Mitochondrial-derived compartments (MDCs) regulate the mitochondrial proteome in response to nutrient stress. English et al. now show that the ERMES complex and the conserved GTPase Gem1 promote MDC biogenesis at sites of contact between the endoplasmic reticulum and mitochondria.
Abstract
Mitochondria are dynamic organelles with essential roles in signaling and metabolism. We recently identified a cellular structure called the mitochondrial-derived compartment (MDC) that is generated from mitochondria in response to amino acid overabundance stress. How cells form MDCs is unclear. Here, we show that MDCs are dynamic structures that form and stably persist at sites of contact between the ER and mitochondria. MDC biogenesis requires the ER–mitochondria encounter structure (ERMES) and the conserved GTPase Gem1, factors previously implicated in lipid exchange and membrane tethering at ER–mitochondria contacts. Interestingly, common genetic suppressors of abnormalities displayed by ERMES mutants exhibit distinct abilities to rescue MDC formation in ERMES-depleted strains and are incapable of rescuing MDC formation in cells lacking Gem1. Thus, the function of ERMES and Gem1 in MDC biogenesis may extend beyond their conventional role in maintaining mitochondrial phospholipid homeostasis. Overall, this study identifies an important function for ER–mitochondria contacts in the biogenesis of MDCs.
Introduction
Mitochondria are metabolic and signaling organelles that act centrally in ATP production, heme and iron-sulfur cluster synthesis, and the metabolism of lipids, nucleotides, amino acids, and other metabolites (Rutter and Hughes, 2015). Dysfunctional mitochondria are linked to many age-related and metabolic disorders (Nunnari and Suomalainen, 2012; Wallace, 2005). As such, cells are adept at maintaining mitochondrial homeostasis under a variety of metabolic and environmental stress conditions. Well-characterized systems used by cells to maintain mitochondrial homeostasis include mitochondrial fission and fusion (Labbé et al., 2014), mitochondrial-localized proteases (Quirós et al., 2015), mitophagy (Pickles et al., 2018), the ubiquitin-proteasome system (Karbowski and Youle, 2011), mitochondrial-derived vesicles (Sugiura et al., 2014), and the mitochondrial unfolded protein response (Shpilka and Haynes, 2018). These pathways operate in coordination to maintain mitochondrial health, and failure of these systems is linked to a host of human disorders (Nunnari and Suomalainen, 2012; Wallace, 2005).
We recently discovered a new mitochondrial protein remodeling pathway in budding yeast, Saccharomyces cerevisiae, called the mitochondrial-derived compartment (MDC) pathway (Hughes et al., 2016). MDCs are distinct subdomains of mitochondria that are generated in response to defects in vacuolar acidification. These structures selectively incorporate the outer mitochondrial membrane (OMM) import receptor Tom70 and inner mitochondrial membrane (IMM) metabolite carriers of the SLC25 family while leaving the remainder of the mitochondrial proteome intact. After formation, MDCs are removed from mitochondria by fission and degraded in the lysosome-like vacuole by autophagy. We recently found that MDC formation is activated in response to a variety of insults that acutely elevate intracellular amino acid pools, including impairment of vacuolar amino acid storage, inhibition of protein translation, exposure to amino acid–derived aldehydes, and inhibition of the mechanistic target of rapamycin pathway (Schuler et al., 2020 Preprint). We also discovered that MDC formation is coordinated with metabolite transporter control on other cellular membranes and that the MDC pathway cooperates with the vacuole and the multivesicular body/endosomal sorting complexes required for transport pathway (Henne et al., 2011) to maintain cell health during amino acid excess (Schuler et al., 2020 Preprint). Despite the emerging importance of this pathway in maintaining mitochondrial homeostasis and cell health, it is unclear how cells generate MDCs.
Here, we sought to illuminate the mechanisms and machinery underlying MDC formation. Using superresolution imaging, we show that MDCs are dynamic structures generated from mitochondria at sites of contact with the ER. MDC biogenesis at ER–mitochondria contacts requires the ER–mitochondria encounter structure (ERMES) and GTPase activity by the conserved GTPase Gem1. Interestingly, common genetic suppressors of ERMES complex mutants differ in their ability to rescue MDC formation and are unable to restore MDC formation in cells lacking Gem1. Thus, it appears that the function of ERMES and Gem1 in MDC biogenesis is complex and may extend beyond the well-characterized role of these proteins in maintaining mitochondrial phospholipid homeostasis. Overall, these results identify a new function for ER–mitochondria contacts in the biogenesis of MDCs during amino acid overabundance stress.
Results and discussion
MDCs are dynamic structures
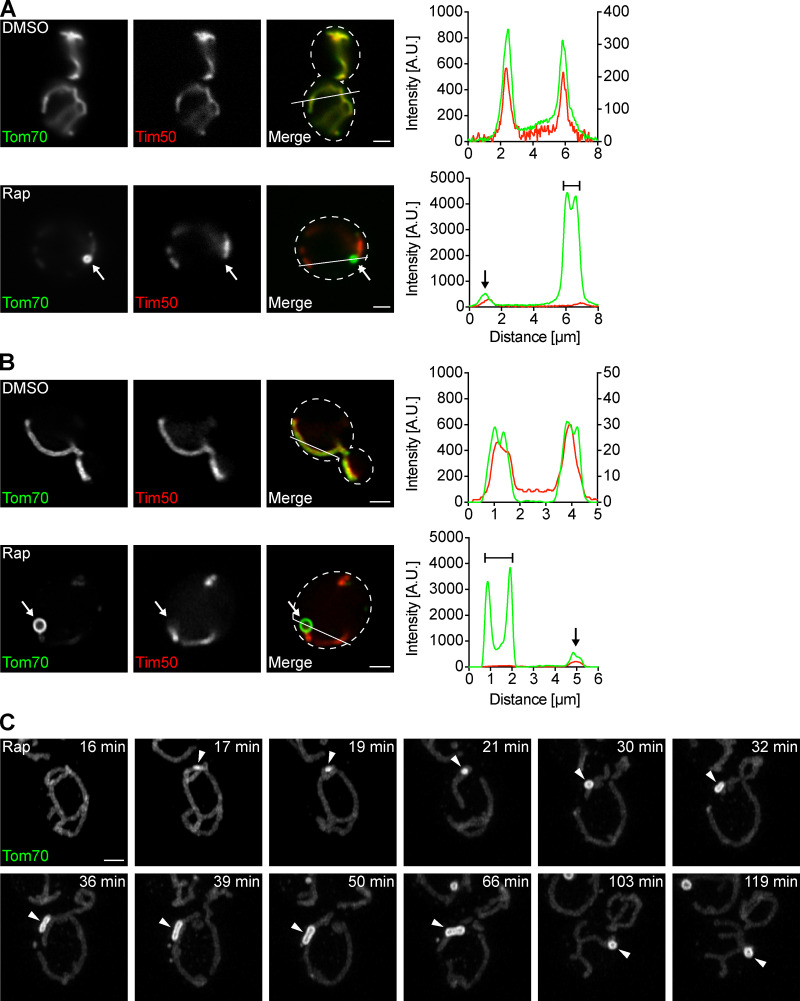
We previously identified MDCs as foci associated with mitochondria in cells with dysfunctional vacuoles (Hughes et al., 2016). In a recent study, we found that MDCs form in response to high levels of intracellular amino acids (Schuler et al., 2020 Preprint). We showed that rapamycin, a potent inhibitor of the mechanistic target of rapamycin (Heitman et al., 1991), increases intracellular amino acids and activates MDC formation (Schuler et al., 2020 Preprint). Rapamycin-induced MDCs are cargo selective and are identified by their enrichment of Tom70, an OMM import receptor required for the import of mitochondrial carriers (Söllner et al., 1990), and exclusion of other mitochondrial proteins, including a subset of IMM proteins such as Tim50 (Fig. 1 A; Yamamoto et al., 2002), as well as soluble matrix proteins (Hughes et al., 2016). To understand how MDCs form, we captured superresolution images of MDCs generated in response to rapamycin and found that they are large, circular structures commonly reaching ∼1 µm in diameter (Fig. 1 B). Using superresolution time-lapse imaging to visualize MDC biogenesis, we observed that MDC formation typically began with accumulation of Tom70 at a distinct site on a mitochondrial tubule (Fig. 1 C, 17 min). Over time, the Tom70 focus grew into a large, round structure and remained associated with mitochondrial tubules for extended periods of time (Fig. 1 C and Video 1). MDCs exhibited dynamic behavior and frequently appeared elongated (Fig. 1 C, 32–66 min; and Video 1).
Figure 1.
MDCs are dynamic structures. (A) Widefield images and line scan analyses of yeast expressing Tom70-GFP and Tim50-mCherry treated with DMSO or rapamycin. White arrows mark MDC. Lines mark fluorescence intensity profile position. Left and right y axes (top line scan graph) correspond to Tom70-GFP and Tim50-mCherry fluorescence intensity, respectively. The single y axis (bottom line scan graph) corresponds to Tom70-GFP and Tim50-mCherry fluorescence intensity. Bracket marks MDC. Black arrow marks a mitochondrial tubule. Images show single focal planes. Scale bars = 2 µm. (B) Superresolution images and line scan analyses of yeast expressing Tom70-GFP and Tim50-mCherry treated with DMSO or rapamycin. White arrows mark MDCs. Lines mark fluorescence intensity profile position. Left and right Y axes (top line scan graph) correspond to Tom70-GFP and Tim50-mCherry fluorescence intensity, respectively. The single Y axis (bottom line scan graph) corresponds to Tom70-GFP and Tim50-mCherry fluorescence intensity. Bracket marks an MDC. Black arrow marks mitochondrial tubule. Images show single focal planes. Scale bars = 2 µm. (C) Superresolution time-lapse images of yeast expressing Tom70-GFP treated with rapamycin. Arrowheads mark MDCs. Images show maximum intensity projections. Scale bars = 2 µm. See also Video 1. A.U., arbitrary units; Rap, rapamycin.
Video 1.
MDCs form from a single site and are dynamic (related to Fig. 1). Maximum intensity projection video of yeast expressing Tom70-GFP treated with rapamycin. Images were taken every minute and are shown at two frames per second.
