Abstract
In poultry, viral infections (e.g., Marek's disease virus, avian leukosis virus, influenza A virus, and so on) can cause devastating mortality and economic losses. Because viruses are solely dependent on host cells to propagate, they alter the host intracellular microenvironment. Thus, understanding the virus-host interaction is important for antiviral immunity and drug development in the poultry industry. MicroRNAs are crucial posttranscriptional regulators of gene expression in a wide spectrum of biological processes, including viral infection. Recently, microRNAs have been identified as key players in virus-host interactions. In this review, we will discuss the intricacies involved in the virus-host cross-talk mediated by host and viral microRNAs in poultry (i.e., chicken and ducks), as well as recent trends and challenges in this field. These findings may provide some insights into the rapidly developing area of research regarding viral pathogenesis and antiviral immunity in poultry production.
Key words: cross-talk, poultry animal, microRNA, viral infection, virus-host
Introduction
Many types of viruses will cause high mortality rates and enormous economic losses to the poultry industry. For example, Marek's disease virus (MDV) and avian leukosis virus can induce neoplastic diseases in chickens. Moreover, influenza A virus (IAV) is highly variable and can spread across species, thereby resulting in a threat to global public health. Therefore, the cross-talk between the host (i.e., chickens) and viruses during infections is critical to understanding viral pathogenesis and antiviral immunity in chicken production. Over the past decade, studies have attempted to depict the cross-talk between the host and virus from different aspects (e.g., microRNA [miRNAs] perspective) and have made progress in this area (Li et al., 2014a, Skalsky and Cullen, 2010).
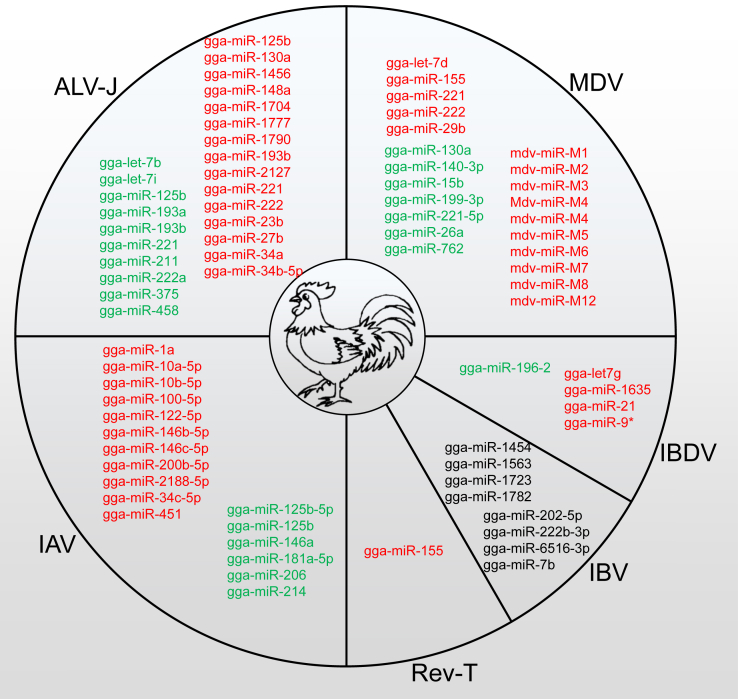
miRNAs Are short noncoding 18- to 25-nucleotide-long RNAs that regulate gene expression at the posttranscriptional level. Substantial evidence demonstrates that a single miRNA can have extensive and crucial effects on a range of fundamental biological processes, and miRNA dysregulation can result in biological dysfunction and diseases (Bartel, 2004, Ha and Kim, 2014). Pathogen-induced phenotypic changes in hosts are always accompanied by remarkable alterations at the transcriptional level (Jenner and Young, 2005). The miRNA expression profile is profoundly influenced by viral infection and contributes to the repertoire of virus-host interactions. Differential miRNA expression can be attributed to both host antiviral defenses and alterations in cellular environment that are affected by viral factors (Scaria et al., 2007, Skalsky and Cullen, 2010, tenOever, 2013, Harwig et al., 2014, Powdrill et al., 2016, Nejad et al., 2018). Moreover, miRNAs have been reported to be biomarkers of viral infections (Wang et al., 2014) and have roles in viral reactivation from latency (Hicks et al., 2019). Therefore, the identification of deregulated miRNAs in virally infected chickens will provide novel insight into understanding virus-host interaction. In this review, we will summarize recent advances into the critical role of virally influenced poultry and viral miRNAs in the cross-talk between the host and virus to help understand viral pathogenesis and antiviral immunity in the poultry industry. Figure 1 shows the deregulation of miRNAs in chickens and viruses, infected by various viruses including MDV, avian leukosis virus subgroup J (ALV-J), IAV, infectious bursal disease virus (IBDV), infectious bronchitis virus (IBV), or reticuloendotheliosis virus strain T (Rev-T). Chicken (Gallus gallus) miRNAs started with “gga-miR” while the MDV miRNAs started with “MDV-miR”.
Figure 1.
Summary of deregulated miRNAs in viral infected chickens. Red: upregulated microRNAs. Green: downregulated microRNAs. Black: expressed microRNAs. Chicken (Gallus gallus) microRNAs started with “gga-miR” while the MDV microRNAs started with “MDV-miR”. Abbreviations: ALV-J, avian leukosis virus subgroup J; IAV, influenza A virus; IBDV, infectious bursal disease virus; IBV, infectious bronchitis virus; MDV, Marek's disease virus; Rev-T, reticuloendotheliosis virus strain T.
Marek's disease virus
MDV is a highly contagious alpha herpesvirus and the causative agent of T-cell lymphoma and Marek's disease in chickens. The chickens and MDV-encoded miRNAs have been reported to have regulatory roles during MDV oncogenesis (Table 1).
Table 1.
miRNAs have been reported to have regulatory roles during MDV infection in chickens.
