Abstract
Samsoniella species have been found on lepidopteran larvae or pupae buried in soil or leaf litter. Three new species, Samsoniella hymenopterorum, S. coleopterorum and S. lepidopterorum, parasitic on hymenopteran larvae, coleopteran larvae and lepidopteran pupae, respectively, are reported. Morphological comparisons with extant species and DNA-based phylogenies from analysis of a multigene (ITS, RPB1, RPB2 and TEF) dataset supported the establishment of the new species. Unusually, all three new species have mononematous conidiophores. The new species are clearly distinct from other species in Samsoniella occurring in separate subclades.
Keywords: Isaria-like, morphology, nutritional preference, phylogeny
Introduction
The genus Isaria Pers. was introduced for entomogenous fungi with mononematous or synnematous conidiophores, usually consisting of several verticillate branches, each bearing a dense whorl of phialides characters. The phialides consist of a cylindrical or swollen basal portion, terminating in a thin, often long neck and produce divergent conidial chains (Samson 1974). However, entomogenous species, morphologically similar to Isaria, can be found distributed throughout the Hypocreales (Luangsa-ard et al. 2004).
Kepler et al. (2017) proposed the rejection of Isaria in favour of Cordyceps, owing to the confusion surrounding the application of Isaria and combined 11 species into Cordyceps. Mongkolsamrit et al. (2018) described some Isaria-like species and proposed the new genus Samsoniella Mongkols., Noisrip., Thanakitp., Spatafora & Luangsa-ard. The typical characteristics of Samsoniella are oval to fusiform conidia and bright red-orange teleomorphic stromata and anamorphic synnemata. Samsoniella species inhabit lepidopteran larvae and pupae in leaf litter or soil. Currently, Samsoniella consists of three species, S. alboaurantia (G. Sm.) Mongkols., Noisrip., Thanakitp., Spatafora & Luangsa-ard, S. aurantia Mongkols., Noisrip., Thanakitp., Spatafora & Luangsa-ard and S. inthanonensis Mongkols., Noisrip., Thanakitp., Spatafora & Luangsa-ard.
Three infected insect specimens were collected during a survey of entomogenous fungi in south-western China. Morphological and molecular phylogenetic analyses suggested that these isolates represented three new species, which are described here as Samsoniella hymenopterorum sp. nov., S. coleopterorum sp. nov. and S. lepidopterorum sp. nov.
Materials and methods
Specimen collection and identification
Three fungus-infected insect specimens were collected from Xishui County (28°29'56.70"N, 106°24'31.04"E) (A1950 and A1952) and Dali, Rongjiang County (26°01'58.70"N, 108°24'48.06"E) (DL1007), Guizhou Province, on 20 July and 1 October 2018, respectively. Isolation of the fungi was done as described by Chen et al. (2019). The surface of the specimens was rinsed with sterile water, followed by surface sterilisation with 75% ethanol for 3–5 sec. A part of the insect body was cut off and inoculated with haemocoel on potato dextrose agar (PDA) and PDA, to which 1% w/v peptone (PDAP) had been added. Fungal colonies emerging from specimens were isolated and cultured at 22 °C for 14 d under 12 h light/12 h dark conditions following protocols described by Zou et al. (2010). Accordingly, strains A19501, A19502, A19521, A19522, DL10071 and DL10072 were obtained. The specimens and the isolated strains were deposited in the Institute of Fungus Resources, Guizhou University (formally Herbarium of Guizhou Agricultural College; code, GZAC), Guiyang City, Guizhou, China.
Macroscopic and microscopic morphological characteristics of the fungi were examined and growth rates determined from PDA cultures incubated at 25 °C for 14 d. Hyphae and conidiogenous structures were mounted in lactophenol cotton blue or 20% lactate solution and observed with an optical microscope (OM, DM4 B, Leica, Germany).
DNA extraction, PCR amplification and nucleotide sequencing
DNA extraction was carried out in accordance with Liang et al. (2011). The extracted DNA was stored at −20 °C. Translation elongation factor 1 alpha (TEF) and RNA polymerase II largest subunit 2 (RPB2) genes were amplified using 983F/2218R and RPB2-5F/RPB2-7Cr primers, according to van den Brink et al. (2012). The RNA polymerase II largest subunit 1 (RPB1) gene was amplified with the primer pair CRPB1 and RPB1-Cr (Castlebury et al. 2004). The internal transcribed spacer (ITS) region was amplified by PCR using ITS4/ITS5, which was described by White et al. (1990). PCR products were purified using the UNIQ-10 column PCR products purification kit (no. SK1141; Sangon Biotech (Shanghai) Co., Shanghai, China) in accordance with the manufacturer’s protocol and sequenced at Sangon Biotech (Shanghai) Co. The resulting sequences were submitted to GenBank.