MDCs form and stably persist at ER–mitochondria contacts
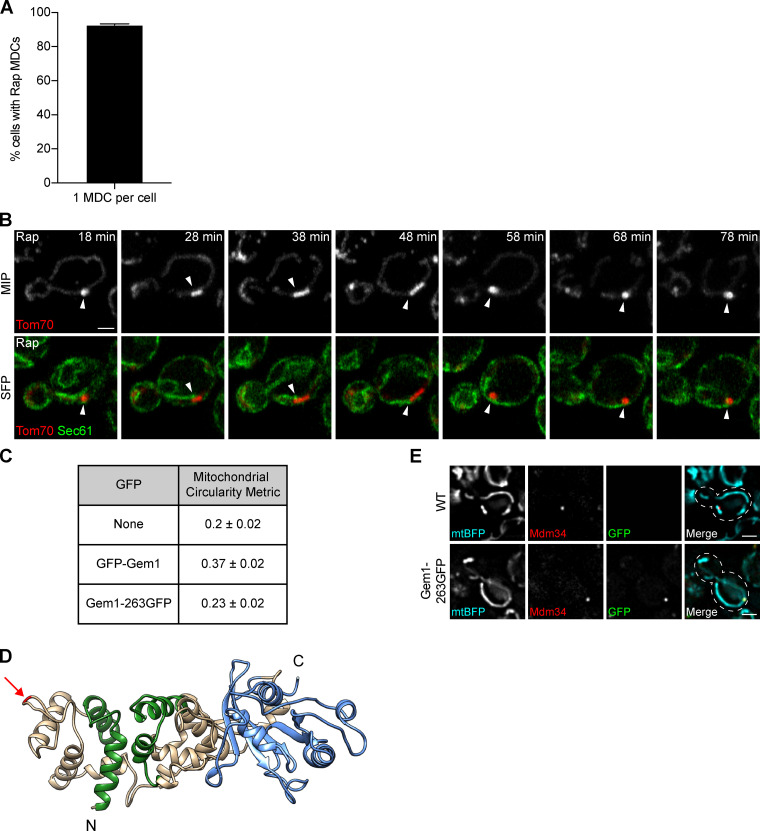
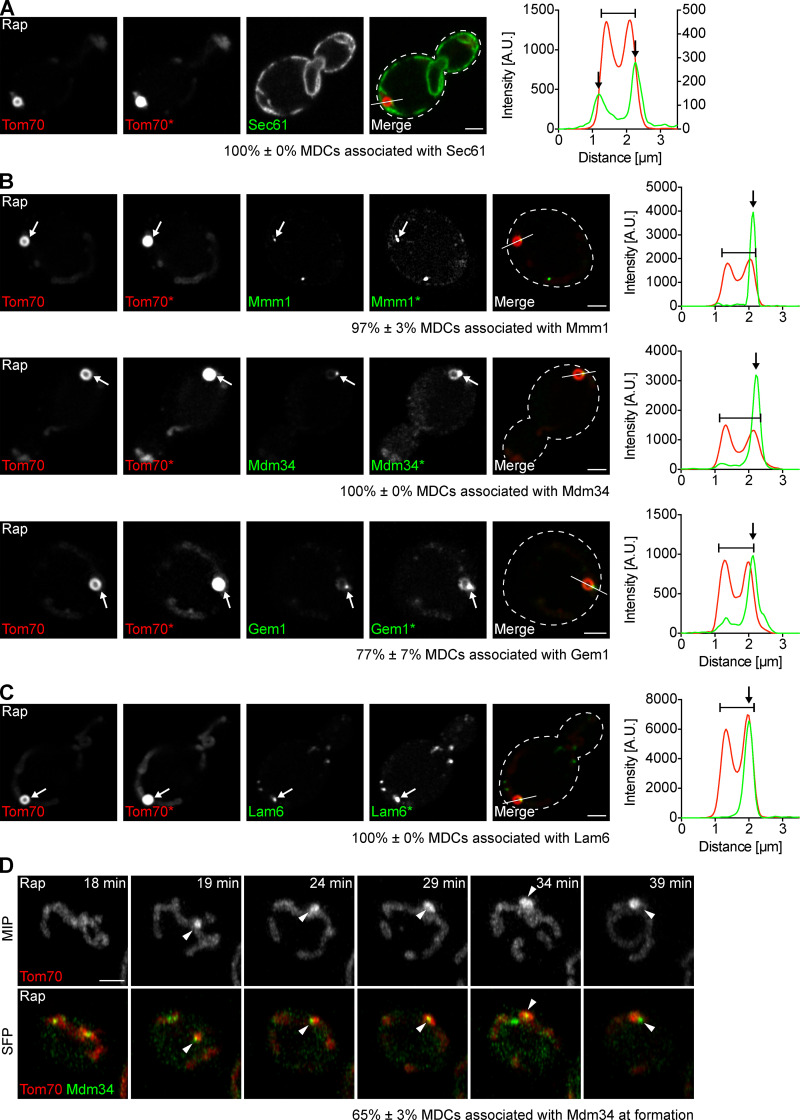
Although the mitochondrial network extends throughout the cell, we typically observed only one large MDC per cell upon treatment with rapamycin (Fig. S1 A), suggesting that MDCs are spatially linked to a distinct subcellular location. Events that take place at discrete sites on mitochondria often do so at points of contact with other organelles, such as the ER. We imaged MDCs in cells expressing the ER marker Sec61-GFP and found that 100% of MDCs were associated with ER tubules (Fig. 2 A and Fig. S1 B). MDCs primarily localized adjacent to the peripheral ER (77% ± 3% of MDCs, mean ± SEM) and often simultaneously associated with cytoplasmic ER tubules (57% ± 7% of peripheral ER-associated MDCs, mean ± SEM; Fig. 2 A and Fig. S1 B).
Figure S1.
MDCs stably associate with the ER (related to Fig. 2). (A) Percentage of MDC-positive cells with a single large MDC per cell upon treatment with rapamycin. Error bar shows mean ± SEM of three replicates, n = 50 cells with at least one MDC per replicate. (B) Superresolution time-lapse images of yeast expressing Tom70-mCherry and Sec61-GFP treated with rapamycin. Arrowheads mark Sec61 associated with MDCs. Images show maximum intensity projections (MIP) and single focal planes (SFP). Scale bar = 2 µm. (C) Gem1 fusion protein functionality was assessed by quantifying the shape of segmented mitochondria using the circularity metric (4π × area/perimeter2, where 1 is a perfect circle) described in Materials and methods. WT (none) mitochondria have a circularity of 0.2 ± 0.02 (mean ± SEM), and mitochondria of yeast expressing N-terminally tagged Gem1 (GFP-Gem1) rose to 0.37 ± 0.02. Mitochondria of yeast expressing internally tagged Gem1 (Gem1-263GFP) had a circularity of 0.23 ± 0.02, indicating near-WT shape and functionality of the fusion protein. (D) 3D model of Gem1. Green marks EF hands and blue marks the C-terminal GTPase domain (N-terminal GTPase domain not shown). Arrow marks the insertion site of GFP after amino acid 262 (Gem1-263GFP). Model displays up to amino acid 614 (full-length Gem1 is 662 amino acids). Homology modeling of Gem1 was performed using the SWISS-MODEL server (Waterhouse et al., 2018) and Miro structure (PDB accession no. 5KSZ; Klosowiak et al., 2016) as a template. 3D protein structures were rendered using UCSF Chimera (Pettersen et al., 2004). (E) Widefield images of yeast expressing no GFP (WT) or GFP inserted after amino acid 262 of Gem1 (Gem1-263GFP). Both strains express mitochondrial matrix-targeted BFP (mtBFP) and Mdm34-mCherry. Mitochondria of yeast expressing Gem1-263GFP display WT-like shape. Images show single focal planes. Scale bars = 2 µm. Rap, rapamycin.
Figure 2.
MDCs form and stably persist at ER–mitochondria contacts. (A) Superresolution images and line scan analysis of yeast expressing Tom70-mCherry and Sec61-GFP treated with rapamycin. Asterisk (Tom70*) indicates that the fluorescence intensity was increased. Line marks fluorescence intensity profile position. Left and right y axes of the line scan graph correspond to Tom70-mCherry and Sec61-GFP fluorescence intensity, respectively. Bracket marks an MDC. Black arrows mark Sec61 associated with MDCs. Images show a single focal plane. Percent associated shows mean ± SEM of three replicates, n = 10 MDCs per replicate. Scale bar = 2 µm. (B) Superresolution images and line scan analyses of yeast expressing Tom70-mCherry and Mmm1-GFP, Mdm34-GFP or Gem1-263GFP treated with rapamycin. Gem1 was internally tagged with GFP after amino acid 262 to create Gem1-263GFP, a functional fusion protein (see Fig. S1, C–E, and Materials and methods for details). Asterisks (Tom70*, Mmm1*, Mdm34*, and Gem1*) indicate that the fluorescence intensity was increased. Lines mark fluorescence intensity profile position. The y axis of the line scan graph corresponds to Tom70-mCherry and Mmm1-GFP, Mdm34-GFP, or Gem1-263GFP fluorescence intensity. Brackets mark MDCs. White and black arrows mark Mmm1, Mdm34, or Gem1 associated with MDCs. Images show single focal planes. Percent associated shows mean ± SEM of three replicates, n = 10 MDCs per replicate. Scale bars = 2 µm. (C) Superresolution images and line scan analysis of yeast expressing Tom70-mCherry and Lam6-GFP treated with rapamycin. Asterisk (Tom70*) indicates that the fluorescence intensity was increased. Line marks fluorescence intensity profile position. The y axis of the line scan graph corresponds to Tom70-mCherry and Lam6-GFP fluorescence intensity. Bracket marks an MDC. White and black arrows mark Lam6 associated with MDC. Images show single focal plane. Percent associated shows mean ± SEM of three replicates, n = 10 MDCs per replicate. Scale bar = 2 µm. (D) Superresolution time-lapse images of yeast expressing Tom70-mCherry and Mdm34-GFP treated with rapamycin. Arrowheads mark Mdm34 associated with MDCs. Images show maximum intensity projections (MIP) and single focal planes (SFP). Percent associated shows mean ± SEM of three replicates, n = 23 MDCs total. Scale bar = 2 µm. See also Fig. S1 and Video 2. A.U., arbitrary units; Rap, rapamycin.
In yeast, mitochondria are tethered to the ER by the ER–mitochondria encounter structure (ERMES), a complex composed of mitochondrial subunits Mdm10 and Mdm34, cytosolic subunit Mdm12, and ER subunit Mmm1 (Kornmann et al., 2009; Stroud et al., 2011). ERMES is regulated by Gem1, a conserved GTPase that colocalizes with the complex (Kornmann et al., 2011; Stroud et al., 2011). In addition, mitochondria and the ER are also connected by the interaction of OMM proteins Tom70 and Tom71 with Lam6, an ER-resident membrane protein (Elbaz-Alon et al., 2015; Murley et al., 2015). To test if MDCs associate with ER–mitochondria contacts, we visualized MDCs in cells expressing fluorescently tagged ER–mitochondria contact site proteins, all of which appear as discrete foci simultaneously associated with the ER and mitochondria (Elbaz-Alon et al., 2015; Kornmann et al., 2009, 2011; Murley et al., 2015). Because N-terminal GFP tagging of Gem1 yields partial functionality, we used an internally GFP-tagged Gem1 fusion protein that appears functional based on its lack of impact on mitochondrial morphology (Fig. 2 B; and Fig. S1, C–E; see Materials and methods for details). MDCs associated with the contact site markers Mmm1 (97% ± 3% of MDCs, mean ± SEM), Mdm34 (100% ± 0% of MDCs, mean ± SEM), Gem1 (77% ± 7% of MDCs, mean ± SEM), and Lam6 (100% ± 0% of MDCs; mean ± SEM; Fig. 2, B and C), indicating that MDCs localize to ER–mitochondria contacts. Interestingly, although cells treated with rapamycin harbored multiple ER–mitochondria contacts marked by ERMES (10 ± 1 Mdm34-GFP foci per cell [mean ± SEM], n = 30 cells), most MDCs associated with only a single Mdm34 focus (60% ± 1% [mean ± SEM], n = 30 MDCs), though we observed MDCs associated with up to four Mdm34 foci simultaneously. Furthermore, the mitochondria-localized contact site proteins Mdm34 and Gem1, but not the ER-localized proteins Mmm1 and Lam6, were present at low levels in MDCs (Fig. 2, B and C). This observation likely stems from the fact that, in addition to contact site foci, we detected low levels of Mdm34 and Gem1, but not Mmm1 and Lam6, distributed along mitochondrial tubules (Fig. 2, B and C).
