Abstract
Salmonella screening is a key to ensure food safety in poultry supply chains. Currently available Salmonella detection methods including culture, polymerase chain reaction and enzyme-linked immuno-sorbent assay could not achieve rapid, sensitive, and in-field detection. In this study, different strategies for separation and detection of Salmonella were proposed, compared, and improved based on our previous studies on immunomagnetic separation and impedance biosensor. First, the coaxial capillary for immunomagnetic separation of target bacteria was improved with less contamination, and 3 strategies based on the improved capillary and immunomagnetic nanoparticles were compared to separate the target bacteria from sample and form the magnetic bacteria. The experimental results showed that the strategy of capture in tube and separation in capillary was the most suitable with separation efficiency of approximately 88%. Then, the immune gold nanoparticles coated with urease were used to label the magnetic bacteria, resulting in the formation of enzymatic bacteria, which were injected into the capillary. After the urea was catalyzed by the urease on the enzymatic bacteria in the capillary, different electrodes were compared to measure the impedance of the catalysate and the screen-printed electrode with higher sensitivity and better stability was the most suitable. This impedance biosensor-based bacterial detection strategy was able to detect Salmonella as low as 102 CFU/mL in 2 h without complex operations. Compared to the gold standard culture method for practical screening of Salmonella in poultry supply chains, this proposed strategy had an accuracy of approximately 90% for 75 real poultry samples.
Key words: Salmonella screening, poultry supply chains, impedance biosensor, immunomagnetic separation, food safety
Introduction
China is the third largest producer and consumer of poultry meat and eggs in the world. According to the report of World Health Organization on February 20, 2018, 550 million people fall ill each year, including 220 million children under the age of 5 years due to diarrheal diseases (World Health Organization, 2018). Salmonella is 1 of the 4 key global causes of diarrheal diseases, which is very often associated with poultry supply chains. China National Center for Food Safety Risk Assessment (2015) estimated that the incidence of food poisoning caused by Salmonella reached over 3 million person-times annually, and around 50% was associated with chicken meats. China is now faced with great challenges in poultry product safety due to large variation and uncertain factors in poultry supply chains, and in-field screening of Salmonella in whole poultry supply chains is a key to ensure poultry product safety.
Currently available methods for bacterial detection mainly include traditional culture plating (Culture, gold standard method) (Rodriguez et al., 2018), polymerase chain reaction (PCR, recommended rapid method) (Xu et al., 2018, Hyeon et al., 2019), enzyme-linked immunosorbent assay (ELISA, recommended rapid method), etc. However, these methods either require long time (3 to 4 D) to obtain final results (culture) (Bouguelia et al., 2013, Wang and Salazar, 2016, Xu et al., 2016) or need complex nucleic acid extraction procedures and expensive instruments (PCR) (Zhu et al., 2018, Cao et al., 2019, Li et al., 2019) or lack sufficient sensitivity (ELISA) (Kanayeva et al., 2012, Zhang et al., 2016, Wang et al., 2019). Therefore, many efforts have been made by researchers worldwide in the past decades to develop new methods for rapid and sensitive detection of foodborne pathogens. As alternatives, various biosensors were reported with shorter time, higher sensitivity, simpler operation, and lower cost using nanomaterials, enzymes, or chemical reagents as signal amplifiers or carriers (Yu et al., 2018, Ye et al., 2019, Zhang et al., 2019). Impedance biosensors, which rely on electrochemical changes on the electrode’s surface during the detection of an analyte, have shown advantages over traditional bacterial detection methods, such as compact design, low cost, and rapid response. Interdigitated microelectrodes (IMEs) are often used as transducer in the development of impedance biosensors to achieve higher sensitivity and faster detection since IMEs are featured with low ohmic drop, rapid reaction kinetics, high signal-to-noise ratio, and miniature size (Fysun et al., 2019, Ibau et al., 2019, Jasim et al., 2019, Poghossian et al., 2019). Recently, some microfluidic electrochemical biosensors were reported for biochemical analysis, which offered shorter detection time, less sample and reagent consumption, automatic operation, and less cross-contamination (Li et al., 2018, Nguyen et al., 2018, Singh et al., 2018, Soares et al., 2018, Wang et al., 2018). In these microfluidic biosensors, the biological recognition elements were generally immobilized on the surface of the electrodes (Pursey et al., 2017, Singh et al., 2017, Weng et al., 2017, Ghrera et al., 2018), which could capture the targets or enhance the signals resulting in more impedance changes on the electrode–solution interface. However, their reproducibility was often unsatisfied due to the complicated electrode modification procedures, and the binding efficiency was often relatively low due to solid-liquid phase reaction, which limited their practical applications. Besides, the volume of the bacterial sample for the microfluidic biosensors is very small, generally at microliter or nanoliter level. Even if the biosensors could detect the bacteria as low as one cell, i.e., their lower detection limits were 1 CFU/μL or 1 CFU/nL, equal to 103 CFU/mL or 106 CFU/mL, the biosensors still were not sensitive enough for foodborne bacteria detection because most of these pathogenic bacteria are not allowed to be detectable in many food samples.
