Abstract
We studied the microbial profiles of the duodenum, jejunum, and ileum during different developmental stages in the duck using high-throughput sequencing of the bacterial 16S rRNA gene. We also investigated the differences in the microbiota in the duodenum, jejunum, and ileum at different developmental times. A correlation analysis was performed between the most abundant bacterial genera and the development of the small intestine. An analysis of alpha diversity indicated different species richness and bacterial diversity in the different small intestinal segments and at different development times. A beta diversity analysis indicated differences in the bacterial community compositions across time. In a weighted UniFrac principal coordinates analysis, the samples clustered into two categories, 2 to 4 wk and 6 to 10 wk, in the duodenum, jejunum, and ileum. Our results show that the small intestine is predominantly populated by the phyla Firmicutes, Bacteroidetes, and Proteobacteria throughout the developmental stages of the duck. The duodenum, jejunum, and ileum shared most of the bacterial phyla and genera present, although they showed significant differences in their relative abundances in the intestinal segments and developmental stages. They shared different bacterial taxa during development times and among different segments when the intergroup differences were analyzed. The genera Bacillus, Corynebacterium 1, Lactococcus, Sphingomonas, and Haliangium correlated moderately positively with the increase in bodyweight and the lengths and weights of the duodenum, jejunum, and ileum, and these genera may be considered important markers when assessing the heath of the intestinal microbiota in ducks. This study provides a foundation upon which to extend our knowledge of the diversity and composition of the duck microbiota and a basis for further studies of the management of the small intestinal microbiota and improvements in the health and production of ducks.
key words: microbiota, diversity and composition, small intestine, duck
INTRODUCTION
The integrity, function, and health of the animal gut depend on many factors, including the animal's environment, feed, and gut microbiota. The gut microbiota is important to the health and production of the bacterial host. The symbiotic interactions between the host and its gut microbes are fundamental to animal health and production. It is increasingly recognized that the gut microbiota plays an important role in a variety of physiological processes, including metabolic homeostasis (Shin et al., 2014; Chen et al., 2019; Lazar et al., 2019), immune function (Ichinohe et al., 2011; Ivanov and Littman, 2011; Kuss et al., 2011), and tissue development (Diaz Heijtz et al., 2011). A better understanding of the animal gut microbiota and gut function will provide new opportunities for the improvement of animal health and production.
The composition and distribution of the gut microbiota in different animals have been extensively investigated in recent years (Kim and Isaacson, 2015; Ellegaard and Engel, 2019; Pourabedin and Zhao, 2017; Tanca et al., 2017; Vatsos, 2017). Different animals display significantly different microbiotal compositions and diversity, even in different sections of the gut (Feng et al., 2019). Less information is available on the microbiota in the small intestine than in the large intestine, especially in poultry and waterfowl. The small intestine includes the duodenum, jejunum, and ileum. The different sections of the small intestine perform different functions, so we can infer that the microbes in the intestinal sections also differ. In humans, many kinds of enzyme help to break down food in the duodenum, and then most nutrients are absorbed in the jejunum. In the ileum, Peyer's lymph node is the hub of a large number of lymphatic processes in the intestinal tract. The number of microbes in every 1 g of intestinal contents increases from the duodenum, to the jejunum, and the ileum. Thus, among the small-intestinal segments in humans, the number of microbes is highest in the ileum (El Aidy et al., 2015). The microbiota of the small intestine may also have profound effects on various aspects of the host's physiology, including the immune, metabolic, and endocrine functions, in different animals. To gain further insight into the composition and function of the gut microbiota, it is crucial to understand the spatial variations in the microbiota across the small intestine.
The intestinal microbiota is always complex and variable, and is in a dynamic and delicate balance with host (Guevarra et al., 2019). We speculated that the composition and diversity of the intestinal microbiota could be affected by the different developmental stages of the domestic duck, and may also differ among its intestinal segments. With advances in the high-throughput sequencing technology, both culture-independent and -dependent microbial communities in the intestinal tract can be studied, and 16S-rRNA-based next-generation sequencing is a powerful tool with which to investigate the biological and ecological roles of the intestinal microbiota in different animals.
In this study, we investigated the structural composition and the predicted functions of the microbial communities in the intestinal segments (duodenum, jejunum, and ileum) in the Gaoyou duck (Anas platyrhynchos) at different developmental stages. Our results show that the microbiota has a slightly different composition and high diversity among the duodenum, jejunum, and ileum in different developmental periods. These findings provide a new insight into the membership of the microbiota along the small intestine, although its functions are yet to be investigated in the duck.
MATERIAL AND METHODS
Animal Breeding and Sample Selection
One hundred Gaoyou ducks were raised on ground. The experimental diets were formulated to meet the nutritional needs of the ducks, according to the National Research Council of China (NRC, 2010). Eight ducks were randomly selected for sampling at 2, 4, 6, and 10 wk. The bodyweights and lengths and weights of the duodenal, jejunal, and ileal segments were recorded. The contents of the intestinal segments were collected with elbow tweezers along the outer walls of the intestines and placed in 5 mL Eppendorf tubes. The samples were kept frozen at −80°C until they were sent to Novogene Bioinformatics Technology Co., Ltd (Tianjin, China) for 16S rRNA amplification and sequencing.
These animal experiments were approved by the Committee of Animal Care at Jiangsu Institute of Poultry Science (CAC-JIPS01342, Yangzhou, China).
DNA Extraction, Amplification, and Sequencing
Eight DNA samples were extracted from the contents of the duodenal, jejunal, and ileal segments for each time group. Total genomic DNA was extracted with the CTAB method (Tang et al., 2008). The quality and quantity of the DNA was verified with a NanoDrop™ 2000 spectrophotometer (Thermo Scientific, MA, USA) and agarose gel electrophoresis. The extracted DNA was diluted to a concentration of 1 ng/μl and stored at −20°C. The diluted DNA was used as the template for the PCR amplification of the bacterial 16S rRNA genes, with barcoded primers and HiFi HotStart ReadyMix (KAPA Biosystems, MA, USA). For the bacterial diversity analysis, the V3-V4 variable regions of the 16S rRNA genes were amplified with the universal primers 343F and 798R (343F: TACGGRAGGCAGCAG; 798R: AGGGTATCTAATCCT). The amplicon quality was checked by visualization with gel electrophoresis, and they were purified with AMPure XP beads (Agencourt, CA, USA). Equal amounts of purified amplicon were pooled for subsequent sequencing.
