Abstract
The dynamic development of the animal intestine with a concurrent succession of microbiota and changes in microbial community and metabolite spectrum can exert far-reaching effects on host physiology. However, the precise mechanism of mutual response between microbiota and the gut is yet to be fully elucidated. Broilers with varying developmental degrees of intestinal wall thickness were selected, and they were divided into the thick group (H type) and the thin group (B type), using multiomics data integration analysis to reveal the fundamental regulatory mechanisms of gut–microbiota interplay. Our data showed, in broilers with similar body weight, the intestinal morphological parameters were improved in H type and the diversity of microbial communities is distinguishable from each other. The beneficial bacteria such as Bifidobacterium breve was increased whereas avian endogenous retrovirus EAV-HP was decreased in the H type compared with the B type. Furthermore, microbial metabolic potentials were more active, especially the biosynthesis of folate was improved in the H type. Similarly, the consolidation of absorption, immunity, metabolism, and development was noticed in the thick group. Correlation analysis indicated that the expression levels of material transport and immunomodulatory-related genes were positively correlated with the relative abundance of several probiotics such as B. breve, Lactobacillus saerimneri, and Butyricicoccus pullicaecorum. Our findings suggest that the chickens with well-developed ileal thickness own exclusive microbial composition and metabolic potential, which is closely related to small intestinal morphogenesis and homeostasis.
Key words: broiler, intestinal thickness, microbiota, ileal transcriptome, immune regulation
Introduction
The gastrointestinal tract is an indispensable multifunctional system in the animal body. The surface of the intestinal mucosa is covered with a single layer of columnar epithelial cells forming the villus–crypt axis, which serves as the foundational unit of the intestinal structure and function. The epithelial type of the villus–crypt axis mainly includes absorptive cells that are responsible for transporting and metabolizing nutrients, goblet cells that secrete mucin to form a chemical barrier, and enterochromaffin cells that produce and secrete serotonin at the villus (Crosnier et al., 2006, Gross et al., 2016). LGR5-positive stem cells fuel intestinal self-renewal capacity and paneth cells secrete antibiotic peptides and lysozyme; both types of cells are located at the bottom of Lieberkühn crypt. Besides, paneth cells can maintain stem cell developmental signals in the same niche (Sato et al., 2011). The epithelial cells not only maintain the intestinal morphological structure and physiological functions such as absorption but also orchestrate the core of the mucosal barrier and homeostasis. Importantly, several immune cells are also present that coordinate to tolerate commensal bacteria and prevent pathogen invasion in the intestine (Peterson et al., 2007).
The intestinal lumen of vertebrates including birds harbors innumerable microorganisms, and the metagenome of the microbial community involving ample metabolic pathways is several times bigger than the size of the host genome (Hanning and Diaz-Sanchez, 2015). Interestingly, it was found that not only microbes could exist in digesta but also some special commensal bacteria, for instance, segmented filamentous bacteria (SFB), could adhere to the apical surface of ileal epithelial cells (Chichlowski et al., 2007). In recent years, plentiful studies on chicken intestinal microbiota had emerged; however, the primary focus of these research studies has been on the modulating feasibility of breed, age, feed composition, intestinal segment (mainly cecum), and pathogenic infection on the bacterial community (Gong et al., 2007, Schokker et al., 2015, Awad et al., 2016). In addition to bacteria, symbionts in the digestive tract also include other taxonomic microorganisms such as fungi and even viruses. Most previous studies used the amplicon high-throughput sequencing technology that is known to better clarify the bacterial community composition than the traditional culture-dependent methods. But some of these studies did not deeply explore microbial metabolism. Furthermore, the selection of the 16S rDNA variable region and the use of amplification primers directly affect the outcome of bacterial diversity (Sun et al., 2013). The metagenomic approach can more accurately unravel the changes in the species composition and metabolic functions of the gut microbiome. However, it is not frequently used in the small intestine microecosystem of broilers.
It has been demonstrated that disturbing the composition of gut microbiota can shape gene expression of epithelial cells (Yin et al., 2010). The genetic transcription levels of mammalian enterocytes in the state of proliferation, differentiation, and inflammation were impacted by various transcription factors such as CDX2, NFKB1, and STAT3 (Gao et al., 2009, Jostins et al., 2012). However, the host gene transcriptional regulatory network is more elaborate and complex; therefore, some degree of divergence between different species may also involve other uncharted transcription factors. At the same time, noncoding RNA and DNA methylation can also impair gene expression of intestinal epithelial cells in varied dimensions leading to a shift in tissue development, immune reaction, and even microbial diversity (Jenke et al., 2013, Liu et al., 2016). To our knowledge, there is no report available on gene expression on the intestinal thickness of normal broilers.
Intestinal health has an important influence on animal growth, disease prevention, and product harvest. To fully comprehend the intricate phenomenon of health and disease, a single technique may not suffice to resolve sophisticated biological events. A comprehensive analysis of host transcriptomics and microbiome may contribute to health management with subsequent disease diagnosis and treatment (Karczewski and Snyder, 2018). Although it has been noted that the small intestinal thickness of birds at the same age had a distinct developmental degree, the detailed mechanism of mysterious alteration of intestinal histomorphology still remains unclear. Therefore, we hypothesized that the modification of intestinal microbiota and gene profile might be responsible for the divergence in intestinal thickness during broiler growth. Hence, we used a multiomics integration approach to compare the gut–microbe reciprocities of broilers with dissimilar intestinal thicknesses and tried to elucidate the mechanisms of microbiota modulation to promote gut health and production performance in future.
Materials and methods
Birds, Diet, and Ethics Guidelines
A total of 400 one-day-old male Arbor Acres broilers were obtained from a local commercial hatchery and were reared in 40 cages. A corn–soybean meal–basal diet was formulated to meet the nutrient requirements recommended by the feeding standards for broilers in China (Table 1), and the diet was in mash form. All the birds were allowed ad libitum access to feed and water throughout the experimental period. All the experimental procedures and sample collection methods in this study were approved by the Animal Care and Use Committee of China Agricultural University.
Table 1.
Composition and nutrient levels of the experimental basal diet.1
| Items | Composition |
|---|---|
| Ingredients, % | |
| Corn | 51.38 |
| Soybean oil | 3.75 |
| Soybean meal | 40.71 |
| Calcium hydrogen phosphate | 1.86 |
| Limestone | 1.24 |
| Sodium chloride | 0.35 |
| DL-Methionine (98%) | 0.20 |
| Vitamin premix2 | 0.03 |
| Trace mineral premix3 | 0.20 |
| Choline chloride (50%) | 0.25 |
| Antioxidant | 0.03 |
| Total | 100 |
| Calculated nutrient level4 | |
| Metabolizable energy, MJ/kg | 12.31 |
| Crude protein, % | 22.00 |
| Lysine, % | 1.21 |
| Methionine, % | 0.52 |
| Calcium, % | 1.00 |
| Available phosphorus, % | 0.45 |
Diets were in mash form.
Vitamin premix (1 kg) contained the following ingredients: vitamin A, 50 MIU; vitamin D3, 12 MIU; vitamin K3, 10 g; vitamin B1, 10 g; vitamin B2, 32 g; vitamin B12, 0.1 g; vitamin E, 0.2 MIU; biotin, 0.5 g; folic acid, 5 g; pantothenic acid, 50 g; niacin, 150 g.
Trace mineral premix (1 kg) contained the following ingredients: copper, 4 g; zinc, 90 g; iron, 38 g; manganese, 46.48 g; selenium, 0.1 g; iodine, 0.16 g; cobalt, 0.25 g.
Calculated value based on the analyzed data for the experimental diets.
Experimental Design and Sample Collection
At day 28, all the birds were weighed individually, and 120 birds with similar body weight were selected and euthanized with an injection of 100 mg/kg of pentobarbital sodium (Solarbio, Beijing, China) via the wing vein. The contents of the terminal half of the ileum (the region of the small intestine extending from Meckel's diverticulum to 2 cm cranial to the ileocecal junction) were collected within 3 min of euthanasia, immediately placed in cryogenic vials, snap frozen in liquid nitrogen, delivered to the laboratory, and stored at −80°C until DNA extraction.
Intestinal Histomorphology and Sample Selection
At day 28, the middle segment of the ileum (1 to 2 cm in length) from each bird was excised, fixed in 4% paraformaldehyde immediately after sacrifice, and then embedded in paraffin. Transverse 5-μm sections were stained with hematoxylin and eosin, and histomorphological parameters were examined using an Olympus optical microscope and ProgRes Capture Pro Software (version 2.7, Jenoptik, Jena, Germany). Afterward, the thickness of the ileal wall (including the mucosa, submucosa, muscularis, and serosa) was measured. The birds were ranked from thickest to thinnest by the measured thickness. The chyme samples from broilers with thick ileal walls (H type; S9, S15, S24, S41, S71, S84, S86, S87, S93, and S98) and broilers with thin ileal walls (B type; S13, S18, S33, S34, S37, S39, S40, S56, S59, S79, S90, and S95) were used for subsequent analysis.
DNA Extraction
Genomic DNA was isolated from approximately 200 mg of ileal digesta using a commercial kit (QIAamp DNA Stool Mini Kit, Qiagen Inc., Valencia, CA). The DNA concentration and purity were determined using the NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, MA) or the Qubit (Thermo Fisher Scientific, MA). For in-depth microbial depiction, 2 strategies were adopted: (i) 16S rDNA sequencing for diversity characterization and (ii) metagenomic sequencing for species composition and metabolic function.
16S rDNA Sequencing
Purified genomic DNA at a normalized concentration (20 ng/μl) was used as a template to analyze the microbial communities. The V3-V4 region of 16S rDNA was amplified using universal eubacterial primers 341F: 5′- CCTAYGGGRBGCASCAG-3′ and 806R: 5′-GGACTACNNGGGTATCTAAT-3′. The PCR products were purified using a Qiagen Gel Extraction Kit (Qiagen, Germany) and then sequenced on the HiSeq2500 platform (Illumina, San Diego, CA) at Novogene, Beijing, China.