| miRNA | Species | Source | Pattern | Targets | Cell function | Reference |
|---|---|---|---|---|---|---|
| let-7d | Chicken | Skin | Up | - | - | Heidari and Delekta, 2017 |
| miR-130a | Chicken | Spleens | Down | HOXA3, MDFIC | Inhibit MD lymphoma cell proliferation and migration | Han et al., 2016 |
| miR-140-3p | Chicken | Spleens | Down | - | Inhibit MDV-transformed lymphoid cells proliferation; MD lymphoma transformation | Lian et al., 2015 |
| miR-155 | Chicken | Cell lines, peripheral blood leukocytes | Up | GPM6B, RREB1, c-Myb, MAP3K7IP2, PU.1, C/EBP | - | Muylkens et al., 2010 |
| miR-155 | Chicken | Spleens | Up | - | - | Li, Zhang, Li, Zheng, Zheng and Liu, 2014c |
| miR-15b | Chicken | Spleens | Down | ATF2 | - | Tian et al., 2012 |
| miR-199-3p | Chicken | Spleens | Down | - | Inhibit MDV-transformed lymphoid cells proliferation; MD lymphoma transformation | Lian et al., 2015 |
| miR-221 | Chicken | Cell lines | Up | p27(Kip1) | - | Lambeth et al., 2009 |
| miR-221-5p | Chicken | Spleens | Down | - | Inhibit MDV-transformed lymphoid cells proliferation; MD lymphoma transformation | Lian et al., 2015 |
| miR-222 | Chicken | Cell lines | Up | p27(Kip1) | - | Lambeth et al., 2009 |
| miR-26a | Chicken | Spleens | Down | NEK6 | Suppress MD lymphoma cell proliferation | Li, Lian, Zhang, Qu and Yang, 2014b |
| miR-29b | Chicken | Spleens | Up | - | - | Li, Zhang, Li, Zheng, Zheng and Liu, 2014c |
| miR-762 | Chicken | Spleens | Down | - | - | Li, Zhang, Li, Zheng, Zheng and Liu, 2014c |
| miR-12 | MDV | Cell lines, kidney or spleen tissues | Up | - | Potential biomarkers for MDV-transformed tumors and lymphoblastoid cell lines. | Xu et al., 2008 |
| miR-4 | MDV | Cell lines, kidney or spleen tissues | Up | - | Potential biomarkers for MDV-transformed tumors and lymphoblastoid cell lines. | Xu et al., 2008 |
| miR-8 | MDV | Cell lines, kidney or spleen tissues | Up | - | Potential biomarkers for MDV-transformed tumors and lymphoblastoid cell lines. | Xu et al., 2008 |
| miR-M1 | MDV | Cell lines, spleens | Up | - | Burnside et al., 2006 | |
| miR-M12-3p | MDV | Spleens | Up | - | - | Li, Zhang, Li, Zheng, Zheng and Liu, 2014c |
| miR-M1-5p | MDV | Spleens | Up | - | - | Li, Zhang, Li, Zheng, Zheng and Liu, 2014c |
| miR-M2 | MDV | Cell lines, spleens | Up | - | Burnside et al., 2006 | |
| miR-M2*/3p | MDV | Spleens | Up | - | - | Morgan et al., 2008 |
| miR-M2-5p | MDV | Spleens | Up | - | - | Li, Zhang, Li, Zheng, Zheng and Liu, 2014c |
| miR-M3 | MDV | Cell lines, spleens | Up | - | Burnside et al., 2006 | |
| miR-M3-5p | MDV | Cell line | Up | - | - | Yao et al., 2008 |
| miR-M4 | MDV | Spleens | Up | - | - | Morgan et al., 2008 |
| miR-M4 | MDV | Embryo fibroblast cultures | Up | - | Variants within its seed region were sufficient to inhibit the induction of lymphomas. | Zhao et al., 2011 |
| miR-M4 | MDV | Cell lines, spleens | Up | - | Burnside et al., 2006 | |
| miR-M4 | MDV | Spleens | Up | - | - | Burnside and Morgan, 2011 |
| miR-M4-5p | MDV | Cell lines | Up | - | - | Yao et al., 2008 |
| miR-M4-5p | MDV | Spleens | Up | - | - | Li, Zhang, Li, Zheng, Zheng and Liu, 2014c |
| miR-M4-5P | MDV | Cell lines, peripheral blood | Up | GPM6B, RREB1, c-Myb, MAP3K7IP, PU.1, C/EBP, UL28, UL32 | - | Muylkens et al., 2010 |
| miR-M5 | MDV | Cell lines, spleens | Up | - | - | Burnside et al., 2006 |
| miR-M5-3p | MDV | Cell lines | Up | - | - | Yao et al., 2008 |
| miR-M6-5p | MDV | Cell lines | Up | - | - | Yao et al., 2008 |
| miR-M7-5p | MDV | Cell lines | Up | - | - | Yao et al., 2008 |
| miR-M8-3p | MDV | Spleens | Up | - | - | Li, Zhang, Li, Zheng, Zheng and Liu, 2014c |
“-” Means no targets of the microRNA in the reference.
Abbreviations: MD, Marek's disease; MDV, Marek's disease virus; miRNA, microRNA.
The dysregulation of chicken miRNAs was observed in MDV-infected samples and may play a crucial role in Marek's disease tumorigenesis. The overexpression of gga-miR-221/-222 decreased the levels of cell cycle regulatory protein p27(Kip1) in the MDV-transformed lymphoblastoid cell line MSB-1 (Lambeth et al., 2009). The level of gga-miR-29b and gga-miR-155 expression increased and that of gga-miR-762 decreased in MDV-infected chicken splenic tumors when compared with nontumorous tissues (Li et al., 2014c). Several miRNAs are also related to the proliferation or migration of MDV-transformed lymphoid cells. It was found that gga-miR-130a targets HOXA3 and MDFIC and inhibits MSB-1 cell proliferation and migration (Han et al., 2016). The dysregulation of gga-miR-199-3p, gga-miR-140-3p, and gga-miR-221-5p was observed during the early tumor transformation phase and demonstrated a suppressive role on cell proliferation (Lian et al., 2015). Moreover, gga-miR-26a was also downregulated in MDV-infected spleens and suppressed the proliferation of MSB-1 cells (Li et al., 2014b). MDV infection affects the miRNA expression levels not only in spleens but also in the skin tissues, such as gga-let-7d, which was upregulated in the skin of MDV-infected chickens (Heidari and Delekta, 2017).
In the MDV-infected chickens, MDV-encoded miRNAs were found to be stably expressed to enable MDV pathogenesis and contribute to MDV-induced transformation and oncogenesis. Mdv-miR-M4, which flanks the Meq oncogene, was highly expressed in MDV-infected cells and MDV-induced tumors compared with noninfected samples and targeted both chicken and viral genes (Burnside et al., 2006, Xu et al., 2008, Yao et al., 2008, Muylkens et al., 2010, Burnside and Morgan, 2011, Li et al., 2014c). In particular, in the MDV-induced tumors, the level of mdv-miR-M4 was threefold higher in very virulent plus MDV-induced tumors than in less virulent samples (Morgan et al., 2008). Furthermore, mutations in mdv-miR-M4 were sufficient to inhibit the induction of lymphomas (Zhao et al., 2011). In addition to mdv-miR-4, mdv-miR-M1 to -M8 were also upregulated in the MDV-infected samples, among which miR-1 was mapped downstream of the Meq gene, mdv-miR-M2 to -M5 were all found to be located upstream of the Meq promoter region, and mdv-miR-M6 to -M8 was mapped to the latency-associated transcript region (Burnside et al., 2006, Xu et al., 2008, Yao et al., 2008, Li et al., 2014c). Moreover, the level of mdv-miR-M2*/3p was more than six-fold higher in very virulent plus MDV-induced tumors than in less virulent samples (Morgan et al., 2008). Mdv-miR-12 was also found to be located upstream of Meq and expressed at high levels in the spleen and kidney tumor tissues compared with normal tissues (Xu et al., 2008, Li et al., 2014c).
Avian leukosis virus subgroup J
Avian leukosis viruses are a group of avian retroviruses that can induce neoplastic diseases in chickens. Specially, an infection with ALV-J can result in serious damage and significant economic loss compared with other virus subgroups because of sporadic or widespread infection (Payne et al., 1991, Gao et al., 2012, Payne and Nair, 2012).
