Abstract
Background
Bacteremia is a leading cause of mortality in developing countries, however, etiologic evaluation is infrequent and empiric antibiotic use not evidence-based. Here, we evaluated the patterns of ESBL resistance in children enrolled into a surveillance study for community acquired bacteremic syndromes across health facilities in Central and Northwestern Nigeria.
Method
Blood culture was performed for children aged less than 5 years suspected of having sepsis from Sept 2008-Dec 2016. Blood was incubated using the BACTEC00AE system and Enterobacteriacea identified to the species level using Analytical Profile Index (API20E®). Antibiotic susceptibility profile was determined by the disc diffusion method. Real time PCR was used to characterize genes responsible for ESBL production.
Result
Of 21,000 children screened from Sept 2008-Dec 2016, 2,625(12.5%) were culture-positive. A total of 413 Enterobacteriaceae available for analysis were screened for ESBL. ESBL production was detected in 160 Enterobacteriaceae, high resistance rates were observed among ESBL-positive isolates for Ceftriaxone (92.3%), Aztreonam (96.8%), Cefpodoxime (96.3%), Cefotaxime (98.8%) and Trimethoprim/sulfamethoxazole (90%), while 87.5%, 90.7%, and 91.9% of the isolates were susceptible to Imipenem, Amikacin and Meropenem respectively. Frequently detected resistance genes were blaTEM—83.8% (134/160), and, blaCTX-M 83.1% (133/160) followed by blaSHVgenes 66.3% (106/160). Co-existence of blaCTX-M, blaTEM and blaSHV was seen in 94/160 (58.8%), blaCTX-M and blaTEM in 118/160 (73.8%), blaTEM and blaSHV in 97/160 (60.6%) and blaCTX-M and blaSHV in 100/160 (62.5%) of isolates tested.
Conclusion
Our results indicate a high prevalence of bacteremia from ESBL Enterobacteriaceae in this population of children. These are resistant to commonly used antibiotics and careful choice of antibiotic treatment options is critical. Further studies to evaluate transmission dynamics of resistance genes could help in the reduction of ESBL resistance in these settings.
Introduction
Bacterial blood stream infection (BSI) is a major public-health concern especially in developing countries as it is one of the leading causes of death. In Nigeria as in most developing countries, sub-standard laboratory methods contribute to improper diagnosis of BSI [1]. The absence of accurate etiologic diagnosis and standard clinical microbiology laboratories results in patients being treated with broad spectrum antibiotics. This unguided approach to clinical care is an important factor which promotes the development and spread of antibiotic resistant bacteria.
Trends of increasing antibiotic resistance of blood pathogens to commonly used antibiotics have been reported in Nigeria [2]. Antibiotic resistance has become a concern worldwide and in Enterobacteriaceae, the production of β-lactamase remains the most important mechanism of β- lactam resistance [3].
β-Lactamases are a group of bacterial enzymes that hydrolyze β-lactam antibiotics [4]. The first β-lactamases discovered are the broad spectrum TEM-1, TEM-2, and SHV-1. The product of mutations of the genes that encode these enzymes gave rise to current extended-spectrum β-lactamases (ESBLs), [4]. The ESBL enzymes were initially recognized in clinical isolates in the 1980s; they are derived mainly from the TEM or SHV types of β-lactamases, by point mutations in the parent enzymes which did not possess extended-spectrum β-lactam substrate activity [5, 6]. More than 200 ESBLs have been identified so far, apart from the TEM, SHV and CTX-M types, other clinically relevant types of ESBLs include the VEB, PER, GES, TLA, IBC, SFO-1, BES-1 and BEL-1 types [6].
Extended-spectrum β-lactamases (ESBLs) are a rapidly evolving group of β-lactamases which hydrolyze the extended-spectrum cephalosporins, penicillins, and aztreonam, but not carbapenems [5]. These ESBL-producing bacteria may also be multiply resistant to other class of antimicrobial agents such as aminoglycosides, trimethoprim/sulfamethoxazole, and quinolones. These Multidrug-resistant (MDR) strains cause infections which are difficult to treat [3, 7, and 8]. The World Health Organization (WHO) has declared infections caused by MDR bacteria as an emerging global health problem of major public health concern [9].
The prevalence of ESBL-producing bacteria has been reported worldwide [10–15], while there are a number of publications on ESBL-producing bacteria causing clinical infections [16–21] in Nigeria, report on the characterization of invasive isolates from infants and children is sparse and because antimicrobial resistance varies greatly among geographical settings, it is crucial to formulate empiric therapy of severe infections such as BSI on comprehensive location specific knowledge of the prevalence and antimicrobial resistance patterns of locally isolated bacteria.
