Abstract
Mycoplasma gallisepticum (MG) and Mycoplasma synoviae (MS) represent the most important avian Mycoplasma species in the poultry industry, causing considerable economic losses. In Italy, the presence of MG or MS has been investigated especially in commercial poultry farms. To our knowledge, no systematic investigations on MG or MS presence using highly specific diagnostic assays have been performed in backyard poultry. The aim of this study was to detect and molecularly characterize MG and MS strains in 11 backyard poultry flocks located in different regions of Italy. Tracheal swabs were collected and DNA was extracted. For MS, a PCR targeting a vlhA gene fragment was performed, and typing and subtyping was attempted. The presence of MG was investigated by a screening PCR, then MG typing by gene-targeted sequencing (GTS). All the amplicons were sequenced, then MG and MS dendrograms were constructed. All the flocks examined resulted Mycoplasma positive: 5 out of 11 (45.45%) were MG and MS positive, 3 (27.27%) were MG positive, and the remaining 3 (27.27%) were MS positive. The MS detections were assigned to types C, D, and F. All strains of type D belonged to subtype D1 and 2 unknown subtypes were identified. A MS sequence showed peculiar characteristics, which did not allow assignment to a known MS type or subtype. MG GTS analysis identified 6 MG strains belonging to 5 subclusters circulating in Italian backyards chicken flocks. The results of this study provide evidence of a risk for commercial poultry farms, especially in areas where backyard and commercial farms are close, suggesting the implementation of biosecurity measures.
key words: backyard poultry, Italy, Mycoplasma gallisepticum, Mycoplasma synoviae
INTRODUCTION
Mycoplasma gallisepticum (MG) and Mycoplasma synoviae (MS) represent the most important avianMycoplasma species worldwide in the poultry industry, causing considerable economic losses (Raviv and Ley, 2013). MG causes chronic respiratory disease in chickens and sinusitis in turkeys. MG disease is characterized by respiratory rales, coughing, nasal discharge and conjunctivitis, and infraorbital sinusitis in turkeys. Increased carcass and downgrading condemnation caused by aereosacculitis, decreased growth and egg production and increased medication costs, make MG one of costliest infection diseases (Raviv and Ley, 2013).
MS is mostly considered to occur as a subclinical upper respiratory infection, which can progress to respiratory disease with air sac lesions, when combined with Newcastle disease or infectious bronchitis, and to infectious synovitis when it becomes systemic (Lockaby et al., 1999). In the last decade, the importance of MS seems to be increasing. The emergence of MS strains that affect egg shell quality with typical eggshell apex abnormalities and decreased egg production (Feberwee et al., 2009; Catania et al., 2010, 2016a) as well as arthropathic and amyloid-inducing strains (Kleven et al., 1975; Landman and Feberwee, 2001) have been reported. Control of Mycoplasma has generally been based on eradication of the organisms from breeder flocks and the maintenance of a Mycoplasma-free status in the breeders and their progeny by implementation of biosecurity.
In Italy, the presence of MG or MS has been investigated especially in commercial poultry farms (Catania et al., 2010, 2016a). A case of MS infection in the lesser flamingo (Phoeniconaias minor) in a zoo of Northern Italy was also reported (Catania et al., 2016b). To our knowledge, no systematic investigations on MG or MS presence using highly specific diagnostic assays have been performed in backyard poultry.
The aim of this study was to detect and molecularly characterize MG and MS strains in backyard poultry flocks in different regions of Italy.
MATERIALS AND METHODS
Samples
Between January 2016 and December 2017, eleven backyard poultry flocks (1 to 11) located in different areas of Italy (Northern, Central and Southern Italy) were investigated for MG and MS presence (Table 1). The main breeds were Sussex, Robusta Lionata, and Cocincine Millefiori, reared for ornamental purposes or consumption of meat. Anamnesis reported a low or moderate morbidity of respiratory signs (sneezing, nasal discharge, and foam in the eye or rattle breathing) in adult birds, but this was severe in chicks with mortality up to 100%. None of these flocks were vaccinated against MG or MS, except flock 10, where a killed MG vaccine was used.
