ABSTRACT
Methylglyoxal (MG), a cytotoxic oxygenated short aldehyde, is a by-product of various metabolic reactions in plants, including glycolysis. The basal level of MG in plants is low, whereby it acts as an essential signaling molecule regulating multiple cellular processes. However, hyperaccumulation of MG under stress conditions is detrimental for plants as it inhibits multiple developmental processes, including seed germination, photosynthesis, and root growth. The evolutionarily conserved glyoxalase system is critical for MG detoxification, and it comprises of two-enzymes, the glyoxalase-I and glyoxalase-II. Here, we report the functional characterization of six putative glyoxalase-I genes from date palm (Phoenix dactylifera L.) (PdGLX1), by studying their gene expression under various environmental stress conditions and investigating their function in bacteria (Escherichia coli) and yeast (Saccharomyces cerevisiae) mutant cells. The putative PdGLX1 genes were initially identified using computational methods and cloned using molecular tools. The PdGLX1 gene expression analysis using quantitative PCR (qPCR) revealed differential expression under various stress conditions such as salinity, oxidative stress, and exogenous MG stress in a tissue-specific manner. Further, in vivo functional characterization indicated that overexpression of the putative PdGLX1 genes in E. coli enhanced their growth and MG detoxification ability. The putative PdGLX1 genes were also able to complement the loss-of-function MG hypersensitive GLO1 (YML004C) yeast mutants and promote growth by enhancing MG detoxification and reducing the accumulation of reactive oxygen species (ROS) under stress conditions as indicated by flow cytometry. These findings denote the potential importance of PdGLX1 genes in MG detoxification under stress conditions in the date palm.
KEYWORDS: Functional characterization, glyoxalase-I, date palm, methylglyoxal, ROS, salinity
Introduction
Plants being sessile organisms are directly affected by various abiotic and biotic factors, which determine their growth and development, thereby affecting the potential yields of crop plants.1,2 Date palm (Phoenix dactylifera L.) is a monocotyledon, dioecious plant, which is an important socioeconomic and abiotic stress-tolerant plant mainly grown in the hot and arid regions of the middle east and north Africa.3 Currently, this tree is suffering excessive salinity, which may be attributed to many environmental factors such as lack of precipitation, overexploitation of underground water, high evaporation rate, and the rising sea levels brought about by climate change.4 To overcome the adverse effects of stress conditions, plants have developed multiple mechanisms to alter their morphological, physiological, biochemical, and molecular processes.5–7 These alterations help in reducing the stress, thereby enabling the plants to grow and develop under various adverse environmental conditions.8 A vast number of plant-stress interaction pathways have been reported to be involved in stress tolerance and adaptation.9 Interestingly, it has been noted that various stress conditions lead to the accumulation of toxic aldehydes, of which methylglyoxal (MG), an α, β-dicarbonyl ketoaldehyde by-product of various metabolic processes, acts as an essential signaling molecule,10 and functions as a Hill oxidant and catalyzes the photoreduction of O2 to superoxide (O2.) in the photosystem-I.11 Therefore, MG could be used as a biomarker for plant stress tolerance.12
It has been previously reported that there is a 2- to 6- fold increase in the accumulation of MG in response to various stress.13 This increase leads to an upregulation in the reactive oxygen species (ROS) production, a decrease in the glutathione (GSH) levels and a disruption in the antioxidant enzyme function in plants.8,14 Various works have shown that MG regulatory function is governed in a concentration-dependent manner, whereby a lower concentration (0.1 mM) of MG did not inhibit germination but inhibited root elongation, however, at a higher concentration (1 mM MG) it inhibited seed germination in Arabidopsis.15 It was also observed that exogenous application of MG in Arabidopsis at a lower concentration (≤1 mM) induced stomatal closure via an extracellular oxidative burst and an elevation of [Ca2+]cyt without reducing the viability of guard cells, however, at a higher concentration (≥1 mM), MG was cytotoxic.16 In addition, MG has also been shown to prevent light-induced stomatal opening by inhibiting inward potassium (K+) channels into the guard cells of Arabidopsis.16 A gene chip microarray analysis of rice plants exposed to 10 mM MG revealed that 966 genes were upregulated, and 719 genes were down-regulated, of which 18% and 14% are genes related to signal transduction, respectively.17 These findings indicate a regulatory role of MG on signal transduction, thus signifying its possible involvement in plant stress signaling.
Further, MG has also been reported to inactivate the antioxidant enzyme, cytosolic ascorbate peroxidase (NtcAPX) in Nicotiana tabacum by modification of the amino acids of the enzyme.18 Therefore, to prevent the hyperaccumulation of MG in the cell under stress conditions, various detoxification pathways are involved in MG breakdown,19 of which the universally conserved glyoxalase system is the most efficient.20 Within the glyoxalase pathway, MG, along with GSH is first non-enzymatically converted to hemithioacetate, after which the enzyme glyoxalase-I (also known as lactoylglutathione lyase) catalyzes the breakdown to S-lactoylglutathione. The glyoxalase-II further hydrolyzes S-lactoylglutathione to D-lactate and recycles GSH. The MG-scavenging glyoxalase pathway is believed to have diverse physiological functions in the plant. However, its role in stress tolerance is the most widely studied.21 In addition, various reports have observed an increase in the expression of the MG detoxification glyoxalase genes in plants under stress conditions, thus suggesting that these enzymes play an essential role in the plant’s stress adaptation.22–25 Furthermore, transgenic overexpression of the MG-detoxification glyoxalase enzymes has been shown to enhance tolerance to various stress conditions and also to lower MG levels in the plants.26–29
The aim of this project was to functionally characterize glyoxalase-I (EC 4.4.1.5) genes encoded by the date palm genome (PdGLX1), study their expression profile under various stress conditions, and determine their function in E. coli and GLO1 knockout yeast cells using a functional complementation assay. Here, we report six putative PdGLX1 genes in the genome of the date palm, namely PdGLX1:1, PdGLX1:2, PdGLX1:3, PdGLX1:4, PdGLX1:5, and PdGLX1:6. The expression of these genes varies under various stress treatments suggesting that their expression is induced in a stress-dependent manner. Further, heterologous expression of the PdGLX1 in BL21 (DE3) E. coli and yeast mutant cells enhanced stress tolerance and significantly reduced MG accumulation in the cells. Our research highlights the importance of the PdGLX1 gene family in stress tolerance in date palm.
Materials and methods
Plant growth conditions and treatments
Date palm seeds (cv. Khalas) were soaked overnight in distilled water at 37ºC. Debris was washed away from the seeds with tap water. A 70% ethanol solution was then used to surface-sterilize the seeds, followed by rinsing them twice using sterile distilled water. The seeds were then germinated at 37ºC on moist sterile vermiculite for ten days as previously described.30,31 The germinated seedlings were potted in multipurpose compost (Bulrush Horticulture Ltd, UK). The pots were maintained in the greenhouse at 30ºC, 350 μmol.m−2.s−1 light intensity and a photoperiod of 16 h/8 h light/dark cycle. The pots were irrigated on a weekly basis with distilled water to field capacity for about three weeks. Subsequently, the pots were segregated according to the irrigation solutions (treatments): control (distilled water), salt stress (300 mM NaCl), oxidative stress (10 mM H2O2), and MG stress (10 mM MG). The plants were harvested after six weeks of treatment.
The concentration of the NaCl and MG used in the following experiments was selected empirically. The highest non-lethal level the NaCl and MG was selected to trigger the tolerance/detoxification process in the used cells. In general, the proper highest concentration used in these experiments was the one which can significantly affect the growth of the wildtype organism but did not show a negative effect on the transgenic microorganisms used in this project. The date palm, as well as yeast cells, are NaCl tolerant organisms by nature; however, bacteria cells are less tolerant; therefore, a lower concentration of NaCl was used in the corresponding experiments.
