Abstract
Applying broiler litter containing extended-spectrum beta-lactamase (ESBL)–producing Escherichia coli (E. coli) to arable land poses a potential risk for humans to get colonized by contact with contaminated soil or vegetables. Therefore, an inactivation of these bacteria before land application of litter is crucial. We performed 2 short-term litter storage trials (one in summer and winter, respectively), each covering a time span of 5 D to investigate the effectiveness of this method for inactivation of ESBL-producing E. coli in chicken litter. Surface and deep litter samples were taken from a stacked, ESBL-positive chicken litter heap in triplicates in close sampling intervals at the beginning and daily for the last 3 D of the experiments. Samples were analyzed quantitatively and qualitatively for ESBL-producing E. coli, total E. coli, and enterococci. Selected isolates were further characterized by whole-genome sequencing (WGS). In the depth of the heap ESBL-producing E. coli were detected quantitatively until 72 h and qualitatively until the end of the trial in winter. In summer detection was possible quantitatively up to 36 h and qualitatively until 72 h. For surface litter samples a qualitative detection of ESBL-producing E. coli was possible in all samples taken in both trials. In the deep samples a significant decrease in the bacterial counts of over 2 Log10 was observed for total E. coli in the winter and for total E. coli and enterococci in the summer. Genetic differences of the isolates analyzed by WGS did not correlate with survival advantage. In conclusion, short-term storage of chicken litter stacked in heaps is a useful tool for the reduction of bacterial counts including ESBL-producing E. coli. However, incomplete inactivation was observed at the surface of the heap and at low ambient temperatures. Therefore, an extension of the storage period in winter as well as turning of the heap to provide aerobic composting conditions should be considered if working and storage capacities are available on the farms.
Key words: antibiotic resistance, ESBL, E. coli, broiler litter, environment
Introduction
Extended-spectrum beta-lactamases (ESBL) are enzymes occurring in Enterobacteriaceae. Their ability to hydrolyze the β-lactam ring of a variety of β-lactam antibiotics including extended-spectrum cephalosporins of the third and fourth generation leads to an inactivation of antibiotic properties. Cephalosporins of the third and fourth generation have a broad-spectrum activity against Gram-positive and Gram-negative bacteria and are often used for the treatment of infections in intensive care units. The emergence of resistance against these drugs limits therapeutic options (Remschmidt et al., 2017).
ESBL-producing Escherichia coli (E. coli) are commonly found in broiler production with a prevalence of up to 100% in fattening farms (Dierikx et al., 2010, Dierikx et al., 2013, Laube et al., 2013, Blaak et al., 2015, Hering et al., 2016, Daehre et al., 2018). Furthermore, ESBL-producing E. coli have been detected in the vicinity of broiler barns, and an airborne and waterborne dissemination have been described (Laube et al., 2014, Blaak et al., 2015). In Germany, 600 million broiler chickens were slaughtered in 2017 (Statistisches Bundesamt, 2017) contributing to the 1.1 million metric tons of poultry litter that are spread to arable land in Germany annually (Statistisches Bundesamt, 2016). This presents a possible important emission source of resistant bacteria from the barns to the environment.
Blaak et al. (2015) reported that ESBL-producing E. coli were found in the soil at a distance of 1-5 m of litter storage areas with up to 2.0 × 104 cfu/kg. Additionally, it was shown that ESBL-producing E. coli can be transferred from animal husbandry to soil and are able to survive on the fields for at least 1 y (Hartmann et al., 2012). A cross-transmission of ESBL-producing E. coli and mobile genetic elements encoding for the production of ESBL between animals, including chickens, humans, and the environment is hypothesized (Leverstein-van Hall et al., 2011, Huijbers et al., 2014). The spread of litter containing ESBL-producing Enterobacteriaceae to the environment poses a potential risk for humans to be colonized with these bacteria after contact with contaminated soils or via contaminated vegetables. Hence, inactivation of these resistant bacteria before land application is crucial.
Storing the litter in piles after removal from the barns could be a useful and cost efficient tool for the reduction of resistant bacteria in litter. Studies that investigated the reduction of nonresistant E. coli in chicken litter by storage (anaerobic conditions) and composting (active aeration) under practical conditions were performed previously (Erickson et al., 2010, Wilkinson et al., 2011). Considering practicability and economic sustainability short-term storage of litter presents the most advantageous method of litter treatment. The decline of ESBL-producing E. coli in chicken litter under field conditions has not been investigated so far. For a more detailed assessment of bacterial inactivation in short-term chicken litter storage, concentrations of nonresistant E. coli and enterococci were additionally monitored in this study. Enterococci are approved gram-positive indicator microorganisms present in feces and have a higher tenacity compared to E. coli. Two short-term storage trials each covering a time span of 5 D were performed. One trial was performed in the summer (summer trial) and one in the winter (winter trial) to explore climatic influences on the decline of these bacteria.
Materials and methods
Experimental Design
An initial screening of 40 different barns of a large broiler farm in Germany was performed to select barns with high quantities of ESBL-producing E. coli in the litter. Screening consisted of one boot swab and one composite litter sample per barn and was done for both the summer trial and the winter trial. The winter trial was carried out in early March, and the summer trial in early June. The time span between the screening and the litter storage trial was 2 wk for the winter trial and 1 wk for the summer trial.
In all barns, 1.5 kg/m2 of wood pellets were used as bedding material. The litter (approximately 15 metric tons for each trial) was removed from the barns with a front-end loader and piled up on a concrete surface behind the barn directly after the chickens were housed out.
Storing the litter behind the barns for several days is not unusual in broiler production. Although on the farm where the trials were performed, the litter is removed as fast as possible if working capacities are available. Under suitable conditions, it is used directly for fertilization or otherwise transported to further storage areas.
The first samples were taken immediately after the litter heap was stacked. For both trials, the litter heaps were sampled at 6 points in time: 12 h, 24 h, 36 h, 48 h, 72 h, and 96 h after storage begin. We expected a faster reduction of the bacterial counts in the summer because of the higher ambient temperatures. Hence, in the summer trial, additional samplings were performed at 1 h, 3 h, and 6 h after the begin of storage. At each point in time, 3 surface and 3 deep samples were taken from the litter heap.
For the surface litter samples, approximately 50 g of litter from the heap's surface were collected in sterile 120 mL specimen containers (VWR, Radnor, PA)
For the deep litter samples, custom-built steel sample containers were used. These containers are cylindrical, 9.9 cm long, have a diameter of 4.4 cm, and drill holes with a diameter of 7 mm, ensuring the same environmental conditions in the sampling container and the surrounding litter heap. These sterilized containers were filled with litter from the heap and were placed in the litter heap at a depth of 50 to 55 cm at the start of the experiment. Wires were attached to the containers allowing quick retraction and sample collection at each point in time.
The ALMEMO 2490 device (AHLBORN, ZA9020-FS and FH A696-GF1 Holzkirchen, Germany) was used to record temperature and moisture at each sampling spot immediately after sampling.
The weather data for the trial periods were obtained from the closest weather station located approximately 20 km from the sampling site (Archive of the German Meteorological Office)
pH Value Analysis
The pH value was measured for all litter samples. Samples were diluted with purified water at a ratio of 1:10 and homogenized for 30 s with a vortex mixer. The pH value was measured with the handheld measurement instrument AL10 (AQUALYTIC, Dortmund, Germany).
Microbiological Analyses
All boot swabs and litter samples from the screenings were analyzed quantitatively and qualitatively for ESBL-producing E. coli. Litter samples from the litter storage trials were analyzed quantitatively and qualitatively for total E. coli and ESBL-producing E. coli. Additionally, enterococci were analyzed quantitatively.
All litter samples were mixed with Luria/Miller-broth (LB) (Roth, Karlsruhe, Germany) in stomacher bags at a ratio of 1:10. Boot swabs were put in stomacher bags, and 200 ml of LB medium was added. The samples were homogenized using a Stomacher 400 Circulator (Seward Limited, West Sussex, UK) for 2 min at 200 rpm.