Table 1.
Examined flocks with MG and MS PCR results.
| Year | Flock | Region | MS (type-subtype) | MG |
|---|---|---|---|---|
| 2016 | 1 | Lombardy | +(F2) | + |
| 2016 | 2 | Lombardy | − | + |
| 2016 | 3 | Emilia Romagna | − | + |
| 2016 | 4 | Tuscany | +(D1) | + |
| 2017 | 5 | Sicily | +(D1) | + |
| 2017 | 6 | Campania | +(n.a.) | + |
| 2017 | 7 | Emilia Romagna | +(n.a.) | − |
| 2017 | 8 | Friuli Venezia Giulia | + (C7) | − |
| 2017 | 9 | Lazio | +(n.k.) | + |
| 2017 | 10 | Umbria | − | + |
| 2017 | 11 | Lazio | +(n.a.) | − |
n.a.: not available.
n.k.: not known.
In total, 100 birds were randomly selected and sampled (8 to 10 per flock). Tracheal swabs were collected from only the symptomatic birds and then stored at −20°C until processing as a pool for each flock.
DNA Extraction
Pools of tracheal swabs were eluted in 2 mL of sterile PBS. Total DNA was extracted from elutions using the DNA Mini kit (Qiagen, Germany) according to the manufacturer's instructions, then tested using PCR for MS and MG.
Detection and Typing of Mycoplasma Synoviae
With respect to MS, a PCR targeting a 430 bp fragment of the vlhA gene coding for an abundant immunodominant surface protein was performed. The target region included tandem repeats that encode proline-rich repeats (PRR) and the highly polymorphic RIII region, allowing the typing and subtyping of MS strains (Benčina et al., 2001). The forward primer vlhA-F (5′-GATGCGTAAAATAAAAGGAT-3′) (Moscoso et al., 2004) and the reverse primer vlhA R2 (5′-AGTAACCGATCCGCTTAATGC-3′) (Hammond et al., 2009) were used. A previously characterized MS sample was included as positive control in all PCRs.
Detection and Typing of Mycoplasma Gallisepticum
The presence of MG was investigated using a PCR targeting a 237 to 303 bp fragment of the proline-rich domain of the mgc 2 (pr-mgc2) adhesin-encoding gene (Moscoso et al., 2004). In order to type DNA samples, the gene-targeted sequencing (GTS) assay targeting portions of putative cytadhesin genes (pvpA, gapA and mgc2) and an uncharacterized hypothetical surface lipoprotein-encoding gene (designated coding DNA sequence MGA_0319) (Ferguson et al., 2005) was performed.
In particular, 590, 824, and 332 bp fragments of the MGA_0319, mgc 2, and gap A genes were amplified according to Ferguson et al. (2005). With regard to the pvp A gene, forward pvpA3F (5′-GCCAMTCCAACTCAACAAGCTGA-3′) and reverse pvpA4R primers (5′-GGACGTSGTCCTGGCTGGTTAGC-3′) (Ferguson et al., 2005) or modified forward pvpA3F (5′-GGYAGTCCTAAGTTATTWGGTC-3′) (Liu et al.,2001) and pvpA4R (Ferguson et al., 2005) primers, amplifying a 497 or 702 bp fragment, respectively, were used. Ts-11 vaccine strain was included as positive control in all PCRs.
Sequencing and Phylogenetic Analysis
The vlhA MS and the GTS MG amplicons were sequenced in both directions by a commercial sequencing service (Macrogen Europe). The obtained sequences were edited and assembled using Bioedit software. Nucleotide identity of the vlhA sequences was determined using the nucleotide BLAST algorithm with GenBank database (http://www.ncbi.nlm.nih.gov) (Johnson et al., 2008).