Sequences were excluded if they did not meet the default QIIME quality criteria. The sequence data were denoised using the denoise_wrapper.py command within QIIME. The chimeras were identified using the UPARSE software (v7.0.1001; Tiburon, CA) and removed. Then, the effective tags were finally obtained and used for further analyses. UPARSE was used to identify operational taxonomic units at 97% similarity on the Bio-Linux platform (NEBC, Oxford, United Kingdom). Representative sequences were processed using the QIIME pipeline. Alignment-based lowest common ancestor (LCA) assignment was carried out based on the SILVA database. Resampling according to the minimum sequence numbers across all samples was performed before the downstream analyses. The community composition provides the classification information at different taxonomic levels. Alpha diversity and beta diversity were analyzed using QIIME (version 1.7.0.355). The principal coordinate analysis was performed based on the weighted Unifrac distance and analysis of similarities used the anosim function of the R vegan package (version 2.15.3; R Foundation, Vienna, Austria) and ggplot2 software packages. Linear discriminant analysis effect size (LEfSe) analyses were performed using the LEfSe tool (http://huttenhower.sph.harvard.edu/lefse/).
Metagenomic Sequencing
DNA library construction was performed following the manufacturer's instructions, and libraries were sequenced using the Illumina Hiseq X Ten instrument. During the experimental process, the S39 and S56 were found to be unqualified and deleted for subsequent analysis. High-quality reads were obtained by filtering low-quality reads, adapter contamination, or DNA contamination (Gallus gallus, Zea mays, and Glycine max) from the raw Illumina data. We used SOAP de novo 2 (v2.0) (Luo et al., 2012) and MetaGene Mark (v.3.38) (Zhu et al., 2010) to perform de novo assembly and gene prediction, respectively, with the high-quality reads. All predicted genes were aligned using CD-HIT (identity > 95% and coverage > 90%) (Fu et al., 2012) to obtain a catalog of nonredundant genes. To obtain the relative abundance of each gene, the high-quality reads from each sample were aligned against the gene catalog by SOAP2 (identity > 95%). We aligned the gene catalog against the NCBI-nr database using DIAMOND (v0.8.22.84) (Buchfink et al., 2015) and performed taxonomic binning by assigning genes in the NCBI Taxonomy database using the LCA algorithm (Huson et al., 2011). MetaPhlAn2 (v.2.0) (Truong et al., 2015) was used to construct species matrices and analyze microbial community composition, and HUMAnN2 (Franzosa et al., 2018) was used to construct metabolic pathway matrices and annotate microbial functions and metabolic capabilities. Samples were clustered at the species level and visualized by principal component analysis implemented in the ade4 R package (R Foundation, Lyon, France). Welch's t-test was executed between the 2 groups of microbial species composition and metabolic pathways using STAMP version 2.1, and the significance analysis was performed using Welch's t-test and Bonferroni correction.
Ileal Transcriptome Analysis
To construct the cDNA library, a total of 20 ileal RNA (H type; S9, S15, S24, S41, S71, S83, S86, S87, S93, and S98; B type; S13, S18, S33, S34, S37, S40, S59, S79, S90, and S95) were extracted using TRIzol (Invitrogen, Carlsbad, CA). The libraries were sequenced on the HiSeq X Ten platform (Illumina). After removing sequencing adapters and trimming consecutive low-quality bases (quality < 20) from both the 5′ and 3′ end of the reads, high-quality RNA-seq reads were aligned to the G. gallus genome (assembly Gallus gallus 5.0) using HISAT2 (v2.0.5) with default parameters. Then, gene expression abundance was quantified by FPKM (Fragments Per Kilobase of exon per Million fragments mapped) using String Tie (version 1.3.1c), and the differentially expressed genes (DEG) between 2 groups were identified using DESeq2 (v.1.20.0) with a P value < 0.05 and fold change > 2 (Rapaport et al., 2013), and the false discovery rate (0.05) was used. The functional protein associations were identified using String (version 11.0) and the interaction network was visualized using Cytoscape (version 3.7.1). The KEGG pathway enrichment analysis of DEG was performed using R software (version 2.15.3).
Correlation Analysis
The gene transcript matrix and the species composition matrix were used for Spearman correlation analysis. First, the Spearman correlation coefficient values of microbial species abundance and intestinal gene expression were calculated using the corr.test function of the psych package (v1.8.12) in R (R Foundation, Evanston, IL), and the significance was tested and then visualized using the pheatmap package. The species abundance as a conditional factor was used to explore the mutual response between the gut and microbe by applying the rda function in the vegan package, and then, it was used to perform sorting analysis. The R (0.8 < |R| < 1) and P value (P value ≤ 0.001) of the influence of condition factors on sample distribution were calculated using the envfit function, and then, the conditional factors with significant effects were selected to perform redundancy analysis (RDA).
Statistical Analysis
Differences were analyzed for the histomorphological parameters and body weight in H group and B group by the independent samples t-test (SPSS 25.0; IBM Corp., Armonk, NY). Data are presented as mean ± SEM. The alpha diversity index between groups was analyzed using the Mann–Whitney U test (SPSS 25.0). The results were shown as median and quartile.
Results
Histological Structure of Ileal Wall Thickness Alienates in Chickens
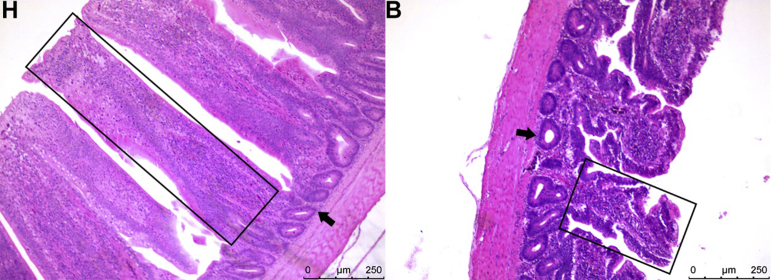
Previous data from our laboratory indicated the heterogeneity in ileal morphology of the normal broiler population, and the Pearson correlation analysis found that there was a significant positive correlation between the ileal wall thickness and body weight (R = 0.575, P = 0.001) at 28 d of age (Supplementary Table 1). As shown in Table 2, there was no significant difference in body weight between the 2 phenotypes, but the ileal thickness in the H type was significantly greater (P < 0.05) than in the B type. In addition, the ileal mucosal thickness, villus height, and crypt depth of the H type were also significantly higher (P < 0.05) than those of the B type. All experimental birds did not have visible pathologic changes. Under the light microscope, it was observed that the villus of the H type was thick, the crypts were closely arranged, and the structural integrity of enterocytes was intact. In contrast, the B type had short villus, dilated crypts, and loose connections between the epithelium (Figure 1).
Table 2.
Histomorphological parameter and body weight of birds with different phenotype at day 28.1
| Item | Ileal thickness, μm | Mucosal thickness, μm | Villus height, μm | Crypt depth, μm | Body weight, kg |
|---|---|---|---|---|---|
| H type | 1214.20 | 986.73 | 829.81 | 156.92 | 1.42 |
| B type | 822.75*** | 662.90*** | 542.47*** | 120.43** | 1.41 |
| SEM | 51.15 | 41.41 | 37.15 | 6.39 | 0.03 |
| P-value | <0.001 | <0.001 | <0.001 | 0.002 | 0.953 |
**P < 0.01, and ***P < 0.001, for B type vs. H type.
Values are means with SEM (n = 10–12).
Figure 1.
Observation of intestinal wall morphology in broilers. Representation of the ileal wall of the H type and B type under a light microscope ( × 40) at day 28, the rectangular frame indicating the villus and the arrow indicating the crypt. Sections were stained with hematoxylin and eosin. Scale bar = 250 μm.
The Microbial Community Diversity Diminishes in H-Type Birds
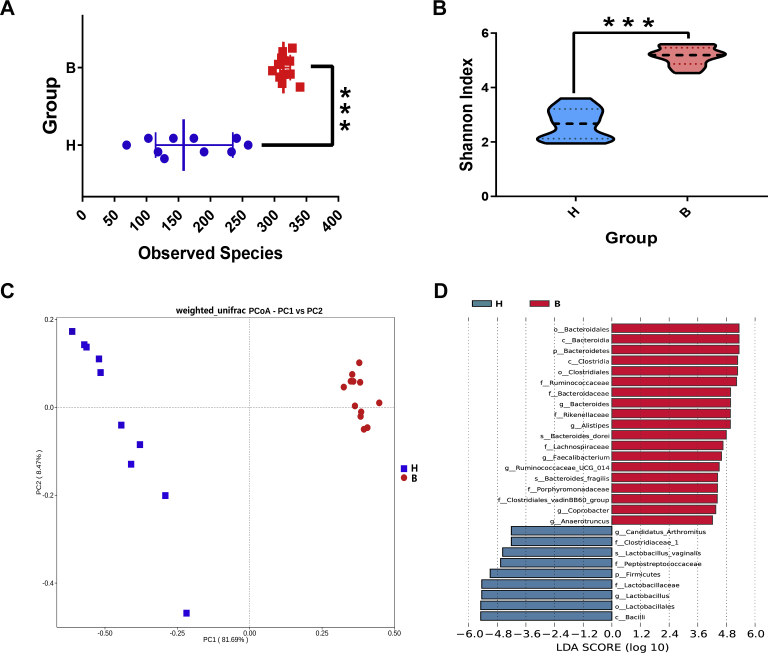
To evaluate the diversity of ileal microbiota in the 2 groups, the observed species number and the Shannon index were calculated using 16S rDNA analysis (Figures 2A, 2B). The results showed that the observed species number (P = 3.09E-06) and the Shannon index (P = 3.09E-06) in the B type were significantly higher than those in the H type. Principal coordinate analysis was used to distinguish the microbial communities, and the results illustrated that the PCo1 contribution value was 81.69% and the PCo2 contribution value was 8.47%. The 2 phenotypes could be separated from each other on the PCo1 axis, and samples from the B type were closely clustered; conversely, H-type samples were more discrete (Figure 2C). Similarly, analysis of similarities (R = 1, P = 0.001) indicated a striking distinctness in the similarity of the microbial community between the groups.