For the quantitative analyses aliquots of the suspensions were taken, and triplicates of 100 μL were streaked on specific agar plates after serial dilution. For E. coli, MacConkey agar No. 3 (Oxoid, Wesel, Germany) was used. To detect ESBL-producing E. coli, 1 mg/L cefotaxime (AppliChem, Darmstadt, Germany) was added as suggested by the EFSA (2011). For enterococci, we used Bile Aesculin Azide agar (Merck, Darmstadt, Germany). We did not set a minimum number of cfu per plate for the evaluation of the undiluted samples, resulting in a quantitative detection limit of 3.3 × 101 cfu/g of litter.
For qualitative testing, the homogenized samples were incubated in LB medium for 20 to 24 h at 37°C. Subsequently, 10 μL were streaked on MacConkey agar with and without the addition of cefotaxime, respectively, with an inoculation loop.
MALDI-TOF Mass Spectrometry (MALDI Microflex LT and Biotyper database, Bruker Daltonics, Bremen, Germany) was used for species confirmation of colonies which were phenotypically suspected to be E. coli or enterococci.
Real-Time PCR and Sanger Sequencing
Real-time qPCR as described by Roschanski et al. (2014) was used to detect the most important beta-lactamase genes blaSHV, blaTEM, blaCTX-M, and the CIT-type AmpC blaCMY-2 in isolates of all samples.
For both trials, the ESBL-gene of 8 isolates were sequenced by Sanger sequencing to identify the present ESBL-variants. Two isolates from the litter and one isolate from a surface and deep litter sample for day 1, 3, and 5 were chosen for sequencing for each trial, respectively. All isolates that showed a blaTEM resistance gene in addition to the predominant resistance gene were also chosen for sequencing.
The DNA was isolated, and PCR was performed as published previously by Projahn et al. (2017). The purified PCR products were sent to LGC Genomics (Berlin, Germany) who provided the sequences. Nucleotide sequences were analyzed using DNASTAR Lasergene (Madison, WI) and compared with the reference sequences of GenBank (https://www.ncbi.nlm.nih.gov/genbank/) according to the accession numbers of the lahey database (https://www.lahey.org/studies/).
Phylotyping
All isolates were analyzed for their phylogenetic group as published by Clermont et al. (2013) with modified PCR conditions according to Projahn et al. (2017). Isolates that could not be assigned to a phylogroup because of unspecific band patterns were declared as a combined phylogroup.
Whole Genome Sequencing
Forty-four ESBL-producing E. coli isolates were selected for whole genome sequencing and recultivated on Brain-Heart-Infusion agar (Roth, Karlsruhe, Germany). DNA was isolated using the Qiagen Blood and Tissue Kit (Qiagen, Hilden, Germany), and sequencing libraries were prepared using the Nextera XT protocol with modifications (Baym et al., 2015, Steglich et al., 2018). The libraries were sequenced on a NextSeq machine with a NextSeq 500/550 mid output v2 kit (Illumina, San Diego) to ≥ 50-fold sequencing coverage.
Short-read sequencing data were uploaded to the online platform Enterobase (https://enterobase.warwick.ac.uk), where they were assembled. Resulting contigs were quality-controlled and subjected to classification by 7-gene multilocus sequence typing (MLST), core-genome multilocus sequence typing (cgMLST), and cgMLST-based hierarchical clustering. Clusters at the level HC1100 (Hierarchical Cluster 1,100, that is, chains of genomes differing pairwise by maximally 1,100 cgMLST alleles) represent major genetic populations within the species E. coli, largely congruent with sequence-type complexes based on legacy 7-gene MLST (Zhou et al., 2019). In addition, EnteroBase used genome sequence information to predict phylogroups according to Clermont typing (Zhou et al., 2019) based on algorithms by Beghain et al. (2018) and Waters et al. (2018).
Genome sequences were screened for antibiotic resistance genes by using the tools Resfinder (Zankari et al., 2012), AMRFinder (Feldgarden et al., 2019), and CARD (Jia et al., 2017), as implemented in ABRicate (https://github.com/tseemann/abricate). Genome sequencing data were submitted to the European Nucleotide Archive (https://www.ebi.ac.uk/ena) under study number PRJEB34161.
Statistical Analysis
The software SPSS, version 25, (SPSS, Inc., Chicago, IL) was used for statistical analysis. The data on microbial counts had no normal distribution. We used log transformation to achieve log normal distribution, and geometric means were calculated as proposed by Bland and Altman (1996). The upper and lower 95% confidence intervals were calculated. The winter trial and summer trial were analyzed separately.
Results
Environmental Conditions During the Litter Storage Trials Conducted in Summer and Winter
The relevant weather data for the period of both trials are summarized in Table 1.
Table 1.
Environmental conditions during the short-term litter storage trials as provided by the weather station closest to the trial site (German meteorological office).
| Trial | Day | Daily air temperature (°C) |
RH (%)1 | Sunshine (h) | Rainfall (mm) | ||
|---|---|---|---|---|---|---|---|
| Minimum | Maximum | Mean | |||||
| Winter | 1 | −13.1 °C | −2.0 °C | −7.2 °C | 58.8% | 9.5 h | 0.0 mm |
| 2 | −12.8 °C | −0.6 °C | −6.1 °C | 56.5% | 9.4 h | 0.0 mm | |
| 3 | −7.7 °C | 7.4 °C | −0.5 °C | 54.7% | 5.1 h | 1.1 mm | |
| 4 | −3.8 °C | 11.9 °C | 4.4 °C | 74.5% | 7.9 h | 0.0 mm | |
| 5 | −5.5 °C | 5.2 °C | 1.0 °C | 82.6% | 0.9 h | 6.9 mm | |
| Summer | 1 | 13.6 °C | 28.8 °C | 21.6 °C | 75.6% | 10.0 h | 20.4 mm |
| 2 | 13.6 °C | 30.4 °C | 23.5 °C | 69.9% | 12.3 h | 0.0 mm | |
| 3 | 14.9 °C | 27.0 °C | 21.6 °C | 78.8% | 6.5 h | 0.2 mm | |
| 4 | 14.4 °C | 21.5 °C | 19.2 °C | 86.1% | 0.3 h | 0.1 mm | |
| 5 | 13.7 °C | 25.2 °C | 20.5 °C | 76.0% | 5.4 h | 0.0 mm | |
Relative humidity.
Temperature, Moisture, and pH Value of the Litter
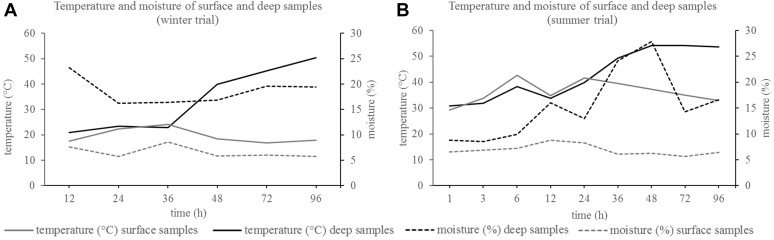
In both trials, the temperature of the litter increased continuously at a depth of 50 to 55 cm compared with the surface of the litter heap. In the winter trial, 50.4°C were reached at the end of the trial period (96 h). In the summer trial, temperatures over 50°C were already reached after 36 h, and the maximum temperature measured was 58.5°C at the end of the trial (96 h). The temperature in the surface samples was lower for both trials ranging from 16.8°C to 24.2°C in the winter trial and 29.3°C to 42.7°C in the summer trial.
In the winter trial, moisture of the deep samples ranged from 16.2 to 23.2%. In the summer trial, moisture of the deep samples increased from approximately 9.0% at the beginning of the experiment to 27.9% at the end of the experiment. Surface samples from both trials showed lower moisture levels with values ranging from 5.8 to 8.6% in the winter trial and 5.9 to 8.8% in the summer trial. Temperature and moisture development for both trials is shown in Figure 1.
Figure 1.
Development of mean sample temperatures and moistures in the winter trial (A) and summer trial (B).
