Abstract
The gene coding for antimicrobial peptides produced by probiotic Lactobacillus reuteri KUB-AC5 located on the cloned DNA fragment I-C46 containing 2 open reading frames I-C46-F2.1 and I-C46-F2.2 were designed for strain-specific primers P1 and P2, respectively, and assessed by real-time quantitative polymerase chain reaction. According to the obtained results, primer P1 has limited strain specificity. Primer P2 exhibited high efficacy and specificity at annealing temperature of 70°C while P1 annealed at 57°C causing nonspecific bands. Hence, P2 was selected for quantitative polymerase chain reaction assay by isothermal annealing and extension reaction at high temperature of 70°C resulting in linearity for its DNA sequences ranging from 102 to 107 target copy numbers per assay, and displaying a detection limit of 6.17 log cfu/g of cecal digesta. Using spike testing, this system was able to detect 7.88 ± 0.06 to 11.78 ± 0.06 log copy number/g of digesta, higher than cultivation assay at about 1 log cfu/g, with good correlation of 0.99. These results suggested possible detection of strain KUB-AC5 in the gastrointestinal tract of chicken to evaluate the efficacy and persistence of a probiotic strain which requires correct inclusion rates in the feed.
Key words: Lactobacillus reuteri KUB-AC5, antimicrobial peptide, strain-specific primer, quantitative polymerase chain reaction
INTRODUCTION
With the recent rapid increase in global population, the food and agro-industry must expand and respond to increased consumer demands for boiler chicken and poultry products. Infection by pathogens such as Escherichia coli and Salmonella sp. causes severe losses in chicken production. Antibiotics have been used as growth promoters to solve these problems; however, many countries have banned the use of antibiotics in animal feed due to drug residues which promote the emergence of antibiotic-resistant strains as obstacles in the treatment of various bacterial infections. Lactic acid bacteria (LAB) are increasingly applied in the food and feed industry as probiotics and producers of metabolites such as small antimicrobial peptides (AMPs). In our previous study, Lactobacillus reuteri KUB-AC5 isolated from chicken intestine exerted antimicrobial substances exhibiting broad-spectrum activity against both Gram-positive and Gram-negative bacterial strains including Escherichia coli and Salmonella sp. (Nitisinprasert et al., 2000; Nakphaichit et al., 2011). This strain also promoted growth of chicken supplied with feed supplement of 105 cfu/g for a week and suppressed five pathogenic genera in chicken ilia comprising Klebsiella, Chryseobacterium, Citrobacter, Aeromonas, and Acinetobacter in the order Campylobacterales. Abundance of several species of lactobacilli including Lb. reuteri increased as a result of probiotic treatment (Nakphaichit et al., 2011).
However, it proved difficult to assess and quantify the concentration of specific spiked probiotic strains, since Lactobacilli strains are closely related with high sequence homology in variable regions of the 16S rRNA gene sequence (Yeung et al., 2002). To evaluate the efficacy and persistence of a probiotic strain requires correct inclusion rates in the feed, with the ability to trace the introduced strain through the gastrointestinal tract (GIT). Methods to monitor the probiotic should be strain-specific. Colony polymerase chain reaction—the random amplified polymorphic DNA has been applied to determine the survivability of Lb. reuteri KUB-AC5 from various compartments of GIT; however, this method is tedious and time consuming (Rodklongtan et al., 2014). Quantitative polymerase chain reaction (qPCR) is a technique that has been used to detect several bacterial species in food (De Martin et al., 2007) or feces (Rintrila et al., 2004). It is highly sensitive and enables quantification of microorganisms with low abundance within an environmental sample.
The challenge in qPCR development involves primer design that specifically targets species or strains of interest, despite the presence of closely related bacteria. To date, only a few studies have reported the use of qPCR to identify and quantify probiotics in vivo. Sattler et al. (2014) developed a quantification method for the probiotic Lb. reuteri DSM 16,350 in poultry feed and intestine by designing primers based on a genomic sequence of the strain derived from suppression subtractive hybridization with the type strain Lb. reuteri DSM 20,016. However, to date, no primers designed from specific AMPs have been reported. As mentioned above, the probiotic strain KUB-AC5 has also been isolated from chicken intestine and can produce an interesting AMP with wide inhibitory activity. Tangthong (2012) has successfully cloned the gene encoding AMP from Lb. reuteri KUB-AC5 into pNZ307and expressed in Escherichia coli DH5α to obtain the recombinant strain E. coli ACE-C46 exerting extracellular inhibitory activities of 15 mm against indicator strain Salmonella Enteritidis S003. Its recombinant plasmid contained an insert DNA I-C46 (ID number 2,253,028) which consisted of 2 open reading frames (ORF) of a small ORF I-C46-F2.1 (153 bp) and big ORF I-C46-F2.2 (630 bp). Based on the molecular weight of active AMP of 4.7 kDa analysed by MALDITOF Mass, the structural gene coding for AMP potentially located on the ORF-I-C46-F2.1. Moreover, the nucleotide sequence of both ORFs showed low similarity to other bacterial genes in the NCBI database. Therefore, here, a specific primer sequence from the gene coding for AMP was designed to evaluate strain KUB-AC5 in broiler chickens using qPCR. Presence and amount of probiotic strain KUB-AC5 were also determined in a compartment of chicken GIT.