In this study we limited our population to children who were enrolled for community acquired bacteremia surveillance in young Nigerian children. Children are at a greater risk of acquiring bloodstream infections compared to adult due to their immature immune system.[22] We thus aimed to investigate the genetic profile of Extended spectrum betalactamase Enterobacteriaceae (ESBL-E) from pediatric BSI patients.
Methods
Isolate collection
The study was conducted at seven hospitals in Federal Capital Territory (FCT) and three in Kano Nigeria as previously described from Sept 2008—Dec 2016 [1, 23]. Children aged less than five years were enrolled at the different sites, blood specimens of 1–3 ml were collected using a vacutainer set, after aseptically cleansing the skin with alcohol swab and povidone-iodine, the specimen was collected directly into an aerobic blood culture bottle (BD BactecPeds Plus/F culture vials; Becton Dickinson, Ireland), and incubated in an automated Bactec® 9050 machine. All positive bottles were sub cultured onto MacConkey, Sheep blood and chocolate agar plates at 36°C for 24 h.
A total number of 887 culture-positive Enterobacteriaceae were obtained. Of the 887 isolates, 474 salmonella species including Typhi which have been reported by Obaro et al. 2015 were excluded [23], therefore 413 including Escherichia coli, Klebsiella species, Enterobacter species, Serratia marcescens, Pantoea species, Salmonella Typhi and Citrobacter species from September 2008 to December 2016 were included. The isolates were stored in 10% skim milk glycerol at -80°C.
Phenotypic screening and confirmation of ESBL
Susceptibility was determined using the disk diffusion method on Mueller Hinton agar as recommended by the Clinical and Laboratory Standard Institute [24]. Susceptibility was tested against amoxicillin/clavunate (20/10μg), cefoxitin (30μg), trimethoprim-Sulfamethoxazole (1.25/23.75μg), ciprofloxacin (5μg), ceftriaxone (30μg), amikacin (30μg), cefpodoxime (10μg), ceftazidime (30μg), imipenem (10μg) and meropenem (10μg), cefotaxime (30μg), piperacillin tazobactam (110μg), aztreonam (30μg) and tigecycline (15μg), (Oxoid Ltd, Basingstoke, Hampshire, England) and interpretation of breakpoint was according to CLSI, 2015 guideline.
Enterobacteriaceae were identified to species level using Analytical Profile Index (API 20E) identification strip (Biomeriux Inc, France). Phenotypic ESBL production was confirmed with the combination disc diffusion test with clavulanic acid. Confirmatory test was considered positive when the inhibition zone produced by the discs in combination clavulanate increased ≥5 mm than the disks without the clavulanate [24].
Molecular identification of ESBL genes
All confirmed ESBL bacteria using the combined disk method were analyzed by Real time PCR for the presence of genes encoding TEM, SHV, CTX-M. Primers and Probes were designed for ESBL producing genes by LGC, Biosearch, USA based on primers used by Roschanski et al., 2014 [25].
Genomic DNA was extracted using Maxwell 16 cell DNA purification kit (Promega) on an automated DNA extraction machine (Maxwell 16 extraction system, USA). Real time PCR assay was performed on AriaMx system (Agilent Inc, USA) using 25 μL PCR reaction mixture containing 12.5 μL Perfecta master mix low ROX kit (Quanta Bioscience Inc, USA), 1 μL of 10 μM primers, 1 μL of probes, 7.5 μL Nuclease free water (Sigma-Aldrich, USA) and 2 μL DNA template. The thermal conditions were as follows: denaturation at 95°C for 15 min, then 30 cycles consisting of a denaturation step at 95°C for 15 seconds, annealing at 50°C for 15 seconds and extension at 70°C for 20 seconds.
After completion of the run, a cycle threshold (Ct) was calculated by determining the signal strength at which the fluorescence exceeded a threshold limit. This value was analyzed using the AriaMx system software version 3.1.
Statistical analysis
Two-way ANOVA was performed using SPSS V 21 SPSS Inc., (Chicago, Ill., USA) for statistical analysis of data. A p<0.05 was considered statistically significant.
Ethical consideration
Consent was obtained from the International Foundation against Infectious Disease in Nigeria (IFAIN), Abuja to gain access to the stored Enterobacteriaceae obtained between 2009 and 2016 from blood of children. IFAIN has the written approval by the Research Ethics Committee of the FCT, National Hospital Abuja, Zankli Medical Center, Federal Medical Center Keffi, Aminu Kano Teaching Hospital. Informed consent was obtained from the parent or guardian of the children. All data/sample were fully anonymized.
