Abstract
Skeletal muscle dysfunction in survivors of pneumonia disproportionately affects older individuals in whom it causes substantial morbidity. We found that skeletal muscle recovery was impaired in old compared with young mice after influenza A virus‐induced pneumonia. In young mice, recovery of muscle loss was associated with expansion of tissue‐resident skeletal muscle macrophages and downregulation of MHC II expression, followed by a proliferation of muscle satellite cells. These findings were absent in old mice and in mice deficient in Cx3cr1. Transcriptomic profiling of tissue‐resident skeletal muscle macrophages from old compared with young mice showed downregulation of pathways associated with phagocytosis and proteostasis, and persistent upregulation of inflammatory pathways. Consistently, skeletal muscle macrophages from old mice failed to downregulate MHCII expression during recovery from influenza A virus‐induced pneumonia and showed impaired phagocytic function in vitro. Like old animals, mice deficient in the phagocytic receptor Mertk showed no macrophage expansion, MHCII downregulation, or satellite cell proliferation and failed to recover skeletal muscle function after influenza A pneumonia. Our data suggest that a loss of phagocytic function in a CX3CR1+ tissue‐resident skeletal muscle macrophage population in old mice precludes satellite cell proliferation and recovery of skeletal muscle function after influenza A pneumonia.
Keywords: influenza, macrophages, pneumonia, skeletal muscle
A schematic illustrating how impaired phagocytic function in CX3CR1+ tissue‐resident skeletal muscle macrophages prevents muscle recovery after influenza A virus‐induced pneumonia in old mice
1. INTRODUCTION
As is evident from the ongoing coronavirus disease 2019 (COVID‐19) pandemic, elderly individuals are at increased risk of developing and dying from pneumonia (Zhou et al., 2020). Nevertheless, most elderly patients with access to modern healthcare survive (Jain et al., 2015; Thannickal et al., 2015). Elderly pneumonia survivors are at increased risk of developing age‐related disorders including persistent lung dysfunction (Mittl et al., 1994), skeletal muscle atrophy‐limiting mobility (Herridge et al., 2003), myocardial infarction (Corrales‐Medina Vicente et al., 2012), chronic kidney disease (Murugan et al., 2010), dementia (Tate et al., 2014), and cognitive impairment (Girard et al., 2018), raising significant concerns about these sequelae in elderly survivors of COVID‐19. In particular, skeletal muscle dysfunction develops in the majority of survivors of pneumonia and may persist for years after hospital discharge, where it is a major driver of morbidity (Chan et al., 2018; Dos Santos et al., 2016; Herridge et al., 2003, 2011; Walsh, Batt, Herridge, & Dos Santos, 2014). Persistent skeletal muscle weakness in survivors of severe pneumonia disproportionately affects elderly patients, resulting in reduced quality of life including an increased risk of hospitalizations, long‐term disability, and loss of independence (Barreiro, Sznajder, Nader, & Budinger, 2015; Falsey, Hennessey, Formica, Cox, & Walsh, 2005; Gozalo, Pop‐Vicas, Feng, Gravenstein, & Mor, 2012; Herridge et al., 2011; Pfoh et al., 2016).
Infection with the influenza A virus is among the most common causes of pneumonia, and like pneumonia induced by the severe acute respiratory syndrome–coronavirus‐2 (SARS‐CoV‐2), the mortality attributable to influenza A pneumonia disproportionately impacts the elderly (Jain et al., 2015; Kanegai et al., 2016; Ortiz et al., 2013; Zhou et al., 2020). Influenza A virus‐induced pneumonia in mice is an attractive model to study the effects of aging on pneumonia‐induced muscle function as it recapitulates the disproportionate skeletal muscle dysfunction reported in older survivors of pneumonia (Bartley et al., 2016). Furthermore, as the influenza A virus is trophic for the lung epithelium, there is little if any viremia (Radigan et al., 2019). In previous work foundational to the current study, we demonstrated that influenza A infection induces the expression of the muscle‐specific E3 ligase FBXO32 (atrogin‐1) and that Fbxo32 −/− mice were resistant to influenza A‐induced muscle loss. Furthermore, we found that the administration of tocilizumab, an antibody targeting the IL‐6 receptor, to mice, prevented the induction of atrogin‐1 and the loss of muscle function after influenza A infection without affecting the severity of the lung injury or viral clearance. We concluded that the release of IL‐6 and possibly other endocrine signals from the injured lung induce the active degradation of skeletal muscle during acute influenza A‐induced pneumonia. Notably, tocilizumab and sarilumab are being tested to treat organ injury induced by the cytokine storm in patients with COVID‐19 pneumonia (Di Giambenedetto et al., 2020; Lu, Chen, & Chang, 2020).
Because skeletal muscle fibers are post‐mitotic, muscle myofibers lost during injury must be replaced through activation, proliferation, and differentiation of muscle satellite cells, a muscle‐specific progenitor cell population (Brack & Rando, 2012; Wang & Rudnicki, 2011). After injury, muscle satellite cells undergo rapid proliferation, expand in number, and fuse with damaged myofibers, restoring the number of myonuclei, myofiber content, and volume (Munoz‐Canoves & Michele, 2013; Sala et al., 2019; Tierney et al., 2014). The process of muscle repair has been studied in models of direct injury, for example, freezing, trauma, and toxin injection, but the mechanisms by which muscle fibers are regenerated after pneumonia are not known.
Given this background, the disproportionate loss of skeletal muscle mass in older pneumonia survivors could be attributable to enhanced muscle loss during the acute infection, impaired recovery, or both. We report that when the dose of influenza A administered to young and old mice is titrated to induce equivalent mortality, old and young mice suffer a nearly proportionate loss of skeletal muscle size and function. However, while young mice fully recovered muscle function to pre‐pneumonia levels 60 days after injury, old mice did not. In young mice, a population of CX3CR1‐expressing tissue‐resident macrophages in the skeletal muscle expanded and downregulated MHCII expression after influenza A infection in the absence of recruitment of monocytes from the bone marrow. This was followed by proliferation of muscle satellite cells. All of these findings were absent in old mice and mice deficient in Cx3cr1. Transcriptomic profiling suggested a loss of phagocytic function in tissue‐resident macrophages from the skeletal muscle of old mice, which was confirmed using in vitro assays. Knockout of Mertk, a tyrosine kinase involved in macrophage phagocytosis, phenocopied the molecular and physiologic findings in old mice. Our findings suggest phagocytosis‐induced signaling in CX3CR1+ tissue‐resident skeletal muscle macrophages is necessary for satellite cell proliferation during muscle recovery after influenza A virus‐induced pneumonia.
2. RESULTS
2.1. Old mice do not recover skeletal muscle function after influenza A‐induced pneumonia
In mice, the course of influenza A follows a predictable and consistent pattern in which replicating viruses are detectable by plaque assay over the first 2–5 days of the infection accompanied by the recruitment of neutrophils and monocyte‐derived alveolar macrophages (Peteranderl et al., 2016). Mice then develop progressive hypoxemia, respiratory failure, and death (Traylor, Aeffner, & Davis, 2013), or begin a long process of lung repair, which is incomplete even months after the injury (Kanegai et al., 2016) (Figure 1a). During the acute phase of viral replication, blood cytokines including IL‐6 are dramatically increased coinciding with the loss of muscle mass. We reported that the IL‐6‐dependent activation of FBXO32 in the skeletal muscle was necessary for the acute loss of muscle mass during IAV infection (Radigan et al., 2019).
Figure 1.
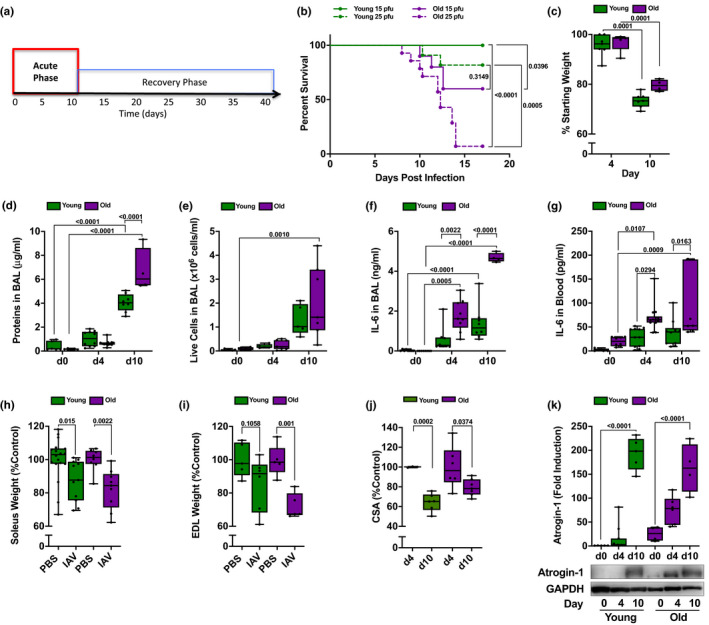
Acute influenza A virus (IAV) infection is worse in old compared with young mice. (a) Timeline of influenza A‐induced pneumonia in mice. Data in this figure focus on the acute phase (red). (b) Young (4 months old; green lines) and old (20–24 months old; purple lines) mice were infected with either 15 pfu (solid lines) or 25 pfu (dotted lines) of IAV (A/WSN/33). A Kaplan–Meier survival curve is shown (n = 9–14 mice per group). Log‐rank (Mantel–Cox) tests were performed, and p values are shown on graph. Each dot represents a single animal. For all data (c–k), young (4 months old; green bars) and old (20–24 months old; purple bars) mice were infected with IAV (25 pfu for young mice, 15 pfu for old mice). Mice were harvested for analysis at 0, 4, and 10 days post‐infection. A two‐way ANOVA with Tukey's post hoc corrections for comparison with more than three groups was used to determine statistical differences, or Student's t test for comparisons between two groups, p values are shown on graph. Box plot center lines are median, and box limits are upper and lower quartiles. Each dot represents an individual animal. (c) Percentage of starting total body weight is shown for day 4 and day 10 post‐IAV infection (n = 5–7 mice per group). (d) Total protein in bronchoalveolar lavage (BAL) fluid was measured by Bradford assay (n = 4–12 mice). (e) Number of live cells in the BAL was counted using hemocytometer, and dead cells were excluded using trypan blue (n = 5–7 mice). (f) IL‐6 levels in the BAL were measured by ELISA (n = 5–7 mice). (g) IL‐6 levels in the serum were measured by ELISA (n = 4–8 mice). (h) Soleus muscles were excised, and soleus wet muscle weights were determined. Values are presented as a percent of naive age‐matched controls (n = 8–18 mice). (i) Extensor digitorum longus (EDL) muscles were excised, and EDL wet muscle weights were measured. Values are presented as a percent of naive age‐matched controls (n = 5–7 mice). (j) Soleus cross‐sectional areas were determined using transverse cross sections immunostained for laminin. Values are normalized to naive age‐matched controls (n = 4–7 mice). (k) Quantification (top panel) and representative Western blot (bottom panel) of atrogin‐1 expression in the mouse tibia anterior muscle. Values are normalized to GAPDH (loading control) (n = 5–11 mice)
Critically ill elderly patients with pneumonia have been reported to have both enhanced muscle loss during their ICU stay and prolonged muscle dysfunction that persists more than a year after injury. Modeling this in mice poses challenges, as the LD50 for influenza A virus is dramatically different in old mice compared with young mice (Figure 1b). Accordingly, we developed a model in which we measured muscle loss and recovery in young and old mice treated with a dose of virus that resulted in similar (~25%) mortality and weight loss in each group (25 pfu per mouse for young mice and 15 pfu per mouse for old mice, Figure 1b,c). Despite the reduced dose of virus, old mice still had worse lung injury as measured by bronchoalveolar lavage protein content 10 days after infection (Figure 1d, e) and increased levels of IL‐6 in bronchoalveolar lavage fluid (Figure 1f) and in the plasma (Figure 1g) compared with young mice. The moderate age‐related increase in systemic inflammation corresponds with injury to the hind limb muscles that is only slightly worse in old compared with young mice. The soleus muscle wet weight decreased similarly in both young and old mice (Figure 1h), but the loss of wet weight in the extensor digitorum longus (EDL) muscle was more pronounced in old mice (Figure 1i). In the soleus muscle, the average fiber cross‐sectional area (CSA) was significantly reduced in both young and old mice after influenza A virus‐induced pneumonia (Figure 1j), although the proportional reductions were smaller in old mice due to a lower starting volume (Figure S1). Consistent with the higher levels of plasma IL‐6, the increase in skeletal muscle atrogin‐1 occurred more rapidly in old animals, but was similar 10 days after infection in young and old mice (Figure 1k).