MATERIALS AND METHODS
Bacterial Strains and Culture Conditions
Recombinant strain E. coli ACE-C46 containing recombinant plasmid was used as a source of the target gene encoded AMP to design a specific primer for polymerase chain reaction (PCR) and as a positive control for PCR analysis. Real-time PCR or qPCR analysis was performed using the probiotic strain Lb. reuteri KUB-AC5 as a tester. Fifteen other bacterial strains including Lb. fermentum KUB-D18, Lb. johnsonii KUB-NN19, Lb. plantarum (KL-1 and KUB-SP1-3), Lb. reuteri (KUB-AC16 and KUB-D28), Lb. salivarius (KL- D4, KUB-AC21 and KUB-I49), Lb. sakei JCM1157, Lactococcus lactis JCM5805, Pediococcus acidilactici KUB-M6, Pd. pentosaceus JCM5885T, E. coli E010 and S. Enteritidis S003, were used for specificity testing. All strains were grown in MRS agar (Difco Laboratories, USA) at 37°C for 24 h, except for E. coli E010 and S. Enteritidis S003 which were cultivated in nutrient broth (Merck, Germany) with agitation of 200 rpm for 24 h. All bacterial strains were kept in the culture collection of the Department of Biotechnology, Faculty of Agro-Industry, Kasetsart University and maintained as frozen stocks at −80°C in the presence of 20% glycerol.
Primer Design
Two sets of strain-specific primers were synthesized by 1st Base Laboratories Sdn Bhd, Malaysia and designed based on the nucleotide sequence of gene encoding for AMP of Lb. reuteri KUB-AC5. The specificity of primers was verified in silico with FASTA and the NCBI BLAST programs (http://blast.ncbi.nlm.nih.gov/Blast.cgi), with their qualities checked by OligoAnalyzer 3.1 program (http://sg.idtdna.com/calc/analyzer). All primers used are listed in Table 1.
Table 1.
Primer properties.
| Primers | Oligonucleotide sequence (5′-3′) | %GC | Tm (°C) | Length (bp) | Amplicon size (bp) |
|---|---|---|---|---|---|
| P1 (from ORF-1) | 169 | ||||
| Forward primer P1-F | CATGCCATGGATGATTATGTTGAACGGTTTTG | 40.6 | 60.2 | 32 | |
| Reverse primer P1-R | CGGCTCGAGTGAATCAGGCCAACAAACAA | 51.7 | 64.2 | 29 | |
| P2 (from ORF-2) | 189 | ||||
| Forward primer P2-F | TCGCTCACGGCTGTTAGGACA | 54.5 | 60.6 | 21 | |
| Reverse primer P2-R | AGCACTCCACGTTGCCACA | 57.9 | 60.0 | 19 |
DNA Extraction
Total DNA was isolated from pure cultures of various test strains using QIAamp Stool Mini Kit (Qiagen, Hilden, Germany) in combination with the bead-beating method (Sakamoto et al., 2011) as a template to determine specificity of designed primers and qPCR analysis. In brief, bacterial cells re-suspended in 900 µL of phosphate buffer saline solution (PBS; pH 8.0) were transferred in 2 mL screw-cap polypropylene tubes containing 0.3 g of zirconia beads (0.1 mm in diameter; Biospec, USA) and 300 µ L of PCI (Phenol Chloroform Isoamyl; 25:24:1) and cells were broken by a minibead beater 3110BX (Biospec, USA) for 3 min at 4800 rpm. Then, the broken cells suspension was kept on ice for further DNA extraction according to the protocol of QIAamp Stool Mini Kit. Total DNA solution obtained was kept at −80°C until required for use. Extraction of plasmid DNA from recombinant ACE-C46 was performed using a NucleoSpin Plasmid Mini Kit (NucleoSpin® Plasmid/Plasmid (NoLid), Berlin, Germany).
PCR and qPCR Assays
Two primer sets were tested for their specificity and efficiency by an annealing temperature gradient PCR with total DNA of each target strain or recombinant plasmid pACE-C46 used as a template. Total reaction volume of 10 µL contained 50 ng of DNA template, 0.2 µL; 10 µmol of each primer, 0.4 μL; 2.5 mM each dNTP, 1 μL; 2.5 U/µL of i-TaqTM Plus DNA Polymerase (JH Science), 0.1 µL; 10X Plus Buffer with MgSO4, 1 µL with MilliQ water added to obtain final volume of 10 μL. Amplification conditions consisted of initial denaturation at 94°C for 2 min followed by 34 cycles of 94°C for 30 s, gradient annealing temperature at 54 to 62°C and 55 to 70°C for 30 s for primers P1 and P2, respectively, and extension at 72°C for 20 s, followed by a final extension step at 72°C for 5 min. Amplified fragments were analyzed by electrophoresis in a 1% agarose gel and 1X TAE buffer at 100 volts for 30 min. DNA was visualized under a UV light tray Gel Doc System (Bio-Rad Laboratories, Hercules, CA, USA) using 100 bp plus DNA Ladder (Thermo Scientific) as a marker.
Efficiency of the primers was tested using a serial dilution of genomic DNA from the strain KUB-AC5 by qPCR assay (LightCyclerR 480, Roche, Germany). Total reaction volume of 20 µL contained 50 ng/µl of DNA template, 2 µL; 10 µmol of each primer, 0.4 µL; LightCycler® 480 SYBR Green master (Roche, Germany), 10 µL and free nuclease water added to obtain the final volume of 20 µL. Amplification conditions of primer P2 consisted of initial denaturation at 95°C for 5 min followed by 45 cycles of 95°C for 10 s, annealing and extension step combined at 70°C for 45 s and elongation at 72°C for 5 min.