Finally, we used superresolution time-lapse imaging in cells expressing Mdm34-GFP and Tom70-mCherry to determine whether MDCs form at ER–mitochondria contacts or if association with the ER occurs at a later stage of MDC biogenesis. We found that 65% ± 3% (mean ± SEM) of MDCs associated with Mdm34 at the time of formation (Fig. 2 D and Video 2), and the other 35% ± 3% (mean ± SEM) associated with Mdm34 within 4 min of initiation. This latter observation suggests that our inability to detect MDC–Mdm34 association in some instances may result from technical limitations. Together, our data indicate that MDC biogenesis is spatially linked to ER–mitochondria contacts and that MDCs remain stably associated with these sites after formation.
Video 2.
MDCs form at ER–mitochondria contacts (related to Fig. 2). Maximum intensity projection video of yeast expressing Tom70-mCherry and Mdm34-GFP treated with rapamycin. Images were taken every minute and are shown at two frames per second.
MDC formation requires ERMES and Gem1
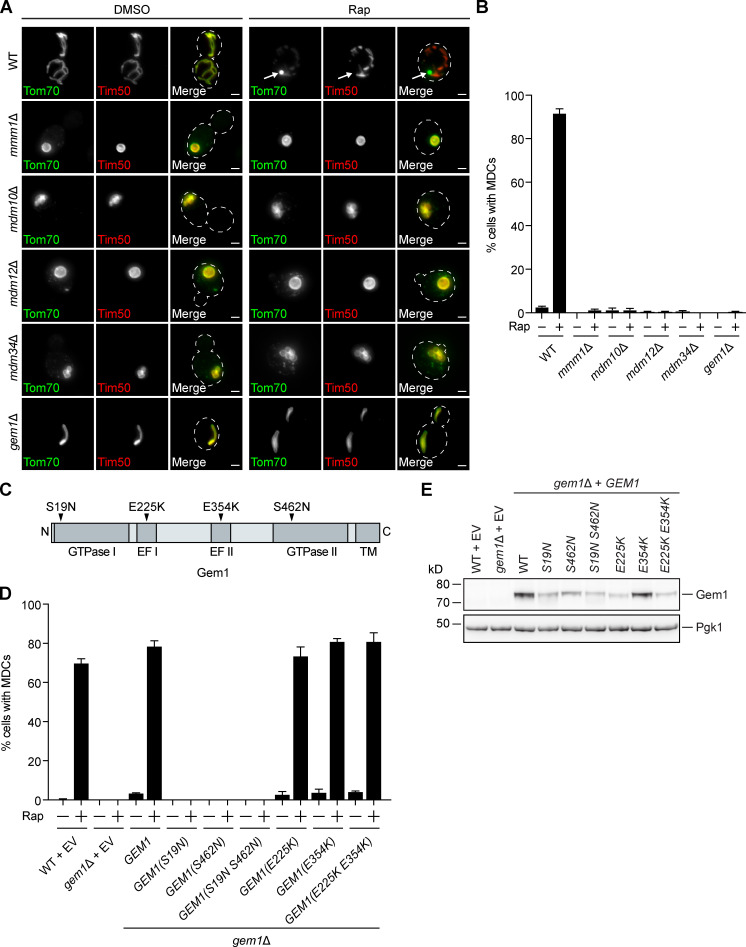
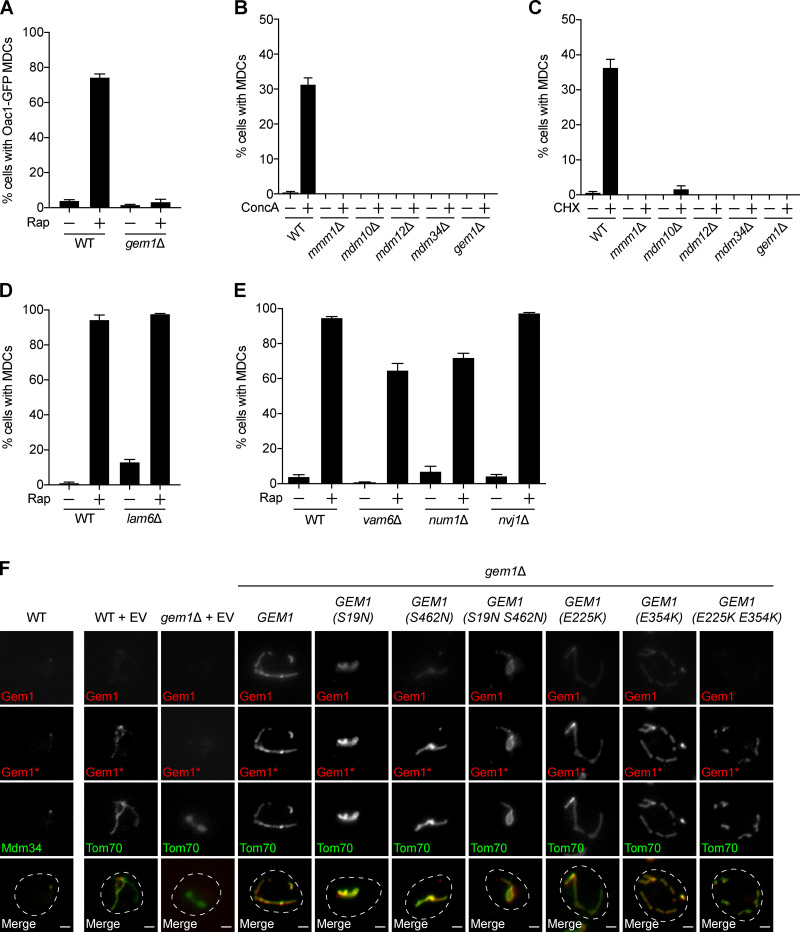
We next tested the requirement of ER–mitochondria contact site machinery for MDC formation. We individually deleted genes encoding ERMES subunits Mmm1, Mdm10, Mdm12, and Mdm34, which prevent complex assembly (Kornmann et al., 2009) and produce spherical mitochondria (Berger et al., 1997; Sogo and Yaffe, 1994). We found that these mutants failed to form rapamycin-induced MDCs (Fig. 3, A and B). Similarly, MDC formation was also inhibited in cells lacking GEM1 (Fig. 3, A and B). In the absence of Gem1, the ERMES complex remains intact, though gem1Δ cells display fewer and larger ERMES foci by fluorescence microscopy (Kornmann et al., 2011). We obtained similar results with other MDC cargo (Oac1; Fig. S2 A) and other well-characterized MDC inducers, including concanamycin A, which alters vacuolar storage of amino acids by disrupting the activity of the vacuolar H+-ATPase (Dröse et al., 1993), and cycloheximide, which increases amino acid pools by inhibiting protein translation (Fig. S2, B and C; Beugnet et al., 2003). In contrast to ERMES/Gem1, MDC formation was unaltered in lam6Δ cells (Fig. S2 D). MDC biogenesis triggered by rapamycin was also largely unaffected by the loss of other organelle contacts, including vacuole and mitochondria patches (vam6Δ; Elbaz-Alon et al., 2014; Hönscher et al., 2014), mitochondria–ER–cortex anchors (MECAs, num1Δ; Lackner et al., 2013), and nucleus–vacuole junctions (nvj1Δ; Fig. S2 E; Pan et al., 2000). These results identify a specific role for ERMES/Gem1 in the biogenesis of MDCs.
Figure 3.
MDC formation requires ERMES and Gem1. (A) Widefield images of WT and the indicated mutant yeast expressing Tom70-GFP and Tim50-mCherry treated with DMSO or rapamycin. Arrows mark MDCs. Images show maximum intensity projections. Scale bars = 2 µm. (B) Quantification of A. Error bars show mean ± SEM of three replicates, n = 100 cells per replicate. (C) Schematic of Gem1 and mutants used in D. (D) Quantification of rapamycin-induced MDC formation in WT and gem1Δ cells expressing EV or overexpressing GEM1 or the indicated GEM1 mutant. Error bars show mean ± SEM of three replicates, n = 100 cells per replicate. (E) Western blot analysis of Gem1 in WT and gem1Δ cells expressing EV or overexpressing GEM1 or the indicated GEM1 mutant. Pgk1 was used as a loading control. Note that the Gem1 antibody did not detect endogenous Gem1 (WT + EV). See also Fig. S2. Rap, rapamycin; TM, transmembrane.
Figure S2.
MDC formation requires ERMES and Gem1, but not other organelle contact site proteins (related to Fig. 3). (A) Quantification of rapamycin-induced MDC formation in WT and gem1Δ cells. Oac1-positive, Tim50-negative MDCs were quantified. Error bars show mean ± SEM of three replicates, n = 50–100 cells per replicate. (B) Quantification of concanamycin A–induced MDC formation in WT and the indicated mutant yeast. Error bars show mean ± SEM of three replicates, n = 100 cells per replicate. (C) Quantification of cycloheximide-induced MDC formation in WT and the indicated mutant yeast. Error bars show mean ± SEM of three replicates, n = 100 cells per replicate. (D) Quantification of rapamycin-induced MDC formation in WT and lam6Δ cells. Error bars show mean ± SEM of three replicates, n = 100 cells per replicate. (E) Quantification of Rap-induced MDC formation in WT and the indicated mutant yeast. Error bars show mean ± SEM of three replicates, n = 100 cells per replicate. (F) Indirect immunofluorescence of cells expressing EV or overexpressing GEM1 or the indicated GEM1 mutant and Mdm34-GFP or Tom70-GFP. Top row images (Gem1) were acquired with the same settings and are nonadjusted. Asterisk (Gem1*) indicates that the fluorescence intensity was increased. Images show single focal planes. Merged images show Gem1* and Mdm34-GFP or Tom70-GFP. Scale bars = 2 µm. CHX, cycloheximide; ConcA, concanamycin A; Rap, rapamycin.
