Abstract
The Chaetosphaeriaceae are a diverse group of pigmented, predominantly phialidic hyphomycetes comprised of several holomorphic genera including Chaetosphaeria, the most prominent genus of the family. Although the morphology of the teleomorphs of the majority of Chaetosphaeria is rather uniform, their associated anamorphs primarily exhibit the variability and evolutionary change observed in the genus. An exception from the morphological monotony among Chaetosphaeria species is a group characterised by scolecosporous, hyaline to light pink, multiseptate, asymmetrical ascospores and a unique three-layered ascomatal wall. Paragaeumannomyces sphaerocellularis, the type species of the genus, exhibits these morphological traits and is compared with similar Chaetosphaeria with craspedodidymum- and chloridium-like synanamorphs. Morphological comparison and phylogenetic analyses of the combined ITS-28S sequences of 35 isolates and vouchers with these characteristics revealed a strongly-supported, morphologically well-delimited clade in the Chaetosphaeriaceae containing 16 species. The generic name Paragaeumannomyces is applied to this monophyletic clade; eight new combinations and five new species, i.e. P. abietinussp. nov., P. eleganssp. nov., P. granulatussp. nov., P. sabinianussp. nov. and P. smokiensissp. nov., are proposed. A key to Paragaeumannomyces is provided. Using morphology, cultivation studies and phylogenetic analyses of ITS and 28S rDNA, two additional new species from freshwater and terrestrial habitats, Codinaea paniculatasp. nov. and Striatosphaeria castaneasp. nov., are described in the family. A codinaea-like anamorph of S. castanea forms conidia with setulae at each end in axenic culture; this feature expands the known morphology of Striatosphaeria. A chaetosphaeria-like teleomorph is experimentally linked to Dendrophoma cytisporoides, a sporodochial hyphomycete and type species of Dendrophoma, for the first time.
Keywords: molecular phylogeny, phialidic conidiogenesis, scolecosporous, systematics, wood-inhabiting fungi, 15 new taxa
Introduction
The family Chaetosphaeriaceae (Réblová et al. 1999) is a speciose, diverse group of pigmented, predominantly phialidic fungi some of which possess known teleomorphs (sexual and asexual morphs, hereafter teleomorph and anamorph respectively). Members of the family have a world-wide geographical distribution. They are essential components of biodiversity and play a role in decomposition of woody and herbaceous material and leaf litter, occur in soil, and some exhibit an endophytic lifestyle and have been isolated from living herbs and trees (e.g. Gams and Holubová-Jechová 1976; Hughes and Kendrick 1968; Réblová and Gams 1999; Réblová and Seifert 2003; Réblová 2004; Fernández and Huhndorf 2005; Huhndorf and Fernández 2005; Crous et al. 2012; Hashimoto et al. 2015; Yang et al. 2018; Lin et al. 2019; Luo et al. 2019).
Sexually reproducing fungi encompassed in the Chaetosphaeriaceae are perithecial ascomycetes that share several morphological traits such as similar anatomy of the brittle, melanised ascomatal wall, persistent paraphyses, unitunicate, thin-walled asci with a refractive, non-amyloid apical annulus, transversely septate ascospores that germinate by germ tubes and phialidic conidiogenesis. Several species produce both ascospores and conidia, ascomata are often associated with conspicuous conidiophores arranged in the juxtaposition. Most representatives of the family reproduce only asexually and are known as “anamorphic holomorphs” (Seifert et al. 2011). They either permanently lost the ability to sexually reproduce and do not develop the teleomorph, or the latter remains to be discovered.
Most of the sexually reproducing fungi in the family were classified in Chaetosphaeria (Tulasne & Tulasne, 1863), a prominent genus of the family. Chloridium botryoideum has long been known to be a part of the life cycle of Ch. innumera, the generic type (Tulasne and Tulasne 1863; Gams and Holubová-Jechová 1976). Using ITS and 28S DNA sequence data, Ch. innumera was resolved as unrelated to other Chaetosphaeria and chaetosphaeria-like species associated with morphologically different anamorphs (Réblová and Winka 2000; Fernández et al. 2006; Lin et al. 2019). Following the “one fungus, one name” concept (Hawksworth 2011, 2012; Hawksworth et al. 2011), some of the former Chaetosphaeria linked with different anamorphs now belong in the respective anamorphic genera based on priority, for example, Catenularia (Berkeley and Broome 1871; Hughes 1965a; Holubová-Jechová 1982), Cacumisporium (Réblová and Gams 1999), Chloridium (Gams and Holubová-Jechová 1976; Réblová et al. 2016), Exserticlava (Hino 1961; Matsushima 1985; Réblová and Seifert 2003; Fernández and Huhndorf 2005), Menispora (Booth 1957, 1958; Holubová-Jechová 1973; Réblová and Seifert 2008), Sporoschisma (Müller et al. 1969, Réblová et al. 2016), Tainosphaeria (Fernández and Huhndorf 2005) and Zanclospora (Hughes and Kendrick 1965b). Other Chaetosphaeria that form natural units, characterised primarily by the morphological traits of their anamorphs, will form the basis of generic classification in the family and, thus, need to be re-examined based on phylogenetic studies.
The majority of species accommodated in Chaetosphaeria possess ellipsoidal, fusiform to cylindrical-fusiform, 1–5-septate, hyaline, symmetrical ascospores with their length generally ranging from 6 to 40 μm. Ascomata are brown to black, papillate, often glossy with a two-layered ascomatal wall; the outer layer consisting of several rows of brick-like cells with dark brown, opaque walls. The transfer of a scolecosporous Lasiosphaeria raciborskii (Carroll and Munk 1964) with a three-layered ascomatal wall to Chaetosphaeria by Miller and Huhndorf (2004) expanded the concept of the genus. Huhndorf and Fernández (2005) introduced another four morphologically similar species based on ITS sequence data, i.e. Ch. ellisii (= Ch. longispora), Ch. lapaziana, Ch. panamensis and Ch. rubicunda, characterised by unique ascomatal wall anatomy, multiseptate scolecosporous ascospores and occurrence on decaying wood. Their ascomatal wall is composed of three layers. The typical chaetosphaeriaceous outer layer is present as the middle layer, while the outer layer consists of thin-walled, mostly globose cells. The ascospores are hyaline, cylindrical-filiform (up to 150 μm long), 7–16-septate, usually asymmetrical with a bluntly rounded apical end and tapering towards the basal end. These species were experimentally linked with a craspedodidymum-like anamorph, and some also form a chloridium-like synanamorph in axenic culture (Huhndorf and Fernández 2005). Atkinson et al. (2007) and Perera et al. (2016) introduced another three Chaetosphaeria matching the diagnostic characters of this group. Among the known ascomycetes, the monotypic genus Paragaeumannomyces (Matsushima 2003), based on P. sphaerocellularis, is remarkably similar to these scolecosporous species of Chaetosphaeria in features of ascomata, asci and ascospores and ecology.
Our sampling of saprobic lignicolous fungi in terrestrial biotopes in various localities in Europe, New Zealand and North America revealed several species whose morphological characters best match those of the genus Paragaeumannomyces and other scolecosporous Chaetosphaeria, i.e. Ch. albida (Atkinson et al. 2007), Ch. longispora (Barr 1993; Huhndorf and Fernández 2005) and five unknown species. We also collected additional specimens that represent new species, an unknown Codinaea (Maire 1937; Hughes and Kendrick 1968) on submerged wood and leaves in France and United Kingdom and an undescribed Striatosphaeria (Samuels and Müller 1978) on decaying bark of a woody liana in French Guiana. Codinaea, typified by C. aristata, comprises fungi forming tufts of fertile or sterile setae accompanied by conidiophores terminating into a phialide and hyaline, aseptate, falcate conidia with setulae at both ends. Striatosphaeria is well distinguishable from other members of the family by brown, 1-septate ascospores with longitudinal ridges and furrows running the entire length of the ascospore and a codinaea-like anamorph with brown, 1-septate conidia.
Dendrophoma (Saccardo 1880; Crous et al. 2012) is characterised by superficial, stromatic, stipitate, cupulate conidiomata, phialidic conidiogenous cells arranged in terminal whorls and naviculate to botuliform, aseptate, hyaline conidia with polar appendages. Using DNA sequence data, Crous et al. (2012) confirmed its systematic placement in the Chaetosphaeriaceae. However, its teleomorph-anamorph relationship remains unknown. A collection of a chaetosphaeria-like species with glabrous, dark, erumpent, aggregated ascomata sometimes in caespitose clusters, stipitate asci with inconspicuous apical annulus and fusiform, hyaline, 1-septate ascospores was encountered in the cracks of the bark of twigs of Buxus sempervivens in Germany. The axenic culture derived from ascospores yielded an anamorph similar to Dendrophoma. A BLASTn search (Zhang et al. 2000) for possible relatives in GenBank (Sayers et al. 2019) suggested our isolate is similar to Dendrophoma cytisporoides, the type species of the genus.
The present study provides new data that improve our understanding of morphological and genetic diversity of the Chaetosphaeriaceae and its pleomorphism. Our longer term goals focus on identification of monophyletic, morphologically well-delimited natural lineages and the life history of species currently assigned to the family. To assess phylogenetic relationships of our isolates, we based the study on morphological and cultivation studies along with the analysis of DNA sequence data from the nuc rDNA internal transcribed spacer region (ITS1-5.8S-ITS2 = ITS) and nuclear large subunit 28S ribosomal DNA gene (28S).
Materials and methods
Herbarium material and fungal strains
Material for this study was collected in north temperate regions of Europe (France, Germany and Ukraine) and North America (North Carolina, Tennessee), south subtropical and temperate climate zones of New Zealand, and in the neotropical regions of the Caribbean (Puerto Rico) and South America (French Guiana). An additional living culture was obtained from BCCM/MUCL Agro-food & Environmental Fungal Collection (MUCL), Université catholique de Louvain, Louvain, Belgium. Representative strains and ex-type strains were deposited at Westerdijk Fungal Biodiversity Institute (CBS), Utrecht, the Netherlands. Holotypes and other herbarium material (as dried voucher specimens) were deposited in the Fungarium of the Illinois Natural History Survey (ILLS), Champaign, Illinois, USA, the New Zealand Fungarium (PDD), Auckland, New Zealand and Herbarium of the Institute of Botany (PRA), Czech Academy of Sciences, Průhonice, Czech Republic. Isolates and specimens, their sources and GenBank accession numbers for ITS and 28S sequences generated in this study, are listed in Table 1.
Table 1.
Taxa, isolate information and GenBank accession numbers for new sequences (in bold) determined for this study.
| Taxon | Specimen | Status | Country | Host | Substrate | GenBank accessions | Reference | |
|---|---|---|---|---|---|---|---|---|
| ITS | 28S | |||||||
| Codinaea paniculata | CBS 145098 | T | France | unidentified | submerged decaying wood | MT118230 | MT118201 | This study |
| CBS 126573 | France | Alnus glutinosa | submerged decaying wood | MT118231 | MT118202 | This study | ||
| CBS 127692 | France | Fraxinus excelsior | submerged decaying wood | MT118232 | MT118203 | This study | ||
| MUCL 34876 | United Kingdom | unidentified | submerged dead leaf | MT118233 | MT118204 | This study | ||
| Dendrophoma cytisporoides |
CBS 144107 IMI 506817 |
Germany | Buxus sempervivens | decaying periderm of a twig | MT118234 | MT118205 | This study | |
| Paragaeumannomyces abietinus | CBS 145351 | T | France | Abies alba | decaying wood | MT118235 | MT118206 | This study |
| Paragaeumannomyces albidus | PDD 118738 | New Zealand | unidentified | decaying wood | MT876579 | – | This study | |
| Paragaeumannomyces elegans | PDD 118740 | T | New Zealand | unidentified | decaying wood | MT876580 | – | This study |
| Paragaeumannomyces granulatus | ICMP 15133 | T | New Zealand | unidentified | decaying wood | MT876575 | MT876577 | This study |
| PDD 118745 | New Zealand | unidentified | decaying wood | MT876576 | MT876578 | This study | ||
| Paragaeumannomyces lapazianus | S.M.H. 2182 | Costa Rica | unidentified | decaying wood | AY906945 | MT118207 | Huhndorf and Fernández (2005), this study | |
| S.M.H. 2900 | Puerto Rico | unidentified | decaying wood | AY906946 | MT118208 | Huhndorf and Fernández (2005), this study | ||
| S.M.H. 3043 | Puerto Rico | unidentified | decaying wood | AY906947 | MT118209 | Huhndorf and Fernández (2005), this study | ||
| Paragaeumannomyces longisporus | A.N.M. 1269 | USA, Tennessee | unidentified | decaying wood | MT118239 | MT118210 | This study | |
| ILLS00121385 | USA, Tennessee | unidentified | decaying wood | MT118237 | MT118211 | This study | ||
| ILLS00121386 | USA, Tennessee | unidentified | decaying wood | MT118238 | MT118212 | This study | ||
| S.M.H. 2519 | USA, Indiana | unidentified | decaying wood | AY906939 | MT118213 | Huhndorf and Fernández (2005), this study | ||
| S.M.H. 2758 | USA, North Carolina | unidentified | decaying wood | AY906940 | MT118214 | Huhndorf and Fernández (2005), this study | ||
| S.M.H. 3805 | USA, North Carolina | unidentified | decaying wood | MT118236 | MT118215 | This study | ||
| S.M.H. 3809 | USA, North Carolina | unidentified | decaying wood | AY906942 | MT118216 | Huhndorf and Fernández (2005), this study | ||
| S.M.H. 3860 | USA, South Carolina | unidentified | decaying wood | AY906944 | MT118217 | Huhndorf and Fernández (2005), this study | ||
| Paragaeumannomyces panamensis | S.M.H. 3596 | T | Panama | unidentified | decaying wood | AY906948 | MT118218 | Huhndorf and Fernández (2005), this study |
| Paragaeumannomyces sp. 1 | S.M.H. 2025 | Puerto Rico | unidentified | decaying wood | MT118241 | MT118219 | This study | |
| S.M.H. 3014 | Puerto Rico | unidentified | decaying wood | AY906952 | MT118222 | Huhndorf and Fernández (2005) (as ‘raciborskii’), this study | ||
| Paragaeumannomyces sp. 2 | S.M.H. 2036 | Puerto Rico | unidentified | decaying wood | AY906950 | MT118220 | Huhndorf and Fernández (2005) (as ‘raciborskii’), this study | |
| S.M.H. 2132 | Puerto Rico | unidentified | decaying wood | AY906951 | MT118221 | Huhndorf and Fernández (2005) (as ‘raciborskii’), this study | ||
| Paragaeumannomyces rubicundus | S.M.H. 2881 | PT | Puerto Rico | unidentified | decaying wood | AY906954 | MT118223 | Huhndorf and Fernández (2005), this study |
| S.M.H. 3221 | T | Costa Rica | unidentified | decaying wood | MT118242 | MT118224 | This study | |
| Paragaeumannomyces sabinianus | ILLS00121384 | T | USA, Tennessee | unidentified | decaying wood | MT118243 | MT118225 | This study |
| S.M.H. 3807 | USA, North Carolina | unidentified | decaying wood | AY906941 | MT118226 | Huhndorf and Fernández (2005), this study | ||
| S.M.H. 3824 | USA, North Carolina | unidentified | decaying wood | AY906943 | MT118227 | Huhndorf and Fernández (2005), this study | ||
| Paragaeumannomyces smokiensis | ILLS00121398 | T | USA, Tennessee | unidentified | decaying wood | MT118240 | MT118228 | This study |
| Striatosphaeria castanea | CBS 145352 | T | French Guinea | woody liana | decaying periderm | MT118244 | MT118229 | This study |
| Striatosphaeria codinaeophora | S.M.H. 1524 | Puerto Rico | Nectandra turbacensis | decaying wood | MT118245 | AF466088 | Huhndorf et al. (2004), this study | |
Note: T and PT denote ex-type and ex-paratype strains.
Morphological characterisation
Morphological characteristics were obtained from fungi growing on natural substrate and growth media. Descriptions in the key are based on fungi growing on natural substrate. Herbarium material was rehydrated with tap water and examined with an Olympus SZX12 dissecting microscope (Olympus America, Inc., Melville, USA). Hand-sectioned ascomata and centrum material (asci, ascospores and paraphyses), conidiophores and conidia were mounted in 90 % lactic acid, Melzer’s reagent, and lactophenol with cotton blue. All measurements were in Melzer’s reagent. Means ± standard deviation (SD) based on a minimum of 20–25 measurements are given for dimensions of asci, ascospores and conidia. Micromorphological observations were made using an Olympus BX51 compound microscope with differential interference contrast (DIC) and phase contrast (PC) illumination. Images of microscopic structures were captured with an Olympus DP70 camera operated by Imaging Software Cell^D (Olympus). Macroscopic images of colonies were documented using a Canon EOS 77D digital camera with Canon EF 100mm f/2.8L Macro IS USM objective with daylight spectrum 5500K 16W LED lights (Canon Europe Ltd., Middlesex, United Kingdom). All images were processed with Adobe Photoshop CS6 (Adobe Systems, San Jose, USA).
For comparative purposes, strains were inoculated in triplicate on cornmeal dextrose agar (CMD) [17 g of cornmeal agar (Oxoid Limited, Hampshire, United Kingdom), 2 g of dextrose, 1 L of distilled water, sterilized for 15 min at 121 C], Modified Leonian’s agar (MLA) (Malloch 1981), oat-meal agar (OA) modified after Gooding and Lucas (1959) (30 g of oatmeal cooked in 1 L of distilled water for 15–30 min, filtered through cheesecloth, the filtrate was brought back to volume with distilled water, 15 g of agar, sterilized for 60 min at 121 C) and potato-carrot agar (PCA) (Crous et al. 2019). To induce sporulation, strains were also inoculated on CMA (Crous et al. 2019) with sterile stems of Urtica dioica. Descriptions of colonies are based on 4 wk old cultures grown in darkness at 22–23 C.
DNA extraction and amplification
Methods for the DNA extraction and amplification of samples with A.N.M., ILLS and S.M.H. prefixes followed Huhndorf et al. (2004) and Hustad and Miller (2015). Other samples were processed according to the following protocols. Total genomic DNA was extracted from mycelium removed from 3-wk-old cultures grown on MLA using the DNeasy UltraClean Microbial Kit (Qiagen GmbH, Germany) following the manufacturer’s protocol for filamentous fungi. All PCR amplifications were carried out in 25 μL volume reactions using a Q5 High Fidelity DNA polymerase kit (New England Biolabs Inc., United Kingdom) according to the manufacturer’s protocol. Primers used for the amplification included: V9G/LR8 (de Hoog and Gerrits van den Ende 1998; Vilgalys unpublished) for the internal transcribed spacers (ITS) of the nuclear rRNA cistron and D1, D2 and D3 domains (approx. 1900 bp of the 5' end) of the 28S rDNA gene.