Sequence alignment and phylogenetic analyses
The DNA sequences, generated in this study, were assembled and edited using Lasergene software (version 6.0 DNASTAR). Generated ITS, RPB1, RPB2 and TEF sequences were aligned with those published by Mongkolsamrit et al. (2018) and others selected on the basis of BLAST algorithm-based searches in GenBank (Table 1). Purpureocillium lilacinum (Thom) Luangsa-ard, Houbraken, Hywel-Jones & Samson (isolates CBS 284.36 and CBS 431.87) and Beauveria bassiana (Bals.-Criv.) Vuill. (ARSEF 1564) were chosen as outgroup taxa for the analysis of Samsoniella in Cordycipitaceae and Samsoniella species and closely-related species, respectively. Multiple datasets of ITS, RPB1, RPB2 and TEF were aligned using MAFFT v7.037b (Katoh and Standley 2013) and alignments were edited with MEGA6 (Tamura et al. 2013). Sequences were concatenated with SequenceMatrix v.1.7.8 (Vaidya et al. 2011). The partition homogeneity test in PAUP4.0b10 (Swofford 2002) was undertaken by using the command ‘hompart’.
Table 1.
Taxa included in the phylogenetic analyses.
Maximum Likelihood (ML) analyses were constructed with RAxMLGUI (Silvestro and Michalak 2012). The GTRGAMMA model was used for all partitions, in accordance with recommendations in the RAxML manual against the use of invariant sites. For Bayesian Inference (BI), a Markov Chain Monte Carlo (MCMC) algorithm was used to generate phylogenetic trees with Bayesian probabilities using MrBayes v.3.2 (Ronquist et al. 2012) for the combined sequence datasets. The selection of the best-fit nucleotide substitution model for each locus was calculated by the Akaike Information Criterion (AIC) with jModelTest 2 (Darriba et al. 2012). The TIM+I+G model was selected for the concatenated ITS+RPB1+RPB2+TEF sequences. The Bayesian analysis resulted in 20,001 trees after 10,000,000 generations. The first 4,000 trees, representing the burn-in phase of the analyses, were discarded, while the remaining 16,001 trees were used for calculating posterior probabilities in the majority rule consensus tree. After the analysis was finished, each run was examined using the programme Tracer v1.5 (Drummond and Rambaut 2007) to determine burn-in and confirm that both runs had converged. The final alignment is available from TreeBASE under submission ID: 24710 (http://www.treebase.org).
Results
Phylogenetic analyses
The phylogenetic tree of Samsoniella in Cordycipitaceae (Fig. 1) and Samsoniella species and closely related species (Fig. 2) were generated from the ML and BI analysis, based on a combined data set of ITS, RPB1, RPB2 and TEF sequence data. Statistical support (≥ 50%/0.5) is shown at the nodes for ML bootstrap support/BI posterior probabilities (Figs 1, 2). The strain numbers are noted after each species’ name. The concatenated sequences of analysis 1 and analysis 2 included 67 and 17 taxa, and consisted of 2,152 (ITS: 528, RPB1: 488, RPB2: 442 and TEF: 694) and 2,194 (ITS: 477, RPB1: 565, RPB2: 473 and TEF: 679) characters with gaps, respectively.
Figure 1.