Gem1 GTPase activity is required for MDC formation
Gem1 is anchored by its C-terminus in the OMM (Frederick et al., 2004) and harbors two GTPase domains that hydrolyze GTP and two EF hands that are capable of binding calcium ions (Fig. 3 C; Koshiba et al., 2011). To determine if these functional domains are important for MDC formation, we generated well-characterized mutations in the GTPase motifs and EF hands of Gem1 and overexpressed them in gem1Δ cells (Fig. 3 C; Koshiba et al., 2011). We found that mutations that disrupted Gem1’s ability to bind calcium ions had no effect on MDC formation (Fig. 3 D). However, MDC formation was abolished when the ability of Gem1 to hydrolyze GTP via either GTPase domain was compromised (Fig. 3 D). Interestingly, we observed no MDCs when a single GTPase domain was mutated, although single GTPase mutants have reduced, not completely impaired, GTP hydrolysis (Koshiba et al., 2011). Importantly, and consistent with previous observations (Koshiba et al., 2011), mutations in the GTPase motifs (S19N, S462N, and S19N S462N) and the first EF hand (E225K and E225K E354K) resulted in decreased, yet detectable, Gem1 protein levels compared with the WT (Fig. 3 E). Of note, the steady-state levels of all overexpressed Gem1 constructs were higher than endogenous Gem1 (which was undetectable by Western blot with our antibody; Fig. 3 E). Additionally, although endogenous Gem1 localizes to distinct foci, WT and mutant Gem1, when overexpressed, localized similarly along mitochondrial tubules, as assessed by indirect immunofluorescence, indicating that overexpressed Gem1 likely saturates binding sites on ERMES (Fig. S2 F). Thus, the ability of a given Gem1 mutant to promote MDC formation did not correlate with its level of expression or ability to localize to mitochondrial tubules. These results suggest that Gem1 GTPase activity is required for MDC formation, while calcium binding is dispensable.
Disruption of select phospholipid biosynthesis pathways does not impact MDC formation
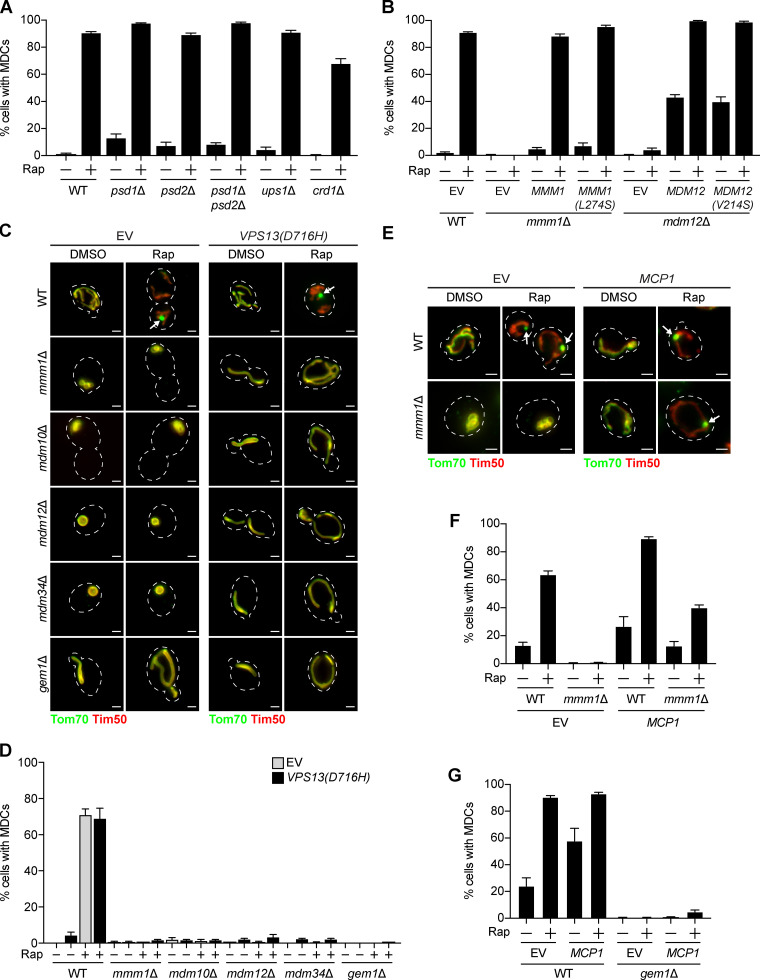
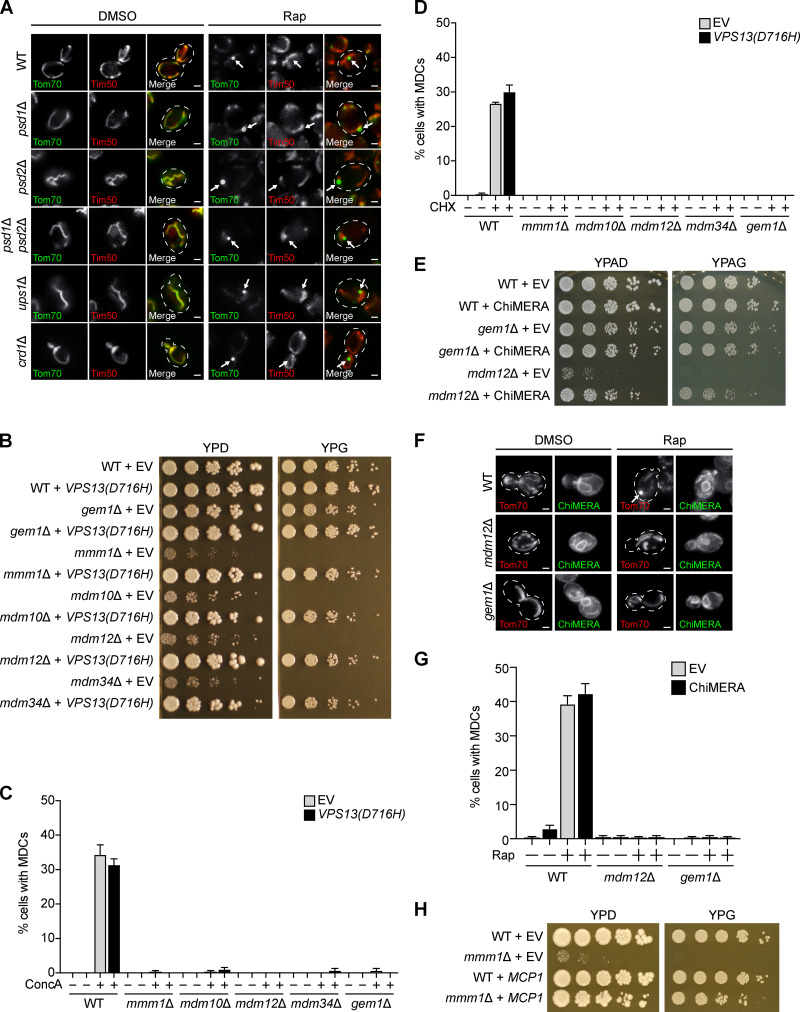
Next, we aimed to identify the function of ERMES/Gem1 in MDC formation. The central role of the ERMES complex is to facilitate phospholipid transport between mitochondria and the ER (Kornmann et al., 2009). Phospholipid synthesis takes place at mitochondria and the ER, and many phospholipid intermediates are transported between the two organelles. Specifically, phosphatidylserine is generated at the ER and can be transported to the IMM, where it is converted to phosphatidylethanolamine by a mitochondria-localized pool of the phosphatidylserine decarboxylase Psd1 (Clancey et al., 1993; Friedman et al., 2018; Trotter et al., 1993). Similarly, phosphatidic acid is trafficked from the ER to the IMM by Ups1 (Connerth et al., 2012) and ultimately converted to cardiolipin by Crd1 (Chang et al., 1998; Tuller et al., 1998). Yeast cells lacking ERMES have abnormal phospholipid levels (Kornmann et al., 2009), and the ERMES complex subunits Mmm1, Mdm12, and Mdm34 possess lipid-binding domains (AhYoung et al., 2015; Jeong et al., 2016, 2017; Kawano et al., 2018; Kopec et al., 2010). Therefore, we considered it a possibility that ERMES is required to achieve a proper mitochondrial phospholipid composition necessary for MDC formation. To test this, we assessed MDC formation in various phospholipid synthesis mutants, including psd1Δ, ups1Δ, and crd1Δ. Because yeast contain Psd2, an additional phosphatidylserine decarboxylase that localizes to the Golgi and vacuole (Trotter and Voelker, 1995), we also examined MDC formation in psd2Δ and psd1Δ psd2Δ cells. In contrast to the MDC defects we observed in ERMES/Gem1 mutants, MDC formation was unaffected in phospholipid synthesis mutants (Fig. 4 A and Fig. S3 A). Consistent with these observations, we also found that MDCs persisted in cells expressing MMM1(L274S) and MDM12(V214S), mutants reported to impair lipid trafficking by the ERMES complex (Fig. 4 B; Kawano et al., 2018).
Figure 4.
Genetic suppressors of ERMES mutants differ in their ability to restore MDC formation. (A) Quantification of rapamycin-induced MDC formation in WT and the indicated mutant yeast. Error bars show mean ± SEM of three replicates, n = 100 cells per replicate. (B) Quantification of Rap-induced MDC formation in WT, mmm1Δ, and mdm12Δ cells expressing EV or overexpressing MMM1, MMM1(L247S), MDM12, or MDM12(V214S). Error bars show mean ± SEM of three replicates, n = 100 cells per replicate. (C) Widefield images of WT and the indicated mutant yeast expressing Tom70-GFP and Tim50-mCherry and EV or VPS13(D716H) treated with DMSO or rapamycin. Arrows mark MDCs. Images show maximum intensity projections of merged Tom70-GFP and Tim50-mCherry. Scale bars = 2 µm. (D) Quantification of C. Error bars show mean ± SEM of three replicates, n = 100 cells per replicate. (E) Widefield images of WT and mmm1Δ yeast expressing Tom70-GFP and Tim50-mCherry and EV or overexpressing MCP1 treated with DMSO or Rap. Arrows mark MDCs. Images show maximum intensity projections of merged Tom70-GFP and Tim50-mCherry. Scale bars = 2 µm. (F) Quantification of E. Error bars show mean ± SEM of three replicates, n = 100 cells per replicate. (G) Quantification of rapamycin-induced MDC formation in WT and gem1Δ cells expressing EV or overexpressing MCP1. Error bars show mean ± SEM of three replicates, n = 100 cells per replicate. See also Fig. S3. Rap, rapamycin.
Figure S3.
Genetic suppressors of ERMES mutants differ in their ability to restore MDC formation (related to Fig. 4). (A) Widefield images of WT and the indicated mutant yeast expressing Tom70-GFP and Tim50-mCherry treated with DMSO or rapamycin. Arrows mark MDCs. Images show single focal planes. Scale bars = 2 µm. (B) Serial dilutions of WT and the indicated mutant yeast expressing EV or VPS13(D716H) spotted on media with glucose (YPD) or glycerol (YPG). (C) Quantification of concanamycin A–induced MDC formation in WT and the indicated mutant yeast expressing EV or VPS13(D716H). Error bars show mean ± SEM of three replicates, n = 100 cells per replicate. (D) Quantification of cycloheximide-induced MDC formation in WT and the indicated mutant yeast expressing EV or VPS13(D716H). Error bars show mean ± SEM of three replicates, n = 100 cells per replicate. (E) Serial dilutions of WT and the indicated mutant yeast expressing EV or ChiMERA spotted on media with glucose (YPAD) or glycerol (YPAG). (F) Widefield images of WT and the indicated mutant yeast expressing Tom70-mCherry and ChiMERA (GFP) treated with DMSO or rapamycin. Arrow marks an MDC. Tom70-mCherry images show maximum intensity projections and ChiMERA images show single focal planes. Scale bars = 2 µm. (G) Quantification of rapamycin-induced MDC formation in WT and the indicated mutant yeast expressing empty EV or ChiMERA. Error bars show mean ± SEM of three replicates, n = 100 cells per replicate. (H) Serial dilutions of WT and mmm1Δ cells expressing EV or overexpressing MCP1 spotted on media with glucose (YPD) or glycerol (YPG). CHX, cycloheximide; ConcA, concanamycin A; Rap, rapamycin.