Dendrograms were generated by the neighbor-joining method using Maximum Likelihood model, by Molecular Evolutionary Genetics Analysis (MEGA 6) (Tamura et al., 2013). Bootstrap values were calculated based on 1,000 replicates and considered significant when >70. In addition to the sequences obtained in this study, published corresponding sequences of 34 MS reference strains and previously pubblished Italian strains were included in the phylogenetic analysis (Supplementary Table 1). For MG sequences, the dendrogram was constructed with only MG samples for which all four genes have been successfully sequenced. The MG sequences in GTS analysis were compared with published strains including reference and vaccine strains (Supplementary Table 2).
RESULTS AND DISCUSSION
All flocks examined were Mycoplasma positive when detected using PCR. Of 11 flocks, 5 (45.45%) were MG and MS positive, 3 (27.27%) were MG positive, and the remaining 3 (27.27%) were MS positive (Table 1).
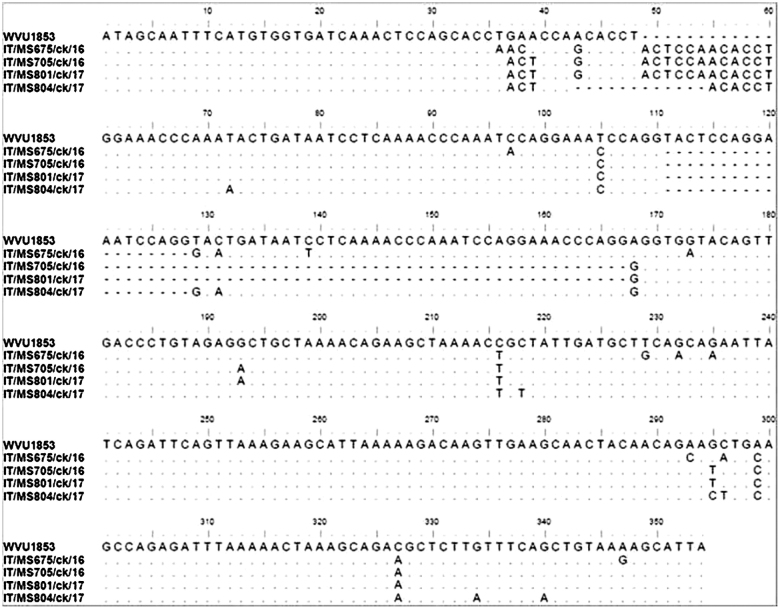
With respect to MS sequence analysis, the vlhA amplicons were successfully sequenced except those from flocks 6, 7, and 11. The MS strains analyzed were named as follows: IT/MS675/ck/16 (flock 1), IT/MS705/ck/16 (flock 4), IT/MS801/ck/17 (flock 5), IT/MS804/ck/17 (flock 8), IT/MS879/ck/17 (flock 9). The nucleotide sequences were submitted to the GenBank database (Table 2). The comparison of the sequences with GenBank showed a percentage of similarity ranging from 92 to 100%. According to the length of the PRR region (Benčina et al., 2001), three known MS types, C (96 nt), D (69 nt), and F (108 nt), were detected in flocks 1 (F), 4 (D), 5 (D), and 8 (C) (Figure 1). Interestingly, the vlha sequence of the IT/MS879/ck/17 strain (flock 9) showed mutations in the PRR region, which meant this flock could not be assigned to a known MS type or subtype (Figure 2). With regards to the RIII region, the known subtype D1 was detected in flocks 4 and 5, and two unknown subtypes, termed C7 and F2, were identified in flocks 8 and 1 (Figure 1). Based on the dendrogram, vlha sequences of MS strains detected in the study clustered with Spanish, Japanese (IT/MS675/ck/16) and Italian MS strains (IT/MS705/ck/16 and IT/MS801/ck/17) except for the vlha IT/MS879/ck/17 sequence, which formed a completely separate branch from other Italian and reference strains (Figure 3).
Table 2.
Accession numbers of MS strains submitted to GeneBank database.