Figure 2.
Bacterial community variation in intestinal thickness. (A) The effect of intestinal thickness on the observed species number in ileal microbiota (97% similarity rate). The values are shown as median and quartile. (B) The effect of intestinal thickness on the Shannon index in ileal microbiota. The values are shown as median and quartile; ***, P < 0.001 (H type, n = 10; B type, n = 12). (C) Principal coordinate analysis (PCoA) plot based on the weighted Unifrac distance for all samples, on which the intestinal thickness (PCo1) made the 2 communities distinctly separated (H type, n = 10; B type, n = 12). (D) The histogram displaying 28 microorganisms with different intestinal thicknesses obtained by linear discriminant analysis effect size. Abbreviation: LDA, linear discriminant analysis.
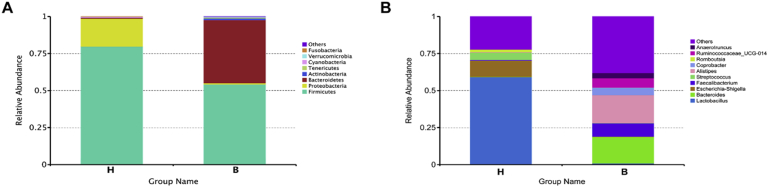
At the phylum level (Supplementary Figure 1A), the ileal microbiota of broilers was mainly composed of Firmicutes, Proteobacteria, and Bacteroides. The proportion of Firmicutes (79.76%) was highest in the H type, followed by Proteobacteria (18.91%) and Bacteroides (0.56%). Interestingly, the proportion of Bacteroidetes (42.55%) in the B type was dramatically high, whereas Firmicutes (54.23%) and Proteobacteria (0.84%) was lower. At the genus level (Supplementary Figure 1B), the percentage of Lactobacillus (59.14%) was highest in the H type, followed by Escherichia–Shigella (11.02%), Streptococcus (4.89%), and Bacteroides (0.22%). However, compared with the H type, the percentage of Alistipes (18.88%), Bacteroides (17.80%), and Faecalibacterium (8.90%) was largely increased, whereas Lactobacillus (0.98%), Escherichia–Shigella (0.28%), and Streptococcus (0.21%) were markedly diminished in the B type. A total of 28 taxonomic microorganisms (LDA > 4) were identified in the ileal microbial communities by using LEfSe, including 9 in the H type and 19 in the B type (Figure 2D). Contrary to the B type, the abundance of Bacilli, Lactobacillus, and Candidatus Arthromitus (SFB) in the ileal microbial community of the H type was extremely elevated, which could be used as preliminary biomarkers for this group.
The Proportion of Beneficial Bacteria Expands in H-Type Birds
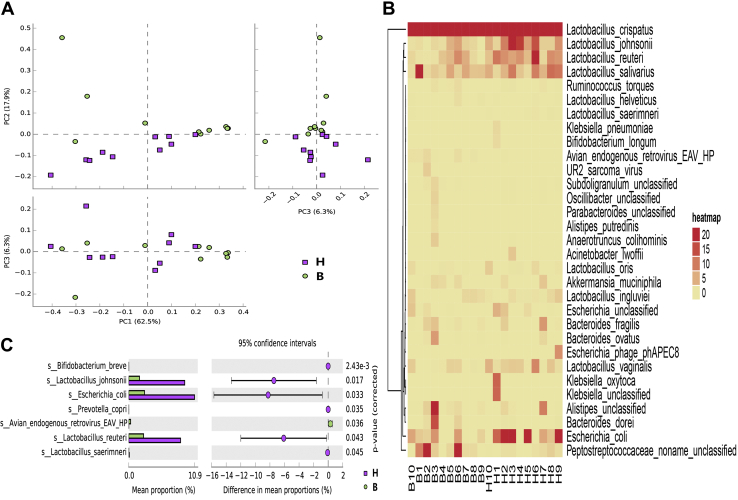
To further clarify alterations in the ileal microbiome, the community compositions of groups were analyzed by metagenomic sequencing. Principal component analysis was used to compare the community compositions at the species level. The results showed that the first principal component contribution was 62.5%, the second principal component contribution was 17.9%, and the third principal component contribution was 6.3%. It was also observed that the samples of the H type were tight, whereas B-type samples exhibited large heterogeneity. In addition, the 2 groups were able to manifest valid separation on the PC2 axis (Figure 3A). It was further found that the ileal contents of broilers primarily consisted of Lactobacillus (80.45% in the H type vs. 79.42% in the B type), and Lactobacillus crispatus as the dominant species had the highest relative abundance (52.98% in the H type vs. 68.44% in the B type) at 28 d of age (Figure 3B). In addition, 8 viruses were detected at the species level (0.80% in the H type vs. 0.54% in the B type) including Escherichia phage phAPEC8, avian myelocytomatosis virus, UR2 sarcoma virus, avian endogenous retrovirus EAV-HP (EAV-HP), and Staphylococcus phage ROSA (Supplementary Figure 2). Subsequently, comparing the ileal microbiota between differential types demonstrated that there were 7 kinds of species with meaningful distinctions (q value ≤ 0.05). Among them, the relative abundance of Bifidobacterium breve, Lactobacillus johnsonii, Escherichia coli, Prevotella copri, Lactobacillus reuteri, and Lactobacillus saerimneri was increased, whereas the relative abundance of EAV-HP was reduced in the H type (Figure 3C).
Figure 3.
The alterations of species composition in intestinal thickness. (A) Principal component analysis demonstrated all samples at the species level (H type, n = 10; B type, n = 10). (B) The clustered heat map of representative species in all samples. (C) STAMP analyzed the ileal microbiome and indicated a total of 7 species with a significant difference (H type, n = 10; B type, n = 10).
Commensal Bacteria Increase the Anabolism Pathway in H-Type Birds
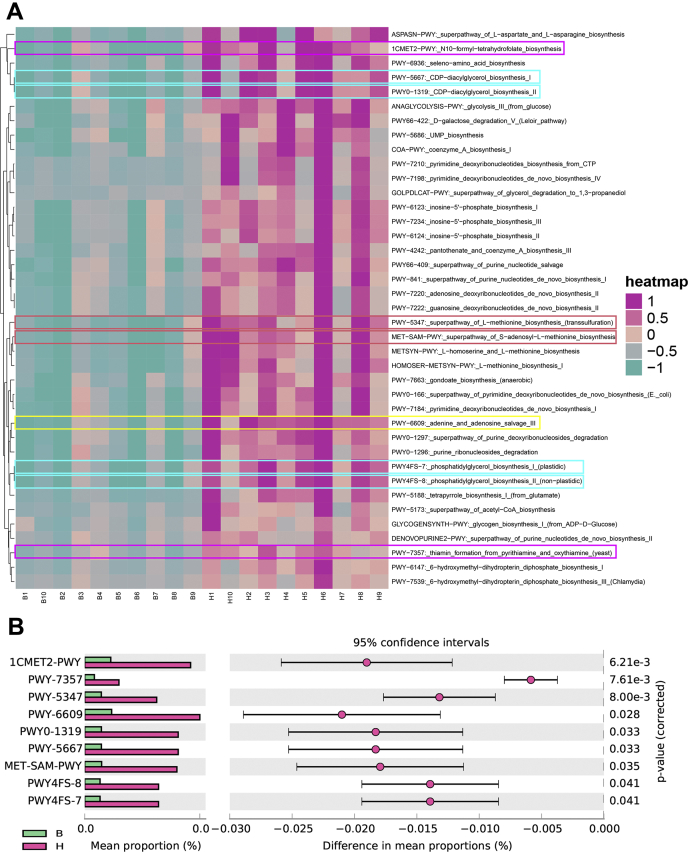
The metagenomic sequencing of microbial function potential revealed that the biosynthesis pathway of ileal microbiota in the H type was significantly more improved than in the B type (Figure 4A). Comparing the microbial metabolic capabilities between phenotypes, a total of 9 metabolic pathways were obtained (q value ≤ 0.05). The pathways involved in vitamin anabolism were N10-formyl-tetrahydrofolate biosynthesis (1CMET2-PWY; q value = 6.21e-3) and thiamine formation from pyrithiamine and oxythiamine (yeast) (PWY-7357; q value = 7.61e-3). The 2 pathways related to methionine anabolism were superpathway of L-methionine biosynthesis (transsulfuration) (PWY-5347; q value = 8.00e-3) and superpathway of S-adenosyl-L-methionine biosynthesis (MET-SAM-PWY; q value = 0.035). One adenine metabolism–related pathway was adenine and adenosine salvage III (PWY-6609; q value = 0.028). Besides, the 4 pathways involved in acylglycerol synthesis were CDP-diacylglycerol biosynthesis I (PWY-5667; q value = 0.033), CDP-diacylglycerol biosynthesis II (PWY0-1319; q value = 0.033), phosphatidylglycerol biosynthesis II (nonplastidic) (PWY4FS-8; q value = 0.041), and phosphatidylglycerol biosynthesis I (plastidic) (PWY4FS-7; q value = 0.041) (Figure 4B).
Figure 4.
The switches of microbial metabolic potentiality with intestinal thickness. (A) The clustered heat map of microbial metabolic pathways in all samples, in which the color boxes indicate distinctive metabolic pathways. (B) STAMP analyzed the core metabolic potentialities of the ileal microbiome in chickens. (H type, n = 10; B type, n = 10).