Genetic suppressors of ERMES mutants differ in their ability to restore MDC formation
As an alternative approach to test whether the loss of MDCs in ERMES mutants was due to disrupted phospholipid synthesis, we assayed the ability of three different genetic suppressors of ERMES mutants to restore MDC formation. The first suppressor is a well-characterized dominant substitution mutation in the endosomal protein Vps13 (Lang et al., 2015). Cells lacking subunits of the ERMES complex characteristically display spherical mitochondria (Berger et al., 1997; Sogo and Yaffe, 1994), loss of mitochondrial DNA (Hobbs et al., 2001), minimal growth on fermentable media, and no growth on nonfermentable media (Berger et al., 1997; Burgess et al., 1994; Youngman et al., 2004). Expression of the VPS13(D716H) allele was reported to suppress the characteristic abnormalities exhibited by ERMES mutants, importantly, without restoring complex assembly (Lang et al., 2015). Three lines of evidence suggest that VPS13(D716H) suppresses ERMES mutant–associated defects by restoring mitochondrial lipid homeostasis. First, Vps13 localizes to vacuole and mitochondria patches (John Peter et al., 2017), contacts between mitochondria and vacuoles that expand in the absence of ERMES to funnel lipids to mitochondria (Elbaz-Alon et al., 2014; Hönscher et al., 2014). Second, loss of VPS13 and ERMES is synthetically lethal, suggesting that Vps13 functions redundantly with ERMES (Lang et al., 2015). Third, Vps13 has the capacity to transport lipids between organelles (Kumar et al., 2018). We expressed VPS13(D716H) in cells lacking individual ERMES subunits or Gem1 and observed tubular mitochondrial morphology (Fig. 4 C) and near-WT growth on fermentable and nonfermentable media (Fig. S3 B), consistent with previous findings (Lang et al., 2015; Park et al., 2016). We then assessed MDC formation in these strains and found that although mitochondrial health was restored, MDCs failed to form in response to rapamycin, concanamycin A, or cycloheximide (Fig. 4, C and D; and Fig. S3, C and D). Although we did not detect MDCs in ERMES/Gem1 mutants expressing VPS13(D716H), we considered it possible that the cells were able to execute the initial step of MDC biogenesis, accumulation of Tom70 of increased fluorescence intensity versus the mitochondrial tubule (Fig. 1 C). However, we observed no obvious enrichment of Tom70 on mitochondria and no clear difference between untreated and rapamycin-treated cells (Fig. 4 C), suggesting that ERMES mutants expressing VPS13(D716H) are unable to initiate MDC formation.
Next, we tested whether artificially tethering mitochondria to the ER could restore the ability of ERMES/Gem1 mutants to generate MDCs. We used ChiMERA (construct helping in mitochondria-ER association), a synthetic ER–mitochondria tether that suppresses the growth deficiencies and abnormal mitochondrial morphology displayed by some ERMES mutants (Fig. S3, E and F; Kornmann et al., 2009). Like the VPS13(D716H) suppression experiment, we were unable to observe MDCs in mdm12Δ or gem1Δ cells expressing ChiMERA, despite substantial restoration of the tubular mitochondrial morphology (Fig. S3, F and G).
Finally, we used a third well-characterized method to suppress the cellular defects associated with loss of ERMES, overexpression of the OMM protein Mcp1 (Tan et al., 2013). It was recently shown that MCP1 overexpression recruits Vps13 to mitochondria, facilitating bypass of the ERMES complex (John Peter et al., 2017). As previously reported, we found that overexpression of MCP1 restored the mitochondrial morphology and growth of cells lacking the ERMES complex (Fig. 4 E and Fig. S3 H). Interestingly, in contrast to our VPS13(D716H) and ChiMERA results, MDC formation was partially rescued by MCP1 overexpression in strains lacking the ERMES subunit Mmm1 (Fig. 4, E and F). However, overexpression of MCP1 did not restore MDCs in cells lacking Gem1 (Fig. 4 G). Taken together, these results suggest that the role of ERMES and Gem1 in MDC formation is potentially separable and may extend beyond the function of these proteins in the maintenance of mitochondrial phospholipid homeostasis.
Overall, the data presented here provide a step forward in our understanding of the molecular underpinnings of how cells form MDCs, structures with important implications in the maintenance of mitochondrial homeostasis during metabolic excess. Using superresolution imaging, we now show that MDCs are dynamic structures that stably persist at contacts with the ER. We also demonstrate that MDC biogenesis is spatially linked to ER–mitochondria contacts and that MDCs mature at these sites over time. Finally, we identify ERMES and Gem1 as important factors for MDC biogenesis.
These results raise several important questions for future consideration. Chief among them is: What is the role of ERMES/Gem1 and the ER in MDC formation? To date, the best-characterized function of ERMES is to facilitate phospholipid transport between mitochondria and the ER. However, we find that MDCs persist in numerous phospholipid synthesis mutants and in cells expressing Mmm1 and Mdm12 mutant proteins with impaired lipid transfer activities. We also show that common suppressors of ERMES mutant–associated defects, expression of a synthetic ER–mitochondria tether or restoration of mitochondrial lipid homeostasis by expression of VPS13(D716H), do not rescue MDCs in cells lacking ERMES/Gem1. These results suggest that regulation of mitochondrial phospholipid homeostasis by ERMES/Gem1 may not play a role in MDC formation.
In apparent contradiction with this conclusion, however, we found that MCP1 overexpression partially restores MDCs in cells lacking the ERMES complex. What could explain this difference? One possibility is that overexpressed MCP1 is more effective at suppressing the defects associated with loss of ERMES than expression of VPS13(D716H). However, we have no evidence to support this hypothesis, and our data show that each of these suppressors restored mitochondrial morphology and growth in ERMES mutants to a similar extent. Alternatively, MCP1 overexpression may bypass the requirement of ERMES in MDC formation through an unknown mechanism not related to its function in the ERMES mutant rescue pathway. It is interesting to note that while both ERMES and Gem1 are required for MDC formation, loss of ERMES can be bypassed by MCP1 overexpression, while loss of Gem1 cannot. This result suggests that Gem1 may play a different or additional role than the ERMES complex in MDC formation. One possibility is that the ERMES complex is required for optimal Gem1 positioning or recruitment and that this requirement can somehow be bypassed by MCP1 overexpression.
If the function of ERMES and Gem1 in MDC formation does extend beyond their conventional role in maintaining mitochondrial phospholipid homeostasis, then what might these factors do? One option is that MDC formation requires the recruitment of cytoskeletal machinery to mitochondria. The ERMES complex has previously been implicated in the attachment of mitochondria to the actin cytoskeleton (Boldogh et al., 1998, 2003), and it is likely that the formation of a large cellular structure like the MDC requires a cytoskeletal-based force. Consistent with this idea, the mammalian Gem1 orthologues Miro1 and Miro2 are well known to connect mitochondria to microtubules for movement (Fransson et al., 2006; Glater et al., 2006; Guo et al., 2005). Additionally, similar to MDC formation, Gem1 GTPase activity was demonstrated to be important for the separation of mitochondrial tubules after division, possibly due to a potential role in the transport of mitochondria (Murley et al., 2013). Though Gem1 has not been shown to join mitochondria and the actin cytoskeleton or microtubules, this similarity raises the possibility that Gem1 may perform a similar function for MDC formation and mitochondrial division. Future experiments will be aimed at addressing all of these exciting possibilities for the role of ERMES and Gem1 in MDC biogenesis.
In addition to the questions surrounding the function of ERMES and Gem1 in MDC formation, the exact membrane topology of the MDC remains elusive, as does the sorting mechanism that governs the selection of cargo into the MDC. What has become clear through this work and another recently published study from our laboratory is that MDCs are generated in response to distinct signaling cues, such as an increase in intracellular amino acids (Schuler et al., 2020 Preprint). How metabolic alterations are relayed to the MDC pathway remains a mystery, one that will likely be solved by the identification of additional biogenesis and sorting machinery. As with the discovery of any new cellular system, much remains to be resolved regarding MDCs, and addressing some of these key outstanding questions will be important for elucidating the role of the MDC pathway in the maintenance of cellular amino acid homeostasis.
Materials and methods
Yeast strains and culture
All yeast strains are derivatives of S. cerevisiae S288C (BY; Brachmann et al., 1998) and are listed in Table S1. Strains expressing fluorescently tagged TOM70, TIM50, SEC61, MMM1, MDM34, and LAM6 were created by one-step PCR-mediated C-terminal endogenous epitope tagging using standard techniques and oligo pairs listed in Table S2. Plasmid templates for fluorescent epitope tagging were from the pKT series of vectors (Sheff and Thorn, 2004). Correct integrations were confirmed by a combination of colony PCR across the chromosomal insertion site and correctly localized expression of the fluorophore by microscopy. Because N-terminal tagging of Gem1 yields only partially functional proteins, yEGFP-tagged GEM1 was created by PCR-mediated internal endogenous yEGFP tagging after amino acid 262 using the Gauss toolbox (Gauss et al., 2005). Functionality of the fusion protein was assessed by quantifying the shape of segmented mitochondria using the circularity metric (4π × area/perimeter2, where 1 is a perfect circle). While WT mitochondria have a circularity of 0.2 ± 0.02 (mean ± SEM), those of yeast expressing N-terminally tagged Gem1 rose to 0.37 ± 0.02. Mitochondria of yeast expressing internally tagged Gem1 had a circularity of 0.23 ± 0.02, indicating near-WT shape and thus functionality of the fusion protein. Though the mitochondrial morphology of cells expressing Gem1-263GFP is near WT, we do not know if the fusion protein is fully functional; however, it does permit MDC formation. Deletion strains were created by one-step PCR-mediated gene replacement using the previously described pRS series of vectors (Brachmann et al., 1998; Sikorski and Hieter, 1989) and oligo pairs listed in Table S2. Correct gene deletions were confirmed by colony PCR across the chromosomal insertion site. For strains bearing deletion of MMM1, MDM10, MDM12, or MDM34, one copy of the gene was deleted to create a heterozygous diploid, which was subsequently sporulated to obtain haploid mutants. For Fig. 3, D and E and Fig. S2 F, strains expressing empty vector (EV), GEM1, GEM1(S19N), GEM1(S462N), GEM1(S19N S462N), GEM1(E225K), GEM1(E354K), or GEM1(E225K E354K) from a GPD promoter integrated into an empty region of chromosome I (chr I; 199;456–199;457) were constructed by transformation of parental yeast strains with NotI-digested pAG306GPD-empty chr I, pAG306GPD-GEM1 chr I, pAG306GPD-GEM1(S19N) chr I, pAG306GPD-GEM1(S462N) chr I, pAG306GPD-GEM1(S19N S462N) chr I, pAG306GPD-GEM1(E225K) chr I, pAG306GPD-GEM1(E354K) chr I, or pAG306GPD-GEM1(E225K E354K) chr I, respectively, as previously described (Hughes and Gottschling, 2012). For Fig. 4 B, strains expressing EV, MMM1, MMM1(L274S), MDM12, or MDM12(V214S) from a GPD promoter integrated into an empty region of chr I (199,456–199,457) were constructed by transformation of parental yeast strains with NotI-digested pAG306GPD-empty chr I, pAG306GPD-MMM1 chr I, pAG306GPD-MMM1(L274S) chr I, pAG306GPD-MDM12 chr I, or pAG306GPD-MDM12(V214S) chr I, respectively, as previously described (Hughes and Gottschling, 2012). All strains in Fig. 4 B also express VPS13(D716H) to restore the abnormalities associated with loss of the ERMES complex. For Fig. 4, E–G, strains expressing EV or MCP1 from a GPD promoter integrated into an empty region of chr I (199,456–199,457) were constructed by transformation of parental yeast strains with NotI-digested pAG306GPD-empty chr I or pAG306GPD-MCP1 chr I as previously described (Hughes and Gottschling, 2012).
