Abstract
Wheat dwarf virus (WDV) is considered as one of the most common viruses on cereal crops. Recently, severe outbreaks of WDV have been observed especially on winter wheat in southwestern part of Poland. Moreover, the presence of genetically different WDV-barley-specific and WDV-wheat-specific forms (WDV-B and WDV-W, respectively) was confirmed. In this study, a loop-mediated isothermal amplification assay (LAMP) was developed for the first time for efficient and rapid detection of WDV-B and WDV-W in infected plants. The reaction was performed using a set of three primer pairs: WDVF3/WDVB3, WDVFIB/WDVBIP and WDVLoopF/WDVLoopB specific for coat protein coding sequence. The amplified products were analyzed by direct staining of DNA, gel electrophoresis and real-time monitoring of the amplification curves. The sensitivity of optimized reaction was tenfold higher in comparison with conventional PCR. LAMP assay developed here is a useful and practical method for the rapid detection of different WDV isolates and can be implemented by phytosanitary services.
Keywords: WDV-W, WDV-B, Wheat, Barley, Identification, LAMP technique
Introduction
Wheat dwarf virus (WDV) is a member of the genus Mastrevirus in the family Geminiviridae. The virus is transmitted by leafhoppers Psammotettix alienus (Dahlb.) [1] and P. provincialis [2] and it infects numerous species within the family Poaceae including important cereals (mainly wheat and barley) [3]. WDV induces yellowing of leaves, severe stunting and, in case of early infection, death of infected plants which may lead to yield losses up to 80% [4]. The virus was first reported in Czechoslovakia [5] and subsequently in many countries in Europe, Asia and North Africa [6]. Severe outbreaks of WDV have been reported in Sweden [7], Czech Republic [8], Austria [6] and recently also in Poland [9].
The virus has monopartite single-stranded circular (ss)DNA genome of ~2.7 kb which encodes four proteins: the movement protein (MP), the coat protein (CP) and two others associated with replication (Rep and RepA). Large and small intergenic regions (LIR and SIR, respectively) contain regulatory elements for viral replication and transcription [3]. The recently published results of the phylogenetic studies confirmed division of the global WDV population into two clearly separated WDV-wheat and WDV-barley-specific groups, according to their host. The studies have shown that the crucial regions for this division were MP and LIR [10]. Moreover, six WDV strains (A-E) have been distinguished based on sequence similarity between virus isolates and phylogenetic relationships among them [10].
To date, molecular detection of WDV is based on polymerase chain reaction (PCR), immunocapture (IC-PCR), real-time PCR, PCR and rolling circle amplification (RCA) methods combined with restriction fragment length polymorphism (RFLP) [11–16]. The promising alternative for above mentioned techniques is loop-mediated isothermal amplification of nucleic acids which has been developed for the rapid and efficient detection of many DNA and RNA viruses [17, 18]. LAMP assay amplifies a target sequence with high sensitivity and specificity under isothermal conditions (63–65 °C) and exhibits sensitivity similar to or even higher than the conventional PCR. Taking into account all the advantages of the LAMP assay, our objective was to develop and evaluate LAMP for the detection of WDV isolates collected from different hosts.