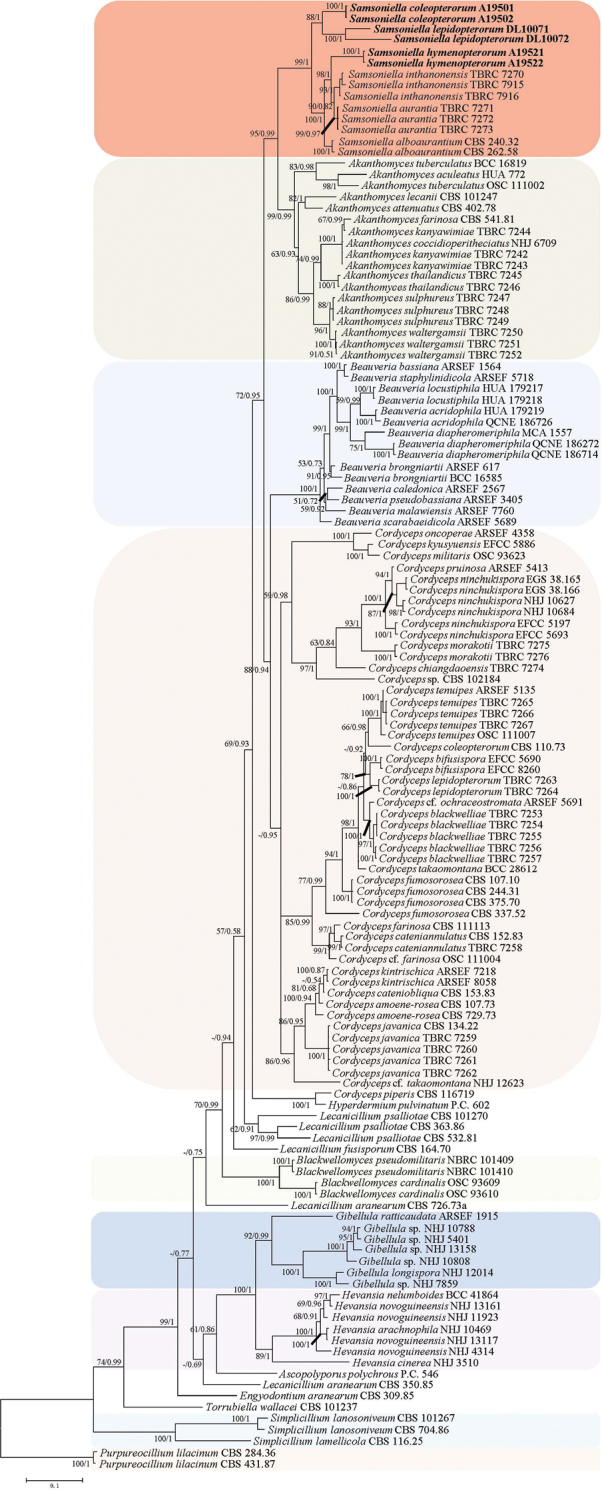
Phylogenetic relationships of the genus Samsoniella in Cordycipitaceae, based on multigene dataset (ITS, RPB1, RPB2 and TEF). Statistical support values (≥ 50%/0.5) are shown at the nodes for ML bootstrap support/BI posterior probabilities.
Figure 2.
Phylogenetic relationships between the genus Samsoniella and closely-related species, based on multigene dataset (ITS, RPB1, RPB2 and TEF). Statistical support values (≥ 50%/0.5) are shown at the nodes for ML bootstrap support/BI posterior probabilities.
Analysis 1: Samsoniella in Cordycipitaceae. The RAxML analysis of the combined dataset (ITS+RPB1+RPB2+TEF) yielded a best scoring tree (Fig. 1) with a final ML optimisation likelihood value of –28,809.222105. Parameters for the GTR model of the concatenated dataset was as follows: estimated base frequencies; A = 0.234094, C = 0.301291, G = 0.260521, T = 0.204093; substitution rates AC = 1.111784, AG = 3.130020, AT = 0.930972, CG = 0.886915, CT = 6.300092, GT = 1.000000; gamma distribution shape parameter α = 0.390179. In the phylogenetic tree (Fig. 1), Samsoniella species were clustered in a clade and resolved into two obvious clades. Samsoniella species have a close relationship with Akanthomyces species.
Analysis 2: Samsoniella species and closely-related species. The RAxML analysis of the combined dataset (ITS+RPB1+RPB2+TEF) yielded a best scoring tree (Fig. 2) with a final ML optimisation likelihood value of –9,722.503130. Parameters for the GTR model of the concatenated data set were as follows: estimated base frequencies; A = 0.233473, C = 0.298686, G = 0.261629, T = 0.206212; substitution rates AC = 1.250081, AG = 2.534760, AT = 0.891128, CG = 0.827805, CT = 5.916085, GT = 1.000000; gamma distribution shape parameter α = 0.674468. In the phylogenetic tree (Fig. 2), Samsoniella species were clustered in a clade and easily distinguished with Akanthomyces species. S. coleopterorum and S. lepidopterorum clustered in a clade (Fig. 2) and formed two independent branches. S. hymenopterorum was phylogenetically close to S. inthanonensis and S. aurantia.
Taxonomy
Samsoniella coleopterorum
W.H. Chen, Y.F. Han & Z.Q. Liang sp. nov.
629653D9-9471-5715-9DB0-8FBB3FDE9DD7
831735
Figure 3.