Identification of GLYOXALASE-I genes (PdGLX1) within P. dactylifera L. genome
The GLYOXALASE-I specific amino acid domain was retrieved from the protein families (Pfam) database and used as a probe for the protein sequence search (KLKNPMLVVTDIDKSVEFYKKVFGLYVIMDFGANKTLTGGLALQTSETYKEFIGTSDISFGGNNFEVYFEEEDFDKFADRLKEYDIEYVHPIIEHSWGQRVVRFYDPDKHIIEV). The BLASTP search tool was then used against the date palm genome (taxid:42345), previously deposited within the National Center for Biotechnology Information (NCBI) database, to identify putative PdGLX1 proteins, which were then analyzed using the Hidden Markov Model (HMM) profile on the Pfam database.32
In silico analysis and subcellular localization prediction of PdGLX1
Computational and biochemical analyses, in addition to subcellular localization prediction, were performed to understand the function of the PdGLX1 proteins in date palm. The pI/MW tool on the ExPASy platform was used to determine the theoretical molecular weight (MW) and the isoelectric point (pI).33 Three subcellular localization prediction tools were utilized with default search parameters: the Wolf PSORT,34 the CELLO v.2.5,35 and the ChloroP localization tools.36 The presence of cis-acting elements within 1.5 kb of the deduced promoter region was determined using PlantCARE.37 The coding region from the genomic DNA was verified using the GSDS 2.0 tool.38 The Multiple Expectation for Motif Elicitation (MEME) tool was utilized for conserved motif prediction using default parameters.39 The phylogenetic relationship between PdGLX1 and other protein orthologues of Arabidopsis thaliana, Medicago sativa, Oryza sativa, Glycine max, and Sourgum bicolor was constructed using the neighbor-joining method and Kimura 2-parameter model implemented within the MEGAX software.40
Cloning of the putative PdGLX1 genes
Total RNA from date palm leaf and root samples was isolated using the MRIP method.41 SuperScript IV Reverse Transcriptase (Invitrogen, California, USA) was utilized for complementary DNA (cDNA) synthesis. DreamTaq Master Mix (ThermoScientific, Massachusetts, USA) and gene-specific primers (Table S1) were used to amplify the different date palm putative PdGLX1 coding regions using the synthesized cDNA. These amplified genes were cloned into the pGEM-T Easy vector system (Promega, Wisconsin, USA), and the sequences were verified using the standard Sanger DNA sequencing method. The attB1 and attB2 sites were integrated using PCR and suitable oligonucleotides (Table S1) at the 5’ and 3’ end of each gene followed by the BP-clonase (Invitrogen, California, USA) reaction to generate the Gateway-entry clone pDONR-Zeo-PdGLX. Subsequently, the LR-clonase reaction (Invitrogen, California, USA) was used to sub-clone the genes into a bacterial expression vector pET-DEST42 (Invitrogen, California, USA) and into a yeast expression vector pYES-DEST52 (Invitrogen, California, USA). The Gateway cassette of pET-DEST42 vector was excised using the BsrGI restriction enzyme (ThermoScientific, Massachusetts, USA) and re-circularized using T4 ligase (NEB, Massachusetts, USA). This procedure has generated an empty vector, which was used as a negative control for subsequent experiments.
Expression profile of the PdGLX1 gene family under stress conditions
The cDNA was synthesized using RNA extracted from the leaf and root tissues of date palm seedlings, as described earlier. This cDNA was used as a template to perform qPCR using Fast-SYBR (Invitrogen, California, USA) and gene-specific primers (Table S1) on the Biorad CFX96 Real-time PCR system (Biorad). The expression data were normalized to the expression of ACTIN as a reference gene42 and analyzed using the 2−ΔΔCT method.43
Abiotic stress tolerance assay in E. coli
The bacterial expression cassettes pET-DEST42-PdGLX and empty pET-DEST42 were genetically transformed into BL21 E. coli cells using the Gene Pulser Xcell electroporation system (Bio-Rad, USA). The transgenic cells were cultured at 37°C in liquid LB containing 1 mg/ml ampicillin until they reach a mid-exponential growth phase (OD600 ~ 0.6). Subsequently, PdGLX gene expression was induced by the addition of 0.5 mM Isopropyl β- d-1-thiogalactopyranoside (IPTG) followed by treatment with the different stress inducers included salinity (250 mM NaCl), oxidative stress (5 mM H2O2), and MG toxicity (0.5 mM MG). The optical density (OD600) was recorded at 2-hour intervals for a total growth period of 12 hours. The cells were then harvested to quantify the MG concentration. All measurements were conducted in three biological replicates.
Abiotic stress tolerance assay in yeast
The yeast expression cassettes pYES-DEST52-PdGLX and the empty pYES-DEST52 were transformed to MG hypersensitive yeast knockout GLO1 cell lines (YML004C; GLO1 BY4741; MAT a; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; YML004C::kanMX4) (HVD Biotech Vertriebs Ges.m.b.H, Austria). The empty vector was also transformed into the wildtype yeast (Saccharomyces cerevisiae) parent strain (BY4741; MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) using a previously described PEG-lithium acetate heat shock method.44 The transformed yeast knockouts cells were then streaked on synthetically defined (SD)-agar plates with different concentrations of MG and incubated at 30ºC for 72 h. Additionally, the transgenic yeast knockouts were cultured in SD glucose media until a mid-exponential growth phase (OD600 ~ 0.8), washed, and an equal number of cells were separately inoculated into SD galactose liquid media supplemented with the different stress conditions: 500 μM MG, 700 μM H2O2 and 500 mM NaCl. The growth of the cells was monitored spectrophotometrically (OD600) at 6-h intervals for a total of 30 h and harvested for MG quantification at the 30-h time point.
MG quantification
The cellular MG level was estimated as previously described.45 Briefly, the cells were harvested from liquid culture and lysed by suspension in 250 μl of 5 M perchloric acid on ice for 15 min. The lysed cells were centrifuged at 11,000 g for 10 min, and the resultant supernatant was neutralized with 1 M Na2HPO4. The neutralized lysate was centrifuged at 11,000 g for 10 min, and 1 ml of the supernatant was transferred to a fresh tube, to which 10 μl of NaN3 was added. The concentration of MG in the cell lysate was quantified using a reaction mixture containing 650 μl of the cell lysate, 250 μl of 7.2 mM 1,2-diaminobenzene, and 100 μl of 5 M perchloric acid. The absorbance was measured spectrophotometrically at 336 nm after 3 h incubation in the dark. The concentration of MG was calculated from an MG standard curve (1–100 μM).
ROS accumulation in yeast cells
To evaluate the consequences of MG stress on ROS accumulation in yeast knockout GLO1 cells genetically transformed with the different expression cassettes (pYES-DEST52-PdGLX1), the transgenic yeast cells were grown until an early exponential stage (OD600 ~ 0.6) and harvested by centrifugation at room temperature. The cells were then treated with 0.5 mM MG for 7 h at 30ºC, followed by staining with a peroxide-specific dye 50 mM 2’,7’-dichlorodihydrofluorescein diacetate (H2DCFAD) (Invitrogen, California, USA). The cells were also stained with a mitochondrial superoxide-specific dye 5 mM MitoSOX™ solution (Invitrogen, California, USA). The cells were stained in the dark for 30 min at 30ºC, washed with phosphate buffer saline, and the ROS levels were determined by flow cytometry using the BD FACSAria™ flow cytometer (BD Biosciences, USA). The FlowJo software46 was utilized to determine the mean fluorescent intensity (MFI) values from 5000 events.
Statistical analysis
One-way analysis of variance (ANOVA) and Dunnett’s T3 post hoc test was used to compare the statistically significant differences between the mean of tested parameters against the control. Statistical analyses were performed using the IBM SPSS statistical package version 21 (IBM Corp. Armonk, NY, USA).
Results
P. dactylifera L. genome encodes six PdGLX1 genes
BLASTP search on the NCBI database against the date palm genome revealed a total of six putative PdGLX1 genes from the genome of P. dactylifera L. Their sequence was then retrieved and further classified as PdGLX1:1, PdGLX1:2, PdGLX1:3, PdGLX1:4, PdGLX1:5 and PdGLX1:6 with the largest being PdGLX1:4 with 1107 bp and the smallest being PdGLX1:6 with 708 bp. Their locus location, gene identification number (id), mRNA identification number (id), protein identification number (id), coding sequences (CDS) length, and protein length are shown in Table S2.
Molecular phylogenetic analysis of the putative PdGLX1 proteins was constructed using the Maximum Likelihood method (ML). The percentage of trees in which the proteins clustered together is shown next to the branches based on 1000 bootstrap replicates (Figure 1a). Further, analyses of the coding region against the corresponding genomic sequence revealed the presence of eight introns in PdGLX1:1, PdGLX1:3, and PdGLX1:4 genes. However, only seven introns were found in PdGLX1:2, PdGLX1:5 and PdGLX1:6 coding sequence in the P. dactylifera L. genome (Figure 1b).
Figure 1.
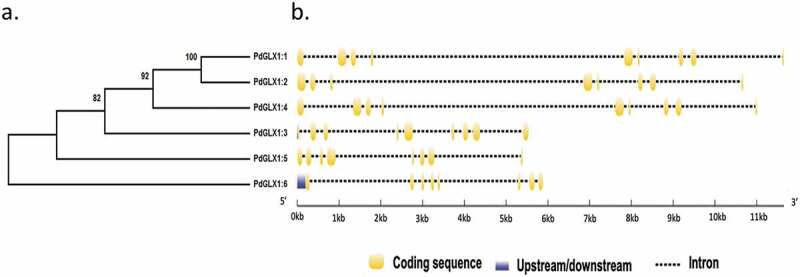
Phylogenetic analysis of the putative PdGLX1 proteins using the Maximum Likelihood (ML) method (a). The percentage of trees in which the proteins clustered together is shown next to the branches based on 1000 bootstrap replicates. Intron-exon map of the different PdGLX1 genes (b)
The retrieved protein sequences were then searched for the presence of specific domains using the Hidden Markov Model (HMM) profile on the Pfam database. The results showed the presence of two Glyoxalase/Bleomycin resistance protein/Dioxygenase superfamily domain (PF00903.25) each for PdGLX1:1–5, which classifies these proteins as Ni2+-dependent GLX1 proteins, whereas PdGLX1:6 has a single GLX1 domain and therefore classified as Zn2+-dependent GLX1 proteins. Together, the presence of these domains confirmed the identity of the retrieved sequences (Figure 2, Table S3).
Figure 2.