Several miRNAs may play a role as tumor-formation relevant genes in chickens. The overexpression of gga-miR-221 has been observed in cells, liver tissues, or liver tumors of ALV-J–infected chickens compared with noninfected samples; moreover, gga-miR-221 promoted the proliferation, migration, and growth of cancer cells and inhibited apoptosis by targeting CDKN1B and BCL-2 modifying factor (BMF) (Li et al., 2012, Wang et al., 2013a, Dai et al., 2015, Ren et al., 2018). However, in ALV-J–infected bone marrow dendritic cells, gga-miR-221 was downregulated (Liu et al., 2016). A similar expression pattern and function of gga-miR-222 was also reported in ALV-J–infected samples (Li et al., 2012, Dai et al., 2015, Liu et al., 2016). In addition, gga-miR-125b and -193b also displayed opposing expression patterns in infected liver tissues and dendritic cells. Their expression level was decreased in infected liver tissues but increased in bone marrow dendritic cells (Wang et al., 2013a, Liu et al., 2016). Gga-miR-34b-5p promoted the proliferation and migration of ALV-J–infected cells, as well as ALV-J replication by suppressing melanoma differentiation–associated gene 5 signaling pathway (Li et al., 2017). Other miRNAs reported to exhibit significant levels of expression between infected and noninfected samples are listed in Table 2.
Table 2.
miRNAs have been reported to play a regulatory role during ALV-J infection in chickens.
| miRNA | Source | Pattern | Targets | Cell function | Reference |
|---|---|---|---|---|---|
| let-7b | Liver | Down | - | - | Li et al., 2012 |
| let-7i | Liver | Down | - | - | Li et al., 2012 |
| miR-125b | Liver | Down | - | - | Li et al., 2012 |
| miR-125b | Bone marrow dendritic cells | Up | - | - | Liu et al., 2016 |
| miR-125b | Cell lines, liver | Down | - | - | Wang, Gao, Ji, Qi, Qin, Gao, Wang and Wang, 2013a |
| miR-130a | Bone marrow dendritic cells | Up | - | - | Liu et al., 2016 |
| miR-1456 | Liver | Up | - | - | Li et al., 2012 |
| miR-148a | Bone marrow dendritic cells | Up | - | - | Liu et al., 2016 |
| miR-1704 | Liver | Up | - | - | Li et al., 2012 |
| miR-1777 | Liver | Up | - | - | Li et al., 2012 |
| miR-1790 | Liver | Up | - | - | Li et al., 2012 |
| miR-193a | Cell lines, liver | Down | - | - | Wang, Gao, Ji, Qi, Qin, Gao, Wang and Wang, 2013a |
| miR-193b | Bone marrow dendritic cells | Up | - | - | Liu et al., 2016 |
| miR-193b | Cell lines, liver | Down | - | - | Wang, Gao, Ji, Qi, Qin, Gao, Wang and Wang, 2013a |
| miR-211 | Bone marrow dendritic cells | Down | - | - | Liu et al., 2016 |
| mir-2127 | Liver | Up | - | - | Li et al., 2012 |
| miR-222 | Liver | Up | - | - | Li et al., 2012 |
| miR-221 | Bone marrow dendritic cells | Down | - | - | Liu et al., 2016 |
| miR-221 | Cell line, liver tissue | Up | CDKN1B | Promote cell proliferation and cell cycle progression | Ren et al., 2018 |
| miR-221 | Liver | Up | - | - | Li et al., 2012 |
| miR-221 | Cell lines, liver | Up | BMF | Promote the proliferation, migration, and growth of cancer cells, and inhibit apoptosis | Dai et al., 2015 |
| miR-221 | Cell lines, liver | Up | - | - | Wang, Gao, Ji, Qi, Qin, Gao, Wang and Wang, 2013a |
| miR-222 | Liver, marrow | Up | BMF | Promote the proliferation, migration, and growth of cancer cells, and inhibit apoptosis | Dai et al., 2015 |
| miR-222a | Bone marrow dendritic cells | Down | - | - | Liu et al., 2016 |
| miR-23b | Spleens | Up | IRF1 | Promote ALV-J replication | Li et al., 2015a |
| miR-27b | Bone marrow dendritic cells | Up | - | - | Liu et al., 2016 |
| miR-34a | Bone marrow dendritic cells | Up | - | - | Liu et al., 2016 |
| miR-34b-5p | Spleens | Up | MDA5 | Promote ALV-J infected cells proliferation and ALV-J replication | Li et al., 2017 |
| mir-375 | Livers | Down | - | - | Li et al., 2012 |
| mir-458 | Livers | Down | - | - | Li et al., 2012 |
Abbreviations: ALV-J, avian leukosis virus subgroup J; BMF, BCL-2 modifying factor; miRNA, microRNA.
Influenza A virus
IAVs are negative-sense single-stranded RNA viruses that can cause significant economic losses to the poultry industry and represent a high threat to global public health because of the potential of overcoming the host barrier, which would allow transmission from animals to humans (Rebmann and Zelicoff, 2012). The rapid mutation rate and evolution of IAVs result in the loss of vaccine optimal efficacy (Shao et al., 2017). Therefore, more effective strategies are required to control AIV. Based on the virus surface glycoproteins, hemagglutinin and neuraminidase, IAVs can be divided into 18 hemagglutinin subtypes and 11 neuraminidase subtypes (Hay et al., 2001). Thus, more effective strategies are required to control IAV. Here, we summarize the regulatory roles of miRNAs in chickens during IAV infection, including H5N3, H5N1, and H9N2 (Table 3).
Table 3.
miRNAs reported to have a regulatory role during IAV infection.
| miRNA | Virus | Species | Source | Pattern | Targets | Reference |
|---|---|---|---|---|---|---|
| miR-100-5p | H9N2 | Chicken | CEF | Up | - | Peng et al., 2015 |
| miR-10a-5p | H9N2 | Chicken | CEF | Up | - | Peng et al., 2015 |
| miR-10b-5p | H9N2 | Chicken | CEF | Up | - | Peng et al., 2015 |
| miR-125b-5p | H9N2 | Chicken | CEF | Down | - | Peng et al., 2015 |
| miR-146c-5p | H9N2 | Chicken | CEF | Up | - | Peng et al., 2015 |
| miR-181a-5p | H9N2 | Chicken | CEF | Down | - | Peng et al., 2015 |
| miR-214 | H9N2 | Chicken | CEF | Down | - | Peng et al., 2015 |
| miR-125b | H5N3 | Chicken | Lung, trachea | Down | - | Wang et al., 2009 |
| miR-146a | H5N3 | Chicken | Lung, trachea | Down | - | Wang et al., 2009 |
| miR-1a | H5N3 | Chicken | Lung, trachea | Up | - | Wang et al., 2009 |
| miR-206 | H5N3 | Chicken | Lung | down | - | Wang et al., 2012 |
| miR-451 | H5N3 | Chicken | Lung | Up | - | Wang et al., 2012 |
| mir-133c | H5N1 | Chicken | Lung | Expressed | PB1, PB1-F2, and N40 | Kumar et al., 2014 |
| mir-146c* | H5N1 | Chicken | Lung | Expressed | PB1, PB1-F2, and N40 | Kumar et al., 2014 |
| miR-1658* | H5N1 | Chicken | Lung | Expressed | NS1 | Asaf et al., 2015 |
| miR-1710 | H5N1 | Chicken | Lung | Expressed | PB1, PB1-F2, and N40 | Kumar et al., 2014 |
| miR-2188-5p | H5N1 | Chicken | Spleen | Up | - | Li, Zhang, Su, Liu, Guo, Zhang, Lu, Xing, Guan, Li, Sun and Zhao, 2015b |
| miR-34c-5p | H5N1 | Chicken | Spleen | Up | - | Li, Zhang, Su, Liu, Guo, Zhang, Lu, Xing, Guan, Li, Sun and Zhao, 2015b |
| miR-200b-5p | H5N1 | Chicken | Spleen | Up | - | Li, Zhang, Su, Liu, Guo, Zhang, Lu, Xing, Guan, Li, Sun and Zhao, 2015b |
| miR-122-5p | H5N1 | Chicken | Spleen | Up | - | Li, Zhang, Su, Liu, Guo, Zhang, Lu, Xing, Guan, Li, Sun and Zhao, 2015b |
| miR-146b-5p | H5N1 | Chicken | Spleen | Up | - | Li, Zhang, Su, Liu, Guo, Zhang, Lu, Xing, Guan, Li, Sun and Zhao, 2015b |
| miR-122-5p | H5N1 | Duck | Spleen | Down | - | Li, Zhang, Su, Liu, Guo, Zhang, Lu, Xing, Guan, Li, Sun and Zhao, 2015b |
Abbreviations: CEF, chicken embryonic fibroblasts; IAV, influenza A virus; miRNA, microRNA.
