Abstract
The physiological roles of thyrotropin-releasing hormone (TRH) are proposed to be mediated by TRH receptors (TRHR), which have been divided into 3 subtypes, namely, TRHR1, TRHR2, and TRHR3, in vertebrates. Although 2 TRH receptors (TRHR1 and TRHR3) have been predicted to exist in birds, it remains unclear whether TRHR3 is a functional TRH receptor similar to TRHR1. Here, we reported the functionality and tissue expression of TRHR3 in chickens. The cloned chicken TRHR3 (cTRHR3) encodes a receptor of 387 amino acids, which shares high-amino-acid identities (63–80%) to TRHR3 of parrots, lizards, Xenopus tropicalis, and tilapia and comparatively lower sequence identities to chicken TRHR1 or mouse TRHR2. Using cell-based luciferase reporter assays and Western blot, we demonstrated that similar to chicken TRHR1 (cTRHR1), cTRHR3 expressed in HEK 293 cells can be potently activated by TRH and that its activation stimulates multiple signaling pathways, indicating both TRH receptors are functional. Quantitative real-time PCR revealed that cTRHR1 and cTRHR3 are widely, but differentially, expressed in chicken tissues, and their expression is likely controlled by promoters located upstream of exon 1, which display strong promoter activities in cultured DF-1 cells. cTRHR1 is highly expressed in the anterior pituitary and testes, while cTRHR3 is highly expressed in the muscle, testes, fat, pituitary, spinal cord, and many brain regions (including hypothalamus). These findings indicate that TRH actions are likely mediated by 2 TRH receptors in chickens. In conclusion, our data provide the first piece of evidence that both cTRHR3 and cTRHR1 are functional TRH receptors, which helps to elucidate the physiological roles of TRH in birds.
Key words: chicken, TRH, TRHR3, TRHR1, pituitary
Introduction
It is generally believed that thyrotropin-releasing hormone (TRH), also known as thyrotropin-releasing factor, is a releasing hormone secreted by the hypothalamus which constitutes an essential part of the hypothalamic–pituitary–thyroid axis in vertebrates. TRH is mainly produced by neurons in the paraventricular nucleus and transported via the hypophysial portal circulation to the pituitary gland (Taylor et al., 1990). The precursor of TRH is a long polypeptide which often contains multiple repeats of Gln-His-Pro-Gly. These repeats are likely cleaved to form the mature form of TRH (Richter et al., 1984, Lechan et al., 1986), which is a tripeptide (pyro-Glu-His-Pro-NH2) that shows a remarkable structural conservation in most vertebrates including mammals, birds, amphibians, reptiles, and fish. TRH was originally named after its ability to promote the release of thyroid-stimulating hormone (TSH) in mammalian pituitaries (Bøler et al., 1969, Burgus et al., 1970). Nevertheless, this function has also been found to be mediated by other hormones in vertebrates, including corticotropin-releasing hormone in nonmammalian vertebrates and glucagon-like peptide (GCGL) in chickens (De Groef et al., 2003, Huang et al., 2014). Furthermore, multiple effects of TRH were found in the central nervous system (CNS) and peripheral tissues in various species. For instance, TRH exerts an antidepressant function in the mammalian CNS (Marangell, 1997). TRH was reported to have a prolactin (PRL)-stimulating effect in cultured rat pituitary cells and chicken pituitary cells (Tashjian et al., 1971, Harvey et al., 1978). It has also been established as a potent growth hormone (GH)–releasing factor in vertebrates. As an example, in chickens, TRH stimulates GH release both in vivo and in vitro (Harvey, 1990).
The physiological roles of TRH are reported to be mediated by thyrotropin-releasing hormone receptor (TRHR). In vertebrates, 3 subtypes of TRHR have been identified or predicted, namely, TRHR1, TRHR2, and TRHR3. All of them belong to G protein–coupled receptor family. Among the three, TRHR1 was the first isolated one, being discovered in mice back in 1990 (Straub et al., 1990). This receptor is widely present across various vertebrate groups and has been reported to mediate the function of TRH in the anterior pituitary of mice (Rabeler et al., 2004). TRHR2 was first identified in rats in 1998 (Itadani et al., 1998). It shares 51% sequence identity with both mouse and rat TRHR1. Unlike TRHR1, TRHR2 has so far been found in frogs, teleosts, and a few mammalian species including rats, mice, and horses (Heuer et al., 2000, O'Dowd et al., 2000). In comparison, studies on TRHR3 are scarce. This receptor has been predicted to exist in a small number of species across different animal kingdoms but is hypothesized to be absent in humans and rodents. Although TRHR3 of Xenopus laevis has been functionally analyzed, the binding affinity of Xenopus TRHR3 to TRH is considerably lower than that of the other 2 TRH receptors, casting doubt on whether TRHR3 is a functional TRH receptor (Lu et al., 2003).
In birds, 2 TRH receptors (TRHR1 and TRHR3) were predicted from the genome. Similar to mammalian TRHR1, chicken TRHR1 has been established to be a functional receptor coupled to Gq proteins and its activation can stimulate intracellular calcium concentration (Sun et al., 1998). Nevertheless, the tissue expression and functionality of TRHR3 in birds remains in the dark. Hence, in this study, we cloned TRHR3 from chickens and characterized its tissue expression and signal transduction, aiming to reveal the functional similarity and differences between cTRHR3 and cTRHR1. These findings not only help to unveil the influence of TRH and its receptors on the physiological functions and traits of chickens, such as pituitary hormone (GH, PRL, TSH) secretion and growth, but also provide clues to explore the functional change of TRHR3 during vertebrate evolution.
Methods and materials
Chemicals, Primers, Peptides, and Antibodies
Chicken TRH used in experiments was synthesized by GL Biochem (Shanghai, China). All primers were synthesized by Beijing Genome Institute (China), and primer sequences are shown in Supplementary Table 1. ERK1/2 and pERK1/2 antibodies were purchased from Cell Signaling Technology (Beverly, MA). All chemicals including H89, MDL-12330A, and 2-aminoethoxydiphenyl borate (2-APB) were obtained from Sigma-Aldrich (St. Louis, MO).
Total RNA Extraction
Adult chickens (aged 6 mo) of both sexes (Lohmann layer) and ducks were purchased from local commercial companies. Six adult chickens (3 males and 3 females) were killed, and different tissues (including the telencephalon, midbrain, cerebellum, hindbrain, hypothalamus, spinal cord, anterior pituitary, heart, duodenum, kidneys, liver, lung, ovary, testes, spleen, pancreas, crop, proventriculus, gizzard, jejunum, ileum, cecum, colon, subcutaneous fat, back skin, and breast muscle) were collected and stored at −80°C before use. In addition, anterior pituitary glands collected from 6 adult male chickens were carefully segregated into the caudal lobe (Ca) and cephalic lobe (Ce) and subjected to RNA extraction. Total RNA was extracted using RNAzol (Molecular Research Center, Cincinnati, OH) in accordance with the manufacturer's instruction. The animal experiment protocol was approved by the Animal Ethics Committee of Sichuan University (Chengdu, China).
Reverse Transcription and Quantitative Real-Time PCR
Two micrograms of total RNA and 0.5 pg of oligodeoxythymide were mixed in a total volume of 5 μL, incubated at 70°C for 10 min and cooled at 4°C for 2 min. The first strand buffer, 0.5 mmol each deoxynucleotide triphosphate, and 100 U Moloney murine leukemia virus reverse transcriptase (Takara) were then added into the reaction mix in a total volume of 10 μL. Reverse transcription (RT) was performed at 42°C for 90 min. In accordance with our previously established method (Cai et al., 2015), quantitative real-time RT-PCR was performed on the CFX96 Real-time PCR Detection System (Bio-Rad, Hercules, CA) to examine the mRNA levels of target genes in chicken tissues.