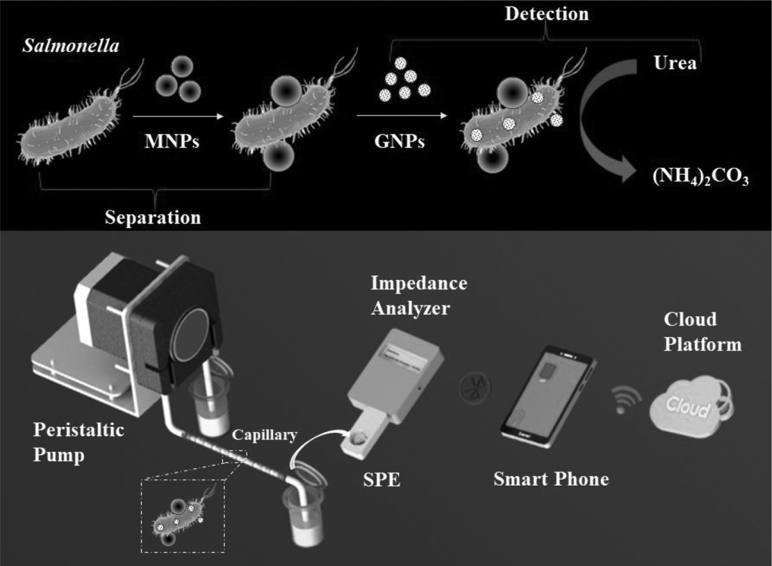
In our previous studies (Chen et al., 2015, Chen et al., 2016, Wang et al., 2017a, Wang et al., 2017b), we have successfully developed some impedance biosensors for detection of foodborne pathogenic bacteria and some immunomagnetic separators for separation of target bacteria from complex food matrix. However, we have not systematically compared, improved, and combined them to set up the most suitable strategy based on these immunomagnetic separators and impedance biosensors for in-field detection of foodborne pathogenic bacteria in real food supply chains. Thus, different strategies based on the improved immunomagnetic separators and impedance biosensors were investigated in this study. As shown in Figure 1, the anti-Salmonella capture antibody-conjugated magnetic nanoparticles (MNPs) were first used to specifically separate the target Salmonella cells and efficiently concentrate them in small volume of phosphate-buffered saline (PBS) solution, resulting in the formation of magnetic bacteria. Then, the gold nanoparticles (GNPs), which were modified with the anti-Salmonella detection antibodies and the urease, were used to label the magnetic bacteria, resulting in the formation of enzymatic bacteria, and injected into the improved coaxial capillary with less cross-contaminations. Finally, the enzymatic bacteria were used to catalyze urea to produce the catalysate, ammonium carbonate, resulting in a decrease on the impedance of the urea-catalysate mixture, and the impedance change was measured by the impedance analyzer and analyzed by the App on the smart phone to determine the concentration of the Salmonella cells. The result was combined with the time and the global positioning system location for collecting the sample, and transmitted to the cloud platform via WIFI for real-time risk assessment.
Figure 1.
Scheme of the biosensor-based strategy for rapid screening of Salmonella in poultry supply chains. The target salmonella is first separated by the immune magnetic nanoparticles (MNPs), then labeled by the gold nanoparticles (GNPs) with urease, and finally used to catalyze urea. The catalysate is measured by the impedance analyzer with the electrode to obtain the result. The result is first transmitted via Bluetooth to the smartphone, then analyzed by the App, and finally uploaded onto the cloud platform for risk assessment. Abbreviation: SPE, screen-printed electrode.
Materials and methods
Bacteria Preparation
Salmonella Typhimurium was used as the target bacteria, while Escherichia coli O157: H7 and Listeria monocytogenes were used as the non-target bacteria. They all were first incubated in Luria-Bertani medium at 37°C with shaking at 180 rpm for 16 to 18 h, and then were serially diluted with the sterile PBS to obtain the bacteria with the concentrations of 101 to 105 CFU/mL. Besides, the chicken/duck meat samples and the whole chicken carcass samples were collected from the breeding link and the slaughtering link in poultry supply chains, respectively, for evaluation of the biosensor-based Salmonella detection strategy.
Nanomaterials Preparation
The immune MNPs are crucial to specific capture, rapid separation and efficient enrichment of Salmonella from complex sample background. The carboxylated MNPs from Allrun (10 mg/mL, size: ∼180 nm, Shanghai, China) were first modified with streptavidin by the EDC method according to our previous study (Lin et al., 2015) to obtain the streptavidin-modified MNPs (2 mg/mL). Then, 2.5 μL of the biotinylated capture antibodies from Abcam (ab69255, 2 mg/mL, Cambridge, UK) and 50 μL of the streptavidin-modified MNPs were added into 500 μL of PBS containing 1% bovine serum albumin (BSA) and incubated at 15 rpm for 45 min. After magnetic separation for 2 min to remove the surplus antibodies, the immune MNPs with the concentration of 2 mg/mL were obtained and stored at 4°C for further use.
The GNPs were modified with the detection antibodies against Salmonella and the urease by electrostatic adsorption for specific labeling of the magnetic bacteria. Briefly, the GNPs with the diameter of approximately 20 nm were first synthesized by adding 0.1% sodium citrate into boiling 0.01% HAuCl4∙3H2O and heating for 10 min. Then, the detection antibodies and the urease at the mass ratio of 1:3 were added into the GNPs and incubated at 15 rpm for 1 h. After the GNPs were blocked by 1% (w/v) PEG 20,000 for 30 min and 10% (w/v) BSA for 30 min, respectively, they were centrifuged at 15,000 rpm and 4°C for 15 min to remove the excessive antibodies and urease. The immune GNPs were finally dissolved in water containing 1% BSA and stored at 4°C for further use.
Capillary Fabrication
The coaxial capillary is the key to separation of target bacteria from sample background. In this study, the capillary was fabricated with some improvements based on our previous designs (Wang et al., 2017b; Huang et al., 2018, Xue et al., 2018). Briefly, it consisted of an inner quartz capillary (inner diameter: 1.0 mm, outer diameter: 1.2 mm) and an outer quartz one (inner diameter: 1.8 mm, outer diameter: 2.0 mm). As shown in Supplementary Figure 1, the inner capillary was filled with an iron ball (diameter: 0.8 mm)-column magnet (diameter: 0.8 mm, length: 1 mm, Grade: N42, Material: neodymium) staggered chain, and both ends of this capillary were connected with 2 3D printed cylinder aligners to ensure that these 2 capillaries were concentric. Four symmetric half-cylinders were removed from the aligner for allowing the solutions to flow through the coaxial capillary, and 1 triangle cone (diameter: 1.8 mm, height: 0.6 mm) was attached to the aligner for avoiding the residuals.