Results
From Sept 2008-Dec 2016, 21,000 children were screened for bacteremia with a culture positivity rate of 12.5% (2,625/21,000). Four hundred and thirteen (413) Enterobacteriaceae were available for analysis, comprising Klebsiella species 141(34.14%), Escherichia coli 96(23.24%), Enterobacter species 42 (10.16%), Salmonella typhi 100 (24.21%) and others 34 (8.23%) (Serratia, Pantoea, Citrobacter, Proteus and Kluyvera species) (Table 1).
Table 1. Frequency of ESBL and Non ESBL producing isolates from blood culture.
| Isolates | No of isolates (%) | No of ESBL producers (%) | No of non ESBL producers (%) |
|---|---|---|---|
| Escherichia coli | 96(23.24) | 22 (13.75) | 74 (29.24) |
| Klebsiellaspp | 141 (34.14) | 105 (65.62) | 36 (14.22) |
| Enterobacter spp | 42 (10.16) | 21 (13.12) | 23 (9.09) |
| Serratia species | 8 (1.93) | 4 (2.5) | 4 (1.58) |
| Pantoeaspp | 14 (3.38) | 7 (4.37) | 7 (2.76) |
| Citrobacters pp | 8 (1.93) | 1 (0.62) | 7 (2.76) |
| Proteus mirabilis | 3(0.72) | 0(0) | 3(1.18) |
| Kluyveraspp | 1(0.24) | 0(0) | 1(0.39) |
| Salmonella typhi | 100 (24.21) | 0(0) | 100(39.52) |
| Total (%) | 413 | 160 (38.74) | 253 (61.25) |
Prevalence of ESBLs
The overall prevalence of ESBL producers observed in this study was 160/413 (38.74%). Overall, the prevalence of ESBL-producing isolates was greater among male enrollees 53.12% as compared to females 33.12%. Gender was not specified in 13.75%. ESBL-producing Klebsiella pneumoniae, Enterobacter species, Pantoea species and Serratia species occurred most frequently in children ≤1 month of age, while Escherichia coli was isolated more from age range 2 days -12 months.
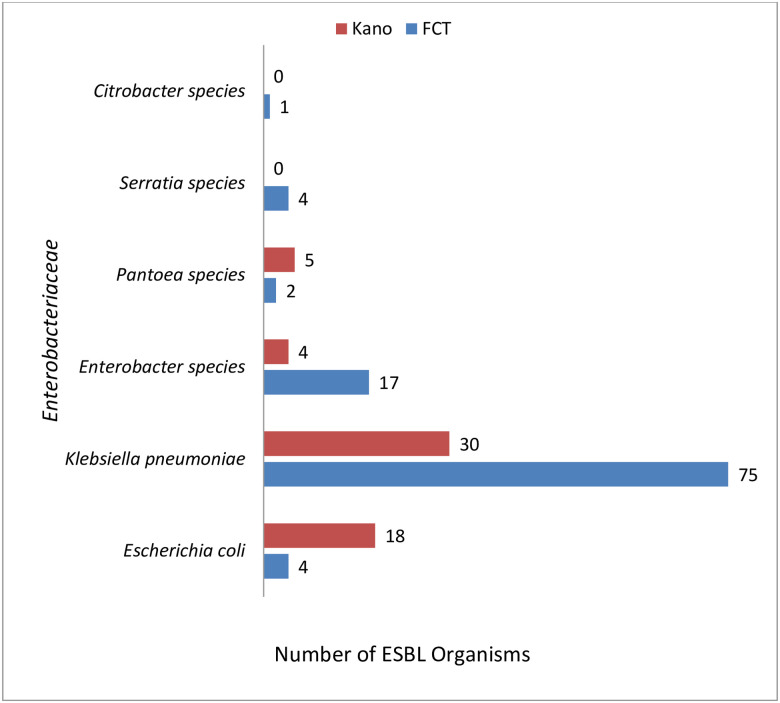
There was no significant difference (p = 0.813; p>0.05) Fig 1, between ESBL distribution in patients from Kano and those from FCT (FCT 103/160(64.37%) and Kano 57/160 (35.62%)).
Fig 1. Distribution pattern of ESBL organisms from FCT and Kano.