Cloning of the cDNAs Encoding Chicken TRHR1 and TRHR3 and Duck TRHR1
In accordance with the cDNA sequence of cTRHR1 (NM_204930) and predicted cDNA sequence of dTRHR1 (XM_005022942) and cTRHR3 (XM_004947049) in GenBank, gene-specific primers were designed to amplify the 5′-cDNA and 3′-cDNA ends of cTRHR1 and cTRHR3 from adult chicken or duck brain (or pituitary) using SMART-RACE cDNA amplification Kit (Clontech, Palo Alto, CA). The amplified PCR products were cloned into pTA2 vector (TOYOBO, Japan) and sequenced. Finally, the open reading frames were obtained by assembly of resultant sequences.
Identification of the Promoter Regions of Chicken TRHR1 and TRHR3
To map the promoter regions of chicken TRHR1 and TRHR3, we designed gene-specific primers and amplified their 5′-flanking regions (near exon 1) with high-fidelity Taq DNA polymerase (TOYOBO, Japan). The PCR products were cloned into pGL3-Basic vector (Promega, Madison, WI), and their sequences verified. Then, a series of promoter–luciferase reporter constructs for cTRHR1 (−2097/+127Luc, −933/+127Luc, −371/+127Luc, −137/+127Luc) and cTRHR3 (−2003/+23Luc, −1000/+23Luc, −509/+23Luc, −180/+23Luc) were prepared. In this experiment, the transcription start site on exon 1 of each gene determined by rapid amplification of 5′ cDNA ends (5′-RACE) was designated as “+1,” and the first nucleotide upstream of the transcription start site was designed as “−1.” Finally, the promoter activities of these constructs were examined in cultured DF-1 cells by the Dual-Luciferase Reporter Assay (Promega, Madison, MI), as described in our previous study (Wang et al., 2010).
Functional Characterization of Chicken TRHR1 and TRHR3
The open reading frames of TRHRs were amplified using gene-specific primers based on their cDNA sequences obtained from the adult chicken brain or pituitary using high-fidelity Tag DNA polymerase (TOYOBO, Japan). Then, the PCR products were cloned into the pcDNA3.1(+) expression vector (Invitrogen) and sequenced. In accordance with our previously established methods (Wang et al., 2007, Wang et al., 2012, Mo et al., 2015, Mo et al., 2017), the function of each receptor was examined in HEK 293 cells using the pGL3-NFAT-RE-luciferase reporter system, pGL4-SRE-luciferase reporter system, and pGL3-CRE-1uciferase reporter system.
Western Blot
As described in our previous studies (He et al., 2016, Mo et al., 2017), HEK 293 cells transfected with cTRHR1 or cTRHR3 expression plasmid were cultured on a 24-well plate at 37°C and then treated by TRH (10 nM) for 10 min. Then, cells were lysed and used for Western blot detection of phosphorylated ERK1/2 (pERK1/2) and ERK1/2.
Data Analysis
The mRNA level of the target gene was first normalized by that of β-actin and then shown as a fold difference compared to a selected tissue. The change in luciferase activity of HEK 293 cells expressing TRHR was expressed as a relative fold increase in response to peptide treatment. Data were analyzed by one-way ANOVA followed by Dunnett's test. Dose–response curves were constructed using a nonlinear regression model, and the corresponding half maximal effective concentration (EC50) values were assessed using GraphPad Prism, version 7 (GraphPad, San Diego, CA). The construction of the Maximum Likelihood Tree phylogenetic tree was carried out using MEGA, version 7 (MEGA software, Sudhir Kumar, Arizona State University, Phoenix, AZ). All experiments were repeated at least twice.
Results
Cloning of the Full-Length cDNA of Chicken TRHR3
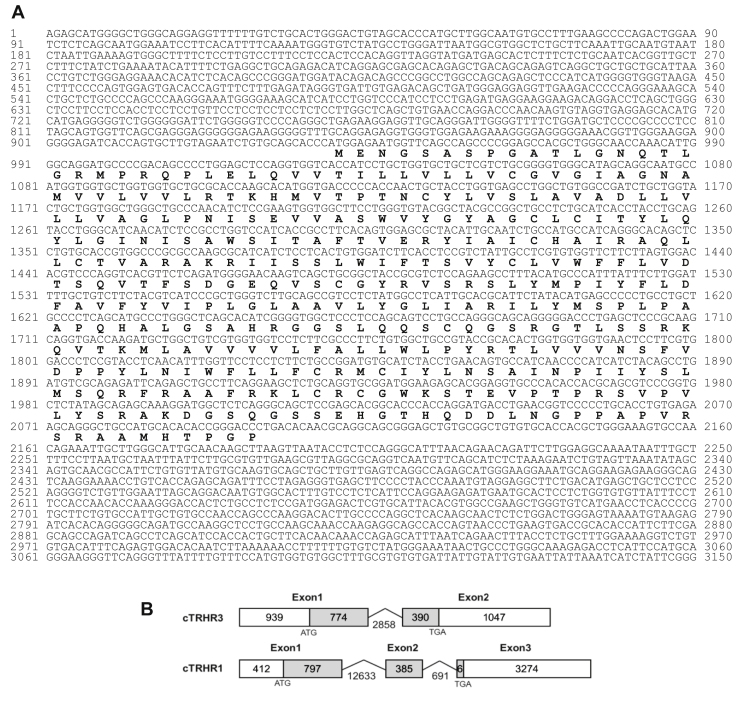
Based on the predicted partial cDNA sequence of chicken TRHR3 (XM_004947049), we cloned the full-length cDNAs of cTRHR3 from adult chicken brain tissue by RACE-PCR. The cloned cTRHR3 cDNA is 3,150 bp in length (accession no. MK138989) and predicted to encode a receptor of 387 amino acids (Figure 1A). Comparison of cTRHR3 cDNA sequence with the chicken genome database (http://www.ensembl.org/Gallus_gallus) shows that cTRHR3 consists of 2 exons. The first exon of cTRHR3 consists of a 774-bp coding sequence (CDS) fragment and a 939-bp 5′-untranslated region (5′-UTR), and the second exon consists of a 390-bp CDS fragment and a 1047-bp 3′-UTR region.
Figure 1.
(A) The full-length cDNA and amino acid sequences of chicken TRHR3. (B) Exon organization of chicken TRHR1 and TRHR3 genes. Numbers in the boxes indicate the size (bp) of the coding region (shaded) and noncoding region. Numbers under the broken lines indicate the size (bp) of the introns.
To compare the structural and functional difference between cTRHR3 and cTRHR1, we also cloned the chicken TRHR1 of 395 amino acids from pituitary. cTRHR1 is composed of 3 exons, and its exon 3 contains a very short CDS region of 6 bp and a long 3′-UTR region of 3,274 bp (Figure 1B).
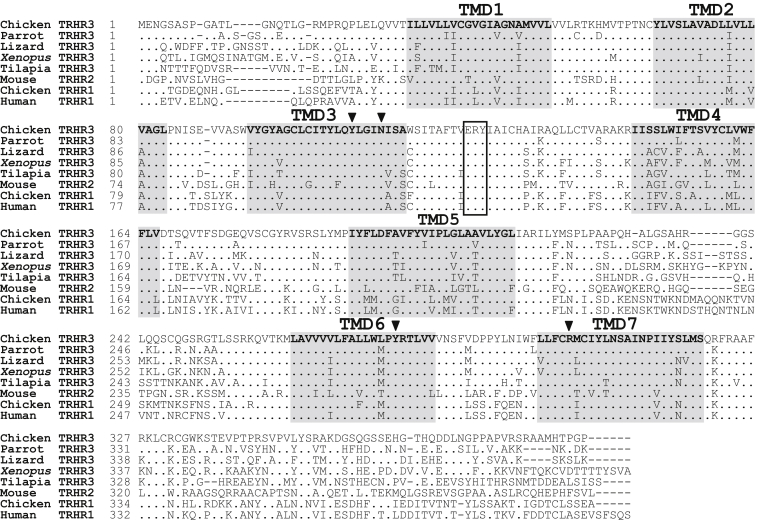
Sequence alignment revealed that cTRHR3 shows high-amino-acid-sequence identities of 80.3%, 72.9%, 66.5%, and 62.6% with TRHR3 of parrots, lizards, Xenopus tropicalis, and tilapia, respectively. Chicken TRHR1 shows high-amino-acid-sequence identity of 81% with human TRHR1 and a much lower identity of 56.2% with chicken TRHR3 (Figure 2, Supplementary Table 2). Both cTRHR1 and cTRHR3 possess a G protein–coupling ERY motif located near the third transmembrane domain and a conserved NP (XX)Y sequence located in the seventh transmembrane domain. In addition, both TRH receptors have the same four-amino-acid residues (Tyr108, Asn112, Tyr277, and Arg301) that were proposed to directly interact with TRH (Sun et al., 1998).