Electrode Fabrication
The electrode is also very important to sensitive and stable detection of the catalysate. In this study, 3 different electrodes were fabricated by different fabrication processes and compared for impedance measurement. As shown in Supplementary Figure 2, the interdigitated microelectrodes (finger width: 15 μm, interfinger space: 15 μm, finger length: 3 mm, finger pair: 25) were fabricated using photolithography and wet etching techniques on the glass substrate. The screen-printed electrodes (SPE, concentric circle, circle width: 200 μm, intercircle space: 200 μm, circle pair: 3) were fabricated using screen-printed technique on the ceramic substrate. The printed circuit board electrodes (PCBE, finger width: 200 μm, interfinger space: 200 μm, finger pair: 10, finger length: 6 mm) were fabricated using gold plating on the printed circuit board.
Bacteria Separation
The separation of the target bacteria from food matrix is the precondition of sensitive detection, which mainly includes the capture of the target bacteria and the separation of the magnetic bacteria. Three different strategies for separation of the target Salmonella cells using the immune MNPs and the improved coaxial capillary were compared in this study, including (1) both capture and separation in tube, (2) capture in tube and separation in capillary, and (3) both capture and separation in capillary.
For the strategy of both capture and separation in tube, 50 μL of the immune MNPs (2 mg/mL) were first washed in the BSA blocking centrifuge tube and resuspended with 1 mL of the bacterial sample, then incubated at 15 rpm for 45 min, and finally separated using a magnetic separator (MS0204, Aibit Biotech, Wuxi, China) with the maximum strength of 1.3 T for 2 min to remove the sample background and resuspended with 1 mL of PBS, followed by culture plating for calculating the separation efficiency.
For the strategy of capture in tube and separation in capillary, 50 μL of the immune MNPs were first washed in the tube and resuspended with 1 mL of the bacterial sample, then incubated at 15 rpm for different time, and finally injected at 1 mL/min into the BSA blocking coaxial capillary and washed with 1 mL of PBS. All the wastes were collected. The original bacterial sample and the waste were determined using culture plating for calculating the separation efficiency.
For the strategy of both capture and separation in capillary, 50 μL of the immune MNPs were first injected at 1 mL/min into the BSA blocking coaxial capillary and washed with 1 mL of PBS. Then, 1 mL of the bacterial sample was injected at 0.36 mL/min into the capillary, resulting in specific capture of the flowing target bacteria. All the wastes were collected. The original bacterial sample and the waste were determined using culture plating for calculating the separation efficiency.
Bacteria Detection
Three different strategies for detection of the target Salmonella cells combining the immune magnetic separation and the impedance biosensor were compared in this study, including (1) immunoreaction and catalysis in tube, (2) immunoreaction in tube and catalysis in capillary, and (3) immunoreaction and catalysis in capillary.
For the strategy of immunoreaction and catalysis in tube, 1 mL of the bacterial sample was first added into 50 μL of the MNPs to form the magnetic bacteria, and then incubated with 10 μL of the GNPs at 15 rpm for 45 min to form the enzymatic bacteria in the centrifuge tube, followed by separation with the magnetic separator to remove the excessive GNPs. After washing with deionized water twice, the enzymatic bacteria were used to catalyze the urea for 30 min and finally detected using the electrodes to determine the bacterial concentration.
For the strategy of immunoreaction in tube and catalysis in capillary, 1 mL of the bacterial sample was first incubated with 50 μL of the MNPs to form the magnetic bacteria and then conjugated with 10 μL of the GNPs to form the enzymatic bacteria in the centrifuge tube. After they were injected at 1 mL/min into the BSA blocking coaxial capillary and washed with 1 mL of deionized water to remove the excessive GNPs and PBS, urea was injected into the capillary and catalyzed by the urease for 30 min. Finally, the catalysate was flushed out and detected using the electrodes to determine the bacterial concentration.
For the strategy of immunoreaction and catalysis in capillary, 50 μL of the MNPs were first injected at 1 mL/min into the BSA blocking coaxial capillary. After 1 mL of the bacterial sample and 10 μL of the GNPs were injected and incubated for 45 min, respectively, 1 mL of deionized water was successively used to wash and remove the excessive GNPs and PBS. After urea was injected into the capillary and catalyzed by the urease for 30 min, the catalysate was finally flushed out and detected using the electrodes to determine the bacterial concentration.
Impedance Measurement
The catalysate (ammonium carbonate) was detected using these 3 electrodes, including the screen-printed electrodes, the printed circuit board electrodes, and the interdigitated microelectrodes. The electrochemical impedance data were collected simultaneously using a self-developed impedance analyzer, a portable LCR meter (886, BK Precision, Yorba Linda, CA), and an electrochemical station (IM6, ZAHNER, Kronach, Bavaria, Germany). For the electrochemical station, the sinusoidal alternating potential with the amplitude of 5 mV, the direct current bias of 0 V, and the frequency range of 100 Hz to 1 MHz was applied on the electrodes. For the LCR meter, the sinusoidal alternating potential with the amplitude of 50 mV, the direct current bias of 0 V, and the fixed frequency of 10 kHz was applied on the electrodes. For the self-developed impedance analyzer, a commercial AD5934 IC was used with an 89C51 microcontroller to measure the impedance, and the sinusoidal potential with the amplitude of 25 mV, the bias of 0 V, and the fixed frequency of 10 kHz was applied on the electrodes. The impedance data of the impedance analyzer were transmitted via Bluetooth to a low-cost smartphone (Honor 9, US $190, Huawei, Shenzhen, China), analyzed by a self-developed App, and uploaded via WIFI onto a food safety monitoring cloud platform for risk assessment and early warning. After each measurement, the electrodes were rinsed with deionized water to remove the residual ions until their impedance returned to the comparable level with deionized water.
