Abstract
Wooden breast (WB) results in significant losses to the broiler industry due to reductions in meat quality. While the etiology of WB is unknown, it is believed to be associated with localized hypoxia and decreased lactate levels in skeletal muscles, indicating the presence of altered lactate metabolism in WB. We hypothesized that the expression levels of the major signaling molecules that control lactate metabolism, including lactate dehydrogenases (LDHA and LDHB) and monocarboxylate transporters (MCT1 and MCT4), were altered in WB. Therefore, the objectives of this study were to evaluate whether there were changes in mRNA and protein levels of LDHA, LDHB, MCT1, and MCT4 in WB compared to normal breast (NB) muscles. Biochemical analysis for LDH enzyme activity in NB and WB muscles was studied. MicroRNA375 (miR-375) expression, known to be inversely associated with LDHB protein expression in human cells, was also investigated. The level of LDHA mRNA was 1.7-fold lower in WB tissues than in NB tissues (P < 0.0001). However, the LDHA protein levels were similar in WB and NB tissues. In contrast, the levels of LDHB mRNA and protein were 8.4-fold higher (P < 0.002) and 13.6-fold higher (P < 0.02) in WB than in NB tissues, respectively. The level of miR-375 was not different between WB and NB muscles. The specific LDH isoenzyme activity that converted lactate to pyruvate was 1.8-fold lower in WB compared to NB tissues (P < 0.01). The level of MCT1 mRNA was 2.3-fold higher in WB than those in NB muscles (P < 0.02). However, this upregulation was not observed with MCT1 protein expression levels. The expression levels of MCT4 mRNA and protein were elevated 2.8-fold (P < 0.02) and 3.5-fold (P < 0.004) in WB compared to NB tissues, respectively. Our current findings suggest the potential roles of LDHB and MCT4 on lactate metabolism and provide a unique molecular elucidation for altered lactate homeostasis in WB muscles of broilers.
Key words: wooden breast, lactate dehydrogenase, monocarboxylate transporters, LDH isoenzyme activity, broiler lactate metabolism
INTRODUCTION
Per capita consumption of broilers has increased 290% over the last 58 yr in the US (National Chicken Council, 2018). To meet the increasing demand for poultry meat, broilers have been selected based on fast growth rate, a high feed conversion ratio, and high meat yields (Zuidhof et al., 2014; Petracci et al., 2015; Tallentire et al., 2018). However, the fast-growing and heavy weights of broilers have increased the incidence of degenerative myopathies, severely affecting meat appearance and quality (Kuttappan et al., 2012; Tijare et al., 2016). One muscle myopathy is wooden breast (WB), which affects the pectoralis muscles in fast-growing commercial broilers (Sihvo et al., 2014). It is considered as one of the manifestations of metabolic syndrome in these fast-growing broilers, which makes the Pectoralis major muscle pale, rigid, and bulge outwards (Sihvo et al., 2014; Bailey et al., 2015; Tasoniero et al., 2016). Histologically, WB is characterized by myofiber necrosis, fibrosis, and high levels of fibrillar collagen in skeletal muscle tissues (Sihvo et al., 2014; Velleman and Clark, 2015; Kuttappan et al., 2016). Sihvo et al. (2014 and 2017) reported that WB starts to develop at approximately 2 wk of age in broiler chickens and estimated that 48 to 73% of commercial broilers had WB. The widespread incidence of WB has caused large economic losses due to poor meat quality, such as low water-holding capacity, reduced product yield, decreased myofibrillar protein content, and poor meat texture (Mazzoni et al., 2015; Mudalal et al., 2015; Aguirre et al., 2018). The causes of WB are still unknown, however, WB is believed to be caused by a combination of genetic and environmental factors (reviewed in Zampiga et al., 2018). Studies have suggested that WB in broilers is associated with genetic background (Macrae et al., 2006; Griffin et al., 2018), environmental factors (Bailey et al., 2015), WB-related gene changes associated localized hypoxia (Mutryn et al., 2015), inflammation and abnormal metabolisms (Abasht et al., 2016), increased oxidative stress (Mutryn et al., 2015; Abasht et al., 2016), and decreased capillary density (Hoving-Bolink et al., 2000).
Due to the presence of localized tissue hypoxia (Mutryn et al., 2015; Sihvo et al., 2018) and decreased lactate levels in WB-affected muscle (Abasht et al., 2016), we propose that the expression of key molecules, including lactate dehydrogenase (LDHA and LDHB) enzymes and 2 major monocarboxylate transporters (MCT1 and MCT4) that regulate the levels of lactate are altered in WB. Lactate dehydrogenase is a cytoplasmic enzyme found in nearly all living animal cells, composed of tetramers derived from two LDH subunits. It is encoded by the LDHA which converts pyruvate to lactate and LDHB gene which converts lactate to pyruvate in the last step of glycolysis (Cahn et al., 1962; Dawson et al., 1964; Pesce et al., 1964). The combination of LDHA and LDHB produce five LDH isoenzymes, categorized as LDH1 (B4), LDH2 (A1 B3). LDH3 (A2 B2), LDH4 (A3 B1), and LDH5 (A4) (Dawson et al., 1964). The altered level of LDH enzyme in human serum indicates damaged cells and tissues and has become a common marker for tissue injuries and disease (Drent et al., 1996; Kato et al., 2006; Ding et al., 2017). Lactates, the end product of anaerobic glycolysis, are transported by MCT1 and MCT4, which are located on the cell membrane. Translocation of lactate in skeletal muscles is facilitated by MCT1, which facilitates uptake of lactate into cells, and MCT4, which exports lactate out of cells (Roth and Brooks, 1990; Halestrap and Price, 1999; Halestrap and Meredith, 2004). Micro RNAs (miRNAs) are small no-coding RNAs that negatively regulate gene expression. Micro RNA-375 has been reported to be inversely associated with LDHB gene expression in cancer cells including human squamous cell carcinoma, esophageal squamous cancer cells, and merkel cell carcinoma (Kinoshita et al., 2012; Isozaki et al., 2012; Kumar et al., 2018). Therefore, the objectives of this study were to evaluate whether there were changes in the mRNA and protein levels of LDHA and LDHB, MCT2 and MCT4, and miR-375 in WB compared to normal breast (NB).
We report here in WB tissues, up-regulation of LDHB mRNA and protein that converts lactate to pyruvate was accompanied by an increase in MCT4 mRNA and protein expression, which act as a lactate efflux transporter. Further, we did not observe the inverse relationship of miR-375 with LDHB in WB tissue. Interestingly, the LDH isoenzyme activity that converts lactate to pyruvate was reduced in WB tissues compared to NB tissues. Future studies will involve isolating individual components of cells from WB tissues which will help us understand the cellular and molecular mechanisms behind WB muscles, leading to the development of new strategies for preventing and/or treating WB condition.
MATERIALS AND METHODS
Sample Collection
A total of 16 commercial high yielding broilers at 49 d of age were used to collect breast tissues, including 8 WB, which were determined independently by three experienced individuals who followed the methods described by Dalle Zotte et al. (2017), and 8 NB. Due to reports that a defective blood supply causes muscle damage and ischemia in the cranial section of WB muscles (Hoving-Bolink et al., 2000), we collected the tissue samples from the cranial part of the pectoralis major muscles. The breast tissue samples were dissected and immediately snap-frozen in liquid nitrogen. All tissues were then transferred and stored at −80°C for later total RNA and protein extractions. The animal use protocol was approved by Texas A&M University Animal Care and Use Committee.