Bioinformatics Analysis
The raw sequencing data were in the FASTQ format. Paired-end reads were preprocessed with the Trimmomatic software (Bolger et al., 2014) to remove ambiguous bases (N) and low-quality sequences, with average quality scores below 20, using a sliding-window trimming approach. After trimming, the paired-end reads were assembled with the FLASH software using the parameters: minimal overlap of 10 bp, maximum overlap of 200 bp, and maximum mismatch rate of 20% (Reyon et al., 2012). The sequences were further denoised with the (QIIME) software (version 1.9.1) (Caporaso et al., 2010), and all sequences with bases above Q20 were retained. Chimeric reads were detected and removed. The reads were compared with the reference database (Silva database, https://www.arb-silva.de/) (Quast et al., 2012) using UCHIME algorithm (http://www.drive5.com/usearch/manual/uchime_algo.html) (Edgar et al., 2011) to detect chimera sequences, and then the chimera sequences were removed (Haas et al., 2011). Then the clean reads finally obtained.
The primer sequences were removed from the clean reads, which were then clustered to generate operational taxonomic units (OTUs) with the Vsearch software, with a similarity cutoff of 97% (Edgar, 2013). A representative read for each OTU was selected with the QIIME package. All representative reads were annotated and searched with BLAST against the Silva database release 123 using the Ribosomal Database Project classifier (confidence threshold of 70%) (Wang et al., 2007).
The alpha diversity metrics (Chao1, observed species, Shannon's index, and Simpson's index) were calculated with the QIIME software (Version 1.9.1) and displayed with R package (version 2.15.3). The differences in the alpha diversity indices were analyzed with R project and parameter and nonparameteric tests were performed groups. For the beta diversity metrics, the weighted UniFrac distance matrices were calculated by QIIME software (version 1.9.1). The principal coordinates analysis (PCA) figures were generated based on FactoMineR package and ggplot 2 package in R software (Version 2.15.3). The LEfSe analysis was performed with LEfSe software, and the screening value, the linear discriminant analysis (LDA) score, was 7. R project was used for the Metastats analysis, and the permutation test between groups under phylum and genus classification level to obtain the p values. The Benjamini and Hochberg false discovery rate was used to revise the p values (White et al., 2009). Analysis of similarities (ANOSIM) was performed in the R vegan package (2.15.3) and the analysis of molecular variance with the mothur software (1.33.3). The species analysis, to identify significant differences between groups, was performed with the T_test in the R package. The correlations between the bacterial genera in the small intestine and duck development were investigated with Pearson's correlation analysis.
Statistical Analysis
The bodyweights and the lengths and weights of the duodenal, jejunal, and ileal segments were assessed with analysis of variance. Differences in alpha diversity and in the relative abundances of the bacterial phyla, genera, and species between groups were analyzed with the Mann–Whitney U test. All analyses were performed with IBM SPSS v. 20.0 (SPSS Inc., Chicago, IL, USA). For all tests, values of P < 0.05 were considered to indicate statistically significant differences.
RESULTS
Diversity of the Bacterial Communities in Duck Small Intestine
The microbiota was analyzed based on 96 sequenced samples (three intestinal segments from each of eight Gaoyou ducks collected at 2, 4, 6, and 10 wk after the start of the experiment). Each sample included an average of 36,315 sequences, and a total of 4,025 OTUs were detected, based on 97% nucleotide sequence identity between the reads in an OTU. Individual-segment-based rarefaction curves were generated to assess whether the sampling of each segment of the small intestine was sufficient for the analysis. Good's coverage ranged from 98 to 99% for all samples (data not shown).
Chao1, the observed species, Shannon's index, and Simpson's index indicated that there were significant differences among the microbiota in the duodenal, jejunal, and ileal segments and in the different age groups (Table 1). The Chao1 values and observed species were higher at 2 and 4 wk than at 6 and 10 wk in the duodenum (P < 0.05). In jejunum, the Chao1 values showed no difference among development periods (2, 4, 6, and 10 wk) (P > 0.05), the observed species at 10 wk were significant less than that at 2 and 4 wk in jejunum (P < 0.05). In ileum, the Chao1 values and observed species showed no significant difference between 2 and 4 wk or 6 and 10 wk, and these values at 6 wk were significant less than that at 2 and 4 wk (P < 0.05). Shannon's and Simpson's indices showed similar trends in duodenal, the values at 2 and 4 wk were significantly higher than that at 6 wk (P < 0.05). Shannon's indices in jejunum and Simpson's indices in jejunum and ileum showed no difference among 2, 4, 6, and 10 wk. These data suggest that the communities differed significantly as the age of the ducks increased and among the intestinal segments. The community richness in the duodenum, jejunum and ileum was less at 6 and 10 wk than at 2 and 4 wk. The microbiota also showed less diversity at 6 and 10 wk in the duodenum and jejunum.
Table 1.
Alpha-diversity in the duodenum, jejunum, and ileum at different development stages.
| 2 wk | 4 wk | 6 wk | 10 wk | ||
|---|---|---|---|---|---|
| Chao1 | duodenum | 1084.53a ± 18.33 | 1000.55a ± 154.29 | 399.62b ± 112.42 | 637.74b ± 204.21 |
| jejunum | 849.55 ± 177.31 | 1012.33 ± 94.27 | 637.51 ± 219.17 | 482.14 ± 262.18 | |
| ileum | 1144.13a ± 42.22 | 952.75a,b ± 137.96 | 331.72d ± 124.69 | 539.39b-d ± 137.12 | |
| Observed species | duodenum | 1004.00a ± 9.79 | 912.4a ± 128.37 | 323.83b ± 90.01 | 512b ± 131.28 |
| jejunum | 728.00a,b ± 120.09 | 870.20a ± 75.10 | 486.25b,c ± 115.79 | 365.00c ± 182.23 | |
| ileum | 957.80a ± 37.32 | 701.00a,b ± 134.77 | 217.67c ± 82.23 | 400.33b,c ± 98.19 | |
| Shannon's index | duodenum | 7.5a ± 0.03 | 7.69a ± 0.05 | 5.08b ± 0.58 | 6.18a,b ± 0.56 |
| jejunum | 7.36a ± 0.12 | 7.61a ± 0.03 | 6.42a,b ± 0.42 | 5.32b ± 1.04 | |
| ileum | 6.6552 ± 0.2276 | 5.1151 ± 0.8384 | 3.4425 ± 0.6824 | 5.0676 ± 0.6748 | |
| Simpson' index | duodenum | 0.9719a ± 0.0015 | 0.9777a ± 0.0031 | 0.9136b ± 0.0225 | 0.9549a,b ± 0.0144 |
| jejunum | 0.9726 ± 0.0030 | 0.9775 ± 0.0008 | 0.9630 ± 0.0087 | 0.9093 ± 0.0380 | |
| ileum | 0.9400 ± 0.0124 | 0.8258 ± 0.0806 | 0.7359 ± 0.1109 | 0.8729 ± 0.0559 |
Note: different superscript letters indicate that the alpha-diversity indices differed among the different development stages at P < 0.05. (n = 5–6).