Results and discussions
Comparison of Different Strategies for Magnetic Separation of Target Bacteria
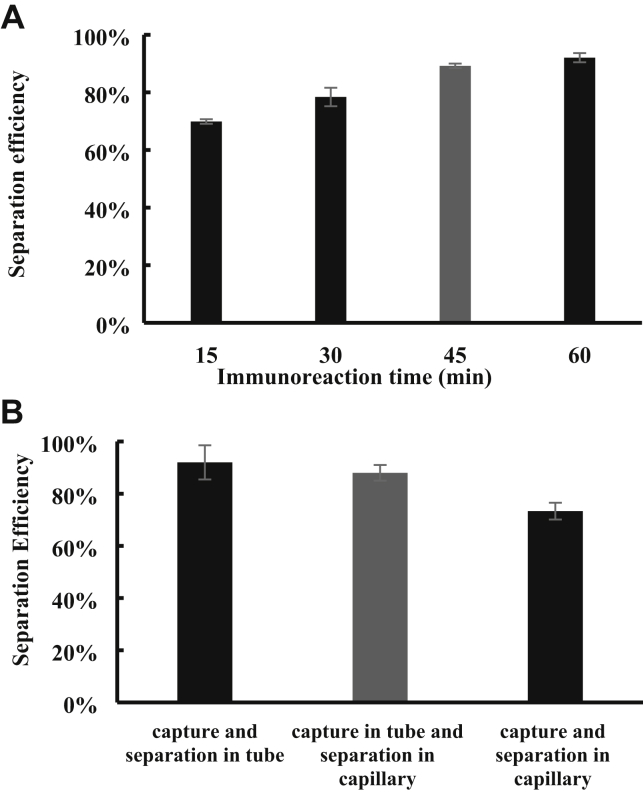
Specific separation and efficient concentration of the target bacteria from complex food samples are vital to their sensitive and accurate detection. Since the incubation time plays an important role in separation of target bacteria, a different time from 15 to 60 min was first used to separate the bacteria for optimization of incubation time. As shown in Figure 2A, the separation efficiency has an obvious increase (approximately 19.3%) from 69.9 to 89.2%, while the incubation time changes from 15 to 45 min, i.e., the increasing rate is approximately 10% per 15 min; however, a further increase in the incubation time to 60 min only results in little change (2.8%) in the separation efficiency, i.e., the increasing rate is less than 3% per 15 min. This is because it needs more time for the immune MNPs to react with the target bacteria when they become less in the same volume. Thus, the optimal incubation time of 45 min was used in the study.
Figure 2.
(A) The results for magnetic separation of Salmonella Typhimurium (1 mL, 103 CFU/mL) by the immune MNPs (50 μL, 2 mg/mL) with different incubation time from 15 to 60 min. The optimal incubation time is selected as 45 min. (B) The results for magnetic separation of Salmonella typhimurium (1 mL, 103 CFU/mL) by the immune MNPs (50 μL, 2 mg/mL) using 3 different strategies (both capture and separation in tube, capture in tube and separation in capillary, and both capture and separation in capillary). The most suitable strategy is capture in tube and separation in capillary after trading off the separation efficiency and operation skills
Three different strategies for magnetic separation of Salmonella were compared, and the experimental results are shown in Figure 2B. The conventional strategy of both capture and separation in tube has a higher separation efficiency of 92.0% than the strategy of capture in tube and separation in capillary (88.0%) and the strategy of both capture and separation in capillary (73.3%). This is because the immune reaction between the antibodies on the MNPs and the target bacteria in the centrifuge tube is liquid-liquid phase reaction and more efficient than those in the capillary, which are solid-phase phase reaction, resulting in the capture of more bacteria in the same time. However, the standard deviation of the strategy of both capture and separation in tube (6.6%) is about 2 times larger than the others, indicating it has bigger variations and requires higher operation skills. To trade off the separation efficiency and operation skills, the strategy of capture in tube and separation in capillary was used in this study. The high separation efficiency of this magnetic separation strategy of Salmonella might be attributed to the following aspects: (1) sufficient reaction between the MNPs and the target Salmonella cells due to high affinity of the capture antibodies, (2) rapid capture of the magnetic bacteria due to high gradient magnetic field in the coaxial capillary, and (3) efficient concentration of the magnetic bacteria in the coaxial channel.
Comparison of Different Electrodes and Detectors for Impedance Measurement
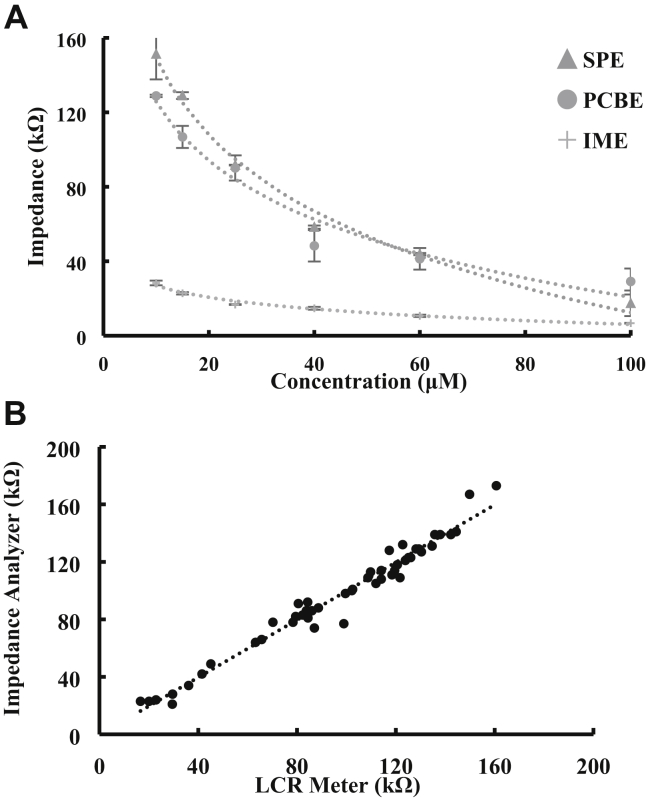
Sensitive and accurate impedance measurement is crucial to the sensitivity and stability of the impedance biosensor-based strategies. Three different electrodes with different sizes and shapes were developed using different fabrication processes and compared in this study. As shown in Figure 3A, these 3 electrodes (IME, SPE, and PCBE) are able to quantitatively measure the impedance of the simulated catalysate (ammonium carbonate) with a linear range from 10 to 100 μM. The slopes of the SPE and PCBE are much larger than that of the IME, indicating that the SPE and the PCBE have higher sensitivity than the IME. However, the standard deviation of the PCBE is obviously larger than those of the SPE and the IME, indicating the PCBE has a higher noise level and a poorer stability. Besides, the cost of the IME (US$ approximately 50 for each) is much higher than those of the PCBE (US$ approximately 0.5 for each) and the SPE (US$ approximately 1 for each). To trade off the sensitivity, the stability and the cost, the SPE was finally used in this study.