Total RNA and Small RNA Extraction
Total RNA was extracted from NB and WB samples utilizing the RNeasy Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer's protocol. Briefly, a small piece of chicken breast was homogenized in buffer RLT using a GEX 130 PB ultrasonic processor (Cole-Parmer, Vernon Hills, IL, USA) at 50% power for 10 s on ice. The lysate was centrifuged at the speed of 2000 × g for 5 min, and the supernatant was collected. Then 1 volume of 70% ethanol was added to partially precipitate total RNA. The partially precipitated total RNA bonded to the RNeasy spin column, and treated with DNase to digest the contamination of genomic DNA. Buffer RPE was used to wash the column. The total RNA was eluted in 50 μL DNase/RNase free water. The quantity and quality of total RNA were evaluated through the NanoDrop 1000 (Thermo Scientific, MA, USA).
Small RNA isolation was performed using the mirVana PARIS kit (Ambion, Austin, TX). The procedure was followed according to the manufacturer's protocol. The concentration of small RNA was confirmed by an Agilent Small RNA kit on the Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA).
Quantitative Reverse Transcription Polymerase Chain Reaction for mRNAs
One microgram of total RNA from each sample was transcribed into cDNA using SuperScript III First-Strand Synthesis System (Invitrogen, Carlsbad, CA) following the manufacturer's protocol. Briefly, first strand cDNA synthesis was prepared by mixing 1 μg of total RNA, 1 μL of 50 ng/μl random hexamers, and 1 μL of 10 mM dNTPs mix. The mixture was incubated at 65°C for 5 min to denature RNA, and then placed on ice for at least 1 min. Ten microliters cDNA synthesis mix (2 μL 10 × RT Buffer, 4 μL 25 mM MgCl2, 2 μL 0.1 M DTT, 1 μL RNase OUT, and 1 μL SuperScript III RT) was added and incubated at 25°C for 10 min, followed by 50°C for 50 min, and terminated at 85°C for 5 min. One microliter RNase H (2 U/μL) was added and incubated at 37°C for 20 min. The cDNA product was stored at −20°C. The cDNA samples were diluted to the concentration of 1 ng/μL, and 8 μL of the diluted cDNA was subjected to a real-time polymerase chain reaction (PCR) reaction with PowerUp SYBR Green Master Mix (ThermoFisher Scientific, Waltham, MA) using an ABI 7900H system (ThermoFisher Scientific, Waltham, MA). The oligonucleotide primers for LDHA, LDHB, MCT1, MCT4, and GAPDH were listed in Table 1. The conditions of real time PCR amplification were 1 cycle at 50°C for 2 min, 95°C for 10 min, 40 cycles at 95°C for 15 s, 60°C for 1 min, followed by a melting curve analysis. The GAPDH was employed as an internal control used to normalize the expression of target transcript (Zambonelli et al., 2016). The values of average cycle threshold (Ct) were determined for each sample, and 2−ΔΔCt values for the comparison of the target gene and GAPDH were used for relative quantification by fold-change (Schmittgen and Livak, 2008). The PCR efficiency test was performed using serial dilutions of cDNA pool and primers. All reactions were performed in triplicates.
Table 1.
Primer Sequences used for qRT-PCR.
| Target gene | Primers | Primer Sequence (5′ to 3′) | Amplicons (bp) | Reference |
|---|---|---|---|---|
| LDHA | Forward | CCATGTCTCTCAAGGATCATCTC | 295 | Imagawa et al. (2006) |
| Reverse | GCACCAGCAGTGACAATGAC | |||
| LDHB | Forward | TTCCCAGCAACAAGATCACCGT | 511 | Imagawa et al. (2006) |
| Reverse | AACACCTGCCACATTAACTCCG | |||
| MCT1 | Forward | AGCAGCATCCTGGTGAACAAG | 59 | Zhang et al. (2017) |
| Reverse | AGGCACCCACCCACGAT | |||
| MCT4 | Forward | GCTGGTCTCAAGTGGGTTAG | 107 | Current study |
| Reverse | CCACCGTAATCGACAGACATAG | |||
| GAPDH | Forward | AGAACATCATCCCAGCGT | 182 | Guo et al. (2016) |
| Reverse | AGCCTTCACTACCCTCTTG |
Quantitative RT-PCR For miR-375
A total of 100 ng small RNA was reverse-transcribed for miRNA detection. Complementary DNA (cDNA) synthesis was performed using the miScript II reverse transcription (RT) kit (Catalog no. 218,161; Qiagen, Valencia, CA, USA) following the manufacturer's instruction. Briefly, the RT reaction was conducted in a 20 μL reaction mixture containing 100 ng of small RNA, 4 μL 5x miScript HiFlex Buffer, 2 μL 10x miScript Nucleics Mix and 2 μL miScript Reverse Transcriptase Mix. The reverse transcription was performed at 37°C for 60 min and 95°C for 5 min, and the cDNA product was stored at −20°C. A total of 200 pg of cDNA was used for quantitative RT-PCR reaction using the miScript SYBR Green PCR kit and Custom miScript Primer Assay for miR-375 (Catalog no. MSC0075989; Qiagen, Valencia, CA, USA) and U6 small nucleolar RNA (Catalog no. MSC0075901; Qiagen, Valencia, CA, USA). The following components were combined in a 10 μL reaction using 384-well plates: 5 μL 2x QuantiTect SYBR Green PCR Master Mix, 1 μL miScript Universal Primer, 1 μL of 10x miR-375 primer, and 200 pg of cDNA with nuclease-free water. The conditions of PCR were comprised of denaturation at 95°C for 10 min, followed by 40 cycles of 94°C for 15 s, 55°C for 30 s, and 70°C for 30 s, followed by the melting curve analysis. Data was analyzed using comparative delta delta Ct method (2−ΔΔCt) (Schmittgen and Livak, 2008). Relative miR-375 expression was normalized to U6 small nucleolar RNA. The efficiency test was performed using serial dilutions of cDNA pool and primers. Melting curve analyses were performed for validating the specificity of amplicons.
Western Blotting Analysis
Frozen tissues were homogenized in radioimmunoprecipitation assay buffer (Thermo Fisher Scientific, Waltham, MA). The protein concentration was quantified by a BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA). Ten micrograms of protein from each sample was loaded onto 4 to 12% SDS-PAGE gradient gels (Bio-Rad, Hercules, CA). After electrophoresis, proteins were transferred to PVDF membranes by a semi-dry electroblotting system (Cat. #: 170–3940; Bio-Rad, Hercules, CA) at 50 mA for 2 h. Then, the PVDF membranes were blocked in a Tris-buffered saline containing 0.1% Tween (TBST) and 5% non-fat milk overnight at 4°C. The membranes were washed 4 times for 20 min in TBST, and then incubated with primary antibody for 4 h. The following primary antibodies were purchased from Abcam, TLC (Cambridge, MA) unless indicated and used at the indicated dilutions: mouse anti-GAPDH (catalog no. ab8245; 1:20,000 dilution), rabbit anti-monocarboxylic acid transporter 1 (MCT1) (catalog no. ab93048; 1:1000 dilution), rabbit anti-LDHA (catalog no. ab135366; 1:1000 dilution), rabbit anti-SLC16A3 (MCT4) (catalog no. ab74109; 1:500 dilution), rabbit anti-LDHB (catalog no. ab75167; 1:1000 dilution). The membrane was washed 4 times in TBST, followed by incubation with a secondary antibody for 1 h at room temperature. The following secondary antibodies were used: HRP-conjugated goat anti-rabbit IgG (catalog no. ab205718; 1:20,000 dilution), HRP-conjugated goat anti-mouse IgG from Life Technology (Carlsbad, CA; catalog no. A16072; 1:5000 dilution). Enhanced chemiluminescence detection reagents (Amersham Pharmacia Biotech, Piscataway, NJ) were employed. The chemiluminescent signals were captured and analyzed by a ChemiDocTM MP Image System (Bio-Rad, Hercules, CA). Immunoblot images were quantified by Quantity One-4.6.1 software (Bio-Rad, Hercules, CA), and target protein signals were normalized to GAPDH and presented as relative band densities.