Because H9N2 is the most common influenza virus in Asia, it has caused large economic losses to chicken production. Moreover, H9N2 has been detected in multiple avian species and can infect humans (Peiris et al., 1999, Xu et al., 2007). Using a deep sequencing platform, Peng et al. measured the expression profiles of miRNAs in H9N2-infected and noninfected chicken embryo fibroblasts and found that 48 miRNAs exhibited significantly different levels of expression between the 2 groups; the miRNAs were predicted to target immune response–related genes (Peng et al., 2015). Among these, the miRNAs that presented with a high abundance (top 10) in the chicken fibroblasts are summarized in Table 3.
H5N1 and H5N3 are highly pathogenic subtypes of avian influenza. Kumar et al. (2014) screened the miRNAs that were expressed in chicken lungs against the H5N1 genome and found that mir-133c, mir-1710, and mir-146c* were predicted to regulate the viral genes PB1, PB1-F2, and N40, while miR-1658* targeted the viral NS1 gene (Kumar et al., 2014, Asaf et al., 2015). Moreover, the miRNA expression pattern in H5N1-infected chickens and ducks diverged substantially, indicating a difference in the miRNA regulatory networks of the 2 species (Li et al., 2015b). For example, miR-122-5p was upregulated in infected chickens and was predicted to target the genes in the B cell receptor signaling pathway, whereas miR-122-5p was downregulated in ducks and predicted to target RASGRP3. In H5N3-infected chicken lungs, miR-1a, miR-206, miR-451, miR-125b, and miR-146a displayed significantly differentially expressed levels compared with the noninfected lungs (Wang et al., 2009, Wang et al., 2012).
Other viruses
IBDV, a member of the genus Avibirnavirus of the family Birnaviridae, is the etiological agent of infectious bursal disease, which is a highly contagious and immunosuppressive disease affecting young chickens (Brandt et al., 2001, Wang et al., 2007). The overexpression of gga-miR-9* was observed in IBDV-infected specific-pathogen-free chickens and significantly promoted IBDV replication by targeting interferon regulatory factor 2 (Ouyang et al., 2015). Similarly, the overexpression of miR-21 in chicken fibroblasts suppressed IBDV replication by downregulating IBDV VP1 expression (Wang et al., 2013b). Based on the genome-wide profiling of IBDV-infected chicken dendritic cells (DCs), Lin et al. identified 3 transcription factor-miRNA networks in IBDV-simulated DCs (CTCF-Let-7 g, CTCF-miR196-2, and CTCF-miR1635), in which let-7g and miR-1635 were upregulated in IBDV-infected samples and miR196-2 was downregulated (Lin et al., 2016).
IBV is a positive-sense RNA virus of the genus Gammacoronavirus that has extensive genetic diversity and a high mutation rate. IBV can cause pathological lesions in the respiratory tract, alimentary tract, kidney, oviduct, and gonads and is the pathogenic agent of avian infectious bronchitis (Cavanagh, 2007). To study the regulation of miRNAs in response to various virulent IBV infections, high-throughput sequencing was performed to determine the level of miRNA expression in the kidneys of noninfected chickens and those infected with attenuated IBV, normal virulent IBV, and highly virulent IBV (Yang et al., 2017). For example, when compared with noninfected groups, the expression of miR-202-5p, miR-222b-3p, and miR-6516-3p was upregulated in all of the 3 virally infected groups; however, miR-7b and miR-1454 were upregulated in the attenuated IBV and normal virulent IBV groups but downregulated in highly virulent IBV.
Rev-T is a highly oncogenic retrovirus that carries the viral oncogene, v-rel (Chen et al., 1981). High levels of miR-155 expression was observed in both Rev-T–infected chicken embryo fibroblast cultures and Rev-T–induced B-cell lymphomas and promoted cellular survival and transformation (Bolisetty et al., 2009, Yao et al., 2017).
To date, there are substantially fewer studies on the expression pattern and function of miRNAs in virus-infected ducks. Duck Tembusu virus (DTMUV) is a single-stranded positive RNA virus responsible for substantial economic losses in the duck industry since it was first isolated (Su et al., 2011). Cui et al. analyzed the miRNA expression profiles in DTMUV-infected and uninfected duck embryonic fibroblasts cells using high-throughput sequencing and validated 9 miRNAs that were differentially expressed in response to DTMUV infection (Cui et al., 2018). Duck embryonic fibroblasts infected with virulent duck enteritis virus, a member of the Herpesviridae family, displayed a unique pattern of viral miRNAs and a novel set of host miRNAs compared with other strains of duck enteritis virus–attenuated vaccines (Wu et al., 2018). The above deregulated miRNAs are listed in Table 4.