Figure 3.
(A) Comparison of 3 electrodes (interdigitated microelectrode [IME], screen-printed electrode [SPE], printed circuit board electrode [PCBE]) for impedance measurement of the catalysate with the concentrations from 10 to 100 μM. The SPE and the PCBE have higher sensitivity and much lower cost than the IME. However, the PCBE has a higher noise level and a poorer stability. The SPE is considered as the most suitable electrode after trading off the sensitivity, the stability, and the cost. (B) Comparison of the impedance analyzer with the LCR meter for impedance measurement of the simulated catalysate with the concentrations from 10 to 100 μM and real poultry samples at the same characteristic frequency of 10 kHz. The impedance values measured by the impedance analyzer and the LCR meter are very close. The impedance analyzer is suitable for impedance measurement.
Besides, accurate impedance measurement is another key to the sensitivity of the impedance biosensor-based strategies. The self-developed impedance analyzer was compared with the commercial 886 LCR meter using both pure ammonium carbonate and real poultry samples. As shown in Figure 3B, the impedance values measured by the impedance analyzer are consistent with those measured by the LCR meter at the same characteristic frequency of 10 kHz, indicating that the impedance analyzer is suitable for accurate impedance measurement.
Comparison of Different Strategies for Detection of Target Bacteria
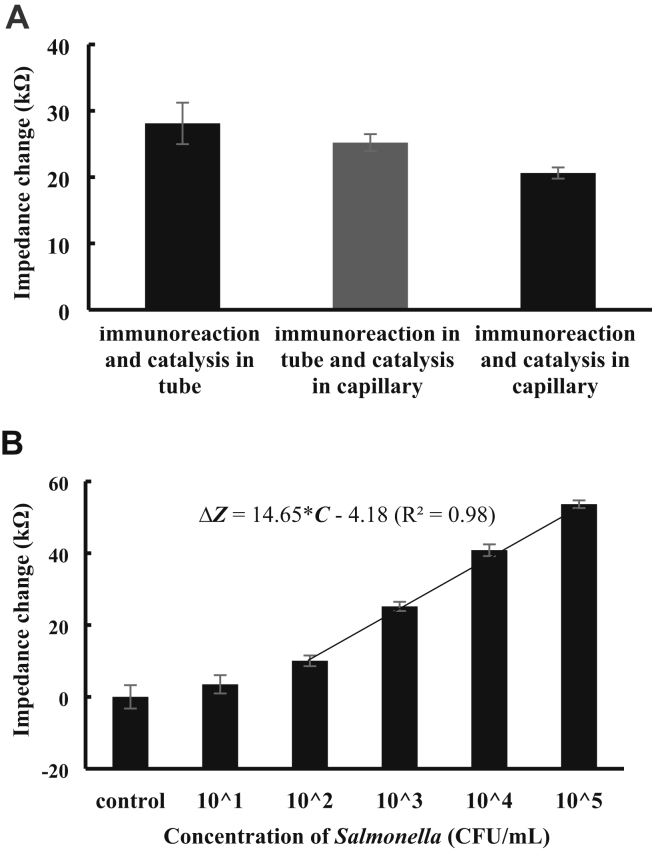
Sufficient sensitivity and easy operation are 2 key criteria for practical application of the biosensor-based bacteria detection strategies. Three different strategies were compared for detection of Salmonella at the concentration of 103 CFU/mL, and the experimental result is shown in Figure 4A. The impedance change for the strategy of immunoreaction and catalysis in tube (28.1 kΩ) is higher than the strategy of immunoreaction in tube and catalysis in capillary (25.2 kΩ) and the strategy of immunoreaction and catalysis in capillary (20.6 kΩ), indicating that the strategy of immunoreaction and catalysis in tube has the strongest detection signal. This might be due to the following reasons: (1) the higher capture efficiency of the target bacteria in the tube than in the capillary, indicating less loss of the target bacteria; (2) the higher labeling efficiency of the magnetic bacteria in the tube than in the capillary, resulting in more GNPs with urease; and (3) the higher catalysis efficiency of the urease in the tube than in the capillary, leading to more catalysate. However, the strategy of immunoreaction and catalysis in tube has the largest variations as well, indicating it might have the poorest stability. This should be attributed to the manual operations. Thus, to trade off the sensitivity and the stability, the optimal strategy of immunoreaction in tube and catalysis in capillary with strong signal and less operation was used in this study.
Figure 4.
(A) Comparison of different strategies (immunoreaction and catalysis in tube, immunoreaction in tube and catalysis in capillary, and immunoreaction and catalysis in capillary) for detection of Salmonella with the concentration of 103 CFU/mL at the optimal conditions (immunoreaction time: 45 min; electrode: screen-printed electrode [SPE]). (B) Calibration curve of the optimal strategy. The target Salmonella with the concentrations from 101 to 105 CFU/mL were detected using this optimal strategy at the optimal conditions (immunoreaction time: 45 min; electrode: SPE).
To determine the unknown concentration of bacterial sample, different concentrations of target bacteria ranging from 101 to 105 CFU/mL were detected using this optimal strategy to build up its calibration model. As shown in Figure 4B, a good linear relationship between the impedance change (ΔZ) and the concentration (C) of target bacteria from 102 to 105 CFU/mL is found and can be expressed as ΔZ = 14.65*C–4.18 (R2 = 0.98). The lower detection limit of this strategy was determined as 102 CFU/mL according to 3 times of signal-to-noise ratio. The high sensitivity of this optimal strategy can be attributed to the following aspects: (1) magnetic separation of the target bacteria from the sample, resulting in purification and enrichment of target bacteria; (2) enzymatic catalysis of non-conductive urea into conductive ammonia carbonate, resulting in great amplification of impedance signal; and (3) semi-automatic operations in the capillary, resulting in the decrease of background noise.