Specific LDH Enzyme Activity Assay
Specific LDH activity (conversion from lactate to pyruvate) was determined using a kit made in-house. Briefly, frozen breast muscle tissue was homogenized in the extraction buffer containing 50 mM of Tris-HCl (pH 8.5), 1 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride, and 1 mM ethylenediaminetetraacetic acid by a GEX 130 PB ultrasonic processor (Cole-Parmer, Vernon Hills, IL, USA) at 50% power for 10 s on ice. The lysate from each sample was centrifugated at 12,000 × g for 5 min at 4°C. The supernatant was decanted, and snap frozen in liquid nitrogen and stored at −80°C. The protein concentration for each sample was measured by BCA Protein Assay Kit. The protein samples were diluted to 1 μg in 30 μL extraction buffer and added to 200 μL LDH assay buffer containing 100 mM lithium lactate, 4 mM NAD+, 3 mM NaHCO3, 166.6 mM NaCl and 8.33 mM Tris-HCl (pH 8.8). The change of NADH absorbance was measured in 51-s intervals for 10 min at 340 nm absorbance at room temperature using a Synergy H1 multi-mode microplate reader (BioTek, Winooski, VT, USA). The specific activity of LDH was calculated as the ratio of the enzyme activity divided by the protein mass for enzymatic assay. One unit of LDH activity was defined as the amount of enzyme that yielded 1 μM NADH per min. Gen5 3.02 Kinetic v.3.5.1 program (BioTek, Winooski, VT) was used to measure enzyme kinetic activity. Lactate dehydrogenase activities were calculated in the formula below and expressed as units per gram of protein.
Statistical Analysis
Statistical analyses were conducted via a student's T test, using JMP Pro 14 (SAS institute Inc., Cary, NC, U.S.A.). All of the data were presented as mean ± standard error of the mean (SEM). A P-value of < 0.05 was considered significant.
RESULTS
Expression of the LDHA and LDHB mRNA and Protein in WB Muscles Compared to NB Muscles
Since skeletal muscle tissues are the major location for lactate production, we first investigated the impact of WB on expression levels of LDHA, which encodes the LDH muscle subunit (LDH M), that converts pyruvate to lactate in the last step of fermentative glycolysis (Summermatter et al., 2013). Relative LDHA mRNA levels decreased 1.7-fold (P < 0.0001) in WB tissues with respect to those from NB tissues (Figure 1A). In contrast, the LDHA protein levels in WB muscles showed no significant change (Figure 1 C and D). We assessed the mRNA and protein expression levels of LDHB, encoding LDH heart subunit (LDH H) that converts lactate to pyruvate (Dawson et al., 1964). Compared to NB, the mRNA level of LDHB was 8.4-fold higher (P < 0.002) in WB tissues (Figure 1B). Western blot analyses revealed that WB exhibited 13.6-fold higher (P < 0.02) LDHB protein levels compared to NB muscles (Figure 1C and E).
Figure 1.
The altered mRNA and protein levels of LDHA and LDHB in WB compared to NB tissues. (A) Relative LDHA mRNA expression in NB and WB tissues determined by qRT-PCR with normalization to the reference GAPDH mRNA levels. NB data is represented by the filled bar while the WB data is represented by the hatched bar. Data are represented as means ± SEM of 8 NB and 8 WB tissues. (B) Relative LDHB mRNA level, measured in the same experiment as in (A), was upregulated in WB tissues (N = 8) compared to NB tissue (N = 8). (C) Representative immunoblots showing protein expression levels of LDHA, LDHB, and GAPDH in lysates of NB and WB tissues. GAPDH band is shown as an internal control for sample loading. (D) The graph of immunoblot data shows that there is no significant difference of the levels of LDHA protein between NB (N = 8) and WB (N = 8) tissues. (E) Graphical representation of LDHB immunoblot data, showing greater levels of LDHB protein in WB tissue compared to NB tissue.
The Levels of miR-375 Expression in WB did not Differ from those of NB Tissues
Levels of miR-375 were reported to be inversely associated with LDHB protein levels in human squamous cell carcinoma (Kinoshita et al., 2012). To test whether the increased LDHB protein levels observed in WB tissue was mediated by miR-375, we performed qRT-PCR analysis on expression of miR-375 in both NB and WB muscles. As shown in Figure 2, WB muscles did not exhibit a significant reduction of miR-375 compared to NB tissues (P = 0.54).
Figure 2.
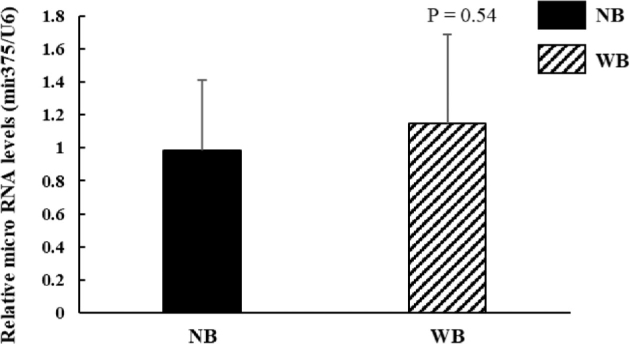
The levels of miR-375 expression in WB did not differ from those of NB tissues. Small RNAs from NB and WB were extracted using the mirVana PARIS kit as described in methods. Relative miR-375 expression was quantitated by qRT-PCR. Chicken U6 was used as an endogenous control for normalization. N = 8 per group.
WB has Decreased Specific LDH Enzymatic Activities that Convert Lactate to Pyruvate
Given the enhanced LDHB protein expression in WB tissues that converts lactate to pyruvate, we also investigated the enzymatic activity of LDH-mediated lactate-to-pyruvatee conversion. In contrast, the special enzymatic activity of LDH that converted lactate to pyruvate was 1.8-fold (P < 0.01) lower in WB tissues than NB tissues (Figure 3).
Figure 3.
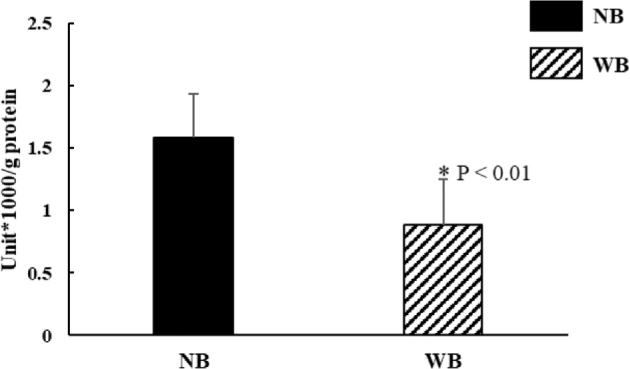
LDH enzyme activity that converts lactate to pyruvate is decreased in WB tissue compared to NB tissues. Results of LDH activity were normalized by using the same amount of total protein in the assay. The LDH enzyme activity of WB was significantly lower than NB. P < 0.05 is significant different, N = 8 per group.