Small Intestinal Functions Strengthen in H-Type Birds
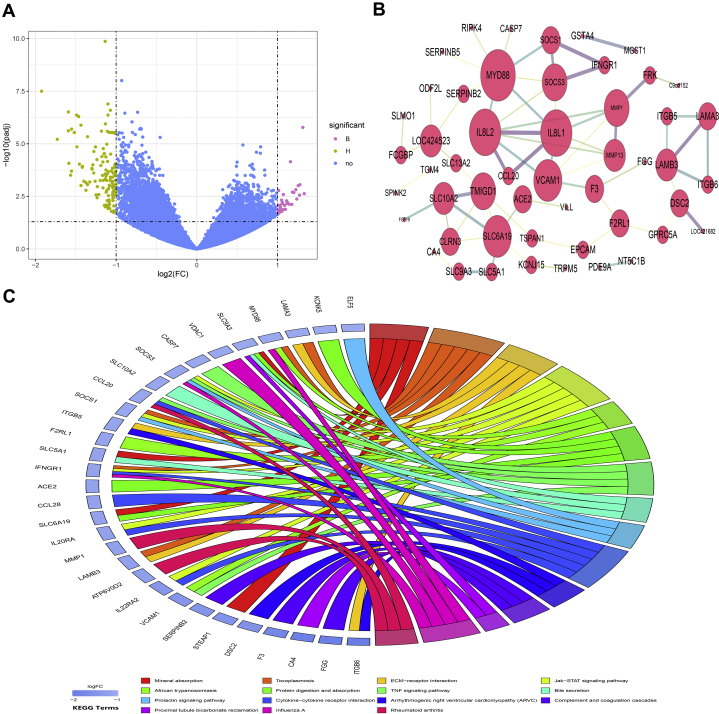
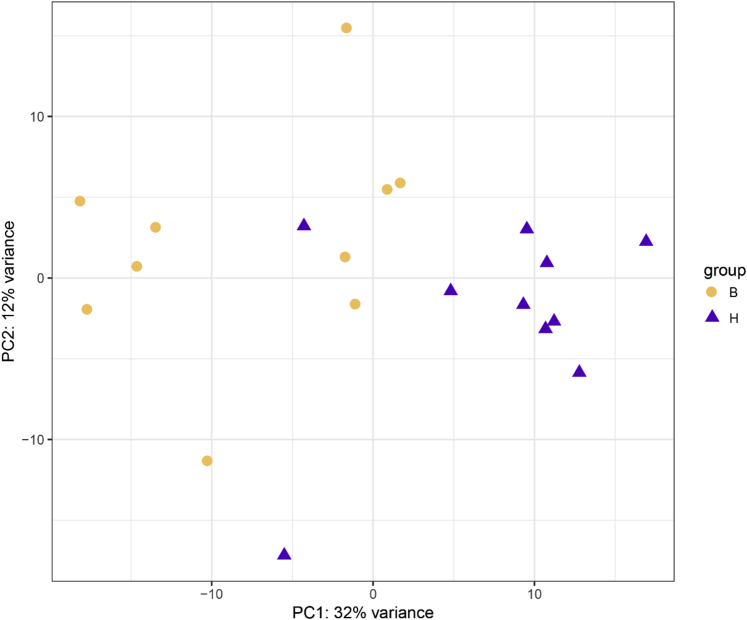
The notable changes in ileal morphology prompted us to explore the underlying molecular mechanisms responsible for differences in intestinal thickness through RNA-seq. Principal component analysis was performed to identify and visualize the similarity of gene expression, with the first principal component contribution being 32% and the second principal component contribution being 12%. The 2 groups could be distinguished from each other, and the H-type samples were closely clustered together (Supplementary Figure 3). A total of 88,839 transcripts were detected in all samples of this study, including 59,470 known annotated transcripts and 29,369 novel transcripts. Using the volcano plot to analyze the DEG (P value < 0.05 and fold change > 2), it was declared that the number of upregulated genes in the H type was 131 and the number of downregulated genes was 35. Intriguingly, most of the downregulated genes were long noncoding RNA or uncharted transcripts (Figure 5A). The protein–protein interaction network showed that there were close interactions between MYD88, SOCS1, SOCS3, IL8L1, and IL8L2 in intestinal tissues. Meanwhile, SLC6A19, SLC5A1, SLC10A2, TMIGD1, and CLRN3 were closely linked (Figure 5B). The KEGG enrichment analysis of differential genes revealed 15 kinds of different pathways (q value < 0.05). Among them, there were 3 pathways related to intestinal absorption in the H type, including mineral absorption (q value = 0.002), protein digestion and absorption (q value = 0.008), and bile secretion (q value = 0.033). Meanwhile, there were also several immunity-related pathways involved in the H type, such as toxoplasmosis (q value = 0.003), JAK-STAT signaling pathway (q value = 0.007), African trypanosomiasis (q value = 0.008), tumor necrosis factor (TNF) signaling pathway (q value = 0.015), and cytokine–cytokine receptor interaction (q value = 0.035). Further connecting the differential KEGG pathway to hub genes, it was discovered that SLC6A19, SLC5A1, and SLC10A2 responsible for absorbing neutral amino acids, glucose, and bile acids were significantly upregulated in the intestinal tract of the H type. It was further found that MYD88, SOCS1, and SOCS3 involved in immune signaling transduction were significantly upregulated in the intestinal tract of the H type (Figure 5C). In addition, the gene expression level of ELF5 was markedly strengthened in the H type compared with the B type.
Figure 5.
Shapes of gene expression in intestinal thickness. (A) The volcano plot shows the differential gene expression in 2 groups, wherein each dot represents 1 gene. The golden dots represent upregulated genes in the H type, the purple dots represent upregulated genes in the B type. (B) The hub protein–protein interaction network, showing 51 nodes and 66 edges. Each node represents a protein, and the links between nodes represent known or predicted functional protein–protein interactions. The degree of a node indicates its number of links to other nodes. (C) Chord diagram mapping the core genes to connect with KEGG pathways.
The Interplay Between Microbial Species and Ileal Gene Expression
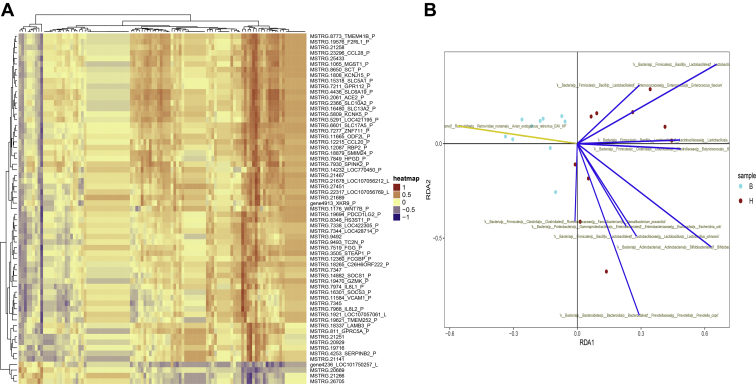
To better understand the relationship between gut microbiota and host gene transcription, we first performed the Spearman correlation analysis on transcriptomic and metagenomic data. A total of 10 microbes were found: B. breve, P. copri, Enterococcus faecium, L. johnsonii, L. saerimneri, Lactobacillus vaginalis, Butyricicoccus pullicaecorum, Faecalibacterium prausnitzii, E. coli, and EAV-HP, which had strongly positive or negative correlation with a total of 63 host gene expressions (P < 0.001) (Figure 6A). Among them, B. breve depicted a significantly positive correlation with 24 genes involved in epithelial function, such as LOC107056212, MSTRG21689, and SLC17A5, and showed a significant negative correlation with MSTRG20669. EAV-HP exhibited a conspicuously negative correlation with 7 genes, such as FGG, PDCD1LG2, and WNT7B, which were probably involved in the regulation of cell fate. Then, using microbial community composition as the conditional factor to observe the response of intestinal gene expression by RDA(m), it was found that the 2 phenotypes of samples could be dispersed on the RDA1 axis. Nine species of bacteria showed a positive correlation with ileal gene expression of H-type samples. However, EAV-HP was the only 1 species that negatively correlated with the H type (Figure 6B).
Figure 6.
The correlational response between gut genes and microbial species. (A) The Spearman correlational heat map of ileal gene expression and microbial species. (B) The abundance of microbial species as a conditional factor for redundancy analysis (RDA)(m). Each line represents a single species of microorganism in the coordinate system. The brown dots represent transcriptomic samples of the H type, and the cyan dots represent transcriptomic samples of the B type.
Discussion
Over the past decade, more and more pieces of evidence about the microbial community compositions and their regulatory influences on host physiology had surfaced. The succession of microbiome usually starts at an early stage of neonates with the villus–crypt axis formation, enterocyte maturation, and gene expression alteration in mammals and birds (Uni et al., 2000, Schokker et al., 2015). Small intestinal epithelial cells could be classified into differentiated types according to cellular morphology and unique gene markers, which mainly perform absorption and barrier functions.
Antibiotics growth promoters (AGP) had long been used as therapeutic or feed additive to prevent bacterial infections and improve growth performance and animal welfare. But the abuse of AGP led to the emergence of resistance and interference with the early colonization, succession, and function of normal microbiota in the gut, which is conceded as a serious threat to public health (Dibner and Richards, 2005). Previously, researchers found that AGP and probiotics as substitutes can obviously increase the body weight of broilers; however, inconsistencies exerted on the small intestinal structure especially on the villus, which became lower than those with probiotics or even basal diets (Salim et al., 2013, Lei et al., 2015). This suggested that changes in microbiome caused by various elements could have dissimilar regulatory patterns on epithelial differentiation and tissue morphogenesis. Moreover, previous chicken studies in our laboratory found that despite the normal clinical features in the populations, a certain proportion of the small intestine, particularly the posterior ileum, became thinner. This hinted that the diversity in the intestinal wall development of broilers might have a tangible connection with microbiota.