PCR was carried out in a BioRad C1000 thermal cycler (Bio-Rad Laboratories Inc., USA) as follows: 98 C for 30 s; 40 cycles of denaturation (98 C for 10 s), annealing (62 C for 30 s) and elongation (72 C for 90 s) and a final extension step at 72 C for 5 min. Amplicons were purified from agarose gels using a NucleoSpin® Gel and PCR Clean-up Kit (Macherey-Nagel GmbH & Co. KG, Germany) following the manufacturer’s instructions, with an elution volume of 25 μL. The DNA concentration was assessed fluorimetrically using Quant-iT PicoGreen dsDNA Assay Kit and Qubit fluorometer (Invitrogen / Thermo Fisher Scientific, USA) to assure required sequencing concentrations adjusted for the length of amplicons/ number of reads required.
Each of the amplicons was sequenced in both directions using the PCR primers and nested primers: ITS5, ITS4, JS1, JS7, JS8 and LR7 for ITS-28S (Vilgalys and Hester 1990; White et al. 1990; Landvik 1996; Vilgalys unpublished). Automated sequencing was carried out by Eurofins GATC Biotech Sequencing Service (Cologne, Germany). Raw sequence data were assembled, examined and edited using Sequencher v.5.4.6 (Gene Codes Corp., Ann Arbor, USA).
Alignments and phylogenetic analyses
Two gene markers, ITS and 28S rDNA, were analysed to assess evolutionary relationships of the unknown fungi with members of the Chaetosphaeriaceae. Consensus secondary structure (2D) models for the ITS1 and ITS2 for members of the Chaetosphaeriaceae were built using the Ppfold program v.3.0 (Sukosd et al. 2012). The obtained 2D consensus models were further improved using the program Mfold (Zuker 2003) and adjusted manually if necessary, based on comparison of homologous positions in the multiple sequence alignment. A predicted 2D model of the 28S of Saccharomyces cerevisiae (Gutell et al. 1993) was used to improve the alignment of this gene. The models were highly consistent in all taxa.
ITS and 28S sequences were aligned manually in Bioedit v.7.1.8 (Hall 1999). GenBank accession numbers for ITS and 28S sequences of members of the Chaetosphaeriaceae retrieved from GenBank and published in other studies are listed in Table 2. Single-locus data sets of the Chaetosphaeriaceae (ITS: 89 sequences/602 characters including gaps, 28S: 86/1176) and Paragaeumannomyces (ITS: 35/489, 28S: 32/1104) were evaluated using PartitionFinder2 (Lanfear et al. 2016), implemented in the CIPRES Science Gateway v.3.3 (http://www.phylo.org) (Miller et al. 2010), to find the best partitioning scheme for our datasets and to select best-fit models under corrected Akaike information criteria. Conflict-free data sets were concatenated into two alignments (deposited in TreeBASE 25964) that were subjected to subsequent phylogenetic analyses.
Table 2.
Taxa, isolate information and accession numbers for sequences retrieved from GenBank.
| Taxon | Strain | Status | Country | Host | Substrate | GenBank accessions | Reference | |
|---|---|---|---|---|---|---|---|---|
| ITS | 28S | |||||||
| Adautomilanezia caesalpiniae | CC-LAMIC 102/12 | T | Brazil | Caesalpina echinata | wood | KX821777 | KU170671 | Crous et al. (2016) |
| Anacacumisporium appendiculatum | HMAS 245593 | T | China, Hainan | broad-leaved tree | dead stems | KP347129 | KT001553 | Ma et al. (2016) |
| Brunneodinemasporium brasiliense | CBS 112007 | T | Brazil | unidentified | decaying leaf | JQ889272 | JQ889288 | Crous et al. (2012) |
| Brunneodinemasporium jonesii | GZCC 16-0050 | T | China | unidentified | decaying wood | KY026058 | KY026055 | Lu et al. (2016) |
| Cacumisporium capitulatum | FMR 11339 | Spain | unidentified | decaying wood | HF677176 | HF677190 | Hernández-Restrepo et al. (2017) | |
| Calvolachnella guaviyunis | CBS 134695 | T | Uruguay | Myrcianthes pungens | bark | KJ834524 | KJ834525 | Crous et al. (2014a) |
| Chaetosphaeria chlorotunicata | S.M.H. 1565 | T | Puerto Rico | unidentified | decaying wood | – | AF466064 | Fernández et al. (2006) |
| Chaetosphaeria innumera | M.R. 1175 | Czech Republic | Fagus sylvatica | decaying wood | AF178551 | AF178551 | Réblová and Winka (2000) | |
| Chaetosphaeria lignomollis | S.M.H. 3015 | T | Puerto Rico | unidentified | decaying wood | EU037896 | AF466073 | Atkinson et al. (2007), Fernández et al. (2006) |
| Chaetosphaeria myriocarpa | CBS 264.76 | The Netherlands | unidentified | decaying wood | AF178552 | AF178552 | Réblová and Winka (2000) | |
| Chaetosphaeria pygmaea | M.R. 1365 | Czech Republic | Fagus sylvatica | decaying wood | AF178545 | AF178545 | Réblová and Winka (2000) | |
| Chloridium caesium | CBS 102339 | Austria | Salix cinerea | decaying wood | AF178564 | AF178564 | Réblová and Winka (2000) | |
| Chloridium gonytrichii | CBS 195.60 | South Africa | unidentified | unknown | MH857954 | MH869503 | Vu et al. (2019) | |
| Chloridium virescens | CBS 152.53 | France | Acer sp. | unknown | MH857142 | MH868678 | Vu et al. (2019) | |
| Codinaea acaciae | CBS 139907 | T | Malaysia, Sarawak | Acacia mangium | leaf spot | KR476732 | – | Crous et al. (2015b) |
| Codinaea lambertiae | CBS 143419 | T | Australia, N.S. Wales | Lambertia formosa | leaves | MG386052 | MG386105 | Crous et al. (2017) |
| Codinaea pini | CBS 138866 | T | Uganda | Pinus patula | dead needles | KP004465 | KP004493 | Crous et al. (2014b) |
| Codinaea simplex | CBS 966.69 | The Netherlands | Quercus sp. | cupule | AF178559 | AF178559 | Réblová and Winka (2000) | |
| Codinaeopsis gonytrichodes | CBS 593.93 | Japan | unidentified | decaying plant material | AF178556 | AF178556 | Réblová and Winka (2000) | |
| Conicomyces pseudotransvaalensis | HHUF 29956 | T | Japan | Machilus japonica | dead twig | LC001710 | LC001708 | Liu et al. (2015) |
| Cryptophiale udagawae | GZCC 18-0047 | China, Guizhou | unidentified | decaying wood | MN104608 | MN104619 | Lin et al. (2019) | |
| Dendrophoma cytisporoides | CBS 223.95 | ET | The Netherlands | Rhododendron sp. | branches and twigs | JQ889273 | JQ889289 | Crous et al. (2012) |
| Dictyochaeta assamica | CBS 242.66 | Guadeloupe | Musa sp. | root | MH858788 | MH870426 | Vu et al. (2019) | |
| Dictyochaeta callimorpha | ICMP 15130 | New Zealand | unidentified | decaying wood | MT454483 | MT454498 | Réblová et al. (2020) | |
| Dictyochaeta cangshanensis | MFLUCC 17-2214 | T | China, Yunnan | unidentified | submerged decaying wood | MK828632 | MK835832 | Luo et al. (2019) |
| Dictyochaeta ellipsoidea | MFLUCC 18-1574 | T | China, Yunnan | unidentified | submerged decaying wood | MK828628 | MK835828 | Luo et al. (2019) |
| Dictyochaeta fuegiana | ICMP 15153 | T | New Zealand | unidentified | decaying wood | MT454487 | EF063574 | Réblová and Seifert (2007), Réblová et al. (2020) |
| Dictyochaeta lignicola | DLUCC 0899 | T | China, Yunnan | unidentified | submerged decaying wood | MK828630 | MK835830 | Luo et al. (2019) |
| Dictyochaeta pandanicola | KUMCC 16-0153 | T | China, Yunnan | Pandanus sp. | decayng leaf | MH388338 | MH376710 | Tibpromma et al. (2018) |
| Dictyochaeta septata | CBS 143386 | ET | Chile | Eucalyptus grandis × urophylla | leaves | MH107889 | MH107936 | Crous et al. (2018a) |
| Dictyochaeta siamensis | MFLUCC 15-0614 | T | Thailand | unidentified | submerged decaying twig | KX609955 | KX609952 | Liu et al. (2016) |
| Dictyochaeta terminalis | GZCC 18-0085 | T | China, Guizhou | unidentified | decaying leaves | MN104613 | MN104624 | Lin et al. (2019) |
| Dinemasporium americanum | CBS 127127 | T | USA, Iowa | n/a | soil of tallgrass prairie | JQ889274 | JQ889290 | Crous et al. (2012) |
| Dinemasporium pseudoindicum | CBS 127402 | T | USA, Kansas | n/a | soil of tallgrass prairie | JQ889277 | JQ889293 | Crous et al. (2012) |
| Ellisembia aurea | CBS 144403 | T | France | Sambucus nigra | decaying wood | MH836375 | MH836376 | Hyde et al. (2019) |
| Ellisembia folliculata | CBS 101317 | France | Salix sp. | decaying wood | – | AF261071 | Réblová and Winka (2001) | |
| Eucalyptostroma eucalypti | CBS 142074 | T | Malaysia | Eucalyptus pellita | leaf spots | KY173408 | KY173500 | Crous et al. (2016) |
| Exserticlava vasiformis | TAMA 450 | Japan, Chiba | unidentified | plant debris | – | AB753846 | Tsuchiya et al., unpublished | |
| Infundibulomyces cupulatus | BCC 11929 | T | Thailand | Lagerstroemia sp. | dead leaf | EF113976 | EF113979 | Plaingam et al. (2003) |
| Infundibulomyces oblongisporus | BCC 13400 | T | Thailand | unidentified, angiosperm | leaf litter | EF113977 | EF113980 | Somrithipol et al. (2008) |
| Kionochaeta castaneae | GZCC 18-0025 | T | China | Castanea mollissima | decaying seed shell | MN104610 | MN104621 | Lin et al. (2019) |
| Kionochaeta microspora | GZCC 18-0036 | T | China, Guizhou | unidentified | decaying wood | MN104607 | MN104618 | Lin et al. (2019) |
| Leptosporella arengae | MFLUCC 15-0330 | T | Thailand | Arenga pinnata | dead rachis | MG272255 | MG272246 | Konta et al. (2017) |
| Leptosporella bambusae | MFLUCC 12-0846 | T | Thailand | bamboo | dead culms | KU940134 | KU863122 | Dai et al. (2016) |
| Menispora ciliata | CBS 122131 | T | Czech Republic | Acer campestre | decaying wood | EU488736 | – | Réblová and Seifert (2008) |
| Menispora tortuosa | DAOM 231154 | unknown | unidentified | unknown | KT225527 | AY544682 | Schoch et al. (2009) | |
| Menisporopsis dushanensis | GZCC 18-0084 | T | China, Guizhou | unidentified | decaying leaves | MN104615 | MN104626 | Lin et al. (2019) |
| Menisporopsis theobromae | MFLUCC 15-0055 | Thailand | unidentified | submerged decaying wood | KX609957 | KX609954 | Liu et al. (2016) | |
| Nawawia filiformis | MFLUCC 17-2394 | Thailand | unidentified | decaying wood | MH758196 | MH758209 | Yang et al. (2018) | |
| Neopseudolachnella acutispora | MAFF 244358 | T | Japan, Aomori | Pleioblastus chino | dead twigs | AB934065 | AB934041 | Hashimoto et al. (2015) |
| Neopseudolachnella magnispora | MAFF 244359 | T | Japan, Aomori | Sasa kurilensis | dead twigs | AB934066 | AB934042 | Hashimoto et al. (2015) |
| Paliphora intermedia | CBS 896.97 | IST | Australia, Queensland | unidentified | leaf litter | MH862682 | EF204501 | Shenoy et al. (2010), Vu et al. (2019) |
| Paragaeumannomyces albidus | PDD 92537 | T | New Zealand | Nothofagus sp. | decaying wood | EU037890 | EU037898 | Atkinson et al. (2007) |
| P. albidus | PDD 92540 | New Zealand | Nothofagus sp. | decaying wood | EU037891 | – | Atkinson et al. (2007) | |
| Paragaeumannomyces bombycinus | PDD 92538 | T | New Zealand | Nothofagus sp. | decaying wood | EU037892 | – | Atkinson et al. (2007) |
| Paragaeumannomyces elegans | PDD 92561 | New Zealand | unidentified | decaying wood | EU037895 | – | Atkinson et al. (2007) | |
| Paragaeumannomyces garethjonesii | MFLUCC 15-1012 | T | Thailand | Fabaceae | seed pod | KY212751 | KY212759 | Perera et al. (2016) |
| Paragaeumannomyces panamensis | MFLUCC 15-1011 | Thailand | Pinus sp. | decaying wood | KY212752 | KY212760 | Perera et al. (2016) | |
| Paragaeumannomyces sp. 3 | S.M.H. 2017 | Puerto Rico | unidentified | decaying wood | AY906949 | AF466078 | Huhndorf and Fernández (2005) (as ‘raciborskii’) | |
| Paragaeumannomyces sp. 4 | S.M.H. 3119 | Puerto Rico | unidentified | decaying wood | AY906953 | AY436402 | Huhndorf and Fernández (2005) (as ‘raciborskii’) | |
| Phialosporostilbe scutiformis | MFLUCC 17-0227 | T | China | unidentified | submerged decaying wood | MH758194 | MH758207 | Yang et al. (2018) |
| Polynema podocarpi | CBS 144415 | T | New Zealand | Podocarpus totara | unknown | MH327797 | MH327833 | Crous et al. (2018b) |
| Pseudodinemasporium fabiforme | CBS 140010 | Malaysia, Sarawak | Acacia mangium | leaf spots | KR611889 | KR611906 | Crous et al. (2015a) | |
| Pseudolachnea fraxini | CBS 113701 | T | Sweden | Fraxinus excelsior | unknown | JQ889287 | JQ889301 | Crous et al. (2012) |
| Pseudolachnea hispidula | MAFF 244365 | Japan, Aomori | Morus bombycis | dead twig | AB934072 | AB934048 | Hashimoto et al. (2015) | |
| Pseudolachnella asymmetrica | MAFF 244366 | Japan, Fukuoka | Phyllostachys nigra var. henonis | dead twig | AB934073 | AB934049 | Hashimoto et al. (2015) | |
| Pseudolachnella scolecospora | MAFF 244379 | Japan, Gifu | Sasa sp. | dead twigs | AB934086 | AB934062 | Hashimoto et al. (2015) | |
| Pyrigemmula aurantiaca | CBS 126743 | T | Hungary | Vitis vinifera | bark | HM241692 | HM241692 | Magyar et al. 2011) |
| Sporoschisma longicatenatum | MFLUCC 16-0180 | T | Thailand | unidentified | submerged decaying wood | KX505871 | KX358077 | Yang et al. (2016) |
| Sporoschisma mirabile | FMR 11247 | Spain | unidentified | dead wood | HF677174 | HF677183 | Hernández-Restrepo et al. (2017) | |
| Striatosphaeria castanea | monte6.2 | Brazil | Encyclia ghillanyi | root | KC928368 | – | Almeida et al., unpublished | |
| Striatosphaeria codinaeophora | M.R. 1230 | Puerto Rico | Dacryodes excelsa | decaying wood | AF178546 | AF178546 | Réblová and Winka (2000) | |
| Tainosphaeria jonesii | GZCC 16-0065 | PT | China, Guangxi | unidentified | submerged decaying wood | KY026060 | KY026057 | Lu et al. (2016) |
| Tainosphaeria siamensis | MFLUCC 15-0607 | T | Thailand | unidentified | submerged decaying wood | KX609956 | KX609953 | Liu et al. (2016) |
| Thozetella nivea | n/a | unknown | unidentified | unknown | EU825201 | EU825200 | Jeewon et al. (2009) | |
| Thozetella tocklaiensis | CBS 378.58 | T | India | Camellia sinensis | decaying flower | MH857817 | MH869349 | Vu et al. (2019) |
| Tracylla aristata | CBS 141404 | ET | Australia, Victoria | Eucalyptus regnans | leaf | KX306770 | KX306795 | Hernández-Restrepo et al. (2016) |
| Tracylla eucalypti | CBS 144429 | T | Colombia | Eucalyptus urophylla | spots on living leaves | MH327810 | MH327846 | Crous et al. (2018b) |
| Zanclospora iberica | CBS 130426 | T | Spain | unidentified | decaying wood | KY853480 | KY853544 | Hernández-Restrepo et al. (2017) |
Note: T, ET, IST and PT denote ex-type, ex-epitype, ex-isotype and ex-paratype strains.
Three analyses were employed to estimate phylogenetic relationships. Bayesian Inference (BI) and Maximum Likelihood (ML) analyses were performed through the CIPRES Science Gateway v.3.3. ML analyses were conducted with RAXML-HPC v.8.2.12 (Stamatakis 2014) with a GTRCAT approximation. Nodal support was determined by non-parametric bootstrapping (BS) with 1 000 replicates. BI analyses were performed in a likelihood framework as implemented in MrBayes v.3.2.6 (Huelsenbeck and Ronquist 2001). Two Bayesian searches were performed using default parameters. The B-MCMCMC analyses lasted until the average standard deviation of split frequencies was below 0.01 with trees saved every 1000 generations. The first 25 % of saved trees, representing the burn-in phase of the analysis, were discarded. The remaining trees were used for calculating posterior probabilities (PP) of recovered branches. Maximum Parsimony (MP) analyses were conducted with PAUP 4.0a167 (Swofford 2003). A heuristic search was performed with the stepwise-addition option with 1000 random taxon addition replicates and TBR branch swapping. Because the secondary structures of the ITS and 28S were carefully studied when aligning the sequences, and regions with incomplete sequences were excluded from the analysis, we treated gaps as a fifth character state; they were given equal weight as the other characters. All characters were unordered. Branch support was estimated on the recovered topologies by performing a heuristic search of 1000 bootstrap replicates consisting of ten random-addition replicates for each bootstrap replicate.
Results
Phylogenetic analyses
Employing the predicted 2D structure of the variable ITS region and 28S enabled us to construct a reliable multiple sequence alignment of homologous positions at both helices and loops, thus eliminating potential ambiguous regions in the alignments. Initially, we compared trees from ML phylogenetic analyses of the two combined data sets (Chaetosphaeriaceae and scolecosporous species of Chaetosphaeria) after alignments were improved with the 2D structure, with and without applying Gblocks (Castresana 2000) using default options, to delimit and remove putative ambiguous regions. The phylogenetic trees based on datasets using Gblocks had lower support for nodes and relationships within and among several clades that could not be resolved (data not shown) compared to trees in which these regions remained. Therefore, the final phylogenies were based on datasets in which Gblocks was not employed.