Standard Curves for Quantification of Strain KUB-AC5
To quantify the number of Lb. reuteri KUB-AC5 in chicken ceca, a standard curve of qPCR was created. Total DNA was extracted from 1 mL of cell suspension (2.99 × 1011 cfu/mL) according to the method of QIAamp Stool Mini Kit combined with bead-beating method mentioned elsewhere and serially diluted in nuclease-free deionized water. Standard DNA was amplified by qPCR in duplicates, applied at the optimal condition as for primer specificity testing and generated by plotting threshold cycle values versus equivalent log DNA concentration of 10 to 107. Amplification efficiency was determined by the slope of the standard curves and calculated based on the equation E = (10 −1/slope) × 100.
Preparation and Quantification of Lb. reuteri KUB-AC5 Contaminated in Cecal Digesta
Lb. reuteri KUB-AC5 free cecum was spiked with strain KUB-AC5 at cell concentration of 2.9 × 105–2.9 × 1010 cfu/g of cecal digesta. Genomic DNA was extracted from about 200 mg digesta by mixing well with PBS (pH 8.0) in a stomacher® 400 circulator (Seward Ltd, Worthing, UK) for 3 min at high speed according to the modified method of Fujimoto et al. (2008) and further performing DNA extraction by the method of QIAamp Stool Mini Kit combined with bead-beating method mentioned. Quantification of the strain KUB-AC5 was assessed from chicken GIT by qPCR as cfu/g according to the optimized condition.
Quantification of Lb. reuteri KUB-AC5 in Chicken GIT Sample
Chicken fed in natural and closed farming systems were purchased from the market and used as sample sources to determine possible detection of the strain KUB-AC5. Determination of strain KUB-AC5 was assessed in triplicate from both ileal and cecal digesta by qPCR analysis according to the optimized condition.
RESULTS
Primer Design
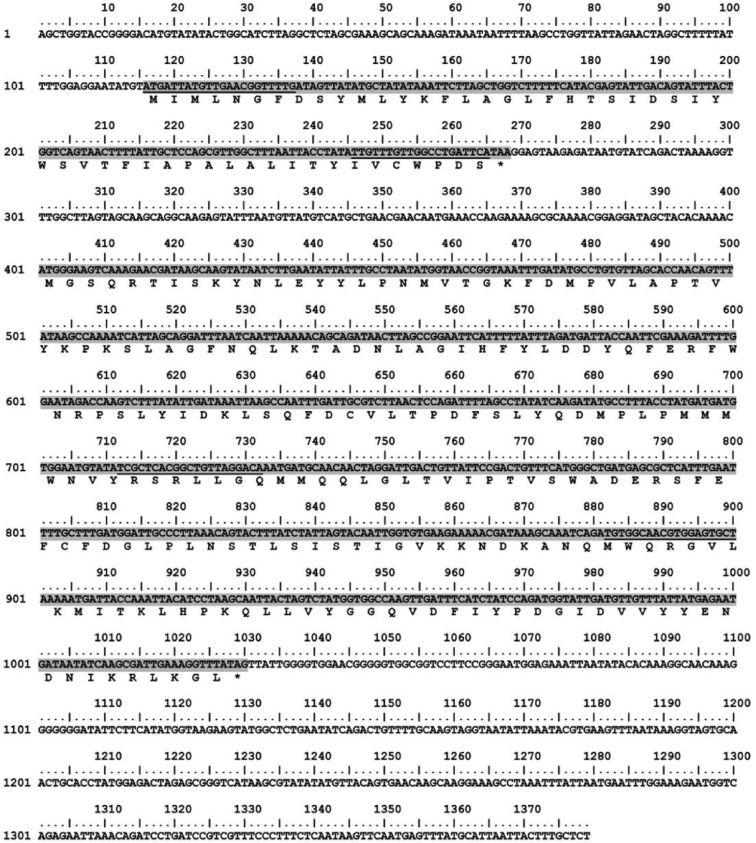
According to an experimental study concerning molecular cloning of gene coding for AMP from Lb. reuteri KUB-AC5, the recombinant clone E. coli ACE-C46 obtained consisted of 2 ORFs as ORF-1 and ORF-2 containing 153 and 630 nucleotides encoding 50 and 209 amino acids, respectively (Figure 1). ORF-1 is a tentative active AMP based on its molecular mass of about 4.7 kDa named. While the output expressed by the gene located on ORF-2 was still unknown. Using NCBI BLAST analysis, the amino acid sequence deduced from ORF-1 showed low identity to the predicted protein accession no. ZP_0,555,3516.1 of Lb. coleohominis 101–4-CHN at 35%, while the nucleotide sequence of ORF-2 showed low identity to bacteria accession No. CP016953.1 at 30%. Primers P1 and P2 were designed based on high variable regions of ORF-1 and ORF-2, respectively. Their characteristics and locations are shown in Table 1 and Figure 1, respectively. Both primers had high annealing temperature at up to 60°C or more, providing suitable annealing temperatures of P1 and P2 at 57 and 55°C, respectively by in silico analysis.
Figure 1.