As a physiologic measure of skeletal muscle function (Figure 2a), we continuously recorded voluntary running distance on monitored exercise wheels in young and old mice over the course of influenza A infection and recovery. Mice adapted to the wheels over 14 days, after which the voluntary distance run on the wheel was consistent and similar between animals (Figure 2b), although on average it was lower in old mice (5.2 ± 1.1 and 2.1 ± 1.2 km/night/mouse in old young and old mice, respectively, Figure S2a). Once baseline running distances were established, mice were infected with influenza A virus and then monitored continuously for up to 60 days. Both young and old mice almost stopped running after the influenza A virus infection, reaching a minimum at day 10 post‐infection (Figure 2b, c). However, by day 20, the distance run returned to pre‐infection levels in young mice. In contrast, old mice failed to return to their pre‐infection activity levels (Figure 2b, c). Similarly, while influenza A virus pneumonia induced a loss of forelimb grip strength in both young and old mice during the acute phase of infection (Figure S2b), old mice failed to recover forelimb grip strength 30 and 60 days after influenza A virus infection (Figure 2d). Hence, when young and old mice are infected with doses of influenza A virus titrated to result in significant but similar weight loss and mortality, old mice have only slightly worse muscle loss. However, while young mice completely recover, old mice do not.
Figure 2.
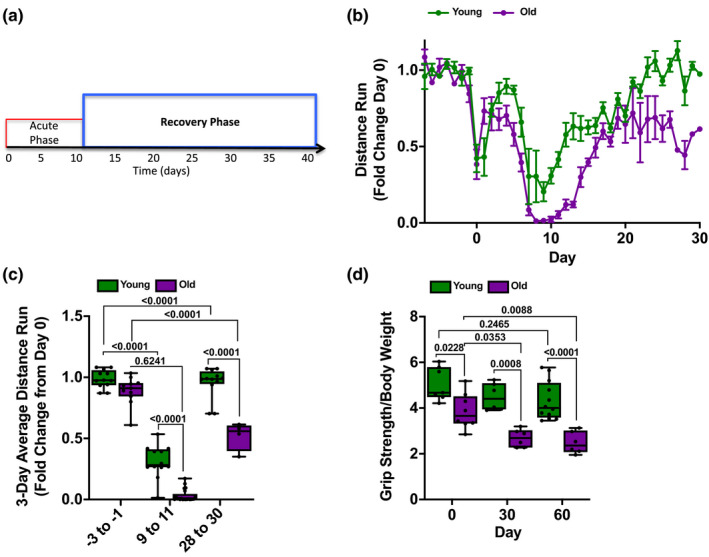
Old mice fail to recover skeletal muscle function after influenza A infection. (a) Timeline of influenza A‐induced pneumonia in mice. Data in this figure focus on the recovery phase (blue). (b) Monitored running wheels were placed in cages, and the voluntary distance run was measured for 14 days before animals were infected with influenza A virus (IAV). At day 0, young (4 months old; green lines) mice were infected with 25 pfu of IAV and old (20–24 months old; purple lines) mice were infected with 15 pfu IAV. Voluntary wheel running was recorded for 30 days, and the 8 p.m. to 6 a.m. average wheel rotation/mouse, normalized to the 10‐day average control, is shown (n = 12 mice per group). A graph of the distance run is shown with means ± SD See also Figure S2. (c) Three‐day average voluntary wheel running distance per cage normalized to the 10‐day average control is shown for young mice (4 months old; green bars) and old mice (20–24 months old; purple bars). The average distance run for days −3 to −1 prior to infection with IAV (25 pfu for young mice, 15 pfu for old mice), days 9–11 post‐infection, and days 28–30 post‐infection is shown (n = 12 mice per group). Box plot center lines are median, box limits are upper and lower quartiles, whiskers are minimal and maximal values, a two‐way ANOVA with Tukey's post hoc corrections for comparison with more than three groups was used to determine statistical differences, and p values are shown on graph. Each dot represents a single animal. (d) Young (4 months old; green bars) and old (20–24 months old; purple bars) mice were infected with IAV at day 0 (25 pfu for young mice, 15 pfu for old mice). Forelimb grip strength was measured at the indicated times (n = 6–12 mice per group). Box plot center lines are median, box limits are upper and lower quartiles, whiskers are minimal and maximal values, a two‐way ANOVA with Tukey's post hoc corrections for comparison with more than three groups was used to determine statistical differences, and p values are shown on graph. Each dot represents a single animal
2.2. Satellite cells and fibroadipogenic progenitors fail to expand in old mice during recovery from influenza A virus‐induced pneumonia
We questioned whether the failure to recover muscle strength and function in old mice was due to a failure to activate regenerative cell populations. We used flow cytometry to simultaneously quantify muscle satellite cells (MuSCs) and fibroadipogenic progenitors (FAPs) as well as lymphoid and myeloid immune cells during muscle recovery after influenza A‐induced lung injury. We identified MuSC as CD45–CD31–CD34+Sca‐1–Itga7+ and FAP as CD45–CD31–CD34+Sca‐1+Itga7+ (Figure 3a). In young, but not in old mice, we observed an expansion of MuSC and FAP after influenza A virus‐induced pneumonia (Figure 3b‐G). This suggests that muscle dysfunction induced by influenza A pneumonia is sufficient to induce regenerative cell populations in young mice, but that this response is impaired in old animals.
Figure 3.
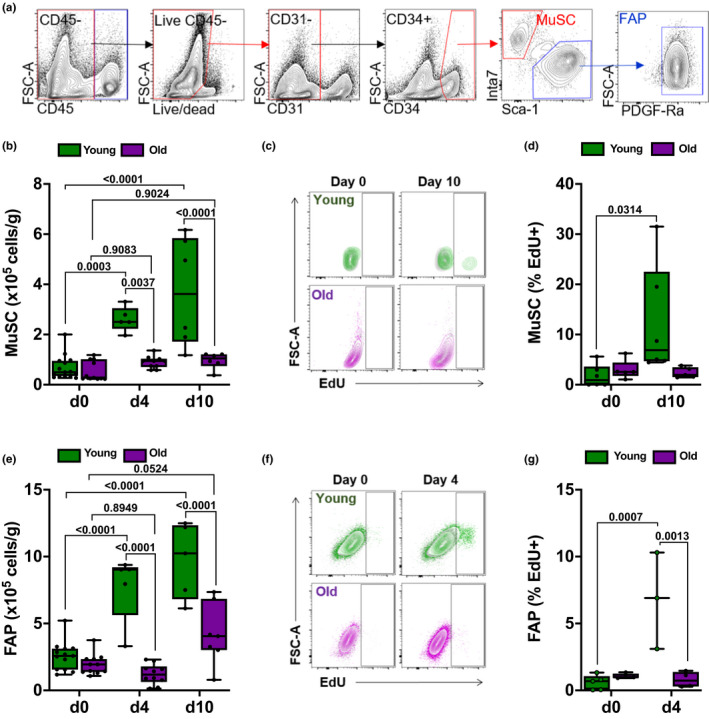
Expansion of muscle satellite cells and fibroadipogenic progenitors is reduced in old compared with young mice after influenza A virus (IAV) pneumonia. For all experiments, young (4 months old; green bars) and old (20–24 months old; purple bars) mice were infected with IAV (25 pfu for young mice, 15 pfu for old mice). Mice were harvested for analysis before the infection (day 0) and 4 and 10 days after infection. A two‐way ANOVA with Tukey's post hoc corrections for comparison with more than three groups was used to show statistical differences. Box plot center lines are median, box limits are upper and lower quartiles, whiskers are minimal and maximal values, p values are shown on the graph. Each dot represents an individual animal. (a) Representative gating strategy to identify muscle satellite cells (MuSCs) and fibroadipogenic progenitors (FAPs). (b) Enumeration of MuSC using flow cytometry. Values shown are per gram of tissue (n = 6–11 mice). (c) Young (top 2 panels; green) or old mice (bottom 2 panels; purple) were injected with 2 mg EdU, 16 hr prior to day 0 or day 10. Representative contour plots showing percentage of EdU‐positive MuSC are shown. Plots from EdU‐injected mice are overlaid against plots of a separate, non‐EdU‐injected control mouse (shown in gray). (d) Box and whiskers plot showing percent EdU‐positive MuSC of total MuSC at days 0 and 10 (n = 5–6 mice). (e) Enumeration of FAP using flow cytometry. Values shown are per gram of tissue (n = 5–13 mice). (f) Young (top 2 panels; green) or old mice (bottom 2 panels; purple) were injected with 2 mg EdU 16 hr prior to day 0 or day 10. Representative contour plots showing percentage of EdU‐positive FAP are shown. Plots from EdU‐injected mice are overlaid against plots of a separate, non‐EdU‐injected control mouse (shown in gray). (g) Box and whiskers plot showing percent EdU‐positive FAP of total FAP at day 0 and day 4 (n = 3–5 mice)
2.3. Tissue‐resident CX3CR1+ skeletal muscle macrophages expand and downregulate MHCII during recovery from influenza A virus‐induced pneumonia
We measured myeloid and lymphoid populations in the skeletal muscle after influenza A infection using flow cytometry (Figures 4a and S3m). In young mice, we observed increases in the number of skeletal muscle macrophages (CD45+Ly6G‐Siglec‐F‐NK1.1‐CD11b+Ly6C‐CD64+) after influenza A infection (Figure 4b). This increase in skeletal muscle macrophages was not observed in old mice (Figure 4b). In contrast, we did not observe influenza A virus‐induced increases in the numbers of neutrophils, eosinophils, natural killer cells, CD11b+ dendritic cells, Ly6C‐high classical monocytes or Ly6C‐low non‐classical monocytes, CD4 and CD8 T cells, regulatory T cells, or B cells in the skeletal muscle of either young or old mice after influenza A infection (Figure S3).
Figure 4.
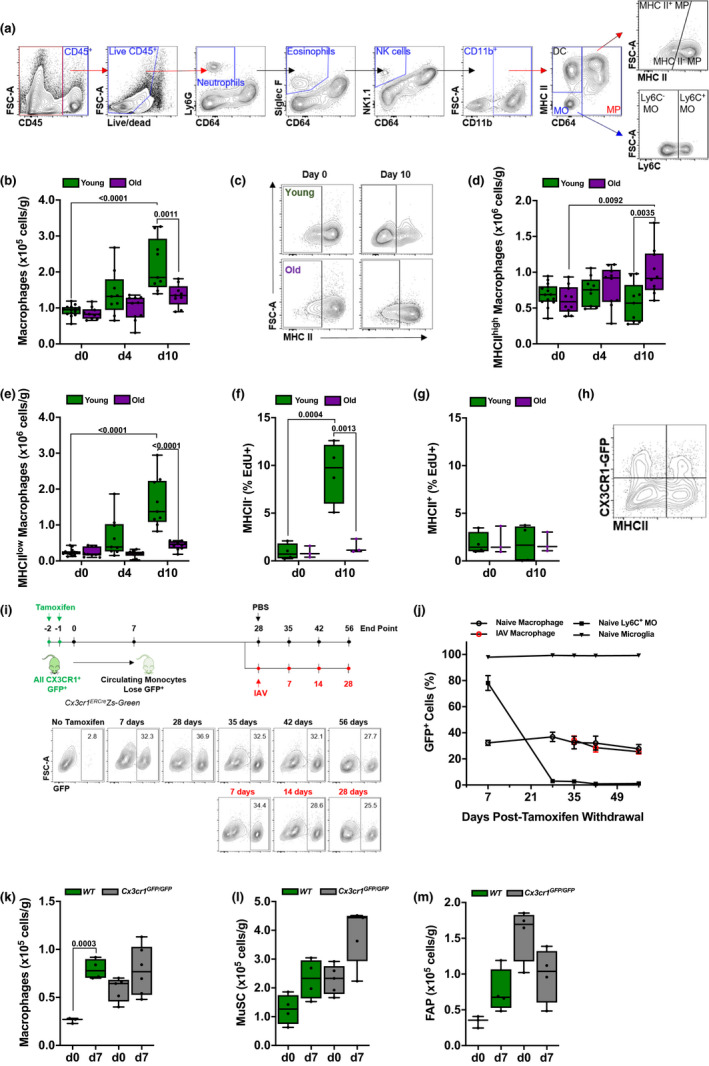
Tissue‐resident skeletal muscle macrophages expand and downregulate MHCII expression after influenza A pneumonia in young, but not in old mice. (a) Gating strategy to identify myeloid cell populations in skeletal muscle tissue using flow cytometry. (b) Young (4 months old; green bars) and old (20–24 months old; purple bars) mice were infected with influenza A virus (IAV) (25 pfu for young mice, 15 pfu for old mice). Graph shows number of macrophages per gram of tissue, before infection (day 0) and 4 and 10 days post‐infection (n = 9–14 mice). (c) Representative flow cytometry showing MHCII staining in skeletal muscle macrophages from young (top two panels) or old (bottom two panels) mice before and 10 days after IAV infection. (d) Young (4 months old; green bars) and old (20–24 months old; purple bars) mice were infected with IAV (25 pfu for young mice, 15 pfu for old mice). Graph shows number of MHCIIhigh macrophages in skeletal muscle per gram of tissue before infection (day 0) and 4 and 10 days after influenza A infection (n = 9–14 mice). (e) Young (4 months old; green bars) and old (20–24 months old; purple bars) mice were infected with IAV (25 pfu for young mice, 15 pfu for old mice). Graph shows number of MHCIIlow macrophages in skeletal muscle per gram of tissue before infection (day 0) and 4 and 10 days after influenza A infection (n = 9–14 mice). (f) Young (4 months old; green bars) and old (20–24 months old; purple bars) mice were infected with IAV (25 pfu for young mice, 15 pfu for old mice). Graph shows percentage of EdU‐positive MHCIIhigh macrophages, of total MHCIIhigh macrophages, in skeletal muscle before infection (day 0) and 10 days after influenza A infection (n = 3–4 mice). (g) Young (4 months old; green bars) and old (20–24 months old; purple bars) mice were infected with IAV (25 pfu for young mice, 15 pfu for old mice). Graph shows percentage of EdU‐positive MHCIIlow macrophages, of total MHCIIlow macrophages, in skeletal muscle before infection (day 0) and 10 days after influenza A infection (n = 3–4 mice). (h) Representative plot of macrophages from Cx3cr1Gfp/WT mice showing heterogeneous expression of MHCII and CX3CR1 (GFP). (i) Cx3cr1ERCre/zsGreen mice were administered tamoxifen (10 mg via oral gavage, twice, 24 hr apart). Mice were harvested 7, 28, 35, 42, and 56 days after the tamoxifen pulse. A separate cohort of mice was infected with 25 pfu IAV 28 days after the tamoxifen pulse and harvested 7, 14, and 28 days later. Representative flow cytometry plots of macrophage GFP expression at each of these time points are shown. (j) Refers to the schematic in (i). Cx3cr1ERCre/zsGreen mice were administered a pulse of tamoxifen (10 mg via oral gavage, twice, 24 hr apart) at day 0. The percentage of GFP+ cells from the indicated cell populations is shown. Some mice were infected with 25 pfu IAV 28 days after the tamoxifen pulse and harvested 7, 14, and 28 days later. Black symbols indicate naïve mice. Red symbols indicate IAV‐infected mice. (k) Young wild‐type mice (4 months old; green bars) and age‐matched Cx3cr1Gfp/Gfp mice (gray bars) were infected with 25 pfu of IAV. Graph represents skeletal muscle macrophage number before infection (day 0) and 7 days after influenza A infection. Values are expressed per gram of tissue (n = 4–5 mice). (l) Young wild‐type mice (4 months old; green bars) and age‐matched Cx3cr1Gfp/Gfp mice (gray bars) were infected with 25 pfu of IAV. Graph represents muscle satellite cell (MuSC) number before infection (day 0) and 7 days after influenza A infection. Values are expressed per gram of tissue (n = 4–5 mice). (m) Young wild‐type mice (4 months old; green bars) and age‐matched Cx3cr1Gfp/Gfp mice (gray bars) were infected with 25 pfu of IAV. Graph represents fibroadipogenic progenitor (FAP) number before infection (day 0) and 7 days after influenza A infection. Values are expressed per gram of tissue (n = 4–5 mice). For all experiments, a two‐way ANOVA with Tukey's post hoc corrections for comparison with more than three groups was used to show statistical differences, and p values are shown on the graph. Each dot represents an individual animal. Also, see Figure S4
As we observed a significant expansion in macrophages after influenza A infection in young mice, we focused our attention on these cells. In naïve mice, the majority of macrophages were represented by an MHCIIhigh population (Figure 4c). During recovery from influenza A infection, the majority of macrophages were MHCIIlow in young, but not in old mice (Figure 4c‐e). Furthermore, incorporation of EdU into MHCIIlow macrophages in after pneumonia‐induced muscle atrophy increased in young but not old mice, indicating an age‐dependent proliferation specific to this population (Figure 4f,g). Using reporter mice in which GFP is knocked into the Cx3cr1 gene (Cx3cr1GFP/+), we found that approximately 25%–30% of skeletal muscle macrophages expressed Cx3cr1 in naïve mice (Figure S4), and both CX3CR1+ and CX3CR1‐ macrophages included populations that were MHCIIhigh and MHCIIlow (Figure 4h).