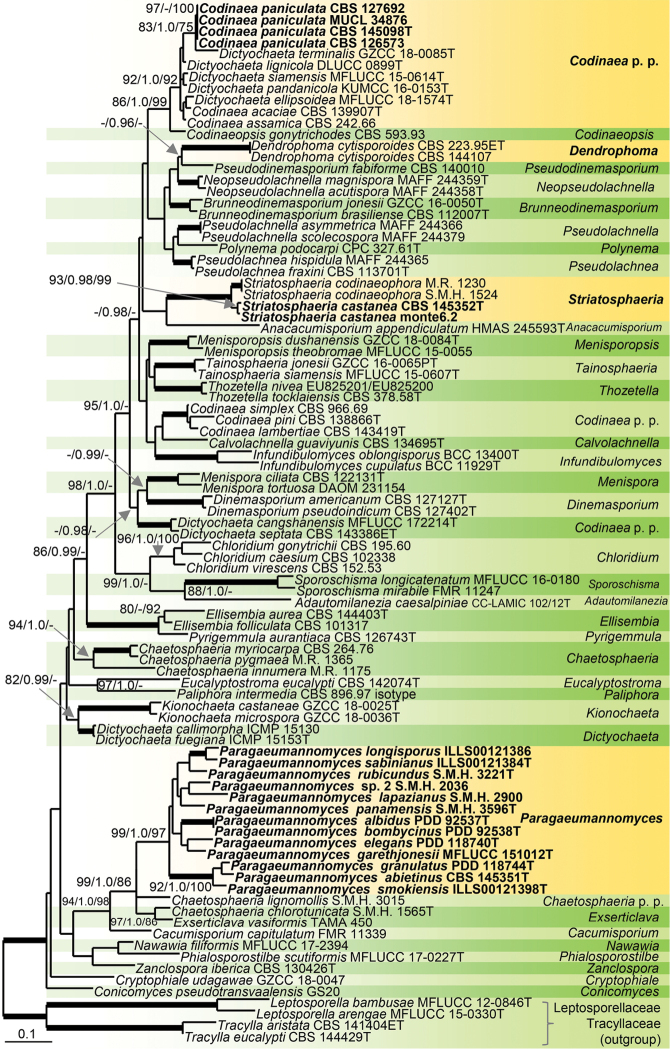
Evolutionary relationships of studied fungi were evaluated in the phylogenetic analysis based on the combined ITS and 28S sequences of 87 representative species of the Chaetosphaeriaceae. Leptosporella arengae, L. bambusae (Leptosporellaceae), and Tracylla eucalypti and T. aristata (Tracyllaceae) were used to root the tree. 76 nucleotides (nt) at the 5'-end and 606 nt at the 3'-end of 28S were excluded from the alignment because of missing data in the majority of sequences. The alignment had 1778 characters including gaps and 882 unique character sites (RAxML). In the MP analysis, 1021 characters were constant (proportion = 57.42 %), 134 variable characters were parsimony-uninformative, 623 characters were parsimony-informative (included); two most parsimonious trees were produced (length = 5066 steps, consistency index = 0.0.298, homoplasy index = 0.702, retention index = 0.631). For the BI analysis, GTR+I+G model was selected for ITS and 28S partitions. The ML tree (RAxML) is shown in Fig. 1. There were no conflicts among the trees generated by the three different phylogenetic analyses. The Chaetosphaeriaceae were resolved as a strongly supported clade; some of the nodes of the backbone tree, which obtained support in the ML and/or BI analyses, were not statistically supported in MP analysis. The 39 identified terminal clades corresponded to individual genera or natural groups of species. Codinaea was resolved as polyphyletic in three subclades. The unknown species was grouped in a clade (92 % MLBS/1.0 PP/92 % MPBS) containing seven Codinaea or Dictyochaeta species, three of which possess the typical Codinaea phenotype, while other morphologically similar species with setulate conidia clustered in the other two subclades, C. lambertiae, C. pini and C. simplex (100/1.0/100) and Dictyochaeta septata and D. cangshanensis (96/1.0/100). The new species Codinaea paniculata, based on four strains, was resolved as a monophyletic clade in all three analyses, although the statistical support varied. In ML and MP analyses the clade obtained 97 % and 100 % support, respectively, in the BI analysis it was weakly supported with 0.77 PP. The intraspecific variability of C. paniculata, based on ITS sequences, varied slightly. Three strains (CBS 145098 ex-type, CBS 126573, MUCL 34876) had identical ITS sequences, strain CBS 127692 differed from them by one base pair. The 28S sequence similarity of all strains of C. paniculata was 100 %. The undescribed Striatosphaeria was nested in the monophyletic Striatosphaeria (100/1.0/100) clade as sister to S. codinaeophora. It clustered in a subclade (93/0.98/99) with an endophytic isolate Striatosphaeria sp. monte6.2; their ITS exhibited 98.5 % sequence similarity. The five unknown chaetosphaeria-like species with scolecosporous ascospores were nested in a strongly-supported monophyletic clade (99/1.0/97). This clade contained eight additional morphologically similar species with scolecosporous ascospores and three-layered ascomatal wall. The clade is introduced as Paragaeumannomyces in this study. A chaetosphaeria-like species grouped with the ex-type strain of Dendrophoma cytisporoidesCBS 223.95 in a monophyletic clade (100/1.0/100). Dendrophoma was resolved as a member of a large, statistically weakly-supported grouping containing six other genera characterised by sporodochial conidiomata.
Figure 1.
Combined phylogeny using ITS and 28S of selected members of the Chaetosphaeriaceae. Species names given in bold are taxonomic novelties; T, ET, IST and PT indicate ex-type, ex-epitype, ex-isotype and ex-paratype strains. Thickened branches indicate branch support with MLBS = 100%, PP values = 1.0 and MP = 100 %. Branch support of nodes ≥ 75 % ML and MPBS, and ≥ 0.95 PP is indicated above branches.
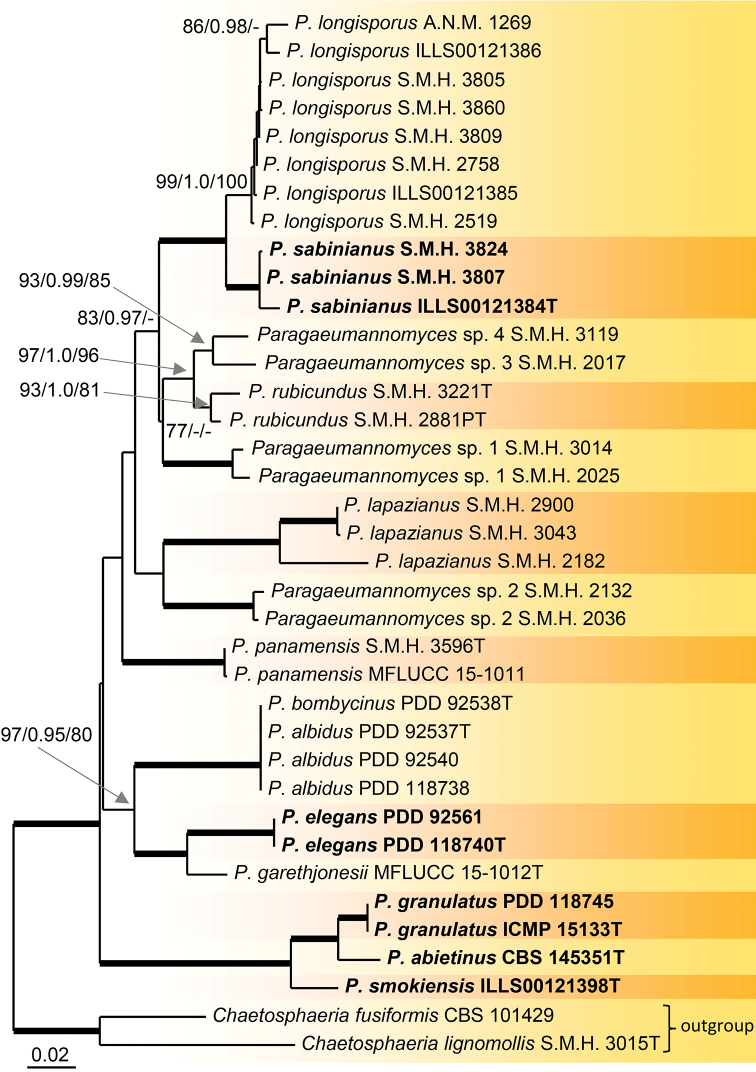
Phylogenetic relationships within the genus Paragaeumannomyces were assessed in the second analysis of the combined ITS-28S loci. Chaetosphaeria fusiformis and Ch. lignomollis (Chaetosphaeriaceae) were used to root the tree, and thus served as outgroup. The analysis included 35 sequences belonging to 16 species. 28 nt at the 5'-end and 714 nt at the 3'-end of 28S were excluded from the alignment due to missing data in the majority of sequences. The alignment had 1593 characters including gaps and 410 unique character sites (RAxML). In the MP analysis, 1247 characters were constant (proportion = 78.28 %), 78 variable characters were parsimony-uninformative, and 268 characters were parsimony-informative (included); 286 most parsimonious trees were produced (length = 832 steps, consistency index = 0.6118, homoplasy index = 0.3882, retention index = 0.8082). For the BI analysis, SYM+G and GTR+I+G models were selected for ITS and 28S partitions, respectively. The ML tree is shown in Fig. 2. There were no conflicts among the trees generated by the three different phylogenetic analyses. In the MP strict consensus tree, branches collapsed within the P. longisporus, P. sabinianus and P. albidus-bombycinus clades. The unknown species from wood of Abies alba clustered in a subclade (100/1.0/100) with two other unknown species from New Zealand and USA. They were introduced as new species sharing similar ascoma morphology, i.e. P. abietinus, P. granulatus and P. smokiensis. Another unknown species from New Zealand with densely setose, brownish-grey ascomata was grouped as a sister to P. garethjonesii and is introduced as P. elegans. The subclade (100/1.0/100) identified initially as Chaetosphaeria ellisiifideHuhndorf and Fernández (2005) [= Chaetosphaeria longisporafideKirk (2014)] was segregated into two well-supported subclades distinguished by ascospore morphology. These subclades represent two species, P. longisporus (99/1.0/100) and the new species, P. sabinianus (100/1.0/100). Paragaeumannomyces raciborskiifideHuhndorf and Fernández (2005) was resolved as polyphyletic forming four subclades accompanied by different anamorph morphology. Because none of these subclades could be designated ‘raciborskii s. str.’, they were labelled Paragaeumannomyces sp. 1–4.
Figure 2.
Combined phylogeny using ITS and 28S of 35 members of Paragaeumannomyces. Species names given in bold are new species; T and PT indicate ex-type and ex-paratype strains. Thickened branches indicate branch support with MLBS = 100%, PP values = 1.0 and MP = 100 %. Branch support of nodes ≥ 75 % ML and MPBS, and ≥ 0.95 PP is indicated above branches.
Taxonomy
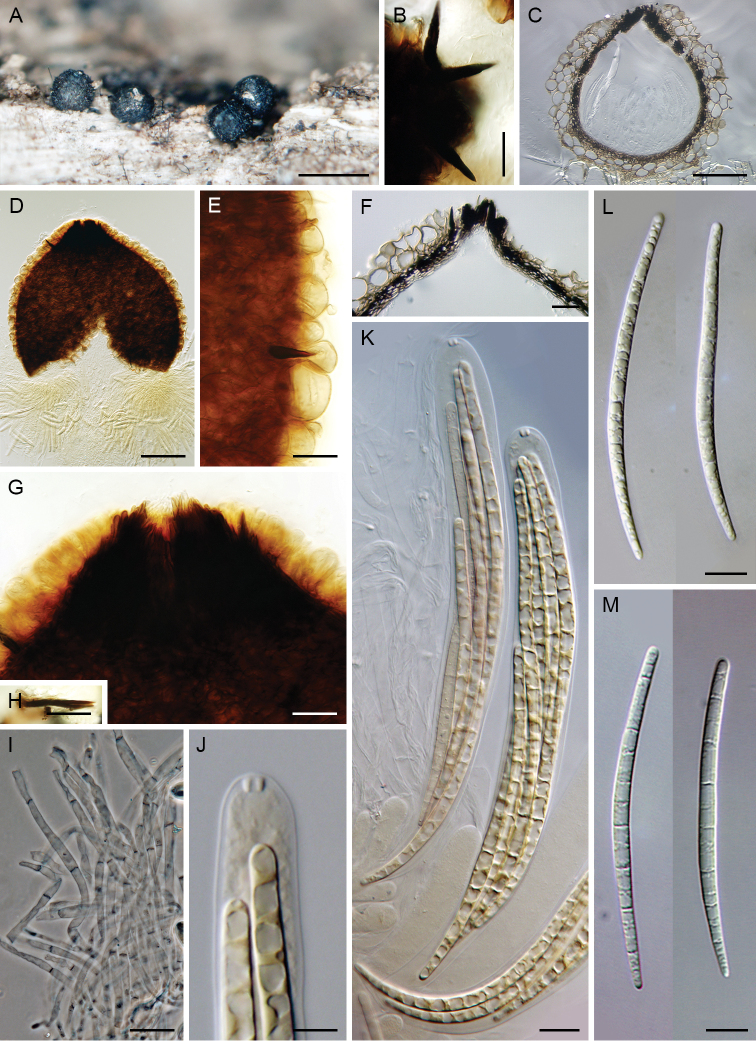
Codinaea paniculata
Réblová & J. Fourn. sp. nov.
900DCFCB-3172-5D7C-9453-8DE8414B9B09
836526
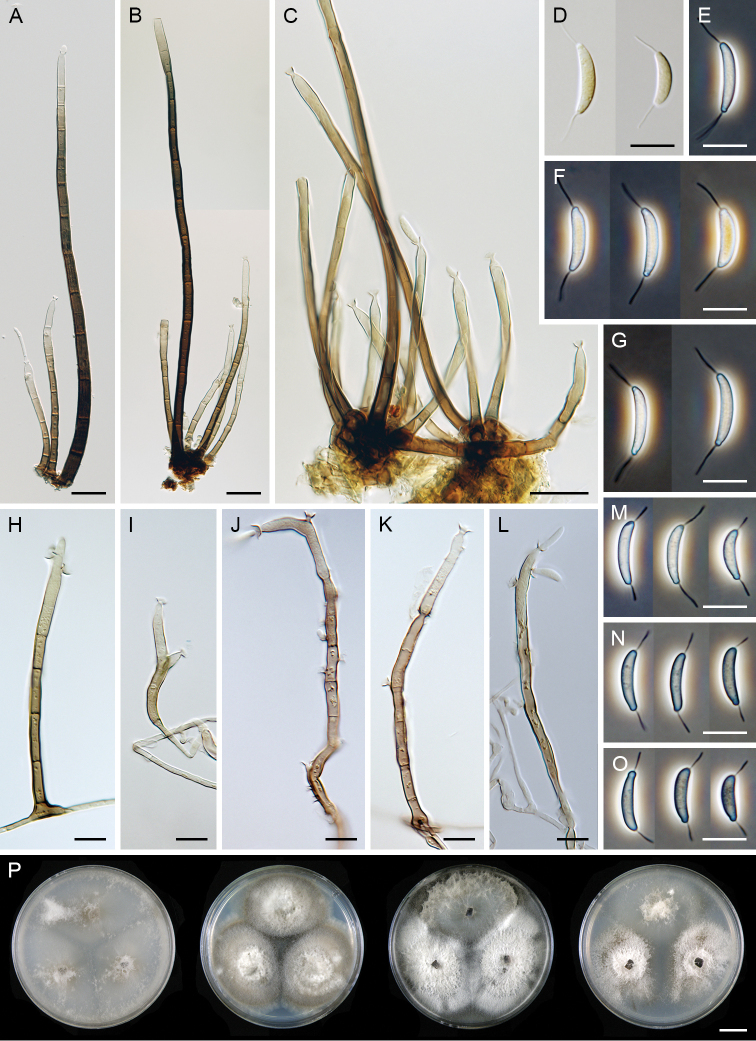
Figure 3.
Codinaea paniculata. A–C setae and conidiophores on nature substrate D–G conidia on nature substrate H–L conidiophores in MLA culture (6 wk) M–O conidia in MLA culture (6 wk) P colonies on CMD, MLA, OA and PCA after 4 wk (from left to right). Images: CBS 145098 (A, B, G–O); CBS 126573 (C); CBS 127692 (D–F). Scale bars: 20 μm (A–C); 10 μm (D–O); 1 cm (P).
Typification.
France – Ariège • Pyrénées Mts., Rimont, La Maille brook; alt. 550 m; 28 May 2018 (incubated in moist chamber for 1 wk); on submerged decaying wood; J. Fournier leg.; M.R. 3950 (holotype: PRA-16319!, ex-type culture CBS 145098).
Etymology.
Panicula (Latin) tuft, referring to the dense groups of setae and conidiophores on the natural substrate.
Description on the natural substrate.
Colonies on the nature substrate effuse, hairy, greyish-brown. Setae erect, straight or slightly flexuous, smooth-walled, dark brown and thick-walled, becoming pale brown to subhyaline and thin-walled towards the apex, 230–290 μm long, 6–7.5 μm wide above the base, tapering gradually towards the apex which almost always develops into a monophialide. Conidiophores macronematous, mononematous, 62–127 × 3.5–4.5 µm, septate, erect, straight or flexuous, arising singly or in groups of 4–6 from hyphal cells associated with the bases of setae, septate, mid-brown to pale brown becoming gradually paler towards the apex. Conidiogenous cells 16.5–30(–38) × 3.5–5 μm, tapering to 1.5–2 μm just below the collarette, integrated, terminal, monophialidic, cylindrical to cylindrical-lageniform, subhyaline or pale brown at the base becoming hyaline to subhyaline towards the apex, smooth-walled; collarettes funnel-shaped, 3.5–4.5 μm wide, 1.5–2.5 μm deep. Conidia in slimy droplets, hyaline in mass, (11.5–)12–17 × (2–)2.5–3(–3.5) µm (mean ± SD = 14.7 ± 1.5 × 2.5 ± 0.3 µm), of two types, narrower and longer, 13.5–17(–17.5) × 2.5–3.5 μm (mean ± SD = 15.2 ± 1.0 × 2.8 ± 0.3 µm), and shorter and usually wider, 11.5–13.5(–14) × 3–3.5(–4) μm, falcate, asymmetrical, rounded at the apical end, with an inconspicuous scar at the basal end, hyaline, aseptate, smooth-walled, with simple, straight or gently curved setulae at both ends, 5–8 μm long; setulae inserted on the concave sides of the conidia.
Description on MLA.
Vegetative hyphae hyaline to pale brown. Setae absent. Conidiophores 95–150(–195) μm long, 3.5–4.5 μm wide, conidiogenous cells 25–35 × 3.5–4(–4.5) μm, tapering to 1.5 µm just below the collarette, integrated, terminal, polyphialidic, usually cylindrical, pale brown to subhyaline, smooth-walled; collarette funnel-shaped, 3.5–4(–4.5) μm wide, 1.5–2 μm deep. Conidia in slimy droplets, of two types, narrower and longer (13–)13.5–15.5(–17) × 2.5–3 µm (mean ± SD = 14.4 ± 0.9 × 2.7 ± 0.2 µm), usually slightly wider and shorter 11–13 × 2.5–3.5 µm (mean ± SD = 12.0 ± 0.7 × 3.1 ± 0.3 µm), falcate, asymmetrical, hyaline, with simple setulae 3.5–5.5(–7.5) μm long at both ends.