Nucleotide and amino acid sequences of ORF-1 and ORF-2 encoded AMP from Lb. reuteri KUB-AC5 and position of primers P1 and P2 designed based on nucleotide sequences of both ORFs; consisting of ORF-1 and ORF-2 at 116 to 268 bp and 401 to 1030 bp labeled in gray, respectively. An underline under both ORFs indicates the position of primers P1 and P2 located on ORF-1 and ORF-2, respectively.
Specificity of Primers P1 and P2
Primers P1 and P2 designed from ORF-1 and ORF-2 were determined for their specificities against 14 LAB strains, including the target strain KUB-AC5 and 2 g negative pathogens, by PCR assay under annealing conditions of 57 and 55°C, respectively. The 11 species tested were the representative species existing in GI-tract of chicken from previous findings except Lb. sakei JCM1157, Lactococcus lactis JCM5805 and Pd. pentosaceus JCM5885T. These 3 latter species which may involve in fermentation process were included in case of other application in the future. Another 2 strains of Lb. reuteri were tested in parallel to confirm primer specificity at strain level. We have included E. coli and Salmonella Enteritidis isolated from intestines of chicken in this experiment as well because these two species were often found and caused contamination in the carcass. For primer P1, a DNA band 169 bp in length appeared when DNA templates from the positive control of the recombinant plasmid DNA pI-C46 and target strain KUB-AC5 were applied. However, nonspecific DNA bands amplified by strains Lb. salivarius KUB-I49, Lb. sakei JCM1157 and Lc. lactis JCM5805 also appeared (data not shown), These results implied that primer P1 showed only 81.25% specificity of total test strains. To solve this problem, a high gradient temperature of 59.3 to 63.9°C was performed. However, the nonspecific band from strain KUB-I49 still appeared.
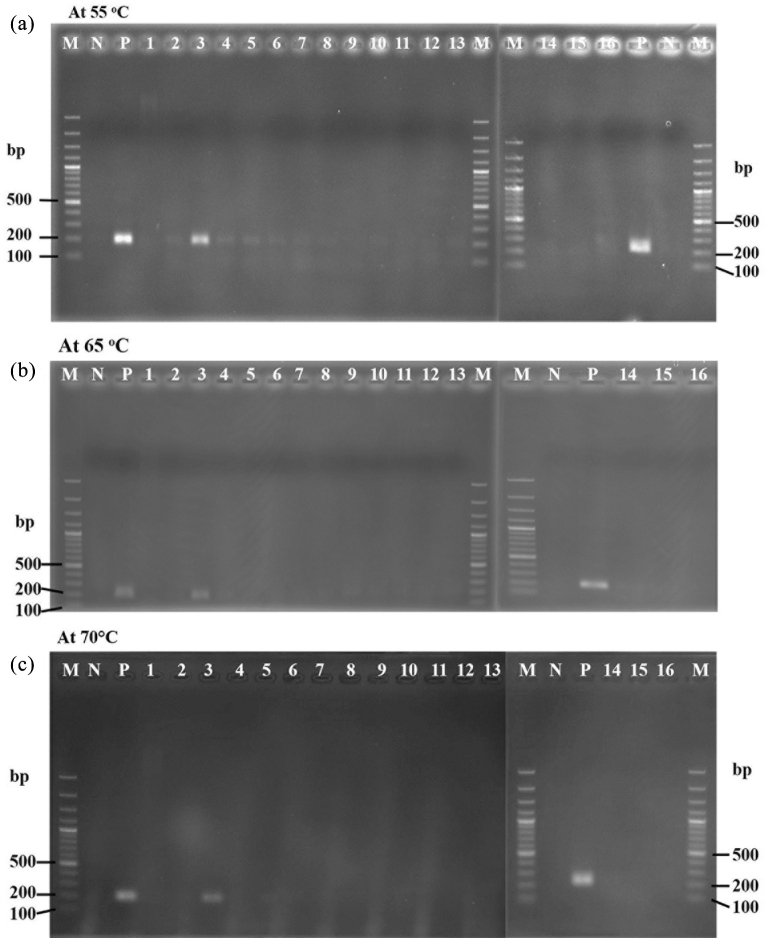
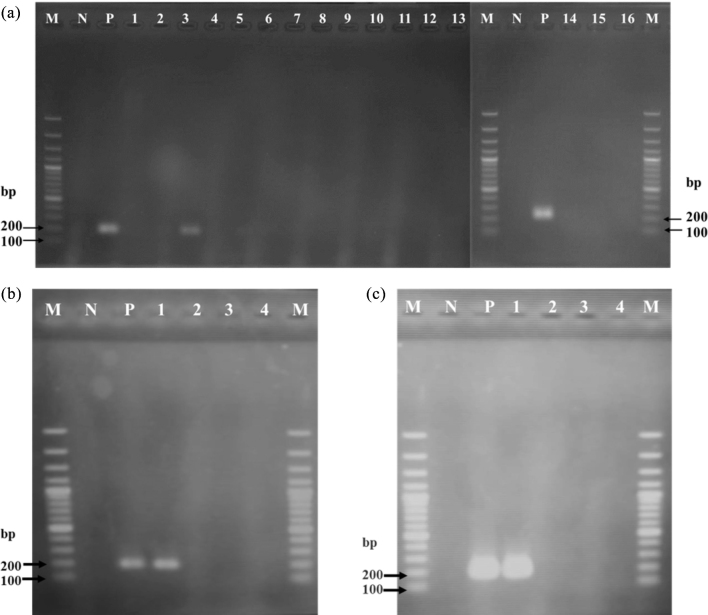
By contrast, primer P2 was specific to strain KUB-AC5, with appearance of strongly specific amplified PCR products of 189 bp at low PCR annealing temperature of 55°C. However, faint bands still occurred against several test strains (Figure 2 a). Therefore, high gradient annealing at 60 to 70°C was performed and resulted in high specificity to only Lb. reuteri KUB-AC5 at 65–70°C (Figure 2b and 2c). Since high annealing temperature of 70°C was close to DNA elongation temperature of 72°C, isothermal amplification of the annealing and extension step for 45 s was tested at 70°C. This resulted in successfully amplified PCR products shown in Figure 3. In addition, the specificity of primer P2 was tested for different strain of Lb. reuteri, KUB-AC5, KUB-AC16, and KUB-D28 by both conventional PCR and qPCR analysis. The strain KUB-AC16 was isolated from the same source of the target strain KUB-AC5 while the strain KUB-D28 was from intestine of duck. By both techniques, it was clearly shown that both strain KUB-AC16 and KUB-D28 showed negative results while only the right PCR products amplified by the target strain KUB-AC5 had occurred as shown in Fig. 3. It elucidated that the primer P2 was specific to only the strain Lb. reuteri KUB-AC5. Therefore, primer P2 was selected for further study.