Table 4.
miRNAs that have been reported to have a regulatory role during IBDV, IBV, Rev-T, DTMUV, or DEV infection.
| miRNA | Virus | Species | Source | Pattern | Targets | Cell function | Reference |
|---|---|---|---|---|---|---|---|
| let-7g | IBDV | Chicken | Bone marrow dendritic cells | Up | - | - | Lin et al., 2016 |
| miR-1635 | IBDV | Chicken | Bone marrow dendritic cells | Up | - | - | Lin et al., 2016 |
| miR-196-2 | IBDV | Chicken | Bone marrow dendritic cells | Down | - | - | Lin et al., 2016 |
| miR-21 | IBDV | Chicken | Cell lines | Up | VP1 | Suppress replication of IBDV | Wang, Ouyang, Pan, Wang, Xia, Bi, Wang and Wang, 2013b |
| miR-9* | IBDV | Chicken | Cell lines | Up | IRF2 | Promote IBDV replication | Ouyang et al., 2015 |
| miR-1454 | IBV | Chicken | Kidney | Up/up/down | - | Associate with the virulence of IBV | Yang et al., 2017 |
| miR-7b | IBV | Chicken | Kidney | Up/up/down | - | Associate with the virulence of IBV | Yang et al., 2017 |
| miR-222b-3p | IBV | Chicken | Kidney | Up/up/up | - | Associate with the virulence of IBV | Yang et al., 2017 |
| miR-1563 | IBV | Chicken | Kidney | Down/up/up | - | Associate with the virulence of IBV | Yang et al., 2017 |
| miR-1723 | IBV | Chicken | Kidney | Down/up/down | - | Associate with the virulence of IBV | Yang et al., 2017 |
| miR-1782 | IBV | Chicken | Kidney | Up/down/down | - | Associate with the virulence of IBV | Yang et al., 2017 |
| miR-202-5p | IBV | Chicken | Kidney | Up/up/up | - | Associate with the virulence of IBV | Yang et al., 2017 |
| miR-6516-3p | IBV | Chicken | Kidney | Up/up/up | - | Associate with the virulence of IBV | Yang et al., 2017 |
| miR-155 | REV-T | Chicken | Cell lines | Up | JARID2 | Promotes cell survival | Bolisetty et al., 2009 |
| miR-155 | REV-T | Chicken | Cell lines | Up | - | - | Yao et al., 2017 |
| miR-7 | DTMUV | Duck | DEF | Up | - | - | Cui et al., 2018 |
| miR-133a-3p | DTMUV | Duck | DEF | Down | - | - | Cui et al., 2018 |
| miR-222a | DTMUV | Duck | DEF | Up | - | - | Cui et al., 2018 |
| miR-206 | DTMUV | Duck | DEF | Down | - | - | Cui et al., 2018 |
| miR-146b-5p | DTMUV | Duck | DEF | Up | - | - | Cui et al., 2018 |
| miR-1662 | DTMUV | Duck | DEF | Down | - | - | Cui et al., 2018 |
| miR-155 | DTMUV | Duck | DEF | Up | - | - | Cui et al., 2018 |
| miR-1a-3p | DTMUV | Duck | DEF | Down | - | - | Cui et al., 2018 |
| miR-221-3p | DTMUV | Duck | DEF | Up | - | - | Cui et al., 2018 |
| miR-133a-3p | DEV | Duck | DEF | Up | - | - | Wu et al., 2018 |
| let-7a-2-3p | DEV | Duck | DEF | Up | - | - | Wu et al., 2018 |
| miR-148a-5p | DEV | Duck | DEF | Up | - | - | Wu et al., 2018 |
| miR-34c-5p | DEV | Duck | DEF | Up | - | - | Wu et al., 2018 |
| miR-1a-3p | DEV | Duck | DEF | Up | - | - | Wu et al., 2018 |
| miR-20b-3p | DEV | Duck | DEF | Down | - | - | Wu et al., 2018 |
| miR-125-2-3p | DEV | Duck | DEF | Down | - | - | Wu et al., 2018 |
| miR-215-5p | DEV | Duck | DEF | Down | - | - | Wu et al., 2018 |
| miR-2954-5p | DEV | Duck | DEF | Down | - | - | Wu et al., 2018 |
| miR-29b-1-5p | DEV | Duck | DEF | Down | - | - | Wu et al., 2018 |
| miR-D25-5p | DEV | DEV | DEF | Up | - | - | Wu et al., 2018 |
| miR-D2-3p | DEV | DEV | DEF | Up | - | - | Wu et al., 2018 |
| miR-D26-5p | DEV | DEV | DEF | Up | - | - | Wu et al., 2018 |
| miR-D27-5p | DEV | DEV | DEF | Up | - | - | Wu et al., 2018 |
| miR-D28-3p | DEV | DEV | DEF | Up | - | - | Wu et al., 2018 |
| miR-D29-5p | DEV | DEV | DEF | Up | - | - | Wu et al., 2018 |
| miR-D30-3p | DEV | DEV | DEF | Up | - | - | Wu et al., 2018 |
| miR-D31-3p | DEV | DEV | DEF | Up | - | - | Wu et al., 2018 |
Abbreviations: DEF, duck embryonic fibroblasts; DEV, duck enteritis virus; DTMUV, Duck Tembusu virus; IBDV, infectious bursal disease virus; IBV, infectious bronchitis virus.
Pathways significantly enriched by the targets of dysregulated miRNAs during viral infection
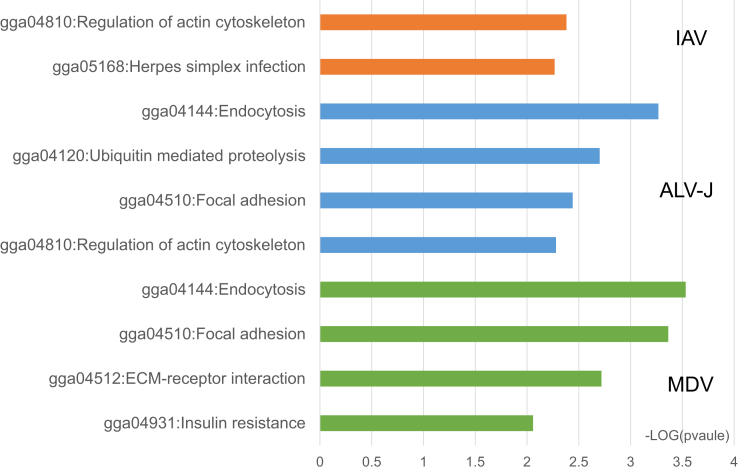
To help understand the mechanisms of how dysregulated miRNAs mediate virus-host interactions, we predicted the targets of the dysregulated miRNAs reported previously during MDV, ALV-J, and IAV infection by miRDB (Liu and Wang, 2019). An enrichment analysis was then subsequently performed by DAVID (Huang da, et al., 2009) to obtain Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways that were significantly enriched by the targets (P < 0.01). As shown in Figure 2, there were some common pathways activated under different viral infections. For example, regulation of the actin cytoskeleton was enriched by the targets of the dysregulated miRNAs during IAV or ALV-J infection, whereas focal adhesion and endocytosis were enriched during ALV-J or MDV infection. However, there are also specific pathways during each type of viral infection (e.g., herpes simplex infection for IAV infection, ubiquitin-mediated proteolysis for ALV-J infection, and insulin resistance and extracellular matrix-receptor interaction pathways for MDV infection). These results indicate that there may be common pathways that are influenced by dysregulated miRNAs during infections with different viruses. There may also be specific pathways activated in response to each viral infection. Thus, different mechanisms may be associated with each infection, which requires further research.
Figure 2.
Significant enriched KEGG pathways for the targets of reported dysregulated miRNAs when IAV, ALV-J, and MDV infection occurs. Abbreviations: ALV-J, avian leukosis virus subgroup J; ECM, extracellular matrix; IAV, influenza A virus; KEGG, Kyoto Encyclopedia of Genes and Genomes; MDV, Marek's disease virus.