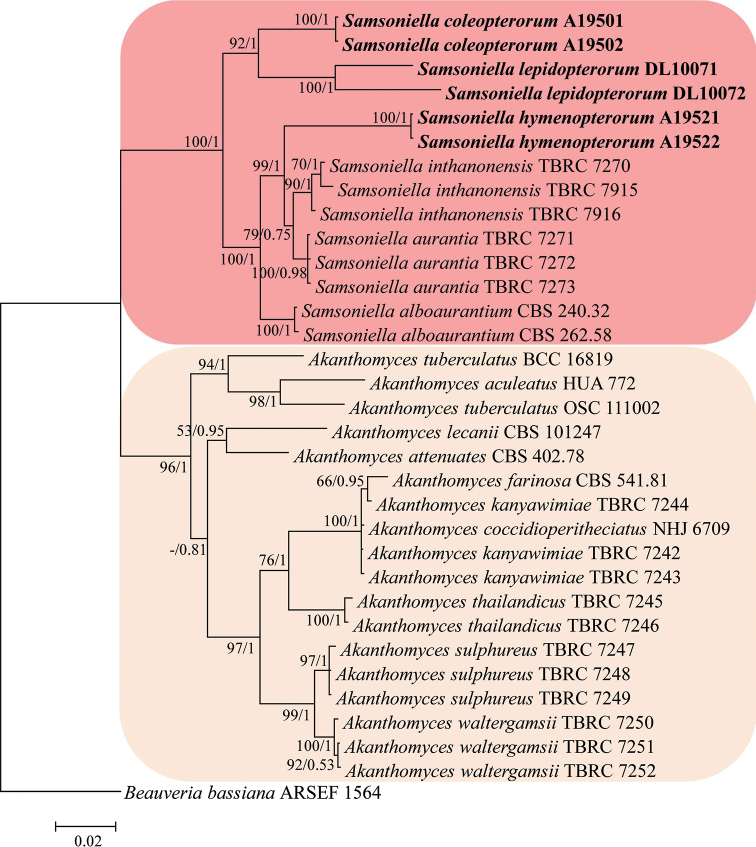
Samsoniella coleopterorumA infected insect (Coleoptera) B, C top (B) and underside (C) of a colony cultured on PDA medium at 14 d D, E, F, G, I phialides and conidia in chains J conidiophore and phialides H conidia. Scale bars: 10 mm (B, C); 10 μm (D–J).
Diagnosis.
Differs from Samsoniella aurantia by having smaller conidia and snout beetle host in the family Curculionidae. Differs from S. lepidopterorum by having cylindrical to ellipsoidal phialides, smaller fusiform to ellipsoidal conidia and a different host.
Type.
China, Guizhou Province, Xishui County (28°29'56.70"N, 106°24'31.04"E), July 2018, Jiandong Liang, holotype GZAC A1950, ex-type culture GZAC A19501. Sequences from isolated strain A19501 have been deposited in GenBank with accession numbers: ITS = MT626376, RPB1 = MT642600, RPB2 = MN101585 and TEF = MN101586.
Description.
Colonies on PDA, 3.6–4.0 cm diam. in 14 d at 25 °C, white, consisting of a basal felt and cottony, floccose hyphal overgrowth, reverse yellowish. Prostrate hyphae smooth, septate, hyaline, 1.1–1.8 μm diam. Erect conidiophores usually arising from aerial hyphae, Isaria-like with phialides in whorls of two to four. Phialides 5.4–9.7 × 1.2–1.8 μm, with a cylindrical to ellipsoidal basal portion, tapering into a short distinct neck. Conidia in chains, hyaline, fusiform, ellipsoidal or subglobose, one-celled, 1.7–2.5 × 1.2–1.8 μm. Chlamydospores and synnemata not observed. Size and shape of phialides and conidia similar in culture and on natural substratum. Sexual state not observed.
Host.
Snout beetle, family Curculionidae.
Distribution.
Xishui County, Guizhou Province, China.
Etymology.
Referring to its insect host, order Coleoptera.
Remarks.
Samsoniella coleopterorum was easily identified as belonging to Samsoniella based on the phylogenetic analyses (Fig. 1). Comparing with the typical characteristics of three species (Table 2), S. coleopterorum has a close relationship with S. aurantia by having cylindrical to ellipsoidal phialides and similar in size. However, it differs from S. aurantia by having shorter conidia and snout beetle host in the family Curculionidae. Based on the combined dataset of ITS, RPB1, RPB2 and TEF sequences, S. coleopterorum has a close relationship with S. lepidopterorum (Fig. 2). However, S. coleopterorum has cylindrical to ellipsoidal phialides, smaller fusiform to ellipsoidal conidia and a different host.