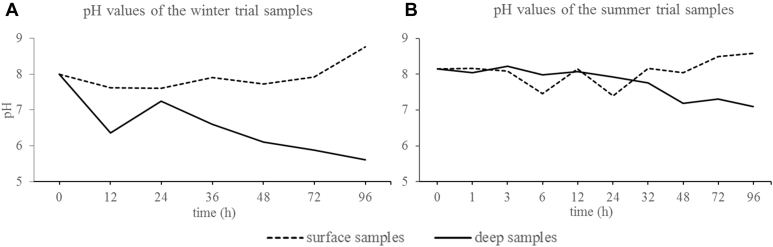
The pH values measured in the chicken litter directly after the removal from the barn was 8.0 in the winter trial and 8.1 in the summer trial.
For both trials, the pH value increased in the surface samples over 5 consecutive day up to a maximum pH value of 8.8 in the winter trial and 8.6 in the summer trial. In the deep samples, the pH value decreased in both trials to a minimum of 5.6 in the winter trial and 6.5 in the summer trial. The mean pH values for both trials are depicted in Figure 2.
Figure 2.
Mean sample pH values in the winter trial (A) and summer trial (B) for each point in time.
Microbiological Status of the Barns in the Initial Screenings
For both trials, the barn with the highest concentration of ESBL-producing E. coli in the litter samples of the initial screenings was chosen for the litter storage trial. The bacterial counts of the investigated microorganisms in the barns are shown in Table 2 for the boot swabs and composite litter samples.
Table 2.
Bacterial counts for the initial screening of the barns in cfu/boot swab and cfu/g of litter.
| Trial | Type of sample | ESBL-producing E. coli in cfu/boot swab or cfu/g | Total E. coli in cfu/boot swab or cfu/g | Enterococci in cfu/boot swab or cfu/g |
|---|---|---|---|---|
| Winter | Boot swab | 1.1 × 106 cfu/boot swab | 2.3 × 107 cfu/boot swab | 2.7 × 108 cfu/boot swab |
| Litter sample | 3.9 × 104 cfu/g | 4 × 105 cfu/g | 2.9 × 107 cfu/g | |
| Summer | Boot swab | 3.6 × 106 cfu/boot swab | 3.6 × 108 cfu/boot swab | 5.9 × 108 cfu/boot swab |
| Litter sample | 6.3 × 105 cfu/g | 1.7 × 107 cfu/g | 7.2 × 107 cfu/g |
Quantitative and Qualitative Detection of ESBL-Producing E. coli in the Litter
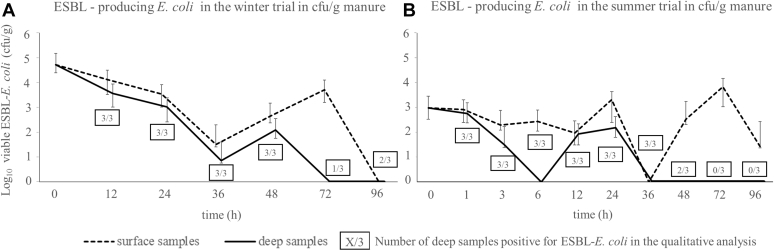
Surface litter samples were firstly taken immediately after the litter was removed from the barns (0 h). The mean number of ESBL-producing E. coli per g of litter was 5.2 × 104 cfu in the winter trial and 9.5 × 102 cfu in the summer trial.
In the winter trial, the mean number of ESBL-producing E. coli was 1.3 × 104 cfu/g of litter at the 12 h sampling point for the surface samples, decreased until 36 h and increased again to 5 × 103 cfu/g after 72 h. At the end of the trial (96 h), ESBL-producing E. coli were not quantitatively detectable.
The deep litter samples in the winter trial showed an ESBL-producing E. coli concentration of 3.6 × 103 cfu/g at 12 h and gradually decreased in the following measurements. For the 72 h and 96 h samples, ESBL-producing E. coli could not be detected quantitatively. A qualitative detection was possible for 33% (n = 1/3) of the samples after 72 h and for 66% (n = 2/3) of the samples after 96 h.
In the summer trial, the number of ESBL–producing E. coli in the surface samples decreased slightly in the first 12 hours (mean at 1 h = 2 × 102 cfu/g, mean at 12 h = 9.3 × 101 cfu/g). At 24 h and 72 h, the concentration of ESBL-producing E. coli was higher than the initial count with up to 6.7 × 103 cfu/g. At 36 h, the number of ESBL-producing E. coli was below the detection limit.
ESBL-producing E. coli were not quantitatively detectable in the deep samples of the summer trial for the 6 h point in time. Additionally, after 36 h for the last 4 points in time (36 h, 48 h, 72 h, and 96 h), ESBL-producing E. coli were constantly under the detection limit. The qualitative analysis was only positive for 67% (n = 2/3) of the samples after 48 h and for 0% (n = 0/3) after 72 h and 96 h, respectively.
A qualitative detection of ESBL-producing E. coli was possible for all surface samples in both trials. The data on ESBL-producing E. coli are shown in Figure 3.
Figure 3.
Results of the quantitative and qualitative analysis of ESBL-producing E. coli in cfu/g of litter for the winter trial samples (A) and the summer trial samples (B). The geometric mean of 3 samples is shown for each point in time. The error bars indicate the upper and lower 95% confidence intervals. The graphs were shifted to improve the visibility of the error bars. ESBL, extended-spectrum beta-lactamase.
Quantitative and Qualitative Detection of Total E. coli in the Litter
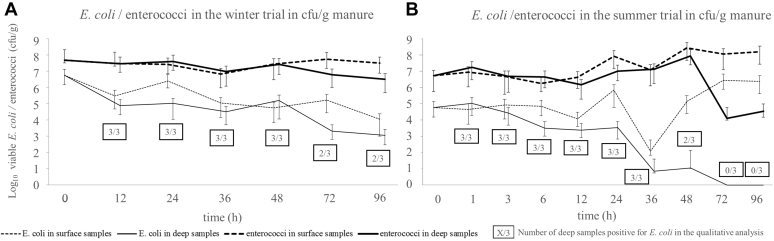
In the litter samples taken directly after the litter was removed from the barns (0 h), the mean number of E. coli per g of litter was 5.6 × 106 cfu for the winter trial and 5.8 × 104 cfu for the summer trial.
In the winter trial, the number of E. coli dropped in both, the surface and the deep samples. After 96 h, the mean amount of E. coli was 1.1 × 104 cfu/g of litter in the surface and 1.1 × 103 cfu/g of litter in the deep samples. Qualitative E. coli detection was possible in 66% (n = 2/3) of the deep samples at 72 h and 96 h.
In the summer trial, on the other hand, the mean number of E. coli in surface litter samples increased from 5.8 × 104 cfu/g of litter at 0 h to a maximum of 2.8 × 106 cfu/g at 72 h and 2.4 × 106 cfu/g at 96 h. Deep litter samples showed a constant decrease in E. coli concentrations with a drop below the detection limit at 72 h. A qualitative detection was possible for 67% (n = 2/3) of samples at 48 h and for 0% (n = 0/3) of samples at 72 h and 96 h. The data for total E. coli are shown in Figure 4.
Figure 4.
Results of the quantitative and qualitative analysis of total E. coli in cfu/g of litter, and the quantitative analysis of enterococci in cfu/g of litter for the winter trial samples (A) and the summer trial samples (B). The geometric mean of 3 samples is shown for each point in time. The error bars indicate the upper and lower 95% confidence intervals. The graphs were shifted to improve the visibility of the error bars.
Quantitative Detection of Enterococci in the Litter
The number of enterococci for the 0 h point in time was 4.8 × 107 cfu/g of litter for the winter trial and 5.5 × 106 cfu/g for the summer trial, respectively.
In the winter trial, the quantity of enterococci was comparatively stable for both, surface and deep samples, ranging from 3.2 × 106 to 5.6 × 107 cfu/g in all samples taken.