Conclusions
miRNAs Regulate various biological processes within cells in poultry, including virus infections. Changes in the levels of poultry and viral miRNA expression after viral infection are clearly an important component of the cross-talk between the host and virus. In this review, we provide an extensive overview of the various viruses that infect poultry and involve miRNAs. However, we are just beginning to understand the underlying mechanisms of how the genes targeted by dysregulated miRNAs mediate virus-host interactions. It is important to note the following points: 1) Similar to MDV-infected chickens, some viruses encode their own miRNAs that are expressed during infection, while the host-encoded miRNAs simultaneously regulate viral genes. Moreover, viral miRNAs can also target host genes (Muylkens et al., 2010), which modify the cell environment during infection and add another layer of cross-talk between the host and virus. For future studies, it will be important to elucidate the molecular mechanisms behind these observations; 2) Coinfection of more than one type of virus in chicken flocks is common and increases the illness severity and threat against poultry health (Sun et al., 2017). The coinfection of viruses in chickens also make the cross-talk between host and virus even more complex. The roles of the miRNA systems in the context of coinfection and pathogenicity require further in-depth research; 3) Recent research has begun to focus on the application of miRNAs to the development of antiviral therapeutics for viruses that threaten human health (e.g., HIV and Ebola) (Janssen et al., 2013, Dunning et al., 2016). However, antiviral therapeutics based on miRNAs in poultry remain limited. Thus, future research tackling these questions are likely to emerge from this exciting area of investigation.
Acknowledgments
This research was funded by the National Natural Science Foundation of China (no.: 61602332, 31600671), and Jiangsu Agri-animal Husbandry Vocational College (NSFPT201727).
Conflicts of Interest: The authors declare no conflict of interest.
Contributor Information
Wenying Yan, Email: wyyan@suda.edu.cn.
Yang Yang, Email: yyang@suda.edu.cn.
References
- Asaf V.N., Kumar A., Raut A.A., Bhatia S., Mishra A. In-silico search of virus-specific host microRNAs regulating avian influenza virus NS1 expression. Theor. Biosci. 2015;134:65–73. doi: 10.1007/s12064-015-0211-9. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bolisetty M.T., Dy G., Tam W., Beemon K.L. Reticuloendotheliosis virus strain T induces miR-155, which targets JARID2 and promotes cell survival. J. Virol. 2009;83:12009–12017. doi: 10.1128/JVI.01182-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt M., Yao K., Liu M., Heckert R.A., Vakharia V.N. Molecular determinants of virulence, cell tropism, and pathogenic phenotype of infectious bursal disease virus. J. Virol. 2001;75:11974–11982. doi: 10.1128/JVI.75.24.11974-11982.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnside J., Bernberg E., Anderson A., Lu C., Meyers B.C., Green P.J., Jain N., Isaacs G., Morgan R.W. Marek's disease virus encodes MicroRNAs that map to meq and the latency-associated transcript. J. Virol. 2006;80:8778–8786. doi: 10.1128/JVI.00831-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnside J., Morgan R. Emerging roles of chicken and viral microRNAs in avian disease. BMC Proc. 2011;5(Suppl 4:):S2. doi: 10.1186/1753-6561-5-S4-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007;38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- Chen I.S., Mak T.W., O'Rear J.J., Temin H.M. Characterization of reticuloendotheliosis virus strain T DNA and isolation of a novel variant of reticuloendotheliosis virus strain T by molecular cloning. J. Virol. 1981;40:800–811. doi: 10.1128/jvi.40.3.800-811.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M., Jia R., Huang J., Wu X., Hu Z., Zhang X., Wang M., Zhu D., Chen S., Liu M., Zhao X., Wu Y., Yang Q., Zhang S., Liu Y., Zhang L., Yin Z., Jing B., Cheng A. Analysis of the microRNA expression profiles in DEF cells infected with duck Tembusu virus. Infect. Genet. Evol. 2018;63:126–134. doi: 10.1016/j.meegid.2018.05.020. [DOI] [PubMed] [Google Scholar]
- Dai Z., Ji J., Yan Y., Lin W., Li H., Chen F., Liu Y., Chen W., Bi Y., Xie Q. Role of gga-miR-221 and gga-miR-222 during tumour formation in chickens infected by subgroup J avian leukosis virus. Viruses. 2015;7:6538–6551. doi: 10.3390/v7122956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning J., Sahr F., Rojek A., Gannon F., Carson G., Idriss B., Massaquoi T., Gandi R., Joseph S., Osman H.K., Brooks T.J., Simpson A.J., Goodfellow I., Thorne L., Arias A., Merson L., Castle L., Howell-Jones R., Pardinaz-Solis R., Hope-Gill B., Ferri M., Grove J., Kowalski M., Stepniewska K., Lang T., Whitehead J., Olliaro P., Samai M., Horby P.W., team R.-T. t. Experimental Treatment of Ebola virus disease with TKM-130803: a single-Arm phase 2 Clinical Trial. PloS Med. 2016;13:e1001997. doi: 10.1371/journal.pmed.1001997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Yun B., Qin L., Pan W., Qu Y., Liu Z., Wang Y., Qi X., Gao H., Wang X. Molecular epidemiology of avian leukosis virus subgroup J in layer flocks in China. J. Clin. Microbiol. 2012;50:953–960. doi: 10.1128/JCM.06179-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- Han B., Lian L., Li X., Zhao C., Qu L., Liu C., Song J., Yang N. Chicken gga-miR-130a targets HOXA3 and MDFIC and inhibits Marek's disease lymphoma cell proliferation and migration. Mol. Biol. Rep. 2016;43:667–676. doi: 10.1007/s11033-016-4002-2. [DOI] [PubMed] [Google Scholar]
- Harwig A., Das A.T., Berkhout B. Retroviral microRNAs. Curr. Opin. Virol. 2014;7:47–54. doi: 10.1016/j.coviro.2014.03.013. [DOI] [PubMed] [Google Scholar]
- Hay A.J., Gregory V., Douglas A.R., Lin Y.P. The evolution of human influenza viruses. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001;356:1861–1870. doi: 10.1098/rstb.2001.0999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidari M., Delekta P.C. Transcriptomic analysis of host immune response in the skin of chickens infected with Marek's disease virus. Viral Immunol. 2017;30:377–387. doi: 10.1089/vim.2016.0172. [DOI] [PubMed] [Google Scholar]