Antibiotic susceptibility data
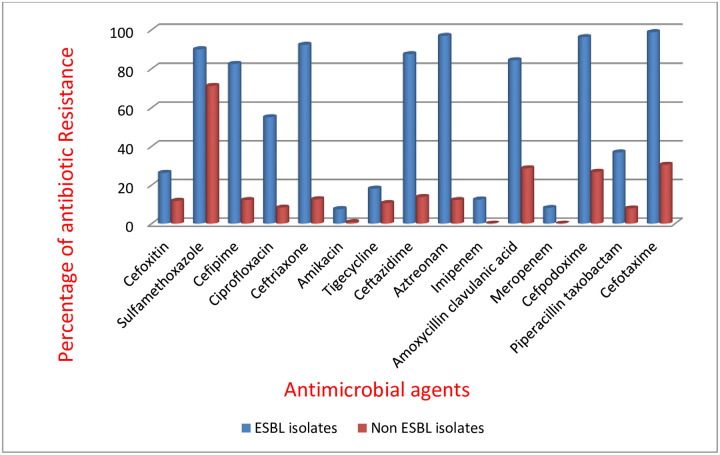
Susceptibility profile of the 160 ESBL-producing isolates revealed high resistance rates for ceftriaxone (92.32%), aztreonam (96.81%), cefpodoxime (96.25%), cefotaxime (98.75%) and sulphamethoxazole- trimethoprim (90%). Over 80% of the isolates were resistant to cefepime, Amoxicillin clavulanic acid and ceftazidime (Fig 2), while 87.5%, 90.63%, and 91.87% of the isolates were susceptible to imipenem, amikacin and meropenem respectively.
Fig 2. Resistance pattern of ESBL and Non ESBL producing Enterobactericeae.
bla gene composition of ESBL-producing strains
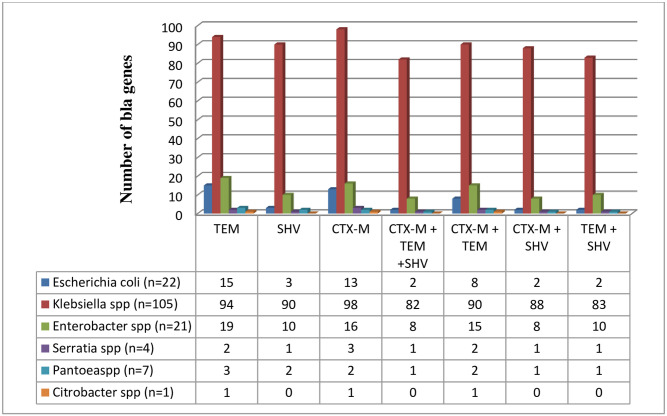
Analysis of the phenotypically confirmed isolates revealed that 134 (62.79%) had the TEM gene, out of which 94(48.14%) were Klebsiella pneumoniae, 15 (25.93%) were Escherichia coli, 19 (22.22%) were Enterobacter species, 2 Serratia species, 3 Pantoea species and 1 strain of Citrobacter species. Also, 133 (34.88%) of the total isolates had the CTX-M gene with 98 (53.33%) being Klebsiella pneumoniae, 13 (6.67%) Escherichia coli, 5 (33.33%), 16 Enterobacter species, 3 were Serratia species, 2 Pantoea species and one isolate of Citrobacter species and 66.25% (106/160) of the total isolates had the SHV gene out of which 90/106 (66.67%) were Klebsiella pneumoniae, 3 were Escherichia coli, 10 Enterobacter species, 2 Pantoea species, and1 Serratia species (Fig 3). Some of the isolates expressed multiple occurrences of genes, the co-existence of blaCTX-M, blaTEM and blaSHV was seen in 94 of the isolates, while blaCTX-M and blaTEM co-existed in 118 of the isolates, blaTEM and blaSHV in 97 of the isolates while blaCTX-M and blaSHV in 100 of the isolates. Two Escherichia coli isolates expressed ESBL phenotype but no blaTEM, blaSHV or blaCTX-M was detected by PCR.
Fig 3. Prevalence of ESBL genes among Enterobacteriaceae isolates.
Discussion
ESBLs have become a widespread serious problem and these enzymes are becoming increasingly expressed by many strains of pathogenic bacteria [26]. The rate of ESBL-producing Enterobacteriacea in our study was 38.74% and the isolates were multi drug resistant. ESBL producing isolates showed a higher degree of antimicrobial resistance as compared to non-ESBL producers. Carbapenems and aminoglycosides were shown to be the most effective antimicrobials for both ESBL and Non ESBL isolates. In comparison with non ESBL, there was a significant difference in the antibiotic resistance pattern (P = 0.0004; p>0.05).
From our study, it was observed that there were more ESBL isolates from FCT compared to Kano; this probably might be due to the affordability and access to third generation cephalosporins in FCT. Few studies have investigated the prevalence and genetic characteristics of ESBL producing Enterobacteriaceae from blood stream infections of pediatric patients in Nigeria. Kasap et al. 2010 from Southwestern Nigeria reported the isolation of SHV 12 from the blood of a 2 year old girl [27], and Aibinu et al., conducted a study in 2003 in Lagos and found that eight out of 40 Enterobacter isolates (20%) investigated were ESBL producers [28]. The limitations in comparison with their studies were the sample size and study period. Their study covered between a period of one month and nine months, while ours had a bigger sample size and a study period of 8 years (September 2008 to December 2016).