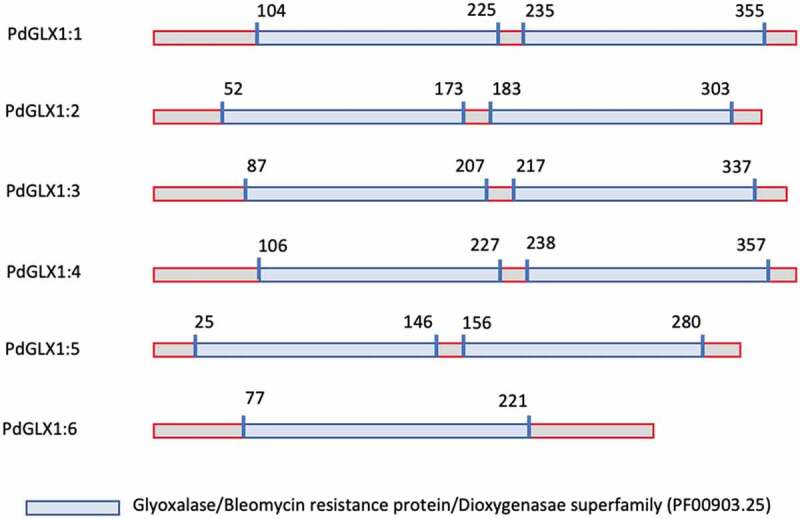
Domain architecture of the different PdGLX1:1–6 proteins
In-silico analysis of the molecular weight revealed that PdGLX1:4 had the largest molecular weight of 40.76 kDa, and PdGLX1:6 had the smallest molecular weight of 26.41 kDa. The results also showed that all the PdGLX1 proteins were acidic except PdGLX1:4, which was almost neutral (pI = 7.09), whereas the most acidic protein was PdGLX1:5 (pI = 5.17) (Table S4). Furthermore, computational subcellular localization prediction showed that all the putative PdGLX1 proteins were localized to the mitochondria, the chloroplast, and the cytosol using the PSORT and CELLO tools. Consistently, chloroplast transit peptides were also identified in the sequences of all the putative PdGLX1 proteins except for PdGLX1:5 proteins using the ChloroP localization tools (Table S4).
Analyses of the putative promoter sequences showed the presence of various cis-acting regulatory elements within 1.5 kb upstream of the PdGLX1 start codon of the gene, including potential environmental stress and phytohormone transcription binding sites, suggesting that different environmental stimuli and phytohormones may regulate the expression of the PdGLX1 genes (Table S5). Further, conserved motifs analysis using the MEME tool predicted the presence of various 7–8 bp conserved motifs with C/G rich regions, which may act as MG-responsive elements (Figure S1).
Multiple sequence analysis of the putative PdGLX1 proteins along with other previously well-characterized GLX1 orthologues from Oryza sativa and Arabidopsis thaliana showed that the GLX1 proteins are conserved. Besides, the Zn2+-dependent GLX1 proteins OsGLX1-8 and PdGLX1:6 had extended sequences which are labeled as A, B, and C (Figure 3). These extra sequence stretches are characteristic of Zn2+-dependent GLY protein. In addition, the metal-binding sites were also identified and marked in triangles (Q/E/H/E), whereas the Ni2+-dependent GLX1 metal-binding sites (Q/E/Q/E) were also identified (Figure 3).
Figure 3.
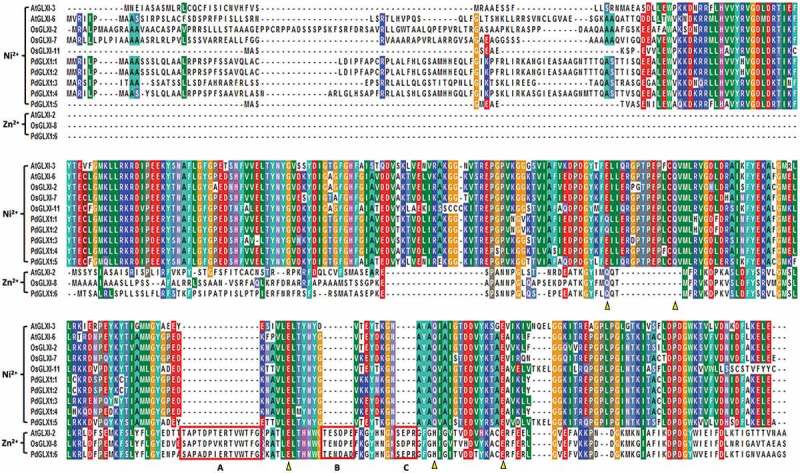
Multiple sequence analysis of the putative PdGLX1 proteins along with other previously well-characterized GLX1 orthologues from Oryza sativa and Arabidopsis thaliana. The Zn2+-dependent protein extended sequences are labeled as A, B, and C. Whereas, the Zn2+ (Q/E/H/E) and Ni2+ (Q/E/Q/E) metal-binding domain are shown in triangles
Conserved motif prediction using the MEME tool revealed the presence of various motifs from the retrieved protein sequences similar to those from Arabidopsis thaliana and Oryza sativa (Figure 4). The results showed that the motif 1 was shared among all the analyzed sequences. However, amino acid motifs 1–8 were common among all the Ni2+ dependent GLX1 proteins, while the Zn2+ dependent GLX1 proteins, AtGLX1-2, OsGLX1-8, and PdGLX1:6, had four identical motifs each. In addition, AtGLX1-6, PdGLX1:1, PdGLX1:2, PdGLX1:3, and PdGLX1:4 had nine identical motifs (Figure 4).
Figure 4.
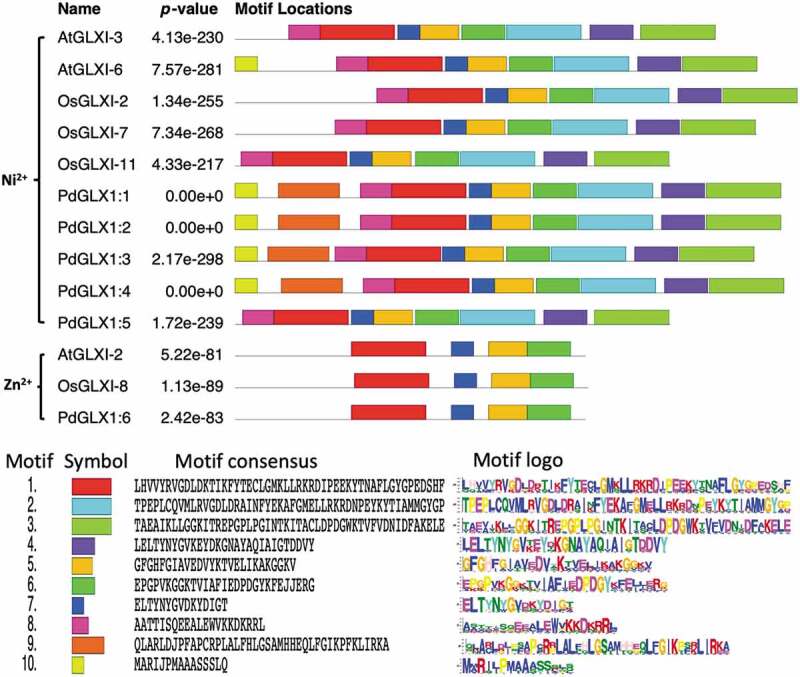
Conserved motifs identified by MEME of GLYOXALASE-I proteins identified from Arabidopsis thaliana, Oryza sativa, and P. dactylifera L. Lengths of motifs of each GLYOXALASE-I protein are displayed proportionally
Phylogenetic analysis of the putative PdGLX1 and other protein orthologues of Arabidopsis thaliana, Medicago sativa, Oryza sativa, Glycine max, and Sourgum bicolor showed that the proteins were clustered into three major groups- Ni2+-dependent proteins (Clads I), Zn2+-dependent (Clade II) and GLX1-like proteins (Clade III). PdGLX1:1–5 proteins were all clustered into clade I, whereas PdGLX1:6 was clustered into clade II (Figure 5).
Figure 5.