Specificity of Optimal Strategy for Detection of Target Bacteria
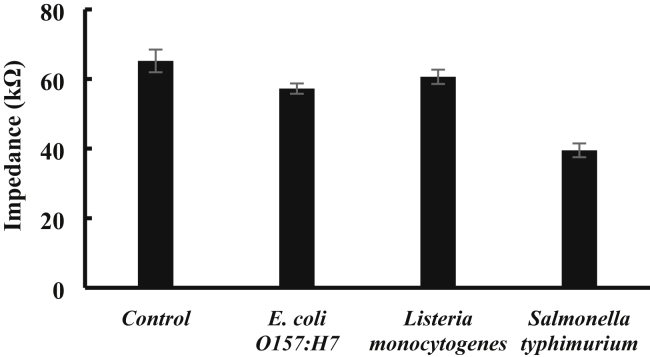
The specificity of this optimal strategy for detection of Salmonella mainly relies on the capture antibodies and the detection antibodies against Salmonella. Two common foodborne pathogenic bacteria at the same concentration (103 CFU/mL), such as E. coli O157: H7 and L. monocytogenes, were used as non-target bacteria to test the specificity of this strategy. The experimental results are shown in Figure 5. Compared to the control (65.2 kΩ), the target Salmonella Typhimurium (39.5 kΩ) has an obviously lower impedance while the non-target E. coli O157: H7 (57.3 kΩ) and L. monocytogenes (60.7 kΩ) have the comparable impedance, indicating this optimal strategy has a good specificity. This should be attributed to the following aspects: (1) the good selectivity of both the capture antibodies and the detection antibodies to the target Salmonella typhimurium; (2) the effective washing in the capillary to minimize the non-specific binding with the non-target bacteria.
Figure 5.
The specificity test of this optimal strategy. The target bacteria (Salmonella typhimurium) and the non-target bacteria (Escherichia coli O157: H7 and Listeria monocytogenes) at the concentration of 103 CFU/mL were detected using this optimal strategy at the optimal conditions (immunoreaction time: 45 min; electrode: screen-printed electrode). Phosphate-buffered saline was used as the negative control.
Detection of Salmonella in Poultry Supply Chains
To verify the feasibility of the optimal strategy for detection of Salmonella in the real samples, 2 different types of real samples, including swab samples from the poultry breeding farm and whole chicken carcass from the poultry slaughtering plant, were collected and detected using this optimal strategy. Each sample was divided into 3 tubes for bacterial test using standard culture plating, real-time PCR (QuanPLEX, Intelligence, Qingdao, China), and this strategy, respectively. The results are shown in Table 1. The detailed protocols to perform the comparative culture plating and real-time PCR are provided in the Supplementary Material. Compared to the culture plating results, this proposed strategy had an accuracy of 89.3% (including 7 false positives and 1 false negative), a little lower than real-time PCR (93.3%, including 4 false positives and 1 false negative), indicating this strategy was suitable for practical applications of Salmonella screening in poultry supply chains. The false positives probably resulted from the interferences of the impurities in the poultry samples, and the false negative might be due to insufficient sensitivity for both the proposed strategy and the commercial PCR.
Table 1.
The results of Salmonella detection in poultry supply chains using the optimal biosensor-based strategy.
| Sample type | Sample number | Impedance biosensor |
Real-time PCR |
Culture plating |
|||
|---|---|---|---|---|---|---|---|
| Negative | Positive | Negative | Positive | Negative | Positive | ||
| Chicken breast | 6 | 5 | 1 | 6 | 0 | 6 | 0 |
| Chicken carcass | 6 | 6 | 0 | 5 | 1 | 6 | 0 |
| Chicken intestine | 6 | 6 | 0 | 6 | 0 | 5 | 1 |
| Chicken leg | 12 | 12 | 0 | 12 | 0 | 12 | 0 |
| Chicken meat | 8 | 6 | 2 | 7 | 1 | 8 | 0 |
| Chicken wing | 17 | 15 | 2 | 15 | 2 | 17 | 0 |
| Duck breast | 8 | 8 | 0 | 8 | 0 | 8 | 0 |
| Duck leg | 4 | 4 | 0 | 4 | 0 | 4 | 0 |
| Duck liver | 4 | 2 | 2 | 4 | 0 | 4 | 0 |
| Duck wing | 4 | 4 | 0 | 4 | 0 | 4 | 0 |
| Total | 75 | 68 | 7 | 71 | 4 | 74 | 1 |
Conclusion
In this study, different strategies based on impedance biosensor and immune magnetic separation were established and compared for separation and detection of Salmonella. The strategy of capture in tube and separation in capillary was the most effective for magnetic separation of Salmonella and was able to separate approximately 88% of Salmonella in 45 min without complex manual operations. The application of the improved coaxial capillary could greatly improve the separation efficiency and reduce the operation skills of the technicians. Compared to other strategies for detection of Salmonella, the strategy of immunoreaction in tube and catalysis in capillary had shown a higher sensitivity and a better stability, and was able to detect Salmonella as low as 102 CFU/mL in 2 h. The proposed biosensor-based strategy could be further improved by integrating the magnetic separation and impedance biosensor onto a chip to achieve fully automatic detection.
Acknowledgments
This study was supported by Walmart Foundation (SA17031161). The authors would like to thank Walmart Food Safety Collaboration Center for its great support.
Footnotes
Supplementary data are available at Poultry Science online.
Supplementary data
Development of the coaxial capillary.
