Abstract
Owing to the practical interest in understanding duration of fertility (DF) to reduce the cost of producing hatching eggs by decreasing the frequency of artificial insemination, as well to uncover the mechanism of the estrogen-gut microbiome axis, elucidating the interaction between the maternal microbiome and the function of sperm storage tubules (SST) has become important for revealing the DF in laying hens. In this study, we investigated the compositional, structural, and functional differences in gut microbiomes between hens with high (HSST, n = 8) and low SST activity (LSST, n = 10) by performing phenotypic selection from approximately 400 individual hens based on their DFs. Their cecal microbial communities were analyzed by sequencing the V4 region of the 16S rRNA gene. The microbiome abundance estimators from the ceca of HSST and LSST hens were not significantly different at the phylum and genus taxonomic levels, although the relative abundances for the phylum Firmicutes and the genus Lactobacillus were higher in the HSST group. Furthermore, some taxonomic levels of bacteria expressing the components of several metabolic pathways differed between the HSST and LSST groups. Moreover, predicting functional microbiomes by Kyoto Encyclopedia of Genes and Genomes (KEGG) revealed that certain pathways, such as the metabolism of carbohydrates and protein, cellular processes, and organismal systems, of the HSST group exhibited higher expression of genes associated with bioactivity and energy biosynthesis than those in the LSST group. Our results may provide insights into hen-microbe interactions with respect to DF and will be useful in establishing a strategy for new research to uncover the functional regulation of SST in laying hens.
Key words: sperm storage tubules, duration of fertility, cecal microbiota, 16S rRNA gene, laying hens
Introduction
For the analysis of sperm storage tubules (SST) performance, the duration of fertility (DF, number of days between insemination and the last fertile egg) in laying hens is a phenotypic variable of practical interest because any reduction in artificial insemination (AI) frequency without a loss of fertility reduces the cost of producing hatching eggs (Liu et al., 2008). Avian females have the ability to store sperm in their reproductive tracts after natural copulation or AI from 1 to 4 wk, within SST and the utero-vaginal junction (UVJ) (Das et al., 2010). SST and UVJ are considered primary and secondary sites for residing sperm, respectively. Sperm are gradually released from the SST, ascend to the anterior end of the oviduct, and fertilize the next ovulated ovum (Bakst, 2011). Some mechanisms have already been suggested for the period of sperm survivability during storage in SST, including enhanced expressions of transforming growth factors β (TGFbs-1 – 5) and TbRs (their receptors), as seen in the UVJ, probably via suppression of antisperm immunoreactions (Chandra Das et al., 2006). Therefore, the series of fertile eggs following a single copulation event or AI reveals the survival of fertile sperm in the SST site and is applicable to analysis using the DF phenotype.
The gut microbiota is an environmental factor that is significantly involved in energy harvest from the host diet (Turnbaugh et al., 2006), mammalian body composition (Tremaroli and Backhed, 2012), and energy storage in adipose tissue (Wang et al., 2017), although the underlying mechanisms and their metabolic pathways remain unclear (Nicholson et al., 2012, Shepherd et al., 2018). The functional interactions between the microbial inhabitants of the gut and host metabolism influence host performance and health. Therefore, many host metabolic pathways are affected by alterations in gut microbial communities (Claesson et al., 2012). Compositional and functional changes in gut microbiota have been linked to productive performance, including growth activity and immune responses; the gut microbiota is significantly involved in energy harvest from the host diet (Turnbaugh et al., 2006), immune system regulation (Tremaroli and Bã¤Ckhed, 2012, Proctor et al., 2019), organ development and morphogenesis (Sommer and Backhed, 2013), and energy storage in tissues (Wang et al., 2017).
In addition, the symbiotic host-microbiota interaction provides host immune responses for regulating a physiological niche in a nutrient-rich environment. Because of this regulation, the microbiota exerts various beneficial functions for the host, such as digestion and vitamin production, as well as protection from pathogens. Recently, the role of the estrogen-gut microbiome axis was discovered to regulate reproductive performance in female mammals through the secretion of the enzyme β-glucuronidase, which deconjugates estrogens into their active forms, and lower microbial composition was estimated to occur in infertile females (Flores et al., 2012, Baker et al., 2017). Furthermore, estrogen-progesterone ratio also contributed to regulating the period of sperm survivability in the SST (Yoshimura et al., 2000). Ito et al. (2011) suggested that progesterone regulates the release of the resident sperm from the SST in the Japanese quail with a contraction-like morphological change of the SST. Consequently, gut microbiota diversity may contribute to regulating SST activity through an effective mechanism involving the estrogen-progesterone ratio and the susceptibility of progesterone receptors in the SST. Precisely, we hypothesized that the longevity period of sperm within storage in SST is regulated by the gut microbiome through the interacting estrogen-gut microbiome axis to determine the DF. Therefore, we conducted this experiment using 2 groups of hens that were selected for high (HSST) and low (LSST) SST activity during the period of sperm storage through detection of the phenotype of DF, assessing the compositional, structural, and functional diversity of the cecal microbiome using 16S RNA amplicon-based metagenomic association analysis.
Materials and Methods
All experimental procedures were reviewed and approved by the College of Animal Science and Veterinary Medicine, Huazhong Agricultural University, China, and hens were handled in accordance with the guidelines described by the Animal Care Committee of Hubei Province, P.R. China.
Experimental Design and Management
A total of 400 crossbreeding hens at 60 wk of age were used in this study to select for HSST and LSST activity. All hens were under similar husbandry conditions and were housed in individual cages inside an enclosed farm during the experimental period. They were fed a standard caloric and nitric breeder diet (2,700 kcal/kg, 13% protein, 1% calcium, and 0.45% phosphorous), and water was provided ad libitum. The DF (maximum number of days for a fertile egg) for HSST and LSST were ended by the 7th and 20th D from insemination, respectively.