In the summer trial, the quantity of enterococci increased in surface samples in the sampling period, reaching a maximum of 2.7 × 108 cfu/g after 48 h. The quantity at the end of the sampling period (96 h) was 1.6 × 108 cfu/g of litter.
In contrast, in the summer trial, deep litter samples showed a significant decrease in enterococci concentration. A mean of 1.3 × 104 and 3.4 × 104 cfu/g were detected after 72 h and 96 h, respectively. The data for enterococci are shown in Figure 4.
Molecular Characterization of ESBL-Producing E. coli
Phylogroups and Sequence Types
In the winter trial, the phylogenetic group was determined for 48 ESBL-producing E. coli isolates using classical gel-based PCR. Forty-seven isolates were allocated to phylogroup F, and one isolate was allocated to phylogroup A/C.
In the summer trial, the number of ESBL-producing E. coli isolates available for phylogenetic analysis was 54 of which 17 isolates belonged to phylogroup B1. One isolate belonged to the groups A and F, respectively, and 35 isolates were allocated to the combined phylogroup D/E.
In Table 3, a comparison of the Clermont phylogroups determined by PCR and the by whole genome sequenced isolates is shown. It also provides information on the 7-gene MLST, cgMLST, and HC1100 clustering from the Enterobase analyses and the predicted O and H antigens for all isolates.
Table 3.
Phylogroups determined with gel-based PCR and phylogroups and sequence types determined via Enterobase.
| Sample ID | Trial | Timepoint (h) | Sampling site | Phylogroup FU1 | Phylogroup Enterobase | ST 7 gene MLST2 | ST cGMLST3 | HC1100 cgST4 | H-Antigen | O-Antigen |
|---|---|---|---|---|---|---|---|---|---|---|
| 7-1EP06 | Summer | 0 h | Barn | D/E | D | 2,309 | 86,589 | 5,033 | H6 | O15 |
| 7-1EP05 | Summer | 0 h | Barn | D/E | D | 2,309 | 86,626 | 5,033 | H6 | O15 |
| 7-1MP03 | Summer | 0 h | Surface | D/E | D | 2,309 | 86,616 | 5,033 | H6 | O15 |
| 7-1MP05 | Summer | 1 h | Depth | D/E | D | 2,309 | 86,618 | 5,033 | H6 | O15 |
| 7-1MP11 | Summer | 3 h | Depth | B1 | B1 | 162 | 86,592 | 138 | H10 | O9 |
| 7-1MP10 | Summer | 3 h | Surface | D/E | D | 2,309 | 86,619 | 5,033 | H6 | O15 |
| 7-1MP16 | Summer | 6 h | Surface | B1 | B1 | 1,304 | 86,593 | 152 | H7 | O91 |
| 7-1MP17 | Summer | 6 h | Depth | D/E | D | 2,309 | 86,608 | 5,033 | H6 | O15 |
| 7-1MP23 | Summer | 12 h | Depth | B1 | D | 2,309 | 86,590 | 5,033 | H6 | O15 |
| 7-1MP22 | Summer | 12 h | Surface | B1 | B1 | 162 | 86,622 | 138 | H10 | O88 |
| 7-1MP31 | Summer | 24 h | Depth | D/E | D | 2,309 | 86,580 | 5,033 | H6 | O15 |
| 7-1MP39 | Summer | 36 h | Depth | D/E | D | 2,309 | 86,577 | 5,033 | H6 | O15 |
| 7-1MP34 | Summer | 36 h | Surface | B1 | B1 | 1,304 | 86,593 | 152 | H7 | O91 |
| 7-1MP38 | Summer | 36 h | Surface | D/E | D | 2,309 | 86,614 | 5,033 | H6 | O15 |
| 7-1MP35 | Summer | 36 h | Depth | D/E | D | 2,309 | 86,620 | 5,033 | H6 | O15 |
| 7-1MP37 | Summer | 36 h | Depth | D/E | D | 2,309 | 86,617 | 5,033 | H6 | O15 |
| 7-1MP41 | Summer | 48 h | Depth | D/E | D | 2,309 | 86,580 | 5,033 | H6 | O15 |
| 7-1MP40 | Summer | 48 h | Surface | D/E | D | 2,309 | 86,601 | 5,033 | H6 | O15 |
| 7-1MP42 | Summer | 48 h | Surface | D/E | D | 2,309 | 86,623 | 5,033 | H6 | O15 |
| 7-1MP43 | Summer | 48 h | Depth | D/E | D | 2,309 | 86,623 | 5,033 | H6 | O15 |
| 7-1MP44 | Summer | 48 h | Surface | D/E | D | 2,309 | 86,580 | 5,033 | H6 | O15 |
| 7-1MP46 | Summer | 72 h | Surface | B1 | B1 | 162 | 86,615 | 138 | H10 | O88 |
| 7-1MP48 | Summer | 72 h | Surface | D/E | D | 2,309 | 86,625 | 5,033 | H6 | O15 |
| 7-1MP56 | Summer | 96 h | Surface | B1 | B1 | 1,304 | 86,593 | 152 | H7 | O91 |
| 7-1MP54 | Summer | 96 h | Surface | D/E | D | 2,309 | 86,624 | 5,033 | H6 | O15 |
| 5-2EP02 | Winter | 0 h | Barn | F | F | 117 | 86,591 | 50 | H4 | O8 |
| 5-2EP01 | Winter | 0 h | Barn | F | F | 117 | 86,578 | 50 | H4 | O8 |
| 5-2MP32 | Winter | 24 h | Surface | A/C | A | 10 | 86,613 | 13 | H48 | O12 |
| 5-2MP33 | Winter | 24 h | Depth | F | F | 117 | 86,579 | 50 | H4 | O8 |
| 5-2MP35 | Winter | 36 h | Depth | F | F | 117 | 86,599 | 50 | H4 | O8 |
| 5-2MP34 | Winter | 36 h | Surface | F | F | 117 | 86,653 | 50 | H4 | O8 |
| 5-2MP41 | Winter | 48 h | Depth | F | F | 117 | 86,647 | 50 | H4 | O8 |
| 5-2MP43 | Winter | 48 h | Depth | F | F | 117 | 86,600 | 50 | H4 | O8 |
| 5-2MP44 | Winter | 48 h | Surface | F | F | 117 | 86,621 | 50 | H4 | O8 |
| 5-2MP42 | Winter | 48 h | Surface | F | F | 117 | 86,632 | 50 | H4 | O8 |
| 5-2MP49 | Winter | 72 h | Depth | F | F | 117 | 86,578 | 50 | H4 | O8 |
| 5-2MP50 | Winter | 72 h | Surface | F | F | 117 | 86,578 | 50 | H4 | O8 |
| 5-2MP48 | Winter | 72 h | Surface | F | F | 117 | 86,578 | 50 | H4 | O8 |
| 5-2MP46 | Winter | 72 h | Surface | F | F | 117 | 86,578 | 50 | H4 | O8 |
| 5-2MP55 | Winter | 96 h | Depth | F | F | 117 | 86,578 | 50 | H4 | O8 |
| 5-2MP54 | Winter | 96 h | Surface | F | F | 117 | 86,627 | 50 | H4 | O8 |
| 5-2MP57 | Winter | 96 h | Depth | F | F | 117 | 86,579 | 50 | H4 | O8 |
| 5-2MP56 | Winter | 96 h | Surface | F | F | 117 | 86,578 | 50 | H4 | O8 |
| 5-2MP52 | Winter | 96 h | Surface | F | F | 117 | 86,612 | 50 | H4 | O8 |
Freie Universität Berlin.
Sequence type 7 gene multilocus sequence type.
Sequence type core genome multilocus sequence type.
Hierarchical cluster 1,100 core genome sequence type.
Resistance Genes
Real time qPCR revealed that all isolates from the winter trial (n = 48) harbored a resistance gene belonging to the blaSHV gene family. The resistance gene was sequenced in 8 isolates, identifying it as blaSHV-12 in all chosen isolates. In the summer trial, all isolates (n = 54) harbored a resistance gene belonging to the blaCTX-M gene family. All 8 blaCTX-M genes sequenced were identified as blaCTX-M-1.