Figure 2.
Amino acid alignment of chicken TRHR3 (MK138989) with parrot TRHR3 (XP_005146871.1), lizard TRHR3 (XP_003220685.1), Xenopus tropicalis TRHR3 (XP_002933212.1), tilapia TRHR3 (XP_003439144.1), mouse TRHR2 (NP_573465.1), chicken TRHR1 (NP_990261.1), and human TRHR1 (NP_003292). The seven transmembrane domains (TMD1-7) are shaded. The ERY motif is boxed, and the 4 conserved amino acid residues (Tyr108, Asn112, Tyr277, Arg301) associated with ligand binding are marked with black arrows. Note: dots indicate the amino acids identical to chicken TRHR3, and dashes denote the gaps in the sequence.
To study the evolutionary relationship between TRHR1, TRHR2, and TRHR3 in vertebrates, phylogenetic analysis was performed using the maximum likelihood method of MEGA software. As shown in Supplementary Figure 1, TRHR family forms 3 clusters, TRHR1, TRHR2, and TRHR3. Among them, TRHR1 and TRHR2 have closer evolutionary relationships, while the TRHR3 is located on a more distant branch. TRHR1 exists in all vertebrate groups; TRHR2 is found in mice, horses, frogs, reptiles, and teleosts (e.g., zebrafish); TRHR3 is present in birds, frogs, fish, and a few mammalian species (e.g., dogs, pigs, and cattle).
Identification of cTRHR1 and cTRHR3 Promoter Regions
In this study, based on the cloned cTRHR3 and cTRHR1 5′-UTR sequences, the potential promoter regions near or upstream exon 1 of the 2 receptor genes were cloned, and their promoter activity were detected in vitro using DF-1 cells.
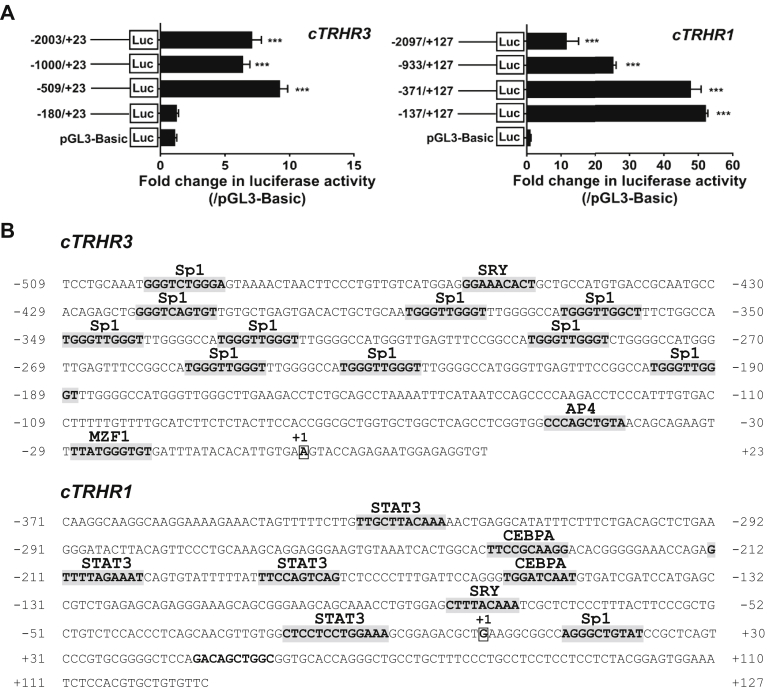
As shown in Figure 3A, the promoter vector (−2003/+23Luc) containing the 5′ flanking sequence of the cTRHR3 gene exhibited strong promoter activity in DF-1 cells. Promoter deletion experiments mapped that the core promoter region of cTRHR3 gene may be located in the −509/+23 region because the fragment near exon 1 (−180/+23Luc) did not show any promoter activity. Similar to cTRHR3, the promoter vector (−2097/+127Luc) containing the 5′ flanking sequence of the cTRHR1 gene exhibited strong promoter activity in DF-1 cells, indicating that this region likely corresponds to the promoter region for this gene. After the deletion experiment, it was preliminarily determined that the core promoter of the cTRHR1 gene is likely located in the −137/+127 region. Using online transcription factor–binding site prediction tools such as JASPAR (jaspar.genereg.net), multiple common transcription factor–binding sites were identified in the promoter region of cTRHR1 (−509/+23) and cTRHR3 (−371/+127) genes. As shown in Figure 3B, the binding sites for many transcription factors such as Sp1, AP4, SRY, and MZF1 were predicted to exist within the core promoter region of cTRHR3, while the binding sites for STAT3, CEBPα, SRY, and Sp1 were predicted to exist within the core promoter region of cTRHR1.
Figure 3.
(A) Detection of promoter activities of the 5′-flanking region of chicken TRHR1 and TRHR3 in cultured DF-1 cells. Various stretches of the 5′-flanking regions of TRHR1 and TRHR3 were cloned into pGL3-Basic vector for the generation of multiple promoter–luciferase constructs. The promoter–luciferase vector and pRL-TK vector were cotransfected into DF-1 cells, and the promoter activities were detected by the dual-luciferase reporter system. All data represent the mean ± SEM of 4 replicates (N = 4). ***P < 0.001 vs. pGL3-Basic vector. The transcriptional start site (A/G, boxed) identified by rapid amplification of 5′ cDNA ends (5′-RACE) was designated as '+1'. (B) Predicted transcription factor binding sites in the promoter regions of cTRHR1 and cTRHR3. The corresponding binding sites are shaded. Sp1, specificity protein 1; SRY, sex-determining region; AP4, activating enhancer–binding protein 4; MZF1, myeloid zinc finger 1; STAT3, signal transducer and activator of transcription 3; CEBPA, CCAAT/enhancer-binding protein alpha.
Functional Analysis of cTRHR3 and cTRHR1
In mammals, it was reported that TRHR can couple to Gq/11 protein upon ligand activation and cause changes in intracellular calcium levels and activate the MAPK/ERK1/2 signaling pathway (Sun et al., 1998, Kanasaki et al., 1999). In this study, the signaling pathways coupled to the newly cloned receptor TRHR3 in chickens were first investigated using the pGL3-NFAT-RE-luciferase reporter system, pGL4-SRE-luciferase reporter system, and pGL3-CRE-luciferase reporter system. In addition, chicken TRHR1 and duck TRHR1 (dTRHR1, MN561700) were examined in parallel to compare their functional similarity and difference with cTRHR3.