Culture characteristics.
On CMD colonies 80–85 mm diam, circular, flat, margin fimbriate, aerial mycelium restricted mainly to the centre and margin of the colony, sparsely lanose, floccose centrally becoming mucoid towards the margin, cobwebby at the margin, colony centre whitish, pale brown to creamy towards the margin, pale brown pigment diffusing from the centre of the colony to the agar; reverse creamy. On MLA colonies 65–70 mm diam, circular, slightly raised, margin filiform, lanose, floccose, colony centre whitish becoming brown-grey towards the margin with a brown outer zone of submerged growth, pale brown pigment diffusing to the agar; reverse dark brown. On OA colonies 89–95 mm diam, circular, raised, margin filiform, aerial mycelium occasionally reduced or absent, colonies similar to those on MLA, lanose, floccose, locally mucoid and smooth or cobwebby, whitish becoming dark grey at the margin, a dark brown to burgundy brown pigment diffusing to the agar; reverse dark grey. On PCA colonies 78–89 mm diam, circular, flat to slightly raised, margin entire to weakly filiform, lanose, floccose, occasionally locally mucoid and smooth or with sparse decumbent aerial hyphae, cobwebby at the margin, whitish becoming brown towards the margin; reverse olivaceous brown. Sporulation on MLA, OA, CMD after 8 wk.
Other specimen examined.
France – Ariège • Pyrénées Mts., Rimont, Le Baup stream, ca. 1.5 km from the village along D18 road; alt. 550 m; 12 Jun. 2009; on submerged wood of Fraxinus excelsior; J. Fournier leg.; J.F. 09153 (PRA-16320, culture CBS 127692) • Ibid.; 23 May 2008; on submerged wood of Alnus glutinosa; J. Fournier & M. Delpont leg.; J.F. 08124 (PRA-16321, culture CBS 126573). United Kingdom • Liverpool, University Campus Liverpool; 1992; on submerged dead leaf in a pool; G.L. Hennebert leg.; (culture MUCL 34876).
Habitat and distribution.
All four isolates analysed in this study originated from the freshwater environment and occurred on decaying wood or leaves of Alnus glutinosa, Fraxinus excelsior and other unidentified hosts. Based on the BLASTn search of the ITS sequence of C. paniculata in GenBank, two isolates from roots of Elymus mollis (ITS: KU838460, KU839605, David et al. 2016), a native beach grass on the USA Pacific Northwest coast, and one environmental soil sample from ancient woodland enclosing a conifer plantation in the United Kingdom (ITS: KM374380, Johnson et al. 2014) showed 100 % sequence similarity. Based on these records, C. paniculata is known from the north temperate region in Europe in France and United Kingdom and North America in USA, Oregon.
Notes.
Among known Codinaea species, C. assamica is similar to C. paniculata, but differs by slightly longer (14.6–16.8 × 2.6–2.8 µm) conidia with longer (9.6–12.8 µm) setulae (Hughes and Kendrick 1968) and formation of polyphialides in vivo. Dictyochaeta terminalis (Lin et al. 2019) matches C. paniculata in monophialidic conidiogenous cells formed in vivo and aseptate conidia, which are slightly longer and wider (14.7–20.7 × 2.9–4.2 µm). The ITS sequences of examined strains of C. paniculata exhibit 99.94–100 % similarity; their comparison with ITS sequences of the closely related C. assamicaCBS 242.66 (MH858788) and D. terminalis GZCC 18-0085 (MN104613) showed 89.7 % and 89.85 % similarity, respectively.
Paragaeumannomyces
Matsush., Matsush. Mycol. Mem. 10: 156. (2003) [2001]. Emend. Réblová & A. N. Miller.
C591DECF-7F55-5BBF-90E8-6BD0078A8660
Type species.
Paragaeumannomyces sphaerocellularis Matsush., Mycol. Mem. 10: 156. (2003) [2001].
Description.
Teleomorph: Ascomata perithecial, non-stromatic, superficial, subglobose to conical, solitary, in small groups or aggregated, sometimes collapsing laterally upon drying, ranging from white, yellow-white, light fawn-grey, ginger-brown, reddish-brown, russet to dark brown, papillate, glabrous or setose, setae dark brown, acute, opaque, scattered over entire ascoma and/or clustered around the ostiole, centrum sometimes pink to pale red. Ostiole periphysate. Ascomatal wall three-layered; outer layer composed of thin-walled, globose, subglobose to polyhedral cells, sometimes containing pale purple pigment when fresh; middle layer composed of brick-like, dark brown cells with opaque walls; inner layer of flattened, thin-walled, subhyaline cells. Paraphyses persistent, branching, tapering. Asci unitunicate, 8-spored, cylindrical-fusiform, stipitate, apex with a non-amyloid apical annulus. Ascospores asymmetrical, cylindrical-filiform, slightly tapering towards the basal end, multiseptate, hyaline, occasionally light pink, with negative or positive dextrinoid reaction in Melzer’s reagent. Synanamorphs: Craspedodidymum-like. Conidiophores mononematous, semi-macronematous to micronematous, brown, septate, unbranched or reduced to single conidiogenous cells. Conidiogenous cells phialidic, obclavate or broadly lageniform, brown, with an apical opening; collarettes flared or cup-shaped. Conidia globose, subglobose, subangular to triangular, unicellular, hyaline, with setulae. Chloridium-like. Conidiophores mononematous, macronematous, brown, septate, unbranched. Conidiogenous cells phialidic, cylindrical, subhyaline, elongating percurrently, with an apical opening; collarette indistinct or flared. Conidia globose, ovoid to clavate, unicellular, hyaline, non-setulate, accumulating in slimy droplets. [Characteristics of the synanamorphs adopted from Huhndorf and Fernández (2005)].
Notes.
The holotype of P. sphaerocellularis (Japan, Schimizu-cho, Wakayama Pref., on decaying twig of unknown broadleaf tree, Apr. 2000, MFC-21077), the type species of Paragaeumannomyces (Matsushima 2003), was not available to us. A comparison of its protologue with our specimens and descriptions of other scolecosporous species of Chaetosphaeria (Carroll and Munk 1964; Huhndorf and Fernández 2005; Atkinson et al. 2007; Perera et al. 2016), combined with phylogenetic analysis of the ITS-28S sequences of 35 isolates, provided sufficient evidence to consider them congeneric. Paragaeumannomyces is proposed as the correct name for this morphologically and phylogenetically well-delimited group of chaetosphaeriaceous fungi. The width of the ascus is sometimes variable even within a single collection depending on the arrangement of ascospores in the sporiferous part, whether they are 2–3-seriate, 4-seriate end-to-end or in a fascicle.
Members of Paragaeumannomyces display a wide geographical distribution pattern; they have a predominantly pantropical distribution in Central America and Asia but were also encountered in the subtropical and temperate climate zones of Europe, Japan, New Zealand and North America.
Key to Paragaeumannomyces species
| 1 | Ascomata almost white, yellowish-white or light fawn-grey when fresh, dark yellow, buff, tawny to dark ginger-brown or yellow-grey when dried, purple pigment absent in cells of the outer ascomatal layer, setae absent | 2 |
| – | Ascomata reddish, reddish-brown, russet or brown, sometimes with red surface crystals, globose cells in the outer ascomatal layer may contain pale purple pigment when fresh, setae present or absent | 3 |
| 2 | Ascomata almost white to yellowish-white, translucent, areolate when fresh, with a distinct black papilla, asci 270–295 × 18.5–20.5 μm, ascospores (3–)5–11-septate, (55–)69–86 × 5–6.5(–7) μm | P. albidus |
| – | Ascomata light fawn-grey, not areolate when fresh, papilla indistinct or absent, asci 215–270 × 11–14 µm, ascospores (7–)11(–13)-septate, 62–88 × 4.5–6 µm | P. bombycinus |
| 3 | Ascomatal setae present only at the apex or absent | 4 |
| – | Ascomatal setae present at the apex and also scattered over entire surface of the ascoma, or setae only scattered over ascoma, occasionally absent | 8 |
| 4 | Ascomata with red surface crystals, ascospores 7-septate, 80–100 × 3.5–4.2 μm, anamorph craspedodidymum-like, conidia globose to subglobose in a vertical position, with setulae | P. rubicundus |
| – | Ascomata without red surface crystals, ascospores with seven or more septa | 5 |
| 5 | Ascomata more than 500 μm diam, setae occasionally absent, ascospores 7-septate, 50–100 × 4.5–6 μm, anamorph craspedodidymum-like, conidia oblate to horizontally oblong with a short abscission scar or frill, without setulae, tropical distribution | P. lapazianus |
| – | Ascomata less than 500 μm diam, setae present at the apex, ascospores with seven or more septa, temperate and subtropical distribution | 6 |
| 6 | Ascospores (7–)11–13-septate, (90–)95–123.5 × 4–5(–5.5) μm, asci 210–295 × (16.5–)17–24.5 μm | P. granulatus |
| – | Ascospores with up to 11 septa, 87 μm and shorter | 7 |
| 7 | Ascospores 9–11-septate, (58–)60.5–80.5 × (3–)3.5–4.5(–5) μm, asci (134–)140–174(–189) × 11–13(–14) μm | P. smokiensis |
| – | Ascospores (5–)7–9(–11)-septate, (62–)65–87 × (3.5–)4–5.5 μm, asci (185–)195–240 × 12–14.5(–15.5) μm | P. abietinus |
| 8 | Setae present at the apex and also scattered over entire surface of ascoma, ascospores 7-septate | 9 |
| – | Setae scattered over entire surface of ascoma, occasionally absent, ascospores with seven or more septa | 10 |
| 9 | Ascospores (50.4–)52.5–68 × 3.5–4.5 μm, craspedodidymum- and chloridium-like synanamorphs, conidia without setulae | P. longisporus |
| – | Ascospores (64.5–)68.5–86.5(–88.5) × (3–)3.5–4.5 μm, anamorph unknown | P. sabinianus |
| 10 | Ascospores 7-septate | 11 |
| – | Ascospores with more than seven septa | 14 |
| 11 | Asci up to 152 μm long | 12 |
| – | Asci 150 μm and longer | 13 |
| 12 | Ascomatal setae 60–75 μm long, ascospores 65–75 × 3–4 µm, asci 123–140 × 10–11 µm, craspedodidymum-like anamorph, conidia without setulae, purple-pigmented aleuriospore-like cells present in culture | P. panamensis |
| – | Ascomatal setae 38–47 μm long, ascospores 63.3–75 × 2.3–3.7 µm, asci 120–152 × 10.7–13.3 µm, anamorph unknown | P. garethjonesii |
| 13 | Ascospores (50–)60–100(–150) × 3–3.75(–4.5) µm, asci (150–)180–250(–350) × 10–20(–27) µm, craspedodidymum-like anamorph, conidia with or without setulae, some strains also with a chloridium-like synanamorph | P. raciborskiis. lat. (fideHuhndorf and Fernández 2005; as Paragaeumannomyces sp. 1–4 in the phylogeny, Fig. 2) |
| – | Ascospores (57.5–)60–73(–75) × (3.5–)4–4.5(–5) μm, asci (152–)174–221(–227) × 10.5–15(–20) μm, anamorph unknown | P. elegans |
| 14 | Ascospores 5–10-septate, 65–90 × 3–4 µm, asci 105–125 × 10–12.5 µm | P. sphaerocellularis |
| – | Ascospores 13–16-septate, 50–65 × 2–4 μm, asci 70–100 × 10–13 μm | P. raciborskiis. str. (fidePenzig and Saccardo 1897; Carroll and Munk 1964) |
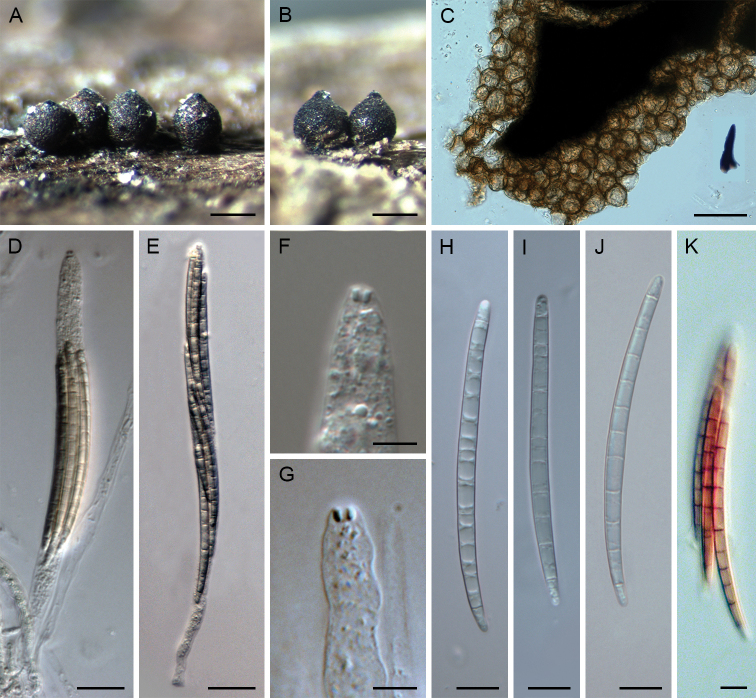
Paragaeumannomyces abietinus
Réblová, J. Fourn. & A.N. Mill. sp. nov.
82B6D25F-DF0B-5E6E-8C85-4882CF07D200
836527
Figure 4.
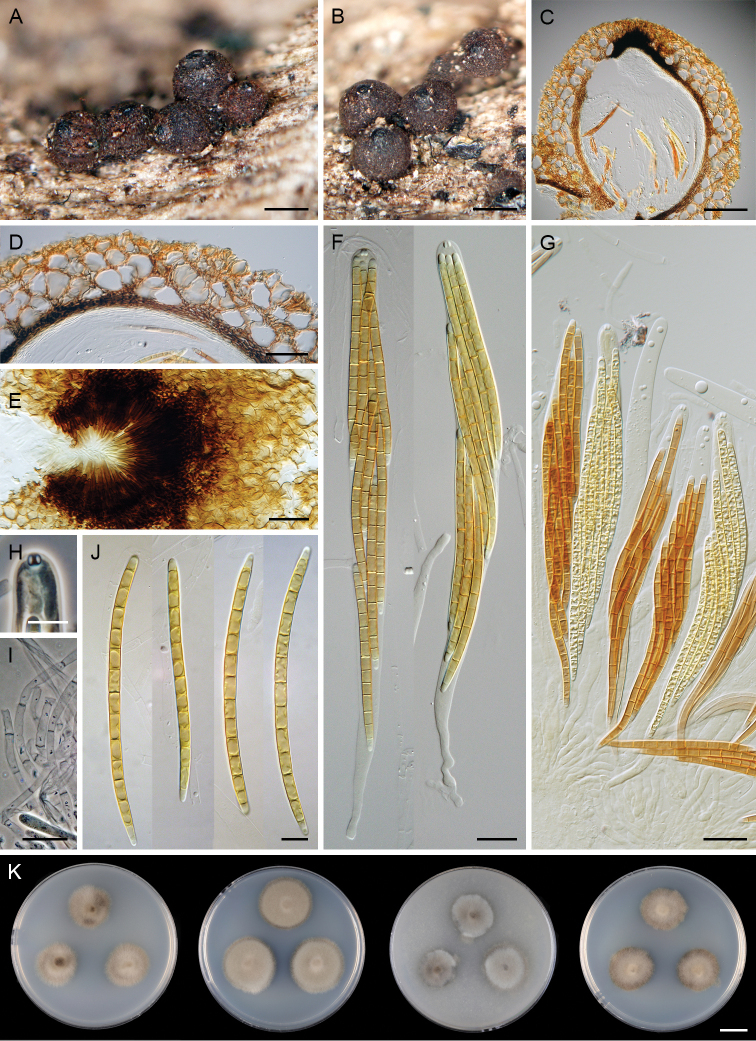
Paragaeumannomyces abietinus. A, B ascomata C, D, F vertical section of ascomal wall E vertical section of the ascomal wall and papilla with apical setae G, H ascospores I, J asci K, L ascal apex with apical ring M paraphyses N colonies on CMD, MLA, OA and PCA after 4 wk (from left to right). Images: PRA-16327 (A); CBS 145351 (B, F–L, N); PRA-16324 (C); PRA-16325 (D, E, M). Scale bars: 250 μm (A, B); 200 μm (C); 50 μm (D–F); 20 μm (I, J, M); 10 μm (G, H, K, L); 1 cm (N).
Typification.
France – Ariège • Pyrénées Mts., Ustou, Cirque de Cagateille, path up to the La Hillette lake, mixed Abies forest; alt. 1550 m; 18 Jul. 2018; on decaying wood of a trunk of Abies alba; J. Fournier leg.; J.F. 18057 (holotype: PRA-16323!, ex-type culture CBS 145351).
Etymology.
Referring to the host Abies alba.
Description on the natural substrate.
Teleomorph: Ascomata perithecial, non-stromatic, superficial, solitary or in small groups, 350–450 μm diam, 360–500 μm high, broadly conical, collapsing laterally upon drying, finely roughened, dark reddish-brown, glabrous except for the black conical papilla, with dark brown, stiff, acute setae, 32–40 × 3–4 μm, densely clustered around the ostiole; centrum pink. Ostiole periphysate. Ascomatal wall leathery, three-layered. Outer layer of textura angularis, 33–58 μm thick, consisting of thin-walled, globose, subglobose to polyhedral, dark ginger-brown to reddish-brown cells, 6.5–11 μm diam, grading into smaller cells towards the exterior. Middle layer of textura prismatica, 18–25 μm thick, composed of thick-walled, polyhedral, dark brown, melanised cells. Inner layer of textura prismatica, 10–15 μm thick, composed of thin-walled, flattened and elongated hyaline cells. Paraphyses abundant, hyaline, sparsely branched, septate, 4.5–7 μm wide, tapering to 2–2.5 μm, longer than the asci. Asci (185–)195–240 × 12–14.5(–15.5) μm (mean ± SD = 209.2 ± 12.0 × 14.1 ± 0.8 μm), (145–)155–205 µm (mean ± SD = 172.6 ± 14.3 μm) long in the sporiferous part, cylindrical-fusiform, stipitate, apically rounded, ascal apex non-amyloid with a distinct apical annulus 3–3.5 μm wide, 2–3 μm high. Ascospores (62–)65–87 × (3.5–)4–5.5 μm, filiform to cylindrical, straight or slightly curved to sigmoid, hyaline, light pink in mass, with dextrinoid reaction in Melzer’s reagent turning reddish-brown except for the end cells which remain hyaline, (5–)7–9(–11)-septate, septa often unevenly distributed, not constricted or slightly constricted at the septa, especially at the septa above and below the middle, asymmetrical, rounded at the apical end, tapering towards the basal end, with one or two guttules in each cell, 2–3-seriate or 4-seriate and partially overlapping or 4-seriate forming two fascicles end to end. Anamorph: Unknown.