In 2012, Alo et al., detected 80% of ESBL production among Klebsiella pneumoniae and Escherichia coli strains isolated from blood samples of hospitalized patients in Ebonyi State University Teaching Hospital [29]. Also a study by Adeyankinnu et al., 2014 reported an ESBL prevalence of 26.4% for all isolates tested, with E. coli having a greater proportion [30]. Their study was restricted to detect the presence of ESBL in Escherichia coli and Klebsiella pneumoniae isolates by phenotypic means only.
The prevalence of ESBL among children in our study was lower when compared to adult studies in Nigeria [20, 31], UK [32], and Spain [33] and pediatric studies in Birmingham [34] and Pakistan [35].
The data from our study have demonstrated that there is a high prevalence of blaTEM, blaCTX-M, and blaSHV ESBL genes in Enterobacteriaceae isolates. A study from South Eastern Nigeria reported blaCTX-M-15 genes from urine, vaginal and wound swabs of out-patients younger than 30 years [36]. Two studies conducted in Western Nigeria by Olowe et al., [37] from blood, wound, HVS, and sputum samples and Raji et al., [38] from blood, Urine and wound samples revealed the isolates harbored blaCTX-M-1 and blaCTX-M-15 genes respectively.
According to our study, blaTEM and blaCTX-M type were the most prevalent ESBL encoding genes, detected in 83% of the ESBL-producing Enterobacteriaceae and the majority were found in Klebsiella isolates, in contrast to our study, Mohammed et al., from North Eastern Nigeria reported blaSHV (36.4%) and blaTEM (31.4%) to be the most prevalent [39], this is likely due to variations in the samples used [39]. The detection of CTX-M, TEM and SHV genes by molecular techniques in ESBL producing bacteria can supply useful data about their epidemiology, association with epidemic clones and risk factors associated with these infections.
The results from our study however, reflect the global trend toward a pandemic spread of CTX-M-type ESBLs in various Enterobacteriaceae. These findings agree with other contemporary studies from around the world that show that ESBL genes of the CTX-M are dominant in Tanzania, Burkina Faso, Texas, Spain, Brazil, Latin America, [40–45]. In similarity to our findings, Ahmed et al. [46] reported that blaTEM was the most frequent β-lactamase-encoding gene in Egypt.
In the present study, it was observed that there were multiple occurrences of genes in some of the isolates, this finding is similar to a study in Peru by Garcia et al., where majority (57.3%) of the ESBL strains harbored 2 or more ESBL genes [47] while in Turkey, Bali et al., observed that about 19.2% ESBL isolates carried more than one type of beta lactamases genes [48].
Two [2] isolates which were screened for ESBL had none of the genes tested for in them from this study; this may be due to the presence of other ESBL genes.
To the best of our knowledge, this is the first report on ESBL resistance patterns from a large surveillance on Bacteremia Pediatric patients from Nigeria.
In conclusion, our findings suggest a high prevalence of ESBL resistance to commonly-used antibiotics in Enterobacteriaceae bacteremia in children in this study. Further studies on the transmission dynamics of resistance genes could help in the control of ESBL resistance in these settings.
Supporting information
(DOCX)
Acknowledgments
We are grateful for the institutional support from administration of local participating sites (University of Abuja Teaching Hospital Gwagwalada, National Hospital Abuja, Nyanya General Hospital, Federal Medical Center Keffi, Aminu Kano Teaching Hospital, Murtala Mohammed Specialist Hospital, and Hasiya Bayero Pediatric Hospitals in Kano).