AI, DF Calculation, and Cecal Content Sampling Assay
To verify the DF per day, the hens were artificially inseminated by pooling the semen from cocks. All cocks were similar in age, genotype, and all management practices. AI was conducted by pooling ejaculates from all cocks without any dilution. Three replicates of AI were applied with 30 μl of pooled semen using a micropipette reinforced with a rubber tube. All handing processes, as well as ejaculate injection, were very similar among the three instances of insemination for calculating the average DF for each hen. After each insemination, the labeled eggs were collected for 25 D and placed in an artificial incubator; the fertility of each egg was determined on the sixth day of incubation using light detection. The average DF in days was calculated for each hen from data from the three inseminations. Finally, according to the DF phenotype, the highest (HSST) and lowest (LSST) individual hens (n = 30) were selected from the experimental population to study fecal microbiome diversity between the groups. Fresh cecal samples from the jejunum (n = 8 and 10 from HSST and LSST, respectively) were obtained after slaughter and placed in separate sterilized tubes and stored immediately at −20°C until DNA extraction.
Microbial DNA Extraction and PCR Amplification
The microbial total DNA extraction of 8 HSST and 10 LSST samples from cecal jejunum content in each group was extracted using a DNA stool mini kit (QIAamp DNA Stool mini Kit; QIAGEN, Hilden, Germany), following the manufacturer's instructions. The DNA concentration and quantity were measured using a Nanodrop device, samples were analyzed by 0.8% agarose gel electrophoresis, and DNA was quantified by UV spectrophotometry. Then, the extracted genomic DNA was used as a template, and 16S amplicons were generated by PCR amplification. For each sample, 16S ribosomal RNA (V3-V4 region) amplicons were amplified using primer (forward 5′ CCTAYGGGRBGCASCAG GNG 3′, reverse 5′ GGACTACNNGGGTATCTAAT 3′), and PCR products were then purified according to Zhao et al. (2013). The PCR conditions were as follows: initial denaturation, annealing, and extension were carried out and repeated at 94°C for 4 min, 94°C for 30 s, 50°C for 45 s, and 72°C for 30 s for 25 cycles. After confirming sufficient quality of PCR products, library construction was conducted.
Sequence Quality Control and Operational Taxonomic Units Calculation
Amplified libraries barcoded V3 and V4 were sequenced using the Illumina Miseq 2000 platform, including 250 bp paired-end reads generated with a 7-cycle index read (sequences with an overlap longer than 10 bp without any mismatch were assembled). Each sequencing process was conducted by Personalbio Co., Ltd., and the results are publicly available in the Penzano Biomicrobiology Group report under opening number MbPL201901330. After removing barcode and primer sequences, the raw tags were merged based on the overlap of 2two reads. Then, clean tags were created by pretreating the raw tags and removing the chimeric sequences to generate effective tags. Quantitative analysis of sequences was performed using QIIME software (Quantitative Insights Into Microbial Ecology, version 1.8.0, http://qiime.org/; Northern Arizona University, Flagstaff, AZ), according to Caporaso et al. (2010). Then, USEARCH (version 5.2.236, http://www.drive5.com/usearch/; University of California, Santa Cruz, CA) was used to detect and eliminate chimeric sequences. Quality sequences were counted for each sample after the removal of sequences shorter than 160 bp, and statistical estimations were created for the distribution of sequence length using R software (version 3.2.2; R Core, University of Auckland, Auckland, New Zealand) to characterize the length distribution of the sequences contained in each sample; the results were presented as a sequence length map. Specaccum analysis was applied to check if all sample sizes and the operational taxonomic units (OTUs) abundance matrix were sufficient to estimate community richness (Supplementary Figure 1). Finally, the phylogeny of OTUs, microbial diversity units that usually refer to the sequences of one or more samples based on a sequence similarity threshold set by an individual, was calculated according to Blaxter et al. (2005).
Identification of Maternal Microbiome Community and OTUs Analysis
For the analysis of bacteria and archaea, the databases for the 16S rRNA gene, Greengenes (Release 13.8, http://greengenes.secondgenome.com/) and RDP (Ribosomal Database Project, Release 11.1, http://rdp.Cme.msu.edu/), were used default to identify the OTU diversity among the samples and between the groups (Desantis et al., 2006, Cole et al., 2009). For fungal identification, the 18S rRNA gene database for fungi and the Silva database (Release 115, http://www.arb-silva.de) were used to label fungi (Christian et al., 2013). The abundances of OTUs with less than 0.001% of the total sample sequencing were removed to ensure reliable and accurate analysis results (Bokulich et al., 2013). Then, the OTU division was carried out, and the classification status identification results were statistically analyzed. At the same time, the R software was used to plot the identification results of each sample at each classification level into a histogram to visually compare the OTUs of different samples. A Venn diagram was constructed to calculate the total number of OTUs by of each sample (group), using the R software.
Annotation of Microbial Composition
Alpha diversity analysis, including the Shannon, Simpson, Chao1, and ACE indices, as well as community uniformity, was applied to find the drivers of variation in microbial community structure among samples (Shannon, 1948, Simpson, 1949, Chao and Shen, 2004). Beta diversity analysis with principal component analysis (PCA), principal coordinate analysis, NMDS, and UPGMA cluster analysis was used to obtain the comparative analysis of intergroup/group differences in UniFrac distance (Lozupone and Knight, 2005, Ramette, 2007). Phylogenetic tree construction, MEGAN Visualization, and KRONA interactive displays were performed to identify the microbial interactive visualization and display the results of species annotation visually between groups according to OTU tags. Heat map analysis was conducted according to the distribution of the top 50 abundances and the degree of similarity between the samples, allowing high-abundance and low-abundance classification units to be distinguished by different color composition gradients that reflect the community composition similarity between samples. The order analyses of partial least squares discriminant analysis, Adonis/PERMANOVA, and ANOSIM were performed to determine the variation in community structure between groups for screening key species.