Culture characteristics.
On CMD colonies 10–11 mm diam, circular, slightly convex, margin entire to weakly fimbriate, lanose, beige-brown with a dark brown outer zone of submerged growth, dark brown pigment diffusing from the colony margin to agar; reverse dark brown to black. On MLA colonies 12–15 mm diam, circular, slightly convex, margin entire, lanose, floccose, cobwebby at the margin, beige-brown with a dark brown outer zone of submerged growth, brown pigment diffusing from the colony margin to agar; reverse dark brown. On OA colonies 8–9 mm diam, circular, convex, margin entire, lanose, beige-brown, with a paler outer ring; reverse brown. On PCA colonies 14–15 mm diam, circular, convex, margin entire, lanose, floccose, cobwebby towards the margin, beige, pale brown towards the margin; reverse brown. Sporulation absent on all media, even after prolonged incubation (> 3 mo).
Other specimen examined.
Ukraine • Carpathian Mts., Kvasi, Bliznica near Rachiv, right bank of the upper flow of the Tisa river; alt. 1000 m; 28 Jun. 1997; on decaying wood of Abies alba; M. Réblová leg.; M.R. 946 (PRA-16324). • Ibid.; M.R. 947 (PRA-16325). • Ibid.; M.R. 959 (PRA-16326). Ukraine • Carpathian Mts., Massif Boržava, Guklivij; 21 Jul. 1998; on decaying wood of Abies alba; M. Réblová leg.; M.R. 1309 (PRA-16327).
Habitat and distribution.
All specimens of P. abietinus occur on decaying wood of Abies alba. The species has been collected in mountain areas and is known in Europe in France and Ukraine.
Notes.
Attempts to cultivate this species were unsuccessful for the Ukrainian specimens; the ascospores germinated over five days with long inflated germ tubes from both ends but did not grow after isolation on agar medium. The axenic culture derived from the ascospore isolate of the French material yielded sterile mycelium only.
Paragaeumannomyces abietinus is similar to P. rubicundus and P. lapazianus in reddish-brown ascomata, the arrangement of setae around the ostiole and distribution in the north temperate region. Paragaeumannomyces rubicundus (Huhndorf and Fernández 2005) can be distinguished from the present species in having 7-septate, longer (80–100 × 3.5–4.2 μm) ascospores and red surface crystals; P. lapazianus has 7-septate ascospores and a broader range of ascospore lengths including shorter and broader ascospores [(45–)50–100(–120) × (3–)4.5–6(–7) µm] and larger ascomata [(400–)500–950 µm diam, 525–825(–1025) µm high]. In the ITS-28S phylogenetic tree (Fig. 2), P. abietinus was clustered with P. granulatus (New Zealand) and P. smokiensis (USA). These species are morphologically highly similar; they share glabrous, dark brown to reddish-brown ascomata except for the black papilla containing short, appressed setae, and ascospores exhibiting a dextrinoid reaction in Melzer’s reagent. Paragaeumannomyces granulatus differs from P. abietinus in longer [(90–)95–123.5 μm], (7–)11–13-septate ascospores, while P. smokiensis is distinguished from the latter species by shorter and slightly narrower asci [(134–)140–174(–189) × 11–13(–14) μm] and ascospores with more septa (9–11-septate).
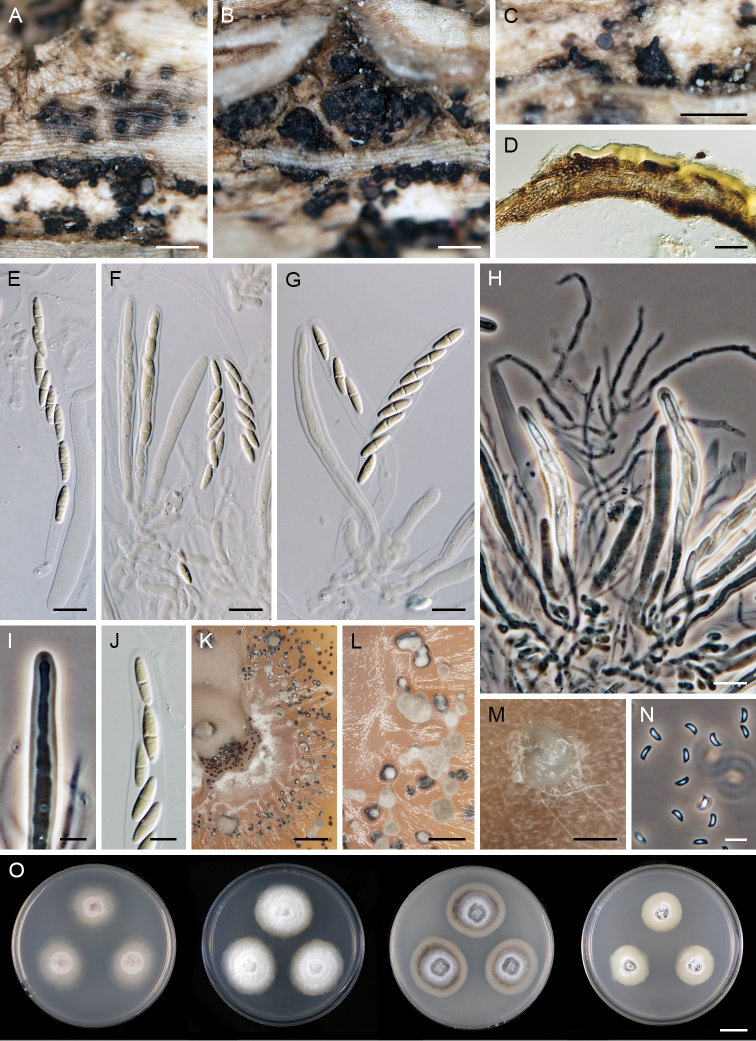
Paragaeumannomyces albidus
(T.J. Atk., A.N. Mill. & Huhndorf) Réblová & A.N. Mill. comb. nov.
6C146A6D-0A83-5132-B176-861389DA2691
836528
Figure 5.
Paragaeumannomyces albidus (PDD 118738). A, B young ascomata C–E mature ascomata F, G asci H paraphyses I, J sporiferous parts of the asci K, L ascospores. Scale bars: 250 μm (A–E); 10 μm (F–L).
Basionym.
Chaetosphaeria albida T.J. Atk., A.N. Mill. & Huhndorf, New Zealand J. Bot. 45: 688. 2007.
Specimens examined.
New Zealand – Tasman • Tasman District, Abel Tasman National Park, Pigeon Saddle point, unpaved road between Tata Beach and Totaranui ca. 10 km NW of Totaranui; 24 Feb. 2003; on decaying wood of Nothofagus sp. buried in soil; M. Réblová leg.; M.R. 2605/NZ 76 (PDD 118737). – West Coast • Westland District, Arthur’s Pass National Park, Kelly Shelter ca. 5 km W of Otira, Cockayane Nature Walk, a podocarp-broadleaf forest; 16 Mar. 2003; on decaying wood of a trunk; M. Réblová leg.; M.R. 2840/NZ 351 (PDD 118738). • Ibid.; 16 Mar. 2003; on decaying wood and bark of a branch; M. Réblová leg.; M.R. 2843/NZ 355 (PDD 118739).
Habitat and distribution.
Paragaeumannomyces albidus has been collected on Metrosideros robusta, Metrosideros sp., Nothofagus sp. and other unidentified hosts and is known from New Zealand (Atkinson et al. 2007; this study).
Notes.
For additional illustrations and description, see Atkinson et al. (2007).
Paragaeumannomyces albidus is the only species of the genus characterised by a wide range of ascoma colours that change when ascomata are young and fresh or mature and dried. Different colours were used by Atkinson et al. (2007) to distinguish P. albidus from closely related P. bombycinus. Paragaeumannomyces albidus differs from the latter species in having distinctly papillate ascomata, which are almost white, yellowish-white, areolate and translucent when young except for the black papilla (Fig. 5A, B). In older specimens and after drying, ascomata often become laterally pinched, dark yellow, buff, tawny to dark ginger-brown (Fig. 5C–E). The ascomatal wall of P. albidus is thicker than that of P. bombycinus, with an outer layer containing an external melanised section. In our material, asci were longer than those in the original description, 270–295 × 18.5–20.5 μm long and 155–225 μm long in the sporiferous part vs 220–260 × 16–20 μm fideAtkinson et al. (2007). The size and septation of ascospores matched those given in the protologue, though being slightly longer in the upper range: (3–)5–11-septate, (55–)69–86 × 5–6.5(–7) μm vs (5–)7(–12)-septate, (47–)60–80 × 5–7 μm fideAtkinson et al. (2007). The ascospores exhibited a dextrinoid reaction in Melzer’s reagent turning reddish-brown except for the tips of the end cells that remain hyaline.
Attempts to isolate our specimens of P. albidus in living culture were not successful. Therefore, the DNA was extracted from herbarium material of all three collections, but only ITS1 of PDD 118738 could be amplified and sequenced. Comparison of the ITS1 sequences of our specimen and the holotype of P. albidus revealed 100 % similarity (Fig. 2).
Paragaeumannomyces bombycinus
(T.J. Atk., A.N. Mill. & Huhndorf) Réblová & A.N. Mill. comb. nov.
7FEEA223-D70E-50BC-B481-31BA7DE9C121
836529
Basionym.
Chaetosphaeria bombycina T.J. Atk., A.N. Mill. & Huhndorf, New Zealand J. Bot. 45: 691. 2007.
Habitat and distribution.
The species has been collected on decaying wood of Nothofagus sp. and is known from New Zealand (Atkinson et al. 2007).
Notes.
For description, illustration and holotype information, see Atkinson et al. (2007). Although P. albidus and P. bombycinus share identical ITS sequences and the size of their ascomata, asci and ascospores considerably overlap, the latter species was distinguished by characters in the ascomatal wall, ascoma appearance and ascospore septation. The ascomata of P. bombycinus are light fawn-grey and non-areolate when fresh, the ascomatal wall lacks the external melanised layer or the melanisation is only weakly present, and the black papilla is lacking or indistinct being covered by the outer layer. The ascospores of P. bombycinus are (7–)11(–13)-septate compared to (5–)7(–12)-septate ascospores of P. albidus. Atkinson et al. (2007) considered the ITS sequence identity uninformative in the light of distinct morphologies between the two species. Another explanation to this peculiar case could be that P. bombycinus is a described anomaly within P. albidus based on a single collection. More specimens of the “bombycinus” phenotype need to be examined, and their ITS and possibly other loci analysed.
Paragaeumannomyces elegans
Réblová & A.N. Mill. sp. nov.
4AAE0649-8AE3-55AD-BEB3-ADA68B12DFBF
836530
Figure 6.
Paragaeumannomyces elegans. A, B ascomata C vertical section of ascomal wall D ascal apex with apical annulus E–G asci H ascospores I paraphyses. Images: PDD 118740 (A, B, D, H, I); PDD 118741 (C, E–G). Scale bars: 250 μm (A, B); 20 μm (C); 10 μm (D–I).
Typification.
New Zealand – West Coast • Westland District, Mount Aspiring National Park, Haast, Roaring Billy track; 22 Mar. 2005; on decaying wood; M. Réblová leg.; M.R. 3295/NZ 566A (holotype: PDD 118740!).
Etymology.
Elegans (L) elegant, referring to elegant and lovely ascomata adorned with setae.
Description on the natural substrate.
Teleomorph: Ascomata perithecial, non-stromatic, superficial, in small groups, often gregarious, 290–350 μm diam, 280–350 μm high, subglobose to slightly conical, dully glossy, brown with a light grey tinge except for the tiny black papilla composed of thick-walled, dark brown cells, ascomata densely setose, setae 28–60 × 4.5–6 μm, stiff, acute, dark brown, thick-walled, opaque. Ostiole periphysate. Ascomatal wall leathery, three-layered. Outer layer of textura angularis, 33–41 μm thick, consisting of thin-walled, globose to subglobose to polyhedral, reddish-brown cells ca. 5–12 μm diam. Middle layer of textura prismatica, 9.5–18 μm thick, composed of thick-walled, polyhedral, elongated, dark brown, melanised cells. Inner layer of textura prismatica, 5–8 μm thick, composed of thin-walled, flattened and elongated hyaline cells. Paraphyses abundant, hyaline, sparsely branched, septate, 3.5–5 μm wide, tapering to ca. 2 μm, longer than the asci. Asci (152–)174–221(–227) × 10.5–15(–20) μm (mean ± SD = 204.8 ± 13.7 × 12.3 ± 1.5 μm), (129–)141–197(–204) μm (mean ± SD = 168.2 ± 17.2 μm) long in the sporiferous part, cylindrical-fusiform, stipitate, apically rounded, ascal apex non-amyloid with a distinct apical annulus 2.5–3 μm wide, 2–2.5 μm high. Ascospores (57.5–)60–73(–75) × (3.5–)4–4.5(–5) μm (mean ± SD = 65.5 ± 3.2 × 4.1 ± 0.2 μm), filiform to cylindrical, straight or slightly curved to sigmoid, hyaline, with negative or very weak dextrinoid reaction in Melzer’s reagent, 7–septate, septa usually obscured by large guttules, not constricted at the septa, asymmetrical, rounded at the apical end, slightly tapering towards the basal end, with one or two guttules in each cell, 2–3-seriate or 3–4-seriate, partially overlapping. Anamorph: Unknown.
Other specimen examined.
New Zealand – Otago • Clutha District, The Catlins, Catlins Coastal Rain Forest Park, MacLennan Range, Lake Wilkie Walk; 17 Mar. 2005; on decaying wood of a branch; M. Réblová leg.; M.R. 3289/NZ 549 (PDD 118742). – West Coast • Westland District, Ship Creek Point, Kahikatea Swamp Forest walk; 8 Mar. 2003; on decaying wood; M. Réblová leg.; M.R. 2819/NZ 329 (PDD 118741). – West Coast • Westland District, Ross, Totara Valley Road, 12 Apr. 2005; on decaying wood; M. Réblová leg.; M.R. 3486/NZ 775 (PDD 118743).
Habitat and distribution.
The present species is a saprobe on decaying wood of Nothofagus sp. and other unidentified hosts, known from New Zealand (Atkinson et al. 2007; this study).
Notes.
Paragaeumannomyces elegans is distinguishable from other members of the genus by densely setose, dull glistening brown ascomata with a light grey tinge, which gives them an almost grey appearance when dried. The new species resembles P. garethjonesii (Perera et al. 2016) and P. panamensis (Huhndorf and Fernández 2005) in 7-septate ascospores and setose ascomata with acute, stiff, opaque setae scattered over the entire surface, but differs from them in larger ascomata, asci and wider ascospores (for a detailed comparison see the key).
Comparison of the ITS sequence of the holotype of P. elegans with available Paragaeumannomyces sequences revealed 100 % sequence similarity with a specimen PDD 92561 (New Zealand, Taupo, Ohakune, ITS: EUO37895) tentatively identified as P. raciborskii (Atkinson et al. 2007) (Fig. 2).
Paragaeumannomyces garethjonesii
(R.H. Perera, Maharachch. & K.D. Hyde) Réblová & A.N. Mill. comb. nov.
2A69DA51-254D-5E22-89E4-1177408D8757
836531
Basionym.
Chaetosphaeria garethjonesii R.H. Perera, Maharachch. & K.D. Hyde, Mycosphere 7: 1308. 2016.
Habitat and distribution.
Paragaeumannomyces garethjonesii was collected on a Fabaceae seed pod and is known from Asia in Thailand (Perera et al. 2016).
Notes.
For description, illustration and holotype information see Perera et al. (2016). Paragaeumannomyces garethjonesii resembles P. panamensis (Huhndorf and Fernández 2005) in size of ascomata, which are the smallest (up to 250 μm diam and 270 μm high) within the genus, setae scattered over the entire ascoma, overlapping length of their asci and 7-septate ascospores, but it differs by shorter (38–47 μm) setae, slightly wider (10.7–13.3 µm) asci and the absence of aleuriospore-like cells in culture (for a detailed comparison see the key).
Paragaeumannomyces granulatus
Réblová & A.N. Mill. sp. nov.
F71FA226-0E8C-56F1-889D-E6C6B4D2D148
836532
Figure 7.
Paragaeumannomyces granulatus (PDD 118744). A, B ascomata C, D vertical section of ascomal wall E papilla and the upper part of ascomal wall in surface view F, G asci H ascal apex with apical annulus I paraphyses J ascospores K colonies on CMD, MLA, OA and PCA after 4 wk (from left to right). Scale bars: 250 μm (A, B); 100 μm (C); 25 μm (G); 20 μm (D–F, I); 10 μm (H, J); 1 cm (K).
Typification.
New Zealand – West Coast • Westland District, Hokitika, Mananui Point, Lake Mahinapua, Swimmers Beach walks; 5 Mar. 2003; on decaying wood; M. Réblová leg.; M.R. 2715/NZ 216 (holotype: PDD 118744!, ex-type culture ICMP 15133).
Etymology.
Granulum (L), granule, small grain, diminutive of granum, referring to the roughened surface of the ascomatal wall composed of globose cells, which appears granulose in the surface view.
Description on the natural substrate.
Teleomorph: Ascomata perithecial, non-stromatic, superficial, solitary or in small groups, 380–495 μm diam, 415–530 μm high, subglobose to conical, finely roughened, dark brown to dark reddish-brown, sometimes with irregular reddish colour except for the black papilla; papilla composed of dark brown, thick-walled, cylindrical to subulate, apically narrowly rounded soft setae; centrum pink. Ostiole periphysate. Ascomatal wall leathery, three-layered. Outer layer of textura angularis, 95–115 μm thick, consisting of thin-walled, globose to subglobose ginger-brown cells ca. 27–33 μm diam, grading into smaller cells 8–16 μm diam. Middle layer of textura prismatica, 14–21 μm thick, composed of thick-walled, polyhedral, elongated, dark brown, melanised cells. Inner layer of textura prismatica, 7–12 μm thick, composed of thin-walled, flattened and elongated hyaline cells. Paraphyses abundant, hyaline, sparsely branched, septate, 3.5–5 μm wide, tapering to 2–2.5 μm, longer than the asci. Asci 210–295 × (16.5–)17–24.5 μm (mean ± SD = 239.7 ± 15.5 × 20.3 ± 2.1 μm), 165–200(–250) μm (mean ± SD = 184.7 ± 10.3 μm) long in the sporiferous part, cylindrical-fusiform, stipitate, apically rounded, ascal apex non-amyloid with a distinct apical annulus 3.5–4 μm wide, 2.5–3(–3.5) μm high. Ascospores (90–)95–123.5 × 4–5(–5.5) μm (mean ± SD = 101.4 ± 10.2 × 4.8 ± 0.4 μm), filiform to cylindrical, straight or slightly curved to sigmoid, hyaline to very light pink, light pink-brown in mass, with dextrinoid reaction in Melzer’s reagent turning reddish-brown except for the end cells which remain hyaline, (7–)11–13-septate, septa often unevenly distributed, not constricted or slightly constricted at the septa, asymmetrical, broadly rounded at the apical end, tapering and narrowly rounded at the basal end, with one or two guttules in each cell, 2–3-seriate or 4-seriate and partially overlapping. Anamorph: Unknown.