The villus is a finger-like structure formed by the protrusion of the intestinal luminal surface during mucosal development and constitutes the central part of the gut wall. This scheme of architecture dramatically increases the mucosal surface area by 10 times, and the villus height is often used as an important parameter in evaluating the absorption capacity of the small intestine in vivo (Walton et al., 2016). Several reports claimed that the probiotics could improve protein utilization and defense capacity by promoting the ileal villus height and the number of neutral goblet cells (Salim et al., 2013, Martinez et al., 2016). This implied that the developmental status of the intestinal wall might improve growth performance in broilers. In this study, to avoid the distinctness in intestinal histomorphology caused by unequal body weight, broilers with similar body weight were selected as subjects. Under the normal physiological circumstances, epithelial cells connect through a variety of tight junction proteins including Zonula Occludens1, Occludin, and Claudins. These proteins interact with cytoskeletal proteins to form the junctional complex and ensure the integrity of the mucosal barrier (Turner, 2009). This, in turn, restricts the entry of microorganisms and food particles from the intestinal lumen into the internal environment. It also regulates the absorption and secretion of fluids and electrolytes (Turner, 2009). However, during intestinal injury, epithelial cells lose their normal morphological characteristics and are detached from the basement membrane, accompanied by massive monocyte infiltration, resulting in loss of the mucosal barrier (Boussenna et al., 2015). The hematoxylin and eosin staining showed that the intestinal architecture and epithelial integrity was intact in the H type, whereas the villus became short, crypts were dilated, and the epithelial cells were disconnected in the B type, indicating that the broilers in the H type had a better physical barrier than the B type. These results illustrated that the difference in intestinal thickness between individuals might closely be related to the ability to absorb nutrients and prevent pathogens.
It is generally believed that richer the alpha diversity and species composition in microbiota, better will be the stability in gut microecosystems (Jackson et al., 2016). Therefore, the alpha diversity can generally be used as an important indicator to judge health status, disease progression, and drug treatment (Nowak et al., 2015). However, our results showed that the diversity in the H type was lower than that in the B type. Previous studies in mammals reported that the species diversity in overweight individuals was reduced, whereas the short-chain fatty acid (SCFA) production, such as acetate and butyrate, and energy acquisition were increased, and it is surmised that “specialized microbiota” may promote weight gain (Turnbaugh et al., 2006). This contradiction may be due to the comparative evaluation of the diversity in varying health status, behavior, and diet, as well as constitutional variance species (Manichanh et al., 2006, Clarke et al., 2014). Microbial composition of the rearing environment and gastrointestinal tract varies considerably in poultry production and can be affected by factors such as age and production batch (Johnson et al., 2018). Therefore, we tried to limit many factors such as age, diet, breed, gender, and house to minimize the fluctuation of the gut microbiome. Principal coordinate analysis and ANOMIS analyses showed that there were overall distinctions in ileal microbiota between the 2 groups and intestinal thickness was the main factor affecting the beta diversity in this experiment, which also indicated the dominant species composition between the 2 types was different. Broilers with less gut microbial species may be more suitable for better feed utilization and weight gain, and the assessment of gut health and productive performance in traditional microbiota diversity during broiler rearing needs to be revised. The ileal bacterial composition of broilers in this experiment was consistent with previous reports and also reflected the existence of “inherent members” during the succession of gut microbiota (Xiao et al., 2017). LEfSe analysis showed that the abundance of Firmicutes and Lactobacillus in the ileum of the H type ascended, whereas the abundance of Bacteroidetes and Bacteroides descended. A sharp increase of Lactobacillus in the ileum may be responsible for the reduction of other species of bacteria and ultimate decrease in the diversity in the H phenotype. It has been reported that obese individuals had a higher number of Firmicutes with less number of Bacteroides, and the overweight phenotype can be obtained by transplanting the microbiota from the obese mammal into germ-free or slim individuals (Ley et al., 2006). As the activity of bile salt hydrolase between bacteria is not the same, it can play a vital role in weight change by regulating key genes involved in lipid metabolism, immune homeostasis, and the circadian rhythm (Joyce et al., 2014). Lactobacillus acts as an essential indigenous probiotic in the animal symbiotic system. It can metabolize and produce multiple organic acids to reduce the pH of chyme; thus, it may inhibit the enterobacterium growth and regulate the gene expression level of epithelial cells. However, it had also been reported that some lactobacilli such as Lactobacillus salivarius can exert negative effects on broiler performance (Torok et al., 2011). We speculated that this inconsistency might be due to changes in dietary ingredients and animal species in each trial. In addition, each microbial genus in the gut includes several species and strains, and there could be distinct metabolic functions between the bacteria. In this study, it was discovered that the abundance of SFB in the B type was significantly increased. Previously, researchers had found that SFB can promote the maturation of B cells and T cells in newborn animals and can also induce cytokines including IL-23 in the intestine to affect the differentiation of Th17 cells in mammals (Schnupf et al., 2015). Liao et al. (2012) reported that the abundance of SFB in the ileum reached its peak at 11 D in chickens and then decreased, while the supplementation of Lactobacillus delbrueckii accelerated the early colonization of SFB. This suggested that there might be differences in the intestinal immunity of the 2 types of broilers. These results indicated that although the diversity of ileal microbiota in the H type decreased, the specialized bacterial members such as Lactobacillus and SFB as microbiomarkers could be beneficial to weight gain and immune development.
The similar microbial species in the 2 groups predicted the presence of specific intrinsic species in the ileal contents of broilers. This was distinct from our 16S rDNA analysis, but it might be due to different technical methods. It was similar to the previous report on the intestinal metagenome of domestic chicken explaining that Lactobacillus is the dominant species in the small intestine, which may competitively inhibit pathogens by forming a negative correlation network with several bacteria such as Staphylococcus (Huang et al., 2018). We also detected numerous viruses belonging to Myoviridae and Retroviridae at the family level. Gut phages generally encode genes beneficial to bacteria helping host bacteria adapt to the external environment, expressing bacterial virulence factors, and maintaining microbiota stability and elasticity (Ogilvie and Jones, 2015). This implies that the phage may affect the composition of beneficial bacteria and pathogenic bacteria in microecosystem in a nonhost immunomodulatory manner in the gut, thereby affecting animal health and disease state. The Retroviridae contains many widely distributed animal viruses. Some exogenous retroviruses can be amplified by a unique replication method using reverse transcriptase after invading the host cell, and the viral genome can be inserted into the host chromosome by integrase. Endogenous retroviruses (ERV) are a class of viruses that integrate their gene fragments into the host genome, including that of domestic chickens, and can replicate, evolve, and coexist with animals (Gifford and Tristem, 2003). EAV-HP is an ancient endogenous transcriptional virus that persists in modern poultry, which is inseparable with the high pathogenic avian leukosis virus subgroup-J. Wragg et al. (2015) analyzed several poultry genomes and found that EAV-HP had approximately 75 integration sites on each bird, and the related genes revealed that it may be involved in immune regulation and cell adhesion. Change in the abundance of ERV such as EAV-HP in the ileum may induce the regulation of gene activity and immune signals in the host cells. Maukonen et al. (2015) reported that the feces of children with inflammatory bowel disease were characterized by a decrease in the number of bifidobacteria and lactobacilli, accompanied by an increase in the expression of proinflammatory cytokines such as IL6. B. breve can activate the TLR2-MYD88 signaling pathway in the intestinal CD103 + dendritic cells to promote IL-10 production, which in turn induces an increase in the number of regulatory T cells (Jeon et al., 2012). Combined with the aforementioned evidence, B. breve in the intestinal tract can boost the production of anti-inflammatory cytokines such as IL-10 and reduce proinflammatory cytokines levels such as those of IL-6, thereby improving the immune microenvironment to maintain the microecological balance. Young et al. (2012) found that when an aged mouse becomes immunodeficient, murine leukemia virus could be activated to infect the host and induce tumorigenesis. Interestingly, the partial or complete absence of gut microbiota may effectively block the ERV infection. Loss of TLR7 which interacts with the viral nucleic acid can directly lead to ERV activation to produce retroviral viremia, while the deletion of TLR3 and TLR9 can promote the integration of ERV into the genome of cells deriving oncogene failure (Yu et al., 2012). Normally, homeostasis is maintained between the gut microbiota and host immune system; however, once the local balance is disturbed, it can lead to dysbiosis and accelerate disease progression. Although we were unable to determine the source of ERV in the intestinal contents (exogenous secretion of enterocytes or exfoliated DNA fragment reprocessing or even external environment), the aforementioned information suggests that the “denormalized” microbiota may suppress immunosurveillance and promote ERV activation in the gut. Our data indicated a bidirectional response between bacteria, viruses, and host immune molecules, forming a sophisticated symbiotic system that can potentially regulate the intestinal epithelial cell development and maturation.
The metabolic functions of gut microbiome in poultry had been reviewed extensively and indicated that the production of vitamins and amino acids was much higher in the hindgut than in the foregut (Ferrario et al., 2017, Huang et al., 2018). In the present study, the ileal microbial synthesis in the H type was enhanced compared with that in the B type, especially the nutrient production pathways such as those of folate and methionine were enhanced, which could be used by the animal. We did not measure the actual amount of metabolites in the ileal chyme; however, the previous study had reported that bifidobacteria could synthesize folate in feces, which could be used as a probiotic to increase the folate levels in serum and the liver for folate-deficient mice, thereby improving the physical health (Pompei et al., 2007). The main animal physiological regulations involving folate are the following: (i) participation in basic purine nucleotide biosynthesis; (ii) anti-inflammation, folate supplementation can reduce the expression of IL1B in the cecal tonsil and increase the level of serum IgG when young hens are stimulated by lipopolysaccharide (Munyaka et al., 2012); (iii) folate produced by microbes might also affect epigenetic modification as a carbon unit (Mischke and Plosch, 2013). In addition, S-adenosylmethionine can also act as a methyl donor to influence the degree of methylation in different DNA regions to regulate transcriptional activity (Mischke and Plosch, 2013). It is quite evident from the aforementioned information that several functional nutrients can be synthesized via the ileal microbiome, which not only can directly ensure physiologically important metabolic reactions in animals but also can have potential regulatory effects at the genetic level. Our data also showed an enhanced adenine metabolism of ileal microbiota in the H type. Huang et al. (2018) showed that the pathways involved in nucleotide metabolism, DNA replication and repair, and transcription were more active in the small intestine than in the large intestine, partly because the nucleotides were essential components for microorganism proliferation and growth. Besides, the microbial metabolites such as SCFA can directly act as signaling molecules to mediate the transformation of epithelial immune activity. A recent study had shown that the columnar epithelium at the tip of the small intestine villus expressed a large number of genes involved in purine metabolism, such as CD73 which acts as an ecto-5′-nucleotidase to convert bacterially derived ATP to adenosine (Moor et al., 2018). This suggested that the microbiome may prevent the excessive activation of local inflammation by reducing bacterial ATP as a danger signal, and this could also promote tolerance to commensal bacteria; sequentially, it could have been of benefit for colonization and growth in the H type. These results indicated that the microbial metabolism which produced a variety of nutrients beneficial for the host in the ileum was enhanced, and it might alter the transcriptional regulation of epithelial cells and affect intestinal morphogenesis and immunologic progress.