One winter trial isolate and 4 summer trial isolates showed an additional blaTEM gene. All 5 blaTEM genes were identified as broad spectrum beta-lactamase resistance gene blaTEM-1.
For all genome-sequenced isolates from the winter trial (n = 19), genome sequencing confirmed the presence of blaSHV-12. In addition, genome sequencing detected the plasmid-mediated quinolone-resistance determinant qnrS1 and mdfA genes in all 19 isolates, and the broad-spectrum beta-lactamase resistance gene blaTEM-1 and the oxytetracycline resistance determinant Tet 34 in one isolate.
In the summer trial, the ESBL-resistance gene blaCTX-M-1 was confirmed or found in all sequenced isolates (n = 25). In addition, the mdfA gene was detected in all isolates, and Tet 34 was detected in 84% (n = 21/25) of the isolates. blaTEM-1 was detected in 4 isolates and the sulII gene in 3 isolates.
Genome-Based Phylogeny
Phylogenetic trees were calculated in Enterobase and can be found in the Supplemental Figures 1 and 2.
While there are genomic differences between isolates from the summer and winter experiments, isolates within each experiment showed little variation with the summer experiment appearing to be slightly more diverse.
Within the 2 main clusters (one for each season), the different sampling sites are distributed equally as well as the time points for sampling. The genetic differences between the 2 experiments is also reflected in the distribution of resistance genes.
Discussion
The aim of our study was to evaluate whether short-term litter storage provides sufficient inactivation of ESBL-producing E. coli present in chicken litter under field conditions in winter and summer.
The most important findings of the study concerning the amount of cultivable ESBL-producing E. coli were that regardless of the season, the inactivation occurs faster in a depth of about 50 cm compared with the surface of the litter pile. Additionally, a seasonal influence concerning the required time span and the effectiveness of the inactivation of ESBL-producing E. coli was shown. Extended-spectrum beta-lactamases–producing E. coli were not detected quantitatively in the deep samples in the winter trial after 72 h and already after 36 h in the summer trial.
Initial concentrations of nonresistant E. coli in the chicken litter were significantly higher than ESBL-producing E. coli for both trials. The proportion of the ESBL-producing subpopulation on the total amount of E. coli was 0.94% in the winter trial and 1.64% in the summer trial for the samples taken at storage begin. A similar proportion of 1.1% ESBL-producing isolates was recently reported by Friese et al. (2019) for turkey-rearing flocks. This is a possible explanation for the extended time in which nonresistant E. coli were detected in the litter compared with the ESBL-producing subpopulation. In the study performed by Erickson et al. (2010), nonresistant E. coli naturally occurring in the litter were not detected quantitatively and qualitatively in surface and deep samples after 4 D in static piles of chicken litter in summer, fall, and winter.
Abiotic Factors Influencing Microbial Counts in the Litter Storage Trials
Litter piles are microbiologically highly heterogeneous and local conditions are influenced by a variety of biological, physical, and chemical factors. In our study, we explored the influence of temperature, pH value, and moisture content of the chicken litter on the bacterial cell count.
In the study performed by Erickson et al. (2010) in the USA, temperatures within static piles of chicken litter were measured at different intervals. The highest mean temperatures reported in a depth of 30 cm from the heaps surface were 54.4°C in the summer after 4 D of storage and 51.8°C in the winter after 3 D of storage. This is in accordance with our findings where a maximum temperature of 58.5°C was measured in the summer and 50.4°C in the winter. Temperatures over 65°C were reported by Wilkinson et al. (2011) for static piles of poultry litter in the first weeks of aging.
In a recently performed laboratory scale anaerobic digestion experiment by Thomas et al. (2019), ESBL-/AmpC-producing E. coli were added in a concentration of over 107 cfu/ml to a mix of chicken litter and an inoculum from a biogas plant. They showed that at a constant temperature of 55°C, ESBL-producing E. coli were quantitatively undetectable by direct count after 2 h of incubation. In our summer trial, temperatures reached levels constantly above 53°C at 48 h. In the subsequent samples (72 h and 96 h), we did not detect ESBL-producing E. coli and nonresistant E. coli quantitatively and qualitatively. We therefore assume that a temperature of 53°C is sufficient for inactivation of all ESBL-producing and nonresistant E. coli under practical conditions of an anaerobic litter storage.
A laboratory scale study by Laport et al. (2003) showed that a 2 h incubation period at 55°C will result in >90% reduction of Enterococcus faecium. Accordingly, at the end of our summer trial enterococci concentrations had decreased by > 99% compared with the initial concentrations in the deep samples.
In the winter trial, a temperature of over 50°C was observed in the deep samples for the last point in time after 96 h only. Extended-spectrum beta-lactamases–producing E. coli were qualitative, and nonresistant E. coli were quantitatively detectable until the end of the trial. The number of enterococci was stable in both the surface and deep samples in the winter trial, ranging from 3.2 × 106 to 5.6 × 107 cfu/g in all samples. Therefore, we assume that the increase in temperature to a maximum of 50.4°C in the winter trial was insufficient for a distinct reduction of the monitored bacteria in chicken litter.
A further factor, which might influence the bacterial counts in the chicken litter, is the pH value. The pH value of the litter at the beginning of the experiment (0 h) was in the alkaline range, a frequent finding in chicken litter (Huang et al., 2017). The pH value drop in the depth of the litter heap was presumably because of anaerobic fermentation and formation of organic acids like propionic acid, butyric acid, and acetic acid (Cornell Waste Management Institute, 1996). The minimum pH value of 5.6 measured in the winter trial and 6.5 in the summer trial is not sufficient to inactivate E. coli. It was shown that E. coli has a high probability of surviving pH values of 1.5 to 4.0 (Takumi et al., 2000).
Insights From Whole Genome Sequencing
Whole genome sequencing revealed that the analyzed isolates for both trials harbored additional resistance genes besides ESBL resistance genes. Identified genes included qnrS1, which may mediate resistance to quinolones (Cerquetti et al., 2009), mdfA, which substantially increases resistance to amphoteric lipophilic compounds (e.g., ethidium bromide, benzalkonium, and tetracycline) (Edgar and Bibi, 1997) and the Tet 34 and sulII resistance genes, which mediate for resistance against oxytetracycline (Nonaka and Suzuki, 2002) and sulfonamides (Radstrom and Swedberg, 1988).
The results also indicate that one strain of ESBL-producing E. coli was predominant in the chicken barns for each of the trials. This finding is especially prominent in the winter trial, where 94.7% (n = 18/19) of the isolates belonged to the same 7 gene MLST (ST117) and HC1100cgST (ST50).
Results of the phylogenetic analyses suggest that genetic differences do not equip the isolates of a certain cluster with a survival advantage.
Advantages and Limitations of the Study
In our study, we did not artificially add ESBL-producing E. coli to the litter. Instead, chicken litter naturally contaminated with these resistant bacteria was used, mirroring true field conditions. Hutchison et al. (2005) pointed out that this is crucial because bacteria are already adapted to their environment, and therefore, bacterial stress is minimized (Wesche et al., 2009). A further advantage over lab-scale studies is that the litter heaps were exposed to environmental conditions which are typical for the winter and summer season in central Europe. For on-farm waste management, similar environmental conditions are likely to appear; therefore, better transferability of the results can be expected when compared with laboratory studies.
A limitation of the study is that the measurements of temperature, humidity, and the litter samples taken for the microbiological analyses cannot cover the conditions in the entire storage mass. It was shown previously that because of increased deposition of fecal droppings on the litter surface, the number of coliform bacteria is significantly higher in the top layer of a chicken litter bed compared with the bottom layers (Barker et al., 2010). Using a front-end loader to remove and stack the litter at the end of the fattening period will not assure complete mixing of the litter, resulting in an inhomogeneous distribution of coliform bacteria. This might explain inconsistencies we saw in the course of the bacterial counts for some points in time. However, through our sampling scheme that covered both surface and deep samples with 3 samples evenly distributed over the litter heap for each point in time, we achieved representative results.