Culture characteristics.
On CMD colonies 14–16 mm diam, circular, convex, margin fimbriate, lanose, grey-brown, reverse dark brown to almost black. On MLA colonies 19–20 mm diam, circular, raised, margin entire to weakly fimbriate, lanose, beige-brown, with a dark brown outer zone, reverse dark brown to almost black. On OA colonies 13–16mm diam, circular, raised, margin weakly fimbriate, lanose, beige-grey becoming grey towards the periphery, reverse dark brown to almost black. On PCA colonies 15–17 mm diam, circular, slightly convex, margin weakly fimbriate, lanose, beige, pale brown at the margin, reverse black. Sporulation absent on all media.
Other specimen examined.
New Zealand – Auckland • Auckland district, Waitakere Ranges Nature Reserve, Anawhata Road; 24 Apr. 2005; on decaying wood; M. Réblová leg.; M.R. 3543/NZ 838 (PDD 118745).
Habitat and distribution.
A saprobe on decaying wood, known from New Zealand.
Notes.
Paragaeumannomyces granulatus most closely resembles P. abietinus in the ascoma appearance, pink content of the ascoma centrum, ascospores with usually more than seven septa and positive dextrinoid reaction in Melzer’s reagent but both species are separated by size of asci and ascospores. The ascospores of P. abietinus are (5–)7–9(–11)-septate and shorter [(62–)65–87 μm] and asci are shorter and narrower [(185–)195–240 × 12–14.5(–15.5) μm].
Paragaeumannomyces lapazianus
(G.C. Carroll & Munk) Réblová & A.N. Mill. comb. nov.
647B4995-143D-5A58-8688-B841BCDEBCC5
836533
≡ Chaetosphaeria lapaziana (G.C. Carroll & Munk) F.A. Fernández & Huhndorf, Fung. Diver. 18: 49. 2005.
Basionym.
Lasiosphaeria lapaziana G.C. Carroll & Munk, Mycologia 56: 90. 1964.
Habitat and distribution.
Paragaeumannomyces lapazianus is common on decaying wood in the neotropics and is known from the Caribbean in Puerto Rico and Jamaica, from Central America in Costa Rica, and from South America in French Guiana (Carroll and Munk 1964; Huhndorf and Fernández 2005).
Notes.
For description, illustration and holotype information see Carroll and Munk (1964) and Huhndorf and Fernández (2005). Paragaeumannomyces lapazianus has 7-septate ascospores and the largest ascomata in the genus, (400–)500–950 μm diam and 525–825(–1025) μm high fideHuhndorf and Fernández (2005), and forms a craspedodidymum-like anamorph in vitro. The anamorph is characterised by inflated, pigmented conidiogenous cells with a flared collarette and oblate to horizontally oblong conidia with a short abscission scar or frill and without setulae.
Paragaeumannomyces longisporus
(Sacc.) Réblová & A.N. Mill. comb. nov.
6E1521E1-E431-5840-BA60-37AE8FC6259A
836534
Figure 8.
Paragaeumannomyces longisporus. A, B ascomata C, D vertical section of ascomal wall E vertical section of ascomal wall and papilla with apical of setae F ascomal wall with setae G globose cells of the outer layer of the ascomal wall H, I asci J paraphyses K–N ascospores. Images: ILLS00121385 (A, B, G); S.M.H. 3860 (C, E, M); S.M.H. 2519 (D); ILLS00121386 (F, H–J, K); S.M.H. 2758 (L); S.M.H. 3809 (N). Scale bars: 250 μm (A–D); 50 μm (E–G); 20 μm (H–J); 10 μm (K–N).
≡ Sphaeria longispora Ellis, Bull. Torrey Bot. Club 6: 135. 1877 non Currey 1859 nec Karsten 1873. (Nom. illegit., Art. 53.1)
≡ Ceratostomella longispora (Sacc.) Cooke, Grevillea 17: 50. 1889.
≡ Chaetosphaeria longispora (Sacc.) P.M. Kirk, Index Fung. 120: 1. 2014.
= Lasiosphaeria ellisii M.E. Barr, Mycotaxon 46: 48. 1993.
≡ Chaetosphaeria ellisii (M.E. Barr) Huhndorf & F.A. Fernández, Fung. Diver. 19: 27. 2005.
Basionym.
Ophioceras longisporum Sacc., Syll. fung. 2: 360. 1883.
Specimens examined.
USA – Tennessee • Cocke Co., Great Smoky Mountains National Park, Cosby, Cosby Nature Trail; alt. 716 m; 23 Mar. 2007; on decaying wood; A.N. Miller, P. Chaudhary & H.A. Raja leg.; A.N.M. 1134 (ILLS00121385). • Ibid.; 19 Jul. 2007; T.J. Atkinson & P. Chaudhary leg.; A.N.M. 1250 (ILLS00121386).
Habitat and distribution.
The species occurs on decaying wood and is known from the north temperate region in the USA (Indiana, New Jersey, North Carolina, South Carolina, Tennessee) (Barr 1993; Huhndorf and Fernández 2005; this study).
Notes.
For description and illustration, refer to Barr (1993), and Huhndorf and Fernández (2005). Our specimens of P. longisporus match well the fungus described and illustrated by Barr (1993) based on the holotype of Sphaeria longispora (Ellis 1877), only the asci are longer and agree with the measurements given by Huhndorf and Fernández (2005). Based on examination of our material (Fig. 8), ascomata are reddish-brown, subglobose to globose, with three-layered wall, setose, setae dark brown, acute, scattered over entire ascomatal surface and also aggregated around the ostiole, asci (140.5–)165–183 × 10.5–12.5 μm and (114–)133–157.5 μm long in the sporiferous part, ascospores (50.5–)52.5–68 × 3.5–4.5 μm, 7-septate, asymmetrical, tapering towards the basal end, with a negative or very weak dextrinoid reaction in Melzer’s reagent.
Sphaeria longispora (Ellis 1877) is a later homonym of S. longispora (Currey 1859) and S. longispora (Karsten 1873). Barr (1993) revised the holotype of S. longispora Ellis (USA, New Jersey, Newfield, on fallen branch of Kalmia latifolia, 20 Jul 1874, J.B. Ellis, NY) and concluded that the fungus is better placed in Lasiosphaeria due to filiform, septate ascospores and setose ascomata and proposed a replacement name, Lasiosphaeria ellisii as a nomen novum. This species was later transferred to Chaetosphaeria by Huhndorf and Fernández (2005) as Ch. ellisii. Kirk (2014) considered the first combination of S. longispora in Ophioceras by Saccardo (1883) as the earliest legitimate name of the taxon in the same rank (Art. 41.3) to replace Sphaeria longispora Ellis. Ophioceras longisporum Sacc. is, therefore, a basionym for all future combinations. Kirk (2014) proposed a new combination Chaetosphaeria longispora but erroneously cited S. longispora as the basionym, which does not affect the valid publication of the new combination (Art. 41.8c).
Paragaeumannomyces panamensis
(Huhndorf & F.A. Fernández) Réblová & A.N. Mill. comb. nov.
A7433647-E7EC-5742-9980-66B243B62468
836535
Basionym.
Chaetosphaeria panamensis Huhndorf & F.A. Fernández, Fung. Diver. 19:33. 2005.
Habitat and distribution.
Paragaeumannomyces panamensis has been collected on decaying wood of Pinus sp. and an unidentified host, and it is known from Asia in Thailand and Central America in Panama (Huhndorf and Fernández 2005; Perera et al. 2016).
Notes.
For description, illustrations and holotype information see Huhndorf and Fernández (2005) and Perera et al. (2016). Paragaeumannomyces panamensis is similar to P. sphaerocellularis in ascoma and is more or less comparable in size of asci, but differs by shorter, always 7-septate ascospores and occurrence in the tropics. For detailed comparison, see notes to P. sphaerocellularis.
Paragaeumannomyces raciborskii
(Penz. & Sacc.) Réblová & A.N. Mill. comb. nov.
65ADA667-339C-51B9-9101-DFEEDA6816E9
836536
≡ Lasiosphaeria raciborskii (Penz. & Sacc.) G.C. Carroll & Munk, Mycologia 56: 91. 1964.
≡ Chaetosphaeria raciborskii (Penz. & Sacc.) F.A. Fernández & Huhndorf, Mycol. Res. 108: 29. 2004.
Basionym.
Ophiochaeta raciborskii Penz. & Sacc., Malpighia 11: 406. 1897.
Habitat and distribution.
Paragaeumannomyces raciborskii has been collected on culms of Chusquea bamboo and other unidentified bamboo species, on palm wood and fruit, and decaying wood of unknown trees. The species has a pantropical geographical distribution and is probably the most commonly encountered species of the genus; it is known from Indonesia in Java and Central America in Costa Rica (Penzig and Saccardo 1897; Carroll and Munk 1964). Other collections published under this name, which may represent different species, originate from Asia in Thailand, the Caribbean in Cuba, Jamaica and Puerto Rico, Central America in Costa Rica and Panama, and South America in Ecuador, French Guiana and Venezuela (Huhndorf and Fernández 2005).
Notes.
For descriptions and illustrations, see Carroll and Munk (1964). The holotype of P. raciborskii (Penzig and Saccardo 1897) originates from decaying wood in Java. In the protologue, the species was described with black, setose ascomata 250 μm diam, short-stipitate asci 130–150 × 9–10 μm and hyaline, multiseptate ascospores 60–70 × 3 μm. Carroll and Munk (1964) redescribed the species based on the holotype and an additional collection from Costa Rica as having setose, reddish-brown ascomata 250–300 μm diam, 13–16-septate ascospores 50–65 × 2–4 μm, and asci 70–100 × 10–13 μm. Huhndorf and Fernández (2005) introduced a broader species concept of P. raciborskii, which was based on numerous specimens of a tropical geographical distribution originating mainly from Central and South America. The species was characterised by setose, reddish, russet or brown ascomata (150–)200–450 μm diam with stiff, dark setae scattered over the entire ascoma or absent in some specimens, 7-septate ascospores (50–)60–100(–150) × 3–3.75(–4.5) μm, and short-stipitate asci (150–)180–250(–350) × 10–20(–27) μm.
Paragaeumannomyces raciborskiifideHuhndorf and Fernández (2005) shows a high degree of ITS sequence variability accompanied by a low phenotypic plasticity, which resulted in the species being polyphyletic and segregated into four lineages labelled Paragaeumannomyces sp. 1–4 (Fig. 2). Two anamorphic craspedodidymum-like morphotypes with and without setulae and a chloridium-like synanamorph have been experimentally linked to several strains of P. ‘raciborskii’ by Huhndorf and Fernández (2005). Although the in vitro anamorphic characters seem promising in becoming another important set of diagnostic features to distinguish species of Paragaeumannomyces, isolated strains often form sterile mycelium in vitro or they are difficult to isolate into living culture.
Paragaeumannomyces rubicundus
(Huhndorf & F.A. Fernández) Réblová & A.N. Mill. comb. nov.
E2E5755E-C058-5925-9B0B-B3EDFA192CA1
836537
Basionym.
Chaetosphaeria rubicunda Huhndorf & F.A. Fernández, Fung. Diver. 19: 39. 2005.
Habitat and distribution.
Paragaeumannomyces rubicundus occurs on decaying wood and is known from the Caribbean in Puerto Rico and from Central America in Costa Rica (Huhndorf and Fernández 2005).
Notes.
For description, illustration and holotype information see Huhndorf and Fernández (2005). Paragaeumannomyces rubicundus is distinguished from other species of the genus by ascomata with red surface crystals not dissolving in water, 3 % KOH or lactophenol. Similar to Paragaeumannomyces sp. 4 (S.M.H. 3119), the craspedodidymum-like anamorph forms conidia with three setulae.
Paragaeumannomyces sabinianus
Réblová & A.N. Mill. sp. nov.
62B7969B-14E2-5D03-97B0-72912621B66F
836538
Figure 9.
Paragaeumannomyces sabinianus. A ascomata. B ascomatal setae C vertical section of ascomal wall D, E ascomal wall F, G upper part of the ascoma with ostiole surrounded by setae H setae from the ostiolar region I paraphyses J ascal apex with apical ring K asci L, M ascospores. Images: ILLS00121384 (A, B, D, E, G–K); S.M.H. 3824 (C, F, L); S.M.H. 3807 (M). Scale bars: 500 μm (A); 20 μm (B, E, G, H); 100 μm (C, D); 25 μm (F); 5 μm (J); 10 μm (I, K–M).
Typification.
USA – Tennessee • Sevier Co., Great Smoky Mountains National Park, Twin Creeks, Twin Creeks Nature Trail, near ATBI plot; alt. 549 m; 11 Oct. 2006; on decaying wood; A.N. Miller & P. Chaudhary leg.; A.N.M. 1011 (holotype: ILLS00121384!).
Etymology.
The species epithet is proposed in honour of Sabine M. Huhndorf for her contribution to mycology and studies in Chaetosphaeria.
Description on the natural substrate.
Teleomorph: Ascomata perithecial, non-stromatic, superficial, usually solitary or in small groups, 250–300(–400) μm diam, 280–320 μm high, subglobose to broadly conical, rarely collapsing laterally upon drying, finely roughened, dark reddish-brown except for a black indistinct papilla, setose, setae 30–41.5 × 4–5 μm, dark brown, stiff, acute, scattered over entire ascoma, shorter and narrower setae 16.5–35 × 2.5–3 μm densely aggregated around the ostiole. Ostiole periphysate. Ascomatal wall leathery, three-layered. Outer layer of textura angularis, 32–53 μm thick, consisting of thin-walled, globose to subglobose, dark orange-brown to reddish-brown cells, 11.5–28 μm diam. Middle layer of textura prismatica, 12–22 μm thick, composed of thick-walled, polyhedral, dark brown, melanised cells. Inner layer of textura prismatica, 3–5 μm thick, composed of thin-walled, flattened and elongated hyaline to subhyaline cells. Paraphyses abundant, hyaline, sparsely branched, septate, 3.5–4.5(–6) μm wide, tapering to 2–2.5 μm, longer than the asci. Asci (154–)161–189 × (11–)12.5–14.5(–15.5) μm (mean ± SD = 174.2 ± 8.7 × 13.0 ± 0.8 μm), (130–)144–165 µm (mean ± SD = 155.2 ± 8.3 μm) long in the sporiferous part, cylindrical-fusiform, stipitate, apically broadly rounded to obtuse, ascal apex non-amyloid with a distinct apical annulus 2.5–3 μm wide, 1.5–2 μm high. Ascospores (64.5–)68.5–86.5(–88.5) × (3–)3.5–4.5 μm, (mean ± SD = 79.1 ± 5.3 × 4.0 ± 0.3 μm), filiform to cylindrical, straight or slightly curved to sigmoid, hyaline, 7-septate, septa often unevenly distributed, not constricted at the septa, asymmetrical, broadly rounded at the apical end and tapering towards the narrowly rounded basal end, with one or two guttules in each cell, 2–3-seriate, rarely 4-seriate, partially overlapping, with a negative or weak dextrinoid reaction in Melzer’s reagent. Anamorph: Unknown.
Other specimen examined.
USA – North Carolina • Macon Co., Coweeta Hydrological Laboratory; 27 Jun. 1998, on decorticated wood; F.A. Fernández leg.; S.M.H. 3807. • Ibid., Horse Cove Drive & Bull Pen Road, alt. 1000 m; 27 Jun. 1998; on decorticated wood; F.A. Fernández leg.; S.M.H. 3824.
Habitat and distribution.
A saprobe on decaying wood, so far known from North America in the USA (North Carolina, Tennessee) (Ellis 1887; Huhndorf and Fernández 2005).
Notes.
Huhndorf and Fernández (2005) reported P. longisporus (as Ch. ellisii) from numerous collections from North America; the phylogenetic analysis of ITS sequences of six specimens resolved this species as a statistically unsupported clade with two strongly supported subclades. Although Huhndorf and Fernández (2005) described P. longisporus with setae scattered over the entire ascoma, in discussion, they admitted the presence of setae also around the ostiole: “In C. ellisii, C. raciborskii and C. panamensis the setae tend to be scattered over the entire surface of the ascomata, however some specimens of C. ellisii may have setae concentrated only at the apex.”
Barr (1993) described the ostiole of the holotype of S. longispora surrounded by a crown of dark brown, stiff setae. We examined three collections tentatively identified as P. longisporus from North America (ILLS00121384, ILLS00121385, ILLS00121386) and in each the ostiole was delimited by densely aggregated acute setae. Apart from the ostiolar setae, additional setae were scattered over the entire ascoma, but they differed by their density among collections. The ascomata and asci of these three collections are comparable in size; the main difference lies in the ascospore length. The specimen ILLS00121384 has longer [(64.5–)68.5–86.5(–88.5) μm] ascospores compared to ILLS00121385 and ILLS00121386, which have shorter [(50.5–)52.5–68 μm] ascospores corresponding to the size given by Barr (1993) for the S. longispora holotype. In the description of P. longisporusfideHuhndorf and Fernández (2005), a wide range of ascospore lengths [(40–)50–75(–80) μm] is given, the upper limit matching the ascospore size of ILLS00121384.
In our ITS-28S phylogeny, P. longisporusfideHuhndorf and Fernández (2005) was resolved as a strongly supported clade (100/1.0/100) with two subclades. The first subclade (100/1.0/100) was introduced as a new species P. sabinianus, including ILLS00121384 (Fig. 9), S.M.H. 3807 (Huhndorf and Fernández 2005: fig. 13) and S.M.H. 3824 (Huhndorf and Fernández 2005: fig. 15) with longer ascospores, distinguished from the second subclade P. longisporus (99/1.0/100) with shorter ascospores (Fig. 8). The anamorph of P. sabinianus is unknown; our specimen was not isolated in axenic culture and the strains S.M.H. 3807 and S.M.H. 3824 formed only sterile mycelium in vitro (Huhndorf and Fernández 2005).
Paragaeumannomyces smokiensis
Réblová & A.N. Mill. sp. nov.
84D781EB-FF49-580A-89E0-73939F49C4D0
836539
Figure 10.
Paragaeumannomyces smokiensis (ILLS00121398). A, B ascomata C globose cells of the outer layer of the ascomal wall and an ostiolar seta D, E asci F, G ascal apex with apical annulus H–K ascospores. Scale bars: 250 μm (A, B); 50 μm (C); 20 μm (D, E); 10 μm (F–K).
Typification.
USA – Tennessee • Sevier Co., Great Smoky Mountains National Park, Greenbrier, alternative side trail to Whaley Cemetery; alt. 549 m; 10 Jul. 2005; on decaying wood; A.N. Miller & A.M. Stchigel leg.; A.N.M. 466 (holotype: ILLS00121398!).
Etymology.
Named after the Great Smoky Mountains National Park from where it was collected.
Description on the natural substrate.