References
- Bouguelia S., Roupioz Y., Slimani S., Mondani L., Casabona M.G., Durmort C., Vernet T., Calemczuk R., Livache T. On-chip microbial culture for the specific detection of very low levels of bacteria. Lab Chip. 2013;13:4024–4032. doi: 10.1039/c3lc50473e. [DOI] [PubMed] [Google Scholar]
- Cao X., Zhao L., Zhang J., Chen X., Shi L., Fang X., Xie H., Chang Y., Wang L. Detection of viable but nonculturable Vibrio parahaemolyticus in shrimp samples using improved real-time PCR and real-time LAMP methods. Food Control. 2019;103:145–152. [Google Scholar]
- Chen Q., Lin J., Gan C., Wang Y., Wang D., Xiong Y., Lai W., Li Y., Wang M. A sensitive impedance biosensor based on immunomagnetic separation and urease catalysis for rapid detection of Listeria monocytogenes using an immobilization-free interdigitated array microelectrode. Biosens. Bioelectron. 2015;74:504–511. doi: 10.1016/j.bios.2015.06.007. [DOI] [PubMed] [Google Scholar]
- Chen Q., Wang D., Cai G., Xiong Y., Li Y., Wang M., Huo H., Lin J. Fast and sensitive detection of foodborne pathogen using electrochemical impedance analysis, urease catalysis and microfluidics. Biosens. Bioelectron. 2016;86:770–776. doi: 10.1016/j.bios.2016.07.071. [DOI] [PubMed] [Google Scholar]
- China National Center for Food Safety Risk Assessment http://www.gov.cn/xinwen/2015-02/04/content_2814121.htm 2015. Accessed July 2019.
- Fysun O., Schmitt A., Auernhammer P.T., Rauschnabel J., Langowski H.C. Electrochemical detection of food-spoiling bacteria using interdigitated platinum microelectrodes. J. Microbiol. Methods. 2019;161:63–70. doi: 10.1016/j.mimet.2019.04.015. [DOI] [PubMed] [Google Scholar]
- Ghrera A.S., Pandey C.M., Malhotra B.D. Multiwalled carbon nanotube modified microfluidic-based biosensor chip for nucleic acid detection. Sens. Actuators B. 2018;266:329–336. [Google Scholar]
- Huang F., Zhang H., Wang L., Lai W., Lin J. A sensitive biosensor using double-layer capillary based immunomagnetic separation and invertase-nanocluster based signal amplification for rapid detection of foodborne pathogen. Biosens. Bioelectron. 2018;100:583–590. doi: 10.1016/j.bios.2017.10.005. [DOI] [PubMed] [Google Scholar]
- Hyeon J.Y., Mann D.A., Wang J., Kim W.K., Deng X. Rapid detection of Salmonella in poultry environmental samples using real-time PCR coupled with immunomagnetic separation and whole genome amplification. Poult. Sci. 2019;98:6973–6979. doi: 10.3382/ps/pez425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibau C., Md Arshad M.K., Gopinath S.C.B., Nuzaihan M.N.M., Fathil M.F.M., Estrela P. Gold interdigitated triple-microelectrodes for label-free prognosticative aptasensing of prostate cancer biomarker in serum. Biosens. Bioelectron. 2019;136:118–127. doi: 10.1016/j.bios.2019.04.048. [DOI] [PubMed] [Google Scholar]
- Jasim I., Shen Z., Mlaji Z., Yuksek N.S., Abdullah A., Liu J., Dastider S.G., El-Dweik M., Zhang S., Almasri M. An impedance biosensor for simultaneous detection of low concentration of Salmonella serogroups in poultry and fresh produce samples. Biosens. Bioelectron. 2019;126:292–300. doi: 10.1016/j.bios.2018.10.065. [DOI] [PubMed] [Google Scholar]
- Kanayeva D.A., Wang R., Rhoads D., Erf G.F., Slavik M.F., Tung S., Li Y. Efficient separation and sensitive detection of Listeria monocytogenes using an impedance immunosensor based on magnetic nanoparticles, a microfluidic chip, and an interdigitated microelectrode. J. Food Prot. 2012;75:1951–1959. doi: 10.4315/0362-028X.JFP-11-516. [DOI] [PubMed] [Google Scholar]
- Li Y., Lai S.N., Zheng B. A microfluidic streaming potential analyzer for label-free DNA detection. Sens. Actuators B. 2018;259:871–877. [Google Scholar]
- Li J., Li J., Xu S., Xiong S., Yang J., Chen X., Wang S., Qiao X., Zhou T. A rapid and reliable multiplex PCR assay for simultaneous detection of fourteen animal species in two tubes. Food Chem. 2019;295:395–402. doi: 10.1016/j.foodchem.2019.05.112. [DOI] [PubMed] [Google Scholar]
- Lin J., Li M., Li Y., Chen Q. A high gradient and strength bioseparator with nano-sized immunomagnetic particles for specific separation and efficient concentration of E. coli O157:H7. J. Magn. Magn. Mater. 2015;378:206–213. [Google Scholar]
- Nguyen N.V., Yang C.H., Liu C.J., Kuo C.H., Wu D.C., Jen C.P. An aptamer-based capacitive sensing platform for specific detection of lung carcinoma cells in the microfluidic chip. Biosensors. 2018;8:E98. doi: 10.3390/bios8040098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poghossian A., Geissler H., Schoning M.J. Rapid methods and sensors for milk quality monitoring and spoilage detection. Biosens. Bioelectron. 2019;140:111272. doi: 10.1016/j.bios.2019.04.040. [DOI] [PubMed] [Google Scholar]
- Pursey J.P., Chen Y., Stulz E., Park M.K., Kongsuphol P. Microfluidic electrochemical multiplex detection of bladder cancer DNA markers. Sens. Actuators B. 2017;251:34–39. [Google Scholar]
- Rodriguez F.I., Procura F., Bueno D.J. Comparison of 7 culture methods for Salmonella serovar Enteritidis and Salmonella serovar Typhimurium isolation in poultry feces. Poult. Sci. 2018;97:3826–3836. doi: 10.3382/ps/pey259. [DOI] [PubMed] [Google Scholar]