- Hicks J.A., Trakooljul N., Liu H.C. Alterations in cellular and viral microRNA and cellular gene expression in Marek's disease virus-transformed T-cell lines treated with sodium butyrate. Poult. Sci. 2019;98:642–652. doi: 10.3382/ps/pey412. [DOI] [PubMed] [Google Scholar]
- Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Janssen H.L., Reesink H.W., Lawitz E.J., Zeuzem S., Rodriguez-Torres M., Patel K., van der Meer A.J., Patick A.K., Chen A., Zhou Y., Persson R., King B.D., Kauppinen S., Levin A.A., Hodges M.R. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- Jenner R.G., Young R.A. Insights into host responses against pathogens from transcriptional profiling. Nat. Rev. Microbiol. 2005;3:281–294. doi: 10.1038/nrmicro1126. [DOI] [PubMed] [Google Scholar]
- Kumar A., Vn M.A., Raut A.A., Sood R., Mishra A. Identification of chicken Pulmonary miRNAs targeting PB1, PB1-F2, and N40 genes of highly pathogenic avian influenza virus H5N1 in silico. Bioinform. Biol. Insights. 2014;8:135–145. doi: 10.4137/BBI.S14631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambeth L.S., Yao Y., Smith L.P., Zhao Y., Nair V. MicroRNAs 221 and 222 target p27Kip1 in Marek's disease virus-transformed tumour cell line MSB-1. J. Gen. Virol. 2009;90:1164–1171. doi: 10.1099/vir.0.007831-0. [DOI] [PubMed] [Google Scholar]
- Li H., Ji J., Xie Q., Shang H., Zhang H., Xin X., Chen F., Sun B., Xue C., Ma J., Bi Y. Aberrant expression of liver microRNA in chickens infected with subgroup J avian leukosis virus. Virus Res. 2012;169:268–271. doi: 10.1016/j.virusres.2012.07.003. [DOI] [PubMed] [Google Scholar]
- Li S., Kong L., Yu X., Zheng Y. Host-virus interactions: from the perspectives of epigenetics. Rev. Med. Virol. 2014;24:223–241. doi: 10.1002/rmv.1783. [DOI] [PubMed] [Google Scholar]
- Li X., Lian L., Zhang D., Qu L., Yang N. gga-miR-26a targets NEK6 and suppresses Marek's disease lymphoma cell proliferation. Poult. Sci. 2014;93:1097–1105. doi: 10.3382/ps.2013-03656. [DOI] [PubMed] [Google Scholar]
- Li Z.J., Zhang Y.P., Li Y., Zheng H.W., Zheng Y.S., Liu C.J. Distinct expression pattern of miRNAs in Marek's disease virus infected-chicken splenic tumors and non-tumorous spleen tissues. Res. Vet. Sci. 2014;97:156–161. doi: 10.1016/j.rvsc.2014.04.003. [DOI] [PubMed] [Google Scholar]
- Li Z., Chen B., Feng M., Ouyang H., Zheng M., Ye Q., Nie Q., Zhang X. MicroRNA-23b promotes avian leukosis virus subgroup J (ALV-J) replication by targeting IRF1. Sci. Rep. 2015;5:10294. doi: 10.1038/srep10294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Zhang J., Su J., Liu Y., Guo J., Zhang Y., Lu C., Xing S., Guan Y., Li Y., Sun B., Zhao Z. MicroRNAs in the immune organs of chickens and ducks indicate divergence of immunity against H5N1 avian influenza. FEBS Lett. 2015;589:419–425. doi: 10.1016/j.febslet.2014.12.019. [DOI] [PubMed] [Google Scholar]
- Li Z., Luo Q., Xu H., Zheng M., Abdalla B.A., Feng M., Cai B., Zhang X., Nie Q., Zhang X. MiR-34b-5p suppresses melanoma differentiation-associated gene 5 (MDA5) signaling pathway to promote avian leukosis virus subgroup J (ALV-J)-Infected cells Proliferaction and ALV-J replication. Front Cell Infect. Microbiol. 2017;7:17. doi: 10.3389/fcimb.2017.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian L., Zhang D., Wang Q., Yang N., Qu L. The inhibitory effects of gga-miR-199-3p, gga-miR-140-3p, and gga-miR-221-5p in Marek's disease tumorigenesis. Poult. Sci. 2015;94:2131–2135. doi: 10.3382/ps/pev175. [DOI] [PubMed] [Google Scholar]
- Lin J., Xia J., Zhang K., Yang Q. Genome-wide profiling of chicken dendritic cell response to infectious bursal disease. BMC Genomics. 2016;17:878. doi: 10.1186/s12864-016-3157-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Dai M., Zhang X., Cao W., Liao M. Subgroup J avian leukosis virus infection of chicken dendritic cells induces apoptosis via the aberrant expression of microRNAs. Sci. Rep. 2016;6:20188. doi: 10.1038/srep20188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Wang X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019;20:18. doi: 10.1186/s13059-019-1629-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan R., Anderson A., Bernberg E., Kamboj S., Huang E., Lagasse G., Isaacs G., Parcells M., Meyers B.C., Green P.J., Burnside J. Sequence conservation and differential expression of Marek's disease virus microRNAs. J. Virol. 2008;82:12213–12220. doi: 10.1128/JVI.01722-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muylkens B., Coupeau D., Dambrine G., Trapp S., Rasschaert D. Marek's disease virus microRNA designated Mdv1-pre-miR-M4 targets both cellular and viral genes. Arch. Virol. 2010;155:1823–1837. doi: 10.1007/s00705-010-0777-y. [DOI] [PubMed] [Google Scholar]
- Nejad C., Stunden H.J., Gantier M.P. A guide to miRNAs in inflammation and innate immune responses. FEBS J. 2018;285:3695–3716. doi: 10.1111/febs.14482. [DOI] [PubMed] [Google Scholar]
- Ouyang W., Wang Y.S., Du X.N., Liu H.J., Zhang H.B. gga-miR-9* inhibits IFN production in antiviral innate immunity by targeting interferon regulatory factor 2 to promote IBDV replication. Vet. Microbiol. 2015;178:41–49. doi: 10.1016/j.vetmic.2015.04.023. [DOI] [PubMed] [Google Scholar]
- Payne L.N., Brown S.R., Bumstead N., Howes K., Frazier J.A., Thouless M.E. A novel subgroup of exogenous avian leukosis virus in chickens. J. Gen. Virol. 1991;72(Pt 4):801–807. doi: 10.1099/0022-1317-72-4-801. [DOI] [PubMed] [Google Scholar]
- Payne L.N., Nair V. The long view: 40 years of avian leukosis research. Avian Pathol. 2012;41:11–19. doi: 10.1080/03079457.2011.646237. [DOI] [PubMed] [Google Scholar]
- Peiris M., Yuen K.Y., Leung C.W., Chan K.H., Ip P.L., Lai R.W., Orr W.K., Shortridge K.F. Human infection with influenza H9N2. Lancet. 1999;354:916–917. doi: 10.1016/s0140-6736(99)03311-5. [DOI] [PubMed] [Google Scholar]
- Peng X., Gao Q.S., Zhou L., Chen Z.H., Lu S., Huang H.J., Zhan C.Y., Xiang M. MicroRNAs in avian influenza virus H9N2-infected and non-infected chicken embryo fibroblasts. Genet. Mol. Res. 2015;14:9081–9091. doi: 10.4238/2015.August.7.17. [DOI] [PubMed] [Google Scholar]
- Powdrill M.H., Desrochers G.F., Singaravelu R., Pezacki J.P. The role of microRNAs in metabolic interactions between viruses and their hosts. Curr. Opin. Virol. 2016;19:71–76. doi: 10.1016/j.coviro.2016.07.005. [DOI] [PubMed] [Google Scholar]