Annotation of Microbial Function
To predict the metabolic functioning of bacteria and archaea based on total genome sequences from the 16S rRNA gene, the functional predictive analysis of Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) was performed (Langille et al., 2013). PICRUSt can predict the association function of 16S rRNA gene sequences with three functional profile databases: Kyoto Encyclopedia of Genes and Genomes (KEGG), COG, and Rfam. Especially useful is the KEGG pathways database (http://www.genome.jp/kegg/pathway.html), which classifies genes into six categories, including Metabolism, Genetic Information Processing, Environmental Information Processing, Cellular Processes, Organismal Systems, and Human Diseases, each of which is further divided into multiple levels. According to the predicted abundance distribution of each functional group in each sample, a histogram was constructed for display, and presentation of shared functional groups was accomplished by Venn diagram. Finally, R software was used for cluster analysis of the top 50 most abundant functional predictions in each sample, presented by the heat map.
Statistical Analyses
The beta diversity analysis includes the PCA and principal coordinate analysis, which were calculated by weighted UniFrac and unweighted UniFrac (Lozupone et al., 2010). Differences of the bacterial taxonomic between the HSST and LSST groups in the DF variable were analyzed, using the LM procedure of R software version 3.2.2, R Core (Team, 2015). The significance of variance was analyzed by one-way ANOVA. The individual hen was considered as the experimental unit, and one fixed effect of the DF was included in the statistical model. All differences were considered significantly different at P < 0.05 and were considered trends when P < 0.10. Pairwise comparisons were performed using Duncan's multiple range test.
Results
OTU Clustering and Annotation
The samples of cecal jejunum from 18 hens, which included 8 HSST and 10 LSST, were collected after insemination. To determine which 16S rRNA variable regions were more suitable to be used to identify taxonomy from cecal communities, specaccum species accumulation curves for the total numbers of OTUs for each sample were constructed using R software, revealing that all sample sizes and the OTU abundance matrix were sufficient to estimate community richness (Supplementary Figure 1). The proportion of common and unique OTUs between HSST and LSST groups was presented in a Venn diagram (Figure 1A).
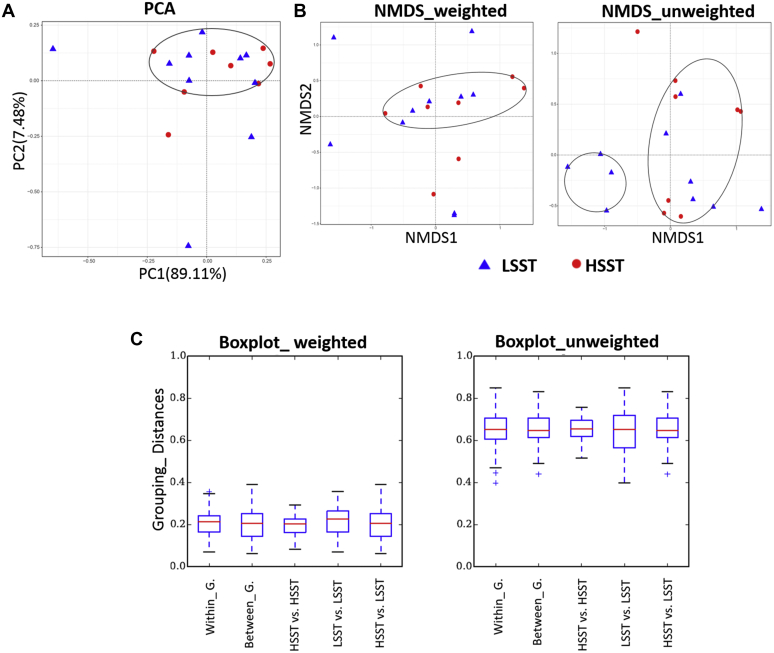
Figure 1.
(A) Principal component analysis (PCA) analysis plot of the natural distribution characteristics between the samples of high activity of sperm storage tubules (HSST) and low activity of sperm storage tubules (LSST). (B) NMDS analysis based on weighted and unweighted UniFrac distance matrices using R software. (C) Multigroup comparison box plot (beta diversity index) of weighted and unweighted UniFrac distance within and between groups of HSST and LSST.
Differences in DF and Alpha and Beta Diversity of the Cecal Microbiotas of LSST and HSST Hens
After obtaining the OTU abundance matrix, a series of analyses was performed for calculating the diversity of each sample community. A total of 644,388 16S rRNA gene sequence reads were generated, with an average of 34,612 ± 1,272.18 reads per hen in the LSST group and 36,479 ± 1,422.34 reads per hen in the LSST group (Table 1). The results of alpha diversity analysis of intestinal microbiota, including observed OTUs, ACE, Chao1, Shannon index, and Simpson index values, are presented in Table 1. The differences in richness values of observed OTUs, ACE, and Chao1 were not significant, although the means were higher for the HSST group than for the LSST group (Table 1). In addition, the Shannon and Simpson diversity indices were also higher for the HSST group than those for the LSST group, although the differences were not significant. In addition, the plots of PCA and NMDS showed overlap between the groups (Figures 1A and 1B). To examine the similarity in community structure between the different samples, beta diversity analysis was applied; there were no significant differences in microbial community within or between HSST and LSST groups of weighted or unweighted UniFrac distance (Figure 1C).
Table 1.
Differences in the duration of fertility and alpha diversity in the cecal microbiome of HSST and LSST groups.
| Parameters | LSST | HSST | SEM | P value2 |
|---|---|---|---|---|
| Duration of fertility (DF)1 | 6.40 | 20.50 | 0.48 | 0.001*** |
| Alpha diversity index | ||||
| Sequence reads | 34,612.00 | 36,479.20 | 1,272.18 | 0.279 |
| Observed OTUs | 2326.62 | 2655.90 | 189.03 | 0.212 |
| ACE | 618.79 | 722.26 | 44.85 | 0.143 |
| Chao1 | 611.12 | 707.56 | 44.13 | 0.164 |
| Shannon | 4.47 | 4.86 | 0.25 | 0.324 |
| Simpson | 0.827 | 0.843 | 0.03 | 0.762 |
All data were expressed as the mean ± SEM.