Figure 2.
Gradient temperature on specificity test of primer P2 by PCR assay. (a), amplicon PCR at 55°C, (b) and (c) amplicon PCR performed by gradient annealing temperature of 65 and 70°C, respectively. Lane M, 100 bp DNA plus; N, negative control of free nuclease DI water; P, positive control of recombinant DNA C46; lane 1, E. coli E010; lane 2, S. Enteritidis S003; lane 3, Lb. reuteri KUB-AC5; lane 4, Lb. reuteri KUB-AC16; lane 5, Lb. salivarius KUB-AC21; lane 6, Lb. salivarius KL-D4; lane 7, Lb. fermentum KUB-D18; lane 8, Lb. reuteri KUB-D28; lane 9, Lb. salivarius KUB-I49; lane 10, Lb. plantarum KL1; lane11, Pd. acidilactici KUB-M6; lane 12, Lb. plantarum KUB-SP1–3; lane 13, Lb. johnsonii KUNN-19–2; lane 14, Lb. sakei JCM1157; lane 15, Lc. lactis JCM5805 and lane 16, Pd. pentosaceus JCM5885T.
Figure 3.
Specificity test of primer P2 by polymerase chain reaction (PCR) assay using optimized condition of combined annealing and extension temperature at 70°C for 45 s. (a), all strain of bacterial test: lane M, 100 bp DNA plus; N, negative control of free nuclease DI water; P, positive control of recombinant DNA C46; lane 1, E. coli E010; lane 2, S. Enteritidis S003; lane 3, Lb. reuteri KUB-AC5; lane 4, Lb. reuteri KUB-AC16; lane 5, Lb. salivarius KUB-AC21; lane 6, Lb. salivarius KL-D4; lane 7, Lb. fermentum KUB-D18; lane 8, Lb. reuteri KUB-D28; lane 9, Lb. salivarius KUB-I49; lane 10, Lb. plantarum KL1; lane 11, Pd. acidilactici KUB-M6; lane 12, Lb. plantarum KUB-SP1–3; lane 13, Lb. johnsonii KUNN-19–2; lane 14, Lb. sakei JCM1157; lane 15, Lc. lactis JCM5805 and lane 16, Pd. pentosaceus JCM5885T. (b) and (c) were specificity test of Lb. reuteri strain by conventional PCR and qPCR assay. Lane M, 100 bp DNA plus; N, negative control of free nuclease DI water; P, positive control of recombinant DNA C46; lane 1, KUB-AC5; lane 2, KUB-AC16; lane 3, KUB-D28; lane 4, Lb. salivarius KUB-AC21 used as negative control.
Efficiency of Primer P2
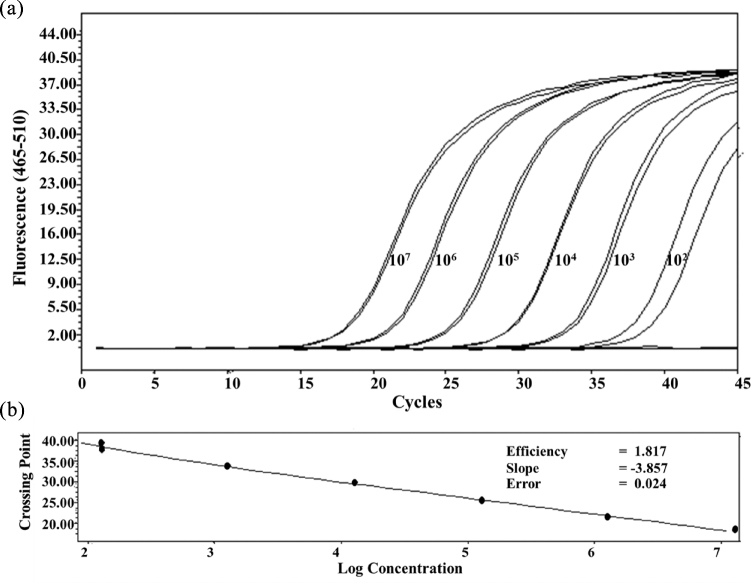
Quantification and detection limits of qPCR assay were determined using purified genomic DNA isolated from the strain KUB-AC5. Amplification reactions were performed for a range of duplicated dilution of DNA equivalent from about 12.9 up to 1.3 × 107 copy number (estimated by a relative genome size of 2.19 MB, analyzed by Illumina de novo HiSeq 4000 platform, Beijing Genomics Institute, China). Logarithms of DNA concentration were plotted against their relative crossing points to obtain the standard curve. In general, PCR efficiency (E) is calculated from the slope of its standard curve by the formula E = (10−1/slope) × 100 to provide the optimal value of 2.0 (where slope of the regression curve is −3.32) which presented 100% doubling with each cycle. Here, the E value calculated from 102 to 107 target copy numbers, was 1.817 or 90.85% doubling with each cycle (Figure 4). Its slope was −3.857 indicating that the assay was suitable to detect the target gene when 100 or more copy numbers were present and therefore could be employed for quantification of Lb. reuteri KUB-AC5. Only one position of gene coding for AMP on a genome was identified by metagenomic analysis which implied that quantification of Lb. reuteri KUB-AC5 cell numbers were equivalent to copy numbers.