- Rebmann T., Zelicoff A. Vaccination against influenza: role and limitations in pandemic intervention plans. Expert Rev. Vaccin. 2012;11:1009–1019. doi: 10.1586/erv.12.63. [DOI] [PubMed] [Google Scholar]
- Ren C., Yu M., Zhang Y., Fan M., Chang F., Xing L., Liu Y., Wang Y., Qi X., Liu C., Zhang Y., Cui H., Li K., Gao L., Pan Q., Wang X., Gao Y. Avian leukosis virus subgroup J promotes cell proliferation and cell cycle progression through miR-221 by targeting CDKN1B. Virology. 2018;519:121–130. doi: 10.1016/j.virol.2018.04.008. [DOI] [PubMed] [Google Scholar]
- Scaria V., Hariharan M., Pillai B., Maiti S., Brahmachari S.K. Host-virus genome interactions: macro roles for microRNAs. Cell Microbiol. 2007;9:2784–2794. doi: 10.1111/j.1462-5822.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- Shao W., Li X., Goraya M.U., Wang S., Chen J.L. Evolution of influenza A virus by mutation and Re-Assortment. Int. J. Mol. Sci. 2017;18:1650. doi: 10.3390/ijms18081650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalsky R.L., Cullen B.R. Viruses, microRNAs, and host interactions. Annu. Rev. Microbiol. 2010;64:123–141. doi: 10.1146/annurev.micro.112408.134243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J., Li S., Hu X., Yu X., Wang Y., Liu P., Lu X., Zhang G., Hu X., Liu D., Li X., Su W., Lu H., Mok N.S., Wang P., Wang M., Tian K., Gao G.F. Duck egg-drop syndrome caused by BYD virus, a new Tembusu-related flavivirus. PLoS One. 2011;6:e18106. doi: 10.1371/journal.pone.0018106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G.R., Zhang Y.P., Zhou L.Y., Lv H.C., Zhang F., Li K., Gao Y.L., Qi X.L., Cui H.Y., Wang Y.Q., Gao L., Pan Q., Wang X.M., Liu C.J. Co-infection with Marek's disease virus and reticuloendotheliosis virus increases illness severity and Reduces Marek's disease vaccine efficacy. Viruses. 2017;9:158. doi: 10.3390/v9060158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- tenOever B.R. RNA viruses and the host microRNA machinery. Nat. Rev. Microbiol. 2013;11:169–180. doi: 10.1038/nrmicro2971. [DOI] [PubMed] [Google Scholar]
- Tian F., Luo J., Zhang H., Chang S., Song J. MiRNA expression signatures induced by Marek's disease virus infection in chickens. Genomics. 2012;99:152–159. doi: 10.1016/j.ygeno.2011.11.004. [DOI] [PubMed] [Google Scholar]
- Wang Y., Brahmakshatriya V., Lupiani B., Reddy S.M., Soibam B., Benham A.L., Gunaratne P., Liu H.C., Trakooljul N., Ing N., Okimoto R., Zhou H. Integrated analysis of microRNA expression and mRNA transcriptome in lungs of avian influenza virus infected broilers. BMC Genomics. 2012;13:278. doi: 10.1186/1471-2164-13-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Brahmakshatriya V., Zhu H., Lupiani B., Reddy S.M., Yoon B.J., Gunaratne P.H., Kim J.H., Chen R., Wang J., Zhou H. Identification of differentially expressed miRNAs in chicken lung and trachea with avian influenza virus infection by a deep sequencing approach. BMC Genomics. 2009;10:512. doi: 10.1186/1471-2164-10-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Gao Y., Ji X., Qi X., Qin L., Gao H., Wang Y., Wang X. Differential expression of microRNAs in avian leukosis virus subgroup J-induced tumors. Vet. Microbiol. 2013;162:232–238. doi: 10.1016/j.vetmic.2012.10.023. [DOI] [PubMed] [Google Scholar]
- Wang Y.S., Ouyang W., Pan Q.X., Wang X.L., Xia X.X., Bi Z.W., Wang Y.Q., Wang X.M. Overexpression of microRNA gga-miR-21 in chicken fibroblasts suppresses replication of infectious bursal disease virus through inhibiting VP1 translation. Antivir. Res. 2013;100:196–201. doi: 10.1016/j.antiviral.2013.08.001. [DOI] [PubMed] [Google Scholar]
- Wang X., Wang H.K., Li Y., Hafner M., Banerjee N.S., Tang S., Briskin D., Meyers C., Chow L.T., Xie X., Tuschl T., Zheng Z.M. microRNAs are biomarkers of oncogenic human papillomavirus infections. Proc. Natl. Acad. Sci. U S A. 2014;111:4262–4267. doi: 10.1073/pnas.1401430111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.S., Wang Z.C., Tang Y.D., Shi Z.L., He K.W., Li Y., Hou J.B., Yao H.C., Fan H.J., Lu C.P. Comparison of four infectious bursal disease viruses isolated from different bird species. Arch. Virol. 2007;152:1787–1797. doi: 10.1007/s00705-007-1022-1. [DOI] [PubMed] [Google Scholar]
- Wu X., Jia R., Zhou J., Wang M., Chen S., Liu M., Zhu D., Zhao X., Sun K., Yang Q., Wu Y., Yin Z., Chen X., Wang J., Cheng A. Virulent duck enteritis virus infected DEF cells generate a unique pattern of viral microRNAs and a novel set of host microRNAs. BMC Vet. Res. 2018;14:144. doi: 10.1186/s12917-018-1468-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Yao Y., Zhao Y., Smith L.P., Baigent S.J., Nair V. Analysis of the expression profiles of Marek's disease virus-encoded microRNAs by real-time quantitative PCR. J. Virol. Methods. 2008;149:201–208. doi: 10.1016/j.jviromet.2008.02.005. [DOI] [PubMed] [Google Scholar]
- Xu K.M., Smith G.J., Bahl J., Duan L., Tai H., Vijaykrishna D., Wang J., Zhang J.X., Li K.S., Fan X.H., Webster R.G., Chen H., Peiris J.S., Guan Y. The genesis and evolution of H9N2 influenza viruses in poultry from southern China, 2000 to 2005. J. Virol. 2007;81:10389–10401. doi: 10.1128/JVI.00979-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Gao W., Liu H., Li J., Chen D., Yuan F., Zhang Z., Wang H. MicroRNA transcriptome analysis in chicken kidneys in response to differing virulent infectious bronchitis virus infections. Arch. Virol. 2017;162:3397–3405. doi: 10.1007/s00705-017-3502-2. [DOI] [PubMed] [Google Scholar]
- Yao Y., Vasoya D., Kgosana L., Smith L.P., Gao Y., Wang X., Watson M., Nair V. Activation of gga-miR-155 by reticuloendotheliosis virus T strain and its contribution to transformation. J. Gen. Virol. 2017;98:810–820. doi: 10.1099/jgv.0.000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Zhao Y., Xu H., Smith L.P., Lawrie C.H., Watson M., Nair V. MicroRNA profile of Marek's disease virus-transformed T-cell line MSB-1: predominance of virus-encoded microRNAs. J. Virol. 2008;82:4007–4015. doi: 10.1128/JVI.02659-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Xu H., Yao Y., Smith L.P., Kgosana L., Green J., Petherbridge L., Baigent S.J., Nair V. Critical role of the virus-encoded microRNA-155 ortholog in the induction of Marek's disease lymphomas. Plos Pathog. 2011;7:e1001305. doi: 10.1371/journal.ppat.1001305. [DOI] [PMC free article] [PubMed] [Google Scholar]