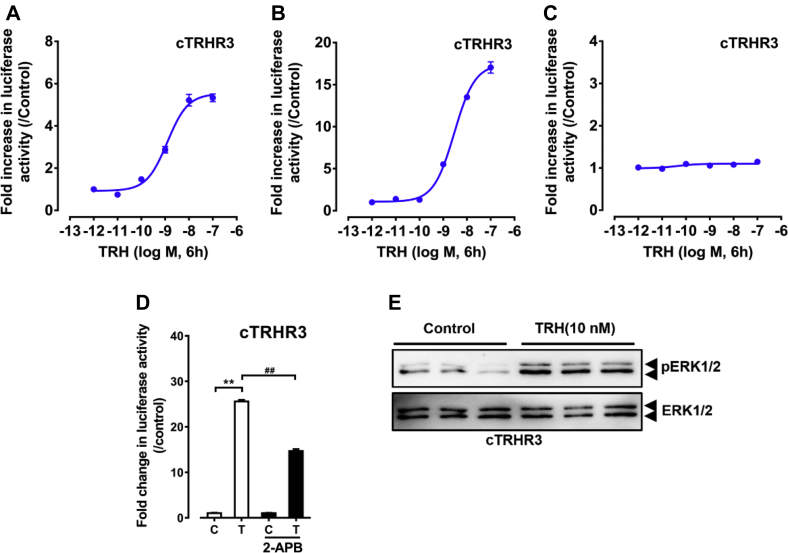
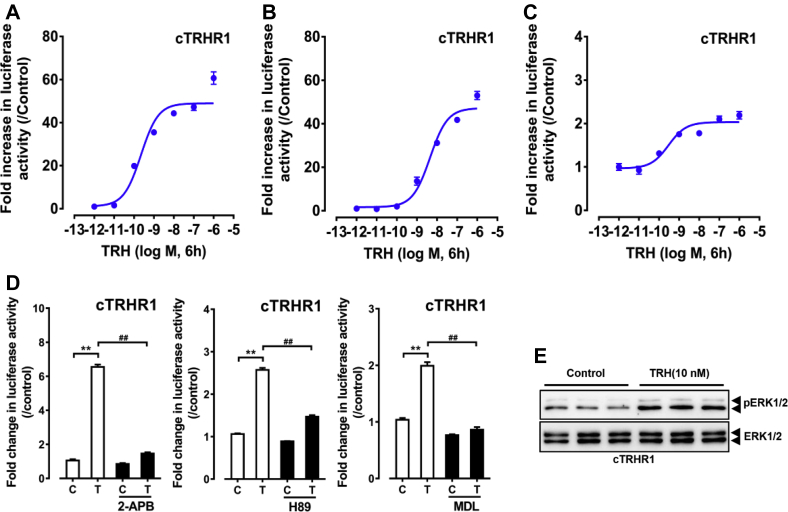
Using the pGL3-NFAT-RE-luciferase reporter assay, we demonstrated that chicken TRHR3 expressed in HEK 293 cells can be activated by cTRH potently with an EC50 value of 1.23 nM, clearly indicating that cTRHR3 is a functional receptor for TRH and its activation can elevate intracellular Ca2+ concentration (Table 1). Furthermore, 2-aminoethoxydiphenyl borate (an inhibitor of inositol triphosphate receptor) could inhibit this induced luciferase activity, supporting that similar to chicken TRHR1, TRHR3 is likely coupled to Gq protein and its activation can induce intracellular calcium mobilization. Using the pGL4-SRE-luciferase reporter assay and Western blot, we also elucidated that TRHR3 activation can activate downstream MAPK/ERK signaling cascade. In contrast, TRHR3 activation cannot stimulate cAMP/PKA signaling pathway at any concentration tested, as monitored by the pGL3-CRE-luciferase reporter system (Figure 4, Table 1).
Table 1.
EC50 values of cTRH in activating different signaling pathways in HEK 293 cells transfected with chicken TRHR1 and TRHR3 and duck TRHR1.
| EC50 values (nM) | |||
|---|---|---|---|
| Receptors | Calcium signaling pathway | MAPK/ERK signaling pathway | cAMP/PKA signaling pathway |
| cTRHR3 | 1.23 | 2.91 | - |
| cTRHR1 | 0.22 | 4.50 | 0.30 |
| dTRHR1 | 0.13 | 2.14 | 0.55 |
‘-’ means that cTRHR3 may not couple to this signaling pathway.
Abbreviations: cTRH, chicken thyrotropin-releasing hormone; EC50, half maximal effective concentration.
Figure 4.
(A–C) Effects of chicken thyrotropin-releasing hormone (TRH) on activating chicken TRHR3, as monitored by the (A) pGL3-NFAT-RE-luciferase reporter system, (B) pGL4-SRE-luciferase reporter system, and (C) pGL3-CRE-luciferase reporter system. All data represent the mean ± SEM of 3 replicates (N = 3). (D) Effects of 2-aminoethoxydiphenyl borate (2-APB) on TRH-induced luciferase activities of HEK 293 cells expressing chicken TRHR3, as monitored by the pGL3-NFAT-RE-luciferase reporter system. 2-APB (100 μM) was added 0.5 h before TRH treatment. T represents TRH treatment (10 nM, 6 h), and C represents control group without TRH treatment. All data represent the mean ± SEM of 3 replicates (N = 3). **P < 0.01 vs. control; ##P < 0.01 between 2 treatment groups. (E) Western blot detection of ERK1/2 phosphorylation (pERK1/2) and total ERK1/2 in HEK 293 cells expressing cTRHR3 treated by cTRH (10 nM) and without TRH (control) for 10 min.
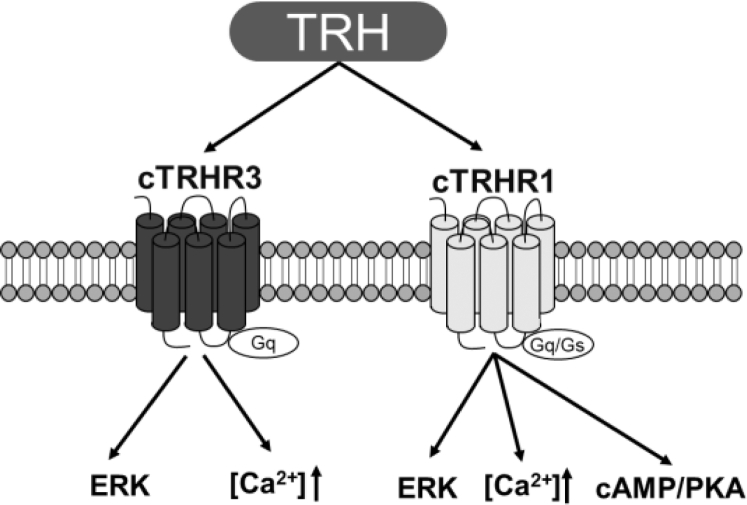
Similar to TRHR3, TRHR1 activation can stimulate intracellular Ca2+ mobilization and MAPK/ERK signaling pathways (Table 1), as monitored by the 2 respective cell-based luciferase reporter systems and Western blot. Interestingly, unlike cTRHR3, cTRHR1 activation can stimulate adenylate cyclase/cAMP/PKA signaling pathway in the pGL3-CRE-luciferase reporter assay, and this stimulatory effect could be inhibited by either an adenylate cyclase inhibitor (MDL-12330A) or a PKA inhibitor (H89) (Figure 5). To further elucidate the functions of bird TRHR1, we cloned and performed the same test with duck TRHR1. The results showed that dTRHR1 can activate multiple signaling pathways including calcium mobilization, MAPK/ERK pathways, and cAMP/PKA pathways (Table 1) similar to cTRHR1 (Supplementary Figure 2), supporting that dTRHR1 is a functional receptor as well. For easy reference, a schematic figure is included to show the similarities and differences of the downstream signaling pathways of cTRHR1 and cTRHR3 when activated by cTRH (Figure 6).
Figure 5.
(A–C) Effects of chicken thyrotropin-releasing hormone (TRH) on activating chicken TRHR1, as monitored by the (A) pGL3-NFAT-RE-luciferase reporter system, (B) pGL4-SRE-luciferase reporter system, and (C) pGL3-CRE-luciferase reporter system. All data represent the mean ± SEM of 3 replicates (N = 3). (D) Effects of 2-aminoethoxydiphenyl borate (2-APB), H89, and MDL-12330A (MDL) on TRH-induced luciferase activities of HEK 293 cells expressing chicken TRHR1, as monitored by the pGL3-NFAT-RE-luciferase reporter system and pGL3-CRE-luciferase reporter system. 2-APB (100 μM), H89 (10 μM) or MDL (20 μM) was added 0.5 h before TRH treatment. T represents TRH treatment (10 nM, 6 h), and C represents control group without TRH treatment. All data represent the mean ± SEM of 3 replicates (N = 3). **P < 0.01 vs. control; ##P < 0.01 between 2 groups. (E) Western blot detection of ERK1/2 phosphorylation (pERK1/2) and total ERK1/2 in HEK 293 cells expressing cTRHR1 treated by TRH (10 nM) and without TRH (control) for 10 min.