Abbreviations: HSST, high activity of sperm storage tubules; LSST, low activity of sperm storage tubules.
DF is the number of days between insemination and the last fertile egg (hens, n = 400 at 60 wk of age), number of artificial insemination (AI, n = 3).
The P values were determined using Welch's t test (***P < 0.001). HSST high activity of SST with long duration of fertility and LSST low activity of SST short duration of fertility (sequences sample, n = 8 for HSST and n = 10 for LSST).
Differences Between the LSST and HSST Hens in the Microbial Taxa Represented in the Cecal Microbiotas
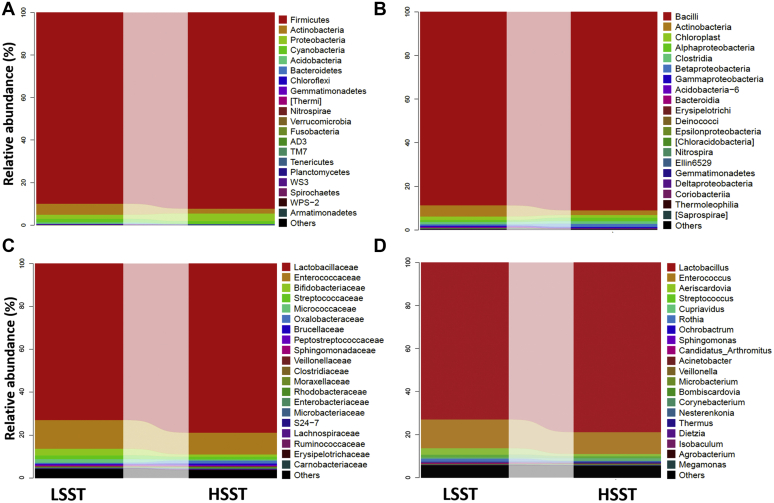
To assess the differences in microbial taxa between HSST and LSST, all the OTUs observed at the 97% similarity level were classified into 21 phyla and 286 genera (Figure 2). Analyses of taxonomic composition at each classification level using the QIIME software, and their abundance differences between groups (HSST vs. LSST) at the five classification levels (class, order, phylum, family, and genus) were obtained as shown in Figure 2. Firmicutes, Actinobacteria, Cyanobacteria, Acidobacteria, Bacteroidetes, and Fusobacteria appeared to be the top phyla in the samples from HSST and LSST groups. In addition, Lactobacillus, Enterococcus, Bifidobacterium, Streptococcus, and Acinetobacter were the most abundant genera in the samples of HSST and LSST groups. The relative abundances of the members of microbial taxa in the LSST and HSST groups indicated that some phyla and genera were represented at significantly different levels in the groups (Table 2). At the phylum level, the abundances of Firmicutes were significantly higher (P < 0.05) in the HSST group than those in the LSST group. On the other hand, the Bacteroidetes was significantly more abundant in the LSST group than in the HSST group. The Firmicutes/Bacteroidetes ratio (FB ratio) was significantly higher (P < 0.05) in the HSST group than that in the LSST group (Table 2). At the genus level, the abundances of Lactobacillus and Bifidobacterium were significantly higher in the cecal microbiotas of the HSST group than those of the LSST group. In contrast, the abundances of Acinetobacter and Clostridium were significantly higher (P < 0.05) in the LSST group than those in the HSST group. Lactococcus and Enterococcus were more abundant in the HSST group than they were in the LSST group, although the difference was not significant (P = 0.384 and 0.223, respectively) as presented in Table 2.
Figure 2.
(A–D) taxonomic composition analysis between groups at each classification level of phylum, class, family, and genus of low activity of sperm storage tubules (LSST) and high activity of sperm storage tubules (HSST) groups.
Table 2.
Differences in the compositional of cecal microbiome of the HSST and LSST groups.
| Taxon | Relative abundance % |
SEM | P value1 | |
|---|---|---|---|---|
| LSST | HSST | |||
| Phylum (%) | ||||
| Firmicutes | 86.90 | 92.24 | 4.75 | 0.048* |
| Bacteroidetes | 2.42 | 1.33 | 0.05 | 0.013* |
| FB ratio | 38.04 | 57.61 | 4.22 | 0.036 |
| Actinobacteria | 5.18 | 2.22 | 1.28 | 0.143 |
| Proteobacteria | 1.98 | 3.68 | 0.86 | 0.204 |
| Cyanobacteria | 1.62 | 1.26 | 0.28 | 0.732 |
| Acidobacteria | 0.62 | 0.03 | 0.24 | 0.134 |
| Genus (%) | ||||
| Lactobacillus | 70.85 | 92.56 | 6.46 | 0.050* |
| Bifidobacterium | 2.69 | 5.87 | 1.87 | 0.047 |
| Lactococcus | 1.43 | 2.39 | 0.69 | 0.384 |
| Enterococcus | 0.61 | 2.03 | 0.70 | 0.223 |
| Streptococcus | 0.57 | 0.32 | 0.03 | 0.345 |
| Sphingomonas | 0.14 | 0.10 | 0.04 | 0.798 |
| Acinetobacter | 1.00 | 2.00 | 0.00 | 0.001** |
| Corynebacterium | 4.50 | 1.74 | 1.95 | 0.058 |
| Thermus | 2.53 | 4.97 | 1.56 | 0.336 |
| Bacteroides | 2.64 | 0.50 | 0.08 | 0.236 |
| Clostridium | 0.13 | 0.66 | 0.03 | 0.316 |
All data were expressed as the mean ± SEM of the percentage of domain bacteria at taxonomic levels (phylum and genus).
Sampling (n = 8 for HSST and n = 10 for LSST).
Abbreviations: HSST, high activity of SST with long duration of fertility; LSST, low activity of SST short duration of fertility.
The P values were determined using Welch's t test (*P < 0.05; **P < 0.01).