Yeast cells were grown exponentially for 15–16 h at 30°C to a final density of 2–7 × 106 cells/ml before the start of all experiments. This period of overnight log-phase growth was performed to ensure vacuolar and mitochondrial uniformity across the cell population and is essential for consistent MDC activation. Unless otherwise indicated, cells were cultured in YPAD medium (1% yeast extract, 2% peptone, 0.005% adenine, and 2% glucose). Cells expressing pRS315 (Sikorski and Hieter, 1989), pVPS13(D716H; Lang et al., 2015), pRS415 (Brachmann et al., 1998), or p415GPD-ChiMERA (Kornmann et al., 2009) were cultured in SD-leucine medium (0.67% yeast nitrogen base without amino acids, 2% glucose, supplemented nutrients 0.072 g/liter each adenine, alanine, arginine, asparagine, aspartic acid, cysteine, glutamic acid, glutamine, glycine, histidine, myo-inositol, isoleucine, lysine, methionine, phenylalanine, proline, serine, threonine, tryptophan, tyrosine, uracil, valine, and 0.007 g/liter para-aminobenzoic acid) for selection of the plasmids and, for MDC assays, were switched to YPAD at the time of drug treatment. Unless otherwise indicated, rapamycin (LC Laboratories; R-5000), concanamycin A (Santa Cruz Biotechnology; sc-202111), and cycloheximide (Sigma-Aldrich; C1988) were added to cultures at final concentrations of 200 nM, 500 nM, and 50 µg/ml, respectively.
Plasmids
To create pAG306GPD-GEM1 chr I, pAG306GPD-MMM1 chr I, pAG306GPD-MDM12 chr I, and pAG306GPD-MCP1 chr I, we used donor Gateway plasmids pDONR221-GEM1, pDONR221-MMM1, pDONR221-MDM12, or pDONR221-MCP1 to insert GEM1, MMM1, MDM12, or MCP1, respectively, into pAG306GPD-ccdB chr I (Hughes and Gottschling, 2012) using LR clonase (Thermo Fisher Scientific; 11791020) according to the manufacturer’s instructions. To create pAG306GPD-GEM1(S19N) chr I, pAG306GPD-GEM1(S462N) chr I, pAG306GPD-GEM1(S19N S462N) chr I, pAG306GPD-GEM1(E225K) chr I, pAG306GPD-GEM1(E354K) chr I, and pAG306GPD-GEM1(E225K E354K) chr I, we used the Q5 Site-Directed Mutagenesis Kit (New England Biolabs; E0554S) and pDONR221-GEM1 to make pDONR221-GEM1(S19N), pDONR221-GEM1(S462N), pDONR221-GEM1(S19N S462N), pDONR221-GEM1(E225K), pDONR221-GEM1(E354K), and pDONR221-GEM1(E225K E354K), respectively. We then used pDONR221-GEM1(S19N), pDONR221-GEM1(S462N), pDONR221-GEM1(S19N S462N), pDONR221-GEM1(E225K), pDONR221-GEM1(E354K), and pDONR221-GEM1(E225K E354K) to insert GEM1(S19N), GEM1(S462N), GEM1(S19N S462N), GEM1(E225K), GEM1(E354K), and GEM1(E225K E354K), respectively, into pAG306GPD-ccdB chr I using LR clonase according to the manufacturer’s instructions. To create pAG306GPD-MMM1(L274S) chr I and pAG306GPD-MDM12(V214S) chr I, we used the Q5 Site-Directed Mutagenesis Kit and pDONR221-MMM1 and pDONR221-MDM12 to make pDONR221-MMM1(L274S) and pDONR221-MDM12(V214S), respectively. We then used pDONR221-MMM1(L274S) and pDONR221-MDM12(V214S) to insert MMM1(L274S) and MDM12(V214S), respectively, into pAG306GPD-ccdB chr I using LR clonase according to the manufacturer’s instructions.
MDC assays
For MDC assays, overnight log-phase cell cultures were grown in the presence of DMSO (Sigma-Aldrich; D2650) or the indicated drug for two hours. After incubation, cells were harvested by centrifugation, and optical z-sections of live yeast cells were acquired with a Zeiss Axio Imager M2 or, for superresolution images, a Zeiss LSM800 with Airyscan or Zeiss LSM880 with Airyscan. The percentage cells with MDCs was quantified in each experiment at the 2-h time point. Unless otherwise indicated, all quantifications show the mean ± SEM from three biological replicates with n = 100 cells per experiment. MDCs were identified as Tom70-positive, Tim50-negative circular structures that were enriched for Tom70 versus the mitochondrial tubule. For Fig. S2 A, MDCs were identified as Oac1-positive, Tim50-negative circular structures.
Microscopy
Optical z-sections of live yeast cells were acquired with a Zeiss Axio Imager M2 equipped with a Zeiss Axiocam 506 monochromatic camera, 100× oil-immersion objective (Plan-Apo, NA 1.4), a Zeiss LSM800 equipped with an Airyscan detector, 63× oil-immersion objective (Plan-Apo, NA 1.4) or a Zeiss LSM880 equipped with an Airyscan detector, 63× oil-immersion objective (Plan-Apo, NA 1.4). Widefield images were acquired with ZEN (Carl Zeiss) and processed with Fiji (Schindelin et al., 2012). Superresolution images were acquired with ZEN (Carl Zeiss) and processed using the automated Airyscan processing algorithm in ZEN (Carl Zeiss) and Fiji. Individual channels of all images were minimally adjusted in Fiji to match the fluorescence intensities between channels for better visualization. Line scan analysis was performed on nonadjusted, single z-sections in Fiji. For Fig. S1 E, yeast were imaged on a DeltaVision MPX microscope (Applied Precision) equipped with a 60× 1.42 NA oil Plan-ApoN or a 100× 1.40 NA oil UplanS-Apo objective lens (Olympus), a multicolor solid state illumination light source, and a CoolSNAP HQ2 camera (Roper Scientific). Acquisition and deconvolution were performed with SoftWoRx software.
Time-lapse imaging
For Figs. 1 C and 2 D, Fig. S1 B, and Videos 1 and 2, overnight log-phase cultures were treated with 1 µM rapamycin for 15 min. Cells were harvested by centrifugation, resuspended in SD medium (0.67% yeast nitrogen base without amino acids, 2% glucose, supplemented nutrients 0.074 g/liter each adenine, alanine, arginine, asparagine, aspartic acid, cysteine, glutamic acid, glutamine, glycine, histidine, myo-inositol, isoleucine, lysine, methionine, phenylalanine, proline, serine, threonine, tryptophan, tyrosine, uracil, valine, 0.369 g/liter leucine, and 0.007 g/liter para-aminobenzoic acid) and 5 µM rapamycin, and pipetted into flow chamber slides as previously described (Fees et al., 2017). Briefly, flow chambers were made using standard microscope slides and coverslips. Strips of Parafilm were used to seal a coverslip to a slide and created the walls of the chamber. Flow chambers were coated with concanavalin A (Sigma-Aldrich; L7647) before loading cells. Melted Vaseline was used to seal the chamber. Optical z-sections of live yeast cells were acquired with a Zeiss Airyscan LSM880 equipped with an environmental chamber set to 30°C. Superresolution time-lapse images were acquired with ZEN (Carl Zeiss) and processed using the automated Airyscan processing algorithm in ZEN (Carl Zeiss) and Fiji. Individual channels of all images were minimally adjusted in Fiji to match the fluorescence intensities between channels for better visualization.
Indirect immunofluorescence
For indirect immunofluorescence, yeast cells were grown exponentially for 15–16 h in YPAD at 30°C to a final density of 4 × 106 cells/ml. Cells were harvested by centrifugation and fixed in 10 ml fixation medium (4% paraformaldehyde in YPAD) for 1 h. Fixed cells were washed with wash buffer (0.1 M Tris, pH 8, and 1.2 M sorbitol) twice and incubated in 2 ml DTT buffer (10 mM DTT in 0.1 M Tris, pH 9.4) at RT for 10 min. Spheroplasts were generated by incubating cells in 2 ml zymolyase buffer (0.1 M KPi, pH 6.5, 1.2 M sorbitol, 0.25 mg/ml zymolyase) at 30°C for 30 min. Spheroplasts were gently diluted 1:40 in wash buffer and attached to glass slides precoated with 0.1% poly-L-lysine (2 mg/ml). Samples were permeabilized in cold 0.1% Triton X-100 in PBS for 10 min at 4°C, briefly dried, and blocked in wash buffer containing 1% BSA at RT for 30 min. After blocking, samples were incubated with primary antibody (affinity-purified anti-Gem1 polyclonal antibody produced in rabbit [Shaw Laboratory], diluted 1:200 in wash buffer containing 1% BSA) for 90 min at RT and secondary antibody (goat anti-rabbit IgG [H+L] cross-adsorbed secondary antibody, Alexa Fluor 647 [Invitrogen; A-21245], diluted 1:300 in wash buffer containing 1% BSA) for 45 min at RT. Samples were washed 10 times after each incubation with PBS containing 1% BSA and 0.1% Tween-20. Slides were washed twice with wash buffer before sealing and were mounted with ProLong Glass Antifade Mountant with NucBlue (Invitrogen; P36981) overnight. Widefield images were acquired as described in Microscopy.