The Chao1 values and observed species did not differ among the duodenum, jejunum, and ileum, whereas Shannon's and Simpson's indices showed that the diversity was lower in the ileum than in the duodenum or jejunum (P < 0.05) (Table 2).
Table 2.
Alpha-diversity in the duodenum, jejunum, and ileum.
| duodenum | jejunum | ileum | |
|---|---|---|---|
| Chao1 | 724.81 ± 51.99 | 646.38 ± 47.52 | 719.28 ± 49.89 |
| Observed species | 655.08 ± 46.51 | 558.12 ± 37.38 | 551.12 ± 41.90 |
| Shannon's index | 7.02a ± 0.14 | 7.04a ± 0.13 | 4.93b ± 0.25 |
| Simpson's index | 0.97a ± 0.00 | 0.97a ± 0.01 | 0.82b ± 0.02 |
Note: different superscript letters indicate that the alpha-diversity indices differ among the duodenum, jejunum, and ileum at P < 0.05. (n = 20–24).
Characterization of the Microbiota in the Duodenum, Jejunum, and Ileum
The differences in the microbial communities in the different parts of the small intestine were measured at the genus level with weighted UniFrac β-diversity measures based on ANOSIM. Considering the similar ecological niches in the 3 segments and their shared location in the small intestine, we assumed that they would share some microbial taxa, and merely differ in the abundances of these taxa. Our results revealed slight group differences among the different developmental stages in the same intestinal segment, and in the different segments at the same developmental stage (P = 0.001, R2 = 0.25–0.5).
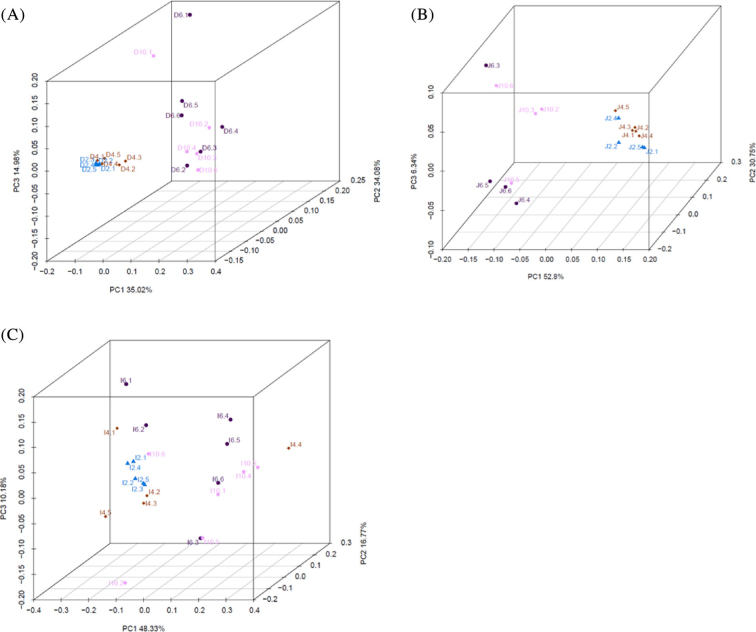
This was further supported by a phylogram of the weighted UniFrac distances, which showed that the bacterial communities in the duodenum, jejunum, and ileum were separated into 2 groups (2 to 4 wk and 6 to 10 wk), and that there was also a very small distance between 2 and 4 wk (Figure 1 A, B, C).
Figure 1.
Phylogram of weighted UniFrac distances in the duodenum (A), jejunum (B), and ileum (C) during duck development. Δ: 2 wk; ⋄: 4 wk; •: 6 wk; ○: 10 wk.
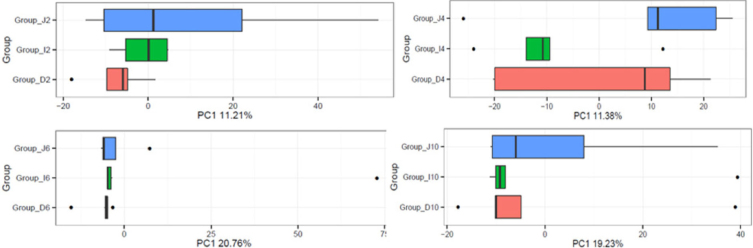
There were no differences among the duodenum, jejunum, and ileum at 6 or 10 wk (P > 0.05), but the jejunal and ileal microbiotas were spatially separated from the duodenal microbiota at 2 wk, and the duodenum and jejunum microbiotas were spatially separated from the ileal microbiota at 4 wk (P < 0.05) (Figure 2). The analysis of the data with a heatmap (details not shown) showed that the distances among the development periods for the same small-intestinal segments (about 0.3) were more than that those between the duodenum, jejunum, and ileum at the same times (about 0.01) (P < 0.05). These results indicate that the group differences across the developmental periods were greater than those among the different intestinal sections.
Figure 2.
Boxplot of weighted UniFrac distances among the duodenum, jejunum, and ileum during duck development (at 2, 4, 6, and 10 wk, respectively). ○: Group of duodenum; ○: Group of jejunum; ○: Group of ileum.