Metastats and Significant Differences Among the Samples From the HSST and LSST Groups
A metastats comparison test using Mothur software package (version 1.33.3; University of Michigan, Ann Arbor, MI) revealed that the differences in sequence quantity of each taxon at the gate of the phylum and genus level between groups of HSST and LSST were 4 and 34, respectively. The statistical values of R using both PERMANOVA and ANOSIM methods at weighted and unweighted UniFrac distances were not significant (Figure 3D), indicating no differences in composition or structure of the microbial populations between HSST and LSST. In addition, a clustering tree of UPGMA was constructed using QIIME software to estimate the similarity between samples, which revealed overlap between the samples from HSST and LSST due to a short UniFrac distance between the samples; consequently, nonsignificant differences in the composition of the microbiota samples were observed between the groups (Supplementary Figure 2). Likewise, the heat map of the top 50 abundances in microbial communities combined with their cluster analysis showed similar microbiome composition between samples from HSST and LSST groups (Supplementary Figure 3).
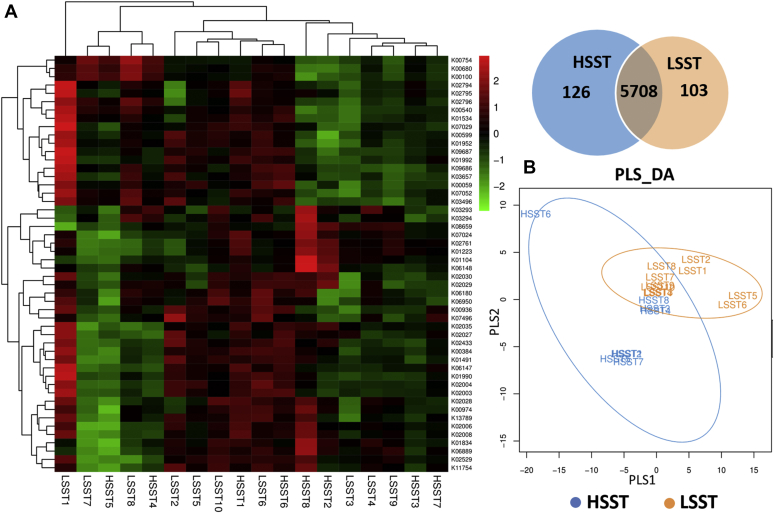
Figure 3.
(A) Heat map of the top 50 predicted functions by Kyoto Encyclopedia of Genes and Genomes (KEGG) based on the similarity of the functional group abundance distributions of cecal samples of high activity of sperm storage tubules (HSST) and low activity of sperm storage tubules (LSST) groups. Venn diagram of the common functional group, (B) Partial least squares discriminant analysis (PLS-DA) discriminant analysis plot showed samples belonging to the same group are closer to each other, and the distance between the points of different groups is farther.
Prediction of Functional Enrichment of Gut Microbiotas
The top 50 functional microbiome groups according to their abundances were analyzed using R software and clustered into a heat map as shown in Figure 3A. The functional microbial communities for the HSST and LSST samples appeared to overlap and belong to the same cluster because of the compositional similarity between them. In addition, the Venn diagram also presented the common and unique aspects of functional microbiomes between groups based on the predicted abundance distribution of each functional group in each sample, which was calculated using R software (Figure 3A). The analysis of partial least squares discriminant analysis based on the coefficient of variable importance in projection for each species also indicated overlap between the HSST and LSST samples (Figure 3B).
Comparison of the Cecal Microbiota KEGG Pathways of the HSST and LSST Groups
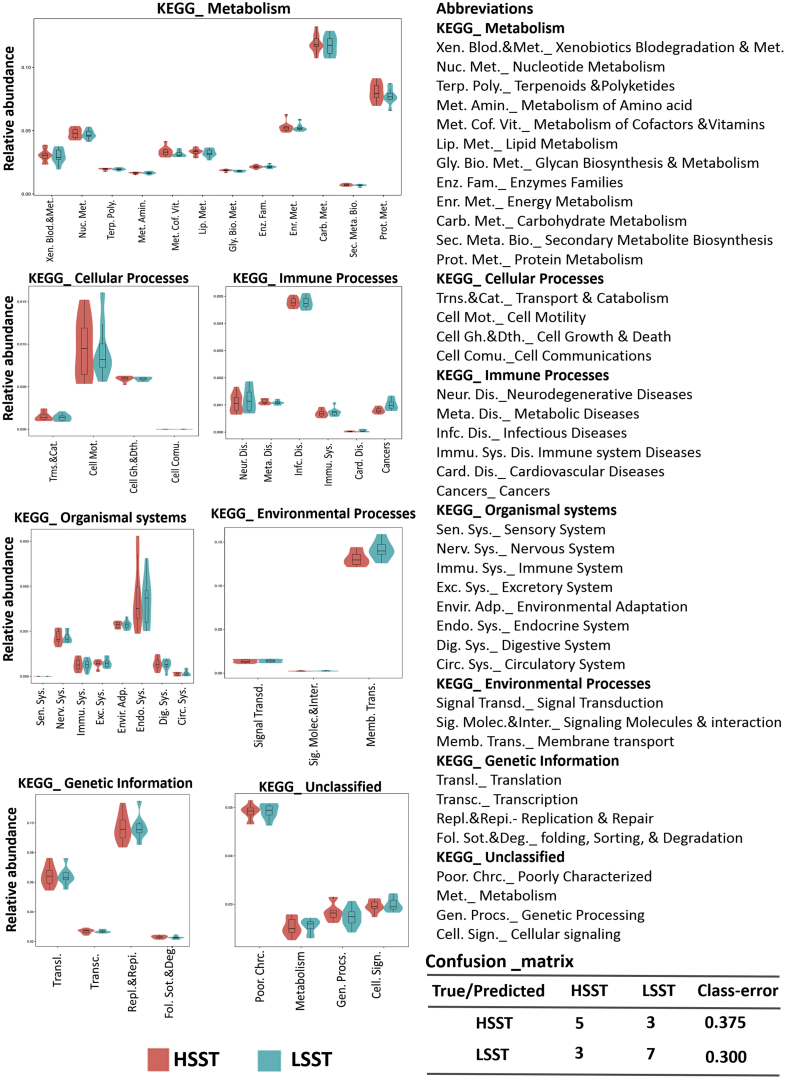
To predict the metabolic functioning of bacteria and archaea, the analysis PICRUSt was performed for functional prediction based on the existing 16S rRNA gene sequencing and known microbial metabolic functions (Figure 4). The functional profile databases of KEGG, COG, and Rfam were applied to predict the metabolic pathways of microbial pathways in the HSST and LSST groups, focusing only on the six KEGG pathways of metabolism, genetic processing, environmental processing, cellular processes, organismal systems, and human diseases. According to the predictions of PICRUSt, the annotation information for each corresponding functional spectrum database of each group is presented in Figure 4. Metabolic pathways of cofactors, vitamins, carbohydrates, protein, and energy were predicted to be expressed at slightly higher levels in the microbiotas of the HSST group, whereas metabolic pathways for nucleotides, lipids, and glycine were predicted to be expressed at higher levels in the LSST group. In addition, the levels of expression of the cell motility and growth and cellular processes pathway, as well as the endocrine system pathway, were predicted to be higher for the HSST group than those for the LSST group (Figure 4).