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (award number R01AI097493); the Bill & Melinda Gates Foundation (grant number OPP1034619); and internal grants from the University of Nebraska Medical Center. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Obaro S., Lawson L., Essen U., Ibrahim K., Brooks K., Otuneye A. Adegbola R. Community acquired bacteremia in young children from central Nigeria—a pilot study. BMC Infectious Diseases, 2011; 11, 137 10.1186/1471-2334-11-137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Medugu N and Iregbu K. C (2016). Trends in profiles of bacteria causing neonatal sepsis in Central Nigeria hospital. African journal of Clinical and experimental Microbiology, 18(1) 1701. [Google Scholar]
- 3.Livermore DM, et al. CTX-M: changing the face of ESBLs in Europe. J. Antimicrob. Chemother.2007; 59:165–174. 10.1093/jac/dkl483 [DOI] [PubMed] [Google Scholar]
- 4.Tooke C. L., Hinchliffe P., Bragginton E. C., Colenso C. K., Hirvonen V., Takebayashi Y., et al. (2019). β-Lactamases and β-Lactamase Inhibitors in the 21st Century. Journal of molecular biology, 431(18), 3472–3500. 10.1016/j.jmb.2019.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pitout J. D. and Laupland K. B., “Extended-spectrum β-lactamase-producing Enterobacteriaceae: an emerging public-health concern,” The Lancet Infectious Diseases, vol. 8, no. 3, pp. 159–166, 2008. [DOI] [PubMed] [Google Scholar]
- 6.Jacoby G.A. and Munoz-Price L.S. The new beta-lactamases. N Engl J Med. 2005; 352: 380–391 10.1056/NEJMra041359 [DOI] [PubMed] [Google Scholar]
- 7.Wilke MS, Lovering AL, Strynadka NCJ. Beta-lactam antibiotic resistance: a current structural perspective. CurrOpinMicrobiol.2005; 8: 525–33. [DOI] [PubMed] [Google Scholar]
- 8.Livermore DM, Woodford N. The beta-lactamase threat in Enterobacteriaceae, Pseudomonas and Acinetobacter. Trends Microbiol.2006; 14: 413–20. 10.1016/j.tim.2006.07.008 [DOI] [PubMed] [Google Scholar]
- 9.World Health Organization 2014: Antimicrobial ResistanceÐGlobal Report on Surveillance
- 10.Demir S, Soysal A, Bakir M, Kaufmann ME, Yagci A. 2008. Extendedspectrum beta-lactamase-producing Klebsiella pneumoniae in paediatric wards: a nested case-control study. J. Paediatr. Child Health 44:548–553. 10.1111/j.1440-1754.2008.01326.x [DOI] [PubMed] [Google Scholar]
- 11.Dhanji H, et al. 2010. Real-time PCR for detection of the O25b-ST131 clone of Escherichia coli and its CTX-M-15-like extended-spectrum betalactamases. Int. J. Antimicrob. Agents 36:355–358. 10.1016/j.ijantimicag.2010.06.007 [DOI] [PubMed] [Google Scholar]
- 12.Hawkey PM. 2008. Prevalence and clonality of extended-spectrum betalactamases in Asia. Clin. Microbiol. Infect. 14(Suppl. 1):159–165. [DOI] [PubMed] [Google Scholar]
- 13.Peirano G, et al. 2010. High prevalence of ST131 isolates producing CTX-M-15 and CTX-M-14 among extended-spectrum-beta-lactamaseproducing Escherichia coli isolates from Canada. Antimicrob. Agents Chemother.54:1327–1330. 10.1128/AAC.01338-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian GB, et al. 2012. Detection of clinically important beta-lactamases in commensal Escherichia coli of human and swine origin in western China. J. Med. Microbiol. 61:233–238. 10.1099/jmm.0.036806-0 [DOI] [PubMed] [Google Scholar]
- 15.Meeta S, Sati P, Preeti S. Prevalence and antibiogram of extended spectrum β-lactamase (ESBL) producing Gram negative bacilli and further molecular characterization of ESBL producing Escherichia coli and KlebsiellaSpecies. J of Clin and Diag Res. 2013;7(10):2173–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egbebi AO, Famurewa O. Prevalence of extended spectrum beta lactamases production among Klebsiella isolates in some parts of South West Nigeria. J Microbiol Biotech Res. 2011;1(2):64–68. [Google Scholar]
- 17.Aibinu IE, Ohaegbulam VC, Adenipekun EA, Ogunsola FT, Odugbemi TO, Mee BJ. Extended spectrum beta lactamase enzymes in clinical isolates of Enterobacter Species from Lagos, Nigeria. J ClinMicrobiol.2003;41:2197–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aboderin OA, Adefehinti O, Odetoyin BW, Olotu AA, Okeke IN, Adeodu OO. Prolonged febrile illness due to CTX-M-15 extended spectrum beta lactamase producing Klebsiella pneumoniae infection in Nigeria. Afr J Lab Med. 2012;1(1):1–4. 10.4102/ajlm.v1i1.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yusha’u MM, Aliyu HM, Kumurya AS, Suleiman L. Prevalence of extended spectrum beta lactamases among Enterobacteriaceae in Murtala Muhammad Specialist Hospital, Kano, Nigeria. Bajopas. 2010;3(1):169–77. [Google Scholar]