Protein preparation and immunoblotting
For Western blot analysis of total protein levels, 2 × 107 overnight log-phase yeast cells were harvested by centrifugation, washed with ddH2O, and incubated in 0.1 M NaOH for 5 min at RT. Pellets were isolated by centrifugation at 14,000 ×g for 10 min at 4°C and incubated for 5 min at 95°C in SDS-lysis buffer (10 mM Tris, pH 6.8, 100 mM NaCl, 1 mM EDTA, 1mM EGTA, and 1% [wt/vol] SDS) plus protease inhibitors (Sigma-Aldrich; 11697498001). Laemmli buffer (63 mM Tris, pH 6.8, 2% [wt/vol] SDS, 10% [vol/vol] glycerol, 1 mg/ml bromophenol blue, and 1% [vol/vol] β-mercaptoethanol) was added to samples and incubated for 5 min at 95°C. Protein concentrations were determined by bicinchoninic acid assay (Thermo Fisher Scientific; 23227). To separate proteins based on molecular weight, equal amounts of protein were subjected to SDS PAGE using Bolt 4–12% Bis-Tris Gels (Thermo Fisher Scientific; NW04125BOX) and transferred to polyvinylidene difluoride membrane by wet transfer. Nonspecific antibody binding was blocked by incubation with TBS containing 5% (wt/vol) dry milk for 1 h at RT. After incubation with primary antibodies (1:500 affinity-purified anti-Gem1 polyclonal antibody produced in rabbit [Shaw Laboratory] or 1:2,000 anti-Pgk1 antibody [Abcam; 22C5D8]) at 4°C overnight, membranes were washed three times with TBS and incubated with secondary antibody (goat-anti-rabbit/mouse HRP conjugated, 1:5,000 in TBS containing 5% dry milk [Jackson ImmunoResearch; 111–035-144/715-035-150]) for 40 min at RT. Membranes were washed three times with TBS, enhanced chemiluminescence solution was applied, and the antibody signal was detected with a Bio-Rad Chemidoc MP system. Blots were exported as TIFFs using ImageLab 6.0 (Bio-Rad) and cropped in Adobe Photoshop CC. Western blots show one representative blot from n = 3 replicates.
Serial-dilution growth assays
Five-fold serial dilutions of exponentially growing yeast cells were in water, and 3 µl of each dilution was spotted onto YPD (1% yeast extract, 2% peptone, and 2% glucose) and YPG (1% yeast extract, 2% peptone, and 3% glycerol) agar or YPAD (1% yeast extract, 2% peptone, 0.005% adenine, and 2% glucose) and YPAG (1% yeast extract, 2% peptone, 0.005% adenine, and 3% glycerol) agar. The total number of cells plated in each dilution spot was 5,000, 1,000, 20, 40, and 8.
Quantification and statistical analysis
The number of replicates, what n represents, and dispersion and precision measures are indicated in the figure legends. In general, quantifications show the mean ± SEM from three biological replicates with n = 100 cells per experiment. In experiments with data depicted from a single biological replicate, the experiment was repeated with the same results.
Online supplemental material
Fig. S1 quantifies the percentage of cells with one MDC per cell, shows time-lapse images of an MDC stably associating with the ER over time, and describes Gem1-263GFP. Fig. S2 demonstrates that Gem1 is required for the formation of MDCs labeled by Oac1 and ERMES/Gem1 are required for concanamycin A and cycloheximide-induced MDC formation. Fig. S3 shows images of MDCs in phospholipid synthesis mutants and demonstrates that ERMES/Gem1 mutants expressing VPS13(D716H) or ChiMERA do not form MDCs in response to concanamycin A, cycloheximide, or rapamycin. Video 1 shows the process of MDC biogenesis. Video 2 shows an MDC forming at and associating with an ER–mitochondria contact site. Table S1 lists the yeast strains used in this study. Table S2 lists the oligonucleotides used in this study.
Supplementary Material
lists the yeast strains used in this study.
shows the oligonucleotides used in this study.
Acknowledgments
We thank members of the A.L. Hughes and J.M. Shaw laboratories for discussion and manuscript comments and L. VanderMeer (Utah) for technical assistance.
This research was supported by the National Institutes of Health (grants AG043095, GM119694, AG061376, and AG055648 to A.L. Hughes; grant T32GM007464 to A.M. English; and grants GM53466 and GM84970 to J.M. Shaw), the American Heart Association (grant 18PRE33960427 to M.-H. Schuler), the Howard Hughes Medical Institute (J.M. Shaw), and the Wellcome Trust (grant 214291/Z/18/Z to B. Kornmann). A.L. Hughes was further supported by an American Federation for Aging Research junior research grant, a United Mitochondrial Disease Foundation early career research grant, a Searle Scholars Program award, and a Glenn Foundation for Medical Research award.
The authors declare no competing financial interests.
Author contributions: Conceptualization, A.M. English, J.M. Shaw, and A.L. Hughes; Methodology, A.M. English, B. Kornmann, J.M. Shaw, and A.L. Hughes; Formal Analysis, A.M. English, M.-H. Schuler, and T. Xiao; Investigation, A.M. English, M.-H. Schuler, and T. Xiao; Writing – Original Draft, A.M. English; Writing – Review & Editing, M.-H. Schuler, T. Xiao, B. Kornmann, J.M. Shaw, and A.L. Hughes; Visualization, A.M. English; Supervision, J.M. Shaw and A.L. Hughes; Funding Acquisition, A.M. English, M.-H. Schuler, B. Kornmann, J.M. Shaw, and A.L. Hughes.
References
- AhYoung, A.P., Jiang J., Zhang J., Khoi Dang X., Loo J.A., Zhou Z.H., and Egea P.F.. 2015. Conserved SMP domains of the ERMES complex bind phospholipids and mediate tether assembly. Proc. Natl. Acad. Sci. USA. 112:E3179–E3188. 10.1073/pnas.1422363112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, K.H., Sogo L.F., and Yaffe M.P.. 1997. Mdm12p, a component required for mitochondrial inheritance that is conserved between budding and fission yeast. J. Cell Biol. 136:545–553. 10.1083/jcb.136.3.545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beugnet, A., Tee A.R., Taylor P.M., and Proud C.G.. 2003. Regulation of targets of mTOR (mammalian target of rapamycin) signalling by intracellular amino acid availability. Biochem. J. 372:555–566. 10.1042/bj20021266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh, I., Vojtov N., Karmon S., and Pon L.A.. 1998. Interaction between mitochondria and the actin cytoskeleton in budding yeast requires two integral mitochondrial outer membrane proteins, Mmm1p and Mdm10p. J. Cell Biol. 141:1371–1381. 10.1083/jcb.141.6.1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh, I.R., Nowakowski D.W., Yang H.-C., Chung H., Karmon S., Royes P., and Pon L.A.. 2003. A protein complex containing Mdm10p, Mdm12p, and Mmm1p links mitochondrial membranes and DNA to the cytoskeleton-based segregation machinery. Mol. Biol. Cell. 14:4618–4627. 10.1091/mbc.e03-04-0225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann, C.B., Davies A., Cost G.J., Caputo E., Li J., Hieter P., and Boeke J.D.. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 14:115–132. [DOI] [PubMed] [Google Scholar]
- Burgess, S.M., Delannoy M., and Jensen R.E.. 1994. MMM1 encodes a mitochondrial outer membrane protein essential for establishing and maintaining the structure of yeast mitochondria. J. Cell Biol. 126:1375–1391. 10.1083/jcb.126.6.1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, S.C., Heacock P.N., Mileykovskaya E., Voelker D.R., and Dowhan W.. 1998. Isolation and characterization of the gene (CLS1) encoding cardiolipin synthase in Saccharomyces cerevisiae. J. Biol. Chem. 273:14933–14941. 10.1074/jbc.273.24.14933 [DOI] [PubMed] [Google Scholar]
- Clancey, C.J., Chang S.C., and Dowhan W.. 1993. Cloning of a gene (PSD1) encoding phosphatidylserine decarboxylase from Saccharomyces cerevisiae by complementation of an Escherichia coli mutant. J. Biol. Chem. 268:24580–24590. [PubMed] [Google Scholar]
- Connerth, M., Tatsuta T., Haag M., Klecker T., Westermann B., and Langer T.. 2012. Intramitochondrial transport of phosphatidic acid in yeast by a lipid transfer protein. Science. 338:815–818. 10.1126/science.1225625 [DOI] [PubMed] [Google Scholar]
- Dröse, S., Bindseil K.U., Bowman E.J., Siebers A., Zeeck A., and Altendorf K.. 1993. Inhibitory effect of modified bafilomycins and concanamycins on P- and V-type adenosinetriphosphatases. Biochemistry. 32:3902–3906. 10.1021/bi00066a008 [DOI] [PubMed] [Google Scholar]
- Elbaz-Alon, Y., Rosenfeld-Gur E., Shinder V., Futerman A.H., Geiger T., and Schuldiner M.. 2014. A dynamic interface between vacuoles and mitochondria in yeast. Dev. Cell. 30:95–102. 10.1016/j.devcel.2014.06.007 [DOI] [PubMed] [Google Scholar]
- Elbaz-Alon, Y., Eisenberg-Bord M., Shinder V., Stiller S.B., Shimoni E., Wiedemann N., Geiger T., and Schuldiner M.. 2015. Lam6 Regulates the Extent of Contacts between Organelles. Cell Rep. 12:7–14. 10.1016/j.celrep.2015.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fees, C.P., Estrem C., and Moore J.K.. 2017. High-resolution Imaging and Analysis of Individual Astral Microtubule Dynamics in Budding Yeast. J. Vis. Exp. 122:55610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransson, S., Ruusala A., and Aspenström P.. 2006. The atypical Rho GTPases Miro-1 and Miro-2 have essential roles in mitochondrial trafficking. Biochem. Biophys. Res. Commun. 344:500–510. 10.1016/j.bbrc.2006.03.163 [DOI] [PubMed] [Google Scholar]
- Frederick, R.L., McCaffery J.M., Cunningham K.W., Okamoto K., and Shaw J.M.. 2004. Yeast Miro GTPase, Gem1p, regulates mitochondrial morphology via a novel pathway. J. Cell Biol. 167:87–98. 10.1083/jcb.200405100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, J.R., Kannan M., Toulmay A., Jan C.H., Weissman J.S., Prinz W.A., and Nunnari J.. 2018. Lipid Homeostasis Is Maintained by Dual Targeting of the Mitochondrial PE Biosynthesis Enzyme to the ER. Dev. Cell. 44:261–270.e6. 10.1016/j.devcel.2017.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauss, R., Trautwein M., Sommer T., and Spang A.. 2005. New modules for the repeated internal and N-terminal epitope tagging of genes in Saccharomyces cerevisiae. Yeast. 22:1–12. 10.1002/yea.1187 [DOI] [PubMed] [Google Scholar]
- Glater, E.E., Megeath L.J., Stowers R.S., and Schwarz T.L.. 2006. Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J. Cell Biol. 173:545–557. 10.1083/jcb.200601067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, X., Macleod G.T., Wellington A., Hu F., Panchumarthi S., Schoenfield M., Marin L., Charlton M.P., Atwood H.L., and Zinsmaier K.E.. 2005. The GTPase dMiro is required for axonal transport of mitochondria to Drosophila synapses. Neuron. 47:379–393. 10.1016/j.neuron.2005.06.027 [DOI] [PubMed] [Google Scholar]
- Heitman, J., Movva N.R., and Hall M.N.. 1991. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 253:905–909. 10.1126/science.1715094 [DOI] [PubMed] [Google Scholar]
- Henne, W.M., Buchkovich N.J., and Emr S.D.. 2011. The ESCRT pathway. Dev. Cell. 21:77–91. 10.1016/j.devcel.2011.05.015 [DOI] [PubMed] [Google Scholar]