Evaluation of Short-Term Litter Storage as an On-Farm Strategy to Prevent the Spread of EBSL-Producing E. coli to the Environment
It was shown by Merchant et al. (2012) that resistant E. coli were detectable in soil fertilized with chicken litter for at least 7 mo. In that study performed in Canada of 295 E. coli isolated from soil, 139 carried either a blaSHV, blaTEM, or blaCMY-2 resistance gene. This highlights the importance of sufficient inactivation of resistant E. coli in litter before land application.
Very low quantities of resistant microorganisms might be able to horizontally transfer mobile genetic elements to microorganisms in the environment, thus potentially contributing to a spread of antibiotic resistance. As a result, the qualitative detection of ESBL-producing E. coli in litter is of particular importance. A recent study by Pornsukarom and Thakur (2017) demonstrated that the application of manure containing Enterobacteriaceae which carry plasmids mediating for antibiotic resistance enriches the environmental resistome. Our results indicate that a storage period of 5 D is sufficient to reduce the amount of ESBL-producing E. coli in the depth of a chicken litter heap in the summer below the detection limit. For very low ambient temperatures, as present in our winter trial, an extension of the storage period should be considered because we observed an incomplete inactivation of EBSL-producing E. coli in a 5-day storage period.
Even if cultivation-based methods are unable to detect resistant bacteria, a transfer of plasmids carrying resistance genes might occur. In a study performed by Le Devendec et al. (2016), chicken manure was stored for 6 wk. After this time span, E. coli were not detected in the manure by cultivation without enrichment. Plasmid capture assays with the stored chicken manure revealed an uptake of plasmids encoding resistance to sulfonamides, aminoglycosides, and streptomycin in recipient strains. This indicates that even if there are no cultivable bacteria left in the litter, the possibility of spread of resistance cannot be discounted because viable plasmids could still be present. In contrast, Guan et al. (2007) stated that composting of chicken manure at high temperatures could help prevent the spread of antibiotic-resistant genes via plasmids in the environment. In their study, neither viable E. coli nor their plasmids could be detected in compost microcosms, which reached temperatures of over 50°C.
The outer edges of litter piles may present a reservoir for bacteria, and turning the litter pile may therefore lead to a recontamination of the interior parts (Pereira-Neto et al., 1986). This is in accordance with our observation in the summer trial, where the number of E. coli and enterococci significantly increased on the surface of the litter heap.
The increased quantity of these bacteria over the course of the trial may be caused by beneficial environmental factors such as rainfall, which influences moisture levels in litter piles and can promote regrowth of enteric bacteria (Gibbs et al., 1997). In the summer trial of our study, rainfall at the end of the first trial day led to increased moisture in the litter heap. Corresponding temperatures on the surface of the litter pile ranged from 29.2°C to 42.7°C, which are known to be sufficiently high for bacterial regrowth (Kumar and Libchaber, 2013).
The survival time of E. coli in manure is significantly longer under anaerobic than under aerobic conditions (Semenov et al., 2011). Additionally, it was shown in the trial by Wilkinson et al. (2011) that temperatures in composted chicken litter piles are higher than in stored piles. As previous research and this study indicate a faster inactivation of ESBL-producing E. coli can be achieved at higher temperatures. It appears therefore that composting litter under aerobic conditions could lead to a faster inactivation of ESBL-producing E. coli compared with storing it in anaerobic conditions. However, increased working and litter-storage capacities are required and not available on all farms.
In conclusion, short-term litter storage is a useful, easily realizable tool leading to an effective reduction of the amount of ESBL-producing E. coli in chicken litter. However, we did not observe a complete inactivation of ESBL-producing E. coli in the depth of the heap in the winter and on the surface of the heap for both trials. An extension of the storage period for low ambient temperatures and stirring the pile one time or composting the litter instead of storing it could increase the effectivity of chicken litter hygienization.
Acknowledgments
The authors would like to thank the staff of the broiler farm for the excellent collaboration. They thank Kerstin Rosen and Benjamin Reichelt (Institute for Animal Hygiene and Environmental Health, Berlin, Germany) for assistance with taking and processing the samples. They also thank Heike Rose, Susann Sellenthin, Sabrina Hansen, Michael Kühl (Institute for Animal Hygiene and Environmental Health, Berlin, Germany), and Vera Junker (DSMZ, Braunschweig, Germany) for excellent technical advice and assistance. They thank Alexander Bartel (Institute for Veterinary Epidemiology and Biostatistics, Berlin, Germany) for statistical advice and Samira Schlesinger (Clinic of Animal Reproduction, Berlin, Germany) for proofreading the manuscript.
The study was funded by the Leibniz Association (grant number: SAW-2017-DSMZ-2)
Footnotes
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.psj.2019.11.043.
Supplementary data
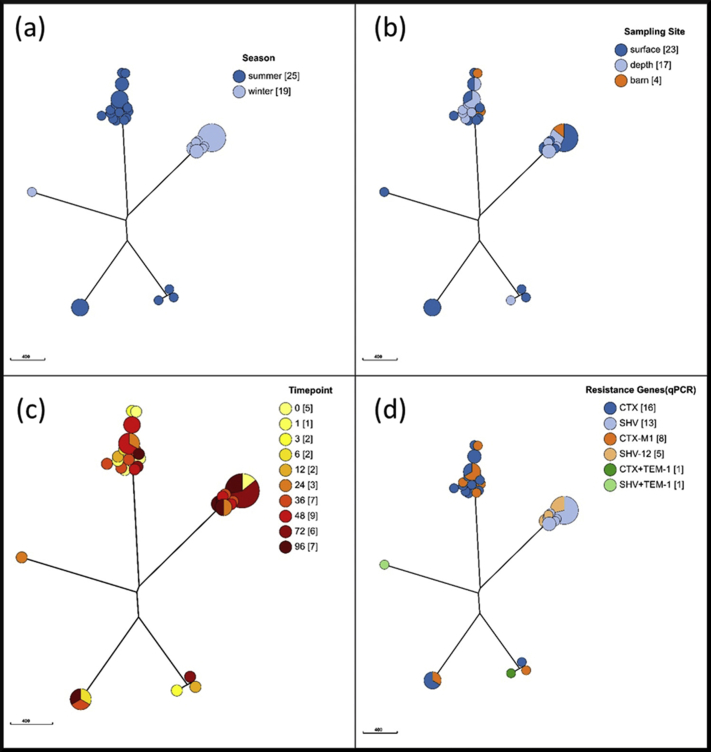
Supplemental Figure 1.
Phylogenetic NJ-Trees. Isolates are colored according to the season in which the experiment was performed (a), the sampling site in litter pile (b), the timepoint of sampling (c) or the resistance genes found by qPCR (d).
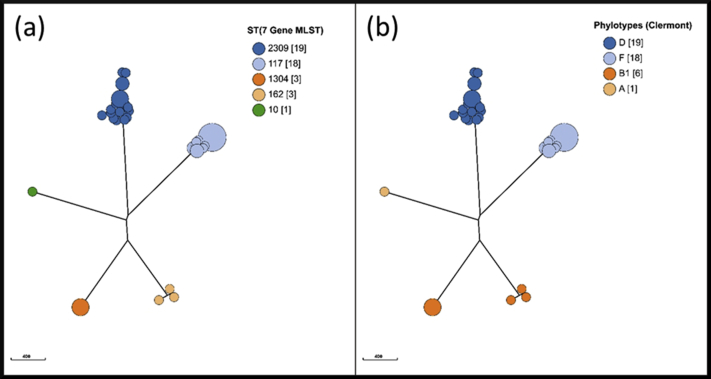
Supplemental Figure 2.