To determine whether these skeletal muscle macrophage populations are maintained by local proliferation or are replenished by monocytes recruited from the circulation, we performed fate‐mapping experiments using Cx3cr1ER‐Cre mice crossed to a ZsGreenLSL reporter. We administered a tamoxifen pulse (two doses of tamoxifen, one day apart via oral gavage) to Cx3cr1ER‐Cre/ZsGreenLSL mice to permanently label all Cx3cr1‐expressing cells with GFP and analyzed the number of GFP‐positive cells in several myeloid populations over the course of 56 days (Figure 4i,j). Microglia, which are tissue‐resident brain macrophages that are not replaced by circulating monocytes over the lifespan of mice and are characterized by constitutive expression of Cx3cr1 (Yona et al., 2013), were used as a positive control. Nearly 100% of microglia were labeled and remained GFP‐positive after the tamoxifen pulse after 56 days (Figure 4j). Seven days after the tamoxifen pulse, 80% of Ly6C+ classical monocytes in the blood were GFP‐positive (Figure 4j). Consistent with the short half‐life of circulating monocytes, almost all GFP‐positive monocytes were replaced by GFP‐negative monocytes 28 days after the tamoxifen pulse (Figure 4j) (Yona et al., 2013). Similarly, 28 days after the tamoxifen pulse nearly all Ly6C‐ non‐classical monocytes, and CD11b+ dendritic cells were replaced by GFP‐negative cells (not shown). Consistent with our findings using Cx3cr1Gfp/WT mice, we found that ~ 30% of skeletal muscle macrophages were GFP‐positive after the tamoxifen pulse (Figure 4j). The number of GFP‐positive skeletal muscle macrophages remained stable 56 days after the tamoxifen pulse. As such, these cells represent a bona fide, long‐lived tissue‐resident macrophage population that maintain their population without input from circulating monocytes with a half‐life of at least several months.
We used the Cx3cr1ER‐Cre/ZsGreenLSL mice to determine whether the increase in skeletal muscle macrophages we observed after influenza A virus‐induced pneumonia was attributable to the recruitment of monocyte‐derived macrophages or the proliferation of tissue‐resident macrophages. We treated Cx3cr1ER‐Cre/ZsGreenLSL with two pulses of tamoxifen one day apart, waited 28 days for GFP‐labeled monocytes to disappear, infected the mice with influenza A virus, and then harvested mice 7, 14, and 28 days after infection (Figure 4i). We did not observe any reduction in the percentage of GFP‐positive macrophages, excluding the possibility that the increase in skeletal muscle macrophages following influenza A infection represented an influx of circulating cells (Figure 4i,j). Further, the finding that the number of macrophages expanded after influenza A infection combined with the constant percentage of GFP‐positive cells suggests both CX3CR1‐positive and CX3CR1‐negative cells expand proportionately after influenza A infection (Figure 4b,j). CX3CR1 is required for the adhesion and migration of leukocytes, and in the microglia, CX3CR1 is necessary for synaptic pruning during development (Paolicelli et al., 2011). We therefore sought to determine whether CX3CR1 is necessary for the expansion of macrophages and recovery of muscle function after influenza A infection. We treated mice deficient in Cx3cr1 (Cx3cr1Gfp/Gfp) with influenza A virus and performed flow cytometry on skeletal muscle during recovery. Mice deficient in Cx3cr1 showed increased numbers of skeletal muscle macrophages at baseline, perhaps reflecting a failure to populate the niche. However, Cx3cr1Gfp/Gfp mice showed no expansion of skeletal muscle macrophages, no change in the proportion of MHCIIlow macrophages, and no proliferation of MuSC or FAP after influenza A virus‐induced pneumonia (Figure 4k–m). This suggests that functional expression of Cx3cr1 on macrophages that reside in skeletal muscle is required for a proliferative response of MuSC and FAP to injury.
2.4. Transcriptomic profiling identifies downregulation of phagocytic and proteostatic pathways and upregulation of inflammatory pathways in tissue‐resident skeletal muscle macrophages from old compared with young mice
To identify possible mechanisms responsible for the lack of a reparative response in tissue‐resident skeletal macrophages from old mice during recovery from influenza A virus‐induced pneumonia, we performed transcriptomic profiling (RNA‐seq) of flow cytometry‐sorted skeletal muscle macrophages. Out of 1,407 differentially expressed genes (FDR q < 0.05), 582 were upregulated in skeletal muscle macrophages from old compared with young animals and 825 were downregulated (Figure 5a). Compared with young mice, skeletal muscle macrophages from naïve old animals demonstrated decreased expression of genes involved lipid storage and metabolism (Ldlrap1, Lrp1, Fabp4, Nr1h3), macrophage activation and cytokine production (Nlrp3, Sphk1, Msr1, Il10), and extracellular matrix remodeling (Spp1, Mmp13, Mmp14, Thbs1, Chil3) (Figure 5a and Tables S1 and S2, list of DEGs and corresponding GO processes). In addition, macrophages from old animals demonstrated decreased expression of the genes involved in response to unfolded protein and cellular stress, particularly molecular chaperones (Dnaja2, Dnajb4, Dnajb11, Hspa9, Hsp90b1, Hspbap1), as well as several molecules involved in positive regulation of phagocytosis (C5ar1, Msr1). In contrast, macrophages from young naïve animals were characterized by decreased expression of the genes involved in negative regulation of cell migration (Sema4a, Sema6d, Plxna3, Sema4c, Sema6a) and antigen processing and presentation (Cd74, H2‐Ab1, H2‐Eb1, H2‐Aa). We also performed transcriptomic profiling of flow cytometry‐sorted skeletal muscle macrophages from young and old mice 60 days after IAV pneumonia. Of 1,054 differentially expressed genes (FDR q < 0.05), 673 genes were upregulated in skeletal muscle macrophages from old mice and were enriched for the genes involved in inflammation (Il6, Tnf, Il10, Il1a, Il1b, Tnfsf14) and proteostatic stress (Hspa9, Hspa5, Hspb8, Hspbap1, Dnaja4, Dnajb4, Dnaja2) (Figure 5b and Tables S3 and S4, list of DEGs and corresponding GO processes). These transcriptomic data suggested impaired phagocytosis and increased inflammatory response in skeletal muscle macrophages from old mice. Consistent with these observations, we found that flow cytometry‐sorted tissue‐resident skeletal muscle macrophages from old mice exhibited reduced uptake of fluorescently labeled polystyrene beads when compared with those from young animals (Figure 5c, d).
Figure 5.
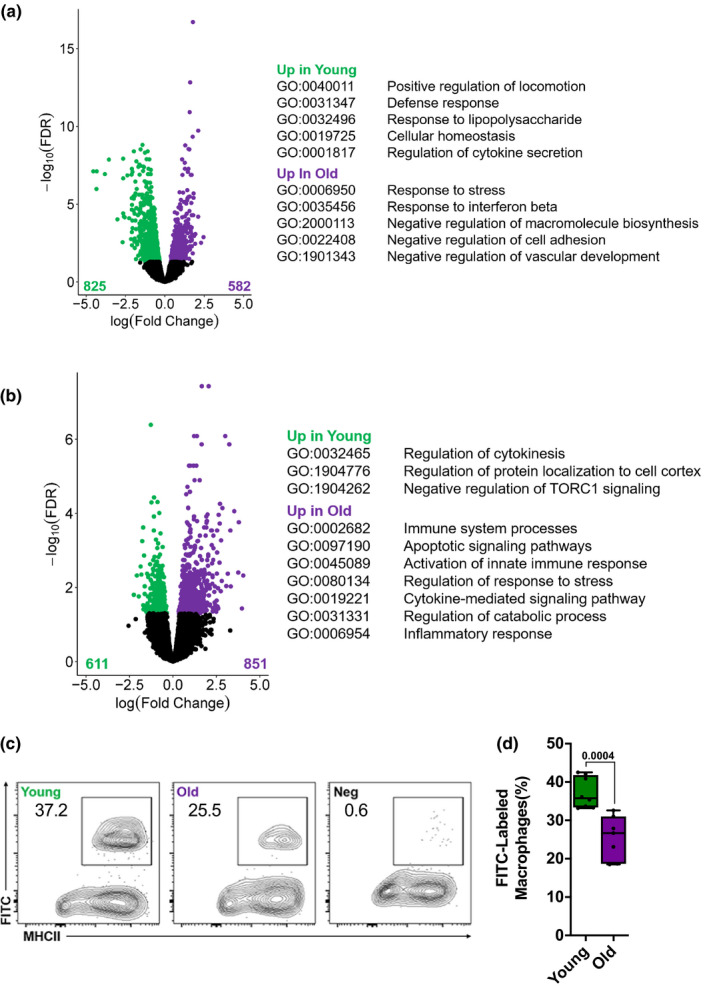
Tissue‐resident macrophages from skeletal muscle from old mice show impaired phagocytosis. (a) Skeletal muscle macrophages were flow cytometry‐sorted from naïve young (4 months old) and old (20–24 months old) mice and analyzed using RNA‐seq (n = 5 mice per group). Differentially expressed genes (FDR q < 0.05) between young (green dots) and old (purple dots) mice are shown with representative genes and GO biological processes (see Tables S1 and S2 for complete list of genes and GO processes). (b) Young (4 months old) and old (20–24 months old) mice were infected with influenza A virus (25 pfu for young mice and 15 pfu for old mice) and harvested 60 days later. Skeletal muscle macrophages were flow cytometry‐sorted and analyzed using RNA‐seq (n = 5 mice per group). Differentially expressed genes (FDR q < 0.05) between young (green dots) and old mice (purple dots) are shown with representative genes and GO biological processes (see Tables S3 and S4 for complete list of genes and GO processes). (c) Single‐cell suspensions prepared from skeletal muscle of young (4 months old) and old (20–24 months old) mice were incubated with antibodies to detect myeloid cells and serum‐opsonized Fluoresbrite microspheres. Uptake of the fluorescently labeled microspheres by macrophages was measured using flow cytometry. Non‐opsonized beads were used as a negative control. Representative contour plots are shown, and numbers indicate percentage from the parent population (gated on macrophages; see Figure 4a for gating strategy). (d) Refers to (c). Graph showing average percent of phagocytosed microspheres per macrophage (n = 7–8 mice). A two‐way ANOVA with Tukey's post hoc corrections for comparison with more than three groups was used to show statistical differences. Box plot center lines are median, box limits are upper and lower quartiles, whiskers are minimal and maximal values, and p values are shown on the graph. Each dot represents an individual animal
2.5. The phagocytic receptor Mertk is necessary for skeletal muscle repair after influenza A‐induced pneumonia
Our transcriptomic data suggested impaired phagocytosis in old compared with young animals. Furthermore, the persistence of MHCII expression in skeletal muscle macrophages from old mice after influenza A infection might reflect impaired phagocytosis (Lemke, 2013). MERTK is a member of the TAM family (Tyro3/Axl/Mer) of tyrosine kinases that is expressed on macrophages, and is involved in recognizing and clearing apoptotic cells through phagocytosis (Lemke, 2013). We found that the level of mRNA encoding Mertk and its phosphatidylserine binding co‐receptor Gas6 were reduced in flow cytometry‐sorted macrophages from old compared with young mice (Figure 6a). Accordingly, we infected Mertk −/− and wild‐type mice with influenza A virus and measured skeletal muscle injury and recovery post‐infection. Wild‐type and Mertk −/− mice developed comparable levels of systemic IL‐6 and atrogin‐1 expression in the skeletal muscle (Figure 6b, c), suggesting a similar level of muscle injury. However, measurements of voluntary distance run on a monitored exercise wheel showed that after an initial transient recovery, Mertk −/− mice failed to maintain their pre‐infection levels of activity after pneumonia (Figure 6d,e). Additionally, we found increased accumulation of age‐related autofluorescent granules representing either lipofuscin or ceroid in the muscle of young Merkt −/− mice compared with young wild‐type mice, and a similar accumulation of lipofuscin or ceroid was observed in old mice (Figures 6f and S5). Similar to Cx3cr1 knockout mice, we observed increased numbers of skeletal muscle macrophages in young Mertk −/− mice at baseline. However, the number of skeletal muscle macrophages in young Mertk −/− mice did not increase during recovery from influenza A virus‐induced pneumonia (Figure 6g) and there was no increase in the proportion of macrophages that were MHCIIlow (Figure 6h), similar to old mice. Consistent with these findings, Mertk −/− mice lacked expansion of MuSC and FAP during recovery from influenza A‐induced pneumonia (Figure 6i,j). These data support a requirement for functional phagocytosis in recovery from influenza A pneumonia‐induced skeletal muscle atrophy.