Table 2.
Morphological comparison of three new species with other Samsoniella species.
| Species | Morphological characteristics | Reference | ||
|---|---|---|---|---|
| Phialide (μm) | Conidia (μm) | Hosts/substrates | ||
| Samsoniella alboaurantium | 5–8 × 2 | ovate to lemon-shaped | soil, lepidopterous pupa | Smith 1957 |
| 2.3–2.5(–3) × 1.5–1.8 | ||||
| S. aurantia | cylindrical to ellipsoidal | fusiform | lepidopterous larvae | Mongkolsamrit et al. 2018 |
| (5–)5.5–8.5(–13) × 2–3 | (2–)2.5–3.5(–4) × (1–)1.5(–2) | |||
| S. inthanonensis | cylindrical | short fusiform | lepidopterous larvae | Mongkolsamrit et al. 2018 |
| (4–)6.5–10(–12) × (1–)1.5–2(–3) | (2–)3(–3.5) × 1.5–2 | |||
| S. coleopterorum | cylindrical to ellipsoidal | fusiform, ellipsoidal or subglobose | snout beetle | this study |
| 5.4–9.7 × 1.2–1.8 | 1.7–2.5 × 1.2–1.8 | |||
| S. hymenopterorum | cylindrical | fusiform to ovoid | bee | this study |
| 6.5–10.6 × 1.2–2.0 | 1.9–2.5 × 1.5–2.1 | |||
| S. lepidopterorum | ellipsoidal | fusiform to subglobose | lepidopterous pupa | this study |
| 5.2–8.5(–13.1) × 1.1–1.7 | 2.0–2.5 × 1.2–2.0 | |||
Samsoniella hymenopterorum
W.H. Chen, Y.F. Han & Z.Q. Liang sp. nov.
F09AE22C-FF9F-59A3-8F34-59DFC6F23B1D
831736
Figure 4.
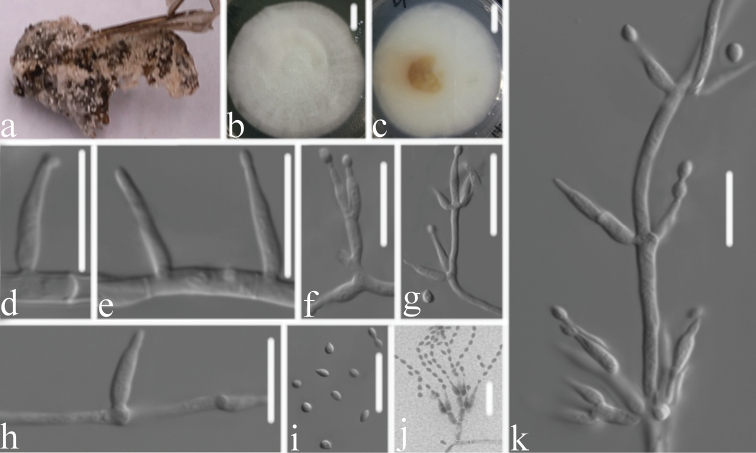
Samsoniella hymenopterorumA infected insect (Hymenoptera) B, C top (B) and underside (C) of a colony cultured on PDA medium at 14 d D–H, J phialides and conidia in chains K conidiophore and phialides I conidia. Scale bars: 10 mm (B, C); 10 μm (D–K).
Diagnosis.
Differs from Samsoniella inthanonensis and S. aurantia by having smaller, fusiform to ovoid conidia and a host in the family Vespidae.
Type.
China, Guizhou Province, Xishui County, at 28°29'56.70"N, 106°24'31.04"E, July 2018, Jiandong Liang, holotype GZAC A1952, ex-type culture GZAC A19522. Sequences from isolated strain A19522 have been deposited in GenBank with accession numbers: ITS = MN128224, RPB1 = MT642603, RPB2 = MT642604 and TEF = MN101588.
Description.
Colonies on PDA, 6.2–6.4 cm diam. in 14 d at 25 °C, white, consisting of a basal felt and cottony, floccose hyphal overgrowth, reverse yellowish. Prostrate hyphae smooth, septate, hyaline, 1.1–1.6 μm diam. Erect conidiophores usually arising from aerial hyphae, Isaria-like with phialides in whorls of three to four. Phialides 6.5–10.6 × 1.2–2.0 μm, with a cylindrical basal portion, tapering to a distinct neck. Conidia in chains, hyaline, fusiform to ovoid, 1-celled, 1.9–2.5 × 1.5–2.1 μm. Chlamydospores and synnemata not observed. Size and shape of phialides and conidia similar in culture and on natural substratum. Sexual state not observed.