Gene Expression of MCT1 and MCT4
Given the enhanced LDHB mRNA and protein levels in WB muscles compared to NB muscles, we also investigated the mRNA and protein expression of MCT1 and MCT4, which are implicated in lactate import and export in muscles, respectively (Juel and Halestrap, 1999; Halestrap and Meredith, 2004; Hashimoto et al., 2006; Ullah et al., 2006). We found that the relative mRNA levels of MCT1 and MCT4 were 2.3-fold and 2.8-fold higher in WB muscles than those in NB muscles (P < 0.02), respectively (Figure 4A and B). However, the protein level of MCT1 in WB was not different from that in NB tissues (Figure 4D). Consistent with the up-regulated MCT4 mRNA levels, the expression levels of MCT4 protein were elevated (3.5-fold, P < 0.004) in WB tissues compared to NB tissues (Figure 4 C and E).
Figure 4.
The altered mRNA and protein levels of MCT1 and MCT4 in WB compared to NB tissues. Relative levels of MCT1 mRNA (A) and MCT4 mRNA (B) in NB and WB tissues determined by qRT-PCR. (C) Western blot analyses were performed on breast muscle lysates prepared from NB and WB tissues. Protein sample (10 μ g) was loaded in each lane. The GAPDH band was shown as internal control for sample loading. (D) Quantification of MCT1 protein level in NB and WB tissues. The protein levels of MCT1 in WB were not different from those in NB tissues. (E) The levels of MCT4 protein were 3.5-fold in WB compared to NB tissues. N = 8 per group.
DISCUSSION
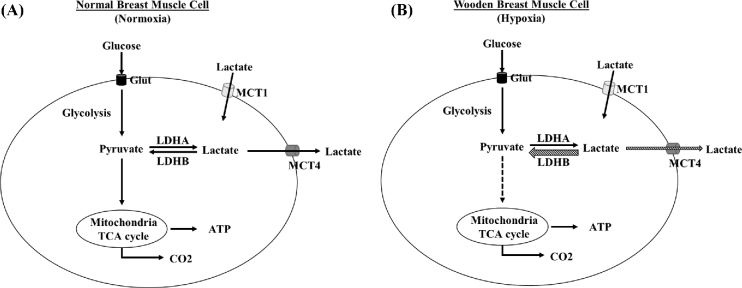
WB myopathy causes notable economic loss in the poultry industry and occurs in about 48% to 73% of commercial broilers (Sihvo et al., 2014). The molecular mechanisms that cause WB myopathy remain unclear. The presence of localized tissue hypoxia (Mutryn et al., 2015; Sihvo et al., 2018) and decreased lactate concentrations in WB (Abasht et al., 2016) indicate the potential alteration of genes involved in lactate metabolism. Identification of specific molecules that regulate lactate levels will improve our understanding of the mechanisms causing WB myopathy and potentially facilitate us to the development of useful therapeutic targets to eliminate the incidence of WB in broilers. In this study, we tested the hypothesis that the gene expression of the LDHA, LDHB, MCT1, and MCT4, involved in lactate metabolism, in WB muscles would change compared to their levels in NB muscles. Through a combination of qRT-PCR and immunoblotting assays, we have now shown that WB tissues indeed show significant up-regulation of both LDHB and MCT4 mRNA and protein levels, 2 key players that decrease lactate levels in WB tissues by oxidizing lactate to pyruvate and enhancing lactate efflux, separately (Figure 5).
Figure 5.
Schematic representation of the lactate metabolism pathway of broiler muscle affected with wooden breast (WB) myopathy (B) compared to normal breast muscle (A). Glucose is transported to the cell cytosol by the glucose transporter (Glut), and then is converted to pyruvate, the end product of glycolysis. Under normoxic conditions (Figure 5A), pyruvate enters the mitochondria and produces 38 ATPs per oxidized glucose molecule, including glycolysis, tricarboxylic acid cycle (TCA cycle), and oxidative phosphorylation. In WB muscle, oxygen delivery is limited (hypoxia, Figure 5B). The activity of TCA cycle is decreased and the pyruvate switches to the fermentation pathway, resulting in the production of lactate by the action of lactate dehydrogenase A (LDHA). The lactate can be re-oxidized to pyruvate by LDHB activity, or it can be transported out of the cell by the monocarboxylate transporter 4 (MCT4). On the other hand, MCT1 uptakes the lactate from the circulatory system into cells. Hatched arrows denote the possible fates for lactate in WB cells. In this model, increased levels of LDHB and MCT4 are the principal proteins that regulate lactate concentrations in the WB cells (Figure 5B). Glut: glucose transporter. LDH: Lactate dehydrogenase. MCT: Monocarboxylate transporter. TCA: tricarboxylic acid.
Although transcript levels of 2 genes, such as LDHA and MCT1, were downregulated and upregulated separately in WB muscles, we did not observe a concomitant reduction of LDHA or increase of MCT1 protein abundances. Both protein levels of LDHA and MCT1 did not change in WB tissues compared to NB tissues, even though their transcript levels significantly differed in WB muscles. Numerous reports have indicated that about 30 to 40% of the changes in mRNA levels are not correlated to the corresponding protein levels due to multiple factors, including differential turnover rates of transcripts or the presence of post-transcriptional or post-translational control mechanisms (Vogel and Marcotte, 2012; Payne, 2015; Karbownik et al., 2016).
High-throughput technologies such as RNA-seq (Mutryn et al., 2015; Papah et al., 2018; Marchesi et al., 2019) and proteomics (Kuttappan et al., 2017; Cai et al., 2018) have been used to identify differentially expressed molecular components in WB. In accordance with our report, Marchesi et al. (2019) showed the upregulation of LDHB mRNA levels and the reduction of LDHA mRNA levels in a different skeletal muscle myopathy known as white striping. However, there are several conflicting reports describing LDHA protein expression in WB tissues (Soglia et al., 2016; Zambonelli et al., 2016; Kuttappan et al., 2017; Cai et al., 2018). Lactate dehydrogenase A protein levels of WB muscles have been reported to be decreased 2-fold compared to the NB in a proteomic study (Kuttappan et al., 2017). The combination of 2-dimensional gel electrophoresis and mass spectrometry by Cai et al. (2018) only identified 8 differentially expressed proteins in WB compared to NB muscles, and LDH was not one of them. In fact, sarcoplasmic LDH protein levels in WB have been reported to be higher (ranging from 14 to 22%) in WB than that of NB using an SDS-PAGE method (Soglia et al. 2016; Zambonelli et al., 2016). The positive identification of LDH protein in Soglia et al. (2016) and Zambonelli et al. (2016) was confirmed by expected molecular weight of the protein in SDS-PAGE gel, although it is not known which LDH subunit was involved since immunoblotting methods using specific antibodies were not employed. This discrepancy about the levels of LDHA protein expression could reflect differences in the choice of sample extraction, preparation, and separation methods.