Figure 6.
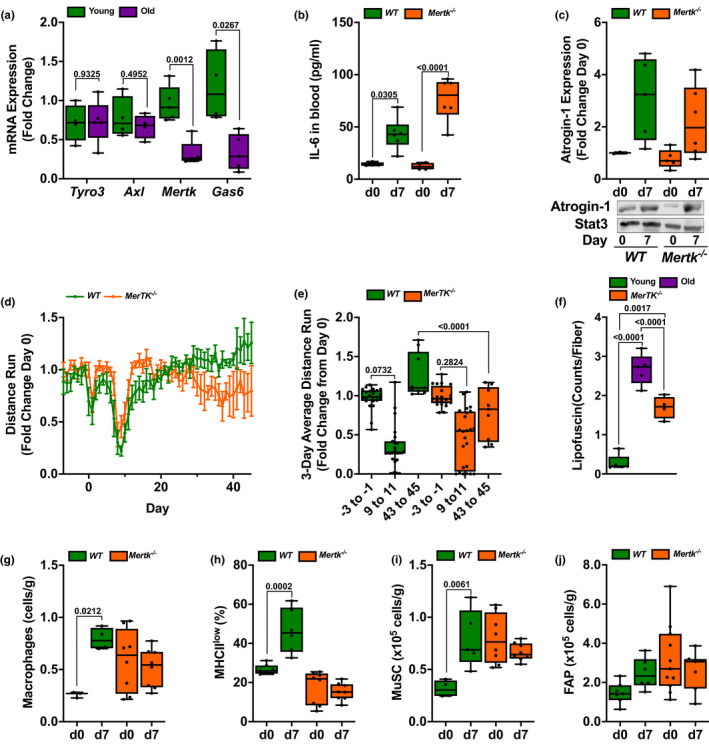
Mertk is necessary for recovery of skeletal muscle function after influenza A‐induced pneumonia. (a) Skeletal muscle macrophages were isolated from naïve young (4 months: green bars) and old (20–24 months: purple bars) mice, and RNA was analyzed for TAM family members via real‐time qPCR with SYBR Green (n = 5 mice). (b) Young (4 months old: orange bars) Mertk −/− mice and age‐matched wild‐type control mice (green bars) were infected with influenza A virus (IAV) (25 pfu) and harvested before the infection (day 0) and 7 days after the infection. IL‐6 was measured by ELISA in the serum (n = 4–6 mice). (c) Young (4 months old: orange bars) Mertk −/− mice and age‐matched wild‐type control mice (green bars) were infected with IAV (25 pfu) and harvested before the infection (day 0) and 7 days after the infection. Quantification (top panel) and representative Western blot (bottom panel) of atrogin‐1 expression in the mouse tibialis anterior muscle. GAPDH was used as a loading control (n = 3–6 mice). (d) Prior to being infected with IAV, young (4 months) Mertk −/− mice (orange lines) and age‐matched wild‐type control mice (green lines) were exposed to voluntary wheel running for 14 days. They were then infected with 25 pfu of IAV, and voluntary wheel running was recorded for 50 days. The 8 p.m. to 6 a.m. average wheel rotation/mouse, corrected to the initial 10‐day average control, is shown (n = 15 mice per group). Graph shows distance run with means ± SD (e) Mice were treated as in (d). The 3‐day average voluntary wheel running per cage is shown for days −3 to −1 prior to IAV, days 9–11 post‐infection, and days 43 to 45 post‐infection (n = 15 mice per group). (f) Graph depicting number of lipofuscin granules per fiber cross section counted manually in Image J. Five mice per group (an average of 3 fields of view per mouse). (g) Young (4 months old, orange bars) Mertk −/− mice and age‐matched wild‐type control mice (green bars) were infected with IAV (25 pfu) and harvested before the infection (day 0) and 7 days after the infection. Skeletal muscle macrophages were enumerated using flow cytometry (n = 4–8 mice). Values are expressed per gram of tissue. (h) Young (4 months old, orange bars) Mertk −/− mice and age‐matched wild‐type control mice (green bars) were infected with IAV (25 pfu) and harvested before the infection (day 0) and 7 days after the infection. MHCIIlow skeletal muscle macrophages were enumerated using flow cytometry (n = 4–8 mice). Values are expressed as a percent of total macrophages. (i) Young (4 months old, orange bars) Mertk −/− mice and age‐matched wild‐type control mice (green bars) were infected with IAV (25 pfu) and harvested before the infection (day 0) and 7 days after the infection. Skeletal muscle satellite cells (MuSCs) were enumerated using flow cytometry (n = 4–8 mice). Values are expressed per gram of tissue. (j) Young (4 months old, orange bars) Mertk −/− mice and age‐matched wild‐type control mice (green bars) were infected with IAV (25 pfu) and harvested before the infection (day 0) and 7 days after the infection. Skeletal muscle fibroadipogenic progenitor (FAP) cells were enumerated using flow cytometry (n = 4–8 mice). Values are expressed per gram of tissue. For all experiments, a two‐way ANOVA with Tukey's post hoc corrections for comparison with more than three groups was used to show statistical differences. Box plot center lines are median, box limits are upper and lower quartiles, whiskers are minimal and maximal values, and p values are shown on the graph. Each dot represents an individual animal
3. DISCUSSION
As vividly illustrated by the COVID‐19 pandemic, pneumonia disproportionately affects the elderly (Zhou et al., 2020). While mortality from pneumonia is higher in older compared with younger individuals, most elderly patients with access to modern healthcare survive their pneumonia (Jain et al., 2015; Ortiz et al., 2013). It is well established that skeletal muscle dysfunction is common in survivors of severe pneumonia requiring intensive care (Herridge et al., 2003). This skeletal muscle dysfunction after pneumonia is disproportionately prolonged and severe in the elderly when compared to younger individuals; however, the mechanisms that underlie this age‐related susceptibility have not been elucidated (Barreiro et al., 2015). In foundational work in advance of this study, we used a murine model of influenza A pneumonia to show the release of IL‐6 from the injured lung signaled the expression of the muscle‐specific E3 ligase FBXO32, actively inducing skeletal muscle degradation. Here, we recapitulate previous findings that old compared with young mice suffer increased mortality when infected with the same dose of influenza A virus. To compare the mechanisms of skeletal muscle dysfunction in young and old mice, we reduced the dose of virus administered to old mice to cause similar mortality and weight loss when compared with young mice. Even at this reduced dose, the levels of IL‐6 and other inflammatory cytokines were increased in old compared with young mice and the severity of acute muscle loss was worse in old compared with old mice. While the differences in acute muscle loss in old compared with young mice were subtle, the differences in recovery were striking. Young animals completely recovered skeletal muscle function after pneumonia, while old animals did not. We used a combination of genetic lineage tracing, flow cytometry, transcriptomics, and genetic loss‐of‐function approaches to show that a loss of phagocytic function in tissue‐resident CX3CR1+ skeletal muscle macrophages contributes to the failure of skeletal muscle recovery in old mice after influenza A infection. Our findings take on new importance during the COVID‐19 pandemic where therapies targeting the IL‐6 receptor are being widely used to target the systemic inflammatory response induced by the SARS‐CoV‐2 coronavirus (Giamarellos‐Bourboulis et al., 2020).
While a subpopulation of macrophages expressing the fractalkine receptor CX3CR1 in the skeletal muscle has been previously reported, nothing was known regarding the function or ontogeny of these cells (Arnold et al., 2015; Jin, Warunek, & Wohlfert, 2018; Zhao, Lu, Wang, Ransohoff, & Zhou, 2016). In young mice recovering from influenza A pneumonia, we observed expansion and downregulation of MHCII in skeletal muscle macrophages followed by muscle satellite cell proliferation. These findings were absent in mice deficient in Cx3cr1. Because macrophages are the only cell type in the skeletal muscle that expresses Cx3cr1 (Schaum et al., 2018), these findings genetically link CX3CR1‐expressing skeletal muscle macrophages with the proliferation of muscle satellite cells (MuSCs) after influenza A‐induced pneumonia. Moreover, we used a lineage tracing system and flow cytometry to show that these skeletal muscle macrophages represent a bona fide tissue‐resident macrophage population maintained in the skeletal muscle via local proliferation independent of recruitment of monocytes or monocyte‐derived macrophages in the steady state and after influenza A pneumonia. Further, unlike other models of skeletal muscle injury and recovery induced by freezing or cardiotoxin administration, we found no evidence of infiltration by other myeloid cell populations or lymphoid populations in response to influenza A pneumonia. While the precise function of CX3CR1 in the skeletal muscle is not known, it may guide macrophages to areas of muscle loss induced by influenza A infection. For example, signaling through CX3CR1 guides monocytes and macrophages to areas of injury or inflammation, and during brain development, CX3CR1 is necessary to guide microglia to synapses where they are pruned from living neurons via a process referred to as trogocytosis (Paolicelli et al., 2011; Weinhard et al., 2018).
We used transcriptomic profiling to compare tissue‐resident skeletal muscle macrophages in young and old mice under naïve conditions and during recovery from influenza A infection. These data suggested the hypothesis that skeletal muscle macrophages from old mice lose phagocytic function, which was confirmed by reduced uptake of serum‐opsonized polystyrene beads in flow cytometry‐sorted skeletal muscle macrophages from old compared with young mice. MERTK is a member of the TAM family of scavenger receptors (MERTK, AXL, and TYRO3), which are activated during injury by interactions with Protein S or Gas6 bound to Annexin V on the surface of apoptotic cells (Lemke, 2013). Ligation of MERTK in macrophages results in the induction of phagocytosis through protein kinase C and Rac‐mediated regulation of the cytoskeleton and the activation of AKT, leading to cellular proliferation and survival (Lemke & Rothlin, 2008). In addition, the activation of MERTK results in its association with the type I interferon receptor (IFNAR) to induce the Jak1/Stat1‐mediated activation of suppressor of cytokine signaling 1 and suppressor of cytokine signaling 3 (SOCS1 and SOC3) (Lemke, 2013). These transcription factors inhibit inflammatory mediators and further reduce expression of MHC class II molecules on the cell surface (Dalpke, Opper, Zimmermann, & Heeg, 2001). We found that during recovery from influenza A infection, young Mertk −/− mice largely phenocopied old mice, in that they failed to expand tissue‐resident macrophages, downregulate MHCII on tissue‐resident macrophages, or expand MuSC and they failed to muscle function after influenza A infection. Our findings are consistent with previous reports that MERTK is necessary for cardiac repair after myocardial infarction and that mesenchymal stem cells promote cardiac repair by activating phagocytic pathways in cardiac macrophages, strengthening the causal association between tissue‐resident skeletal muscle macrophage function and muscle satellite cell proliferation (DeBerge et al., 2017; Vagnozzi et al., 2020; Zhang et al., 2019).