Host.
Bee, family Vespidae.
Distribution.
Xishui County, Guizhou Province, China.
Etymology.
Referring to its insect host, order Hymenoptera.
Remarks.
Samsoniella hymenopterorum was identified as belonging to Samsoniella, based on the phylogenetic analyses (Fig. 1). Comparing with the typical characteristics of the three species (Table 2), S. hymenopterorum has a close relationship with S. inthanonensis by a having cylindrical basal in phialide and similar in size. However, it is distinguished from S. inthanonensis by having smaller, fusiform to ovoid conidia and a host in the family Vespidae. Based on combined dataset of ITS, RPB1, RPB2 and TEF sequences, S. hymenopterorum is phylogenetically close to S. aurantia and S. inthanonensis (Fig. 2). However, S. hymenopterorum has smaller fusiform to ovoid conidia and a different host.
Samsoniella lepidopterorum
W.H. Chen, Y.F. Han & Z.Q. Liang sp. nov.
09A7269B-030B-5978-86BA-22D4B6378101
831737
Figure 5.
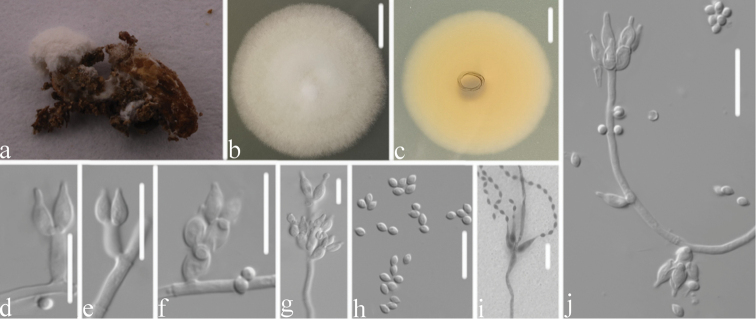
Samsoniella lepidopterorumA infected insect pupa (Lepidoptera) B, C top (B) and underside (C) of a colony cultured on PDA medium at 14 d D–G, I phialides and conidia in chains H conidia J conidiophore and phialides. Scale bars: 10 mm (B, C); 10 μm (D–J).
Diagnosis.
Differs from Samsoniella coleopterorum by having larger, ellipsoidal phialide conidia and a host in the order Lepidoptera.
Type.
China, Guizhou Province, Rongjiang County (26°01'56.13"N, 108°24'48.06"E), October 2018, Wanhao Chen, holotype GZAC DL1007 = RJ1807, ex-type culture GZAC DL10071 = RJ18071. Sequences from isolated strain DL10071 have been deposited in GenBank with accession numbers: ITS = MN128076, RPB1 = MN101592, RPB2 = MN101593 and TEF = MN101594.
Description.
Colonies on PDA, 3.7–3.8 cm diam. in 14 d at 25 °C, white, consisting of a basal felt and cottony, floccose hyphal overgrowth, reverse yellowish. Prostrate hyphae smooth, septate, hyaline, 1.1–2.2 μm diam. Erect conidiophores usually arising from aerial hyphae, Isaria-like with phialides in whorls of two to four. Phialides 5.2–8.5 (–13.1) × 1.1–1.7 μm, with an ellipsoidal basal portion, tapering into a distinct neck. Conidia in chains, hyaline, fusiform to subglobose, 1-celled, 2.0–2.5 × 1.2–2.0 μm. Chlamydospores and synnemata not observed. Size and shape of phialides and conidia similar in culture and on natural substratum. Sexual state not observed.
Host.
Pupa, order Lepidoptera
Distribution.
Rongjiang County, Guizhou Province, China
Etymology.
Referring to its insect host, order Lepidoptera
Remarks.
Samsoniella lepidopterorum was easily identified as belonging to Samsoniella, based on the phylogenetic analyses (Fig. 1). Based on the combined dataset of ITS, RPB1, RPB2 and TEF sequences (Fig. 2) and the typical characteristics of Samsoniella species (Table 2), S. lepidopterorum has a close relationship with S. coleopterorum. However, S. lepidopterorum has larger, ellipsoidal phialide conidia and its pupa host is in the order Lepidoptera.