Figure 4.
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) predicted Kyoto Encyclopedia of Genes and Genomes (KEGG) at the second level distribution map between the high activity of sperm storage tubules (HSST) and low activity of sperm storage tubules (LSST) groups; the width reflects the number of samples corresponding to the abundance. Confutation matrix for the true/predicted HSST and LSST groups.
Discussion
DF ranged widely, from 0 to 21 D, even though AI was performed in the same optimal conditions, suggesting that genotype and environmental variables may be involved. Indeed, the genetic variance of DF is relatively high, supporting environmental variables as candidate contributions. The association between the DF phenotype and the performance of SST in hens was positive, suggesting that DF should be used to assess the performance of SST in female chickens (Liu et al., 2008). Recently, the pattern of host-gut microbiota interaction (symbiotic or opportunistic), based on the drivers of structural, commotional, and functional microbial community structure, was regarded as an internal environmental variable that regulates productive performance and immune responses in livestock (Tremaroli and Bã¤Ckhed, 2012, Li and Guan, 2017, Gopalakrishnan et al., 2018).
In this study, we examined structural and functional differences of gut microbiomes influencing the DF, characteristics of the gut microbiome known to interact with host immune responses, and SST activity in which sperm is stored for quite a long period. We first assessed the phenotype for DF to identify the HSST and LSST groups of hens and then detected cecal microbiome diversity using 16S RNA sequencing, noting that collectively both communities of the HSST and LSST groups were similar in structural, commotional, and functional aspects of their cecal microbiomes. The host genotype and diet are primary selective forces for bacterial colonization factors that shape community membership and functionality (Melanie Lee et al., 2013, Sonnenburg et al., 2016). Atikuzzaman et al. (2015) suggested that immune responses and pH regulation may participate in differentially expressed mucosal genes of the SST segment, speculating that microbiome modulation might also contribute to this regulation. Therefore, further research is required to explore the roles of each of the different gut microbiomes regarding cross-talk between spermatozoa and SST performance to determine the DF of mated hens.
In this study, composition analysis showed that the most abundant phyla identified in the cecal jejunum samples were, Firmicutes, Actinobacteria, Proteobacteria, Acidobacteria, Bacteroidetes, and Fusobacteria, consistent with previous studies (Ferrario et al., 2017). However, the highest differences between the HSST and LSST groups entailed that Firmicutes and Proteobacteria, which include a wide variety of beneficial and pathogenic genera, respectively (Tong et al., 2018), were more diverse in the functional pathways for the microbiome contents of the HSST than those for the LSST group, similar to the results of the study by Ding et al. (2016). Cecal microbiotas of HSST and LSST groups were remarkably similar in composition and structure; however, some functional predictions differed between groups. Although the differences between the groups in the composition of microbiomes were not significant, the functional prediction for the metabolic pathways of the cofactors, vitamins, carbohydrates, protein, and energy was at slightly higher levels in the HSST group, whereas the prediction of the expression levels of the metabolic pathways of the nucleotides, lipids, and glycine were higher in the LSST group. In addition, the predicted levels of expression of the cell motility and cell growth (within cellular processes) pathways, as well as the endocrine system pathways, were higher in the HSST than they were in the LSST group. Wallace et al. (2018) reported that the composition of the maternal gut microbial profiles may be associated with reproductive status and endocrine changes in the female oviduct. In addition, compared with mammals, the gut microbiota regulates estrogen (the estrogen-gut microbiome axis) through the secretion of β-glucuronidase, an enzyme that deconjugates estrogen into its active forms; thus, lower microbial composition is expected in infertile females (Flores et al., 2012, Baker et al., 2017).
In this study, although 16S rRNA amplicon sequencing was the primary method used to analyze compositional and structural aspects of microbial diversity, we also used the computation tool PICRUSt to predict microbial community functions. Functional prediction results revealed that the metabolic pathways of proteins, lipids, carbohydrates, cofactors, and vitamins were more abundant in the HSST than they were in the LSST group, and this outcome was consistent with the probabilities of improved energy harvest from diet and TGF profile. These findings were expected with our previous research revealing that the TGFβ transcription factors, TGFβ1, TGFβ2, and TGFβ3 (TGFb genes), contributed to long DF (Gu et al., 2017). In addition, several studies have shown that immune transcription factors and their signaling pathways are involved in the regulation of SST functions (Das et al., 2005, Chandra Das et al., 2006). The oviduct immune process plays an important role in fertility, which enhances SST ability to store sperm (Kirk et al., 1989, Bakst, 2011). Interestingly, our microbial function results suggested that, compared to the LSST, immune process signal pathways of gut microbiotas in the HSST had higher functional performance, and this outcome was consistent with our previous work regarding the differential expression of lncRNAs in UVJ transcriptomes from hens with long DF (Adetula et al., 2018). According to our understanding, we showed for the first time in the present study that characterizing the composition, structure, and function of gut microbiotas in HSST and LSST hens can predict the DF after the postinsemination of hens. The precise roles of the interaction of the gut microbiota and the efficiency of SST activity are not clearly known, considering the complexity of the gut microbiota, and are associated with energy harvest from the host diet, immune system regulation, and energy storage in tissues.