Our findings fit into an emerging body of literature implicating impaired phagocytosis in tissue‐resident macrophages with aging phenotypes. For example, Pluvinage performed a CRISPR screen to identify genes involved in microglia phagocytosis (Pluvinage et al., 2019). They found that CD22 was upregulated in aging microglia and impaired their phagocytic function. When they administered an antibody directed against CD22, cognitive function in old mice improved. Future studies will answer whether improving phagocytic function in tissue‐resident macrophages in the skeletal muscle will be sufficient to revert influenza A pneumonia‐induced muscle dysfunction in old mice. Furthermore, our results support a role for tissue‐resident macrophages in promoting cell proliferation. For example, co‐culture of monocyte/macrophage populations with muscle progenitor cells induces their proliferation via the release of soluble factors in rodent and avian systems (Cantini & Carraro, 1995; Cantini et al., 2002; Chazaud et al., 2003; Merly, Lescaudron, Rouaud, Crossin, & Gardahaut, 1999; Saclier et al., 2013). In our model, however, we cannot determine whether impaired MuSC proliferation is related to the loss of paracrine signals induced in macrophages upon uptake of cellular debris or an inhibition of cellular proliferation by the debris that is not cleared by macrophages.
Our molecular findings are consistent with the studies of muscle biopsies from patients with myopathy after critical illness. In these biopsies, investigators observed MuSC proliferation in the absence of inflammatory cell infiltration (Dos Santos et al., 2016). This differs from many murine models used to study muscle recovery, most of which involve direct injury to the muscle including trauma, freezing, or cardiotoxin injection. In all of these injuries, inflammatory cells are recruited to the injured muscle including neutrophils, monocytes, monocyte‐derived macrophages, regulatory T cells (Tregs), and other T‐cell populations (Arnold et al., 2007; Heredia et al., 2013; Kuswanto et al., 2016; Teixeira et al., 2003; Varga et al., 2016; J. Zhang et al., 2014). Inflammatory cell recruitment in those models precluded the specific study of tissue‐resident skeletal muscle macrophages. For example, two independent groups reported that deletion of monocyte‐derived macrophages recruited in response to cardiotoxin‐induced injury resulted in impaired satellite cell proliferation. In both of these studies, the strategies used to deplete monocyte‐derived macrophages (CD11b‐DTR and an antibody to M‐CSF) would target both monocyte‐derived macrophages and the skeletal muscle tissue‐resident macrophages we identified in this study (Arnold et al., 2007; Segawa et al., 2008). While Tregs have been reported to be important for skeletal muscle recovery after cardiotoxin injury (Kuswanto et al., 2016), we found that Treg number was reduced in both young and old mice after influenza A infection. Nevertheless, Treg proliferation was higher in young compared with old mice and it is possible that factors released from resident tissue Tregs are important for recovery.
Our study has some limitations. First, while voluntary wheel running in mice is an intuitive and compelling measure of the skeletal muscle dysfunction that impairs mobility in survivors of pneumonia, it is a composite measure that may be altered by factors other than muscle function. Second, while Mertk −/− mice recapitulated the lack of cellular responses in old animals, they transiently regained muscle function after injury. This may be explained by the high basal levels of MuSC in Mertk −/− relative to wild‐type mice, which allows short term, but unsustainable recovery of muscle function. Third, while we observed expansion of FAPs during recovery from muscle injury in young, old, and Mertk‐deficient mice, their role in muscle recovery is not certain. FAP cells have been described as important facilitators of MuSC behaviors, supporting their myogenic differentiation (Fiore et al., 2016), yet FAP cell persistence has been shown to have a detrimental effect on muscle tissue by contributing to fibrosis (Gonzalez et al., 2017) and ectopic fat accumulation (Uezumi, Fukada, Yamamoto, Takeda, & Tsuchida, 2010). Thus, if tissue‐resident macrophages contribute to FAP clearance, as has been reported for inflammatory, monocyte‐derived macrophages (Lemos et al., 2015) some of the effects of aging or Mertk loss in macrophages might be attributed to the persistence of these cells. Finally, while the expression of Cx3cr1 in the skeletal muscle is limited to macrophages, Cx3cr1 is also expressed on monocytes and LYVE1‐expressing macrophages associated with the vasculature and nerve fibers (Lim et al., 2018; Schaum et al., 2018). We therefore cannot completely exclude off target effects related to global deletion of Cx3cr1 in our system.
Skeletal muscle represents approximately 40% of the total body mass (Janssen, Heymsfield, Wang, & Ross, 2000), and maintaining its function is critical for posture, breathing, locomotion, nutrient storage, and overall well‐being. As a result, loss of muscle function and mass with age‐related sarcopenia is a major driver of frailty (Janssen et al., 2000; Topinkova, 2008). Our findings highlight the importance of impaired repair in the persistent, sometimes life‐long loss of muscle function that develops in elderly survivors of pneumonia. Specifically, we suggest that a loss of phagocytic function in tissue‐resident skeletal muscle macrophages during aging impairs their ability to promote muscle satellite cell proliferation necessary for skeletal muscle recovery. Given the prevalence of influenza A, COVID‐19, and other respiratory viral infections, it is likely that our findings have broad implications for elderly survivors of pneumonia.
| Reagent or resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| CD45 clone 30‐F11 PE‐Cy7 | BD Biosciences | 552848; RRID: 5114808 |
| CD31 clone MEC 13.3 PE‐CF594 | BD Biosciences | 563616; RRID: 5163928 |
| CD34 clone RAM34 FITC | BD Biosciences | 553733; RRID: 6019764 |
| CD34 clone RAM34 Alexa Fluor 700 | eBiosciences | 56‐0341‐82; RRID: 4289831 |
| Sca‐1 clone D7 BV421 | BioLegend | 108128; RRID: B254500 |
| Int‐a7 clone 3C12 APC | Miltenyi | 130‐102‐717; RRID: 5190509033 |
| PDGF‐Ra clone APA5 PE | BioLegend | 135906; RRID: B203475 |
| CD64 clone X54‐5/7.1 PE | BioLegend | 139304; RRID: B191540 |
| Ly6G clone 1A8 Alexa Fluor 700 | BD Biosciences | 561236; RRID: 6252780 |
| Siglec‐F clone E50‐2440 PE‐CF594 | BD Biosciences | 562757; RRID: 7128860 |
| NK1.1 clone PK136 BUV395 | BD Biosciences | 564144; RRID: 8155628 |
| CD11b clone M1/70 FITC | BD Biosciences | 561688; RRID: 5357544 |
| CD11b clone M1/70 APC | BioLegend | 101212; RRID: B247959 |
| MHCII M5/114.15.2 eFluor 450 | eBiosciences | 48‐5321‐82; RRID: 4289694 |
| Ly6C clone HK1.4 APC/Cy7 | BioLegend | 128018; RRID: B232708 |
| CD19 clone 1D3 PE‐CF594 | BD Biosciences | 562329; RRID: 5336548 |
| CD3e clone 145‐2C11 FITC | BD Biosciences | 553061; RRID: E00061‐1634 |
| CD3e clone 145‐2C11 APC/Cy7 | BioLegend | 100300; RRID: B239433 |
| CD4 clone RM4‐5 eFluor 450 | eBiosciences | 48‐0042; RRID: 4281988 |
| CD8 clone 53‐6.7 BUV395 | BD Biosciences | 563786; RRID: 7096603 |
| CD25 clone PC61.5 APC | eBiosciences | 17‐0251‐82; RRID: 4300565 |
| FoxP3 clone FJK‐16s PE | eBiosciences | 12‐5773‐82; RRID: 4279536 |
| Fbx32 (atrogin 1) | Abcam | ab168372; RRID: GR322135−8 |
| GAPDH | Cell Signaling | D16H11; RRID: |
| Laminin | Sigma | L9393; RRID: 015M4881V |
| Alexa Fluor 568 secondary antibody | Life Technologies | A‐11011; RRID: 2088068 |
| Chemicals, peptides, and recombinant proteins | ||
| AutoMACS Buffer | Miltenyi | 130‐091‐221 |
| BD Pharm Lyse | BD Biosciences | 555899 |
| eFluor 506 Viability Dye | Thermo Fisher | 65‐0866‐18 |
| 123count eBeads | eBioscience | 01‐1234 |
| Anti‐mouse CD16/CD32 | Invitrogen | 14‐0141‐85 |
| Fluoresbrite YG 1.0 Micron Microspheres | Polysciences, Inc | 16662 |
| RNase‐free DNase Set | Qiagen | 79254 |
| Critical commercial assays | ||
| Skeletal Muscle Dissociation Kit | Miltenyi | 130‐098‐305 |
| Mouse IL‐6 ELISA Kit | Life Technologies | KMC0061C |
| FoxP3 Staining Buffer Set | eBiosciences | 00‐5523 |
| Click‐iT Plus EdU Assay Kit | Invitrogen | C10632 |
| Arcturus PicoPure RNA Isolation Kit | Thermo Fisher | KIT0204 |
| iScript cDNA Synthesis Kit | Bio‐Rad | 1725034 |
| iTaq SYBR One‐Step RT‐PCR kit | Bio‐Rad | 1725151 |
Deposited data
Experimental Models: Organisms/Strains
| C57Bl/6 9–11 weeks/18 months | NIA NIH | |
| B6.129P2(Cg)‐Cx3cr1tm1Litt/J | The Jackson Laboratory | Stock #005582 |
| B6.129P2(C)‐Cx3cr1tm2.1(cre/ERT2)Jung/J | The Jackson Laboratory | Stock #020940 |
| B6.Cg‐Gt(ROSA)26Sortm6(CAG‐ZsGreen1)Hze/J | The Jackson Laboratory | Stock #007906 |
| B6;129‐Mertktm1Grl/J | The Jackson Laboratory | Stock #011122 |
Oligonucleotides
| Gapdh FW | IDT | GCA CAG TCA AGG CCG AGA AT |
| Gapdh RV | IDT | GCC TTC TCC ATG GTG GTG AA |
| MerTK FW | IDT | CGT CTG TCC TAA CCG TAC CT |
| MerTK RV | IDT | GTA CTG TTG AGG ATA TGG ACT |
| Axl FW | IDT | GGT CCC CCT GAG AAC GTT AG |
| Axl RV | IDT | CAT AAG TAC CTC GGG GGT GT |
| Tyro3 FW | IDT | AGT GGA ACG GTC TGA TGC TG |
| Tyro3 RV | IDT | AGA ATG GCA CAC CTT CGA CA |
| Gas6 FW | IDT | ATG GGT GCA TGA GGA GTT GG |
| Gas6 RV | IDT | TGT TCG GGT GTA GTT GAG CC |
Software and Algorithms
| FlowJo V10.1 | Treestar, FlowJo, Ashland, Oregon | https://www.flowjo.com/solutions/flowjo/downloads |
| GraphPad Prism 7.02 | GraphPad Software, California | https://www.graphpad.com/scientific-software/prism/ |
| bcl2fastq 2.17.1.14 | Illumina, California | https://support.illumina.com/sequencing/sequencing_software/bcl2fastq-conversion-software.html |
| Trimmomatic 0.36 | Usadel Lab, RWTH Aachen University, Institute for Biology I | http://www.usadellab.org/cms/?page=trimmomatic |
| STAR 2.6.0 | Alexander Dobin, Cold Spring Harbor Laboratory | https://github.com/alexdobin/STAR |
| HTSeq 0.7.1 | Genome Biology Unit, EMBL Heidelberg | https://htseq.readthedocs.io/en/release_0.10.0/ |
| edgeR 3.20.9 | Bioconductor | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| Gorilla | Eden, Navon, Steinfeld, Lipson, Yakhini, 2009 | http://cbl-gorilla.cs.technion.ac.il/ |
| Other | ||
| Dulbecco's modified Eagle's medium (DMEM) | Fisher Scientific | MT‐10‐013‐CM |
| 2‐Mercaptoethanol | Sigma‐Aldrich | M6250 |
| Bio‐Rad protein assay dye reagent concentrate | Bio‐Rad Laboratories | 500‐0006 |
4. CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contacts: Jacob Sznajder (j‐sznajder@northwestern.edu) and Alexander Misharin (a‐misharin@northwestern.edu).
5. EXPERIMENTAL MODEL AND SUBJECT DETAILS
5.1. Mouse models
All mouse procedures were approved by the Institutional Animal Care and Use Committee at Northwestern University (Chicago, IL, USA). All strains including wild‐type mice are bred and housed at a barrier‐ and specific pathogen‐free facility at the Center for Comparative Medicine at Northwestern University. All experiments were performed with littermate controls. Number of animals per group was determined based on our previous publications. Mice were housed at the Center for Comparative Medicine at Northwestern University, in microisolator cages, with standard 12‐hr light/darkness cycle, ambient temperature 23°C, and were provided standard rodent diet (Envigo/Teklad LM‐485) and water ad libitum. Four‐ and eighteen‐month‐old C57BL/6J mice were obtained from Jackson Laboratories or NIA colony. The C57BL/6J, Cx3cr1ER‐Cre mice (Yona et al., 2013) and ZsGreen reporter (Madisen et al., 2010) mice were obtained from Jackson laboratories (Jax stocks 000664, 020940, and 007906, correspondingly) as were Cx3cr1gfp/gfp mice (Jax stock 005582).