Teleomorph: Ascomata perithecial, non-stromatic, superficial, solitary or in small groups, 270–390 μm diam, 320–400 μm high, subglobose to broadly conical, finely roughened, dark reddish-brown, glabrous except for the black papilla with dark brown, stiff, acute setae, 9.5–13.5 × 2–2.5 μm, densely aggregated around the ostiole. Ostiole periphysate. Ascomatal wall leathery, three-layered; outer layer of textura angularis consisting of globose to subglobose, reddish-brown cells; middle layer composed of brick-like, brown cells; inner layer of flattened, thin-walled, subhyaline cells. Paraphyses abundant, hyaline, longer than the asci, tapering. Asci (134–)140–174(–189) × 11–13(–14) μm (mean ± SD = 158.2 ± 19.4 × 12.3 ± 0.9 μm), cylindrical-fusiform, stipitate, apically narrowly rounded, ascal apex non-amyloid with a distinct apical annulus 2–2.5 μm wide, 1–1.5 μm high. Ascospores (58–)60.5–80.5 × (3–)3.5–4.5(–5) μm (mean ± SD = 69.8 ± 6.3 × 4.0 ± 0.5 μm), filiform to cylindrical, straight or slightly curved, hyaline, 9–11-septate, septa often unevenly distributed, not constricted at the septa, asymmetrical, rounded at the apical end, tapering towards the basal end, with one or two guttules in each cell, 2–3-seriate or 4-seriate and partially overlapping, seldom in a single fascicle. Anamorph: Unknown.
Habitat and distribution.
A saprobe on decaying wood, known only from the USA.
Notes.
The present species is most similar to P. abietinus, the only member of the genus known from Europe. They share dark reddish-brown, glabrous ascomata with short setae surrounding the ostiole. Although the size of ascospores of both species overlap, P. abietinus differs from P. smokiensis by longer [(185–)195–240 × 12–14.5(–15.5) μm] asci and slightly longer and wider ascospores [(62–)65–87 × (3.5–)4–5.5 μm] with usually less septa [(5–)7–9(–11)].
Paragaeumannomyces sphaerocellularis
Matsush., Matsush. Mycol. Mem. 10: 156. (2003) [2001].
893CEAFD-B654-5BC5-A916-AD8E431B547F
Habitat and distribution.
The species was described from dead twigs of an unknown broadleaf tree and is so far known only from the subtropical climate zone of the northern hemisphere in Japan, Wakayama Prefecture (Matsushima 2003).
Notes.
Paragaeumannomyces sphaerocellularis is similar to P. panamensis (Huhndorf and Fernández 2005; Perera et al. 2016) in the morphology of reddish-brown, setose ascomata with acute, dark, opaque setae scattered over the entire surface and hyaline ascospores, but differs from it in larger ascomata 200–350 × 300–425 μm vs 185–235 × 190–270 μm, slightly shorter asci 105–125 μm vs 123–140 µm, and 5–10-septate ascospores longer in their upper range 65–90 × 3–4 μm vs always 7-septate, shorter ascospores 65–75 × 3–4 of P. panamensis. Although the size of the asci may vary, often dependent on the arrangement of ascospores, shorter ascospores with the constant occurrence of seven septa of P. panamensis is considered an important character. In other Paragaeumannomyces species with 7-septate ascospores, such as P. elegans, P. lapazianus and P. rubicundus, the number of seven septa remains constant and is considered a diagnostic feature. While P. sphaerocellularis was collected only once in Japan, two collections of P. panamensis originating from Panama and Thailand suggest that this species has a pantropical distribution. Although the two species are remarkably similar, without molecular evidence we prefer to consider them as separate. For a detailed comparison, see the key.
Striatosphaeria castanea
Réblová & J. Fourn. sp. nov.
E1BF3D38-8FE2-5CFB-941E-87B1ECD83ABF
836540
Figure 11.
Striatosphaeria castanea (CBS 145352). A, B ascomata C asci D ascospores E ascal apex with apical annulus F colonies on CMA with an Urtica dioica stem after 8 wk G–J conidia K–Q conidiophores R colonies on CMD, MLA, OA and PCA after 4 wk (from left to right). Scale bars: 500 μm (A, B); 20 μm (C, E); 10 μm (D, G–Q); 1 cm (F, R).
Typification.
French Guiana • Maripasoula, Saül, sentier des gros arbres, disturbed secondary rainforest; alt. 200 m; 25 Aug. 2018; on the bark of decaying woody liana on the ground associated with Xylaria papillatoides; C. Lechat leg.; GY.J.F. 18140-1 (holotype: PRA-16328!, ex-type culture CBS 145352).
Etymology.
Castanea (Latin) chestnut-coloured, referring to the colour of conidia.
Description on the natural substrate.
Teleomorph: Ascomata perithecial, non-stromatic or formed on rudimentary basal stroma, superficial, solitary or in small groups or dense clusters, 160–200 μm diam, 170–220 μm high, subglobose to broadly conical, dark brown, glabrous, papillate. Ostiole periphysate. Ascomatal wall fragile, carbonaceous, 20–27 μm thick, two-layered. Outer layer of textura prismatica, consisting of brown, polyhedral cells with opaque walls. Inner layer of textura prismatica, consisting of several rows of thin-walled, hyaline, flattened cells. Paraphyses sparse, partially disintegrating at maturity, septate, 3–5 μm wide, tapering to ca. 2.5 μm, longer than the asci. Asci (75–)78–97(–102) × (10.5–)11–14.5 μm (mean ± SD = 87.3 ± 5.0 × 12.5 ± 1.1 µm), (56.5–)65–77(–82.5) μm (mean ± SD = 70.0 ± 4.9 µm) long in the sporiferous part, cylindrical-fusiform, stipitate, apically obtuse, ascal apex with a shallow, non-amyloid apical annulus 3.5–4.5 μm wide, 1–1.5 μm high. Ascospores (10.5–)11–13.5(–14.5) × (5.5–)6–7.5 μm (mean ± SD = 12.2 ± 0.5 × 6.7 ± 0.5 µm), ellipsoidal-fusiform, dark brown to chestnut brown, 1-septate; septum median, dark brown, with a central pore, not constricted or slightly constricted at the septum, with longitudinally arranged darker brown ridges alternating with lighter brown furrows, ascospores uniseriate or obliquely uniseriate in the ascus. Anamorph: Codinaea-like, not present on the nature substrate.
Description on CMA with sterile stems of Urtica dioica.
Colonies effuse, vegetative hyphae hyaline, branched, 2.5–3.5 μm wide. Conidiophores macronematous, mononematous, 22–66 μm long, 3.5–5 μm wide near the base, erect, straight, cylindrical, several-septate, brown, paler towards the apex, unbranched, smooth-walled, or reduced to single conidiogenous cells. Conidiogenous cells (9.5–)12.5–25(–33) × 4–4.5 μm, tapering to 2.5–3.5 μm just below the collarette, monophialidic, integrated, terminal, cylindrical to cylindrical-lageniform, pale brown, subhyaline towards the apex, smooth-walled; collarettes funnel-shaped, 4–6.5 μm wide, (1.5–)2–2.5 μm deep. Conidia (10–)11–13.5 × 4–5.5 μm (mean ± SD = 11.9 ± 0.8 × 4.8 ± 0.4 µm), reniform to ellipsoidal, straight or slightly curved, asymmetrical, narrowly rounded at the apical end, truncate at the basal end, brown, 1-septate, not constricted or slightly constricted at the septum, with 1–2.5 μm long, hyaline setulae at each end, smooth-walled, in slimy droplets, dark brown in mass.
Culture characteristics.
On CMD colonies 23–25 mm diam, circular, flat, margin entire, lanose, floccose, funiculose at the centre, cobwebby towards the periphery, whitish with irregular pale brown spots due to pigmented funiculose mycelium, with an isabelline outer zone of submerged growth; reverse beige. On MLA colonies 22–25 mm diam, circular, raised, margin entire, lanose, floccose, zonate, with grey, brown and white zones, with an isabelline outer zone of submerged growth; reverse dark grey. On OA colonies 31–33 mm diam, circular, flat, margin entire, sparsely lanose, floccose, cobwebby at the margin, zonate, whitish, colony centre with irregular dark brown spots due to pigmented submerged mycelium, pale brown towards the margin, with an olivaceous outer zone of submerged growth, dark brown pigment diffusing to agar at the colony centre; reverse olivaceous grey. On PCA colonies 17–19 mm diam, circular, slightly convex centrally, margin entire, lanose, floccose becoming cobwebby towards the periphery, isabelline to light beige with irregular brown spots due to pigmented mycelium; reverse light beige. Sporulation abundant on CMD, CMA with Urtica stems and PCA, sparse on MLA and OA.
Other specimen.
Brazil • Bahia; isolated from roots of Encyclia ghillanyi; isolate monte6.2; GenBank (ITS): KC928368, unpublished. (Specimen not available).
Habitat and distribution.
Striatosphaeria castanea occurs on the bark of woody liana and as an endophyte of Encyclia ghillanyi. It is known from South America in Brazil and French Guiana.
Notes.
Striatosphaeria codinaeophora closely resembles S. castanea, but differs in having larger [(130–)140–160(–170) × 25–35(– 40) μm] asci, [(17–)19–23(–26) × (6–)7–9(–10) μm] ascospores and (15–20 × 4.5–6 μm) conidia (Samuels and Müller 1978). Based on the present phylogeny (Fig. 1) and comparison of ITS sequences, S. castanea has also been recorded as an endophyte, isolated from roots of Encyclia ghillanyi, a rupiculous orchid inhabiting rock surfaces in semiarid areas in the Bahia state of northern Brazil (strain monte6.2, ITS: KC928368, Almeida et al. unpublished).
Dendrophoma cytisporoides
Sacc., Michelia 2: 4. 1880.
5DE41B6C-6613-5499-A8FF-47B85A62A63E
Figure 12.
Dendrophoma cytisporoides (CBS 144107). A–C ascomata D vertical section of the ascomal wall with remnants of the periderm E–G asci H paraphyses, asci and ascogenous hyphae I, J ascal apex K, L sterile primordia of sporodochia on MLA after 6 mo M sporodochium on OA after 10 wk N conidia O colonies on CMD, MLA, OA and PCA after 4 wk (from left to right). Scale bars: 300 μm (A–C, K); 20 μm (D); 10 μm (E–H); 5 μm (I, J, N); 100 μm (L, M); 1 cm (O).
≡ Phoma cytisporoides Sacc., Michelia 1: 522. 1879.
≡ Phoma cytisporoides subsp. punicina Sacc., Michelia 2: 273. 1881.
≡ Dendrophoma cytisporoides var. punicina (Sacc.) Sacc., Syll. fung. 3: 180. 1884.
≡ Dendrophoma cytisporoides var. pruni-virginianae Sacc., Riv. Accad. di Padova 33: 169. 1917.
= Dendrophoma punicina (Sacc.) Sacc., Rabenh. Krypt.-Fl., Edn. 2, 1(6): 409. 1901.
Description on the natural substrate.
Teleomorph: Ascomata perithecial, non-stromatic, immersed becoming erumpent, in small groups or dense caespitose clusters on the bark of the host, 120–150 μm diam, 180–200 μm high, subglobose to broadly conical, dark brown, glabrous, papillate. Ostiole periphysate. Ascomatal wall fragile, carbonaceous, 22–28 μm thick, two-layered. Outer layer of textura prismatica, consisting of brown, polyhedral cells with opaque walls. Inner layer of textura prismatica, consisting of several rows of thin-walled, hyaline, flattened cells. Paraphyses sparse, persistent, septate, anastomosing, 2–2.5 μm wide, tapering to ca. 1–1.5 μm, longer than the asci. Asci (66–)69–88.5(–92) × 6.5–8 μm (mean ± SD = 77.5 ± 5.9 × 7.3 ± 0.5 µm), (35.5–)43.5–63(–69) μm (mean ± SD = 53.9 ± 5.7 µm) long in the sporiferous part, unitunicate, arising from densely branched, short ascogenous hyphae, 8-spored, cylindrical to cylindrical-clavate, stipitate, apically broadly rounded, ascal apex with an indistinct, non-amyloid apical annulus visible only with the PC illumination, ca. 1.5 μm wide, 1.5–2 μm high. Ascospores 8–10.5(–11) × 2.5–3.5 μm (mean ± SD = 9.5 ± 0.6 × 3.0 ± 0.3 µm), fusiform, hyaline, 1-septate, not constricted at the median septum, uniseriate, obliquely uniseriate or partially biseriate in the ascus. Anamorph: Not observed.
Description on OA.
Colonies effuse, vegetative hyphae hyaline, 2–3 μm wide. Conidiomata stromatic, globose becoming cupulate, up to 350 μm diam. Setae absent. Conidiophores macronematous, septate, branched or unbranched, up to 60 μm long, hyaline. Conidiogenous cells 7–13 × 1.5 μm, monophialidic, integrated and terminal or discrete and lateral, subcylindrical, single or in terminal whorls, hyaline, tapering to ca. 1 μm; collarettes indistinct. Conidia 2.5–3.5 × 1–1.5 (mean ± SD = 3.1 ± 0.3 × 1.2 ± 0.1 µm), naviculate to botuliform, with 0.5–1 μm long setulae at each end, aseptate, smooth-walled, in slimy droplets, hyaline in mass.
Culture characteristics.
On CMD colonies 18–20 mm diam, circular, flat, margin entire, velvety-lanose, mucoid at the margin, white, isabelline towards the periphery, reverse white. On MLA colonies 18–21 mm diam, slightly convex, circular, margin entire to fimbriate, lanose, floccose, cobwebby at the margin, white, with an isabelline outer zone of submerged growth, reverse isabelline. On OA colonies 19–21 mm diam, circular, flat, raised margin, margin entire, velvety becoming lanose towards the margin, mucoid at the margin, zonate, colony centre grey to whitish-grey with intermediate white zone becoming brown, beige at the margin, reverse grey. On PCA colonies 13–15 mm diam, circular, flat, slightly convex centrally, margin entire, velvety-lanose, floccose becoming mucoid towards the periphery, white, pale brown centrally, with an isabelline outer zone of submerged growth, reverse isabelline. Sporulation absent on all media after 4 wk; abundant on OA and PCA after 10 wk or 4 mo, respectively, on MLA sterile primordia of conidiomata were formed.
Specimen examined. Germany • 16 Jan. 2012; on bark and wood of a dead twig of Buxus sempervivens; R. Schumacher leg.; (PRA-16322, culture CBS 144107 = IMI 506817).
Habitat and distribution.
Saprobic on decaying wood and bark of Buxus sempervivens, Deutzia scabra, Rhododendron sp., and Ulmus sp. The species is known from Europe in France, Germany and the Netherlands (Saccardo 1879; Sutton 1965; Crous et al. 2012; this study).
Notes.
Our strain sporulated in vitro only after prolonged incubation. On OA it formed globose to pulvinate fertile conidiomata, while on PCA the conidiomata often became confluent. The comparison of ITS and 28S sequences of our strain with those of the epitype strain of D. cytisporoidesCBS 223.95 (Crous et al. 2012) confirmed they are conspecific; the two strains share 100 % sequence similarity.
Discussion
Based on morphology, cultivation studies and phylogenetic analysis of the combined ITS-28S loci, seven new species and eight new combinations are introduced in the Chaetosphaeriaceae. Paragaeumannomyces (Matsushima 2003) is proposed for the monophyletic, strongly supported clade of former Chaetosphaeria species (Figs 1, 2) characterised by scolecosporous, multiseptate, asymmetrical, hyaline to light pink ascospores and unique three-layered ascomatal wall. The unusual wall was first brought to attention by Carroll and Munk (1964). The colour of the outer wall ranges from white, whitish-yellow, ginger-brown to reddish-brown, russet to dark brown and is composed of thin-walled cells of textura angularis, which may contain pale purple pigment when fresh, occasionally with red surface crystals. Setae, if present, are scattered over the entire ascoma or only surround the ostiole. They are stiff, acute, dark brown with opaque walls, arise from the middle, carbonaceous layer and penetrate the outer layer of globose cells. Members of the genus occur on decayed plant material, especially on strongly decayed decorticated wood.
Examination of our material of Paragaeumannomyces revealed that ascospores of four species exhibit a strong dextrinoid reaction in Melzer’s reagent, namely P. abietinus, P. albidus, P. granulatus and P. smokiensis. The ascospores turned reddish-brown except for the end cells, which remained partially hyaline, especially at the tips. Interestingly, these species share glabrous, non-setose ascomata or only minute setae are arranged around the ostiole. The chemical reaction is visible in ascospores without guttules, which otherwise fill the cells and obscure the colour. The ascospores of P. elegans, P. longisporus and P. sabinianus exhibit a negative or weak dextrinoid reaction; some mature ascospores turned light pink-brown. These species share setose ascomata, sometimes with ostioles surrounded by minute setae. Although more species need to be examined to evaluate this character, we hope it is not premature to argue that the dextrinoid reaction of ascospores is species-specific and has the potential to become another diagnostic feature facilitating species identification. Because we did not examine all known species of Paragaeumannomyces, this character has not been used in the key, but it is mentioned in the species descriptions, if known.
Paragaeumannomyces, typified by P. sphaerocellularis, encompasses 18 species. The present phylogenetic tree (Fig. 2) contains 12 of them and four subclades labelled Paragaeumannomyces sp. 1–4, which represent separate, yet undescribed taxa at the species rank. The molecular data of P. raciborskii and P. sphaerocellularis are unavailable. The closest relatives to Paragaeumannomyces are species of Exserticlava and Chaetosphaeria lignomollis with a kylindria-like anamorph, characterised by septate, versicolorous or hyaline ascospores, respectively (Fig. 1).
Members of Paragaeumannomyces are not easily cultivated and only seldom sporulate in vitro. Huhndorf and Fernández (2005) noted that even isolates from the same specimen varied in their ability to produce the anamorph in culture. The craspedodidymum-like anamorph with usually semi-macronematous to micronematous conidiophores, inflated phialides, deeply flared, cup-shaped collarettes and hyaline non-septate conidia, with or without setulae, was reported for P. lapazianus, P. longisporus, P. panamensis, P. rubicundus, and Paragaeumannomyces sp. 1–4, while the chloridium-like synanamorph is known only in P. longisporus and Paragaeumannomyces sp. 2 (Huhndorf and Fernández 2005; Perera et al. 2016). The systematic placement of Craspedodidymum (Holubová-Jechová 1972), typified by C. elatum, is unknown. The genus was erected for a hyphomycete forming effuse colonies on an old petiole of Phoenix canariensis in a green house in the Czech Republic. To date, 15 binomials were introduced in the genus (Index Fungorum). Craspedodidymum elatum differs from Paragaeumannomyces anamorphs by macronematous, dichotomously branched conidiophores and non-septate, dark brown conidia with a basal hilum. In hyaline, unicellular, globose or triangular conidia with setulae, the Paragaeumannomyces anamorphs also resemble Bahusutrabeeja (Subramanian and Bhat 1977) and Nawawia (Marvanová1980), respectively. Bahusutrabeeja and Nawawia are similar to each other but differ in shape of conidia. Bahusutrabeeja, typified with B. dwaya, forms globose conidia on solitary conidiophores, while Nawawia, based on N. filiformis, have conidia triangular, round-tetrahedral or obpyramidal-shaped on conidiophores arising from small stromata. Their molecular data (Yang et al. 2016; Vu et al. 2019) suggest a distant relationship to Paragaeumannomyces.