- Singh C., Ali M.A., Kumar V., Ahmad R., Sumana G. Functionalized MoS2 nanosheets assembled microfluidic immunosensor for highly sensitive detection of food pathogen. Sens. Actuators B. 2018;259:1090–1098. [Google Scholar]
- Singh N., Ali M.A., Rai P., Sharma A., Malhotra B.D., John R. Microporous nanocomposite enabled microfluidic biochip for cardiac biomarker detection. ACS Appl. Mater. Interfaces. 2017;9:33576–33588. doi: 10.1021/acsami.7b07590. [DOI] [PubMed] [Google Scholar]
- Soares A.C., Soares J.C., Rodrigues V.C., Follmann H.D.M., Arantes L., Carvalho A.C., Melendez M.E., Fregnani J., Reis R.M., Carvalho A.L., Oliveira O.N., Jr. Microfluidic-based genosensor to detect human papillomavirus (HPV16) for head and neck cancer. ACS Appl. Mater. Interfaces. 2018;10:36757–36763. doi: 10.1021/acsami.8b14632. [DOI] [PubMed] [Google Scholar]
- Wang D., Chen Q., Huo H., Bai S., Cai G., Lai W., Lin J. Efficient separation and quantitative detection of Listeria monocytogenes based on screen-printed interdigitated electrode, urease and magnetic nanoparticles. Food Control. 2017;73:555–561. [Google Scholar]
- Wang L., Huang F., Cai G., Yao L., Zhang H., Lin J. An electrochemical aptasensor using coaxial capillary with magnetic nanoparticle, urease catalysis and PCB Electrode for rapid and sensitive detection of Escherichia coli O157:H7. Nanotheranostics. 2017;1:403–414. doi: 10.7150/ntno.22079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Salazar J.K. Culture-Independent rapid detection methods for bacterial pathogens and toxins in food matrices. Comp. Rev. Food Sci. Food Safety. 2016;15:183–205. doi: 10.1111/1541-4337.12175. [DOI] [PubMed] [Google Scholar]
- Wang C., Xing K., Zhang G., Yuan M., Xu S., Liu D., Chen W., Peng J., Hu S., Lai W.H. Novel ELISA based on fluorescent quenching of DNA-stabilized silver nanoclusters for detecting E. coli O157:H7. Food Chem. 2019;281:91–96. doi: 10.1016/j.foodchem.2018.12.079. [DOI] [PubMed] [Google Scholar]
- Wang R., Xu Y., Sors T., Irudayaraj J., Ren W., Wang R. Impedimetric detection of bacteria by using a microfluidic chip and silver nanoparticle based signal enhancement. Mikrochim. Acta. 2018;185:184. doi: 10.1007/s00604-017-2645-x. [DOI] [PubMed] [Google Scholar]
- Weng K.-Y., Chang Y.-J., Ho C.-Y., Liou D.U., Huang Y.-T., Chung W.-Y., Chin T.-Y. Measurement of impedimetric ratio of blood cells using microfluidic chip with ZnO nanowires. J. Med. Biol. Eng. 2017;38:150–158. [Google Scholar]
- World Health Organization. https://www.who.int/news-room/fact-sheets/detail/salmonella-(non-typhoidal) 2018. Accessed July 2019.
- Xu Y., Hu Y., Guo Y., Zhou Z., Xiong D., Meng C., Li Q., Geng S., Pan Z., Jiao X. A new PCR assay based on the new gene-SPUL_2693 for rapid detection of Salmonella enterica subsp. enterica serovar Gallinarum biovars Gallinarum and Pullorum. Poult. Sci. 2018;97:4000–4007. doi: 10.3382/ps/pey254. [DOI] [PubMed] [Google Scholar]
- Xu M., Wang R., Li Y. Rapid detection of Escherichia coli O157:H7 and Salmonella Typhimurium in foods using an electrochemical immunosensor based on screen-printed interdigitated microelectrode and immunomagnetic separation. Talanta. 2016;148:200–208. doi: 10.1016/j.talanta.2015.10.082. [DOI] [PubMed] [Google Scholar]
- Xue L., Zheng L., Zhang H., Jin X., Lin J. An ultrasensitive fluorescent biosensor using high gradient magnetic separation and quantum dots for fast detection of foodborne pathogenic bacteria. Sens. Actuators B. 2018;265:318–325. [Google Scholar]
- Ye Y., Yan W., Liu Y., He S., Cao X., Xu X., Zheng H., Gunasekaran S. Electrochemical detection of Salmonella using an invA genosensor on polypyrrole-reduced graphene oxide modified glassy carbon electrode and AuNPs-horseradish peroxidase-streptavidin as nanotag. Anal. Chim. Acta. 2019;1074:80–88. doi: 10.1016/j.aca.2019.05.012. [DOI] [PubMed] [Google Scholar]
- Yu X., Chen F., Wang R., Li Y. Whole-bacterium SELEX of DNA aptamers for rapid detection of E.coli O157:H7 using a QCM sensor. J. Biotechnol. 2018;266:39–49. doi: 10.1016/j.jbiotec.2017.12.011. [DOI] [PubMed] [Google Scholar]
- Zhang X., Li M., Zhang B., Chen K., He K. Development of a sandwich ELISA for EHEC O157:H7 Intimin gamma1. PLoS One. 2016;11:e0162274. doi: 10.1371/journal.pone.0162274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Tian J., Li K., Tian H., Xu W. Label-free visual biosensor based on cascade amplification for the detection of Salmonella. Anal. Chim. Acta. 2019;1075:144–151. doi: 10.1016/j.aca.2019.05.020. [DOI] [PubMed] [Google Scholar]
- Zhu L., Zhang Y., He P., Zhang Y., Wang Q. A multiplex PCR amplification strategy coupled with microchip electrophoresis for simultaneous and sensitive detection of three foodborne bacteria. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2018;1093–1094:141–146. doi: 10.1016/j.jchromb.2018.06.057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Development of the coaxial capillary.