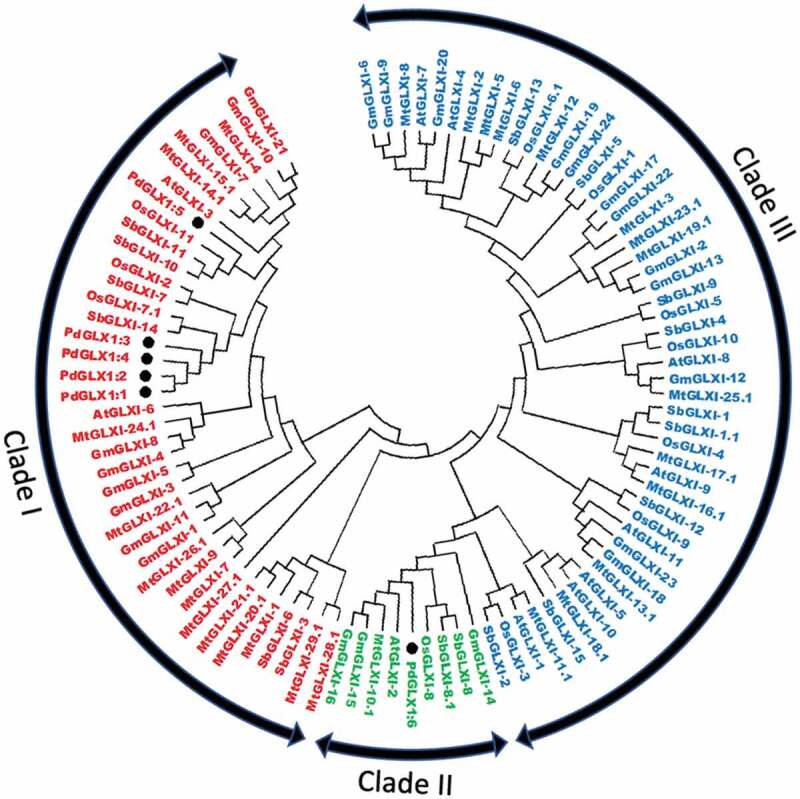
Phylogenetic analysis of Glyoxalase-I proteins identified from P. dactylifera L. and other plant species. Circular tree constructed for the GLXI proteins from P. dactylifera L., Arabidopsis thaliana, Oryza sativa, Sorghum bicolor, Medicago sativa, and Glycine max using Neighbor-Joining method in MEGA X with 1000 bootstrap replicates. The protein sequences are characterized as Clade I for Ni2+-dependent proteins, Clade II for Zn2+-dependent proteins, and Clade III for GLX-like proteins
PdGLX1 genes were induced under different stress conditions
To examine the effect of different environmental stresses including saline (300 mM NaCl), oxidative (10 mM H2O2) and MG toxicity stresses (10 mM MG) on PdGLX1 gene expression in the date palm seedlings, their expression profile was analyzed using qPCR. The results showed that the expression of PdGLX1 genes was altered depending on the stress in the leaf and root tissues (Figure 6). In general, there is a trend of upregulation of all the PdGLX1 genes, with the expression of PdGLX1:1 and PdGLX1:4 being significantly (p 0.05) upregulated under NaCl stress in leaf tissues, compared to the control treatment. However, the expression level of PdGLX1:4 was reduced in the 10 mM H2O2 treated leaves. A similar trend of upregulation was observed in the root tissues of the treated plants, where the PdGLX1:4 was significantly (p 0.05) increased under H2O2 stress treatment. However, the expression of PdGLX1:2, PdGLX1:3, PdGLX1:5, and PdGLX1:6 was not considerably altered under different stress treatments (Figure 6).
Figure 6.
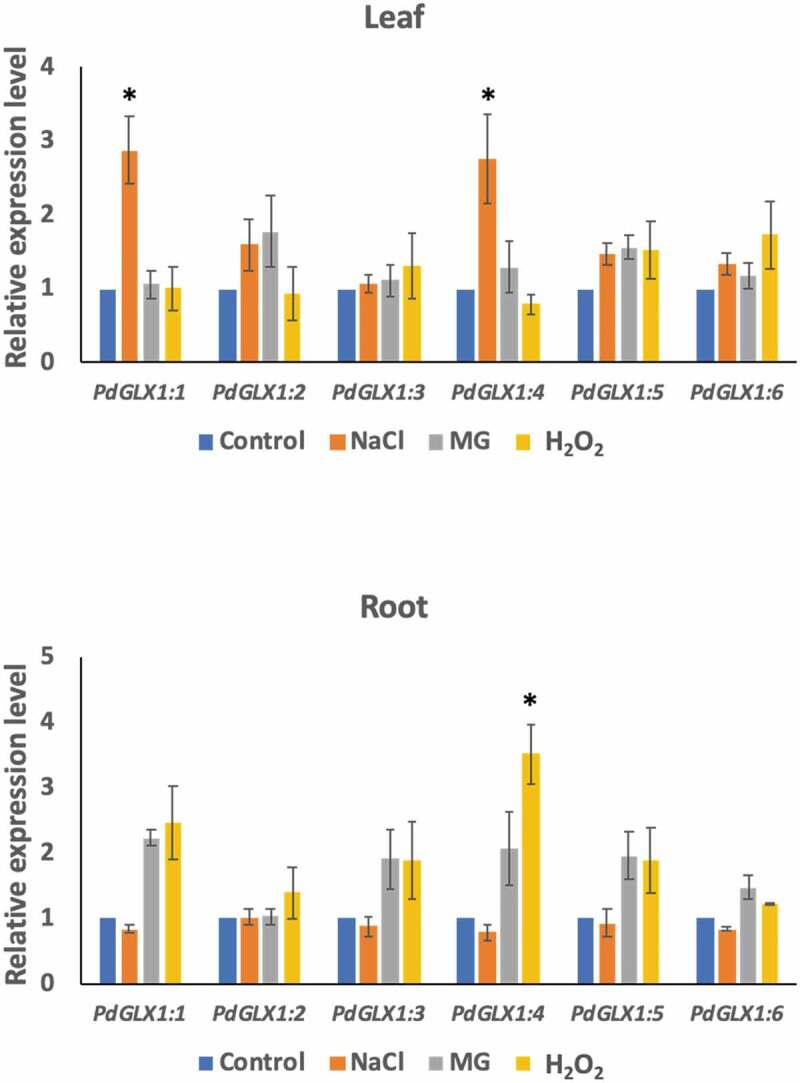
The relative expression level of the different PdGLX1 genes in the leaf (a) and root (b) tissue using qPCR method. Asterisk (*) indicates p 0.05; mean ±SD, n = 3
Heterologous expression of PdGLX1 in E. coli confers tolerance to abiotic stresses
The E. coli BL21 cells transformed with the expression cassettes pET-DEST42-PdGLX1:1–1:6 (Figure 7a) and the empty vector pET-DEST42 (as control) were grown in the presence of different abiotic stresses and the cultures were monitored spectrophotometrically (OD600). The growth of the transgenic E. coli cells increased significantly (p 0.001) when compared with cells transformed with the empty vector under various abiotic stress conditions such as 0.5 mM MG, 5 mM H2O2 and 250 mM NaCl (Figure 7b–d). In addition, the quantification of MG in the cells indicated a significant (p 0.001) increase in the accumulation of MG in the empty control vector as compared to E. coli cells overexpressing the different PdGLX1 genes under 0.5 mM MG (Figure 7b), 5 mM H2O2 (Figure 7c), and 250 mM NaCl (Figure 7d) stresses.
Figure 7.
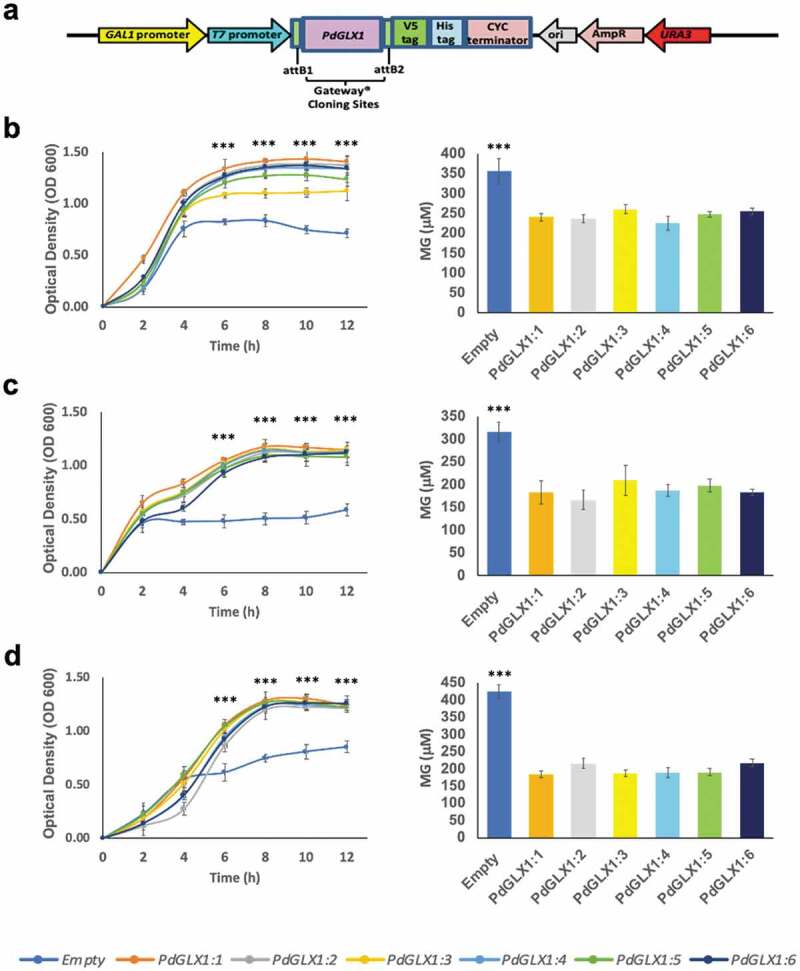
Heterologous expression of PdGLX1 enzymes in E. coli BL21 provides different levels of tolerance to various abiotic stresses. The PdGLX1 genes were cloned into the protein expression vector (pET-DEST52-PdGLX1:1–6) under the control of the T7 promoter (a). The transgenic bacteria were grown in the presence of different abiotic stresses, including MG (0.5 mM MG) (b), salinity (200 mM NaCl) (d), and oxidative stress (5 mM H2O2) (f). The MG concentration in the cells was measured for each treatment. The growth of the cells was spectrophotometrically monitored during the experiment. Asterisks (***) indicates p 0.001; mean ±SD, n = 3
Overexpression of PdGLX1 complements stress tolerance in knockout GLO1 S. cerevisiae
A complementation study of the MG-sensitive GLO1 yeast cells transformed with expression cassettes pYES-DEST52-PdGLX1:1–6 (Figure 8a) and empty vector pYES-DEST52 was conducted. To induce the expression of the PdGLX1 transgene, the transgenic cells were streaked (Figure 8b) on Ura− SD-galactose (Figure 8c) along with different concentrations of MG (Figure 8d–g). To block the expression of the PdGLX1 within the construct and therefore, to verify its effect on the cells under various stresses, the transgenic cells were also streaked on Ura− SD supplemented with glucose (Ura− SD- glucose) (Figure 8h) along with different concentrations of MG (Figure 8i–j). The results of the complementation assays showed that the growth of the control yeast cells (transformed with empty vector) was impaired when Ura− SD-galactose was supplemented with 2 mM MG (Figure 8e). However, the growth was rescued when the yeast cells were transformed with the pYES-DEST52-PdGLX1:1–6 expression cassettes. In addition, only the wild type BY4741 cells and the PdGLX1:6 overexpressing GLO1 yeast knockouts were able to grow at 3 mM MG (Figure 8f) and 4 mM MG (Figure 8g). While the wildtype BY4741 cells were able to grow on the Ura− SD-glucose plates supplemented with 1 mM and 2 mM MG, the knockout cells were unable to grow under the same conditions (Figure 8i–j).