In conclusion, we demonstrated for the first time that functional microbiomes regulate the survival of sperm during residence in SST after insemination. In addition, no significant differences were observed in the structure or composition of gut microbiomes of mated hens; however, some differences in microbiome functions were observed. Thus, possible differences in functional microbiomes may determine the differences in DF between the HSST and LSST hens and could be attributed to the storing ability of SST segments.
Acknowledgments
This study was supported by the National Natural Science Foundation of China, China (No. 31772585), the Fundamental Research Funds for the Central Universities, China (No. 2662018PY088), and Breeding and Reproduction in The Plateau Mountainous Region, Ministry of Education, Guizhou University, China (No. GYSD-K-2018-01). The authors declare that there are no conflicts of interest. All procedures involving of birds handling were approved by Chines Animal Care Committee of Hubei Province and the regulations of College of Animal Science and Veterinary Medicine at Huazhong Agricultural University (protocol no. 2018/0018835). None of the data were deposited in an official repository.
Footnotes
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.psj.2019.10.048.
Supplementary data
References
- Adetula A.A., Gu L., Nwafor C.C., Du X., Zhao S., Li S. Transcriptome sequencing reveals key potential long non-coding RNAs related to duration of fertility trait in the uterovaginal junction of egg-laying hens. Sci. Rep. 2018;8:13185. doi: 10.1038/s41598-018-31301-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atikuzzaman M., Bhai R.M., Fogelholm J., Wright D., Rodriguez-Martinez H. Mating induces the expression of immune- and pH-regulatory genes in the utero-vaginal junction containing mucosal sperm-storage tubuli of hens. Reproduction. 2015;150:473–483. doi: 10.1530/REP-15-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker J.M., Al-Nakkash L., Herbst-Kralovetz M.M. Estrogen-gut microbiome axis: physiological and clinical implications. Maturitas. 2017;103:45–53. doi: 10.1016/j.maturitas.2017.06.025. [DOI] [PubMed] [Google Scholar]
- Bakst M.R. Physiology and endocrinology symposium: role of the oviduct in maintaining sustained fertility in hens. J. Anim. Sci. 2011;89:1323–1329. doi: 10.2527/jas.2010-3663. [DOI] [PubMed] [Google Scholar]
- Blaxter M., Mann J., Thomas T.F., Whitton C., Floyd R., Abebe E. Defining operational taxonomic units using DNA barcode data. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005;360:1935–1943. doi: 10.1098/rstb.2005.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich N.A., Subramanian S., Faith J.J., Gevers D., Gordon J.I., Knight R., Mills D.A., Caporaso J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods. 2013;10:57–59. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Peña A.G., Goodrich J.K., Gordon J.I., Huttley G.A., Kelley S.T., Knights D., Koenig J.E., Ley R.E., Lozupone C.A., McDonald D., Muegge B.D., Pirrung M., Reeder J., Sevinsky J.R., Turnbaugh P.J., Walters W.A., Widmann J., Yatsunenko T., Zaneveld J., Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra Das S., Isobe N., Nishibori M., Yoshimura Y. Expression of transforming growth factor-beta isoforms and their receptors in utero-vaginal junction of hen oviduct in presence or absence of resident sperm with reference to sperm storage. Reproduction. 2006;5:781–790. doi: 10.1530/rep.1.01177. [DOI] [PubMed] [Google Scholar]
- Chao A., Shen T.J. Nonparametric prediction in species sampling. J. Agr. Bio. Env. Sta. 2004;9:253–269. [Google Scholar]
- Christian Q., Elmar P., Pelin Y., Jan G., Timmy S., Pablo Y., J?Rg P., Frank Oliver G.C. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson M.J., Jeffery I.B., Conde S., Power S.E., O’Connor E.M., Cusack S., Harris H.M.B., Coakley M., Lakshminarayanan B., O’Sullivan O. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- Cole J.R., Wang Q., Cardenas E., Fish J., Chai B., Farris R.J., Kulam-Syed-Mohideen A.S., Mcgarrell D.M., Marsh T., Garrity G.M. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:141–145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S.C., Nagasaka N., Yoshimura Y. Changes in the localization of antigen presenting cells and T cells in the utero-vaginal junction after repeated artificial insemination in laying hens. J. Reprod. Dev. 2005;51:683–687. doi: 10.1262/jrd.17027. [DOI] [PubMed] [Google Scholar]
- Das S.C., Naoki I., Yukinori Y. Mechanism of prolonged sperm storage and sperm survivability in hen oviduct: a review. Am. J. Reprod. Immunol. 2010;60:477–481. doi: 10.1111/j.1600-0897.2008.00651.x. [DOI] [PubMed] [Google Scholar]
- Desantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K., Huber T., Dalevi D., Hu P., Andersen G.L. Greengenes: Chimera-checked 16S rRNA gene database and workbenchcompatible in ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J., Zhao L., Wang L., Zhao W., Zhai Z., Leng L., Wang Y., He C., Zhang Y., Zhang H. Divergent selection-induced obesity alters the composition and functional pathways of chicken gut microbiota. Genet. Sel. Evol. 2016;48:93. doi: 10.1186/s12711-016-0270-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrario C., Alessandri G., Mancabelli L., Gering E., Mangifesta M., Milani C., Lugli G.A., Viappiani A., Duranti S., Turroni F. Untangling the cecal microbiota of feral chickens by culturomic and metagenomic analyses. Environ. Microbiol. 2017;11:4771–4783. doi: 10.1111/1462-2920.13943. [DOI] [PubMed] [Google Scholar]