Figure 1.
Nucleotide sequences of the PRR region of vlhA gene of MS strains detected in the study and reference strain WVU1853. Only residues that differ from the sequence of WVU1853 are shown.
Figure 2.
Nucleotide sequences of the PRR region of vlhA gene of IT/MS879/ck/17 strain detected in the study and reference strain WVU1853. Only residues that differ from the sequence of WVU1853 are shown.
Figure 3.
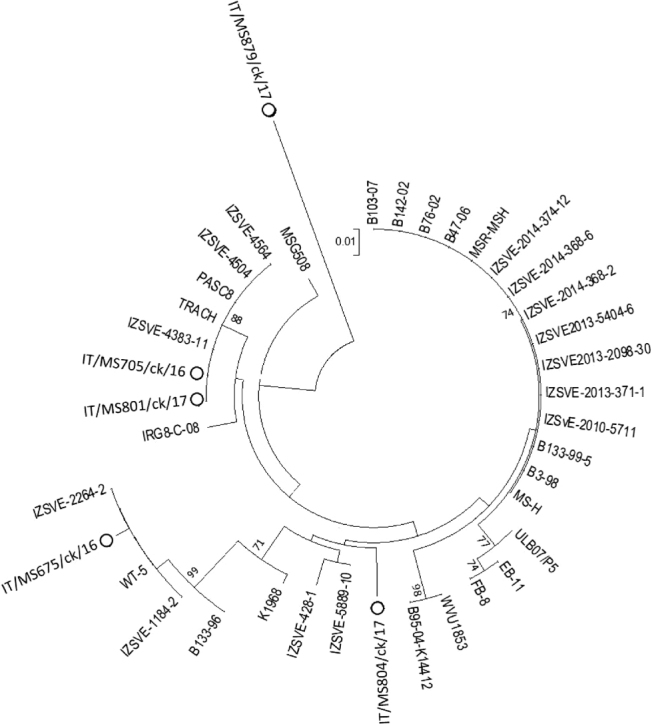
Phylogenetic tree based on the alignment of nucleotide sequences of vlhA genes of MS strains detected in the study (circle) and reference strains. Only bootstrap values >70 are reported.
With respect to the MG sequence analysis, of the 11 samples examined, 8 were pr-mgc 2 positive. All the corresponding GTS amplicons were successfully sequenced, except the MGA_0319 gene from flocks 9 and flock 10 samples. The MG strains analyzed with GTS were named as follows: IT/MG675/ck/16 (flock 1), IT/MG690/ck/16 (flock 2), IT/MG704/ck/16 (flock 3), IT/MG705/ck/16 (flock 4), IT/MG801/ck/17 (flock 5), IT/MG802/ck/17 (flock 6), IT/MG879/ck/17 (flock 9), and IT/MG880/ck/17 (flock 10). Nucleotide sequences were submitted to the GenBank database (Table 3). Since IT/MG879/ck/17 and IT/MG880/ck/17 sequences were not included in the dendrogram, they were analysed using the nucleotide BLAST algorithm with the GenBank database. Sample IT/MG879/ck/17 showed a percentage of nucleotide similarity of 99% with ts-11 vaccine in gapA, 100% with ts-11 vaccine and Australian (K2966) strain in mgc2 and 92% with German 2591/13CK strain in pvpA. Sample IT/MG880/ck/17 showed a percentage of nucleotide similarity of 99% with ts-11 vaccine, Australian (K2966), South Africa, Brazilian (2011/UFMG2) MG strains in mgc2, 99% with ts-11 vaccine, Australian (K2966), South Africa, USA, North America MG strains and Brazilian (2011/UFMG2) MG strains in gapA and 95.3% with German 2591/13CK strain in pvpA.
Table 3.