Figure 6.
Schematic diagram showing the similarities and differences of the downstream signaling pathways of cTRHR1 and cTRHR3 when activated by thyrotropin-releasing hormone (TRH). Both receptors can couple to Gq protein, and their activation stimulates intracellular calcium mobilization and MAPK/ERK signaling pathway. Unlike cTRHR3, cTRHR1 can also couple to Gs protein and stimulate adenylate cyclase/cAMP/PKA signaling pathway.
Tissue Expression of cTRH, cTRHR1, and cTRHR3
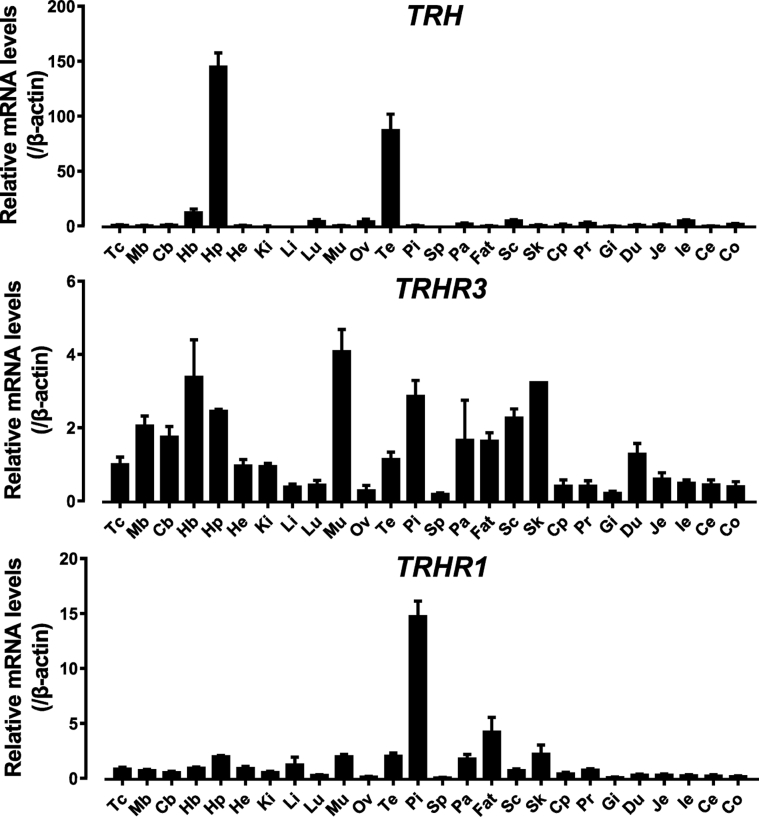
To investigate the tissue expression of TRH and its receptors cTRHR1 and cTRHR3 in chickens, quantitative real-time PCR was used to detect their mRNA expression in adult chickens. As shown in Figure 7, within the CNS, cTRH is highly expressed in the hypothalamus, moderately expressed in the hindbrain, and weakly expressed in other remaining brain regions. In peripheral tissues, only the testes showed a high expression level of TRH. cTRHR1 has the highest expression level in the anterior pituitary and moderate expression levels in other brain regions. In peripheral tissues, besides moderate expression level in the fat, muscle, skin, testes, and intestine, only weak expression of cTRHR1 was detected in other remaining peripheral tissues. Unlike cTRHR1, cTRHR3 has a relatively higher expression level in the muscle, pituitary, skin, spinal cord, midbrain, cerebellum, hindbrain, and hypothalamus, while cTRHR3 is moderately or weakly expressed in other remaining tissues.
Figure 7.
Quantitative real-time PCR assay of cTRH, cTRHR3, and cTRHR1 mRNA expression in adult chicken tissues, including the telencephalon (Tc), midbrain (Mb), cerebellum (Cb), hindbrain (Hb), hypothalamus (Hp), heart (He), kidneys (Ki), liver (Li), lung (Lu), muscle (Mu), ovary (Ov), testes (Te), anterior pituitary (Pi), spleen (Sp), pancreas (Pa), subcutaneous fat (Fat), spinal cord (Sc), skin (Sk), duodenum (Du), crop (Cp), proventriculus (Pr), gizzard (Gi), jejunum (Je), ileum (Ie), cecum (Ce), and colon (Co). The mRNA level of each gene was normalized with the mRNA level of β-actin as an internal control and expressed as the fold difference compared with that of telencephalon (Tc). All data represent the mean ± SEM of 6 individual adult chickens (3 males and 3 females) (N = 6).
Discussion
In this study, the full-length cDNA sequence of TRHR3 was cloned from chickens. Cell-based luciferase reporter assays revealed that cTRHR3 is a novel functional receptor of TRH, and it has a signaling property similar, but not identical, to that of cTRHR1 or duck TRHR1. Quantitative real-time PCR assays showed that both cTRHR3 and cTRHR1 are widely distributed among chicken tissues but that their tissue expression is differentially regulated, probably controlled by their promoters mapped upstream of their respective exon 1. To our knowledge, our study represents the first to characterize the functionality and tissue expression of TRHR3 in avian species.
Structure of cTRHR1 and cTRHR3 and Phylogenetics of Vertebrate TRHR Family
Although 2 TRHRs (TRHR1 and TRHR3) have been predicted from avian genome, the full-length cDNAs of TRHR3 has not been cloned in any avian species. Here, we cloned cTRHR3 from chickens. Sequence comparison reveals that both cTRHRs show considerable conservation in structure, for instance, completely retaining the 4 amino acid residues that were proposed to be important for binding TRH (i.e., Tyr108, Asn112, Tyr277, Arg301). These sites are thought to facilitate ligand binding by altering the conformation of TRHRs to form a more affinitive structure (Sun et al., 1998). TRH was found to initially bind with low affinity to the extracellular region of TRHR, and then the conformation of ligand–receptor complex changes so that TRH binds to the transmembrane region of TRHR with a higher affinity, thus allowing the transduction of downstream signals (Engel and Gershengorn, 2007).
We found that the number of TRHR subtypes varies greatly among vertebrate species, despite their generally conserved structure. To date, only TRHR1 has been identified in the majority of mammalian species including humans, while TRHR2 has only been found in a few mammals such as rats (Duthie et al., 1993, Itadani et al., 1998). In chickens, only TRHR1 and TRHR3 have been identified. By contrast, in lower vertebrates such as amphibians and teleosts, all 3 TRHR subtypes have been found in the majority of these species (Harder et al., 2001, Mekuchi et al., 2011). Taken into account the proposed theory on evolutionary history of vertebrate species and our phylogenetic and synteny analyses on TRHR subtypes shown in Supplementary Figure 1, we hypothesize that multiple gene loss events might have occurred for different subtypes of TRHR in various species during evolution (Kumar and Hedges, 1998, International Chicken Genome Sequencing Consortium, 2004). TRHR2 was likely lost during the evolution of birds as well as many mammals, and TRHR3 might be lost during the evolution of mammals, eventually giving rise to the current repertoire of TRHR subtypes in vertebrates.