Material and methods
The analyses were performed with DNA extracted from infected wheat, barley and triticale plants collected in 2012–2016 from different locations: Dłużec, Kobierzyce and Kondratowice in Dolny Śląsk as well as from Antoniny, Szelejewo and Strzelce in Wielkopolska region of Poland (Table 1). Total DNA was isolated using NucleoSpin® Plant II kit (Macherey–Nagel, Düren, Germany) according to manufacturer’s instructions. The DNA concentration and quality were estimated using a NanoDrop 2000 spectrophotometer (Nonodrop Technologies, Delaware, USA). In preliminary step, complete genome sequences of published, different WDV isolates: WDV-A (AJ783960), WDV-B (FJ620684), WDV-C (JQ647455), WDV-D (JN791096), WDV-E (AM040732) as well as two Polish isolates WDV-W (KM079154) and WDV-B (KM079155) were compared. Based on the result of multiple sequence alignments performed in ClustalW [19] CP region was chosen as a target for amplification in LAMP assay. The set of diagnostic primers (Table 2) were designed using LAMP Designer software (OptiGene, Horsham, UK). The specificity of LAMP primers was examined using 50 ng/µl of total DNA extracted from WDV-infected plants and from healthy wheat cv. Muszelka and barley cv. Bażant plants as the templates for reactions. LAMP assay was performed in total volume of 25 μl. The reaction mixture consisted of 15 μl Isothermal Mastermix ISO-001nd (Novazym, Poznan, Poland), 2 μl of 10 µM WDV FIP and WDV BIP, 1 µl of 10 µM WDV Loop-F and WDV Loop-B, 0.5 μl of 10 µM WDV F3 and WDV B3 primers, 1 µl of template DNA and 2 μl of sterile water. DNA samples from healthy plants and the reaction mix without a template were included as the negative controls. The tubes were incubated at 63 °C for 45 min in a thermoblock (Biometra, Göttingen, Germany). The LAMP products were analysed by electrophoretical separation in 1.5% agarose gel containing Midori Green DNA Stain (NIPPON Genetics Europe GmbH, Düren, Germany) as well as by direct visual inspection of color solution under UV light after the addition of 2 μl of SYBRGreen Dye (Thermo Fisher Scientific, Waltham, MA, USA). The reaction was also carried out using Isothermal Mastermix ISO-001 (Novazym) in a LightCycler 96 Instrument (Roche, Basel, Switzerland) and the results were estimated by analysis of amplification curves. The fluorescence data were obtained on the FAM channel (excitation at 470 nm, detection at 510 nm) in 30 min. The sensitivity of LAMP assay and conventional PCR were estimated and compared. For this purpose, 1 µl of tenfold serial dilutions (from 101 to 10–6) of total DNA of WDV-Strz (adjusted to 50 ng/µl) were used as template for reactions. LAMP assay was performed in conditions described above and PCR was done with Dream Taq Green Master Mix (Thermo Fisher Scientific). The primer pair WDVuniw-F (CGCACTCGGCTTTTCGTGAGTG) and WDVuniw-R (CGCCAGGCGTAGTCGGAGG) were designed by Primer3software [20] based on conservative motifs of fragments of LIR and MP. The potential cross-reactivity of the oligonucleotides as well as their specificity were verified by basic local alignment search tool (BLAST) from website of the National Centre for Biotechnology Information (NCBI) (https://www.ncbi.nim.nih.gov). The PCR was performed in a final volume of 25 µl containing: 1 µl of DNA template with 12.5 µl of 2 × Dream Taq Master mix (Thermo Fisher Scientific), 1 µl of Primer Mix (10 µmol/µl each) and sterile Milli-Q water. Amplification was performed as follows: 94 °C for 2 min, 35 cycles of 94 ºC for 30 s, 55 °C for 30 s, 72 ºC for 30 s and a final cycle of 72 °C for 7 min. The detection limit of compared techniques was determined by electrophoresis in 1.5% agarose gel.
Table 1.
Description of WDV isolates used in this study
| Isolate name | Specification | Geographical origin | Host | Collection date | Accession no |
|---|---|---|---|---|---|
| WDV-Knd1 | WDV-W | Kondratowice | Wheat | 05/2013 | KY781937 |
| WDV-Kob | WDV-W | Kobierzyce | Wheat | 06/2016 | KY781939 |
| WDV-Strz | WDV-W | Strzelce | Wheat | 05/2013 | MT460909 |
| WDV-Ant | WDV-W | Antoniny | Wheat | 04/2014 | KY781933 |
| WDV-Sz2 | WDV-B | Szelejewo | Wheat | 05/2012 | KY781944 |
| WDV-Dl | WDV-B | Dłużec | Barley | 05/2016 | KY781935 |
| WDV-Sz4 | WDV-W | Szelejewo | Triticale | 05/2012 | KY781946 |
Table 2.