Accession numbers of MG strains submitted to GeneBank database.
| Accession numbers |
||||
|---|---|---|---|---|
| Strain | gapA | MGA_0319 | Mgc2 | pvpA |
| IT/MG675/ck/16 | MH807704 | MH807705 | MH807706 | MH807707 |
| IT/MG690/ck/16 | MH807708 | MH807709 | MH807710 | MH814582 |
| IT/MG704/ck/16 | MH807711 | MH807712 | MH807713 | MH807714 |
| IT/MG705/ck/16 | MH807715 | MH814597 | MH814590 | MH814585 |
| IT/MG801/ck/17 | MH814594 | MH814596 | MH814589 | MH814584 |
| IT/MG802/ck/17 | MH814593 | MH814595 | MH814588 | MH814583 |
| IT/MG879/ck/17 | MH814591 | / | MH814586 | MH814580 |
| IT/MG880/ck/17 | MH814592 | / | MH814587 | MH814581 |
The dendrogram obtained from sequences of the MGA_0319, mgc 2, gap A, and pvp A genes showed that MG strains belong to five different subclusters (Figure 4). IT/MG704/ck/16 (flock 3) and IT/MG690/ck/16 (flock 2) clustered with ts-11 and 6/85 vaccine strains, respectively. Similarities observed between IT/MG704/ck/16 and ts-11 was of 92.3% and between IT/MG690/ck/16 and 6/85 was of 94.8%.
Figure 4.
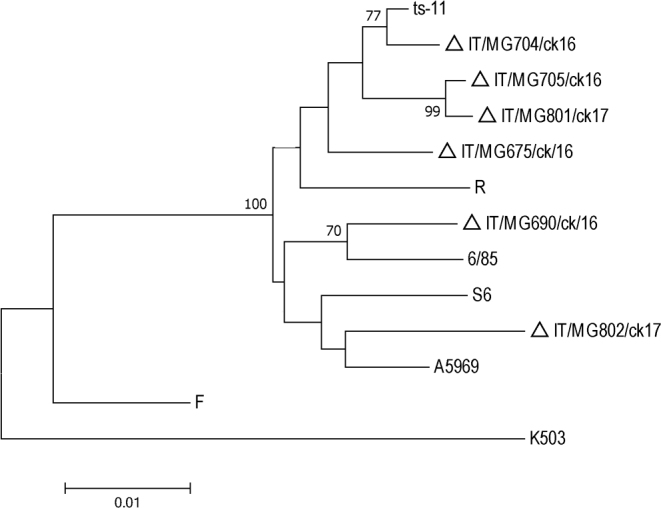
Phylogenetic tree based on the alignment of nucleotide sequences of gap A, MGA_0319, mgc 2, and pvp A genes of MG strains detected in the study (triangle) and reference strains. Only bootstrap values >70 are reported.
IT/MG705/ck/16 (flock 4) and IT/MG801/ck/17 (flock 5), formed a cluster separately from all other analyzed strains, showing a high degree of similarity (99.6%) one with each other. However, the flocks 4 and 5 were geographically distant and no epidemiological link was reported between them.
Despite continued efforts to keep the poultry industry Mycoplasma-free the circulation of MS or MG strains in Italy have been reported in previous works, especially focused on commercial poultry farms (Catania et al., 2010, 2016a),
Our study focused on backyard poultry farms, highlighting MS and MG occurrence in flocks distributed in various Italian regions. MG and MS DNA was detected in birds of different ages and breeds in association with respiratory signs, suggesting a potential clinical role, although other causes of respiratory disease cannot be excluded.
Based on the description of the vlhA fragment, A, C, D, E, F, G, and H types of MS have been detected in Italy (Moronato et al., 2014). In this study, the detection of MS sequences with peculiar molecular characteristics (IT/MS879/ck/17) or new subtypes (C7, F2) evidenced the potential role of backyard chickens as source of new Mycoplasma strains.
Phylogenetic analysis of MS vlhA sequences showed high nucleotide similarity with corresponding sequences of MS strains from Italy and European or extra-European countries, except IT/MS879/ck/17 sequence, which showed only 92% similarity with MS strains from several countries, including Italy. The dendrogram confirmed the peculiarity of IT/MS879/ck/17 being phylogenetically distant from other known MS strains.