Both cTRHR3 and cTRHR1 are Functional Receptors for TRH in Chickens
Although the functionality of TRHR1 has been reported in a variety of species including mice (Gershengorn and Osman, 1996), chickens (Sun et al., 1998), and X. laevis (Bidaud et al., 2002), little is known about the function of its homologous receptor subtype, TRHR3. In previous studies, TRH treatment can increase inositol triphosphate levels of COS-1 cells transfected with chicken/mouse TRHR1 (Sun et al., 1998), and thus, TRHR1 has been proved to couple to Gq/11 and it can activate intracellular signaling pathways, such as activating phospholipase C signaling pathway and inducing intracellular calcium mobilization. In X. laevis, when TRH was applied to HEK 293 cells expressing xTRHR1, an increase in intracellular Ca2+ levels was detected (Bidaud et al., 2002). In our study, we found that similar to mouse TRHR1, chicken TRHR1 is likely coupled to Gq protein and its activation causes intracellular calcium mobilization and activates the MAPK/ERK signaling pathway, as monitored by luciferase reporter assays and Western blot. Interestingly, we also found an increase in pGL3-CRE-luciferase signal in HEK 293 cells transfected with TRHR1 after TRH treatment, suggesting that TRHR1 may also act through the Gs protein which activates the cAMP/PKA signaling pathway. Our finding contrasts a previous report in chickens, in which TRHR1 activation fails to increase the intracellular cAMP levels. This discrepancy is perhaps due to the different experimental approaches used. Nevertheless, our finding in chickens actually concurs with previous report in mammals, in which TRHR1 was found to be coupled to Gs protein in addition to Gq–phospholipase C and able to stimulate adenylate cyclase in rat GH3 cells and contribute to the function of TRH in stimulation of GH release (Paulssen et al., 1992).
Compared with TRHR1, the studies on TRHR3 have been very limited. In this study, we took the first step in characterizing the function of cTRHR3 and found that after TRH treatment of HEK 293 cells transfected with cTRHR3, an increase in intracellular Ca2+ concentration and the activation of the MAPK/ERK signaling pathway were detected. This indicates that chicken TRHR3 is a new functional receptor for TRH. However, this should be noted that cTRHR3 cannot stimulate cAMP/PKA signaling pathway, which suggests that the functionality of the 2 cTRHR receptors is similar but distinct from each other. Our finding, for the first time, indicates that both TRHR1 and TRHR3 can mediate the biological actions of TRH in birds. This contrasts a previous report in X. laevis, in which TRHR3 cannot be activated by TRH effectively, although the other 2 TRH receptors (TRHR1 and TRHR2) can (Bidaud et al., 2002, Lu et al., 2003). Interestingly, in bull frogs, TRHR3 is reported to be a functional receptor for TRH and its activation can increase intracellular calcium level (Nakano et al., 2018). Moreover, TRHR3 is the major receptor subtype expressed in the anterior pituitary and found to be colocalized with PRL cells, which may mediate the PRL-stimulating effect of TRH in this species (Lu et al., 2003, Nakano et al., 2018). These findings strongly suggest that TRHR3 may play distinct roles in different vertebrate classes.
Tissue Expression of TRH, TRHR1, and TRHR3 in Chickens
In this study, we found that cTRH is widely expressed in chicken tissues examined with the highest expression level noted in the hypothalamus and testes. In amphibians (Lamacz et al., 1989), rodents (Segerson et al., 1987), and birds (Péczely and Kiss, 1988, Vandenborne et al., 2005, Aoki et al., 2007), TRH and its precursors are found to be highly expressed in multiple nuclei of the hypothalamus including the supraoptic nucleus, paraventricular nucleus, lateral hypothalamic nucleus, and arcuate nucleus. This highly conserved expression profile of TRH across species in the hypothalamic regions hints that TRH plays an important regulatory role therein, for instance, as a major regulator of TSH synthesis and secretion and secretion of other pituitary hormones such as PRL and GH (Scanes, 1974, Harvey et al., 1978, Harvey, 1990). In addition, the high expression of cTRH in the testis suggests that as in mammals, cTRH may regulate testis function (e.g., testosterone secretion) as an autocrine/paracrine factor in chicken testes (Wilber and Xu, 1998, Geris et al., 2000).
On the other hand, cTRHR1 and cTRHR3 were found to be widely, but differentially, expressed in chicken tissues, and their expression is likely controlled by their promoters upstream of respective exon 1, which contains multiple transcription factor–binding sites. cTRHR1 was detected to be preferentially expressed in the anterior pituitary and weakly or moderately expressed in peripheral tissues such as skin, testis, fat, and muscle. This is consistent with reports in other species, suggesting that cTRHR1 plays important roles in mediating TRH actions on chicken pituitary, such as involvement in the regulation of TSH, GH, and PRL secretion (Scanes, 1974, Harvey et al., 1978, Harvey, 1990). It was reported that in frogs and humans, TRH acts on the skin to regulate wound healing (Meier et al., 2013). In humans, TRH and TRHR1 are coexpressed in hair follicles to regulate hair growth (Gaspar et al., 2010) and have physiological functions that regulate melanin synthesis (Gaspar et al., 2011). The relatively high expression of TRHR1 in chicken testes is consistent with the findings in rats (Satoh et al., 1994). In rat testes, TRHR1 expression levels were detected to be approximately 10% of that in the pituitary gland, which were specifically expressed in mesenchymal cells and shown to regulate testosterone secretion (Wilber and Xu, 1998). Both TRH and TRHR1 genes are highly expressed in porcine fat tissue, which may suggest a paracrine or autocrine role of TRH in this tissue (Jiang et al., 2012). A genome-wide association study on lean body mass variation in humans also identified the functional relevance of TRHR1 in muscle metabolism (Liu et al., 2009). The question whether TRHR1 signaling in chicken plays roles similar to that found in mammals awaits further investigation.
Unlike cTRHR1, cTRHR3 is widely expressed in nearly all tissues examined with a relatively high expression level in various brain regions. This finding, together with the wide distribution of cTRH in chicken tissues, suggests that TRH-TRHR3 signaling may play important autocrine and paracrine roles in the CNS and peripheral tissues such as in testes. Similarly, in Oryzias latipes, TRHR3 was detected by RT-PCR to be widely expressed in various tissues examined, including the anterior pituitary, brain, testes, ovary, kidneys, spleen, various parts of the gastrointestinal tract, and skin (Mekuchi et al., 2011). In X. laevis, TRHR3 expression was detected in tissues such as the brain, heart, and stomach (Bidaud et al., 2002). These findings fall in line with the hypothesis that TRHR3 signaling plays diverse roles in both the CNS and peripheral tissues in nonmammalian vertebrates. The differential expression of both receptors in various chicken tissues, together with the wide tissue expression of cTRH, lead us to hypothesize that TRH plays a wide range of role in chickens and that its action is likely mediated by either TRHR1 or TRHR3, or both receptors in chicken tissues.
In summary, we identified and characterized a new chicken TRH receptor, TRHR3. Our results confirmed that similar to cTRHR1, cTRHR3 is a functional receptor and is able to mediate TRH function through the intracellular Gq protein–related signaling pathway. Furthermore, we found that both TRH receptors are widely, but differentially, expressed in chicken tissues. These findings, together with the wide expression of TRH in chicken tissues, not only support TRH-TRHR1 signaling plays a role in the regulation of pituitary hormone expression and secretion but also suggest that TRH may play diverse roles in extrapituitary tissues such as the testes, hypothalamus, fat, skin, and spinal cord, via TRHR1 and/or TRHR3 in birds.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (31572391, 31771375, and 31772590).
Footnotes
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.psj.2019.10.062.
Contributor Information
Juan Li, Email: lijuanscuhk@163.com.
Yajun Wang, Email: cdwyjhk@gmail.com.