Primers used in LAMP techniques
| Primer’s name | Primer sequence (5′-3′) |
|---|---|
| WDV F3 | GTGAAGAGGAAGTGGTGC |
| WDV B3 | CCACTGACACCACCTCTA |
| WDV FIP | CCGCTACGTAGTTGTAACGAGGGTGAACCTTGTCTCCGATG |
| WDV BIP | GCAAAGGTTTGCGGGTCACCCTTCTTAATGTCGCCTATCTT |
| WDV LoopF | CTTGGATCCGACCTTCTTCC |
| WDV LoopB | ACGGAGTGGATGAACACG |
Results and discussion
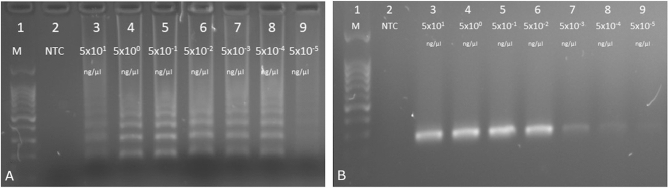
WDV population can be clearly divided into barley and wheat forms [3, 6, 10, 14]. The genomes of these two forms share 84% nucleotide sequence identity however the CP gene is one of the most stable coding regions [21]. The comparative analysis of this region revealed ca. 83% nucleotide sequence identity between different barley- and wheat- infecting WDV isolates whereas up to 98% within the particular group [14, 21]. Taking into account the genetic diversity of the WDV population, the primers for the LAMP assay were designed based on the alignment of CP sequences. LAMP technique developed here was capable of the detection of both WDV-B and WDV-W forms isolated from different hosts in less than hour. In contrast to previously published differentiating methods [11–15] designed set of primers allow for the detection of genetically diverse WDV isolates in single reaction. The amplified products visible as ladder-like DNA fragments were obtained only for tested WDV samples whereas no amplicons were produced for the samples of healthy barley and wheat plants or for the water controls (Fig. 1a). Green color was observed in UV light after adding the dye only to WDV infected samples and no color changes were seen for the negative controls (Fig. 1b). Monitoring of LAMP reaction in real-time revealed rapid and specific detection of WDV in tested samples. The amplification plots was already observed between 10 and 20 min and only for samples with DNA of WDV (Fig. 1c). The specific PCR product (235 bp in size) was detectable up to 5 pg/µl of DNA while LAMP amplicons could be seen even for 500 fg/µl of total DNA indicating that LAMP assay was 10 times more sensitive compared to the conventional PCR (Fig. 2).
Fig. 1.
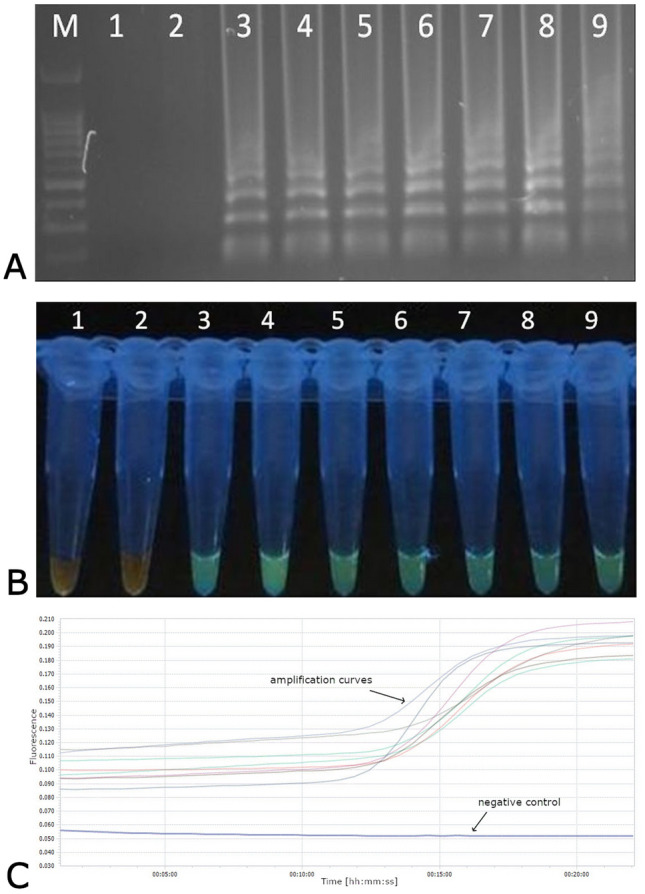
Detection of WDV isolates by LAMP. a, Analysis of LAMP products on agarose gels; lane M–100-bp DNA ladder (Novazym, Poznań, Poland), lane 1-negative control- sap of healthy barley, lane 2–sap of healthy wheat; lane 3- 9- WDV isolates b, Visual detection of LAMP products using SYBRGreen Dye (Thermo Fisher Scientific). Numbers correspond to the gel lanes in panel a. c) Real-time monitoring of LAMP assay of the WDV isolates in LightCycler 96 (Roche)
Fig. 2.