Our results showed that the number of upregulated genes in the H type was more than in the B type, hinting a noteworthy difference in intestinal function. Enrichment analysis declared that there was a disparity in ileal absorption between the groups, and the expression levels of multiple solute carrier (SLC) family members were increased in the H type. Major members of the SLC family can act as cotransporters to assist the movement of vital substances such as inorganic ions, amino acids, lipids, and monosaccharides across the cellular membrane using ionic gradients formed by sodium or hydrion, dysfunction of which can lead to different kinds of diseases (Bai et al., 2017). It had previously been reported that using a microarray to analyze the duodenum with differences in feed utilization efficiency displayed many genes involved in nutrient uptake in broilers (Lee et al., 2015). In contrast, we used RNA sequencing to discover many long noncoding RNA and unknown transcripts, although their biological functions need to be further explored. However, the current information suggested that the alteration of nutrient transports involved in absorption might turn to be one of the main reasons for the distinct growth rate. Interestingly, it was also found that a lot of gene expressions involved in immunity were upregulated in the small intestine of the H type. Although the effects of different immune genes on intestinal microbiota are not completely consistent (Thoene-Reineke et al., 2014), it had been reported that deletion of MYD88 generated a decrease in microbe-induced expression of the colonic antibacterial peptide gene and an excessive increase of SFB in the ileum, but compared with germ-free mice, the colonic antiviral genes in a conventional animal were markedly increased accompanied with norovirus infection (Larsson et al., 2012). As an important negative feedback regulator in the JAK-STAT signaling pathway, SOCS1 can play an important role in inhibiting the excessive immune response. It has been found that SOCS1 did not have complete coincident effect with IL-10 in maintaining intestinal immune tolerance to microorganisms (Chinen et al., 2011). The intestinal tract can reduce the occurrence of excessive inflammation by maintaining remarkable immune surveillance, tolerance, and defense to establish commensalism in the H type. It had been established that ELF5 is an important transcription factor playing a key role in the development of the mammary gland, and a recent study had shown that ELF5 can also act as a core switch to determine the trophoderm stem cell fate and gene regulatory network (Latos et al., 2015). There is barely any detailed evidence about ELF5 on the development of normal intestinal epithelial cells, and we speculated that the intestinal thickness might be related to ELF5 and its interaction network. Intriguingly, apart from MYD88 increase in the Toll-like pathway, we also observed strengthening of the TLR3 expression. Unlike other usual TLR members using MYD88 as an adaptor, TLR3 uses TRIF, in the MYD88-independent manner, as an adaptor to induce interferon beta (INFβ), presenting a unique role in host antiviral defense. TLR3 was also able to sense the dsDNA released from damaged tissue to initiate stem cell regeneration (Nelson et al., 2015). Multiple molecular interactions can play an important role in maintaining epithelium development and immune function. Overall, genes involved in absorption, immunity, and metabolism were upregulated in the ileum of the H type, which seems to be more beneficial for animal health and welfare.
The shifts in the microbial composition induce a valid effect on intestinal gene expression and alterations in the transcriptional activity, which can ultimately shape microbial community. However, the inseparable interaction between the host and microbiota still remains elusive. The correlation analysis in this study indicated that the symbiotic bacteria such as B. breve and Lactobacillus were positively correlated with the genes involved in substance transport and immune regulation, yet these genes had a negative correlation with EAV-HP. SLC17A5 acts as a transporter to efficiently carry sialic acid or glucuronic acid and when mutated can cause severe infant sialic acid storage disease. It was found that salivary gland cells secreted nitrate into saliva through SLC17A5, reducing the nitrate concentration in serum (Qin et al., 2012). The nitrate can be transformed to nitrate via the oral microbiota, which then enters the stomach to produce nitric oxide, and can protect the gastric mucosa from stimulation (Qin et al., 2012). Recent proofs elucidated that the extracellular sialidase could help Bifidobacterium bifidum to enhance carbohydrate utilization and adhesion to enterocytes (Nishiyama et al., 2017). The combination of SLC17A5 and B. breve exhibits a highly positive correlation, suggesting that B. breve may use similar mechanisms to use sialic acid secreted by epithelial cells to increase colonization and growth in intestinal habitats. Alterations in the expression of specific genes such as SLC17A5 in the intestinal epithelium could form a host-selective pressure to remodify the corresponding symbiotic microorganisms, which may initiate feedback to regulate the host transcriptional networks. Taweechotipatr et al. (2009) isolated a variety of lactic acid bacteria from healthy human stool and found that different strains exert inconsistent effects on immune regulation and L. saerimneri can effectively reduce TNFα production to inhibit inflammation. Our data showed that L. saerimneri demonstrated a significant positive correlation with several immune molecules such as SOCS1 and SOCS 3, suggesting that L. saerimneri might play a part in relieving inflammation through SOCS1- and SOCS3-related signaling in broilers. We also found that B. pullicaecorum, an anaerobic indigenous species present in the hindgut contents of chickens, showed a positive correlation not only with transporters such as SLC6A19 and SLC10A2 but also with CCL28. B. pullicaecorum possesses the ability to produce SCFA such as butyrate, which is believed to ameliorate pathological damage caused by Clostridium perfringens and reduce the number of Escherichia–Shigella in the ileum, thereby promoting broiler growth (Eeckhaut et al., 2016). CCL28 not only acts as a chemokine to attract plasma cells to specific mucosal sites but also performs as a broad-spectrum antibacterial molecule against bacteria and Candida albicans (Hieshima et al., 2003). This study confirmed that B. pullicaecorum may transform the mucosal microenvironment by increasing the number of transporters to promote the absorption of neutral amino acids and bile salts and stimulating secretion of immune effectors. The RDA indicated that the microbial community was able to distinguish between host gene expressions. At present, most of the experimental data are being derived from germ-free or knock-out model animals by cross-sectional studies. However, microbial compositions and metabolites are constantly changing during normal animal development. Therefore, it cannot entirely uncover the causal relationship between the host and microbiome without spatiotemporal analysis in the future. In a word, our results indicated that the genes involved in absorption and immunity in the gut had a compact mutual response with the microbial members and metabolites.
In conclusion, broilers with a well-developed ileal thickness have reduced microbiota diversity in the lumen and increased abundance of beneficial bacteria coupled with active community anabolism, thus enhancing the absorption and immune function of epithelial cells. This study provides a theoretical basis for potential probiotic exploitation and application to improve intestinal health and microecological strategies.
Acknowledgments
This study was financially supported by the China Agriculture Research System (grant no. CARS-41-G11). The authors declare that there are no conflicts of interest.
Footnotes
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.psj.2019.10.029.
Supplementary data
Fig S1.
Effect of intestinal thickness on the bacterial composition. (A) The relative abundance of Top10 microbe at the phylum level. (B) The relative abundance of Top10 microbe at the genus level.
Fig S2.
Influence of intestinal thickness on the species composition of ileal microbiota. The histogram shows the relative abundance of the microorganism at the species level.
Fig S3.
Impact of intestinal thickness on the similarity of ileal transcriptome. PCA analyzed all transcriptome samples showing 32% differences in the PC1 axis and 12% differences in the PC2 axis (H type, n = 10; B type, n = 10). PCA, principal component analysis.