Discussion
Phylogenetic analyses, based on the combined datasets of (ITS+RPB1+RPB2+TEF), suggest that the three new species are members of the Cordycipitaceae and belong to the genus Samsoniella (Fig. 1). Mongkolsamrit et al. (2018) noted that the typical characteristics of Samsoniella were oval to fusiform conidia, bright red-orange stromata of the sexual morphs and synnemata of the asexual morphs. The phialides in this genus range from cylindrical to possessing a swollen basal portion. S. coleopterorum, S. hymenopterorum and S. lepidopterorum all have cylindrical phialides and fusiform conidia. However, the three new species had mononematous conidiophores rather than synnemata. Synnematous entomopathogenic fungi (such as Gibellula spp.) can be found on abaxial leaf surfaces of shrubbery, forest floors and shallow soil layers (Hywel-Jones 1996). As air flow under the forest canopy is slow and humid, the dispersal of conidia through airflow diffusion may be difficult. Therefore, these entomopathogenic fungi may employ a particular strategy, such as producing synnemata and sticky conidia, to accommodate various arthropod activities and facilitate conidium spread (Abbott 2002). The three new species were located in the more open portion of the forest and this may favour the dispersal of dry conidia. Thus, we could speculate that the mononematous conidiophores of the three new species may be the result of a convergent evolution to adapt to the ecological environment.
The evolutionary dynamics of fungi and their hosts are usually described either by co-evolution or by host shifts. Shifts often occur to new hosts that are evolutionarily distant, but which occupy a common ecological niche (Vega et al. 2009). Nutrient requirements often determine whether host shifts occur (Vega et al. 2009). Relationships between insects and fungi have been described as biotrophy, necrotrophy and hemibiotrophy, inter alia. The common ancestor of Hypocreaceae and Clavicipitaceae corresponds to a departure from plant-based nutrition to one that specialises on animals and fungi (Spatafora et al. 2007). Prediction of the characteristics and evolutionary placement of any given member should be based on the correlation between molecular-phylogenetic genealogy and nutritional preferences (Spatafora et al. 2007; Vega et al. 2009). Species of Samsoniella were originally found on lepidopteran larvae or pupae buried in soil or leaf litter (Mongkolsamrit et al. 2018). Mongkolsamrit et al. (2018) also reported that the true range of host affiliations of Samsoniella in nature may not be currently represented. Here, we report Samsoniella spp. from coleopteran, hymenopteran larvae and lepidopteran pupae. The presence of different hosts indicates that the nutrient requirements of Samsoniella spp. can change with the environment (Spatafora et al. 2007).
In the present study, a four loci phylogenetic analysis showed that S. coleopterorum, S. lepidopterorum and S. hymenopterorum clustered in separate subclades from other Samsoniella species. They represent new taxa, based on morphological characteristics, nutritional preferences and phylogenetic analyses.
Supplementary Material
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 31860002), High-level Innovative Talents Training Object in Guizhou Province (No. Qiankehepingtairencai [2020]6005), Science and Technology Foundation of Guizhou Province (No. Qiankehejichu [2020]1Y060), National Survey of Traditional Chinese Medicine Resources (No. Caishe [2017]66, 216) and Engineering Research Center of General Higher Education in Guizhou Province (Qianjiaohe (2015) 337). We also thank Lesley Benyon, PhD, from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Citation
Chen W-H, Han Y-F, Liang J-D, Tian W-Y, Liang Z-Q (2020) Morphological and phylogenetic characterisations reveal three new species of Samsoniella (Cordycipitaceae, Hypocreales) from Guizhou, China. MycoKeys 74: 1–15. https://doi.org/10.3897/mycokeys.74.56655
Funding Statement
This work was supported by the National Natural Science Foundation of China (Grant No. 31860002), High-level Innovative Talents Training Object in Guizhou Province (No. Qiankehepingtairencai [2020]6005), Science and Technology Foundation of Guizhou Province (No. Qiankehejichu [2020]1Y060), National Survey of Traditional Chinese Medicine Resources (No. Caishe [2017]66, 216) and Engineering Research Center of General Higher Education in Guizhou Province (Qianjiaohe(2015)337).