- 20.Olowe OA, Aboderin BW. Detection of extended spectrum beta lactamase producing strains of Escherichia coli and Klebsiella Species in a tertiary health centre in Ogun state. Int J of Trop Med. 2010;5(3):62–4. [Google Scholar]
- 21.Soge Olusegun O., Anne Marie Queenan, Ojo Kayode K., Adeniyi Bolanle A., Roberts Marilyn C.; CTX-M-15 extended-spectrum β-lactamase from Nigerian Klebsiella pneumoniae, Journal of Antimicrobial Chemotherapy, Volume 57, Issue 1, 1 January 2006, Pages 24–30, [DOI] [PubMed] [Google Scholar]
- 22.Reddy E. A., Shaw A. V., & Crump J. A. (2010). Community-acquired bloodstream infections in Africa: a systematic review and meta-analysis. The Lancet. Infectious diseases, 10(6), 417–432. 10.1016/S1473-3099(10)70072-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Obaro S. K., Hassan-Hanga F., Olateju E. K., Umoru D., Lawson L., Olanipekun G., et al. (2015). Salmonella Bacteremia Among Children in Central and Northwest Nigeria, 2008–2015. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America, 61 Suppl 4(Suppl 4), S325–31. 10.1093/cid/civ745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clinical and Laboratory Standards Institute. 2015. Performance standards for antimicrobial susceptibility testing. Twenty fifth informational supplement. CLSI document M100-S25. Clinical and Laboratory Standards Institute, Wayne, P
- 25.Roschanski N, Fischer J, Guerra B, Roesler U (2014) Development of a Multiplex Real-Time PCR for the Rapid Detection of the Predominant Beta-Lactamase Genes CTX-M, SHV, TEM and CIT-Type AmpCs in Enterobacteriaceae. PLoS ONE 9(7): e100956 10.1371/journal.pone.0100956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaikh Sibhghatulla, Fatima Jamale, ShaziShakil Syed Mohd. Danish Rizvi, Kamal Mohammad Amjad. Antibiotic resistance and extended spectrum beta-lactamases: Types, epidemiology and treatmentSaudi J Biol Sci. 2015. January; 22(1): 90–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kasap M., Fashae K., Torol S., Kolayli F., Budak F., & Vahaboglu H. (2010). Characterization of ESBL (SHV-12) producing clinical isolate of Enterobacter aerogenes from a tertiary care hospital in Nigeria. Annals of clinical microbiology and antimicrobials, 9, 1 10.1186/1476-0711-9-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aibinu I. E., Ohaegbulam V. C., Adenipekun E. A., Ogunsola F. T., Odugbemi T. O., & Mee B. J. (2003). Extended-spectrum beta-lactamase enzymes in clinical isolates of Enterobacter species from Lagos, Nigeria. Journal of clinical microbiology, 41(5), 2197–200. 10.1128/jcm.41.5.2197-2200.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alo M. N, Anyim C., Igwe J. C. and Elom M (2012). Presence of extended spectrum β-lactamase (ESBL) E. coli and K. pneumonia isolated from blood cultures of hospitalized patients. Advances in Applied Science Research 3 (2):821–825. [Google Scholar]
- 30.Adeyankinnu, F. A., Motayo, B. O., Akinduti, A., Akinbo, J., Ogiogwa, J. I., Aboderin, B. W., et al. (2014). A Multicenter Study of Beta-Lactamase Resistant Escherichia coli and Klebsiella pneumoniae Reveals High Level Chromosome Mediated Extended Spectrum β Lactamase Resistance in Ogun State, Nigeria. Interdisciplinary perspectives on infectious diseases, 819896. [DOI] [PMC free article] [PubMed]
- 31.Afunwa R. A., Odimegwu D. C., Iroha R. I., & Esimone C. O. (2011). Antimicrobial resistance status and prevalence rates of extended spectrum beta-lactamase (ESBL) producers isolated from a mixed human population. Bosnian journal of basic medical sciences, 11(2), 91–96. 10.17305/bjbms.2011.2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McNulty C., Lecky D. M., Xu-McCrae L., Nakiboneka-Ssenabulya D., Chung K. T., Nichols T., et al. (2018). CTX-M ESBL-producing Enterobacteriaceae: estimated prevalence in adults in England in 2014. The Journal of antimicrobial chemotherapy, 73(5), 1368–1388. 