References
- Awad W.A., Mann E., Dzieciol M., Hess C., Schmitz-Esser S., Wagner M., Hess M. Age-related differences in the luminal and mucosa-associated gut microbiome of broiler chickens and shifts associated with Campylobacter jejuni infection. Front. Cell. Infect. Microbiol. 2016;6:154. doi: 10.3389/fcimb.2016.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X., Moraes T.F., Reithmeier R.A.F. Structural biology of solute carrier (SLC) membrane transport proteins. Mol. Membr. Biol. 2017;34:1–32. doi: 10.1080/09687688.2018.1448123. [DOI] [PubMed] [Google Scholar]
- Boussenna A., Goncalves-Mendes N., Joubert-Zakeyh J., Pereira B., Fraisse D., Vasson M.P., Texier O., Felgines C. Impact of basal diet on dextran sodium sulphate (DSS)-induced colitis in rats. Eur. J. Nutr. 2015;54:1217–1227. doi: 10.1007/s00394-014-0800-2. [DOI] [PubMed] [Google Scholar]
- Buchfink B., Xie C., Huson D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- Chichlowski M., Croom W.J., Edens F.W., McBride B.W., Qiu R., Chiang C.C., Daniel L.R., Havenstein G.B., Koci M.D. Microarchitecture and spatial relationship between bacteria and ileal, cecal, and colonic epithelium in chicks fed a direct-fed microbial, PrimaLac, and salinomycin. Poult. Sci. 2007;86:1121–1132. doi: 10.1093/ps/86.6.1121. [DOI] [PubMed] [Google Scholar]
- Chinen T., Komai K., Muto G., Morita R., Inoue N., Yoshida H., Sekiya T., Yoshida R., Nakamura K., Takayanagi R., Yoshimura A. Prostaglandin E2 and SOCS1 have a role in intestinal immune tolerance. Nat. Commun. 2011;2:190. doi: 10.1038/ncomms1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke S.F., Murphy E.F., O'Sullivan O., Lucey A.J., Humphreys M., Hogan A., Hayes P., O'Reilly M., Jeffery I.B., Wood-Martin R., Kerins D.M., Quigley E., Ross R.P., O'Toole P.W., Molloy M.G., Falvey E., Shanahan F., Cotter P.D. Exercise and associated dietary extremes impact on gut microbial diversity. Gut. 2014;63:1913–1920. doi: 10.1136/gutjnl-2013-306541. [DOI] [PubMed] [Google Scholar]
- Crosnier C., Stamataki D., Lewis J. Organizing cell renewal in the intestine: stem cells, signals and combinatorial control. Nat. Rev. Genet. 2006;7:349–359. doi: 10.1038/nrg1840. [DOI] [PubMed] [Google Scholar]
- Dibner J.J., Richards J.D. Antibiotic growth promoters in agriculture: history and mode of action. Poult. Sci. 2005;84:634–643. doi: 10.1093/ps/84.4.634. [DOI] [PubMed] [Google Scholar]
- Eeckhaut V., Wang J., Van Parys A., Haesebrouck F., Joossens M., Falony G., Raes J., Ducatelle R., Van Immerseel F. The probiotic Butyricicoccus pullicaecorum reduces feed conversion and protects from potentially harmful intestinal microorganisms and necrotic enteritis in broilers. Front. Microbiol. 2016;7:1416. doi: 10.3389/fmicb.2016.01416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrario C., Alessandri G., Mancabelli L., Gering E., Mangifesta M., Milani C., Lugli G.A., Viappiani A., Duranti S., Turroni F., Ossiprandi M.C., Hiyashi R., Mackie R., van Sinderen D., Ventura M. Untangling the cecal microbiota of feral chickens by culturomic and metagenomic analyses. Environ. Microbiol. 2017;19:4771–4783. doi: 10.1111/1462-2920.13943. [DOI] [PubMed] [Google Scholar]
- Franzosa E.A., McIver L.J., Rahnavard G., Thompson L.R., Schirmer M., Weingart G., Lipson K.S., Knight R., Caporaso J.G., Segata N., Huttenhower C. Species-level functional profiling of metagenomes and metatranscriptomes. Nat. Methods. 2018;15:962–968. doi: 10.1038/s41592-018-0176-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L., Niu B., Zhu Z., Wu S., Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao N., White P., Kaestner K.H. Establishment of intestinal identity and epithelial-mesenchymal signaling by Cdx2. Dev. Cell. 2009;16:588–599. doi: 10.1016/j.devcel.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford R., Tristem M. The evolution, distribution and diversity of endogenous retroviruses. Virus Genes. 2003;26:291–316. doi: 10.1023/a:1024455415443. [DOI] [PubMed] [Google Scholar]
- Gong J., Si W., Forster R.J., Huang R., Yu H., Yin Y., Yang C., Han Y. 16S rRNA gene-based analysis of mucosa-associated bacterial community and phylogeny in the chicken gastrointestinal tracts: from crops to ceca. FEMS Microbiol. Ecol. 2007;59:147–157. doi: 10.1111/j.1574-6941.2006.00193.x. [DOI] [PubMed] [Google Scholar]
- Gross S., Garofalo D.C., Balderes D.A., Mastracci T.L., Dias J.M., Perlmann T., Ericson J., Sussel L. The novel enterochromaffin marker Lmx1a regulates serotonin biosynthesis in enteroendocrine cell lineages downstream of Nkx2.2. Development (Cambridge, England) 2016;143:2616–2628. doi: 10.1242/dev.130682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanning I., Diaz-Sanchez S. The functionality of the gastrointestinal microbiome in non-human animals. Microbiome. 2015;3:51. doi: 10.1186/s40168-015-0113-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieshima K., Ohtani H., Shibano M., Izawa D., Nakayama T., Kawasaki Y., Shiba F., Shiota M., Katou F., Saito T., Yoshie O. CCL28 has dual roles in mucosal immunity as a chemokine with broad-spectrum antimicrobial activity. J. Immunol. 2003;170:1452–1461. doi: 10.4049/jimmunol.170.3.1452. [DOI] [PubMed] [Google Scholar]
- Huang P., Zhang Y., Xiao K., Jiang F., Wang H., Tang D., Liu D., Liu B., Liu Y., He X., Liu H., Liu X., Qing Z., Liu C., Huang J., Ren Y., Yun L., Yin L., Lin Q., Zeng C., Su X., Yuan J., Lin L., Hu N., Cao H., Huang S., Guo Y., Fan W., Zeng J. The chicken gut metagenome and the modulatory effects of plant-derived benzylisoquinoline alkaloids. Microbiome. 2018;6:211. doi: 10.1186/s40168-018-0590-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D.H., Mitra S., Ruscheweyh H.J., Weber N., Schuster S.C. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011;21:1552–1560. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M.A., Jeffery I.B., Beaumont M., Bell J.T., Clark A.G., Ley R.E., O'Toole P.W., Spector T.D., Steves C.J. Signatures of early frailty in the gut microbiota. Genome Med. 2016;8:8. doi: 10.1186/s13073-016-0262-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenke A.C., Postberg J., Raine T., Nayak K.M., Molitor M., Wirth S., Kaser A., Parkes M., Heuschkel R.B., Orth V., Zilbauer M. DNA methylation analysis in the intestinal epithelium-effect of cell separation on gene expression and methylation profile. PLoS One. 2013;8:e55636. doi: 10.1371/journal.pone.0055636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon S.G., Kayama H., Ueda Y., Takahashi T., Asahara T., Tsuji H., Tsuji N.M., Kiyono H., Ma J.S., Kusu T., Okumura R., Hara H., Yoshida H., Yamamoto M., Nomoto K., Takeda K. Probiotic Bifidobacterium breve induces IL-10-producing Tr1 cells in the colon. PLoS Pathog. 2012;8:e1002714. doi: 10.1371/journal.ppat.1002714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T.J., Youmans B.P., Noll S., Cardona C., Evans N.P., Karnezos T.P., Ngunjiri J.M., Abundo M.C., Lee C.W. A consistent and predictable commercial broiler chicken bacterial microbiota in antibiotic-free production displays strong correlations with performance. Appl. Environ. Microbiol. 2018;84:e00362-18. doi: 10.1128/AEM.00362-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jostins L., Ripke S., Weersma R.K., Duerr R.H., McGovern D.P., Hui K.Y., Lee J.C., Schumm L.P., Sharma Y., Anderson C.A., Essers J., Mitrovic M., Ning K., Cleynen I., Theatre E., Spain S.L., Raychaudhuri S., Goyette P., Wei Z., Abraham C., Achkar J.P., Ahmad T., Amininejad L., Ananthakrishnan A.N., Andersen V., Andrews J.M., Baidoo L., Balschun T., Bampton P.A., Bitton A., Boucher G., Brand S., Buning C., Cohain A., Cichon S., D'Amato M., De Jong D., Devaney K.L., Dubinsky M., Edwards C., Ellinghaus D., Ferguson L.R., Franchimont D., Fransen K., Gearry R., Georges M., Gieger C., Glas J., Haritunians T., Hart A., Hawkey C., Hedl M., Hu X., Karlsen T.H., Kupcinskas L., Kugathasan S., Latiano A., Laukens D., Lawrance I.C., Lees C.W., Louis E., Mahy G., Mansfield J., Morgan A.R., Mowat C., Newman W., Palmieri O., Ponsioen C.Y., Potocnik U., Prescott N.J., Regueiro M., Rotter J.I., Russell R.K., Sanderson J.D., Sans M., Satsangi J., Schreiber S., Simms L.A., Sventoraityte J., Targan S.R., Taylor K.D., Tremelling M., Verspaget H.W., De Vos M., Wijmenga C., Wilson D.C., Winkelmann J., Xavier R.J., Zeissig S., Zhang B., Zhang C.K., Zhao H., International I.B.D.G.C., Silverberg M.S., Annese V., Hakonarson H., Brant S.R., Radford-Smith G., Mathew C.G., Rioux J.D., Schadt E.E., Daly M.J., Franke A., Parkes M., Vermeire S., Barrett J.C., Cho J.H. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce S.A., MacSharry J., Casey P.G., Kinsella M., Murphy E.F., Shanahan F., Hill C., Gahan C.G. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc. Natl. Acad. Sci. U S A. 2014;111:7421–7426. doi: 10.1073/pnas.1323599111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski K.J., Snyder M.P. Integrative omics for health and disease. Nat. Rev. Genet. 2018;19:299–310. doi: 10.1038/nrg.2018.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson E., Tremaroli V., Lee Y.S., Koren O., Nookaew I., Fricker A., Nielsen J., Ley R.E., Backhed F. Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88. Gut. 2012;61:1124–1131. doi: 10.1136/gutjnl-2011-301104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latos P.A., Sienerth A.R., Murray A., Senner C.E., Muto M., Ikawa M., Oxley D., Burge S., Cox B.J., Hemberger M. Elf5-centered transcription factor hub controls trophoblast stem cell self-renewal and differentiation through stoichiometry-sensitive shifts in target gene networks. Genes Dev. 2015;29:2435–2448. doi: 10.1101/gad.268821.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Karnuah A.B., Rekaya R., Anthony N.B., Aggrey S.E. Transcriptomic analysis to elucidate the molecular mechanisms that underlie feed efficiency in meat-type chickens. Mol. Genet. Genomics. 2015;290:1673–1682. doi: 10.1007/s00438-015-1025-7. [DOI] [PubMed] [Google Scholar]