To our knowledge, this is the first report of up-regulation of LDHB protein expression in WB muscles. We confirmed more than 13-fold higher LDHB protein expression in WB tissues than those in NB tissues using western blot analysis. Recent studies on WB have indicated the changes in several glycolytic proteins (Soglia et al., 2016; Zambonelli et al., 2016; Cai et al., 2018). However, none of the studies found the dramatic change in levels of LDHB protein in WB. These studies provide potential candidate signaling molecules and pathways in WB, however, the challenges with finding a low-abundance of regulatory protein remains to be solved by classical proteomics (Cho, 2007; Chandramouli and Qian, 2009). In fact, LDHA is the most abundant subtype of LDH in pectoralis major muscles of chickens (Dawson et al., 1964; Heinova et al., 1999). Lactate dehydrogenase B, a very low-abundance protein in pectoralis major muscles, may not be detectable without proper pretreatment and handling (Andrecht and Hagen, 2008). Using an immunoblotting method with the specific antibody has enabled us to detect the low-concentration of LDHB in our study.
Despite the lower abundance of LDHB protein compared to LDHA protein in normal skeletal muscle, it is not clear why LDHB protein levels were upregulated more than 13-fold in WB muscles compared to NB muscles. One possible mechanism for upregulating LDHB in WB tissue involves the conversion of lactate to pyruvate. To investigate if LDHB decreases lactate levels in WB, we mined published metabolomic datasets from well-characterized WB and NB of different broiler breeds (Supplementary 2 files in Abasht et al., 2016). We found that upregulation of LDHB protein levels in WB is correlated with the reduction of lactate levels. The WB muscles showed 0.66 (P = 0.17), 0.72 (P < 0.002), and 0.56-fold (P < 0.005) reduction in lactate levels from two genetically distinct purebred lines and a commercial breed line, respectively. These data suggest that elevated LDHB protein expression in WB muscles (this study) may contribute to decreased levels of lactate in WB muscles from different broiler breeds (Abasht et al., 2016). The reduction of lactate contents is correlated with upregulation of LDHB gene expression in WB muscle cells, resulting in an increase in cellular pH value. Tasoniero et al. (2016) reported that the breasts with both white striping and WB exhibited the highest pH values than the normal and white striping groups. Various studies also confirmed increased pH values in WB muscles (Petracci et al., 2013; Dalle Zotte et al., 2014, 2017; Mudalal et al., 2015; Zambonelli et al., 2016). The other possibility for up-regulation of LDHB is through the downregulation of micro RNA, miR-375, which is involved in regulating lactate metabolism in several cancer cells including skin, lung, rectal, and gastric cancers by targeting the LDHB gene (reviewed in Haziapostolou et al., 2013). Isozaki et al. (2012) previously revealed that there was an inverse relationship between miR-375 and LDHB using the esophageal squamous cancer cells. In addition, Kinoshita et al. (2012) also found the inverse correlation of miR-375 and LDHB with human squamous cell carcinoma. In the present study, we found that WB muscles had a higher LDHB mRNA expression and the similar expression pattern of miR-375 compared to NB muscles, which is not consistent with previous reports of inverse relationship of LDHB and miR-375 from human studies. This indicates that other miRNAs may be involved directly targeting LDHB mRNA in WB tissues. We are currently investigating this issue.
There is limited information regarding the distribution of isoenzymes of LDH in adult chicken breast tissue. As reviewed by Gallo et al. (2015), LDH isoforms are present in all tissues at different ratios, so that the isoenzymatic profile is tissue-specific. The compositions of LDH isoenzymes in chicken breast muscles have been reported to be regulated depending on the developmental stages (Lindsay, 1963) and pathological conditions (Kaplan and Cahn, 1962). Kaplan and Cahn (1962) reported the complete changes in LDH isoform enzymes in the chicken breasts from LDH1 (four LDHB subunits) in early embryos to the LDH5 (all 4 LDHA subunits) in adult chicken. In contrast, Lindsey (1963) indicated the presence of small amounts of LDH4 (3A1B) and that there may be undetectable levels of LDH3 (2A2B) in adult chicken breast muscles due to the poor sensitivity of the method they used. We studied LDH isoenzyme patterns of WB and NB tissues using an electrophoresis of native polyacrylamide gel, which have LDH5 almost exclusively, but levels of LDH5 isoenzyme between WB and NB tissues were undistinguishable (Data not shown). Similar results were reported by Dawson et al. (1964), with a different unknown breed of chicken, indicated that LDH5 enzymatic activity was at least 4,300 times greater than LDH1 in breast muscles. We subsequently evaluated the specific isoenzyme activity of LDH-mediated lactate to pyruvate and found that it was significantly reduced in WB muscles compared to the NB muscles, which agreed with Wieme and Herpol's (1962) report that total LDH activity was decreased in dystrophic human and chicken muscles (Wilson et al., 1988). However, the organization of the LDH subunits in the chicken dystrophic skeletal muscle cells have been changed, resulting in the up-regulation of LDH1 (four LDHB subunits) and the reduction of LDH5 (all four LDHA subunits) compared to NB muscles (Kaplan and Cahn, 1962). The majority of studies of LDH enzyme activity in WB muscles of broilers have been focused on the changes of the enzyme activity of LDH-mediating pyruvate-to-lactate conversion in serum or plasma from broilers affected by breast muscle myopathy. For example, serum LDH activity in white striping breast muscles of broilers was elevated (Kuttappan et al., 2013). Recent studies suggested the association of increased plasma total LDH enzyme activity with the severity of WB (Meloche et al., 2018a and b). However, the distribution of LDH isoenzymes in broiler blood are totally different from those in breast muscles, containing mixing populations of LDH isoenzymes (66%:23%:6%:5%:3% of LDH5, LDH4, LDH3, LDH2, and LDH1) in serum and an exclusive LDH5 in chicken breast muscles, separately (Heinova et al., 1999). Our data suggests that broiler breast muscles contain a majority of LDH5 isoenzyme (all four LDHA subunits) and a very small amount of isoenzyme containing LDHB subunits that function in oxidizing lactate to pyruvate.
In skeletal muscles, MCT1 and MCT4 are the 2 major MCT isoforms that help the import and export lactate through the cell membrane, respectively (Garcia et al., 1994; Wilson et al., 1998). Little is known about differences of MCT1 and MCT4 protein expression in WB tissues of broilers compared to NB tissues. We found mRNA expression of MCT1 was higher in WB muscles compared to NB muscles. However, the protein levels of MCT1 were not different between WB and NB tissues. In humans, MCT1 gene expression increased in muscles after short-term training (Bonen et al., 1998). Short-term training induced fermentative glycolysis in the skeletal muscle, which is similar to hypoxia in broilers with WB myopathy. Monocarboxylate transporter 4 is highly expressed in white muscles and plays an important role exporting lactate out of cells (Dimmer et al., 2000). Chronic hypoxia has been reported to enhance protein levels of MCT4, not MCT1, in rat skeletal muscles (Py et al., 2005). This selection of upregulation of MCT4 protein, not MCT1 in WB muscles, allows the increased export of lactate out of muscle cells during hypoxia conditions.
The findings of this study present a novel mechanism in which WB muscles of broilers exhibit the reduction of lactate contents by upregulating both LDHB and MCT4 mRNA and protein levels. Further studies are needed to determine the association between LDHB and WB development and whether high expression of LDHB in WB tissues can be a prognostic biomarker for WB myopathy. Development of agents that target LDHB and MCT4 protein may provide a potential therapeutic treatment for reducing the incidence of WB myopathy.
ACKNOWLEDGEMENTS
The authors thank the Agriculture Women Excited to Share Opinions, Mentoring and Experiences (AWESOME) faculty group in the College of Agriculture and Life Sciences at Texas A&M University for assistance with editing the manuscript.