With respect to MG sequences, GTS analysis allowed observation of 6 MG strains in a restricted geographical area such as Italy, indicating the ability of this method to differentiate among Italian MG strains.
The presence of the MS and MG sequences showing high nucleotide similarity with corresponding sequences from Italian or foreign Mycoplasma strains is not surprising considering the features of the farms tested, which commonly introduce animals from Italian farms and from European or extra-European countries, and participate in Italian or foreign exhibitions.
Haesendonck et al. (2014) and Derksen et al. (2018) reported that the backyard poultry flocks could act as reservoir or amplifier for poultry respiratory diseases serving as a continuous source of infection for industrial chickens. The present study detected the circulation of MS and MG in backyard poultry farms, confirming the potential role of this type of breeding to spread pathogens to commercial poultry production, especially in densely poultry-populated areas where backyard and commercial farms are close. Whereas the principles and practices of on-farm biosecurity may be familiar to commercial farmers, hobbyists and backyard farmers may not be aware of the steps required to keep infectious diseases out of their flock and prevent their spread to close farms.
The results of this study suggest that backyard chickens should be checked periodically to investigate the status of Mycoplasma infection. Moreover, the implementation of biosecurity measures in backyard poultry farms are needed. More exhaustive studies including attempts at isolation, in vivo pathogenicity studies and molecular analysis may be useful to better investigate the molecular profile and the potential epidemiological role of Mycoplasma strains circulating in backyard poultry farms.
SUPPLEMENTARY DATA
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.psj.2019.12.020.
Supplementary Table S1. Accession numbers of MS sequences retrieved from GenBank used in the analysis.
Supplementary Table S2. Accession numbers of MG sequences retrieved from GenBank used in the analysis.
Footnotes
The nucleotide sequences data reported in this paper have been submitted to GenBank nucleotide sequence database and have been assigned the following accession numbers: MH727587, from MH807700 to MH807703, from MH807704 to MH807715, from MH814580 to MH814597.
Supplementary Materials
REFERENCES
- Benčina D., Drobnič‐Valič M., Horvat S., Narat M., Kleven S.H., Dovč P. Molecular basis of the length variation in the N-terminal part of Mycoplasma synoviae hemagglutinin. FEMS Microbiol. Letters. 2001;203:115–123. doi: 10.1111/j.1574-6968.2001.tb10829.x. [DOI] [PubMed] [Google Scholar]
- Catania S., Bilato D., Gobbo F., Granato A., Terregino C., Iob L., Nicholas R.A.J. Treatment of Eggshell Abnormalities and Reduced Egg Production Caused by Mycoplasma synoviae Infection. Avian Dis. 2010;54:961–964. doi: 10.1637/9121-110309-Case.1. [DOI] [PubMed] [Google Scholar]
- Catania S., Gobbo F., Bilato D., Gagliazzo L., Moronato M.L., Terregino C., Bradbury J.M., Ramírez A.S. Two strains of Mycoplasma synoviae from chicken flocks on the same layer farm differ in their ability to produce eggshell apex abnormality. Vet. Microbiol. 2016;193:60–66. doi: 10.1016/j.vetmic.2016.08.007. [DOI] [PubMed] [Google Scholar]