- Lei X., Piao X., Ru Y., Zhang H., Peron A., Zhang H. Effect of Bacillus amyloliquefaciens-based direct-fed microbial on performance, nutrient utilization, intestinal morphology and cecal Microflora in broiler chickens. Asian-Australas J. Anim. Sci. 2015;28:239–246. doi: 10.5713/ajas.14.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R.E., Turnbaugh P.J., Klein S., Gordon J.I. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Liao N., Yin Y., Sun G., Xiang C., Liu D., Yu H.D., Wang X. Colonization and distribution of segmented filamentous bacteria (SFB) in chicken gastrointestinal tract and their relationship with host immunity. FEMS Microbiol. Ecol. 2012;81:395–406. doi: 10.1111/j.1574-6941.2012.01362.x. [DOI] [PubMed] [Google Scholar]
- Liu S., da Cunha A.P., Rezende R.M., Cialic R., Wei Z., Bry L., Comstock L.E., Gandhi R., Weiner H.L. The host shapes the gut microbiota via fecal MicroRNA. Cell Host Microbe. 2016;19:32–43. doi: 10.1016/j.chom.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J., He G., Chen Y., Pan Q., Liu Y., Tang J., Wu G., Zhang H., Shi Y., Liu Y., Yu C., Wang B., Lu Y., Han C., Cheung D.W., Yiu S.M., Peng S., Xiaoqian Z., Liu G., Liao X., Li Y., Yang H., Wang J., Lam T.W., Wang J. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichanh C., Rigottier-Gois L., Bonnaud E., Gloux K., Pelletier E., Frangeul L., Nalin R., Jarrin C., Chardon P., Marteau P., Roca J., Dore J. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez E.A., Babot J.D., Lorenzo-Pisarello M.J., Apella M.C., Chaia A.P. Feed supplementation with avian Propionibacterium acidipropionici contributes to mucosa development in early stages of rearing broiler chickens. Benef Microbes. 2016;7:687–698. doi: 10.3920/BM2016.0077. [DOI] [PubMed] [Google Scholar]
- Maukonen J., Kolho K.L., Paasela M., Honkanen J., Klemetti P., Vaarala O., Saarela M. Altered fecal microbiota in paediatric inflammatory bowel disease. J. Crohns Colitis. 2015;9:1088–1095. doi: 10.1093/ecco-jcc/jjv147. [DOI] [PubMed] [Google Scholar]
- Mischke M., Plosch T. More than just a gut instinct-the potential interplay between a baby's nutrition, its gut microbiome, and the epigenome. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2013;304:R1065–R1069. doi: 10.1152/ajpregu.00551.2012. [DOI] [PubMed] [Google Scholar]
- Moor A.E., Harnik Y., Ben-Moshe S., Massasa E.E., Rozenberg M., Eilam R., Bahar Halpern K., Itzkovitz S. Spatial reconstruction of single enterocytes uncovers broad zonation along the intestinal villus axis. Cell. 2018;175:1156–1167.e15. doi: 10.1016/j.cell.2018.08.063. [DOI] [PubMed] [Google Scholar]
- Munyaka P.M., Tactacan G., Jing M., O K., House J.D., Rodriguez-Lecompte J.C. Immunomodulation in young laying hens by dietary folic acid and acute immune responses after challenge with Escherichia coli lipopolysaccharide. Poult. Sci. 2012;91:2454–2463. doi: 10.3382/ps.2012-02381. [DOI] [PubMed] [Google Scholar]
- Nelson A.M., Reddy S.K., Ratliff T.S., Hossain M.Z., Katseff A.S., Zhu A.S., Chang E., Resnik S.R., Page C., Kim D., Whittam A.J., Miller L.S., Garza L.A. dsRNA released by tissue damage activates TLR3 to drive skin regeneration. Cell Stem Cell. 2015;17:139–151. doi: 10.1016/j.stem.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama K., Yamamoto Y., Sugiyama M., Takaki T., Urashima T., Fukiya S., Yokota A., Okada N., Mukai T. Bifidobacterium bifidum extracellular sialidase enhances adhesion to the mucosal surface and supports carbohydrate assimilation. mBio. 2017;8:e00928-17. doi: 10.1128/mBio.00928-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak P., Troseid M., Avershina E., Barqasho B., Neogi U., Holm K., Hov J.R., Noyan K., Vesterbacka J., Svard J., Rudi K., Sonnerborg A. Gut microbiota diversity predicts immune status in HIV-1 infection. AIDS. 2015;29:2409–2418. doi: 10.1097/QAD.0000000000000869. [DOI] [PubMed] [Google Scholar]
- Ogilvie L.A., Jones B.V. The human gut virome: a multifaceted majority. Front. Microbiol. 2015;6:918. doi: 10.3389/fmicb.2015.00918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson D.A., McNulty N.P., Guruge J.L., Gordon J.I. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- Pompei A., Cordisco L., Amaretti A., Zanoni S., Matteuzzi D., Rossi M. Folate production by bifidobacteria as a potential probiotic property. Appl. Environ. Microbiol. 2007;73:179–185. doi: 10.1128/AEM.01763-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L., Liu X., Sun Q., Fan Z., Xia D., Ding G., Ong H.L., Adams D., Gahl W.A., Zheng C., Qi S., Jin L., Zhang C., Gu L., He J., Deng D., Ambudkar I.S., Wang S. Sialin (SLC17A5) functions as a nitrate transporter in the plasma membrane. Proc. Natl. Acad. Sci. U S A. 2012;109:13434–13439. doi: 10.1073/pnas.1116633109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport F., Khanin R., Liang Y., Pirun M., Krek A., Zumbo P., Mason C.E., Socci N.D., Betel D. Comprehensive evaluation of differential gene expression analysis methods for RNA-seq data. Genome Biol. 2013;14:R95. doi: 10.1186/gb-2013-14-9-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim H.M., Kang H.K., Akter N., Kim D.W., Kim J.H., Kim M.J., Na J.C., Jong H.B., Choi H.C., Suh O.S., Kim W.K. Supplementation of direct-fed microbials as an alternative to antibiotic on growth performance, immune response, cecal microbial population, and ileal morphology of broiler chickens. Poult. Sci. 2013;92:2084–2090. doi: 10.3382/ps.2012-02947. [DOI] [PubMed] [Google Scholar]
- Sato T., van Es J.H., Snippert H.J., Stange D.E., Vries R.G., van den Born M., Barker N., Shroyer N.F., van de Wetering M., Clevers H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnupf P., Gaboriau-Routhiau V., Gros M., Friedman R., Moya-Nilges M., Nigro G., Cerf-Bensussan N., Sansonetti P.J. Growth and host interaction of mouse segmented filamentous bacteria in vitro. Nature. 2015;520:99–103. doi: 10.1038/nature14027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schokker D., Veninga G., Vastenhouw S.A., Bossers A., de Bree F.M., Kaal-Lansbergen L.M., Rebel J.M., Smits M.A. Early life microbial colonization of the gut and intestinal development differ between genetically divergent broiler lines. BMC Genomics. 2015;16:418. doi: 10.1186/s12864-015-1646-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D.L., Jiang X., Wu Q.L., Zhou N.Y. Intragenomic heterogeneity of 16S rRNA genes causes overestimation of prokaryotic diversity. Appl. Environ. Microbiol. 2013;79:5962–5969. doi: 10.1128/AEM.01282-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taweechotipatr M., Iyer C., Spinler J.K., Versalovic J., Tumwasorn S. Lactobacillus saerimneri and Lactobacillus ruminis: novel human-derived probiotic strains with immunomodulatory activities. FEMS Microbiol. Lett. 2009;293:65–72. doi: 10.1111/j.1574-6968.2009.01506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoene-Reineke C., Fischer A., Friese C., Briesemeister D., Gobel U.B., Kammertoens T., Bereswill S., Heimesaat M.M. Composition of intestinal microbiota in immune-deficient mice kept in three different housing conditions. PLoS One. 2014;9:e113406. doi: 10.1371/journal.pone.0113406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torok V.A., Hughes R.J., Mikkelsen L.L., Perez-Maldonado R., Balding K., MacAlpine R., Percy N.J., Ophel-Keller K. Identification and characterization of potential performance-related gut microbiotas in broiler chickens across various feeding trials. Appl. Environ. Microbiol. 2011;77:5868–5878. doi: 10.1128/AEM.00165-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong D.T., Franzosa E.A., Tickle T.L., Scholz M., Weingart G., Pasolli E., Tett A., Huttenhower C., Segata N. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Methods. 2015;12:902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- Turnbaugh P.J., Ley R.E., Mahowald M.A., Magrini V., Mardis E.R., Gordon J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Turner J.R. Intestinal mucosal barrier function in health and disease. Nat. Rev. Immunol. 2009;9:799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- Uni Z., Geyra A., Ben-Hur H., Sklan D. Small intestinal development in the young chick: crypt formation and enterocyte proliferation and migration. Br. Poult. Sci. 2000;41:544–551. doi: 10.1080/00071660020009054. [DOI] [PubMed] [Google Scholar]
- Walton K.D., Freddo A.M., Wang S., Gumucio D.L. Generation of intestinal surface: an absorbing tale. Development (Cambridge, England) 2016;143:2261–2272. doi: 10.1242/dev.135400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wragg D., Mason A.S., Yu L., Kuo R., Lawal R.A., Desta T.T., Mwacharo J.M., Cho C.Y., Kemp S., Burt D.W., Hanotte O. Genome-wide analysis reveals the extent of EAV-HP integration in domestic chicken. BMC Genomics. 2015;16:784. doi: 10.1186/s12864-015-1954-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Xiang Y., Zhou W., Chen J., Li K., Yang H. Microbial community mapping in intestinal tract of broiler chicken. Poult. Sci. 2017;96:1387–1393. doi: 10.3382/ps/pew372. [DOI] [PubMed] [Google Scholar]
- Yin Y., Lei F., Zhu L., Li S., Wu Z., Zhang R., Gao G.F., Zhu B., Wang X. Exposure of different bacterial inocula to newborn chicken affects gut microbiota development and ileum gene expression. ISME J. 2010;4:367–376. doi: 10.1038/ismej.2009.128. [DOI] [PubMed] [Google Scholar]
- Young G.R., Eksmond U., Salcedo R., Alexopoulou L., Stoye J.P., Kassiotis G. Resurrection of endogenous retroviruses in antibody-deficient mice. Nature. 2012;491:774–778. doi: 10.1038/nature11599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P., Lubben W., Slomka H., Gebler J., Konert M., Cai C., Neubrandt L., Prazeres da Costa O., Paul S., Dehnert S., Dohne K., Thanisch M., Storsberg S., Wiegand L., Kaufmann A., Nain M., Quintanilla-Martinez L., Bettio S., Schnierle B., Kolesnikova L., Becker S., Schnare M., Bauer S. Nucleic acid-sensing Toll-like receptors are essential for the control of endogenous retrovirus viremia and ERV-induced tumors. Immunity. 2012;37:867–879. doi: 10.1016/j.immuni.2012.07.018. [DOI] [PubMed] [Google Scholar]
- Zhu W., Lomsadze A., Borodovsky M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 2010;38:e132. doi: 10.1093/nar/gkq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.