Figure 4.
Quantitative polymerase chain reaction (qPCR) amplification of genomic DNA serial dilution from Lb. reuteri KUB-AC5 equivalent to 102 to 107 target copy numbers. (a), amplification plot of template DNA concentrations duplicated by primer P2; (b), standard curve of serial dilution of genomic DNA.
Evaluation of Lb. reuteri KUB-AC5 in Cecal Digesta Environment by qPCR Assay
Primer P2 was applied to analyze the required number of Lb. reuteri KUB-AC5 spiked into the cecum to create contaminated KUB-AC5 in cecal digesta at various concentrations of 105 to 1010 cfu/g. Both the control (without Lb. reuteri KUB-AC5 supplementation) and minimum cell concentrations of 105 cfu/g could not be determined by qPCR since they were less than the detection limit of 3.08 log copy number/assay. Cells spiked at 106 to 1010 cfu/g were detected at higher copy numbers or cell numbers of each relative cell concentration, with correlation (r2) equal to 0.9996 (Figure 5). Comparison of cell numbers analyzed by viable cell count and qPCR assay showed that level of Lb. reuteri KUB-AC5 as cfu/g of cecum was about 1 log higher when analyzed by qPCR. However, spike testing of the KUB-AC5 strain, specific to the qPCR system, indicated that quantification did not significantly interfere with the presence of closely related species due to its linearity and r2 values were satisfactory over 106 to 1010 cfu/g.
Figure 5.
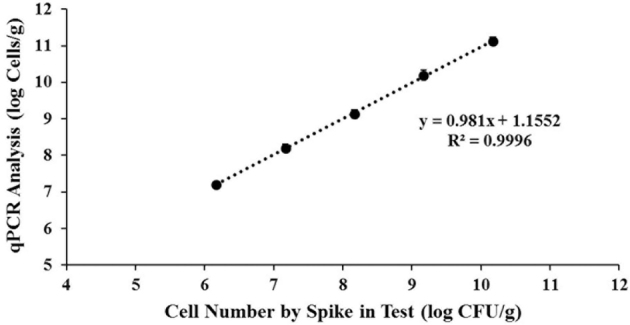
Correlation of cell concentration of Lb. reuteri KUB-C5 analyzed by viable cell count and qPCR.
Possible Detection of Lb. reuteri KUB-AC5 in Chicken GIT from Different Sources by qPCR Assay
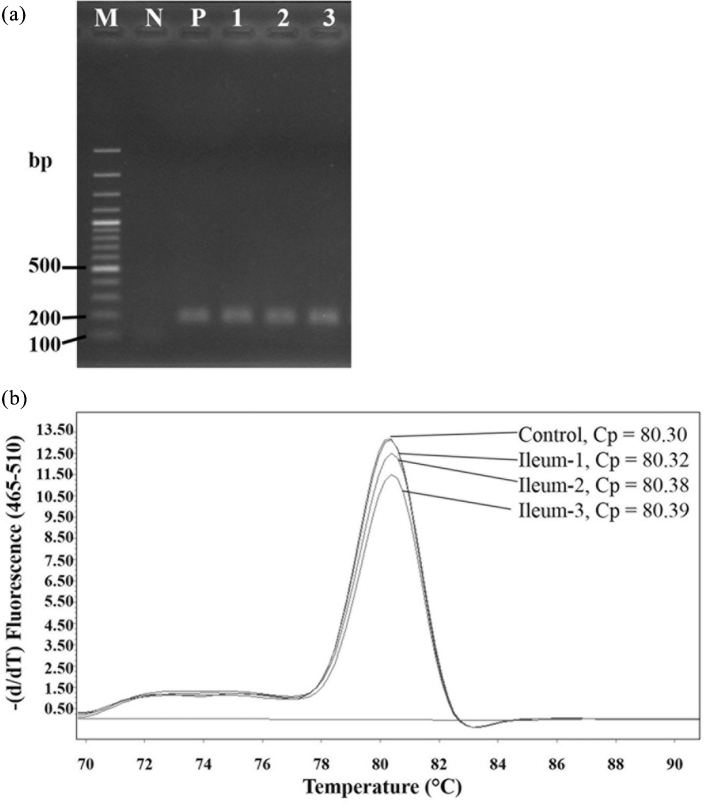
Primer P2 efficiently detected strain KUB-AC5 under spiking in test condition. This strain could not be detected from cecum control treatment (without spiking). In general, LAB belonging to families Enterococcaceae, Lactobacillaceae and Streptococcaceae were abundant in the ileum whereas Lachnospiraceae, Ruminococcaceae and Erysipelotrichaceae were more enriched in the cecum (Nakphaichit et al., 2019). Due to the high abundance of Lactobacillaceae in the ileum but low levels in the cecum, it was possible to detect this strain from the ileum using this technique. More intestinal samples of both ileum and cecum from seven chickens of natural and closed farming systems were tested. Only one ileum sample-1 from natural farming systems was detected at 8.09 log copy number/g while samples of other organs were undetectable. To confirm this positive result, agarose gel electrophoresis and melting curve analysis of amplified PCR products from natural farming system samples 1, 2, and 3 were analyzed and presented in Figure 6. Sizes of amplified PCR products from three samples were similar to the positive control while Ct, shown by the melting curve of sample-1 (80.32), was close to the control (80.30) but different from the other two samples (80.38 and 80.39). Therefore, the strain showing positive results had a profile very similar to strain KUB-AC5, suggesting that it might be identical.