6. METHODS’ DETAIL
6.1. Influenza A infection
Murine‐adapted, influenza A virus (A/WSN/33 [H1N1]) was provided by Robert Lamb, Ph.D., Sc.D., Northwestern University, Evanston, IL. Mice were anesthetized with isoflurane, their lungs were intubated orally with a 20‐gauge Angiocath (Franklin Lanes, NJ), and then instilled with either sterile, phosphate‐buffered saline (S), or mouse‐adapted influenza A virus (IAV) as previously described (Radigan et al., 2019). Various doses in a range of 10‐100pfu were tested to find an optimal sublethal dose matched to the age of the animal. Based on survival curves, we determined that 25 pfu was optimal for young animals, but a lower dose was required for old animals, as a 25 pfu dose resulted in excessive mortality of old mice (Figure S1). We continuously observed mice infected with influenza A virus for signs of distress (slowed respiration, failure to respond to cage tapping, failure of grooming, huddling, and fur ruffling). Mice that developed these symptoms were sacrificed, and the death was recorded as an influenza A‐induced mortality. Weight was measured prior to influenza infection and prior to harvest at 4 or 10 days post‐infection, and recorded as percentage of weight loss from baseline (Radigan et al., 2019).
6.2. Measurement of muscle dysfunction
Immediately prior to muscle harvest, forelimb skeletal muscle strength was assessed using a digital grip strength meter (Columbus Instruments) as described (Radigan et al., 2019). Grip strength was measured in each animal six successive times, and the average of the highest four values for each mouse was used. The same operator performed all tests. The mice were then terminally anesthetized with EUTHASOL (pentobarbital sodium/phenytoin sodium). The soleus and EDL muscles were excised, and tendon was trimmed under a microscope to assure optimal accuracy for weight measurement. The muscles were then blotted dry and weighed. Muscles were either frozen in liquid nitrogen‐cooled isopentane for cryosectioning or snap‐frozen in liquid nitrogen for protein extraction. Alternatively, muscles were minced for flow cytometric analysis as described below.
6.3. Voluntary wheel running
Mice, housed three per cage, were provided access to a trackable, low‐profile saucer wheels (Med Associates, Inc.) 24 hr per day for 14–28 days to establish consistent running patterns. After establishing baseline, all mice were infected with IAV at an age‐appropriate dose (Figure S1). Wheel rotations were gathered in one‐minute increments via wireless hub and analyzed using wheel manager software (Med Associates, Inc.). Distances travelled per mouse per night (8 p.m.‐6 a.m.) were calculated for each night. Data were collected as the average of three mice per cage for six cages each, and running distance is expressed related to average nightly runs over 10 days pre‐Influenza A.
6.4. Immunohistochemistry and fiber size and type assessment
Soleus and EDL serial transverse cryosections (8 μm) were obtained from the Northwestern University Mouse Histology and Phenotyping Laboratory and mounted on glass slides. Sections were fixed in 4% formaldehyde, permeabilized, and blocked. Immunostaining was performed with laminin primary antibody (1:50 dilution; Sigma; Catalog: L9393) followed by Alexa Fluor 568‐conjugated secondary antibody (1:200 dilution; Life Technologies; Catalog: A‐11011). Images were acquired with a Zeiss LSM 510 confocal microscope using a 40x objective (Northwestern University Center for Advanced Microscopy) and analyzed using Zeiss LSM5 Image Browser software. Fiber size was studied by measuring the fibers' minimal inner diameter (at least 100 fibers per muscle), defined as the minimum diameter from inner border to inner border, passing through the center of the muscle fiber. This parameter has been shown to be very insensitive to deviations from the “optimal” cross‐sectioning profile, as compared with direct measure of fiber CSA (Radigan et al., 2019). CSA was calculated using this diameter, and results were expressed as mean CSA ± SE
6.5. Bronchoalveolar lavage
Saline fluid (0.8 ml) was lavaged and then aspirated from the lungs of mice through tracheal angiocatheter. Cell counts were measured by trypan blue exclusion and the automated countess system. To measure the cell differential, 200 μl of BAL fluid was placed in cytospin funnels and spun 450 rcf 5 min onto a glass slide using Shandon Cytospin 3 centrifuge. Cells were stained by Wright–Giemsa and manually counted and identified.
6.6. IL‐6 measurements
IL‐6 expression assays were performed using a kit from Life technologies (KMC0061C) according to the manufacturer's instructions. Assays were performed on either BAL fluids (acquired as described above) or from 50 μl/well blood plasma drawn from posterior vena cava using a 23‐gauge needle, centrifuged to remove cells, and stored at −80°C. Analysis for each mouse was done in triplicate, and a minimum of five mice were analyzed/condition.
6.7. Flow Cytometry and Cell Sorting
Mice were fully perfused through the right ventricle with 20 ml of PBS, and hind limb muscle was removed. Tissue was cut into small pieces with scissors, transferred into C‐tubes (Miltenyi), and processed with a Skeletal Muscle Dissociation Kit according to manufacturer's instructions (Miltenyi). To achieve a single‐cell suspension, two rounds of dissociation were performed using a GentleMACS dissociator (Miltenyi), followed by three rounds of filtration through Falcon 100‐μm, 70‐μm, and then 40‐μm nylon mesh filter units (Thermo Fisher #352340, 352350, and 352360, respectively) into polypropylene 50‐ml Falcon tubes, followed by centrifugation at 300 rcf in an Eppendorf 5810R centrifuge for 15 min. Any remaining red blood cells were lysed by resuspending the pellet in 1ml/tube BD Pharm Lyse (BD Biosciences), and were transferred to flow tubes, pelleted by low‐speed centrifugation 5 min, and resuspended in 0.5ml/tube HBSS containing 0.5 μl/tube viability dye eFluor 506 (eBioscience). Cells were stained in viability dye for 15 min in the dark at room temperature, followed by 2X washes in 1ml MACS buffer (Miltenyi). After live/dead cell staining, pellets are resuspended in 150 μl/tube FcBlock Anti‐mouse CD16/CD32 (Invitrogen) for 30 min in the dark at 4°C to block non‐specific binding. After blocking, cells were divided into three separate tubes at 50 μl each, then stained with a mixture of fluorochrome‐conjugated antibodies in 50 μl/tube, and incubated for an additional 30 min at 4°C in the dark. After antibody incubation, the cells were washed 2X in MACS buffer, and then fixed in 2% paraformaldehyde for 15 min at room temperature in the dark, washed 2X in HBSS, resuspended in 200 μl HBSS, and then stored at 4°C overnight. Data were acquired on a BD LSR Fortessa II flow cytometer using BD FACSDiva software, and compensation and data analyses were performed using FlowJo software (Treestar). Prior to cytometric analysis, 50 μl 123count eBeads (Invitrogen) were added to 200 μl each sample to allow accurate cell counts/gm of tissue. “Fluorescence minus one” controls were used when necessary. Cell populations were identified using sequential gating strategy (Figures 3a,4a,5a). For Foxp3 detection, a staining kit for intracellular antigens was used (eBiosciences). Cells were fixed and permeabilized according to directions, before incubation with Foxp3 antibody prior to analysis. Cell sorting was performed at Northwestern University RLHCCC Flow Cytometry core facility on SORP FACSAria III instrument (BD) with the same optical configuration as LSR II, using a 100‐µm nozzle and 40 psi pressure.
To measure proliferation, EdU was diluted in sterile saline from 100 mg/ml stock in DMSO, and injected intraperitoneally at 2 mg/mouse, 16 hr prior to tissue harvest (Misharin et al., 2017). Single‐cell suspensions were divided into thirds and stained with antibody panels as described in (Figures 3a,4a,5a) with FITC replaced in each panel. After surface antibody staining, cells are fixed and undergo a Click‐IT EdU detection reaction using an Alexa‐Flour 488 picolyl azide and Click‐IT Plus EdU Flow Cytometry Assay Kit (Invitrogen).
6.8. Quantitative RT‐PCR
Ten thousand skeletal muscle macrophages were sorted on a FACSAria 6‐Laser Sorter, into 200 μl PicoPure lysis buffer, and stored at −80°C. RNA was harvested following manufacturer's directions for Arcturus PicoPure RNA Isolation Kit (Applied Biosystems). Total RNA was quantified using a NanoDrop 2000c (Thermo Scientific). cDNA was generated using iScript cDNA Synthesis Kit (Bio‐Rad) in a Bio‐Rad T100 thermal cycler, followed by qPCR with SYBR Green (Bio‐Rad) and a Bio‐Rad CFX Connect real‐time system. The following primers were used:
Gapdh (F‐GCA CAG TCA AGG CCG AGA AT, R‐GCC TTC TCC ATG GTG GTG AA)
MerTK (F‐CGT CTG TCC TAA CCG TAC CT, R‐GTA CTG TTG AGG ATA TGG ACT)
Axl (F‐GGT CCC CCT GAG AAC GTT AG, R‐CAT AAG TAC CTC GGG GGT GT)
Tyro3 (F‐AGT GGA ACG GTC TGA TGC TG, R‐AGA ATG GCA CAC CTT CGA CA)
Gas6 (F‐ATG GGT GCA TGA GGA GTT GG, R‐TGT TCG GGT GTA GTT GAG CC)
6.9. Western blot analysis
Quadriceps muscle tissue was homogenized with a Tissue Tearor (BioSpec Products, Inc) for 1 min in ice‐cold lysis buffer (Nonidet P‐40 1%, glycerol 10%, NaCl 137 mM, Tris‐HCl pH 7.5 20 mM) containing protease (1 Complete Mini, EDTA‐free tablet, Roche) and phosphatase (sodium fluoride 30 mM, β‐glycerophosphate 250 mM, sodium orthovanadate 1mM) inhibitors. Samples were centrifuged at 9,000 g for 10 min at 4°C, and the supernatant was collected. Protein concentrations were determined with Protein Assay Dye (Bio‐Rad) in a 96‐well plate measured against BSA protein standard dilutions at 595 nm with a Bio‐Rad iMark microplate reader (Bio‐Rad, Hercules CA). Equal amount of protein (40–80 total μg) was loaded on a 9% SDS‐PAGE gel, and Western blot analysis was performed as described previously (Radigan et al., 2019). Incubation with primary antibodies was performed overnight at 4°C. Immunoblots were quantified by densitometry using Image J 1.46r (National Institutes of Health) or Image Studio Software (LI‐COR Inc.). The following antibodies were used: rabbit monoclonal to Fbx32 (Abcam; catalog: ab168372; 1:1,000) or rabbit monoclonal to GAPDH (D16H11) (Cell Signaling; catalog: 5174s; 1:1,000).
6.10. Transcriptome profiling via mRNA‐Seq
Ten thousand skeletal muscle macrophages were sorted on a FACSARIA 6‐Laser Sorter, into 200 μl PicoPure lysis buffer, and stored at −80°C. RNA was harvested following manufacturer's directions for Arcturus PicoPure RNA Isolation Kit (Applied Biosystems). RNA quality was assessed with the 4200 TapeStation System (Agilent). Samples with an RNA integrity number (RIN) lower than 7 were discarded. RNA‐seq libraries were prepared from 0.5 ng total RNA using the SMARTer Stranded Total RNA‐Seq Kit v2–Pico Input Mammalian Kit. Libraries were quantified and assessed using the Qubit Fluorimeter (Invitrogen) and 4,200 TapeStation. Libraries were sequenced on NextSeq 500 instrument (Illumina) at 75‐bp length, single‐end reads. The average reading depth across all experiments exceeded 6x106 per sample and over 94% of reads had a Q‐score greater than 30. For RNA‐seq analysis, reads were demultiplexed using bcl2fastq (version 2.17.1.14). Read quality was assessed with FastQC. Low‐quality base calls were trimmed using Trimmomatic (version 0.36). Reads were then aligned to the Mus musculus reference genome (with mm10 assembly) using the STAR aligner (version 2.6.0). Counts tables were generated using HTSeq (version 0.6.1). Raw counts were processed in R (version 3.4.4) using edgeR (version 3.20.9) to generate normalized counts. Negative binomial likelihood with default setting followed by generalized linear models fitting was used to estimate differentially expressed genes. FDR q‐values were used to correct for multiple comparisons, and a value of 0.05 was used as a threshold to indicate statistical significance (see online code). Gene Ontology analysis was performed using GOrilla on two unranked gene lists. The RNA‐seq datasets are available at GEO: GSE142580.