References
- Abbott SP. (2002) Insects and other arthropods as agents of vector-dispersal in fungi. http://www.thermapure.com/pdf/AbbottInsectdispersal-2.pdf
- Castlebury LA, Rossman AY, Sung GH, Hyten AS, Spatafora JW. (2004) Multigene phylogeny reveals new lineage for Stachybotrys chartarum, the indoor air fungus. Mycological Research 108: 864–872. 10.1017/S0953756204000607 [DOI] [PubMed] [Google Scholar]
- Chen WH, Liu C, Han YF, Liang JD, Tian WY, Liang ZQ. (2019) Three novel insect-associated species of Simplicillium (Cordycipitaceae, Hypocreales) from Southwest China. MycoKeys 58: 83–102. 10.3897/mycokeys.58.37176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. (2012) jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9(8): 772–772. 10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A, Rambaut A. (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology 7: e214. 10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed]
- Hywel-Jones N. (1996) Akanthomyces on spiders in Thailand. Mycological Research 9: 1065–1070. 10.1016/S0953-7562(96)80214-0 [DOI] [Google Scholar]
- Katoh K, Standley DM. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30(4): 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepler RM, Luangsa-ard JJ, Hywel-Jones NL, Quandt CA, Sung GH, Rehner SA, Aime MC, Henkel TW, Sanjuan T, Zare R, Chen M, Li Z, Rossman AY, Spatafora JW, Shrestha B. (2017) A phylogenetically-based nomenclature for Cordycipitaceae (Hypocreales). IMA Fungus 8: 335–353. 10.5598/imafungus.2017.08.02.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang JD, Han YF, Zhang JW, Du W, Liang ZQ, Li ZZ. (2011) Optimal culture conditions for keratinase production by a novel thermophilic Myceliophthora thermophila strain GZUIFR-H49-1. Journal of Applied Microbiology 110: 871–880. 10.1111/j.1365-2672.2011.04949.x [DOI] [PubMed] [Google Scholar]
- Luangsa-ard JJ, Hywel-Jones NL, Samson RA. (2004) The order level polyphyletic nature of Paecilomyces sensu lato as revealed through 18S-generated rRNA phylogeny. Mycologia 96: 773–780. 10.1080/15572536.2005.11832925 [DOI] [PubMed] [Google Scholar]
- Mongkolsamrit S, Noisripoom W, Thanakitpipattana D, Wutikhun T, Spatafora JW, Luangsa-ard J. (2018) Disentangling cryptic species with Isaria-like morphs in Cordycipitaceae. Mycologia 110: 230–257. 10.1080/00275514.2018.1446651 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RA. (1974) Paecilomyces and some allied hyphomycetes. Studies in Mycology 6: 1–119. [Google Scholar]
- Silvestro D, Michalak I. (2012) raxmlGUI: a graphical front-end for RAxML. Organisms Diversity & Evolution 12(4): 335–337. 10.1007/s13127-011-0056-0 [DOI] [Google Scholar]
- Smith G. (1957) Some new and interesting species of micro-fungi. Transactions of the British Mycological Society 40(4): 481–488. 10.1016/S0007-1536(57)80054-0 [DOI] [Google Scholar]
- Spatafora JW, Sung GH, Sung JM, Hywel-Jones NL, White JF. (2007) Phylogenetic evidence for an animal pathogen origin of ergot and the grass endophytes. Molecular Ecology 16: 1701–1711. 10.1111/j.1365-294X.2007.03225.x [DOI] [PubMed] [Google Scholar]
- Swofford DL. (2002) PAUP* 4.0b10: phylogenetic analysis using parsimony (*and other methods). Sunderland, MA, Sinauer.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidya G, Lohman DJ, Meier R. (2011) SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27(2): 171–180. 10.1111/j.1096-0031.2010.00329.x [DOI] [PubMed] [Google Scholar]
- van den Brink J, Samson RA, Hagen F, Boekhout T, de Vries RP. (2012) Phylogeny of the industrial relevant, thermophilic genera Myceliophthora and Corynascus. Fungal Diversity 52: 197–207. 10.1007/s13225-011-0107-z [DOI] [Google Scholar]
- Vega FE, Goettel MS, Blackwell M, Chandler D, Jackson MA, Keller KM, Maniania KN, Monzón A, Ownley BH, Pell JK, Rangel DEN, Roy HE. (2009) Fungal entomopathogens: new insights on their ecology. Fungal Ecology 2: 149–159. 10.1016/j.funeco.2009.05.001 [DOI] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (Eds.) PCR protocols: a guide to methods and applications.Academic Press, New York, 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI]
- Zou X, Liu AY, Liang ZQ, Han YF, Yang M. (2010) Hirsutella liboensis, a new entomopathogenic species affecting Cossidae (Lepidoptera) in China. Mycotaxon 111(1): 39–44. 10.5248/111.39 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.