Composition of/and Differences in Bacterial Phyla in the Duodenum, Jejunum, and Ileum
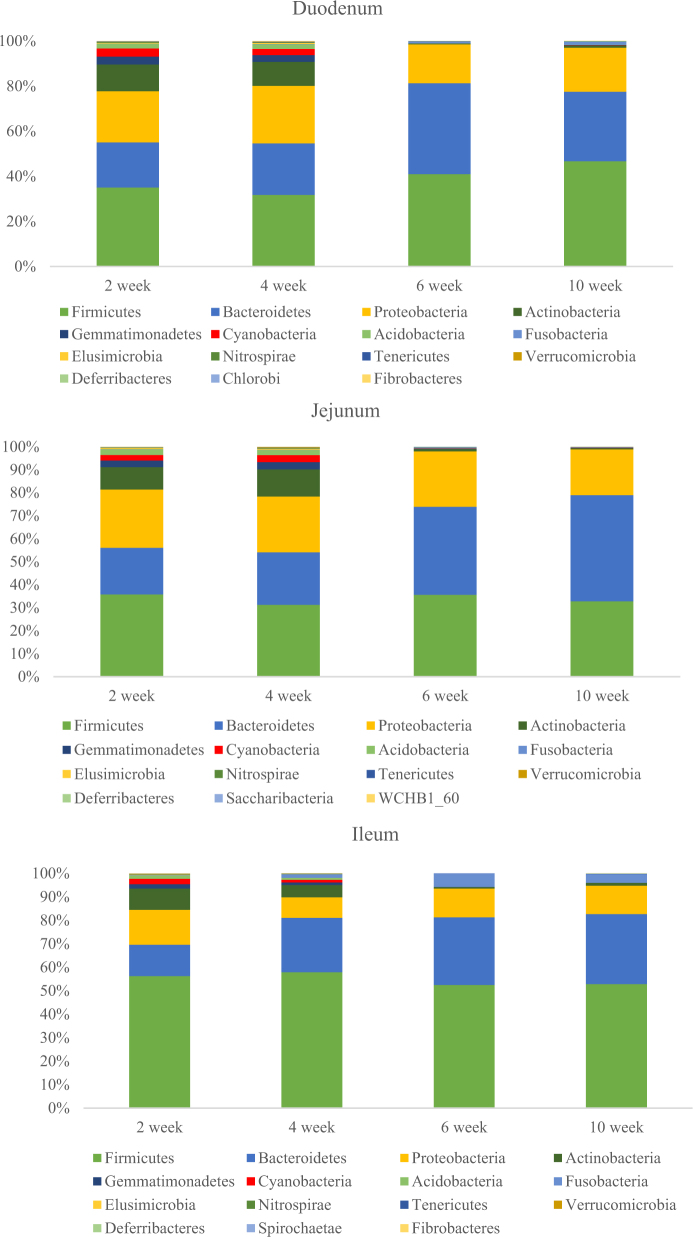
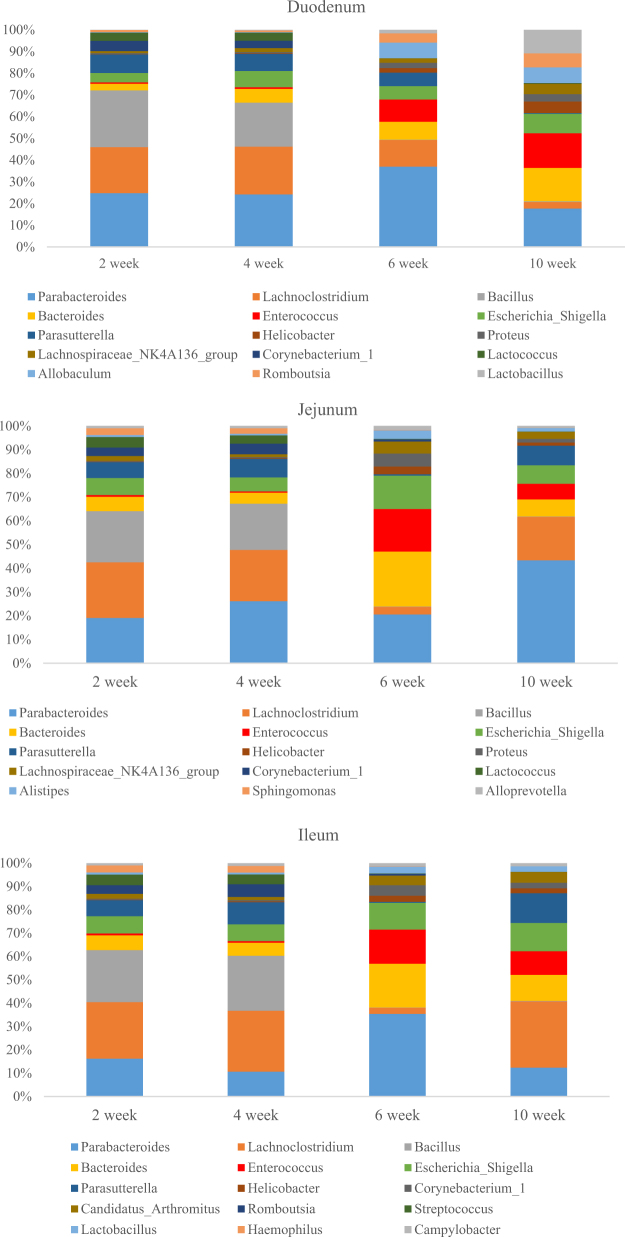
Most of the dominant phyla (the 15 most abundant phyla) were shared among the different intestinal segments, and these 15 most abundant phyla represent 99.14, 99.07, and 99.55% of total microbiome in the duodenum, jejunum, and ileum, respectively. Among those 15 most abundant phyla, Chlorobi was only detected in the duodenum, Saccharibacteria, WCHBI_60 was only detected in the jejunum, and Spirochaetae was only detected in the ileum. The other members of the most abundant 15 phyla occurred in the duodenum, jejunum, and ileum, and only the abundances of these bacterial phyla differed among the segments and developmental periods. The abundances of several bacterial phyla were similar at 2 and 4 wk, and the abundances at 6 and 10 wk were also similar in the duodenum, jejunum, and ileum. Also Actinobacteria, Gemmatimonadetes, Cyanobacteria, Acidobacteria, and Nitrospirae showed higher relative abundances at 2 and 4 wk than that at 6 and 10 wk (P < 0.05) in 3 segments. The details are shown in Figure 3. Phyla Firmicutes, Bacteroidetes, and Proteobacteria were dominant in all the segments, constituting about 92.95 to 97.83% of the microbiome at 6 and 10 wk, and a little less (76.74 to 89.39%) at 2 and 4 wk. However, there were also significant differences among the 3 segments. The ileum contained a significantly larger proportion of Firmicutes (about 50%) than the duodenum or jejunum (each about 35%) (Pileum VS duodenum = 0.001; Pileum VS jejunum = 0.000; Pduodenum VS jejunum = 0.297). Inversely, the duodenum or jejunum contained a significantly larger proportion of Proteobacteria (about 20%) than the ileum (about 11%) (Pileum VS duodenum = 0.000; Pileum VS jejunum = 0.000; Pduodenum VS jejunum = 0.190). Bacteroidetes also showed similar trends in all the segments, the relative abundance at 6 and 10 wk was higher than that at 2 and 4 wk. Furthermore, the abundance of Bacteroidetes at 6 wk were significantly higher than that at 2 and 4 wk (P < 0.05) in duodenum content, the Bacteroidetes abundance at 6 and 10 wk also significantly higher than that at 2 and 4 wk (P < 0.05) in jejunum content. There was no significant detected among different development times in the ileum content.