A comparison of sensitivity of LAMP and PCR methods for WDV detection. Detection limit of LAMP assay (a) and PCR technique (b). Lanes: 1–100-bp DNA ladder (Novazym); 2–NTC negative control (water); 3–9– correspond to serial tenfold dilutions of total DNA
Obtained results and sensitivity of virus detection correspond with those presented by Kuan et al. [22] for Squash leaf curl virus (SLCV) belonging to Begomovirus genus in Geminiviridae family or with that reported by Kill et al. [18] for three virus species of Curtovirus genus also classified in Geminiviridae family. In conclusion, the results confirmed that LAMP assay is a high specific and suitable technique, faster and more sensitive than standard PCR. Moreover, it does not require specialized equipment and the reaction can be carried out with a simple heater or in water bath. All the features mentioned above confirm that optimised in this study LAMP assay is useful diagnostic tool for rapid identification of WDV infection in cereals. Furthermore, according to our knowledge it is the first report presenting the use of the LAMP assay for WDV detection.
Author contributions
Author KT conducted the molecular analyses and wrote the manuscript. Author BH designed the set of diagnostic primers and revised the manuscript. The authors read and approved the final version of the manuscript.
Funding
Supported by the Polish Ministry of Science and Higher Education, statutory activity project: WIB01.
Data availability
The data of this study are available from the corresponding author upon request.
Compliance with ethical standards
Conflict of interest
Author A declares that she has no conflict of interest. Author B declares that she has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Vacke J (1962) Some new findings on wheat dwarf virus. In: Plant Virology, Proceedings of the 5th Conference of the Czechoslovak Plant Virologists, Publishing House of the Czechoslovak Academy of Sciences, Prague, Czechoslovakia, p 331-334
- 2.Ekzayez AM, Kumari SG, Ismail I. First report of wheat dwarf virus and its vector (Psammotettixprovincialis) affecting wheat and barley crops in Syria. Plant Dis. 2011;95:76. doi: 10.1094/PDIS-09-10-0628. [DOI] [PubMed] [Google Scholar]
- 3.Vacke J, Kvarnheden A, Lindbland M, Lindsten K. Wheat Dwarf. In: Lapierre H, Signoret PA, editors. Viruses and virus diseases of poaceae (Gramineae) Paris: INRA; 2004. pp. 590–593. [Google Scholar]
- 4.Lindblad M, Sandgren M, Sigvald R (1999) Epidemiology and control of wheat dwarf. In: Proccedings of VIIth International Plant Virus Epidemiology Symposium. Plant Virus Epidemiology Committee of the International Society of Plant Pathology, Aguadulce, Spain, p 11-16
- 5.Vacke J. Wheat dwarf virus disease. Biol Plant. 1961;3:228–233. doi: 10.1007/BF02933566. [DOI] [Google Scholar]
- 6.Schubert J, Habekuß A, Wu B, Thieme T, Wang X. Analysis of complete genomes of isolates of the Wheat dwarf virus from new geographical locations and descriptions of their defective forms. Virus Genes. 2014;48:133–139. doi: 10.1007/s11262-013-0989-9. [DOI] [PubMed] [Google Scholar]
- 7.Lindsten K, Lindsten B. Wheat dwarf–an old disease with new outbreaks in Sweden. J Plant Dis Protect. 1999;106:325–332. [Google Scholar]
- 8.Kroutil P, Markytánová J. The occurrence of dwarf viruses in cereal crops before the 2005 harvest. Rostlinolékař. 2005;6:9–10. [Google Scholar]