10.1093/jac/dky007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Díaz-Agero Pérez C., López-Fresneña N., Rincon Carlavilla A. L., Hernandez Garcia M., Ruiz-Garbajosa P., Aranaz-Andrés J. M., et al. (2019). Local prevalence of extended-spectrum beta-lactamase (ESBL) producing Enterobacteriaceae intestinal carriers at admission and co-expression of ESBL and OXA-48 carbapenemase in Klebsiella pneumoniae: a prevalence survey in a Spanish University Hospital. BMJ open, 9(3), e024879 10.1136/bmjopen-2018-024879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benner K. W., Prabhakaran P., & Lowros A. S. (2014). Epidemiology of infections due to extended-spectrum Beta-lactamase-producing bacteria in a pediatric intensive care unit. The journal of pediatric pharmacology and therapeutics: JPPT: the official journal of PPAG, 19(2), 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abrar S., Hussain S., Khan R. A., Ul Ain N., Haider H., & Riaz S. (2018). Prevalence of extended-spectrum-β-lactamase-producing Enterobacteriaceae: first systematic meta-analysis report from Pakistan. Antimicrobial resistance and infection control, 7, 26 10.1186/s13756-018-0309-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iroha I R, Esimone C O, Neumann S et al. “First description of Escherichia coli producing CTX-M-15- extended spectrum betalactamse (ESBL) in out- patients from South Eastern Nigeria. Annals of Clinical Microbiology and Antimicrobials 2012:11–19. 10.1186/1476-0711-11-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olowe O A, Oladipo G O, Makanjuola O A, and Olaitan J O. “Prevalence of Extended Spectrum Betalactamases (ESBLs) Carrying Genes in Klebsiellaspp from Clinical Samples at Ile-Ife, South Western Nigeria,” International Journal of Pharma Medicine and Biological Sciences, Vol.1, No.2, pp.129–138, October 2012. [Google Scholar]
- 38.Raji M A, Jamal W, Ojemeh O and Rotimi V O (2015). “Sequence Analysis of genes Mediating Extended Spectrum Beta Lactamase (ESBL) production in isolates of Enterobacteriaceaein a Lagos Teaching Hospital, Nigeria. BMC Infectious Diseases 15:259 10.1186/s12879-015-1005-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohammed Y, Gadzama GB, Zailani SB, Aboderin AO 2016. Characterization of Extended-Spectrum Beta-lactamase from Escherichia coli and Klebsiella Species from North Eastern Nigeria. Journal of Clinical and Diagnostic Research: JCDR. 10(2):DC07–DC10. 10.7860/JCDR/2016/16330.7254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moremi N, Claus H, Vogel U, Mshana SE. Faecal carriage of CTX-M extended-spectrum beta-lactamase-producing Enterobacteriaceae among street children dwelling in Mwanza city, Tanzania. PLoS ONE (2017) 12(9): e0184592 10.1371/journal.pone.0184592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ouedraogo Abdoul-Salam et al. “High Prevalence of Extended-Spectrum SS-Lactamase Producing Enterobacteriaceae among Clinical Isolates in Burkina Fas.” BMC Infectious Diseases 16 (2016): 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chandramohan Lakshmi, and Revell Paula A. “Prevalence and Molecular Characterization of Extended-Spectrum-Β-Lactamase-Producing Enterobacteriaceae in a Pediatric Patient Population.” Antimicrobial Agents and Chemotherapy 569 (2012): 4765–4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Díaz Miguel A. et al. “Diversity of Escherichia Coli Strains Producing Extended-Spectrum Β-Lactamases in Spain: Second Nationwide Study.” Journal of Clinical Microbiology 488 (2010): 2840–2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keite da Silva Nogueira, Danieli Conte, Maia Fernanda Valverde, Dalla-Costa Libera Maria. Distribution of extended-spectrum β-lactamase types in a Brazilian tertiary hospital. Rev. Soc. Bras. Med. Trop. 2015. April 48 (2): 162–169 10.1590/0037-8682-0009-2015 [DOI] [PubMed] [Google Scholar]
- 45.Pallecchi L, Bartoloni A, Fiorelli C, et al. Rapid Dissemination and Diversity of CTX-M Extended-Spectrum β-Lactamase Genes in Commensal Escherichia coli Isolates from Healthy Children from Low-Resource Settings in Latin America. Antimicrobial Agents and Chemotherapy. 2007;51(8):2720–272 10.1128/AAC.00026-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahmed SH, Daef EA, Badary MS, Mahmoud MA, Abd-Elsayed AA. Nosocomial blood stream infectionin intensive care units at Assiut University Hospitals (Upper Egypt) with special reference to extendedspectrum beta-lactamase producing organisms. BMC Res Notes.2009; 2: 76 10.1186/1756-0500-2-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.García C, Astocondor L, Rojo-Bezares B, Jacobs J, Sáenz Y. Molecular Characterization of Extended-Spectrum β-Lactamase-Producer Klebsiella pneumoniae Isolates Causing Neonatal Sepsis in Peru. The American Journal of Tropical Medicine and Hygiene. 2016;94(2):285–288. 10.4269/ajtmh.15-0373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bali BE, Acik L, Sultan N. Phenotypic and molecular characterization of SHV, TEM, CTX-M and extended spectrum beta lactamases produced by Escherichia coli, Acinobacter baumannii and Klebsiella isolates in a Turkish Hospital. Afr J of Res. 2010;4(8):650–54. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.