Figure 3.
Relative abundances of sequences belonging to different bacterial phyla (15 most abundant phyla) in the duck duodenum, jejunum, and ileum.
Composition of/and Differences in Bacterial Genera in the Duodenum, Jejunum, and Ileum
Like the bacterial phyla, most bacterial genera (9/15) occurred in all three intestinal segments, and their relative abundances in the intestinal contents differed with time. Bacillus showed similar trends in all the segments, the relative abundance at 2 and 4 wk was higher than that at 6 and 10 wk (P < 0.05). Corynebacterium_1 in jejunum and ileum showed significant higher abundance at 2 and 4 wk compared to that at 6 and 10 wk (P < 0.05). Lactococcus (in duodenal),Romboutsia (in jejunum) and Proteus (in ileum) showed significant higher abundance at 2 and 4 wk (P < 0.05), respectively. Both duodenal and jejunal content contained larger abundance of Bacteroides at 6 and 10 wk, and the abundance of Bacteroides was significant higher at 10 wk compared to that at 2 and 4 wk (P < 0.05) in duodenum; while the abundance of Bacteroides at 6 wk was significant higher than that at 2 and 4 wk (P < 0.05) in jejunum.
Analysis of Intergroup Differences in Bacterial Taxa
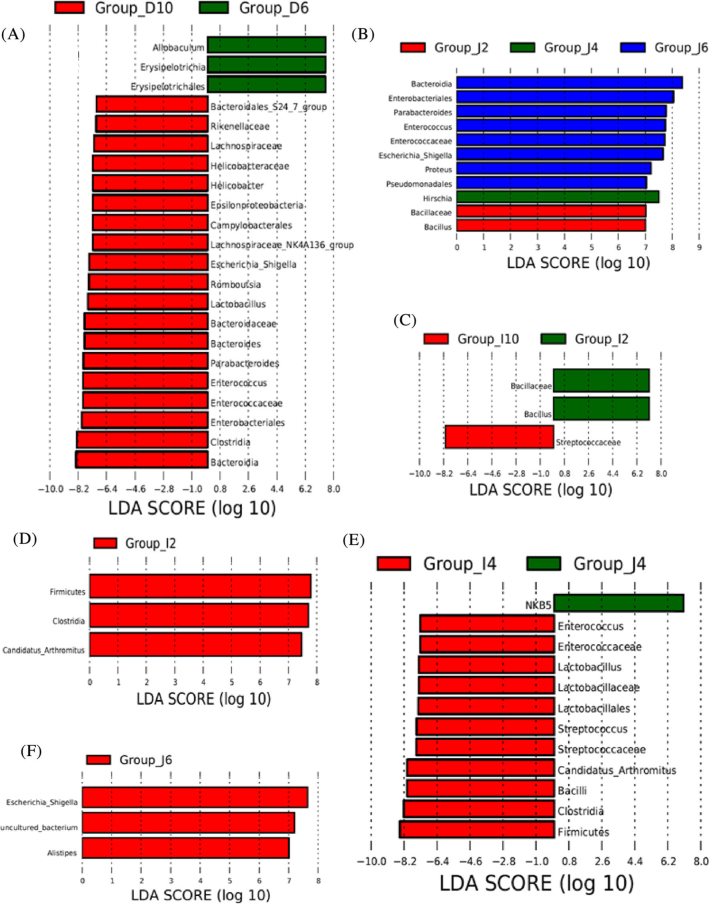
Significant differences were detected in the bacterial taxa present in each segment during the developmental periods (Figure 5 A-C). There are 5 family (f_Bacteroidaceae, f_Rikenellaceae, f_Enterococcaaceae, f_Lachnospiraceae, and f_Helicobacteraceae) detected at 10 wk (Figure 5 A) in duodenum. When we compared the different bacterial genera during the development times in duodenum, the results showed that g. Allobaculum has larger abundance at 6 wk, and 6 genera including g. Bacillus, g. Bacteroides, g. Enterococcus, g. Escherichia_Shigella, g. Helicobacter, and g. Lactobacillus were detected at 10 wk (Figure 5 A). Only f. Bacillaceae and g. Bacillus showed higher abundance at 2 wk, the abundance of g. Hirschia was high at 4 wk, while there are 4 genera including g. Parabacteroides, g. Enterococcus, g. Escherichia Shigella, g. Proteus detected at 6 wk in jejunum (Figure 5 B). There are f_Bacillaceae, g. Bacillus detected at 2 wk and f. Streptococcaceae detected at 10 wk in ileum during the development times (Figure 5 C). The results revealed that the duodenum shared a large number of different bacterial taxa at 6 and 10 wk with the jejunum and ileum.
Figure 5.
Histogram of the linear discriminant analysis (LDA) values showing the statistically significant differences in duodenum (A), jejunum (B), and ileum (C) during development periods and significant differences at 2 wk (D), 4 wk (E), and 6 wk (F) among duodenum, jejunum, and ileum segments. The LDA score was set at 7; the length of the column represents the influence of size. D2: duodenum samples at 2 wk, D4: duodenum samples at 4 wk, D6: duodenum samples at 6 wk and D10: duodenum samples at 10 wk; J2: jejunum at 2 wk, J4: jejunum at 4 wk, J6: jejunum at 6 wk and J10: jejunum at 10 wk; I2: ileum at 2 wk, I4: ileum at 4wk, I6: ileum at 6 wk and I10: ileum at 10 wk.
Figure 4.
Relative abundances of sequences belonging to different bacterial genera (15 most abundant genera) in the duck duodenum, jejunum, and ileum.
Then we compared different bacterial taxa at each development time point among duodenum, jejunum and ileum. A larger number of bacteria taxa were occurred at 4 wk, while there was no bacteria taxa detected at 10 wk (Figure 5 D-F). p. Firmicutes, c. Bacilli, c. Clostridia, o. Lactobacillales, f. Enterococcaceae, f. Lactobacillaceae, f. Streptococcaceae, g. Enterococcus, g. Lactobacillus, g. Streptococcus, and g. Candidatus Arthromitus in jejunum content differed significantly from those in duodenum and ileum at 4 wk (Fig. 6E).