- 9.Trzmiel K. The occurrence of Wheat dwarf virus and Barley yellow dwarf virus species in Poland in the spring of 2019. J Plant Prot Res. 2020 doi: 10.24425/jppr.2020.133959. [DOI] [Google Scholar]
- 10.Wu B, Shang X, Shubert J, Habekuß A, Elena SF, Wang X. Global-scale computational analysis of genomic sequences reveals the recombination pattern and coevolution dynamics of cereal-infecting geminiviruses. Sci Rep. 2015;5:e8153. doi: 10.1038/srep08153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Commandeur U, Huth W. Differentiation of strain of wheat dwarf virus in infected wheat and barley plants by means of polymerase chain reaction. J Plant Dis Protect. 1999;106:550–552. [Google Scholar]
- 12.Kundu JK, Gadiou S, Červená G. Discrimination and genetic diversity of Wheat dwarf virus in the czech republic. Virus Genes. 2009;38:468–474. doi: 10.1007/s11262-009-0352-3. [DOI] [PubMed] [Google Scholar]
- 13.Trzmiel K, Klejdysz T. Detection of barley- and wheat-specific forms of Wheat dwarf virus in their vector Psammotettix alienus by duplex PCR assay. J Plant Prot Res. 2018;58:54–57. doi: 10.24425/119118. [DOI] [Google Scholar]
- 14.Trzmiel K. Detection and discrimination of barley- and wheat-specific forms of Wheat dwarf virus in Poland. Cereal Res Commun. 2018;46(2):300–309. doi: 10.1556/0806.46.2018.11. [DOI] [Google Scholar]
- 15.Gadiou S, Ripl J, Jaňourová B, Jarošová J, Kundu JK. Real-time PCR assay for the discrimination and quantifiacation of wheat and barley strains of Wheat dwarf virus. Virus Genes. 2012;44:349–355. doi: 10.1007/s11262-011-0699-0. [DOI] [PubMed] [Google Scholar]
- 16.Schubert J, Habekuß A, Kazmaier K, Jeske H. Surveying cereal-infecting geminiviruses in Germany-diagnostic and direct sequencing using rolling circle amplification. Virus Res. 2007;127:61–70. doi: 10.1016/j.virusres.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 17.Hasiów-Jaroszewska B, Borodynko N. Detection of pepino mosaic virus isolates from tomato by one-step reverse transcription loop-mediated isothermal amplification. Arch Virol. 2013;158:2153–2156. doi: 10.1007/s00705-013-1706-7. [DOI] [PubMed] [Google Scholar]
- 18.Kil EJ, Cho SH, Byun HS, Kim J, Hwang HS, Auh CK, Heo NY, Shin YG, Lee S. Differential identification of three species of Curtovirus using loop-mediated isothermal amplification. ActaVirol. 2014;58:160–166. doi: 10.4149/av_2014_02_160. [DOI] [PubMed] [Google Scholar]
- 19.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W In: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choise. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosen S, Skaletski HJ. Primer3 on the WWW for general use and for biologist programmers. In: Krawetz S, Misenes S, editors. Bioinformatics methods and protocols: methods in molecular biology. Totova/New Jersey: Humana Press; 2000. pp. 365–386. [Google Scholar]
- 21.Köklü G, Ramsell JNE, Kvarnheden A. The complete genome sequence for a Turkish isolate of Wheat dwarf virus (WDV) from barley confirms the presence of two distinct WDV strains. Virus Genes. 2007;34:359–366. doi: 10.1007/s11262-006-0029-0. [DOI] [PubMed] [Google Scholar]
- 22.Kuan CP, Wu MT, Lu YL, Huang HC. Rapid detection of squash leaf curl virus by loop-mediated isothermal amplification. J Virol Methods. 2010;169:61–65. doi: 10.1016/j.jviromet.2010.06.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data of this study are available from the corresponding author upon request.