Phylogenetic NJ-Trees. The isolates are colored according to their 7 gene MLST sequence type (a) or according to their Phylogroup determined via Enterobase (b)
References
- Barker K.J., Purswell J.L., Davis J.D., Parker H.M., Kidd M.T., McDaniel C.D., Kiess A.S. Distribution of bacteria at different poultry litter depths. Int. J. Poult. Sci. 2010;9:10–13. [Google Scholar]
- Baym M., Kryazhimskiy S., Lieberman T.D., Chung H., Desai M.M., Kishony R. Inexpensive Multiplexed library Preparation for Megabase-Sized genomes. PLoS One. 2015;10:1–15. doi: 10.1371/journal.pone.0128036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beghain J., Bridier-Nahmias A., Le Nagard H., Denamur E., Clermont O. ClermonTyping: an easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb. Genomics. 2018;4:1–8. doi: 10.1099/mgen.0.000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaak H., Van Hoek A.H.A.M., Hamidjaja R.A., Van Der Plaats R.Q.J., Kerkhof-De Heer L., De Roda Husman A.M., Schets F.M. Distribution, numbers, and diversity of ESBL-producing E. coli in the poultry farm environment. PLoS One. 2015;10:1–23. doi: 10.1371/journal.pone.0135402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bland J.M., Altman D.G. Statistics notes: Transformations, means, and confidence intervals. BMJ. 1996;312:1079. doi: 10.1136/bmj.312.7038.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerquetti M., García-ferna A., Giufre M., Fortini D., Accogli M., Graziani C., Luzzi I., Caprioli A., Carattoli A. First Report of plasmid-mediated quinolone resistance determinant qnrS1 in an Escherichia coli strain of animal Origin in Italy. Antimicrob. Agents Chemother. 2009;53:3112–3114. doi: 10.1128/AAC.00239-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clermont O., Christenson J.K., Denamur E., Gordon D.M. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 2013;5:58–65. doi: 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- Cornell Waste Management Institute, C. U. 1996. http://compost.css.cornell.edu/science.html science @ compost.css.cornell.edu. Accessed Feb. 2020.
- Daehre K., Projahn M., Semmler T., Roesler U., Friese A. Extended-sapectrum beta-lactamase-/AmpC beta-lactamase-producing enterobacteriaceae in broiler farms: transmission dynamics at farm level. Microb. Drug Resist. 2018;24:511–518. doi: 10.1089/mdr.2017.0150. [DOI] [PubMed] [Google Scholar]
- Le Devendec L., Mourand G., Bougeard S., Léaustic J., Jouy E., Keita A., Couet W., Rousset N., Kempf I. Impact of colistin sulfate treatment of broilers on the presence of resistant bacteria and resistance genes in stored or composted manure. Vet. Microbiol. 2016;194:98–106. doi: 10.1016/j.vetmic.2015.11.012. [DOI] [PubMed] [Google Scholar]
- Dierikx C., van Essen-Zandbergen A., Veldman K., Smith H., Mevius D. Increased detection of extended spectrum beta-lactamase producing Salmonella enterica and Escherichia coli isolates from poultry. Vet. Microbiol. 2010;145:273–278. doi: 10.1016/j.vetmic.2010.03.019. [DOI] [PubMed] [Google Scholar]
- Dierikx C., van der Goot J., Fabri T., van Essen-Zandbergen A., Smith H., Mevius D. Extended-spectrum-β-lactamase- and AmpC-β-lactamase-producing Escherichia coli in Dutch broilers and broiler farmers. J. Antimicrob. Chemother. 2013;68:60–67. doi: 10.1093/jac/dks349. [DOI] [PubMed] [Google Scholar]
- Edgar R., Bibi E. MdfA , an Escherichia coli Multidrug resistance Protein with an Extraordinarily broad spectrum of drug Recognition. J. Bacteriol. 1997;179:2274–2280. doi: 10.1128/jb.179.7.2274-2280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA. Scientific Opinion on the public health risks of bacterial strains producing extended-spectrum β -lactamases and/or AmpC β -lactamases in food and food producing strains. EFSA J. 2011;9:1–95. [Google Scholar]
- Erickson M.C., Liao J., Boyhan G., Smith C., Ma L., Jiang X., Doyle M.P. Fate of manure-borne pathogen surrogates in static composting piles of chicken litter and peanut hulls. Bioresour. Technol. 2010;101:1014–1020. doi: 10.1016/j.biortech.2009.08.105. [DOI] [PubMed] [Google Scholar]
- Feldgarden M., Vyacheslav B., Hafta D.H., Prasada A.B., Slotta D.J., Tolstoya I., Tysonb G.H., Zhaob S., Hsub C.-H., McDermottb P.F., Tadesseb D.A., Morales C., Simmons M., Tillman G., Wasilenko J., Folsterd J.P., Klimke W. 2019. Using the NCBI AMRFinder tool to determine antimicrobial resistance genotype-phenotype correlations within a collection of NARMS isolates. Accessed Feb. 2020. https://www.biorxiv.org/content/10.1101/550707v1.article-info. [Google Scholar]
- Friese A., Lu H.M., Merle R., Roesler U. Extended-spectrum beta-lactamase and AmpC beta-lactamase-producing Enterobacteriaceae in Turkey fattening farms: a cross-sectional study. Berl. Munch. Tierarztl. Wochenschr. 2019;132:352–359. [Google Scholar]
- Gibbs R.A., Hu C.J., Ho G., Unkovich I. Regrowth of faecal coliforms and salmonellae in stored biosolids and soil amended with biosolids. Water Sci. Technol. 1997;35:269–275. [Google Scholar]
- Guan J., Wasty A., Grenier C., Chan M. Influence of temperature on survival and conjugative transfer of multiple antibiotic-resistant plasmids in chicken manure and compost microcosms. Poult. Sci. 2007;86:610–613. doi: 10.1093/ps/86.4.610. [DOI] [PubMed] [Google Scholar]
- Hartmann A., Locatelli A., Amoureux L., Depret G., Jolivet C., Gueneau E., Neuwirth C. Occurrence of CTX-M producing Escherichia coli in soils, Cattle, and farm environment in France (Burgundy region) Front. Microbiol. 2012;3:83. doi: 10.3389/fmicb.2012.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hering J., Frömke C., von Münchhausen C., Hartmann M., Schneider B., Friese A., Rösler U., Kreienbrock L., Hille K. Cefotaxime-resistant Escherichia coli in broiler farms-A cross-sectional investigation in Germany. Prev. Vet. Med. 2016;125:154–157. doi: 10.1016/j.prevetmed.2016.01.003. [DOI] [PubMed] [Google Scholar]
- Huang J., Yu Z., Gao H., Yan X., Chang J., Wang C., Hu J., Zhang L. Chemical structures and characteristics of animal manures and composts during composting and assessment of maturity indices. PLoS One. 2017;12:1–16. doi: 10.1371/journal.pone.0178110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijbers P.M.C., Graat E.A.M., Haenen A.P.J., van Santen M.G., van Essen-Zandbergen A., Mevius D.J., van Duijkeren E., van Hoek A.H.A.M. Extended-spectrum and AmpC β-lactamase-producing Escherichia coli in broilers and people living and/or working on broiler farms: prevalence, risk factors and molecular characteristics. J. Antimicrob. Chemother. 2014;69:2669–2675. doi: 10.1093/jac/dku178. [DOI] [PubMed] [Google Scholar]
- Hutchison M.L., Walters L.D., Avery S.M., Moore A. Decline of zoonotic agents in livestock waste and bedding heaps. J. Appl. Microbiol. 2005;99:354–362. doi: 10.1111/j.1365-2672.2005.02591.x. [DOI] [PubMed] [Google Scholar]