Correlation Analysis of Microbes and Duck Development
In this study, we measured their bodyweights and the duodenal, jejunal, and ileum lengths, and weights. The 30 most abundant bacterial genera were selected for a correlation analysis. The results revealed the abundances of Bacillus, Corynebacterium 1, Lactococcus, Sphingomonas, and Haliangium correlated significantly with bodyweight, the weights, and lengths of the duodenum and ileum (Table 3).
Table 3.
Correlation analysis of bacterial genera in the small intestine and duck development.
| Body W | Duodenum L | Duodenum W | Jejunum L | Jejunum W | Ileum L | Ileum W | |
|---|---|---|---|---|---|---|---|
| Bacillus | 0.82 | 0.8 | 0.65 | 0.57 | 0.68 | 0.79 | 0.69 |
| Corynebacterium_1 | 0.79 | 0.77 | 0.63 | 0.5 | 0.64 | 0.75 | 0.66 |
| Lactococcus | 0.79 | 0.76 | 0.61 | 0.55 | 0.65 | 0.77 | 0.65 |
| Sphingomonas | 0.77 | 0.75 | 0.59 | 0.51 | 0.62 | 0.74 | 0.63 |
| Haliangium | 0.7 | 0.68 | 0.51 | 0.47 | 0.52 | 0.66 | 0.55 |
Note: W, weight; L, length; Green-yellow-red color gradation was used to indicate the correlation values in the table grids. All the P values were less than 0.01. Five-six samples were used to calculate correlation.
DISCUSSION
In this study, we detected a shift in the alpha diversity of bacteria between the small intestinal segments in the duck, as indicated by Chao1 and Shannon's index. The Chao1 estimator and observed species were higher at 2 and 4 wk than at 6 or 10 wk, indicating a decrease in richness with developmental age. The ileum showed lower richness and diversity than the duodenum or jejunum, which is consistent with previous studies in pigs and other livestock (Salonen and de Vos., 2014; Mao et al., 2015). In dairy cattle, the Chao1 estimator and Shannon index showed significant lower than that in duodenum and jejunum (Mao et al., 2015; Shang et al., 2018). Bacterial diversity differed with both the intestinal segment and the developmental stage.
In this study, it shows that the representative taxonomic groups within the duck small intestine were the phyla Firmicutes, Bacteroidetes, and Proteobacteria. Thirty-eight phyla were detected in the combined profiles of the duodenum, jejunum, and ileum in duck, which exceeds the number in broiler chickens or Guinea fowl (14 phyla in total), whereas all these 14 phyla were present in the profiles of the duck small intestine (Bhogoju et al., 2018). These results indicate that the microbial phylum profiles in the duck do not differ greatly from those reported for avian hosts by other researchers (Oakley et al., 2014; Waite and Taylor, 2015; Bhogoju et al., 2018). Importantly, the 3 small-intestinal segments shared most of the 30 most abundant phyla, and especially the 15 most abundant phyla. We examined whether the spatial continuity of the duodenum, jejunum, and ileum causes these small-intestinal segments to share the same profiles of bacterial phyla.
In chicken, the phylum Firmicutes became the dominant phylum as the chick matured and its gut environment became anaerobic, which could constitute a kind of developmental change (Lim et al., 2015). This result is consistent, to some extent, with Best's report that Firmicutes became the dominant phylum in the cecal contents after 4 d of age in duck (Best et al., 2017). Firmicutes was also the dominant phylum in duck small-intestinal contents in this study. There is about 31.32 to 46.58% in duodenum, 30.81 to 35.52% in jejunum, and 52.07 to 57.66% in ileum. And there was significantly more Firmicutes (about 50%) in the ileum than in the duodenum or jejunum (P < 0.05). The abundances of bacteria genera differed between the development periods 2 to 4 wk and 6 to 10 wk, which is consistent with the changes in beta diversity.
The differences in the abundances of bacteria in the different intestinal segments could be affected by the different dietary formulae consumed during the development periods. The percentage crude protein (CP %) was 20% at 0 to 4 wk but 16% at 5 to 10 wk. There was also a slight difference in the percentage of sulfur-containing amino acids (which decreased to about 0.02% at 5 to 10 wk, 0.40 vs. 0.38%) and percentage of calcium (which decreased to about 0.1% at 5 to 10 wk, 0.8 to 1.3% vs. 0.7 to 1.2%). The formula of the diet could change the composition of the microbiota. In another study, there was a five-fold greater amount of Lactobacillaceae spp.(Firmicutes) and a three-fold greater amount of Clostridia spp.(Bacteroidetes) in sows than in milk-formula-fed piglets, whereas the number of Enterobacteriaceae spp. (Proteobacteria) was five-fold greater in the milk-formula-fed piglets (Yeruva et al., 2016). In yet another study, a low-CP diet altered the bacterial communities, increasing the counts of short-chain-fatty-acid-producing bacteria (Clostridium cluster IV, Clostridium cluster XIVa, Bacteroidetes), reducing the counts of E. coli (Proteobacteria) in pig cecum, and markedly reducing the protein fermentation products (Zhang et al., 2016a). In the present study, the reduced CP% may have affected the abundances of several bacterial taxa. The relative abundance of Firmicutes, Bacteroidetes and Proteobacteria at 6 and 10 wk were a little more than that at 2 and 4 wk in all intestinal segments. Similar results was also detected in genus level, such as g. Bacillus, which abundance at 2 and 4 wk was significantly higher than that at 6 and 10 wk (P < 0.05).
Therefore, the patterns of gut microbial diversity and abundances varied across the different development stages in the host as the bacterial communities adapted to their rapidly changing environments. Ding reported that in various developmental periods (embryo, chick, and hen), chickens shared 65 genera, called the “core microbiota,” and that 42 and 62% of the gut microbial genera in the embryo were found in the maternal hen and chick, respectively. There was a moderate correlation (0.40) between the embryonic and maternal microbiotas, and a correlation of 0.52 between the embryonic and chick microbiotas at the family level (Ding et al., 2017). These results suggest that the number of bacterial taxa decreases as the gut tissue matures, whereas the abundances of the persistent bacteria changed. The stable bacteria in the mature individuals live symbiotically with the host, and they are mutually beneficial. In the present study, the abundances of several of the most abundant bacteria changed significantly between the breeding period and finishing period in the duck.