Figure 6.
Characters of amplified PCR products of ileum samples 1, 2, and 3 from chicken fed by natural farming system. (a), size of amplified PCR product analyzed by gel electrophoresis, M, 100 bp DNA plus; N, negative control of free nuclease dI water; P, positive control of genomic DNA from Lb. reuteri KUB-AC5; 1, Ileum-1; 2, Ileum-2; 3, Ileum-3; (b), melting curve analysis by real time PCR.
DISCUSSION
Lb. reuteri KUB-AC5 anti-pathogenic activity has been previously tested in chicken both in vitro (Nitisinprasert et al., 2000) and in vivo (Nakphaichit et al., 2011). To follow up the existence of this strain in the GIT, DNA sequence coding for AMP, unique for the genome of probiotic strain KUB-AC5 was designed for primers P1 and P2 based on their high variable regions. Primer P2-targeted PCR was superior to P1 due to its higher specificity at annealing temperature of 70°C, providing isothermal annealing and extension PCR reaction. Specificity of primer P2 was shown in both conventional and qPCR assay when DNA isolated from pure bacterial culture was used.
High efficiency of this assay was proposed using primer P2 for identification of Lb. reuteri at strain level. The assay fluorescence signal was good to detect bacteria in intestinal digesta matrix contents of an adult chicken which exhibited up to 1013 bacteria/g (Apajalahti and Kettunen, 2006) with high bacterial diversity of 640 different species (Apajalahti et al., 2004). Linear detection range of Lb. reuteri KUB-AC5 specific real-time PCR was 6 orders of magnitude, ranging from 102 to 107 copy numbers per PCR assay in the intestinal digesta samples. Detection limit was 106 cfu/g of intestinal digesta, with lower sensitivity than previously reported at 6.69 × 105 cfu/g (Radulovic et al., 2012). Requena et al. (2002) developed a technique to detect human Bifidobacterium sp. by PCR assay using a primer designed from 16S rRNA and gene coding for protein transaldolase. They found that 16S rDNA-targeted PCR was superior to PCR-targeting of the transaldolase gene due to its high gene-targeted abundance. Therefore, reduced sensitivity of AMP-targeted PCR was probably due to the low copy number of only one per genome.
Using this optimized method to examine intestine samples confirmed strain specificity of the assay, since Lb. reuteri KUB-AC5 was not found in cecum samples with low concentration of these lactobacilli. However, Lb. reuteri were detected at high concentration of 8.09 log copy number/g in an ileum sample from chicken fed by a natural farming system but not by closed or evaporation system. Bjerrum et al. (2006) proposed that abundance of lactobacilli in ilea of conventional and organic broilers was higher than in the ceca. Therefore, it is possible that strain KUB-AC5 is a common member of the gut microbiota in chicken at low abundance (Guan et al., 2003) and its concentration remains under the detection level of cecal and also some ileal digesta. These findings suggested that this strain is a normal flora in chicken and showed high abundance in chicken ilea, especially from natural farming systems.
Comparison of conventional plating and quantitative PCR methods revealed similar trends with both giving similar final conclusions. Real-time PCR revealed detection of about 1 log higher numbers of Lb. reuteri KUB-AC5 cfu/g of digesta samples. DNA may also have been isolated from dead cells, while single colonies do not necessarily originate from single bacterial cells by plate count and this probably caused the difference in concentrations.
In conclusion, we were able to develop a strain-specific qPCR primer using the P2 primer derived from AMP to detect and enumerate Lb. reuteri KUB-AC5 in intestinal digesta at limiting level of 106 cfu/g or more. This technique with a specific primer from typical AMP is unique to identify strain level and can replace other genotypic identification techniques that include time consuming isolation of the target strain using selective culturing. Therefore, a KUB-AC5 specific qPCR assay has potential for use in feeding trials to ensure the accurate inclusion of feed supplementation and monitor its uptake into the GIT of chicken.
ACKNOWLEDGEMENTS
This work was partially supported by the Research and Researchers Funds for Industries and Betagro Public Co., Ltd. for a Ph.D. scholarship, Special thanks are due to Professor Kenji Sonomoto, Associate Professor Jiro Nakayama and Assistant Professor Takeshi Zendo for providing assistance with performing the DNA sequencing.