Figure 8.
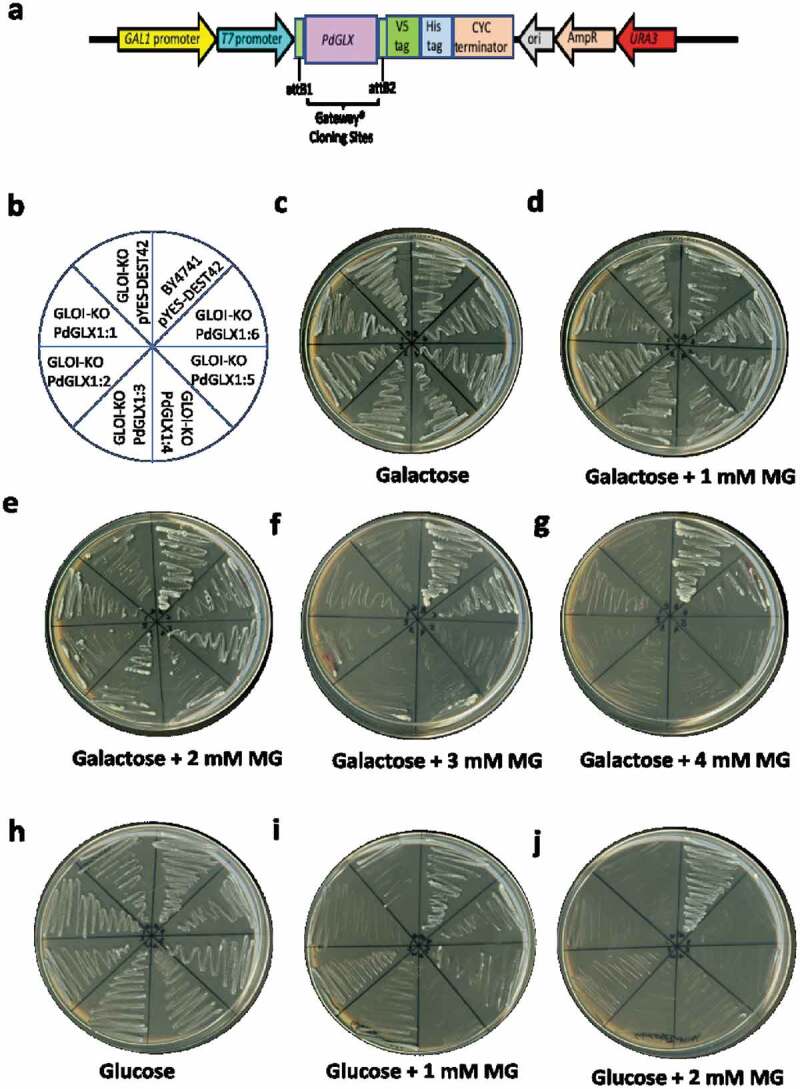
Mutant GLO1 yeast cells genetically transformed with the yeast expression cassette pYES-DEST52-PdGLX1:1–6 (a). The pictorial representation of the different constructs are shown (b), the transgenic cells were then streaked on Ura− SD galactose media (c), along with the different concentrations of MG; 1 mM MG (d), 2 mM MG (e), 3 mM MG (f), 4 mM MG (g), the transgenic cells were also streaked on different Ura− SD glucose media (h), along with different concentrations of MG; 1 mM MG (i), 2 mM MG (j)
Furthermore, the transgenic yeast cells were cultured in liquid SD-galactose along with the different abiotic stress inducers, and their growth was monitored spectrophotometrically (OD600) at 6 h intervals. The growth of the transgenic cells expressing the PdGLX1 genes increased significantly (p 0.001) compared to the cells transformed with an empty vector under various abiotic stress conditions including 500 μM MG (Figure 9a), 700 μM H2O2 (Figure 9b) and 500 mM NaCl (Figure 9c). The cells transformed with the empty vector also indicated a significantly (p 0.001) higher cellular accumulation of MG as compared to cells overexpressing the different PdGLX1 genes under 500 μM MG (Figure 9a), 700 μM H2O2 (Figure 9b) and 500 mM NaCl conditions (Figure 9c).
Figure 9.
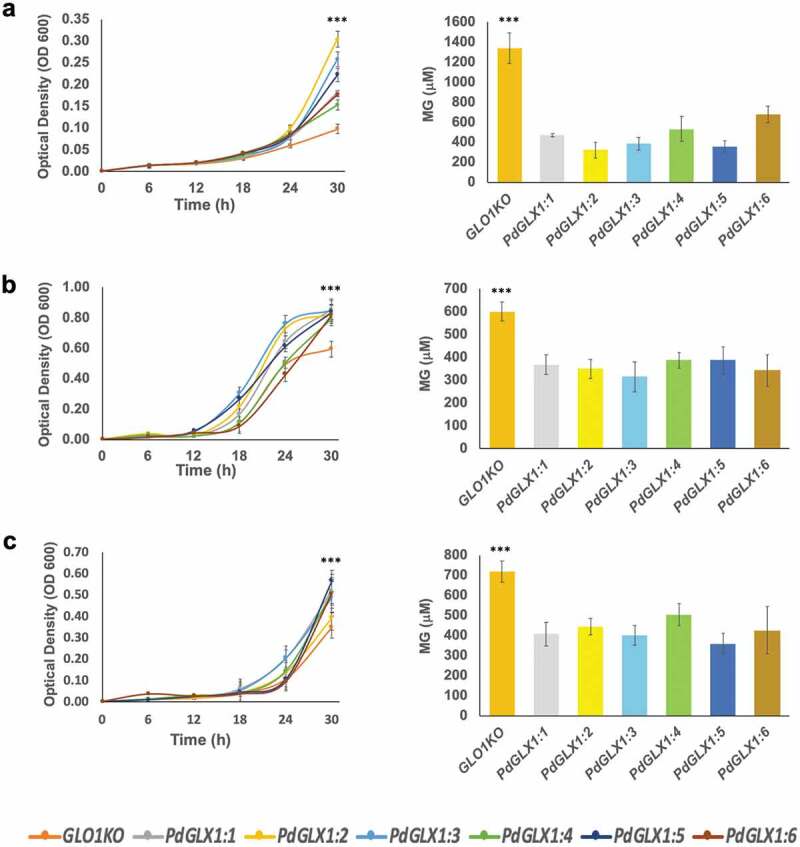
The growth of yeast mutant cells and the estimation of their MG content. The mutant GLO1 yeast cells genetically transformed with the yeast expression cassette pYES-DEST52-PdGLX1:1–6 and the empty vector pYES-DEST52 were grown in liquid Ura− SD galactose media in the presence of 500 µM MG (a), 700 µM H2O2 (b) and 500 mM NaCl (c). Asterisks (***) indicates p 0.001; mean ±SD, n = 3
Overexpression of PdGLX1 in yeast reduced ROS accumulation
The accumulation of ROS in the transgenic yeast GLO1 knockout cell lines in response to the presence of 0.5 mM MG for seven hours was quantified by flow cytometry. The results revealed a significant reduction in cytosolic ROS accumulation in the transgenic yeast cells with PdGLX1:1, PdGLX1:4, PdGLX1:5 (p 0.001) and PdGLX1:6 (p 0.05) genes as compared to knockout yeast cells transformed with the empty vector (Figure 10a). In addition, yeast cells transformed with PdGLX1:2, PdGLX1:3, and PdGLX1:6 genes had a significant (p 0.05) reduction in superoxide molecules in their mitochondria (Figure 10b).
Figure 10.