- Jia B., Raphenya A.R., Alcock B., Waglechner N., Guo P., Tsang K.K., Lago B.A., Dave B.M., Pereira S., Sharma A.N., Doshi S., Courtot M., Lo R., Williams L.E., Frye J.G., Elsayegh T., Sardar D., Westman E.L., Pawlowski A.C., Johnson T.A., Brinkman F.S.L., Wright G.D., McArthur A.G. Card 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017;45:D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Libchaber A. Pressure and temperature Dependence of Growth and Morphology of Escherichia coli : experiments and Stochastic model. Biophys. J. 2013;105:783–793. doi: 10.1016/j.bpj.2013.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laport M.S., Ramos M., Silva C.C., De Freire C., Giambiagi-deMarval M. Heat-resistance and Heat-Shock Response in the Nosocomial pathogen Enterococcus faecium. Curr. Microbiol. 2003;46:313–317. doi: 10.1007/s00284-002-3828-0. [DOI] [PubMed] [Google Scholar]
- Laube H., Friese A., von Salviati C., Guerra B., Käsbohrer A., Kreienbrock L., Roesler U. Longitudinal monitoring of extended-spectrum-beta-lactamase/ampC-producing Escherichia coli at German broiler chicken fattening farms. Appl. Environ. Microbiol. 2013;79:4815–4820. doi: 10.1128/AEM.00856-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laube H., Friese A., von Salviati C., Guerra B., Rösler U. Transmission of ESBL/AmpC-producing Escherichia coli from broiler chicken farms to surrounding areas. Vet. Microbiol. 2014;172:519–527. doi: 10.1016/j.vetmic.2014.06.008. [DOI] [PubMed] [Google Scholar]
- Leverstein-van Hall M.A., Dierikx C.M., Stuart J.C., Voets G.M., van den Munckhof M.P., van Essen-Zandbergen A., Platteel T., Fluit A.C., van de Sande-Bruinsma N., Scharinga J., Bonten M.J.M., Mevius D.J. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin. Microbiol. Infect. 2011;17:873–880. doi: 10.1111/j.1469-0691.2011.03497.x. [DOI] [PubMed] [Google Scholar]
- Merchant L.E., Rempel H., Forge T., Kannangara T., Bittman S., Delaquis P., Topp E., Ziebell K.A., Diarra Moussa S. Characterization of antibiotic-resistant and potentially pathogenic Escherichia coli from soil fertilized with litter of broiler chickens fed antimicrobial-supplemented diets. Can. J. Microbiol. 2012;58:1084–1098. doi: 10.1139/w2012-082. [DOI] [PubMed] [Google Scholar]
- Nonaka L., Suzuki S. New Mg 2+ -Dependent oxytetracycline resistance determinant Tet 34 in Vibrio isolates from Marine fish Intestinal contents. Antimicrob. Agents Chemother. 2002;46:1550–1552. doi: 10.1128/AAC.46.5.1550-1552.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira-Neto J., Stentiford E.I., Smith D.V. Survival of faecal indicator micro-organisms in refuse/sludge composting using the aerated static pile system. Waste Manag. Res. 1986;4:397–406. [Google Scholar]
- Pornsukarom S., Thakur S. Crossm horizontal dissemination of antimicrobial resistance determinants in multiple Salmonella Serotypes following isolation from the Commercial swine Operation environment after manure. Appl. Environ. Microbiol. 2017;83:1–14. doi: 10.1128/AEM.01503-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Projahn M., Daehre K., Roesler U., Friese A. Extended-spectrum-beta-lactamaseand plasmid-encoded cephamycinaseproducing enterobacteria in the broiler hatchery as a potential mode of pseudovertical transmission. Appl. Environ. Microbiol. 2017;83 doi: 10.1128/AEM.02364-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radstrom P., Swedberg G. RSF1010 and a conjugative plasmid Contain sulII , one of two known genes for plasmid-borne sulfonamide resistance Dihydropteroate Synthase. Antimicrob. Agents Chemother. 1988;32:1684–1692. doi: 10.1128/aac.32.11.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remschmidt C., Schneider S., Meyer E., Schroeren-Boersch B., Gastmeier P., Schwab F. Surveillance of antibiotic use and resistance in intensive care units (SARI) a 15-year cohort study. Dtsch. Arztebl. Int. 2017;114:858–865. doi: 10.3238/arztebl.2017.0858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschanski N., Fischer J., Guerra B., Roesler U. Development of a multiplex real-time PCR for the rapid detection of the predominant beta-lactamase genes CTX-M, SHV, TEM and CIT-type AmpCs in enterobacteriaceae. PLoS One. 2014;9:e100956. doi: 10.1371/journal.pone.0100956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenov A.V., Van Overbeek L., Termorshuizen A.J., Van Bruggen A.H.C. Influence of aerobic and anaerobic conditions on survival of Escherichia coli O157 : H7 and Salmonella enterica serovar Typhimurium in Luria-Bertani broth , farm-yard manure and slurry. J. Environ. Manage. 2011;92:780–787. doi: 10.1016/j.jenvman.2010.10.031. [DOI] [PubMed] [Google Scholar]
- Statistisches Bundesamt. 2016. Wirtschaftsdünger Tierischer Herkunft in Landwirtschaftlichen Betrieben/Agrarstrukturerhebung. Fachserie 3 R. 2.2.2 2030222169; pp. 2–74. [Google Scholar]
- Statistisches Bundesamt. Bundesamt (Destatis), 2017. Land- und Forstwirtschaft, Fischerei – Wirtschaftsdünger tierischer Herkunft in landwirtschaftlichen Betrieben/Agrarstrukturerhebung. 2017. https://www.destatis.de/DE/Themen/Branchen-Unternehmen/Landwirtschaft-Forstwirtschaft-Fischerei/Produktionsmethoden/Publikationen/Downloads-Produktionsmethoden/wirtschaftsduenger-2030222169005.html Accessed Feb. 2020.
- Steglich M., Hofmann J.D., Helmecke J., Sikorski J., Spröer C., Riedel T., Bunk B., Overmann J. Convergent Loss of ABC transporter genes from Clostridioides difficile genomes is Associated with Impaired Tyrosine uptake and p -Cresol production. Front. Microbiol. 2018;9:1–12. doi: 10.3389/fmicb.2018.00901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takumi K., De Jonge R., Havelaar A. Modelling inactivation of Escherichia coli by low pH: application to passage through the stomach of young and elderly people. J. Appl. Microbiol. 2000;89:935–943. doi: 10.1046/j.1365-2672.2000.01193.x. [DOI] [PubMed] [Google Scholar]
- Thomas C., Idler C., Ammon C., Herrmann C., Amon T. Inactivation of ESBL-/AmpC-producing Escherichia coli during mesophilic and thermophilic anaerobic digestion of chicken manure. Waste Manag. 2019;84:74–82. doi: 10.1016/j.wasman.2018.11.028. [DOI] [PubMed] [Google Scholar]
- Waters N., Brennan F., Holmes A., Abram F., Pritchard L. 2018. Easily Phylotyping E. coli via the EzClermont Web App and Command-Line Tool.https://www.biorxiv.org/content/10.1101/317610v1.article-info Accessed Feb. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesche A.M., Gurtler J.B., Marks B.P., Ryser E.T. Stress, Sublethal Injury, Resuscitation, and Virulence of bacterial Foodborne Pathogenst. J. Food Prot. 2009;72:1121–1138. doi: 10.4315/0362-028x-72.5.1121. [DOI] [PubMed] [Google Scholar]
- Wilkinson K.G., Tee E., Tomkins R.B., Hepworth G., Premier R. Effect of heating and aging of poultry litter on the persistence of enteric bacteria. Poult. Sci. 2011;90:10–18. doi: 10.3382/ps.2010-01023. [DOI] [PubMed] [Google Scholar]
- Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., Aarestrup F.M., Larsen M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Alikhan N., Mohamed K., Group S., Achtman M., Road G.H., Kingdom U., Chattaway M., Delahay R., Hutton S., Graz H., Petrovska L., Agency P.H., Tyne W., Fan Y., Williams N., Health P. 2019. The User's Guide to Comparative Genomics with EnteroBase. Three Case Studies: Micro-clades within Salmonella enterica Serovar Agama, Ancient and Modern Populations of Yersinia pestis, and Core Genomic Diversity of All Escherichia. Accessed Feb. 2020. https://www.biorxiv.org/content/10.1101/613554v1.article-info. [Google Scholar]