REFERENCES
- Abasht B., Mutryn M.F., Michalek R.D., Lee W.R. Oxidative stress and metabolic perturbations in wooden breast disorder in chickens. PLoS One. 2016;11 doi: 10.1371/journal.pone.0153750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre M.E., Owens C.M., Miller R.K., Alvarado C.Z. Descriptive sensory and instrumental texture profile analysis of woody breast in marinated chicken. Poult. Sci. 2018;97:1456–1461. doi: 10.3382/ps/pex428. [DOI] [PubMed] [Google Scholar]
- Andrecht S., von Hagen . In: General aspects of sample preparation for comprehensive proteomics analysis in proteomic sample preparation. von Hagen J., editor. WILEY-VCH Verlag; Weinheim: 2008. pp. 5–20. [Google Scholar]
- Bailey R.A., Watson K.A., Bilgili S.F., Avendano S. The genetic basis of pectoralis major myopathies in modern broiler chicken lines. Poult. Sci. 2015;94:2870–2879. doi: 10.3382/ps/pev304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonen A., McCullagh K.J., Putman C.T., Hultman E., Jones N.L., Heigenhauser G.J. Short-term training increases human muscle MCT1 and femoral venous lactate in relation to muscle lactate. Am. J. Physiol. Endocrinol. Metab. 1998;274:102–107. doi: 10.1152/ajpendo.1998.274.1.E102. [DOI] [PubMed] [Google Scholar]
- Cahn R.D., Zwilling E., Kaplan N.O., Levine L. Nature and development of lactic dehydrogenase: the two major types of this enzyme from molecular hybrids which change in makeup during development. Science. 1962;136:962–969. doi: 10.1126/science.136.3520.962. [DOI] [PubMed] [Google Scholar]
- Cai K., Shao W., Chen X., Campbell Y.L., Nair M.N., Suman S.P., Beach C.M., Guyton M.C., Schilling M.W. Meat quality traits and proteome profile of woody broiler breast (pectoralis major) meat. Poult. Sci. 2018;97:337–346. doi: 10.3382/ps/pex284. [DOI] [PubMed] [Google Scholar]
- Chandramouli K., Qian P.Y. Proteomics: challenges, techniques and possibilities to overcome biological sample complexity. Hum Genomics Proteomics. 2009;2009 doi: 10.4061/2009/239204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W.C. Proteomics technologies and challenges. Genomics Proteomics Bioinformatics. 2007;5:77–85. doi: 10.1016/S1672-0229(07)60018-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalle Zotte A., Cecchinato M., Quartesan A., Bradanovic J., Tasoniero G., Puolanne E. How does “Wooden Breast” myodegeneration affect poultry meat quality? 60th Int. Congr. Meat Sci. Technol., Punta Del Este, Uruguay. 2014:476–479. [Google Scholar]
- Dalle Zotte A., Tasoniero G., Puolanne E., Remignon H., Cecchinato M., Catelli E., Cullere M. Effect of “Wooden Breast” appearance on poultry meat quality, histological traits, and lesions characterization. Czech J. Anim. Sci. 2017;62:51–57. [Google Scholar]
- Dawson D.M., Goodfriend T.L., Kaplan N.O. Lactic dehydrogenases: functions of the two types rates of synthesis of the two majorforms can be correlated with metabolic differentiation. Science. 1964;143:929–933. doi: 10.1126/science.143.3609.929. [DOI] [PubMed] [Google Scholar]
- Dimmer K.S., Friedrich B., Lang F., Deitmer J.W., Bröer S. The low-affinity monocarboxylate transporter MCT4 is adapted to the export of lactate in highly glycolytic cells. Biochem J. 2000;350:219–227. [PMC free article] [PubMed] [Google Scholar]
- Ding J., Karp J.E., Emadi A. Elevated lactate dehydrogenase (LDH) can be a marker of immune suppression in cancer: Interplay between hematologic and solid neoplastic clones and their microenvironments. Cancer Biomark. 2017;19:353–363. doi: 10.3233/CBM-160336. [DOI] [PubMed] [Google Scholar]
- Drent M., Cobben N.A., Henderson R.F., Wouters E.F., van Dieijen-Visser M. Usefulness of lactate dehydrogenase and its isoenzymes as indicators of lung damage or inflammation. Eur. Respir. J. 1996;9:1736–1742. doi: 10.1183/09031936.96.09081736. [DOI] [PubMed] [Google Scholar]
- Gallo M., Sapio L., Spina A., Naviglio D., Calogero A., Naviglio S. Lactic dehydrogenase and cancer: an overview. Front Biosci. (Landmark Ed) 2015;20:1234–1249. doi: 10.2741/4368. [DOI] [PubMed] [Google Scholar]
- Garcia C.K., Goldstein J.L., Pathak R.K., Anderson R.G., Brown M.S. Molecular characterization of a membrane transporter for lactate, pyruvate, and other monocarboxylates: implications for the Cori cycle. Cell. 1994;76:865–873. doi: 10.1016/0092-8674(94)90361-1. [DOI] [PubMed] [Google Scholar]
- Griffin J.R., Moraes L., Wick M., Lilburn M.S. Onset of white striping and progression into wooden breast as defined by myopathic changes underlying Pectoralis major growth. Estimation of growth parameters as predictors for stage of myopathy progression. Avian Pathol. 2018;47:2–13. doi: 10.1080/03079457.2017.1356908. [DOI] [PubMed] [Google Scholar]
- Guo Y., Zhao P., Guo G., Hu Z., Tian L., Zhang K., Sun Y., Zhang X., Zhang W., Xing M. Effects of arsenic trioxide exposure on heat shock protein response in the immune organs of chickens. Biol. Trace Elem. Res. 2016;169:134–141. doi: 10.1007/s12011-015-0389-1. [DOI] [PubMed] [Google Scholar]
- Halestrap A.P., Meredith D. The SLC16 gene family—from monocarboxylate transporters (MCTs) to aromatic amino acid transporters and beyond. Pflügers Archiv. 2004;447:619–628. doi: 10.1007/s00424-003-1067-2. [DOI] [PubMed] [Google Scholar]
- Halestrap A.P., Price N.T. The proton-linked monocarboxylate transporter (MCT) family: structure, function and regulation. Biochem. J. 1999;343:281–299. [PMC free article] [PubMed] [Google Scholar]
- Hatziapostolou M., Polytarchou C., Iliopoulos D. miRNA link metabolic reprogramming to oncogenesis. Trends Endocrinol. Metab. 2013;24:361–373. doi: 10.1016/j.tem.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Hashimoto T., Hussien R., Brooks G.A. Colocalization of MCT1, CD147, and LDH in mitochondrial inner membrane of L6 muscle cells: evidence of a mitochondrial lactate oxidation complex. Am. J. Physiol. Endocrinol. Metab. 2006;290:1237–1244. doi: 10.1152/ajpendo.00594.2005. [DOI] [PubMed] [Google Scholar]
- Heinova D., Rosival I., Avidar Y., Bogin E. Lactate dehydrogenase isoenzyme distribution and patterns in chicken organs. Res. in Vet. Sci. 1999;67:309–312. doi: 10.1053/rvsc.1999.0317. [DOI] [PubMed] [Google Scholar]
- Hoving-Bolink A.H., Kranen R.W., Klont R.E., Gerritsen C.L.M., de Greef K.H. Fibre area and capillary supply in broiler breast muscle in relation to productivity and ascites. Meat Sci. 2000;56:397–402. doi: 10.1016/s0309-1740(00)00071-1. [DOI] [PubMed] [Google Scholar]