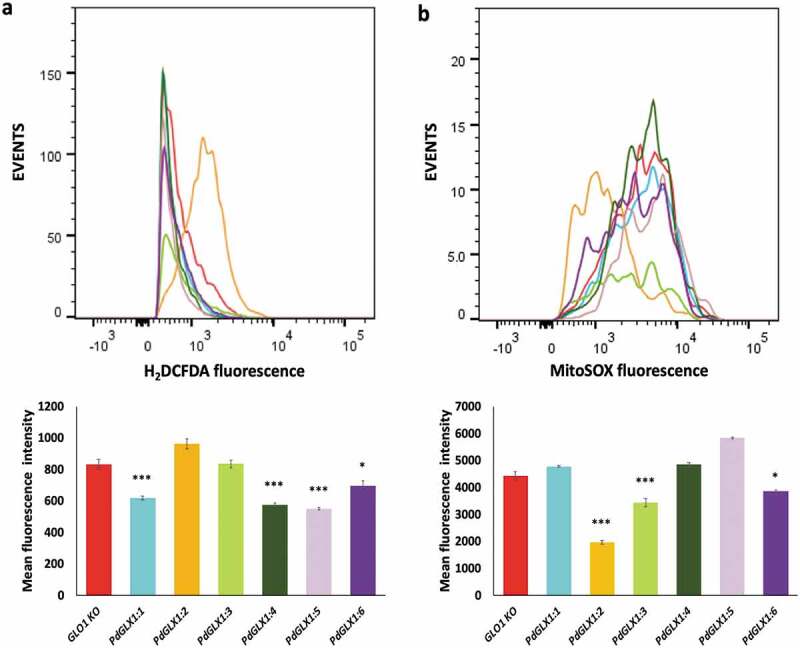
ROS quantification by flow cytometry in GLO1 mutant cells transformed with PdGLX1 genes or the empty negative control vector. The cells were stained to detect cytosolic ROS accumulation using H2DCFAD stain (a) or the mitochondria specific superoxide using the MitoSOX™ stain (b). Graphs indicate the mean fluorescent intensity (5000 events). Asterisks (***) indicates p 0.001, Asterisk (*) indicates p 0.05; mean ±SD, n = 3
Discussion
Plants sense and adapt their morphology, physiology, and gene expression under adverse environmental conditions by altering various stress signaling pathways to bring about cellular stability.47 Different stress-associated signaling molecules have been identified in plants, which includes phytohormones,48 ROS,49 reactive nitrogen species (RNS)50 and reactive carbonyl species (RCN).8,51 Studies have shown that ROS-mediated lipid peroxidation leads to the accumulation of RCN in plants.52,53 Among the RCN, MG has emerged as an essential stress signaling molecule.8 MG is a by-product of numerous enzymatic and non-enzymatic reactions. It is a highly reactive cytotoxic metabolite, which accumulates in the cell under adverse environmental conditions leading to the inactivation of proteins, DNA, RNA, and lipids.12,24,51 To overcome the adverse effects of MG accumulation in the cell, various MG-detoxification mechanisms have been identified, of which the universally conserved glyoxalase system is the most efficient pathway involving two consecutive reactions catalyzed by GLXI and GLXII.10,20
In silico analysis of the subcellular localization of the various putative PdGLX1 proteins showed that the proteins are mainly localized to the cytosol, mitochondria, and chloroplast. In plants, ROS production is found mostly in the chloroplast, mitochondria, and peroxisome. Therefore, these sites may also be prone to ROS-mediated lipid peroxidation, which may lead to the accumulation of MG in these organelles.54 In addition, the localization of the various putative PdGLX1 proteins in the cytosol may help to keep a check on the level of MG produced via glycolysis.55 The localization of the putative PdGLX1 proteins to the chloroplast may suggest a mechanism to control the increasing level of MG originated mainly from the degradation of trios sugars via the Benson-Calvin cycle.56 In the mitochondria, MG disrupts the electron transport chain and increases the generation of superoxide, NO, and peroxynitrite leading to mitochondrial oxidative stress.57 The localization of the various PdGLX1 proteins to the diverse organelles may suggest that the action of the putative PdGLX1 proteins is site-specific, and responds to different developmental stages and metabolic conditions induced by cellular and environmental changes as previously suggested.10,21 Further, promoter region analysis showed the presence of various potential environmental stresses and plant hormone activated transcription factors binding sites, suggesting that different environmental factors and hormones may regulate the expression of the PdGLX1 genes. In addition, the identification of various 7–8 bp C/G rich conserved motifs upstream of the PdGLX1 genes may suggest that these regions act as potential MG-responsive elements, as previously suggested.17
Glyoxalase-I is a metalloenzyme that requires divalent metal ions, such as Zn2+ or Ni2+ for its activity.58 From the date palm genome, we identified both the Zn2+- and the Ni2+-dependent PdGLX1 proteins based on the presence of the conserved domain sequences and the metal-binding domains. Further, the Zn2+- and Ni2+- dependent PdGLX1 also differed in the presence of conserved motifs in their protein sequences. It has been previously suggested that the metal binding property of the GLX1 proteins can be identified based on the domain architecture of the proteins, where Zn2+ dependent proteins have a single larger domain (~140-142 amino acids) as compared to two smaller (~120 amino acids) conserved GLX1 domains found in the Ni2+ dependent GLX1 proteins. This notion is concurrent with our findings that the sequence of Zn2+ dependent PdGLX1:6 has a single conserved GLX1 domain as compared to the other Ni2+-dependent PdGLX1:1–5 proteins which have two conserved GLX1 domains.59,60 Further phylogenetic analysis of putative PdGLX1:1–6 proteins along with previously classified orthologs from Arabidopsis thaliana, Medicago sativa, Oryza sativa, Glycine max and Sourgum bicolor61 show that PdGLX1:1–5 were classified in the (Clade I) Ni2+ dependent GLX1 proteins whereas PdGLX1:6 was grouped into the (Clade II) Zn2+ dependent GLX1 proteins.
Gene expression analysis showed an increasing trend of PdGLX1 expression under the tested stress conditions. The differential expression of these genes appears to be tissue-specific under various environmental stresses since we observed a significant upregulation of the PdGLX1:1 and PdGLX1:4 genes in the leaf tissue in response to 300 mM NaCl stress, whereas there was no change in their expression in the root tissues under the same conditions. However, there was a trend of upregulation of the PdGLX1 genes in the root tissue in response to the H2O2 and MG stress treatment. In line with these observations, various reports have previously shown that there is an increase in the levels of MG and the expression of the MG detoxification GLX1 genes under stress conditions.13,62 Furthermore, the importance of the GLX1 genes in enhancing stress tolerance has been verified by transgenic overexpression of GLX1 genes in numerous crop plants such as tobacco,29,63 black gram,64 rice,65 pumpkin23 and sugar beet.28 Therefore, the overexpression of the putative PdGLX1 genes under the tested conditions implies a stress tolerance mechanism in date palm to maintain MG at a nontoxic level.
Heterologous expression of the putative PdGLX1 genes in E. coli cells significantly enhanced their growth and reduced MG accumulation. This observation can be attributed to the ability of the PdGLX1 proteins to enhance the glyoxalase system in the cells, which is the main route of MG detoxification in bacteria.66 Moreover, it is consistent with previous reports indicating that various stress conditions lead to the accumulation of MG, which in turn hinders the growth of bacteria.67–69 Our conclusions are also supported by previous findings showing that overexpression of the Arabidopsis GLY1 confers stress tolerance to E. coli cells.45
In yeast, the detoxification of MG is controlled by the GLO1 and GRE3 genes. Therefore, mutant yeast cells lacking PdGLX1 genes were sensitive to exogenous application of MG and showed a higher cellular accumulation of MG due to the loss of function of the GLO1 gene.70 In the present study, the yeast complementation assay showed that the PdGLX1 genes were able to complement the loss in function of the MG-sensitive GLO1 knockout yeast cell line genetically transformed with PdGLX1:1–6. These transgenic yeast cells displayed normal growth in Ura−SD-galactose supplemented with high MG concentration, whereas the growth of the empty vector control cells was hindered even at a low MG concentration. The PdGLX1 genes were cloned under the control of the GAL1-promoter, which is activated by galactose and suppressed by glucose. Therefore, the enhanced growth was not observed when the transgenic GLO1 cells were streaked on Ura− SD-glucose supplemented with low concentrations of MG. This test showed that PdGLX1 genes are responsible for enhancing the growth of the GLO1 knockout yeast. Among the different overexpressing PdGLX1:1–6 GLO1 knockout yeast cell lines, the overexpression of the Zn2+-dependent Glyoxalase-I protein PdGLX1:6 showed enhanced survival of the GLO1 knockout lines on Ura−SD-galactose plates supplemented with high MG concentration. This finding may suggest a greater efficiency of the Zn2+-dependent protein than that of the Ni2+-dependent protein in the detoxification of MG, which is consistent with previous studies in other plant species.71 Further, overexpressing the putative PdGLX1 genes enhanced not only the MG-detoxification activity but also the antioxidant activity and, hence the salinity stress tolerance of the GLO1 knockouts mutants.
Cumulatively, our observations imply that the PdGLX1 protein family can catalyze MG detoxification and maintain the cellular redox balance in the date palm, thus enabling its growth under various stress conditions. Future experimental works, including overexpression and localization of PdGLX1 genes in planta, are needed to understand better the MG detoxification and antioxidant mechanisms conferred by PdGLX1 genes in date palm.
Supplementary Material
Acknowledgments
Authors would like to acknowledge the College of Science, Sultan Qaboos University, Oman, for the generous fund number IG/SCI/BIOL/18/01 to MWY. The authors would like to thank Dr. Sirin Adham, Sultan Qaboos University, for her help in the flow cytometry, which was used in this study.
Funding Statement
This work was supported by the Sultan Qaboos University [IG/SCI/BIOL/18/01].
Disclosure of Potential Conflicts of Interest
The authors declare that they have no conflict of interest. All authors revised and approved the final manuscript.