- Catania S., Gobbo F., Ramirez A.S., Guadagnini D., Baldasso E., Moronato M.L., Nicholas R.A.J. Laboratory investigations into the origin of Mycoplasma synoviae isolated from a lesser flamingo (Phoeniconaias minor) BMC Vet. Res. 2016;12:52. doi: 10.1186/s12917-016-0680-1. Available at https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4788927/ (verified 23 February 2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derksen T., Lampron R., Hauck R., Pitesky M., Gallardo R.A. Biosecurity assessment and seroprevalence of respiratory diseases in backyard poultry flocks located close to and far from commercial premises. Avian Dis. 2018;62:1–5. doi: 10.1637/11672-050917-Reg.1. [DOI] [PubMed] [Google Scholar]
- Feberwee A., de Wit J.J., Landman W.J.M. Induction of eggshell apex abnormalities by Mycoplasma synoviae: field and experimental studies. Avian Pathol. 2009;38:77–85. doi: 10.1080/03079450802662772. [DOI] [PubMed] [Google Scholar]
- Ferguson N.M., Hepp D., Sun S., Ikuta N., Levisohn S., Kleven S.H., García M. Use of molecular diversity of Mycoplasma gallisepticum by gene-targeted sequencing (GTS) and random amplified polymorphic DNA (RAPD) analysis for epidemiological studies. Microbiology. 2005;151:1883–1893. doi: 10.1099/mic.0.27642-0. [DOI] [PubMed] [Google Scholar]
- Haesendonck R., Verlinden M., Devos G., Michiels T., Butaye P., Haesebrouck F., Pasmans F., Martel A. High Seroprevalence of respiratory pathogens in hobby poultry. Avian Dis. 2014;58:623–627. doi: 10.1637/10870-052314-ResNote.1. [DOI] [PubMed] [Google Scholar]
- Hammond P.P., Ramírez A.S., Morrow C.J., Bradbury J.M. Development and evaluation of an improved diagnostic PCR for Mycoplasma synoviae using primers located in the haemagglutinin encoding gene vlhA and its value for strain typing. Vet. Microbiol. 2009;136:61–68. doi: 10.1016/j.vetmic.2008.10.011. [DOI] [PubMed] [Google Scholar]
- Johnson M., Zaretskaya I., Raytselis Y., Merezhuk Y., McGinnis S., Madden T.L. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36:W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleven S.H., Fletcher O.J., Davis R.B. Influence of strain of Mycoplasma synoviae and route of infection on development of synovitis or airsacculitis in broilers. Avian Dis. 1975;19:126–135. [PubMed] [Google Scholar]
- Landman W.J.M., Feberwee A. Field studies on the association between amyloid arthropathy and Mycoplasma synoviae infection, and experimental reproduction of the condition in brown layers. Avian Pathol. 2001;30:629–639. doi: 10.1080/03079450120092125. [DOI] [PubMed] [Google Scholar]
- Liu T., García M., Levisohn S., Yogev D., Kleven S.H. Molecular variability of the adhesin-encoding gene pvpA among Mycoplasma gallisepticum strains and its application in diagnosis. J. Clin. Microbiol. 2001;39:1882–1888. doi: 10.1128/JCM.39.5.1882-1888.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockaby S.B., Hoerr F.J., Lauerman L.H., Smith B.F., Samoylov A.M., Toivio-Kinnucan M.A., Kleven S.H. Factors associated with virulence of Mycoplasma synoviae. Avian Dis. 1999;43:251–261. [PubMed] [Google Scholar]
- Moronato M.L., Baldasso E., Fincato A., Qualtieri K., Flaminio B., Catania S. Atti Della Società Italiana di Patologia Aviare. LIII Convegno Annuale. Salsomaggiore Terme (PR), 8-9 Maggio 2014. 2014. Circolazione di differenti genotipi di Mycoplasma synoviae nel settore avicolo industriale; pp. 184–186. [Google Scholar]
- Moscoso H., Thayer S.G., Hofacre C.L., Kleven S.H. Inactivation, storage, and PCR detection of mycoplasma on FTA® filter paper. Avian Dis. 2004;48:841–850. doi: 10.1637/7215-060104. [DOI] [PubMed] [Google Scholar]
- Raviv Z., Ley D.H. Mycoplasma gallisepticum infection. In: Swayne D.E., Glisson J.R., McDougald L.R., Nolan L.K., Suarez D.L., Venugopal N., editors. Disease of Poultry. 13th edn. Iowa State Press; Ames: 2013. pp. 877–893. [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.