Supplementary data
References
- Aoki Y., Ono H., Yasuo S., Masuda T., Yoshimura T., Ebihara S., Iigo M., Yanagisawa T. Molecular evolution of prepro-thyrotropin-releasing hormone in the chicken (Gallus gallus) and its expression in the brain. Zoolog. Sci. 2007;24:686–692. doi: 10.2108/zsj.24.686. [DOI] [PubMed] [Google Scholar]
- Bidaud I., Lory P., Nicolas P., Bulant M., Ladram A. Characterization and functional expression of cDNAs encoding thyrotropin-releasing hormone receptor from Xenopus laevis. Eur. J. Biochem. 2002;269:4566–4576. doi: 10.1046/j.1432-1033.2002.03152.x. [DOI] [PubMed] [Google Scholar]
- Bøler J., Enzmann F., Folkers K., Bowers C.Y., Schally A.V. The identity of chemical and hormonal properties of the thyrotropin releasing hormone and pyroglutamyl-histidyl-proline amide. Biochem. Biophys. Res. Commun. 1969;37:705–710. doi: 10.1016/0006-291x(69)90868-7. [DOI] [PubMed] [Google Scholar]
- Burgus R., Dunn T.F., Desiderio D., Ward D.N., Vale W., Guillemin R. Characterization of ovine hypothalamic hypophysiotropic TSH-releasing factor. Nature. 1970;226:321–325. doi: 10.1038/226321a0. [DOI] [PubMed] [Google Scholar]
- Cai G., Mo C., Huang L., Li J., Wang Y. Characterization of the two CART genes (CART1 and CART2) in chickens (Gallus gallus) PLoS One. 2015;10:e0127107. doi: 10.1371/journal.pone.0127107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Groef B., Goris N., Arckens L., Kuhn E.R., Darras V.M. Corticotropin-releasing hormone (CRH)-induced thyrotropin release is directly mediated through CRH receptor type 2 on thyrotropes. Endocrinology. 2003;144:5537–5544. doi: 10.1210/en.2003-0526. [DOI] [PubMed] [Google Scholar]
- Duthie S.M., Taylor P.L., Anderson L., Cook J., Eidne K.A. Cloning and functional characterisation of the human TRH receptor. Mol. Cell. Endocrinol. 1993;95:R11–R15. doi: 10.1016/0303-7207(93)90043-j. [DOI] [PubMed] [Google Scholar]
- Engel S., Gershengorn M.C. Thyrotropin-releasing hormone and its receptors--a hypothesis for binding and receptor activation. Pharmacol. Ther. 2007;113:410–419. doi: 10.1016/j.pharmthera.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Gaspar E., Hardenbicker C., Bodo E., Wenzel B., Ramot Y., Funk W., Kromminga A., Paus R. Thyrotropin releasing hormone (TRH): a new player in human hair-growth control. FASEB J. 2010;24:393–403. doi: 10.1096/fj.08-126417. [DOI] [PubMed] [Google Scholar]
- Gaspar E., Nguyen-Thi K.T., Hardenbicker C., Tiede S., Plate C., Bodo E., Knuever J., Funk W., Biro T., Paus R. Thyrotropin-releasing hormone selectively stimulates human hair follicle pigmentation. J. Invest. Dermatol. 2011;131:2368–2377. doi: 10.1038/jid.2011.221. [DOI] [PubMed] [Google Scholar]
- Geris K.L., Meeussen G., Kühn E.R., Darras V.M. Distribution of somatostatin in the brain and of somatostatin and thyrotropin-releasing hormone in peripheral tissues of the chicken. Brain Res. 2000;873:306–309. doi: 10.1016/s0006-8993(00)02550-6. [DOI] [PubMed] [Google Scholar]
- Gershengorn M.C., Osman R. Molecular and cellular biology of thyrotropin-releasing hormone receptors. Physiol. Rev. 1996;76:175–191. doi: 10.1152/physrev.1996.76.1.175. [DOI] [PubMed] [Google Scholar]
- Harder S., Dammann O., Buck F., Zwiers H., Lederis K., Richter D., Bruhn T.O. Cloning of two thyrotropin-releasing hormone receptor subtypes from a lower vertebrate (Catostomus commersoni): functional expression, gene structure, and evolution. Gen. Comp. Endocrinol. 2001;124:236–245. doi: 10.1006/gcen.2001.7709. [DOI] [PubMed] [Google Scholar]
- Harvey S. Thyrotrophin-releasing hormone: a growth hormone-releasing factor. J. Endocrinol. 1990;125:345–358. doi: 10.1677/joe.0.1250345. [DOI] [PubMed] [Google Scholar]
- Harvey S., Scanes C.G., Chadwick A., Bolton N.J. The effect of thyrotropin-releasing hormone (TRH) and somatostatin (GHRIH) on growth hormone and prolactin secretion in vitro and in vivo in the domestic fowl (Gallus domesticus) Neuroendocrinology. 1978;26:249–260. doi: 10.1159/000122831. [DOI] [PubMed] [Google Scholar]
- He C., Zhang J., Gao S., Meng F., Bu G., Li J., Wang Y. Molecular characterization of three NPY receptors (Y2, Y5 and Y7) in chickens: gene structure, tissue expression, promoter identification, and functional analysis. Gen. Comp. Endocrinol. 2016;236:24–34. doi: 10.1016/j.ygcen.2016.04.019. [DOI] [PubMed] [Google Scholar]
- Heuer H., Schfer M.K.H., O'Donnell D., Walker P., Bauer K. Expression of thyrotropin-releasing hormone receptor 2 (TRH-R2) in the central nervous system of rats. J. Comp. Neurol. 2000;428:319–336. [PubMed] [Google Scholar]
- Huang G., He C., Meng F., Li J., Zhang J., Wang Y. Glucagon-like peptide (GCGL) is a novel potential TSH-releasing factor (TRF) in Chickens: I) Evidence for its potent and specific action on stimulating TSH mRNA expression and secretion in the pituitary. Endocrinology. 2014;155:4568–4580. doi: 10.1210/en.2014-1331. [DOI] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Itadani H., Nakamura T., Itoh J., Iwaasa H., Kanatani A., Borkowski J., Ihara M., Ohta M. Cloning and characterization of a new subtype of thyrotropin-releasing hormone receptors. Biochem. Biophys. Res. Commun. 1998;250:68–71. doi: 10.1006/bbrc.1998.9268. [DOI] [PubMed] [Google Scholar]
- Jiang X.L., Wang Y., Chen Z., Cai Z.W., Zhang L.F., Zhou H.M., Xu N.Y. Polymorphisms in porcine TRH and TRHR gene and associations with growth and fatness traits. Livest. Sci. 2012;144:67–73. [Google Scholar]
- Kanasaki H., Fukunaga K., Takahashi K., Miyazaki K., Miyamoto E. Mitogen-activated protein kinase activation by stimulation with thyrotropin-releasing hormone in rat pituitary GH3 cells. Biol. Reprod. 1999;61:319–325. doi: 10.1095/biolreprod61.1.319. [DOI] [PubMed] [Google Scholar]
- Kumar S., Hedges S.B. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- Lamacz M., Hindelang C., Tonon M.C., Vaudry H., Stoeckel M.E. Three distinct thyrotropin-releasing hormone-immunoreactive axonal systems project in the median eminence-pituitary complex of the frog ran a ridibunda. Immunocytochemical evidence for co-localization of thyrotropinreleasing hormone and mesotocin in fibers innervating pars intermedia cells. Neuroscience. 1989;32:451–462. doi: 10.1016/0306-4522(89)90093-6. [DOI] [PubMed] [Google Scholar]
- Lechan R., Wu P., Jackson I., Wolf H., Cooperman S., Mandel G., Goodman R. Thyrotropin-releasing hormone precursor: characterization in rat brain. Science. 1986;231:159–161. doi: 10.1126/science.3079917. [DOI] [PubMed] [Google Scholar]
- Liu X.G., Tan L.J., Lei S.F., Liu Y.J., Shen H., Wang L., Yan H., Guo Y.F., Xiong D.H., Chen X.D., Pan F., Yang T.L., Zhang Y.P., Guo Y., Tang N.L., Zhu X.Z., Deng H.Y., Levy S., Recker R.R., Papasian C.J., Deng H.W. Genome-wide association and replication studies identified TRHR as an important gene for lean body mass. Am. J. Hum. Genet. 2009;84:418–423. doi: 10.1016/j.ajhg.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Bidaud I., Ladram A., Gershengorn M.C. Pharmacological studies of thyrotropin-releasing hormone (TRH) receptors from Xenopus laevis: is xTRHR3 a TRH receptor? Endocrinology. 2003;144:1842–1846. doi: 10.1210/en.2002-221074. [DOI] [PubMed] [Google Scholar]