6.11. In Vitro phagocytosis
After enzymatic digestion, 50 μl of the single‐cell suspensions were incubated with 35 μl of myeloid antibody cocktail with 15 μl/tube Fluoresbrite YG 1.0 micron microspheres (Polysciences Inc., Warrington, PA) that had been opsonized for 30 min at 37°C in by mixing 1 drop beads with 1ml 50% FBS/HBSS, followed by 2X wash/resuspension in HBSS 1 ml (non‐opsonized beads were used as a negative control). Cell suspensions were allowed to uptake beads for 30 min at 37°C in the dark with shaking, followed by two washes in HBSS to remove any unbound beads, and fixation in 2% paraformaldehyde, followed by flow cytometry with a FITC negative myeloid cocktail to detect bead uptake in myeloid cell populations.
7. QUANTIFICATION AND STATISTICAL ANALYSIS
7.1. Statistical analysis
Differences between groups were determined according to ANOVA. When ANOVA indicated a significant difference, individual differences were examined using t tests with a Tukey correction for multiple comparisons, as indicated. All analyses were performed using GraphPad Prism, version 8.4.2 (GraphPad Software, San Diego CA). Data are shown as means ± SEMs. p < .05 was considered statistically significant in all tests.
CONFLICT OF INTEREST
The authors declare no competing financial interests.
AUTHOR CONTRIBUTIONS
Constance Runyan designed the study, performed experiments, analyzed results, and wrote the manuscript. Lynn C. Welch performed experiments, analyzed results, and wrote the manuscript. Emilia Lecuona, Masahiko Shigemura, Luciano Amarelle, Raul Piseaux‐Aillon, Kinola J.N. Williams, Nikita Joshi, Kiwon Nam, Alexandra C. McQuattie‐Pimentel, Yuliya Politanska, Ziyan Lu, Lango Sichizya, and Satoshi Watanabe performed experiments and analyzed results. Hiam Abdala‐Valencia designed the study, performed experiments, and analyzed results. Nikolay Markov performed bioinformatic analysis and wrote manuscript. GR Scott Budinger designed and supervised the study, wrote manuscript, and provided reagents, resources, and funding. Jacob I Sznajder and Alexander V. Misharin designed and supervised the study, performed analysis, wrote the manuscript, and provided funding for the project. All authors discussed the results and commented on the manuscript.
Supporting information
Figure S1–S5
Table S1
Table S2
Table S3
Table S4
ACKNOWLEDGMENTS
Northwestern University Flow Cytometry Facility, Center for Advanced Microscopy, and Pathology Core Facility are supported by NCI Cancer Center Support Grant P30 CA060553 awarded to the Robert H. Lurie Comprehensive Cancer Center. This research was supported in part through the computational resources and staff contributions provided by the Genomics Computing Cluster (Genomic Nodes on Quest) which is jointly supported by the Feinberg School of Medicine, the Center for Genetic Medicine, and Feinberg's Department of Biochemistry and Molecular Genetics, the Office of the Provost, the Office for Research, and Northwestern Information Technology.
Runyan CE, Welch LC, Lecuona E, et al. Impaired phagocytic function in CX3CR1+ tissue‐resident skeletal muscle macrophages prevents muscle recovery after influenza A virus‐induced pneumonia in old mice. Aging Cell. 2020;19:e13180 10.1111/acel.13180
G. R. Scott Budinger, Jacob I. Sznajder, and Alexander V. Misharin are contributed equally.
Funding information
Satoshi Watanabe is supported by MSD Life Science Foundation, Public Interest Incorporated Foundation, Japan, and David W. Cugell and Christina Enroth‐Cugell Fellowship Program at Northwestern University. GR Scott Budinger is supported by NIH grants ES013995, HL071643, and AG049665, and the Veterans Administration Grant BX000201. Jacob I Sznajder is supported by NIH grants HL147070, HL071643, and AG049665. Alexander V Misharin is supported by NIH grants HL135124, AG049665, and AI135964.
Contributor Information
G. R. Scott Budinger, Email: s-buding@northwestern.edu.
Jacob I. Sznajder, Email: j-sznajder@northwestern.edu.
Alexander V. Misharin, Email: a-misharin@northwestern.edu.
DATA AVAILABILITY STATEMENT
The RNA‐seq datasets from this study are openly available at GEO: GSE142580.
REFERENCES
- Arnold, L. , Henry, A. , Poron, Françoise , Baba‐Amer, Y. , van Rooijen, N. , Plonquet, A. , … Chazaud, B. (2007). Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. Journal of Experimental Medicine, 204(5), 1057–1069. 10.1084/jem.20070075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold, L. , Perrin, H. , de Chanville, C. B. , Saclier, M. , Hermand, P. , Poupel, L. , … Combadiere, C. (2015). CX3CR1 deficiency promotes muscle repair and regeneration by enhancing macrophage ApoE production. Nature Communications, 6, 8972 10.1038/ncomms9972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro, E. , Sznajder, J. I. , Nader, G. A. , & Budinger, G. R. S. (2015). Muscle dysfunction in patients with lung diseases. A growing epidemic. American Journal of Respiratory and Critical Care Medicine, 191(6), 616–619. 10.1164/rccm.201412-2189OE [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartley, J. M. , Pan, S. J. , Keilich, S. R. , Hopkins, J. W. , Al‐Naggar, I. M. , Kuchel, G. A. , & Haynes, L. (2016). Aging augments the impact of influenza respiratory tract infection on mobility impairments, muscle‐localized inflammation, and muscle atrophy. Aging (Albany NY), 8(4), 620–635. 10.18632/aging.100882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brack, A. S. , & Rando, T. A. (2012). Tissue‐specific stem cells: Lessons from the skeletal muscle satellite cell. Cell Stem Cell, 10(5), 504–514. 10.1016/j.stem.2012.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantini, M. , & Carraro, U. (1995). Macrophage‐released factor stimulates selectively myogenic cells in primary muscle culture. Journal of Neuropathology and Experimental Neurology, 54(1), 121–128. 10.1097/00005072-199501000-00014 [DOI] [PubMed] [Google Scholar]
- Cantini, M. , Giurisato, E. , Radu, C. , Tiozzo, S. , Pampinella, F. , Senigaglia, D. , … Vitiello, L. (2002). Macrophage‐secreted myogenic factors: A promising tool for greatly enhancing the proliferative capacity of myoblasts in vitro and in vivo. Neurological Sciences, 23(4), 189–194. 10.1007/s100720200060 [DOI] [PubMed] [Google Scholar]
- Chan, K. S. , Mourtzakis, M. , Aronson Friedman, L. , Dinglas, V. D. , Hough, C. L. , Ely, E. W. , … Needham, D. M. (2018). Evaluating muscle mass in survivors of acute respiratory distress syndrome: A 1‐year multicenter longitudinal study. Critical Care Medicine, 46(8), 1238–1246. 10.1097/CCM.0000000000003183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazaud, B. , Sonnet, C. , Lafuste, P. , Bassez, G. , Rimaniol, A.‐C. , Poron, Françoise , … Gherardi, R. K. (2003). Satellite cells attract monocytes and use macrophages as a support to escape apoptosis and enhance muscle growth. The Journal of Cell Biology, 163(5), 1133–1143. 10.1083/jcb.200212046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales‐Medina Vicente, F. , Musher Daniel, M. , Wells George, A. , Chirinos Julio, A. , Chen, L. , & Fine Michael, J. (2012). Cardiac complications in patients with community‐acquired pneumonia. Circulation, 125(6), 773–781. 10.1161/CIRCULATIONAHA.111.040766 [DOI] [PubMed] [Google Scholar]
- Dalpke, A. H. , Opper, S. , Zimmermann, S. , & Heeg, K. (2001). Suppressors of cytokine signaling (SOCS)‐1 and SOCS‐3 are induced by CpG‐DNA and modulate cytokine responses in APCs. The Journal of Immunology, 166(12), 7082–7089. 10.4049/jimmunol.166.12.7082 [DOI] [PubMed] [Google Scholar]
- DeBerge, M. , Yeap, X. Y. , Dehn, S. , Zhang, S. , Grigoryeva, L. , Misener, S. , … Thorp, E. B. (2017). MerTK cleavage on resident cardiac macrophages compromises repair after myocardial ischemia reperfusion injury. Circulation Research, 121(8), 930–940. 10.1161/CIRCRESAHA.117.311327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giambenedetto, S. , Ciccullo, A. , Borghetti, A. , Gambassi, G. , Landi, F. , Visconti, E. , … Gasbarrini, A. (2020). Off‐label use of tocilizumab in patients with SARS‐CoV‐2 infection. Journal of Medical Virology, 76, 36–42. 10.1002/jmv.25897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos, C. , Hussain, S. N. A. , Mathur, S. , Picard, M. , Herridge, M. , Correa, J. , … Batt, J. (2016). Mechanisms of chronic muscle wasting and dysfunction after an intensive care unit stay. A Pilot Study. American Journal of Respiratory and Critical Care Medicine, 194(7), 821–830. 10.1164/rccm.201512-2344OC [DOI] [PubMed] [Google Scholar]
- Eden, E. , Navon, R. , Steinfeld, I. , Lipson, D. , & Yakhini, Z. (2009). GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics, 10, 48 10.1186/1471-2105-10-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsey, A. R. , Hennessey, P. A. , Formica, M. A. , Cox, C. , & Walsh, E. E. (2005). Respiratory syncytial virus infection in elderly and high‐risk adults. New England Journal of Medicine, 352(17), 1749–1759. 10.1056/NEJMoa043951 [DOI] [PubMed] [Google Scholar]
- Fiore, D. , Judson, R. N. , Low, M. , Lee, S. , Zhang, E. , Hopkins, C. , … Lemos, D. R. (2016). Pharmacological blockage of fibro/adipogenic progenitor expansion and suppression of regenerative fibrogenesis is associated with impaired skeletal muscle regeneration. Stem Cell Research, 17(1), 161–169. 10.1016/j.scr.2016.06.007 [DOI] [PubMed] [Google Scholar]
- Giamarellos‐Bourboulis, E. J. , Netea, M. G. , Rovina, N. , Akinosoglou, K. , Antoniadou, A. , Antonakos, N. , … Koutsoukou, A. (2020). Complex immune dysregulation in COVID‐19 patients with severe respiratory failure. Cell Host & Microbe, 27(6), 992–1000.e3. 10.1016/j.chom.2020.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard, T. D. , Self, W. H. , Edwards, K. M. , Grijalva, C. G. , Zhu, Y. , Williams, D. J. , … Jackson, J. C. (2018). Long‐term cognitive impairment after hospitalization for community‐acquired pneumonia: A prospective cohort study. Journal of General Internal Medicine, 33(6), 929–935. 10.1007/s11606-017-4301-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, D. , Contreras, O. , Rebolledo, D. L. , Espinoza, J. P. , van Zundert, B. , & Brandan, E. (2017). ALS skeletal muscle shows enhanced TGF‐beta signaling, fibrosis and induction of fibro/adipogenic progenitor markers. PLoS One, 12(5), e0177649 10.1371/journal.pone.0177649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozalo, P. L. , Pop‐Vicas, A. , Feng, Z. , Gravenstein, S. , & Mor, V. (2012). Effect of influenza on functional decline. Journal of the American Geriatrics Society, 60(7), 1260–1267. 10.1111/j.1532-5415.2012.04048.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heredia, J. E. , Mukundan, L. , Chen, F. M. , Mueller, A. A. , Deo, R. C. , Locksley, R. M. , … Chawla, A. (2013). Type 2 innate signals stimulate fibro/adipogenic progenitors to facilitate muscle regeneration. Cell, 153(2), 376–388. 10.1016/j.cell.2013.02.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herridge, M. S. , Cheung, A. M. , Tansey, C. M. , Matte‐Martyn, A. , Diaz‐Granados, N. , Al‐Saidi, F. , … Slutsky, A. S. (2003). One‐year outcomes in survivors of the acute respiratory distress syndrome. New England Journal of Medicine, 348(8), 683–693. 10.1056/NEJMoa022450 [DOI] [PubMed] [Google Scholar]
- Herridge, M. S. , Tansey, C. M. , Matté, A. , Tomlinson, G. , Diaz‐Granados, N. , Cooper, A. , … Cheung, A. M. (2011). Functional disability 5 years after acute respiratory distress syndrome. New England Journal of Medicine, 364(14), 1293–1304. 10.1056/NEJMoa1011802 [DOI] [PubMed] [Google Scholar]
- Jain, S. , Self, W. H. , Wunderink, R. G. , Fakhran, S. , Balk, R. , Bramley, A. M. , … Finelli, L. (2015). Community‐acquired pneumonia requiring hospitalization among U.S adults. New England Journal of Medicine, 373(5), 415–427. 10.1056/NEJMoa1500245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen, I. , Heymsfield, S. B. , Wang, Z. M. , & Ross, R. (2000). Skeletal muscle mass and distribution in 468 men and women aged 18–88 yr. Journal of Applied Physiology, 89(1), 81–88. 10.1152/jappl.2000.89.1.81 [DOI] [PubMed] [Google Scholar]