- Flores R., Shi J., Fuhrman B., Xu X., Veenstra T.D., Gail M.H., Gajer P., Ravel J., Goedert J.J. Fecal microbial determinants of fecal and systemic estrogens and estrogen metabolites: a cross-sectional study. J. Transl. Med. 2012;10:253. doi: 10.1186/1479-5876-10-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan V., Spencer C.N., Nezi L., Reuben A., Andrews M.C., Karpinets T.V., Prieto P.A., Vicente D., Hoffman K., Wei S.C. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L., Sun C., Gong Y., Yu M., Li S. Novel copy number variation of the TGFβ3 gene is associated with TGFβ3 gene expression and duration of fertility traits in hens. Plos One. 2017;12:e0173696. doi: 10.1371/journal.pone.0173696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Yoshizaki N., Tokumoto T., Ono H., Yoshimura T., Tsukada A., Kansaku N., Sasanami T. Progesterone is a sperm-Releasing factor from the sperm-storage tubules in birds. Endocrinology. 2011;152:3952–3962. doi: 10.1210/en.2011-0237. [DOI] [PubMed] [Google Scholar]
- Kirk T.A., Krey H.P.V., Hulet R.M., Dunnington E.A., Denbow D.M. The relationship of infertility to antibody production in the uterovaginal sperm storage tubules of Turkey breeder hens. Theriogenology. 1989;31:955–961. doi: 10.1016/0093-691x(89)90478-0. [DOI] [PubMed] [Google Scholar]
- Langille M.G.I., Jesse Z., J Gregory C., Daniel M.D., Dan K., Reyes J.A., Clemente J.C., Burkepile D.E., Thurber R.L., Vega, Rob K. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Guan L.L. Metatranscriptomic profiling reveals Linkages between the active Rumen microbiome and feed efficiency in Beef Cattle. Appl. Environ. Microbiol. 2017;83 doi: 10.1128/AEM.00061-17. AEM.00061-00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G.Q., Zhu J.J., Wang Z.Y., Jiang X.P., Dafalla M.M. Analysis of sperm storage ability using duration of fertility in hens. Br. Poult. Sci. 2008;49:770–775. doi: 10.1080/00071660802433131. [DOI] [PubMed] [Google Scholar]
- Lozupone C., Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C., Lladser M.E., Knights D., Stombaugh J., Knight R. UniFrac: an effective distance metric for microbial community comparison. ISME J. 2010;5:169. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melanie Lee S., Donaldson G., Mikulski Z., Boyajian S., Ley K., K Mazmanian S. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;7467:426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson J.K., Holmes E., Kinross J., Burcelin R., Gibson G., Jia W., Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- Proctor L.M., Creasy H.H., Fettweis J.M., Lloyd-Price J., Mahurkar A., Zhou W., Buck G.A., Snyder M.P., Strauss J.F., Weinstock G.M., White O., Huttenhower C., H. M. P. R. N. C. The Integrative The Integrative human microbiome project. Nature. 2019;569:641–648. doi: 10.1038/s41586-019-1238-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramette A. Multivariate analyses in microbial ecology. FEMS Microbiol. Ecol. 2007;62:142–160. doi: 10.1111/j.1574-6941.2007.00375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon C.E. A Mathematical theory of Communication. Bell Sys. Techn. J. 1948;27:623–656. [Google Scholar]
- Shepherd E.S., Deloache W.C., Pruss K.M., Whitaker W.R., Sonnenburg J.L. An exclusive metabolic niche enables strain engraftment in the gut microbiota. Nature. 2018;557:434–438. doi: 10.1038/s41586-018-0092-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson E.H. Measurement of diversity. Nature. 1949;163:688. [Google Scholar]
- Sommer F., Bäckhed F. The gut microbiota — masters of host development and physiology. Nat. Rev. Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- Sonnenburg E.D., Smits S.A., Tikhonov M., Higginbottom S.K., Wingreen N.S., Sonnenburg J.L. Diet-induced extinctions in the gut microbiota compound over generations. Nature. 2016;529:212–215. doi: 10.1038/nature16504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong X., Rehman M.U., Huang S., Jiang X., Zhang H., Jia K.L. Comparative analysis of gut microbial community in healthy and tibial dyschondroplasia affected chickens by high throughput sequencing. Microb. Pathog. 2018;118:133–139. doi: 10.1016/j.micpath.2018.03.001. [DOI] [PubMed] [Google Scholar]
- Tremaroli V., Bäckhed F.J.N. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;71:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- Turnbaugh P.J., Ley R.E., Mahowald M.A., Vincent M., Mardis E.R., Gordon J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Wallace J.G., Potts R.H., Szamosi J.C., Surette M.G., Sloboda D.M. The murine female intestinal microbiota does not shift throughout the estrous cycle. Plos One. 2018;13:e0200729. doi: 10.1371/journal.pone.0200729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Kuang Z., Yu X., Ruhn K.A., Kubo M., Hooper L.V. The intestinal microbiota regulates body composition through NFIL3 and the circadian clock. Science. 2017;357:912–916. doi: 10.1126/science.aan0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura Y., Koike K., Okamoto T. Immunolocalization of progesterone and estrogen receptors in the sperm storage tubules of laying and diethylstilbestrol-injected immature hens. Poul. Sci. 2000;79:94–98. doi: 10.1093/ps/79.1.94. [DOI] [PubMed] [Google Scholar]
- Zhao L., Wang G., Siegel P., He C., Wang H., Zhao W., Zhai Z., Tian F., Zhao J., Zhang H., Sun Z., Chen W., Zhang Y., Meng H. Quantitative genetic background of the host influences gut microbiomes in chickens. Sci. Rep. 2013;3:1163. doi: 10.1038/srep01163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.