In this study, the genera Bacillus, Corynebacterium 1, Lactococcus, Sphingomonas, and Haliangium correlated moderately positively with the increases in bodyweight and the duodenal, jejunal, and ileal lengths and weights. Some species of Bacillus and Lactococcus are widely used as probiotics to improve the intestinal environment and increase animal production (Kumar et al., 2017; Oh et al., 2017; Wang et al., 2017). For example, B. subtilis significantly reduced the dextran sodium sulfate (DSS)-induced colonic mucosal injury and inflammatory factors in mice and improved their levels of short-chain fatty acids (Zhang et al., 2016b). The administration of Lactobacillus and B. cereus in feed to chicks modulated their immunity and intestinal microbiota by increasing the relative weights of the immune organs, significantly increasing Lactobacillus and B. cereus numbers, and reducing E. coli numbers (Li and zhao, 2009; Gong et al., 2018). Lactococcus lactis also exerted a protective effect on a mouse model of DSS-induced colitis (Berlec et al., 2017). These data suggest that these bacterial genera may positively affect the development of ducks before 10 wk old, and may be considered important markers in the management of intestinal microbiotal communities. These differences in abundance could be affected by the changes in diet that occur during the different developmental stages.
This study revealed that both development periods and intestinal segments affected the bacterial diversity and abundance of the microbial biota in ducks. Especially, the small intestinal bacteria can be grouped into categories that represent weeks 2 to 4 and weeks 6 to 10 of duck development. Also the relative abundance of several dominant phyla or genera showed significant differences in the different intestinal segments and developmental periods. Several bacterial genera moderately positively correlated with the increase in bodyweight and the lengths and weights of intestinal segments in the ducks. May these genera be considered important markers when assessing the balance of the intestinal microbiota in the duck? This also need further test.
ACKNOWLEDGMENT
This project was supported by the National Key Research and Development Plan Program of China (2016YFD0500500, 2018YFD0501504), and National Natural Science Foundation of China (31172194).
Contributor Information
Chunhong Zhu, Email: zhuch_1304428@126.com.
Huifang Li, Email: lhfxf_002@aliyun.com.
REFERENCES
- Berlec A., Perše M., Ravnikar M., Lunder M., Erman A., Cerar A., Štrukelj B. Dextran sulphate sodium colitis in C57BL/6 J mice is alleviated by Lactococcus lactis and worsened by the neutralization of Tumor necrosis Factor α. Int. Immunopharmacol. 2017;43:219–226. doi: 10.1016/j.intimp.2016.12.027. [DOI] [PubMed] [Google Scholar]
- Best A.A., Porter A.L., Fraley S.M., Fraley G.S. Characterization of gut microbiome dynamics in developing pekin ducks and impact of management system. Front. Microbiol. 2017;7 doi: 10.3389/fmicb.2016.02125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhogoju S., Nahashon S., Wang X., Darris C., Kilonzo-Nthenge A. A comparative analysis of microbial profile of Guinea fowl and chicken using metagenomic approach. PLoS ONE. 2018;13 doi: 10.1371/journal.pone.0191029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Peña A.G., Goodrich J.K., Gordon J.I., Huttley G.A., Kelley S.T., Knights D., Koenig J.E., Ley R.E., Lozupone C.A., McDonald D., Muegge B.D., Pirrung M., Reeder J., Sevinsky J.R., Turnbaugh P.J., Walters W.A., Widmann J., Yatsunenko T., Zaneveld J., Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.L., Takeda K., Sundrud M.S. Emerging roles of bile acids in mucosal immunity and inflammation. Mucosal Immunol. 2019;12:851–881. doi: 10.1038/s41385-019-0162-4. [DOI] [PubMed] [Google Scholar]
- Diaz Heijtz R., Wang S., Anuar F., Qian Y., Björkholm B., Samuelsson A., Hibberd M.L., Forssberg H., Pettersson S. Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. 2011;108:3047–3052. doi: 10.1073/pnas.1010529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J., Dai R., Yang L., He C., Xu K., Liu S., Zhao W., Xiao L., Luo L., Zhang Y., Meng H. Inheritance and establishment of gut microbiota in chickens. Front. Microbiol. 2017;8 doi: 10.3389/fmicb.2017.01967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- Edgar R.C., Haas B.J., Clemente J.C., Quince C., Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Aidy S., van den Bogert B., Kleerebezem M. The small intestine microbiota, nutritional modulation and relevance for health. Curr. Opin. Biotechnol. 2015;32:14–20. doi: 10.1016/j.copbio.2014.09.005. [DOI] [PubMed] [Google Scholar]
- Ellegaard K.M., Engel P. Genomic diversity landscape of the honey bee gut microbiota. Nat Commun. 2019;10:446. doi: 10.1038/s41467-019-08303-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W., Zhang J., Jakovlić I., Xiong F., Wu S., Zou H., Li W., Li M., Wang G. Gut segments outweigh the diet in shaping the intestinal microbiota composition in grass carp Ctenopharyngodon idellus. AMB Expr. 2019;9:44. doi: 10.1186/s13568-019-0770-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong L., Wang B., Mei X., Xu H., Qin Y., Li W., Zhou Y. Effects of three probiotic Bacillus on growth performance, digestive enzyme activities, antioxidative capacity, serum immunity, and biochemical parameters in broilers. Anim. Sci. J. 2018;89:1561–1571. doi: 10.1111/asj.13089. [DOI] [PubMed] [Google Scholar]
- Guevarra R.B., Lee J.H., Lee S.H., Seok M.J., Kim D.W., Kang B.N., Johnson T.J., Isaacson R.E., Kim H.B. Piglet gut microbial shifts early in life: causes and effects. J. Ani. Sci. Biotechnol. 2019;10:1. doi: 10.1186/s40104-018-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B.J., Gevers D., Earl A.M., Feldgarden M., Ward D.V., Giannoukos G., Ciulla D., Tabbaa D., Highlander S.K., Sodergren E., Methé B., DeSantis T.Z., Human Microbiome Consortium. Petrosino J.F., Knight R., Birren B.W. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinohe T., Pang I.K., Kumamoto Y., Peaper D.R., Ho J.H., Murray T.S., Iwasaki A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I.I., Littman D.R. Modulation of immune homeostasis by commensal bacteria. Curr. Opin. Microbiol. 2011;14:106–114. doi: 10.1016/j.mib.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.B., Isaacson R.E. The pig gut microbial diversity: understanding the pig gut microbial ecology through the next generation high throughput sequencing. Vet. Microbiol. 2015;177:242–251. doi: 10.1016/j.vetmic.2015.03.014. [DOI] [PubMed] [Google Scholar]