Authors’ contributions
GAJ conceived, designed, performed the experiments, analyzed data, and wrote the manuscript, and MWY designed the experiments, supervised the work, edited the manuscript, and contributed reagents/materials/analysis tools.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Assaha DV, Ueda A, Saneoka H, Al-Yahyai R, Yaish MW.. The role of Na+ and K+ transporters in salt stress adaptation in glycophytes. Front Physiol. 2017;8:1. doi: 10.3389/fphys.2017.00509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkinson NJ, Urwin PE.. The interaction of plant biotic and abiotic stresses: from genes to the field. J Exp Bot. 2012;63(10):3523–15. doi: 10.1093/jxb/ers100. [DOI] [PubMed] [Google Scholar]
- 3.Jana GA, Al Kharusi L, Sunkar R, Al-Yahyai R, Yaish MW.. Metabolomic analysis of date palm seedlings exposed to salinity and silicon treatments. Plant Signal Behav. 2019;14(11):1663112. doi: 10.1080/15592324.2019.1663112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yaish MW, Kumar P.. Salt tolerance research in date palm tree (Phoenix dactylifera L.). Front Plant Sci. 2015;6:348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Harrasi I, Patankar HV, Al-Yahyai R, Sunkar R, Krishnamurthy P, Kumar PP, Yaish MW. Molecular characterization of a date palm vascular highway 1-interacting kinase (PdVIK) under abiotic stresses. Genes. 2020;11(5):568. doi: 10.3390/genes11050568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al‑Harrasi I, Jana GA, Patankar HV, Al‑Yahyai R, Rajappa S, Kumar PP, Yaish MW. A novel tonoplast Na+/H+ antiporter gene from date palm (PdNHX6) confers enhanced salt tolerance response in Arabidopsis. Plant Cell Rep. 2020;39(8):1079–1093. doi: 10.1007/s00299-020-02549-5. [DOI] [PubMed] [Google Scholar]
- 7.Jana GA, Yaish MW. Genome-wide identification and functional characterization of glutathione peroxidase genes in date palm (Phoenix dactylifera L.) under stress conditions. Plant Gene. 2020;23:100237. doi: 10.1016/j.plgene.2020.100237. [DOI] [Google Scholar]
- 8.Hoque TS, Hossain MA, Mostofa MG, Burritt DJ, Fujita M, Tran L-SP. Methylglyoxal: an emerging signaling molecule in plant abiotic stress responses and tolerance. Front Plant Sci. 2016;7:1341. doi: 10.3389/fpls.2016.01341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma R, De Vleesschauwer D, Sharma MK, Ronald PC. Recent advances in dissecting stress-regulatory crosstalk in rice. Mol Plant. 2013;6(2):250–260. doi: 10.1093/mp/sss147. [DOI] [PubMed] [Google Scholar]
- 10.Schmitz J, Dittmar IC, Brockmann JD, Schmidt M, Hüdig M, Rossoni AW, Maurino VG. Defense against reactive carbonyl species involves at least three subcellular compartments where individual components of the system respond to cellular sugar status. Plant Cell. 2017;29(12):3234–3254. doi: 10.1105/tpc.17.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saito R, Yamamoto H, Makino A, Sugimoto T, Miyake C. Methylglyoxal functions as Hill oxidant and stimulates the photoreduction of O2 at photosystem I: a symptom of plant diabetes. Plant Cell Environ. 2011;34(9):1454–1464. doi: 10.1111/j.1365-3040.2011.02344.x. [DOI] [PubMed] [Google Scholar]
- 12.Kaur C, Singla-Pareek SL, Sopory SK. Glyoxalase and methylglyoxal as biomarkers for plant stress tolerance. CRC Crit Rev Plant Sci. 2014;33(6):429–456. doi: 10.1080/07352689.2014.904147. [DOI] [Google Scholar]
- 13.Yadav SK, Singla-Pareek SL, Ray M, Reddy MK, Sopory SK. Methylglyoxal levels in plants under salinity stress are dependent on glyoxalase I and glutathione. Biochem Biophys Res Commun. 2005a;337(1):61–67. doi: 10.1016/j.bbrc.2005.08.263. [DOI] [PubMed] [Google Scholar]
- 14.Cai Y-T, Zhang H, Qi Y-P, Ye X, Huang Z-R, Guo J-X, Chen L-S, Yang L-T. Responses of reactive oxygen species and methylglyoxal metabolisms to magnesium-deficiency differ greatly among the roots upper and lower leaves of Citrus sinensis. BMC Plant Biol. 2019;19:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoque TS, Uraji M, Tuya A, Nakamura Y, Murata Y. Methylglyoxal inhibits seed germination and root elongation and up-regulates transcription of stress-responsive genes in ABA-dependent pathway in Arabidopsis. Plant Biol. 2012c;14(5):854–858. doi: 10.1111/j.1438-8677.2012.00607.x. [DOI] [PubMed] [Google Scholar]
- 16.Hoque TS, Okuma E, Uraji M, Furuichi T, Sasaki T, Hoque MA, Nakamura Y, Murata Y. Inhibitory effects of methylglyoxal on light-induced stomatal opening and inward K+channel activity in arabidopsis. Biosci Biotechnol Biochem. 2012b;76(3):617–619. doi: 10.1271/bbb.110885. [DOI] [PubMed] [Google Scholar]
- 17.Kaur C, Kushwaha HR, Mustafiz A, Pareek A, Sopory SK, Singla-Pareek SL. Analysis of global gene expression profile of rice in response to methylglyoxal indicates its possible role as a stress signal molecule. Front Plant Sci. 2015a;6:682. doi: 10.3389/fpls.2015.00682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoque MA, Uraji M, Torii A, Banu MNA, Mori IC, Nakamura Y, Murata Y. Methylglyoxal inhibition of cytosolic ascorbate peroxidase from Nicotiana tabacum. J Biochem Mol Toxicol. 2012a;26(8):315–321. doi: 10.1002/jbt.21423. [DOI] [PubMed] [Google Scholar]
- 19.Kaur C, Sharma S, Singla-Pareek SL, Sopory SK. Methylglyoxal, triose phosphate isomerase, and glyoxalase pathway: implications in abiotic stress and signaling in plants. In: Pandey G. (eds) Elucidation of abiotic stress signaling in plants. Springer; 2015. p. 347–366. 10.1007/978-1-4939-2211-6_13 [DOI] [Google Scholar]
- 20.Álvarez Viveros MF, Inostroza-Blancheteau C, Timmermann T, González M, Arce-johnson P. Overexpression of GlyI and GlyII genes in transgenic tomato (Solanum lycopersicum Mill.) plants confers salt tolerance by decreasing oxidative stress. Mol Biol Rep. 2013;40(4):3281–3290. doi: 10.1007/s11033-012-2403-4. [DOI] [PubMed] [Google Scholar]
- 21.Kaur C, Sharma S, Singla-Pareek SL, Sopory SK. Methylglyoxal detoxification in plants: role of glyoxalase pathway. Indian J Plant Physiol. 2016;21(4):377–390. doi: 10.1007/s40502-016-0260-1. [DOI] [Google Scholar]
- 22.Hossain MA, Hasanuzzaman M, Fujita M. Up-regulation of antioxidant and glyoxalase systems by exogenous glycinebetaine and proline in mung bean confer tolerance to cadmium stress. Physiol Mol Biol Plants. 2010;16(3):259–272. doi: 10.1007/s12298-010-0028-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hossain MA, Hossain MZ, Fujita M. Stress-induced changes of methylglyoxal level and glyoxalase I activity in pumpkin seedlings and cDNA cloning of glyoxalase I gene. Aust J Crop Sci. 2009;3:53. [Google Scholar]
- 24.Li Z-G. Methylglyoxal and glyoxalase system in plants: old players, new concepts. Bot Rev. 2016;82(2):183–203. doi: 10.1007/s12229-016-9167-9. [DOI] [Google Scholar]
- 25.Mustafiz A, Singh AK, Pareek A, Sopory SK, Singla-Pareek SL. Genome-wide analysis of rice and Arabidopsis identifies two glyoxalase genes that are highly expressed in abiotic stresses. Funct Integr Genomics. 2011;11(2):293–305. doi: 10.1007/s10142-010-0203-2. [DOI] [PubMed] [Google Scholar]
- 26.Singla-Pareek S, Reddy M, Sopory S. Genetic engineering of the glyoxalase pathway in tobacco leads to enhanced salinity tolerance. Proc Natl Acad Sci. 2003;100(25):14672–14677. doi: 10.1073/pnas.2034667100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Veena RVS, Sopory SK. Glyoxalase I from Brassica juncea: molecular cloning, regulation and its over‐expression confer tolerance in transgenic tobacco under stress. Plant J. 1999;17(4):385–395. doi: 10.1046/j.1365-313X.1999.00390.x. [DOI] [PubMed] [Google Scholar]
- 28.Wu C, Ma C, Pan Y, Gong S, Zhao C, Chen S, Li H. Sugar beet M14 glyoxalase I gene can enhance plant tolerance to abiotic stresses. J Plant Res. 2013;126(3):415–425. doi: 10.1007/s10265-012-0532-4. [DOI] [PubMed] [Google Scholar]
- 29.Yadav SK, Singla-Pareek SL, Reddy MK, Sopory SK. Transgenic tobacco plants overexpressing glyoxalase enzymes resist an increase in methylglyoxal and maintain higher reduced glutathione levels under salinity stress. FEBS Lett. 2005b;579(27):6265–6271. doi: 10.1016/j.febslet.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 30.Yaish MW, Patankar HV, Assaha DV, Zheng Y, Al-Yahyai R, Sunkar R. Genome-wide expression profiling in leaves and roots of date palm (Phoenix dactylifera L.) exposed to salinity. BMC Genomics. 2017;18(1):246. doi: 10.1186/s12864-017-3633-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yaish MW, Sunkar R, Zheng Y, Ji B, Al-Yahyai R, Sardar FA. A genome-wide identification of the miRNAome in response to salinity stress in date palm (Phoenix dactylifera L.). Front Plant Sci. 2015;6(946). doi: 10.3389/fpls.2015.00946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14(9):755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 33.Bjellqvist B, Hughes GJ, Pasquali C, Paquet N, Ravier F, Sanchez JC, Frutiger S, Hochstrasser D. The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis. 1993;14(1):1023–1031. doi: 10.1002/elps.11501401163. [DOI] [PubMed] [Google Scholar]
- 34.Horton P, Park K-J, Obayashi T, Fujita N, Harada H, Adams-Collier C, Nakai K. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35(suppl_2):W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu CS, Chen YC, Lu CH, Hwang JK. Prediction of protein subcellular localization. Proteins: structure. Proteins. 2006;64(3):643–651. doi: 10.1002/prot.21018. [DOI] [PubMed] [Google Scholar]
- 36.Emanuelsson O, Nielsen H, Von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999;8(5):978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu B, Jin J, Guo A-Y, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2014;31(8):1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bailey TL, Johnson J, Grant CE, Noble WS. The MEME suite. Nucleic Acids Res. 2015;43(W1):W39–W49. doi: 10.1093/nar/gkv416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao Y, Yang Y, Cao H, Fan H, Ma Z, Lei X, Mason AS, Xia Z, Huang X. Efficient isolation of high quality RNA from tropical palms for RNA-seq analysis. Plant Omics. 2012;5:584. [Google Scholar]
- 42.Patankar HV, Assaha DV, Al-Yahyai R, Sunkar R, Yaish MW. Identification of reference genes for quantitative real-time PCR in date palm (Phoenix dactylifera L.) subjected to drought and salinity. PloS One. 2016;11(11):e0166216. doi: 10.1371/journal.pone.0166216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 44.Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16(5–6):339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 45.Jain M, Nagar P, Sharma A, Batth R, Aggarwal S, Kumari S, Mustafiz A. GLYI and D-LDH play key role in methylglyoxal detoxification and abiotic stress tolerance. Sci Rep. 2018;8(1):5451. doi: 10.1038/s41598-018-23806-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Becton D (2019) FlowJo ™ Software. 10.5.3 edn.