- Hobbs, A.E., Srinivasan M., McCaffery J.M., and Jensen R.E.. 2001. Mmm1p, a mitochondrial outer membrane protein, is connected to mitochondrial DNA (mtDNA) nucleoids and required for mtDNA stability. J. Cell Biol. 152:401–410. 10.1083/jcb.152.2.401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hönscher, C., Mari M., Auffarth K., Bohnert M., Griffith J., Geerts W., van der Laan M., Cabrera M., Reggiori F., and Ungermann C.. 2014. Cellular metabolism regulates contact sites between vacuoles and mitochondria. Dev. Cell. 30:86–94. 10.1016/j.devcel.2014.06.006 [DOI] [PubMed] [Google Scholar]
- Hughes, A.L., and Gottschling D.E.. 2012. An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature. 492:261–265. 10.1038/nature11654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, A.L., Hughes C.E., Henderson K.A., Yazvenko N., and Gottschling D.E.. 2016. Selective sorting and destruction of mitochondrial membrane proteins in aged yeast. eLife. 5:e13943. 10.7554/eLife.13943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, H., Park J., and Lee C.. 2016. Crystal structure of Mdm12 reveals the architecture and dynamic organization of the ERMES complex. EMBO Rep. 17:1857–1871. 10.15252/embr.201642706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, H., Park J., Jun Y., and Lee C.. 2017. Crystal structures of Mmm1 and Mdm12-Mmm1 reveal mechanistic insight into phospholipid trafficking at ER-mitochondria contact sites. Proc. Natl. Acad. Sci. USA. 114:E9502–E9511. 10.1073/pnas.1715592114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John Peter, A.T., Herrmann B., Antunes D., Rapaport D., Dimmer K.S., and Kornmann B.. 2017. Vps13-Mcp1 interact at vacuole-mitochondria interfaces and bypass ER-mitochondria contact sites. J. Cell Biol. 216:3219–3229. 10.1083/jcb.201610055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski, M., and Youle R.J.. 2011. Regulating mitochondrial outer membrane proteins by ubiquitination and proteasomal degradation. Curr. Opin. Cell Biol. 23:476–482. 10.1016/j.ceb.2011.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano, S., Tamura Y., Kojima R., Bala S., Asai E., Michel A.H., Kornmann B., Riezman I., Riezman H., Sakae Y., et al. 2018. Structure-function insights into direct lipid transfer between membranes by Mmm1-Mdm12 of ERMES. J. Cell Biol. 217:959–974. 10.1083/jcb.201704119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klosowiak, J.L., Park S., Smith K.P., French M.E., Focia P.J., Freymann D.M., and Rice S.E.. 2016. Structural insights into Parkin substrate lysine targeting from minimal Miro substrates. Sci. Rep. 6:33019. 10.1038/srep33019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopec, K.O., Alva V., and Lupas A.N.. 2010. Homology of SMP domains to the TULIP superfamily of lipid-binding proteins provides a structural basis for lipid exchange between ER and mitochondria. Bioinformatics. 26:1927–1931. 10.1093/bioinformatics/btq326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann, B., Currie E., Collins S.R., Schuldiner M., Nunnari J., Weissman J.S., and Walter P.. 2009. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 325:477–481. 10.1126/science.1175088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann, B., Osman C., and Walter P.. 2011. The conserved GTPase Gem1 regulates endoplasmic reticulum-mitochondria connections. Proc. Natl. Acad. Sci. USA. 108:14151–14156. 10.1073/pnas.1111314108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiba, T., Holman H.A., Kubara K., Yasukawa K., Kawabata S., Okamoto K., MacFarlane J., and Shaw J.M.. 2011. Structure-function analysis of the yeast mitochondrial Rho GTPase, Gem1p: implications for mitochondrial inheritance. J. Biol. Chem. 286:354–362. 10.1074/jbc.M110.180034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, N., Leonzino M., Hancock-Cerutti W., Horenkamp F.A., Li P., Lees J.A., Wheeler H., Reinisch K.M., and De Camilli P.. 2018. VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J. Cell Biol. 217:3625–3639. 10.1083/jcb.201807019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé, K., Murley A., and Nunnari J.. 2014. Determinants and functions of mitochondrial behavior. Annu. Rev. Cell Dev. Biol. 30:357–391. 10.1146/annurev-cellbio-101011-155756 [DOI] [PubMed] [Google Scholar]
- Lackner, L.L., Ping H., Graef M., Murley A., and Nunnari J.. 2013. Endoplasmic reticulum-associated mitochondria-cortex tether functions in the distribution and inheritance of mitochondria. Proc. Natl. Acad. Sci. USA. 110:E458–E467. 10.1073/pnas.1215232110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, A.B., John Peter A.T., Walter P., and Kornmann B.. 2015. ER-mitochondrial junctions can be bypassed by dominant mutations in the endosomal protein Vps13. J. Cell Biol. 210:883–890. 10.1083/jcb.201502105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murley, A., Lackner L.L., Osman C., West M., Voeltz G.K., Walter P., and Nunnari J.. 2013. ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. eLife. 2:e00422. 10.7554/eLife.00422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murley, A., Sarsam R.D., Toulmay A., Yamada J., Prinz W.A., and Nunnari J.. 2015. Ltc1 is an ER-localized sterol transporter and a component of ER–mitochondria and ER-vacuole contacts. J. Cell Biol. 209:539–548. 10.1083/jcb.201502033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari, J., and Suomalainen A.. 2012. Mitochondria: in sickness and in health. Cell. 148:1145–1159. 10.1016/j.cell.2012.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, X., Roberts P., Chen Y., Kvam E., Shulga N., Huang K., Lemmon S., and Goldfarb D.S.. 2000. Nucleus-vacuole junctions in Saccharomyces cerevisiae are formed through the direct interaction of Vac8p with Nvj1p. Mol. Biol. Cell. 11:2445–2457. 10.1091/mbc.11.7.2445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J.-S., Thorsness M.K., Policastro R., McGoldrick L.L., Hollingsworth N.M., Thorsness P.E., and Neiman A.M.. 2016. Yeast Vps13 promotes mitochondrial function and is localized at membrane contact sites. Mol. Biol. Cell. 27:2435–2449. 10.1091/mbc.e16-02-0112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen, E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., and Ferrin T.E.. 2004. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Pickles, S., Vigié P., and Youle R.J.. 2018. Mitophagy and Quality Control Mechanisms in Mitochondrial Maintenance. Curr. Biol. 28:R170–R185. 10.1016/j.cub.2018.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirós, P.M., Langer T., and López-Otín C.. 2015. New roles for mitochondrial proteases in health, ageing and disease. Nat. Rev. Mol. Cell Biol. 16:345–359. 10.1038/nrm3984 [DOI] [PubMed] [Google Scholar]
- Rutter, J., and Hughes A.L.. 2015. Power(2): the power of yeast genetics applied to the powerhouse of the cell. Trends Endocrinol. Metab. 26:59–68. 10.1016/j.tem.2014.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin, J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler, M.-H., English, A.M., Campbell, T.J., Shaw, J.M., and Hughes, A.L.. 2020. Mitochondrial-Derived Compartments Facilitate Cellular Adaptation to Amino Acid Stress. bioRxiv. doi: (Preprint posted March 14, 2020) 10.1101/2020.03.13.991091 [DOI] [PMC free article] [PubMed]
- Sheff, M.A., and Thorn K.S.. 2004. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 21:661–670. 10.1002/yea.1130 [DOI] [PubMed] [Google Scholar]
- Shpilka, T., and Haynes C.M.. 2018. The mitochondrial UPR: mechanisms, physiological functions and implications in ageing. Nat. Rev. Mol. Cell Biol. 19:109–120. 10.1038/nrm.2017.110 [DOI] [PubMed] [Google Scholar]
- Sikorski, R.S., and Hieter P.. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo, L.F., and Yaffe M.P.. 1994. Regulation of mitochondrial morphology and inheritance by Mdm10p, a protein of the mitochondrial outer membrane. J. Cell Biol. 126:1361–1373. 10.1083/jcb.126.6.1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söllner, T., Pfaller R., Griffiths G., Pfanner N., and Neupert W.. 1990. A mitochondrial import receptor for the ADP/ATP carrier. Cell. 62:107–115. 10.1016/0092-8674(90)90244-9 [DOI] [PubMed] [Google Scholar]
- Stroud, D.A., Oeljeklaus S., Wiese S., Bohnert M., Lewandrowski U., Sickmann A., Guiard B., van der Laan M., Warscheid B., and Wiedemann N.. 2011. Composition and topology of the endoplasmic reticulum-mitochondria encounter structure. J. Mol. Biol. 413:743–750. 10.1016/j.jmb.2011.09.012 [DOI] [PubMed] [Google Scholar]
- Sugiura, A., McLelland G.-L., Fon E.A., and McBride H.M.. 2014. A new pathway for mitochondrial quality control: mitochondrial-derived vesicles. EMBO J. 33:2142–2156. 10.15252/embj.201488104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, T., Özbalci C., Brügger B., Rapaport D., and Dimmer K.S.. 2013. Mcp1 and Mcp2, two novel proteins involved in mitochondrial lipid homeostasis. J. Cell Sci. 126:3563–3574. 10.1242/jcs.121244 [DOI] [PubMed] [Google Scholar]
- Trotter, P.J., and Voelker D.R.. 1995. Identification of a non-mitochondrial phosphatidylserine decarboxylase activity (PSD2) in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 270:6062–6070. 10.1074/jbc.270.11.6062 [DOI] [PubMed] [Google Scholar]
- Trotter, P.J., Pedretti J., and Voelker D.R.. 1993. Phosphatidylserine decarboxylase from Saccharomyces cerevisiae. Isolation of mutants, cloning of the gene, and creation of a null allele. J. Biol. Chem. 268:21416–21424. [PubMed] [Google Scholar]
- Tuller, G., Hrastnik C., Achleitner G., Schiefthaler U., Klein F., and Daum G.. 1998. YDL142c encodes cardiolipin synthase (Cls1p) and is non-essential for aerobic growth of Saccharomyces cerevisiae. FEBS Lett. 421:15–18. 10.1016/S0014-5793(97)01525-1 [DOI] [PubMed] [Google Scholar]
- Wallace, D.C. 2005. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 39:359–407. 10.1146/annurev.genet.39.110304.095751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse, A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., et al. 2018. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46(W1):W296–W303. 10.1093/nar/gky427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, H., Esaki M., Kanamori T., Tamura Y., Nishikawa S., and Endo T.. 2002. Tim50 is a subunit of the TIM23 complex that links protein translocation across the outer and inner mitochondrial membranes. Cell. 111:519–528. 10.1016/S0092-8674(02)01053-X [DOI] [PubMed] [Google Scholar]
- Youngman, M.J., Hobbs A.E.A., Burgess S.M., Srinivasan M., and Jensen R.E.. 2004. Mmm2p, a mitochondrial outer membrane protein required for yeast mitochondrial shape and maintenance of mtDNA nucleoids. J. Cell Biol. 164:677–688. 10.1083/jcb.200308012 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists the yeast strains used in this study.
shows the oligonucleotides used in this study.