- Imagawa T., Yamamoto E., Sawada M., Okamoto M., Uehara M. Expression of lactate dehydrogenase-A and -B messenger ribonucleic acids in chick glycogen body. Poult. Sci. 2006;85:1232–1238. doi: 10.1093/ps/85.7.1232. [DOI] [PubMed] [Google Scholar]
- Isozaki Y., Hoshino I., Nohata N., Kinoshita T., Akutsu Y., Hanari N., Mori M., Yoneyama Y., Akanuma N., Takeshita N., Maruyama T., Seki N., Nishino N., Yoshida M., Matsubara H. Identification of novel molecular targets regulated by tumor suppressive miR-375 induced by histone acetylation in esophageal squamous cell carcinoma. Int. J. Oncol. 2012;41:985–994. doi: 10.3892/ijo.2012.1537. [DOI] [PubMed] [Google Scholar]
- Juel C., Halestrap A.P. Lactate transport in skeletal muscle - role and regulation of the monocarboxylate transporter. J. Physiol. 1999;517:633–642. doi: 10.1111/j.1469-7793.1999.0633s.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbownik M.S., Szemraj J., Wieteska Ł., Antczak A., Górski P., Kowalczyk E., Pietras T. Antipsychotic drugs differentially affect mRNA expression of genes encoding the neuregulin 1-downstream ErbB4-PI3K pathway. Pharmacology. 2016;98:4–12. doi: 10.1159/000444534. [DOI] [PubMed] [Google Scholar]
- Kaplan N.O., Cahn R.D. Lactic dehydrogenases and muscular dystrophy in the chicken. Proc. Nat. Acad. Sci. 1962;48:2123–2130. doi: 10.1073/pnas.48.12.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato G.J., McGowan V., Machado R.F., Little J.A., Taylor J.V.I., Morris C.R., Nichols J.S., Wang X., Poljakovic M., Morris S.M., Jr, Gladwin M.T. Lactate dehydrogenase as a biomarker of hemolysis-associated nitric oxide resistance, priapism, leg ulceration, pulmonary hypertension, and death in patients with sickle cell disease. Blood J. 2006;107:2279–2285. doi: 10.1182/blood-2005-06-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T., Nohata N., Yoshino H., Hanazawa T., Kikkawa N., Fujimura L., Chiyomaru T., Kawakami K., Enokida H., Nakagawa M., Okamoto Y., Seki N. Tumor suppressive microRNA-375 regulates lactate dehydrogenase B in maxillary sinus squamous cell carcinoma. Int. J. Oncol. 2012;40:185–193. doi: 10.3892/ijo.2011.1196. [DOI] [PubMed] [Google Scholar]
- Kumar S., Xie H., Scicluna P., Lee L., Björnhagen V., Höög A., Larsson C., Lui W.O. MiR-375 regulation of LDHB plays distinct roles in polyomavirus-positive and -negative Merkel cell carcinoma. Cancers (Basel) 2018;10:E443. doi: 10.3390/cancers10110443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuttappan V.A., Brewer V.B., Apple J.K., Waldroup P.W., Owens C.M. Influence of growth rate on the occurrence of white striping in broiler breast fillets. Poult. Sci. 2012;91:2677–2685. doi: 10.3382/ps.2012-02259. [DOI] [PubMed] [Google Scholar]
- Kuttappan V.A., Hargis B.M., Owens C.M. White striping and woody breast myopathies in the modern poultry industry: a review. Poult. Sci. 2016;95:2724–2733. doi: 10.3382/ps/pew216. [DOI] [PubMed] [Google Scholar]
- Kuttappan V.A., Bottje W., Ramnathan R., Hartson S.D., Coon C.N., Kong B.W., Owens C.M., Vazquez-Añon M., Hargis B.M. Proteomic analysis reveals changes in carbohydrate and protein metabolism associated with broiler breast myopathy. Poult. Sci. 2017;96:2992–2999. doi: 10.3382/ps/pex069. [DOI] [PubMed] [Google Scholar]
- Kuttappan V.A., Huff G.R., Huff W.E., Hargis B.M., Apple J.K., Coon C., Owens C.M. Comparison of hematologic and serologic profiles of broiler birds with normal (NORM) and severe (SEV) degrees of WS in breast fillets. Poult. Sci. 2013;92:339–345. doi: 10.3382/ps.2012-02647. [DOI] [PubMed] [Google Scholar]
- Lindsay D.T. Isozyme patterns and properties of lactate dehydrogenase from developing tissues of the chicken. J. Exp. Zool. 1963;152:75–89. doi: 10.1002/jez.1401520108. [DOI] [PubMed] [Google Scholar]
- Macrae V.E., Mahon M., Gilpin S., Sandercock D.A., Mitchell M.A. Skeletal muscle fibre growth and growth associated myopathy in the domestic chicken (Gallus domesticus) Br. Poult. Sci. 2006;47:264–272. doi: 10.1080/00071660600753615. [DOI] [PubMed] [Google Scholar]
- Marchesi J.A.P., Ibelli A.M.G., Peixoto J.O., Cantão M.E., Pandolfi J.R.C., Marciano C.M.M., Zanella R., Settles M.L., Coutinho L.L., Ledur M.C. Whole transcriptome analysis of the pectoralis major muscle reveals molecular mechanismsinvolved with white striping in broiler chickens. Poult. Sci. 2019;98:590–601. doi: 10.3382/ps/pey429. [DOI] [PubMed] [Google Scholar]
- Mazzoni M., Petracci M., Meluzzi A., Cavani C., Clavenzani P., Sirri F. Relationship between pectoralis major muscle histology and quality traits of chicken meat. Poult. Sci. 2015;94:123–130. doi: 10.3382/ps/peu043. [DOI] [PubMed] [Google Scholar]
- Meloche K.J., Fancher B.I., Emmerson D.A., Bilgili S.F., Dozier W. Effects of quantitative nutrient allocaiton on myopathies of the Pectoralis major muscles in broiler chickens at 32, 43, and 50 days of age. Poult. Sci. 2018;97:1786–1793. doi: 10.3382/ps/pex453. [DOI] [PubMed] [Google Scholar]
- Meloche K.J., Fancher B.I., Emmerson D.A., Bilgili S.F., Dozier W. Effects of reduced dietary energy and amino acid density on Pectoralis major myopathies in broiler chickens at 36 and 49 days of age. Poult. Sci. 2018;97:1794–1807. doi: 10.3382/ps/pex454. [DOI] [PubMed] [Google Scholar]
- Mudalal S., Lorenzi M., Soglia F., Cavani C., Petracci M. Implications of white striping and wooden breast abnormalities on quality traits of raw and marinated chicken meat. Animal. 2015;9:728–734. doi: 10.1017/S175173111400295X. [DOI] [PubMed] [Google Scholar]
- Mutryn M.F., Brannick E.M., Fu W., Lee W.R., Abasht B. Characterization of a novel chicken muscle disorder through differential gene expression and pathway analysis using RNA-sequencing. BMC Genomics. 2015;16:399. doi: 10.1186/s12864-015-1623-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Chicken Council Statistics and research: Per capita consumption of poultry and livestock, 1965 to Estimated 2019, in Pounds. 2018. https://www.nationalchickencouncil.org/about-the-industry/statistics/per-capita-consumption-of-poultry-and-livestock-1965-to-estimated-2012-in-pounds/