- 47.Zhu J-K. Abiotic stress signaling and responses in plants. Cell. 2016;167(2):313–324. doi: 10.1016/j.cell.2016.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verma V, Ravindran P, Kumar PP. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016;16(1):86. doi: 10.1186/s12870-016-0771-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choudhury S, Panda P, Sahoo L, Panda SK. Reactive oxygen species signaling in plants under abiotic stress. Plant Signal Behav. 2013;8(4):e23681. doi: 10.4161/psb.23681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luis A, Sandalio LM, Corpas FJ, Palma JM, Barroso JB. Reactive oxygen species and reactive nitrogen species in peroxisomes. Production scavenging, and role in cell signaling. Plant Physiol. 2006;141:330–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mostofa MG, Ghosh A, Li Z-G, Siddiqui MN, Fujita M, Tran L-SP. Methylglyoxal–a signaling molecule in plant abiotic stress responses. Free Radic Biol Med. 2018;122:96–109. doi: 10.1016/j.freeradbiomed.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 52.Biswas MS, Mano J. Lipid peroxide-derived short-chain carbonyls mediate hydrogen peroxide-induced and salt-induced programmed cell death in plants. Plant Physiol. 2015;168(3):885–898. doi: 10.1104/pp.115.256834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mano J. Reactive carbonyl species: their production from lipid peroxides, action in environmental stress, and the detoxification mechanism. Plant Physiol Biochem. 2012;59:90–97. doi: 10.1016/j.plaphy.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 54.Das K, Roychoudhury A. Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front Environ Sci. 2014;2:53. doi: 10.3389/fenvs.2014.00053. [DOI] [Google Scholar]
- 55.Plaxton WC. The organization and regulation of plant glycolysis. Ann Rev Plant Physiol Plant Mol Biol. 1996;47(1):185–214. doi: 10.1146/annurev.arplant.47.1.185. [DOI] [PubMed] [Google Scholar]
- 56.Takagi D, Inoue H, Odawara M, Shimakawa G, Miyake C. The calvin cycle inevitably produces sugar-derived reactive carbonyl methylglyoxal during photosynthesis: a potential cause of plant diabetes. Plant Cell Physiol. 2014;55(2):333–340. doi: 10.1093/pcp/pcu007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hossain MA, Teixeira da Silva J, Fujita M. Glyoxalase system and reactive oxygen species detoxification system in plant abiotic stress response and tolerance: an intimate relationship. Abiotic Stress. 2011;1:235–266. [Google Scholar]
- 58.Jain M, Batth R, Kumari S, Mustafiz A. Arabidopsis thaliana contains both Ni2+ and Zn2+ dependent glyoxalase I enzymes and ectopic expression of the latter contributes more towards abiotic stress tolerance in E. coli. PLoS One. 2016;11(7):7. doi: 10.1371/journal.pone.0159348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaur C, Sharma S, Hasan MR, Pareek A, Singla-Pareek SL, Sopory SK. Characteristic variations and similarities in biochemical, molecular, and functional properties of glyoxalases across prokaryotes and eukaryotes. Int J Mol Sci. 2017;18(4):250. doi: 10.3390/ijms18040250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li T, Cheng X, Wang Y, Yin X, Li Z, Liu R, Liu G, Wang Y, Xu Y. Genome-wide analysis of glyoxalase-like gene families in grape (Vitis vinifera L.) and their expression profiling in response to downy mildew infection. BMC Genomics. 2019;20(1):362. doi: 10.1186/s12864-019-5733-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bhowal B, Singla-Pareek SL, Sopory SK, Kaur C. From methylglyoxal to pyruvate: a genome-wide study for the identification of glyoxalases and D-lactate dehydrogenases in Sorghum bicolor. BMC Genomics. 2020;21(1):145. doi: 10.1186/s12864-020-6547-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Espartero J, Sánchez-Aguayo I, Pardo JM. Molecular characterization of glyoxalase-I from a higher plant; upregulation by stress. Plant Mol Biol. 1995;29(6):1223–1233. doi: 10.1007/BF00020464. [DOI] [PubMed] [Google Scholar]
- 63.Singla-Pareek SL, Yadav SK, Pareek A, Reddy M, Sopory S. Transgenic tobacco overexpressing glyoxalase pathway enzymes grow and set viable seeds in zinc-spiked soils. Plant Physiol. 2006;140(2):613–623. doi: 10.1104/pp.105.073734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bhomkar P, Upadhyay CP, Saxena M, Muthusamy A, Prakash NS, Pooggin M, Hohn T, Sarin NB. Salt stress alleviation in transgenic Vigna mungo L. Hepper (blackgram) by overexpression of the glyoxalase I gene using a novel Cestrum yellow leaf curling virus (CmYLCV) promoter. Mol Breeding. 2008;22(2):169–181. doi: 10.1007/s11032-008-9164-8. [DOI] [Google Scholar]
- 65.Zeng Z, Xiong F, Yu X, Gong X, Luo J, Jiang Y, Kuang H, Gao B, Niu X, Liu Y. Overexpression of a glyoxalase gene, OsGly I, improves abiotic stress tolerance and grain yield in rice (Oryza sativa L.). Plant Physiol Biochem. 2016;109:62–71. doi: 10.1016/j.plaphy.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 66.Ferguson GP, Tötemeyer S, MacLean M, Booth IR. Methylglyoxal production in bacteria: suicide or survival? Arch Microbiol. 1998;170(4):209–218. doi: 10.1007/s002030050635. [DOI] [PubMed] [Google Scholar]
- 67.Campbell AK, Naseem R, Holland IB, Matthews SB, Wann KT. Methylglyoxal and other carbohydrate metabolites induce lanthanum-sensitive Ca2+ transients and inhibit growth in E. coli. Arch Biochem Biophys. 2007;468(1):107–113. doi: 10.1016/j.abb.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 68.Cooper R. Metabolism of methylglyoxal in microorganisms. Ann Rev Microbiol. 1984;38(1):49–68. doi: 10.1146/annurev.mi.38.100184.000405. [DOI] [PubMed] [Google Scholar]
- 69.Inoue Y, Kimura A. Methylglyoxal and regulation of its metabolism in microorganisms. In: RK. Poole (Ed.) Advances in microbial physiology. Vol. 37. Elsevier; 1995. p. 177–227. 10.1016/S0065-2911(08)60146-0 [DOI] [PubMed] [Google Scholar]
- 70.Aguilera J, Antonio Prieto J. Yeast cells display a regulatory mechanism in response to methylglyoxal. FEMS Yeast Res. 2004;4(6):633–641. doi: 10.1016/j.femsyr.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 71.Batth R, Jain M, Kumar A, Nagar P, Kumari S, Mustafiz A. Zn2+ dependent glyoxalase I plays the major role in methylglyoxal detoxification and salinity stress tolerance in plants. Plos One. 2020;15(5):e0233493. doi: 10.1371/journal.pone.0233493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