- Kumar S., Pattanaik A.K., Sharma S., Gupta R., Jadhav S.E., Dutta N. Comparative assessment of canine-origin Lactobacillus johnsonii CPN23 and dairy-origin Lactobacillus acidophillus NCDC 15 for nutrient digestibility, faecal fermentative metabolites and selected gut health indices in dogs. J. Nutr. Sci. 2017;6:e38. doi: 10.1017/jns.2017.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuss S.K., Best G.T., Etheredge C.A., Pruijssers A.J., Frierson J.M., Hooper L.V., Dermody T.S., Pfeiffer J.K. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science. 2011;334:249–252. doi: 10.1126/science.1211057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar V., Ditu L.M., Pircalabioru G.G., Picu A., Petcu L., Cucu N., Chifiriuc M.C. Gut microbiota, host organism, and diet trialogue in diabetes and obesity. Front. Nutr. 2019;6:21. doi: 10.3389/fnut.2019.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.P., Zhao X.J., Wang J.Y. Synergy of Astragalus polysaccharides and probiotics (Lactobacillus and Bacillus cereus) on immunity and intestinal microbiota in chicks. Poult. Sci. 2009;88:519–525. doi: 10.3382/ps.2008-00365. [DOI] [PubMed] [Google Scholar]
- Lim S., Cho S., Caetano-Anolles K., Jeong S.G., Oh M.H., Park B.Y., Kim H.J., Cho S., Choi S.H., Ryu S., Lee J.H., Kim H., Ham J.S. Developmental dynamic analysis of the excreted microbiome of chickens using Next-Generation sequencing. J. Mol. Microbiol. Biotechnol. 2015;25:262–268. doi: 10.1159/000430865. [DOI] [PubMed] [Google Scholar]
- Mao S., Zhang M., Liu J., Zhu W. Characterising the bacterial microbiota across the gastrointestinal tracts of dairy cattle: membership and potential function. Sci. Rep. 2015;5 doi: 10.1038/srep16116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley B.B., Lillehoj H.S., Kogut M.H., Kim W.K., Maurer J.J., Pedroso A., Lee M.D., Collett S.R., Johnson T.J., Cox N.A. The chicken gastrointestinal microbiome. FEMS Microbiol. Lett. 2014;360:100–112. doi: 10.1111/1574-6968.12608. [DOI] [PubMed] [Google Scholar]
- Oh J.K., Pajarillo E.A.B., Chae J.P., Kim I.H., Yang D.S., Kang D.K. The effects of Bacillus subtilis CSL2 on the composition and functional diversity of the faecal microbiota of broiler chickens challenged with Salmonella gallinarum. J. Anim. Sci. Biotechnol. 2017;8:1. doi: 10.1186/s40104-016-0130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F.O. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucl. Acids Res. 2012;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyon D., Tsai S.Q., Khayter C., Foden J.A., Sander J.D., Joung J.K. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin N.R., Lee J.C., Lee H.Y., Kim M.S., Whon T.W., Lee M.S., Bae J.W. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut. 2014;63:727–735. doi: 10.1136/gutjnl-2012-303839. [DOI] [PubMed] [Google Scholar]
- Salonen A., de Vos W.M. Impact of diet on human intestinal microbiota and health. Annu. Rev. Food Sci. Technol. 2014;5:239–262. doi: 10.1146/annurev-food-030212-182554. [DOI] [PubMed] [Google Scholar]
- Shang Y., Kumar S., Oakley B., Kim W.K. Chicken gut microbiota: Importance and detection technology. Front. Vet. Sci. 2018;5:254. doi: 10.3389/fvets.2018.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanca A., Fraumene C., Manghina V., Palomba A., Abbondio M., Deligios M., Pagnozzi D., Addis M.F., Uzzau S. Diversity and functions of the sheep faecal microbiota: a multi-omic characterization. Microb. Biotechnol. 2017;10:541–554. doi: 10.1111/1751-7915.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J.N., Zeng Z.G., Wang H.N., Yang T., Zhang P.J., Li Y.L., Zhang A.Y., Fan W.Q., Zhang Y., Yang X., Zhao S.J., Tian G.B., Zou L.K. An effective method for isolation of DNA from pig faeces and comparison of five different methods. J. Microbiol. Methods. 2008;75:432–436. doi: 10.1016/j.mimet.2008.07.014. [DOI] [PubMed] [Google Scholar]
- Vatsos I.N. Standardizing the microbiota of fish used in research. Lab Anim. 2017;51:353–364. doi: 10.1177/0023677216678825. [DOI] [PubMed] [Google Scholar]
- Waite D.W., Taylor M.W. Exploring the avian gut microbiota: current trends and future directions. Front. Microbiol. 2015;6:673. doi: 10.3389/fmicb.2015.00673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Ni X., Qing X., Zeng D., Luo M., Liu L., Li G., Pan K., Jing B. Live Probiotic Lactobacillus johnsonii BS15 promotes growth performance and lowers fat deposition by improving lipid metabolism, intestinal development, and gut microflora in broilers. Front. Microbiol. 2017;8 doi: 10.3389/fmicb.2017.01073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.R., Nagarajan N., Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput. Biol. 2009;5 doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeruva L., Spencer N.E., Saraf M.K., Hennings L., Bowlin A.K., Cleves M.A., Mercer K., Chintapalli S.V., Shankar K., Rank R.G., Badger T.M., Ronis M.J. Formula diet alters small intestine morphology, microbial abundance and reduces VE-cadherin and IL-10 expression in neonatal porcine model. BMC Gastroenterol. 2016;16:40. doi: 10.1186/s12876-016-0456-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Yu M., Yang Y., Mu C., Su Y., Zhu W. Effect of early antibiotic administration on cecal bacterial communities and their metabolic profiles in pigs fed diets with different protein levels. Anaerobe. 2016;42:188–196. doi: 10.1016/j.anaerobe.2016.10.016. [DOI] [PubMed] [Google Scholar]
- Zhang H.L., Li W.S., Xu D.N., Zheng W.W., Liu Y., Chen J., Qiu Z.B., Dorfman R.G., Zhang J., Liu J. Mucosa-reparing and microbiota-balancing therapeutic effect of Bacillus subtilis alleviates dextrate sulfate sodium-induced ulcerative colitis in mice. Exp Ther Med. 2016;12:2554–2562. doi: 10.3892/etm.2016.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
Uncited References
- Pourabedin M., Zhao X. Prebiotics and gut microbiota in chickens. FEMS Microbiol. Lett. 2015;362 doi: 10.1093/femsle/fnv122. [DOI] [PubMed] [Google Scholar]