- Papah M.B., Brannick E.M., Schmidt C.J., Abash B. Gene expression profiling of the early pathogenesis of wooden breast disease in commercialbroiler chickens using RNA-sequencing. PLoS One. 2018;13 doi: 10.1371/journal.pone.0207346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne S.H. The utility of protein and mRNA correlation. Trends Biochem. Sci. 2015;40:1–3. doi: 10.1016/j.tibs.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesce A., McKay R.H., Stolzenbach F., Cahn R.D., Kaplan N.O. The comparative enzymology of lactic dehydrogenases: I. properties of the crystalline beef and chicken enzymes. J. Biol. Chem. 1964;239:1753–1761. [PubMed] [Google Scholar]
- Petracci M., Mudalal S., Bonfiglio A., Cavani C. Occurrence of white striping under commercial conditions and its impact on breast meat quality in broiler chickens. Poult. Sci. 2013;92:1670–1675. doi: 10.3382/ps.2012-03001. [DOI] [PubMed] [Google Scholar]
- Petracci M., Mudalal S., Soglia F., Cavani C. Meat quality in fast-growing broiler chickens. World Poult. Sci J. 2015;71:363–374. [Google Scholar]
- Py G., Eydoux N., Lambert K., Chapot R., Koulmann N., Sanchez H., Bahi L., Peinnequin A., Mercier J., Bigard A.X. Role of hypoxia-induced anorexia and right ventricular hypertrophy on lactate transport and MCT expression in rat muscle. Metabolism. 2005;54:634–644. doi: 10.1016/j.metabol.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Roth D.A., Brooks G.A. Lactate transport is mediated by a membrane-bound carrier in rat skeletal muscle sarcolemmal vesicles. Arch Biochem. Biophys. 1990;279:377–385. doi: 10.1016/0003-9861(90)90505-s. [DOI] [PubMed] [Google Scholar]
- Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008;3:1101. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Sihvo H.K., Immonen K., Puolanne E. Myodegeneration with fibrosis and regeneration in the pectoralis major muscle of broilers. Vet. Pathol. 2014;51:619–623. doi: 10.1177/0300985813497488. [DOI] [PubMed] [Google Scholar]
- Sihvo H.K., Lindén J., Airas N., Immonen K., Valaja J., Puolanne E. Wooden breast myodegeneration of pectoralis major muscle over the growth period in broilers. Vet. Pathol. 2017;54:119–128. doi: 10.1177/0300985816658099. [DOI] [PubMed] [Google Scholar]
- Sihvo H.K., Airas N., Lindén J., Puolanne E. Pectoral vessel density and early ultrastructural changes in broiler chicken wooden breast myopathy. J. Comp. Pathol. 2018;161:1–10. doi: 10.1016/j.jcpa.2018.04.002. [DOI] [PubMed] [Google Scholar]
- Soglia F., Mudalal S., Babini E., Di Nunzio M., Mazzoni M., Sirri F., Cavani C., Petracci M. Histology, composition, and quality traits of chicken Pectoralis major muscle affected by wooden breast abnormality. Poult. Sci. 2016;95:651–659. doi: 10.3382/ps/pev353. [DOI] [PubMed] [Google Scholar]
- Summermatter S., Santos G., Pérez-Schindler J., Handschin C. Skeletal muscle PGC-1α controls whole-body lactate homeostasis through estrogen-related receptor α-dependent activation of LDH B and repression of LDH A. Proc. Natl. Acad. Sci. U. S. A. 2013;110:8738–8743. doi: 10.1073/pnas.1212976110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallentire C.W., Leinonen I., Kyriazakis I. Artificial selection for improved energy efficiency is reaching its limits in broiler chickens. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-19231-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasoniero G., Cullere M., Cecchinato M., Puolanne E., Dalle Zotte A. Technological quality, mineral profile, and sensory attributes of broiler chicken breasts affected by White Striping and Wooden Breast myopathies. Poult. Sci. 2016;95:2707–2714. doi: 10.3382/ps/pew215. [DOI] [PubMed] [Google Scholar]
- Tijare V.V., Yang F.L., Kuttappan V.A., Alvarado C.Z., Coon C.N., Owens C.M. Meat quality of broiler breast fillets with white striping and woody breast muscle myopathies. Poult. Sci. 2016;95:2167–2173. doi: 10.3382/ps/pew129. [DOI] [PubMed] [Google Scholar]
- Ullah M.S., Davies A.J., Halestrap A.P. The plasma membrane lactate transporter MCT4, but not MCT1, is up-regulated by hypoxia through a HIF-1alpha-dependent mechanism. J. Biol. Chem. 2006;281:9030–9037. doi: 10.1074/jbc.M511397200. [DOI] [PubMed] [Google Scholar]
- Velleman S.G., Clark D.L. Histopathologic and myogenic gene expression changes associated with wooden breast in broiler breast muscles. Avian Dis. 2015;59:410–418. doi: 10.1637/11097-042015-Reg.1. [DOI] [PubMed] [Google Scholar]
- Vogel C., Marcotte E.M. Insights into the regulation of protein abundance from proteomic and transcroptomic analyses. Nat. Rev. Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieme R.J., Herpol J.E. Origin of the lactate dehydrogenase isoenzyme pattern found in the serum of patients having primary musclular dystrophy. Nature. 1962;194:287–288. doi: 10.1038/194287b0. [DOI] [PubMed] [Google Scholar]
- Wilson B.W., Abplanalp H., Buhr R.J., Entrikin J.R., Hooper M.J., Nieberg P.S. Inbred crosses and inherited muscular dystrophy of the chicken. Poult. Sci. 1988;67:367–374. doi: 10.3382/ps.0670367. [DOI] [PubMed] [Google Scholar]
- Wilson M.C., Jackson V.N., Heddle C., Price N.T., Pilegaard H., Juel C., Bonen A., Montgomery I., Hutter O.F., Halestrap A.P. Lactic acid efflux from white skeletal muscle is catalyzed by the monocarboxylate transporter isoform MCT3. J. Biol. Chem. 1998;273:15920–15926. doi: 10.1074/jbc.273.26.15920. [DOI] [PubMed] [Google Scholar]
- Zampiga M., Flees J., Meluzzi A., Dridi S., Sirri F. Application of omics technologies for a deeper insight into quali-quantitative production traits in broiler chickens: a review. J. Anim. Sci. Biotechnol. 2018;9:61. doi: 10.1186/s40104-018-0278-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambonelli P., Zappaterra M., Soglia F., Petracci M., Sirri F., Cavani C., Davoli R. Detection of differentially expressed genes in broiler pectoralis major muscle affected by White Striping – Wooden Breast myopathies. Poult. Sci. 2016;95:2771–2785. doi: 10.3382/ps/pew268. [DOI] [PubMed] [Google Scholar]
- Zhang S., Saremi B., Gilbert E.R., Wong E.A. Physiological and biochemical aspects of methionine isomers and a methionine analogue in broilers. Poult. Sci. 2017;96:425–439. doi: 10.3382/ps/pew253. [DOI] [PubMed] [Google Scholar]
- Zuidhof M.J., Schneider B.L., Carney V.L., Korver D.R., Robinson F.E. Growth, efficiency, and yield of commercial broilers from 1957, 1978, and 20051. Poult. Sci. 2014;93:2970–2982. doi: 10.3382/ps.2014-04291. [DOI] [PMC free article] [PubMed] [Google Scholar]