- Jin, R. M. , Warunek, J. , & Wohlfert, E. A. (2018). Chronic infection stunts macrophage heterogeneity and disrupts immune‐mediated myogenesis. JCI Insight, 3(18), 10.1172/jci.insight.121549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanegai, C. M. , Xi, Y. , Donne, M. L. , Gotts, J. E. , Driver, I. H. , Amidzic, G. , … Rock, J. R. (2016). Persistent pathology in influenza‐infected mouse lungs. American Journal of Respiratory Cell and Molecular Biology, 55(4), 613–615. 10.1165/rcmb.2015-0387LE [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuswanto, W. , Burzyn, D. , Panduro, M. , Wang, K. K. , Jang, Y. C. , Wagers, A. J. , … Mathis, D. (2016). Poor repair of skeletal muscle in aging mice reflects a defect in local, interleukin‐33‐dependent accumulation of regulatory T cells. Immunity, 44(2), 355–367. 10.1016/j.immuni.2016.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke, G. (2013). Biology of the TAM receptors. Cold Spring Harbor Perspectives in Biology, 5(11), 10.1101/cshperspect.a009076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke, G. , & Rothlin, C. V. (2008). Immunobiology of the TAM receptors. Nature Reviews Immunology, 8(5), 327–336. 10.1038/nri2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos, D. R. , Babaeijandaghi, F. , Low, M. , Chang, C.‐K. , Lee, S. T. , Fiore, D. , … Rossi, F. M. V. (2015). Nilotinib reduces muscle fibrosis in chronic muscle injury by promoting TNF‐mediated apoptosis of fibro/adipogenic progenitors. Nature Medicine, 21(7), 786–794. 10.1038/nm.3869 [DOI] [PubMed] [Google Scholar]
- Lim, H. Y. , Lim, S. Y. , Tan, C. K. , Thiam, C. H. , Goh, C. C. , Carbajo, D. , … Angeli, V. (2018). Hyaluronan receptor LYVE‐1‐expressing macrophages maintain arterial tone through hyaluronan‐mediated regulation of smooth muscle cell collagen. Immunity, 10.1016/j.immuni.2018.06.008 [DOI] [PubMed] [Google Scholar]
- Lu, C.‐C. , Chen, M.‐Y. , Lee, W.‐S. , & Chang, Y.‐L. (2020). Potential therapeutic agents against COVID‐19: What we know so far Journal of the Chinese Medical Association, 83(6), 534–536. 10.1097/JCMA.0000000000000318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen, L. , Zwingman, T. A. , Sunkin, S. M. , Oh, S. W. , Zariwala, H. A. , Gu, H. , … Zeng, H. (2010). A robust and high‐throughput Cre reporting and characterization system for the whole mouse brain. Nature Neuroscience, 13(1), 133–140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merly, F. , Lescaudron, L. , Rouaud, T. , Crossin, F. , & Gardahaut, M. F. (1999). Macrophages enhance muscle satellite cell proliferation and delay their differentiation. Muscle and Nerve, 22(6), 724–732. [DOI] [PubMed] [Google Scholar]
- Misharin, A. V. , Morales‐Nebreda, L. , Reyfman, P. A. , Cuda, C. M. , Walter, J. M. , McQuattie‐Pimentel, A. C. , … Perlman, H. (2017). Monocyte‐derived alveolar macrophages drive lung fibrosis and persist in the lung over the life span. Journal of Experimental Medicine, 214(8), 2387–2404. 10.1084/jem.20162152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittl, R. L. Jr , Schwab, R. J. , Duchin, J. S. , Goin, J. E. , Albeida, S. M. , & Miller, W. T. (1994). Radiographic resolution of community‐acquired pneumonia. American Journal of Respiratory and Critical Care Medicine, 149(3 Pt 1), 630–635. 10.1164/ajrccm.149.3.8118630 [DOI] [PubMed] [Google Scholar]
- Munoz‐Canoves, P. , & Michele, D. (2013). Special issue: Myogenesis: Introduction. FEBS Journal, 280(17), 3979 10.1111/febs.12454 [DOI] [PubMed] [Google Scholar]
- Murugan, R. , Karajala‐Subramanyam, V. , Lee, M. , Yende, S. , Kong, L. , Carter, M. , … Kellum, J. A. (2010). Acute kidney injury in non‐severe pneumonia is associated with an increased immune response and lower survival. Kidney International, 77(6), 527–535. 10.1038/ki.2009.502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz, J. R. , Neuzil, K. M. , Rue, T. C. , Zhou, H. , Shay, D. K. , Cheng, P.‐Y. , … Goss, C. H. (2013). Population‐based incidence estimates of influenza‐associated respiratory failure hospitalizations, 2003 to 2009. American Journal of Respiratory and Critical Care Medicine, 188(6), 710–715. 10.1164/rccm.201212-2341OC [DOI] [PubMed] [Google Scholar]
- Paolicelli, R. C. , Bolasco, G. , Pagani, F. , Maggi, L. , Scianni, M. , Panzanelli, P. , … Gross, C. T. (2011). Synaptic pruning by microglia is necessary for normal brain development. Science, 333(6048), 1456–1458. 10.1126/science.1202529 [DOI] [PubMed] [Google Scholar]
- Peteranderl, C. , Morales‐Nebreda, L. , Selvakumar, B. , Lecuona, E. , Vadász, I. , Morty, R. E. , … Herold, S. (2016). Macrophage‐epithelial paracrine crosstalk inhibits lung edema clearance during influenza infection. J Clin Invest, 126(4), 1566–1580. 10.1172/JCI83931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfoh, E. R. , Wozniak, A. W. , Colantuoni, E. , Dinglas, V. D. , Mendez‐Tellez, P. A. , Shanholtz, C. , … Needham, D. M. (2016). Physical declines occurring after hospital discharge in ARDS survivors: A 5‐year longitudinal study. Intensive Care Medicine, 42(10), 1557–1566. 10.1007/s00134-016-4530-1 [DOI] [PubMed] [Google Scholar]
- Pluvinage, J. V. , Haney, M. S. , Smith, B. A. H. , Sun, J. , Iram, T. , Bonanno, L. , … Wyss‐Coray, T. (2019). CD22 blockade restores homeostatic microglial phagocytosis in ageing brains. Nature, 568(7751), 187–192. 10.1038/s41586-019-1088-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radigan, K. A. , Nicholson, T. T. , Welch, L. C. , Chi, M. , Amarelle, L. , Angulo, M. , … Budinger, G. R. S. (2019). Influenza A virus infection induces muscle wasting via IL‐6 regulation of the E3 ubiquitin ligase atrogin‐1. The Journal of Immunology, 202(2), 484–493. 10.4049/jimmunol.1701433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saclier, M. , Yacoub‐Youssef, H. , Mackey, A. L. , Arnold, L. , Ardjoune, H. , Magnan, M. , … Chazaud, B. (2013). Differentially activated macrophages orchestrate myogenic precursor cell fate during human skeletal muscle regeneration. Stem Cells, 31(2), 384–396. 10.1002/stem.1288 [DOI] [PubMed] [Google Scholar]
- Sala, D. , Cunningham, T. J. , Stec, M. J. , Etxaniz, U. , Nicoletti, C. , Dall’Agnese, A. , … Sacco, A. (2019). The Stat3‐Fam3a axis promotes muscle stem cell myogenic lineage progression by inducing mitochondrial respiration. Nature Communications, 10(1), 1796 10.1038/s41467-019-09746-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaum, N. , Karkanias, J. , Neff, N. F. , May, A. P. , Quake, S. R. , Wyss‐Coray, T. , … Principal, I. (2018). Single‐cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature, 562(7727), 367–372. 10.1038/s41586-018-0590-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segawa, M. , Fukada, S. , Yamamoto, Y. , Yahagi, H. , Kanematsu, M. , Sato, M. , … Miyagoesuzuki, Y. (2008). Suppression of macrophage functions impairs skeletal muscle regeneration with severe fibrosis. Experimental Cell Research, 314(17), 3232–3244. 10.1016/j.yexcr.2008.08.008 [DOI] [PubMed] [Google Scholar]
- Tate, J. A. , Snitz, B. E. , Alvarez, K. A. , Nahin, R. L. , Weissfeld, L. A. , Lopez, O. , … Yende, S. (2014). Infection hospitalization increases risk of dementia in the elderly. Critical Care Medicine, 42(5), 1037–1046. 10.1097/CCM.0000000000000123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira, C. F. , Zamuner, S. R. , Zuliani, J. P. , Fernandes, C. M. , Cruz‐Hofling, M. A. , Fernandes, I. , … Gutierrez, J. M. (2003). Neutrophils do not contribute to local tissue damage, but play a key role in skeletal muscle regeneration, in mice injected with Bothrops asper snake venom. Muscle and Nerve, 28(4), 449–459. 10.1002/mus.10453 [DOI] [PubMed] [Google Scholar]
- Thannickal, V. J. , Murthy, M. , Balch, W. E. , Chandel, N. S. , Meiners, S. , Eickelberg, O. , … Budinger, G. R. S. (2015). Blue journal conference. Aging and susceptibility to lung disease. American Journal of Respiratory and Critical Care Medicine, 191(3), 261–269. 10.1164/rccm.201410-1876PP [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierney, M. T. , Aydogdu, T. , Sala, D. , Malecova, B. , Gatto, S. , Puri, P. L. , … Sacco, A. (2014). STAT3 signaling controls satellite cell expansion and skeletal muscle repair. Nature Medicine, 20(10), 1182–1186. 10.1038/nm.3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topinkova, E. (2008). Aging, disability and frailty. Annals of Nutrition & Metabolism, 52(Suppl 1), 6–11. 10.1159/000115340 [DOI] [PubMed] [Google Scholar]
- Traylor, Z. P. , Aeffner, F. , & Davis, I. C. (2013). Influenza A H1N1 induces declines in alveolar gas exchange in mice consistent with rapid post‐infection progression from acute lung injury to ARDS. Influenza and Other Respiratory Viruses, 7(3), 472–479. 10.1111/j.1750-2659.2012.00414.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uezumi, A. , Fukada, S. , Yamamoto, N. , Takeda, S. , & Tsuchida, K. (2010). Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nature Cell Biology, 12(2), 143–152. 10.1038/ncb2014 [DOI] [PubMed] [Google Scholar]
- Vagnozzi, R. J. , Maillet, M. , Sargent, M. A. , Khalil, H. , Johansen, A. K. Z. , Schwanekamp, J. A. , … Molkentin, J. D. (2020). An acute immune response underlies the benefit of cardiac stem cell therapy. Nature, 577(7790), 405–409. 10.1038/s41586-019-1802-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga, T. , Mounier, R. , Horvath, A. , Cuvellier, S. , Dumont, F. , Poliska, S. , … Chazaud, B. (2016). Highly dynamic transcriptional signature of distinct macrophage subsets during sterile inflammation, resolution, and tissue repair. The Journal of Immunology, 196(11), 4771–4782. 10.4049/jimmunol.1502490 [DOI] [PubMed] [Google Scholar]
- Walsh, C. J. , Batt, J. , Herridge, M. S. , & Dos Santos, C. C. (2014). Muscle wasting and early mobilization in acute respiratory distress syndrome. Clinics in Chest Medicine, 35(4), 811–826. 10.1016/j.ccm.2014.08.016 [DOI] [PubMed] [Google Scholar]
- Wang, Y. X. , & Rudnicki, M. A. (2011). Satellite cells, the engines of muscle repair. Nature Reviews Molecular Cell Biology, 13(2), 127–133. 10.1038/nrm3265 [DOI] [PubMed] [Google Scholar]
- Weinhard, L. , di Bartolomei, G. , Bolasco, G. , Machado, P. , Schieber, N. L. , Neniskyte, U. , … Gross, C. T. (2018). Microglia remodel synapses by presynaptic trogocytosis and spine head filopodia induction. Nature Communications, 9(1), 1228 10.1038/s41467-018-03566-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona, S. , Kim, K.‐W. , Wolf, Y. , Mildner, A. , Varol, D. , Breker, M. , … Jung, S. (2013). Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity, 38(1), 79–91. 10.1016/j.immuni.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Xiao, Z. , Qu, C. , Cui, W. , Wang, X. , & Du, J. (2014). CD8 T cells are involved in skeletal muscle regeneration through facilitating MCP‐1 secretion and Gr1(high) macrophage infiltration. The Journal of Immunology, 193(10), 5149–5160. 10.4049/jimmunol.1303486 [DOI] [PubMed] [Google Scholar]
- Zhang, S. , Weinberg, S. , DeBerge, M. , Gainullina, A. , Schipma, M. , Kinchen, J. M. , … Thorp, E. B. (2019). Efferocytosis fuels requirements of fatty acid oxidation and the electron transport chain to polarize macrophages for tissue repair. Cell Metabolism, 29(2), 443–456.e445. 10.1016/j.cmet.2018.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, W. , Lu, H. , Wang, X. , Ransohoff, R. M. , & Zhou, L. (2016). CX3CR1 deficiency delays acute skeletal muscle injury repair by impairing macrophage functions. The FASEB Journal, 30(1), 380–393. 10.1096/fj.14-270090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, F. , Yu, T. , Du, R. , Fan, G. , Liu, Y. , Liu, Z. , … Cao, B. (2020). Clinical course and risk factors for mortality of adult inpatients with COVID‐19 in Wuhan, China: A retrospective cohort study. Lancet, 395(10229), 1054–1062. 10.1016/s0140-6736(20)30566-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1–S5
Table S1
Table S2
Table S3
Table S4
Data Availability Statement
The RNA‐seq datasets from this study are openly available at GEO: GSE142580.