- Marangell L.B. Effects of intrathecal thyrotropin-releasing hormone (protirelin) in refractory depressed patients. Arch. Gen. Psychiatry. 1997;54:214. doi: 10.1001/archpsyc.1997.01830150034007. [DOI] [PubMed] [Google Scholar]
- Meier N.T., Haslam I.S., Pattwell D.M., Zhang G.Y., Emelianov V., Paredes R., Debus S., Augustin M., Funk W., Amaya E., Kloepper J.E., Hardman M.J., Paus R. Thyrotropin-releasing hormone (TRH) promotes wound re-epithelialisation in frog and human skin. PLoS One. 2013;8:e73596. doi: 10.1371/journal.pone.0073596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekuchi M., Saito Y., Aoki Y., Masuda T., Iigo M., Yanagisawa T. Molecular cloning, gene structure, molecular evolution and expression analyses of thyrotropin-releasing hormone receptors from medaka (Oryzias latipes) Gen. Comp. Endocrinol. 2011;170:374–380. doi: 10.1016/j.ygcen.2010.10.013. [DOI] [PubMed] [Google Scholar]
- Mo C., Cai G., Huang L., Deng Q., Lin D., Cui L., Wang Y., Li J. Corticotropin-releasing hormone (CRH) stimulates cocaine- and amphetamine-regulated transcript gene (CART1) expression through CRH type 1 receptor (CRHR1) in chicken anterior pituitary. Mol. Cell. Endocrinol. 2015;417:166–177. doi: 10.1016/j.mce.2015.09.007. [DOI] [PubMed] [Google Scholar]
- Mo C., Huang L., Cui L., Lv C., Lin D., Song L., Zhu G., Li J., Wang Y. Characterization of NMB, GRP and their receptors (BRS3, NMBR and GRPR) in chickens. J. Mol. Endocrinol. 2017;59:61–79. doi: 10.1530/JME-17-0020. [DOI] [PubMed] [Google Scholar]
- Nakano M., Hasunuma I., Minagawa A., Iwamuro S., Yamamoto K., Kikuyama S., Machida T., Kobayashi T. Possible involvement of thyrotropin-releasing hormone receptor 3 in the release of prolactin in the metamorphosing bullfrog larvae. Gen. Comp. Endocrinol. 2018;267:36–44. doi: 10.1016/j.ygcen.2018.05.029. [DOI] [PubMed] [Google Scholar]
- O'Dowd B.F., Lee D.K., Huang W., Nguyen T., Cheng R., Liu Y., Wang B., Gershengorn M.C., George S.R. TRH-R2 exhibits similar binding and acute signaling but distinct regulation and anatomic distribution compared with TRH-R1. Mol. Endocrinol. 2000;14:183–193. doi: 10.1210/mend.14.1.0407. [DOI] [PubMed] [Google Scholar]
- Paulssen R.H., Paulssen E.J., Gautvik K.M., Gordeladze J.O. The thyroliberin receptor interacts directly with a stimulatory guanine-nucleotide-binding protein in the activation of adenylyl cyclase in GH3 rat pituitary tumour cells. Evidence obtained by the use of antisense RNA inhibition and immunoblocking of the stimulatory guanine-nucleotide-binding protein. Eur. J. Biochem. 1992;204:413–418. doi: 10.1111/j.1432-1033.1992.tb16651.x. [DOI] [PubMed] [Google Scholar]
- Péczely P., Kiss J.Z. Immunoreactivity to vasoactive intestinal polypeptide (VIP) and thyreotropin-releasing hormone (TRH) in hypothalamic neurons of the domesticated pigeon (Columba livia). Alterations following lactation and exposure to cold. Cell Tissue Res. 1988;251:485–494. doi: 10.1007/BF00215858. [DOI] [PubMed] [Google Scholar]
- Rabeler R., Mittag J., Geffers L., Ruther U., Leitges M., Parlow A.F., Visser T.J., Bauer K. Generation of thyrotropin-releasing hormone receptor 1-deficient mice as an animal model of central hypothyroidism. Mol. Endocrinol. 2004;18:1450–1460. doi: 10.1210/me.2004-0017. [DOI] [PubMed] [Google Scholar]
- Richter K., Kawashima E., Egger R., Kreil G. Biosynthesis of thyrotropin releasing hormone in the skin of Xenopus laevis: partial sequence of the precursor deduced from cloned cDNA. EMBO J. 1984;3:617–621. doi: 10.1002/j.1460-2075.1984.tb01857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T., Feng P., Kim U.J., Wilber J.F. Identification of thyrotropin-releasing hormone receptor in the rat testis. Neuropeptides. 1994;27:195–202. doi: 10.1016/0143-4179(94)90070-1. [DOI] [PubMed] [Google Scholar]
- Scanes C.G. Some in vitro effects of synthetic thyrotrophin releasing factor on the secretion of thyroid stimulating hormone from the anterior pituitary gland of the domestic fowl. Neuroendocrinology. 1974;15:1–9. doi: 10.1159/000122287. [DOI] [PubMed] [Google Scholar]
- Segerson T.P., Hoefler H., Childers H., Wolfe H.J., Wu P., Jackson I.M., Lechan R.M. Localization of thyrotropin-releasing hormone prohormone messenger ribonucleic acid in rat brain in situ hybridization. Endocrinology. 1987;121:98–107. doi: 10.1210/endo-121-1-98. [DOI] [PubMed] [Google Scholar]
- Straub R.E., Frech G.C., Joho R.H., Gershengorn M.C. Expression cloning of a cDNA encoding the mouse pituitary thyrotropin-releasing hormone receptor. Proc. Natl. Acad. Sci. U S A. 1990;87:9514–9518. doi: 10.1073/pnas.87.24.9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y.M., Millar R.P., Ho H., Gershengorn M.C., Illing N. Cloning and characterization of the chicken thyrotropin-releasing hormone receptor. Endocrinology. 1998;139:3390–3398. doi: 10.1210/endo.139.8.6133. [DOI] [PubMed] [Google Scholar]
- Tashjian A.H., Barowsky N.J., Jensen D.K. Thyrotropin releasing hormone: direct evidence for stimulation of prolactin production by pituitary cells in culture. Biochem. Biophys. Res. Commun. 1971;43:516–523. doi: 10.1016/0006-291x(71)90644-9. [DOI] [PubMed] [Google Scholar]
- Taylor T., Wondisford F.E., Blaine T., Weintraub B.D. The paraventricular nucleus of the hypothalamus has a major role in thyroid hormone feedback regulation of thyrotropin synthesis and secretion. Endocrinology. 1990;126:317–324. doi: 10.1210/endo-126-1-317. [DOI] [PubMed] [Google Scholar]
- Vandenborne K., Roelens S.A., Darras V.M., Kuhn E.R., Van der Geyten S. Cloning and hypothalamic distribution of the chicken thyrotropin-releasing hormone precursor cDNA. J. Endocrinol. 2005;186:387–396. doi: 10.1677/joe.1.06161. [DOI] [PubMed] [Google Scholar]
- Wang Y., Li J., Wang C.Y., Kwok A.H., Leung F.C. Identification of the endogenous ligands for chicken growth hormone-releasing hormone (GHRH) receptor: evidence for a separate gene encoding GHRH in submammalian vertebrates. Endocrinology. 2007;148:2405–2416. doi: 10.1210/en.2006-1013. [DOI] [PubMed] [Google Scholar]
- Wang Y., Li J., Yan Kwok A.H., Ge W., Leung F.C. A novel prolactin-like protein (PRL-L) gene in chickens and zebrafish: cloning and characterization of its tissue expression. Gen. Comp. Endocrinol. 2010;166:200–210. doi: 10.1016/j.ygcen.2009.10.007. [DOI] [PubMed] [Google Scholar]
- Wang Y., Wang C.Y., Wu Y., Huang G., Li J., Leung F.C. Identification of the receptors for prolactin-releasing peptide (PrRP) and Carassius RFamide peptide (C-RFa) in chickens. Endocrinology. 2012;153:1861–1874. doi: 10.1210/en.2011-1719. [DOI] [PubMed] [Google Scholar]
- Wilber J.F., Xu A.H. The thyrotropin-releasing hormone gene 1998: cloning, characterization, and transcriptional regulation in the central nervous system, heart, and testis. Thyroid. 1998;8:897–901. doi: 10.1089/thy.1998.8.897. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.