REFERENCES
- Apajalahti J., Kettunen A. Avian Gut Function In Health and Disease. CABI Publishing; Wallingford, UK: 2006. Microbes of the chicken gastrointestinal tract; pp. 124–137. [Google Scholar]
- Apajalahti J., Kettunen A., Graham A. Characteristics of the gastrointestinal microbial communities, with special reference to the chicken. World's Poult. Sci. J. 2004;60:223–232. [Google Scholar]
- Bjerrum L., Engberg R.M., Leser T.D., Jensen B.B., Finster K., Pedersen K. Microbial community composition of the ileum and cecum of broiler chickens as revealed by molecular and culture-based techniques. Poult. Sci. 2006;85:1151–1164. doi: 10.1093/ps/85.7.1151. [DOI] [PubMed] [Google Scholar]
- De Martin E.C., Duvall R.E., Hitchins A.D. Real-time PCR detection of 16S rRNA genes speeds most-probable-number enumeration of foodborne Listeria monocytogenes. J. Food. Prot. 2007;70:1650–1655. doi: 10.4315/0362-028x-70.7.1650. [DOI] [PubMed] [Google Scholar]
- Fujimoto J., Matsuki T., Sasamoto M., Tomii Y., Watanabe K. Identification and quantification of Lactobacillus casei strain Shirota in human feces with strain-specific primers derived from randomly amplified polymorphic DNA. Int. J. Food Microbiol. 2008;126:210–215. doi: 10.1016/j.ijfoodmicro.2008.05.022. [DOI] [PubMed] [Google Scholar]
- Guan L.L., Hagen K.E., Tannock G.W., Korver D.R., Fasenko G.M. Detection and identification of Lactobacillus species in crops of broilers of different ages by using PCR-denaturing gradient gel electrophoresis and amplified ribosomal DNA restriction analysis. Appl. Environ. Microbiol. 2003;69:6750–6757. doi: 10.1128/AEM.69.11.6750-6757.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakphaichit M., Thanomwongwattana S., Phraephaisarn C., Sakamoto N., Keawsompong S., Nakayama J., Nitisinprasert S. The effect of including Lactobacillus reuteri KUB-AC5 during post-hatch feeding on the growth and ileum microbiota of broiler chickens. Poult. Sci. 2011;90:2753–2765. doi: 10.3382/ps.2011-01637. [DOI] [PubMed] [Google Scholar]
- Nakphaichit M., Sobanbua S., Siemuang S., Vongsangnak W., Nakayama J., Nitisinprasert S. Protective effect of Lactobacillus reuteri KUB-AC5 against Salmonella Enteritidis challenge in chickens. Bene. Microb. 2019;10:43–54. doi: 10.3920/BM2018.0034. [DOI] [PubMed] [Google Scholar]
- Nitisinprasert S., Nilphai V., Bunyun P., Sukyai P., Doi K., Sonomoto K. Screening and identification of effective thermotolerant Lactic acid bacteria producing antimicrobial activity against Escherichai coli and Salmonella sp. resistant to antibiotics. Kasetsart J. Nat. Sci. 2000;34:387–400. [Google Scholar]
- Radulovic Z., Mirkovic N., Bogovic M.B., Petrusic M., Petrovic T., Manojlovic V., Nedovic V. Quantification of viable spray-dried potential probiotic Lactobacilli using real-time PCR. Arch. Biol. Sci. 2012;64:1465–1472. [Google Scholar]
- Requena T., Burton J., Matsuki T., Munro K., Simon M.A., Tanaka R., Watanabe K., Tannock G.W. Identification, detection and Enumeration of human Bifidobacterium species by PCR targeting the transaldolase gene. Appl. Environ. Microbiol. 2002;68:2420–2427. doi: 10.1128/AEM.68.5.2420-2427.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rintrila T., Kassinen A., Malinen E., Krogius L., Palva A. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J. Appl. Microbiol. 2004;97:1166–1177. doi: 10.1111/j.1365-2672.2004.02409.x. [DOI] [PubMed] [Google Scholar]
- Rodklongtan A., La-ongkham O., Nitisinprasert S., Chitprasert P. Enhancement of Lactobacillus reuteri KUB-AC5 survival in broiler gastrointestinal tract by microencapsulation with alginate-chitosan semi-interpenetrating polymer networks. J. Appl. Microbiol. 2014;117:227–238. doi: 10.1111/jam.12517. [DOI] [PubMed] [Google Scholar]
- Sakamoto M., Imayoshi I., Ohtsuka T., Yamaguchi M., Mori K., Kageyama R. Continuous neurogenesis in the adult forebrain is required for innate olfactory responses. Proc. Natl. Acad. Sci. USA. 2011;108:8479–8484. doi: 10.1073/pnas.1018782108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler A.V., Mohnl M., Klose V. Development of a Strain-Specific Real-Time PCR Assay for enumeration of a probiotic Lactobacillus reuteri in chicken feed and intestine. PLOS One. 2014;9:1–7. doi: 10.1371/journal.pone.0090208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangthong J. Kasetsart University; 2012. Cloning of kac5 gene encoding bacteriocin KAC5 from Lactobacillus reuteri KUB-AC5. M.S Thesis. [Google Scholar]
- Yeung P.S., Sanders M.E., Kitts C.L., Cano R., Tong P.S. Species-specific identification of commercial probiotic strains. J. Dairy. Sci. 2002;85:1039–1051. doi: 10.3168/jds.S0022-0302(02)74164-7. [DOI] [PubMed] [Google Scholar]