The original P. longisporus clade with two strongly supported subclades was recognized as two species, the short-spored P. longisporus and the long-spored P. sabinianus. In general, a high degree of ITS sequence variability and more or less uniform teleomorphic phenotype pose special problems in species identification, especially in P. raciborskii, which was resolved as polyphyletic (Huhndorf and Fernández 2005; this study). Morphology of this species was studied by Huhndorf and Fernández (2005) based on more than 100 species, mostly from the neotropics. The broad species concept of P. raciborskiifideHuhndorf and Fernández (2005) includes specimens with wider ascomata, longer ascospores with less septa and longer asci than reported in the protologue (Penzig and Saccardo 1897) and re-description of this species prepared by Carroll and Munk (1964); for details see above. Although no significant variability among ascomata, asci and ascospores was encountered, Huhndorf and Fernández (2005) reported intraspecific variability regarding setae, which were lacking in some specimens. On the other hand, certain variability at the anamorphic level, typical of many natural groups of the Chaetosphaeriaceae, also occurs in P. raciborskiifideHuhndorf and Fernández (2005) and to some extent delimits the four subclades. One isolate (S.M.H. 3119) produced a craspedodidymum-like anamorph with triangular conidia with setulae, while others with the morphologically similar anamorph produced globose, non-setulate conidia and can be further distinguished by formation of the chloridium-like synanamorph (S.M.H. 2036, S.M.H. 2132), or its absence but are separated by dark, purplish-brown phialides (S.M.H. 2017) or light brown phialides (S.M.H. 3014) (Huhndorf and Fernández 2005). A close comparison of the descriptions of the holotype of P. raciborskii (Penzig and Saccardo 1897; Carroll and Munk 1964) with collections gathered by Huhndorf and Fernández (2005) and identified as P. raciborskii confirms that none of the four subclades inferred in the ITS-28SML tree (Fig. 2) could be delimited as P. raciborskii s. str. From a biogeographical perspective it is more likely that they belong to different species entirely. Therefore, the name P. raciborskii for these strains was rejected in our phylogeny; instead, the four subclades were designated Paragaeumannomyces sp. 1–4.
Codinaea (Maire 1937) is one of the largest genera of the Chaetosphaeriaceae with a turbulent taxonomic history. Based on a cluster analysis of phialidic dematiaceous hyphomycetes, Arambarri and Cabello (1989) considered Codinaea, with usually falcate, septate or non-septate conidia bearing setulae at both end, and Dictyochaeta (Spegazzini 1923) with non-setulate, non-septate, cylindrical and asymmetrical conidia, congeneric. Since then, Dictyochaeta (syn. Codinaea) became a broadly circumscribed genus with more than 100 species and the new morphological concept was followed by many mycologists. Réblová and Seifert (2007) confirmed with DNA sequence data that Dictyochaeta fuegiana, the type species of the genus, is a member of the Chaetosphaeriaceae. Based on the ITS-28S phylogeny (Fig. 1), D. fuegiana is unrelated to morphologically similar species with setulate conidia, classified in Codinaea or Dictyochaeta, and resolved as polyphyletic, which is in agreement with Lin et al. (2019) and Réblová et al. (2020). However, in the absence of molecular DNA data of Codinaea aristata, the generic type, it is difficult to delimit Codinaea phylogenetically. The morphological traits delimiting the new species C. paniculata, i.e. presence of setae, unbranched and shorter conidiophores growing at the base of the setae with monophialidic conidiogenous cells in vivo and falcate, non-septate conidia with setulae at both ends, best match those of Codinaea. In the subclade where C. paniculata was clustered, several species sharing the same Codinaea morphotype were present, namely C. assamica (Hughes and Kendrick 1968), D. siamensis (Liu et al. 2016) and D. terminalis (Lin et al. 2019).
Based on published records, Striatosphaeria is an uncommon lignicolous genus with a known distribution in the neotropics. It was introduced by Samuels and Müller (1978) based on two Brazilian collections of S. codinaeophora. Additional specimens of S. codinaeophora known to us were collected on decaying wood of Dacryodes excelsa and Nectandra turbacensis and unidentified hosts in Costa Rica, French Guiana and Puerto Rico (Réblová and Winka 2000; Fernández et al. 2006; S.M. Huhndorf pers. data). The codinaea-like anamorph develops only in axenic culture. The conidia are asymmetrical, brown, 1-septate with minute, hyaline setulae at each end. Although S. codinaeophora was described with non-setulate conidia (Samuels and Müller 1978), a photograph of conidia with setulae accompanied the S. codinaeophora lineage on a phylogenetic tree (Fernández et al. 2006: fig. 1, 6g). The setulate conidia were also present in the new species, S. castanea. It is probable that setulae are formed later, after conidia detach from the conidiogenous cells. The conidia with setulae at both ends are formally introduced in Striatosphaeria for the first time in this study.
The genus Dendrophoma with a single species, D. cytisporoides, was proposed by Saccardo (1880) for fungi with phoma-like fruit bodies, botuliform hyaline conidia and conidiogenous cells arranged in a verticillate fashion. Sutton (1965) lectotypified Phoma cytisporoides (Saccardo 1879) and reported additional characters not mentioned by Saccardo in the protologue, i.e. dark brown, acute setae accompanying conidiomata and minute, unbranched setulae at both ends of conidia. Sutton (1965) compared D. cytisporoides with Dinemasporium graminum, the type species of the genus, and reduced Dendrophoma to synonymy with Dinemasporium (Léveillé 1846). Using nuclear ribosomal loci, Crous et al. (2012) re-established Dendrophoma and placed the genus in the Chaetosphaeriaceae, where it emerged as a separate lineage from Dinemasporium. The anamorph-teleomorph relationship of Dendrophoma has been established for the first time in this study. The teleomorph is morphologically similar to Chaetosphaeria (Tulasne and Tulasne 1863), but differs in having immersed to erumpent ascomata, densely branched ascogenous hyphae (Fig. 12H), the ascal apex lacking a visible discharge mechanism, which can only be seen as a minute apical ring with PC illumination (Fig. 12I) and a morphologically distinct anamorph forming stromatic, stipitate, cupulate sporodochial conidiomata.
Supplementary Material
Acknowledgements
We thank Sabine Huhndorf for the use of her collections, images and DNA extracts and Alberto Stchigel and Preeti Chaudhary for assistance in our collection. Peter Johnston is thanked for his assistance to M.R. in obtaining the Manaaki Whenua Fellowship and collecting permits for New Zealand. We thank J. Leland Crane for grammatical review of some of the new names. J.F. gratefully acknowledges the Parc Naturel Amazonien de Guyane for initiating, financing and organizing fieldwork in Saül in 2018 and 2019 in connection with the inventorial ABC project, during which a new species of Striatosphaeria castanea was collected. We thank René Schumacher for a collection of Dendrophoma cytisporoides. We thank both reviewers for their valuable comments and suggestions.
This study was supported by the project of the Czech Science Foundation (GAČR 20-14840S), and as long-term research development projects of the Czech Academy of Sciences, Institute of Botany (RVO 67985939) (M.R.) and the University Hospital Hradec Králové MH CZ – DRO (UHHK, 00179906) (J.N.). This study was also supported by a National Science Foundation award (DEB-0515558) to A.N.M. The WM Keck Center at the University of Illinois Urbana-Champaign is thanked for sequencing services. The field work of M.R. in New Zealand was partly supported by Studienstiftung für mykologische Systematik und Ökologie (2003) and Manaaki Whenua Fellowship, Landcare Research Auckland (2005).The authors have declared that no competing interests exist.
Citation
Réblová M, Nekvindová J, Fournier J, Miller AN (2020) Delimitation, new species and teleomorph-anamorph relationships in Codinaea, Dendrophoma, Paragaeumannomyces and Striatosphaeria (Chaetosphaeriaceae). MycoKeys 74: 17–74. https://doi.org/10.3897/mycokeys.74.57824
References
- Arambarri AM, Cabello MN. (1989) A numerical taxonomic study of some phialidic genera of hyphomycetes: cluster analysis. Mycotaxon 34: 679–696. [Google Scholar]
- Atkinson TJ, Miller AN, Huhndorf SM, Orlovich DA. (2007) Unusual new Chaetosphaeria species from New Zealand: intrafamilial diversity and elucidations of the Chaetosphaeriaceae–Lasiosphaeriaceae relationship (Sordariomycetes, Ascomycotina). New Zealand Journal of Botany 45: 685–706. 10.1080/00288250709509744 [DOI] [Google Scholar]
- Barr ME. (1993) Redisposition of some taxa described by J.B. Ellis. Mycotaxon 46: 45–76. [Google Scholar]
- Booth C. (1957) Studies of Pyrenomycetes I. Four Species of Chaetosphaeria, two with Catenularia conidia. II. Melanopsamma pomiformis and its Stachybotrys conidia. Mycological Papers 68: 1–27.
- Booth C. (1958) The genera Chaetosphaeria and Thaxteria in Britain. Naturalist 1958: 83–90. [Google Scholar]
- Berkeley M J, Broome CE. (1871) Notices of British Fungi Annals and Magazine of Natural History, Ser. 4, 7: 425–436. 10.1080/00222937108696408 [DOI]
- Carroll GC, Munk A. (1964) Studies on lignicolous Sordariaceae. Mycologia 56: 77–98. 10.1080/00275514.1964.12018085 [DOI] [Google Scholar]
- Castresana J. (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution 17: 540–552. 10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- Crous PW, Schumacher RK, Wingfield MJ, Akulov AY, Denman S, Roux J, Braun U, Burgess TI, Carnegie AJ, Váczy KZ, Guatimosim E, Schwartsburd PB, Barreto RW, Hernández-Restrepo M, Lombard L, Groenewald JZ. (2018a) New and interesting fungi. 1. Fungal Systematics and Evolution 1: 169–215. 10.3114/fuse.2018.01.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Schumacher RK, Wingfield MJ, Lombard L, Giraldo A, Christensen M, Gardiennet A, Nakashima C, Pereira OL, Smith AJ, Groenewald JZ. (2015a) Fungal systematics and evolution: FUSE 1. Sydowia 67: 81–118. [Google Scholar]
- Crous PW, Shivas RG, Quaedvlieg W, van der Bank M, Zhang Y, Summerell BA, Guarro J, Wingfield MJ, Wood AR, Alfenas AC, Braun U, Cano-Lira JF, García D, Marin-Felix Y, Alvarado P, Andrade JP, Armengol J, Assefa A, den Breeÿen A, Camele I, Cheewangkoon R, De Souza JT, Duong TA, Esteve-Raventós F, Fournier J, Frisullo S, García-Jiménez J, Gardiennet A, Gené J, Hernández-Restrepo M, Hirooka Y, Hospenthal DR, King A, Lechat C, Lombard L, Mang SM, Marbach PAS, Marincowitz S, Marin-Felix Y, Montaño-Mata NJ, Moreno G, Perez CA, Pérez Sierra AM, Robertson JL, Roux J, Rubio E, Schumacher RK, Stchigel AM, Sutton DA, Tan YP, Thompson EH, van der Linde E, Walker AK, Walker DM, Wickes BL, Wong PTW, Groenewald JZ. (2014a) Fungal Planet description sheets: 214–280. Persoonia 32: 184–306. 10.3767/003158514X682395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Verkley GJM, Christensen M, Castañeda-Ruiz RF, Groenewald JZ. (2012) How important are conidial appendages? Persoonia 28: 126–137. 10.3767/003158512X652624 [DOI] [PMC free article] [PubMed]
- Crous PW, Verkleij GJM, Groenewald JZ, Houbraken J. (2019) Fungal biodiversity. CBS laboratory manual series 1. CBS-KNAW Fungal Biodiversity Centre, Utrecht, 1–425.
- Crous PW, Wingfield MJ, Burgess TI, Carnegie AJ, Hardy GEStJ, Smith D, Summerell BA, Cano-Lira JF, Guarro J, Houbraken J, Lombard L, Martín MP, Sandoval-Denis M, Alexandrova AV, Barnes CW, Baseia IG, Bezerra JDP, Guarnaccia V, May TW, Hernández-Restrepo M, Stchigel AM, Miller AN, Ordoñez ME, Abreu VP, Accioly T, Agnello C, Agustin Colmán A, Albuquerque CC, Alfredo DS, Alvarado P, Araújo-Magalhães GR, Arauzo S, Atkinson T, Barili A, Barreto RW, Bezerra JL, Cabral TS, Camello Rodríguez F, Cruz RHSF, Daniëls PP, da Silva BDB, de Almeida DAC, de Carvalho Júnior AA, Decock CA, Delgat L, Denman S, Dimitrov RA, Edwards J, Fedosova AG, Ferreira RJ, Firmino AL, Flores JA, García D, Gené J, Góis JS, Gomes AAM, Gonçalves CM, Gouliamova DE, Groenewald M, Guéorguiev BV, Guevara-Suarez M, Gusmão LFP, Hosaka K, Hubka V, Huhndorf SM, Jadan M, Jurjević Ž, Kraak B, Kučera V, Kumar TKA, Kušan I, Lacerda SR, Lamlertthon S, Lisboa WS, Loizides M, Luangsa-ard JJ, Lysková P, Mac Cormack WP, Macedo DM, Machado AR, Malysheva EF, Marinho P, Matočec N, Meijer M, Mešić A, Mongkolsamrit S, Moreira KA, Morozova OV, Nair KU, Nakamura N, Noisripoom W, Olariaga I, Oliveira RJV, Paiva LM, Pawar P, Pereira OL, Peterson SW, Prieto M, Rodríguez-Andrade E, Rojo De Blas C, Roy M, Santos ES, Sharma R, Silva GA, Souza-Motta CM, Takeuchi-Kaneko Y, Tanaka C, Thakur A, Smith MTh, Tkalčec Z, Valenzuela-Lopez N, Van der Kleij P, Verbeken A, Viana MG, Wang XW, Groenewald JZ. (2017) Fungal Planet description sheets: 625–715. Persoonia 39: 270–467. 10.3767/persoonia.2017.39.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Burgess TI, Hardy GEStJ, Crane C, Barrett S, Cano-Lira JF, Le Roux JJ, Thangavel R, Guarro J, Stchigel AM, Martín MP, Alfredo DS, Barber PA, Barreto RW, Baseia IG, Cano-Canals J, Cheewangkoon R, Ferreira RJ, Gené J, Lechat C, Moreno G, Roets F, Shivas RG, Sousa JO, Tan YP, Wiederhold NP, Abell SE, Accioly T, Albizu JL, Alves JL, Antoniolli ZI, Aplin N, Araújo J, Arzanlou M, Bezerra JDP, Bouchara J-P, Carlavilla JR, Castillo A, Castroagudín VL, Ceresini PC, Claridge GF, Coelho G, Coimbra VRM, Costa LA, da Cunha KC, da Silva SS, Daniel R, de Beer ZW, Dueñas M, Edwards J, Enwistle P, Fiuza PO, Fournier J, García D, Gibertoni TB, Giraud S, Guevara-Suárez M, Gusmão LFP, Haituk S, Heykoop M, Hirooka Y, Hofmann TA, Houbraken J, Hughes DP, Kautmanová I, Koppel O, Koukol O, Larsson E, Latha KPD, Lee DH, Lisboa DO, Lisboa WS, López-Villalba Á, Maciel JLN, Manimohan P, Manjón JL, Marincowitz S, Marney TS, Meijer M, Miller AN, Olariaga I, Paiva LM, Piepenbring M, Poveda-Molero JC, Raj KNA, Raja HA, Rougeron A, Salcedo I, Samadi R, Santos TAB, Scarlett K, Seifert KA, Shuttleworth LA, Silva GA, Silva M, Siqueira JPZ, Souza-Motta CM, Stephenson SL, Sutton DA, Tamakeaw N, Telleria MT, Valenzuela-Lopez N, Viljoen A, Visagie CM, Vizzini A, Wartchow F, Wingfield BD, Yurchenko E, Zamora JC, Groenewald JZ. (2016) Fungal Planet description sheets: 469–557. Persoonia 37: 218–403. 10.3767/003158516X694499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Burgess TI, Hardy GEStJ, Gené J, Guarro J, Baseia IG, García D, Gusmão LFP, Souza-Motta CM, Thangavel R, Adamčík S, Barili A, Barnes CW, Bezerra JDP, Bordallo JJ, Cano-Lira JF, de Oliveira RJV, Ercole E, Hubka V, Iturrieta-González I, Kubátová A, Martín MP, Moreau P-A, Morte A, Ordoñez ME, Rodríguez A, Stchigel AM, Vizzini A, Abdollahzadeh J, Abreu VP, Adamčíková K, Albuquerque GMR, Alexandrova AV, Álvarez Duarte E, Armstrong-Cho C, Banniza S, Barbosa RN, Bellanger J-M, Bezerra JL, Cabral TS, Caboň M, Caicedo E, Cantillo T, Carnegie AJ, Carmo LT, Castañeda-Ruiz RF, Clement CR, Čmoková A, Conceição LB, Cruz RHSF, Damm U, da Silva BDB, da Silva GA, da Silva RMF, de A Santiago ALCM, Déniel F, de Oliveira LF, de Souza CAF, Déniel F, Dima B, Dong G, Edwards J, Félix CR, Fournier J, Gibertoni TB, Hosaka K, Iturriaga T, Jadan M, Jany J-L, Jurjević Ž, Kolařík M, Kušan I, Landell MF, Leite Cordeiro TR, Lima DX, Loizides M, Luo S, Machado AR, Madrid H, Magalhães OMC, Marinho P, Matočec N, Mešić A, Miller AN, Morozova OV, Neves RP, Nonaka K, Nováková A, Oberlies NH, Oliveira-Filho JRC, Oliveira TGL, Papp V, Pereira OL, Perrone G, Peterson SW, Pham THG, Raja HA, Raudabaugh DB, Řehulka J, Rodríguez-Andrade E, Saba M, Schauflerová A, Shivas RG, Simonini G, Siqueira JPZ, Sousa JO, Stajsic V, Svetasheva T, Tan YP, Tkalčec Z, Ullah S, Valente P, Valenzuela-Lopez N, Abrinbana M, Viana Marques DA, Wong PTW, Xavier de Lima V, Groenewald JZ. (2018b) Fungal Planet description sheets: 716–784. Persoonia 40: 240–393. 10.3767/persoonia.2018.40.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Guarro J, Hernández-Restrepo M, Sutton DA, Acharya K, Barber PA, Boekhout T, Dimitrov RA, Dueñas M, Dutta AK, Gené J, Gouliamova DE, Groenewald M, Lombard L, Morozova OV, Sarkar J, Smith MTH, Stchigel AM, Wiederhold NP, Alexandrova AV, Antelmi I, Armengol J, Barnes I, Cano-Lira JF, Ruiz Castañeda RF, Contu M, Courtecuisse PrR, da Silveira AL, Decock CA, de Goes A, Edathodu J, Ercole E, Firmino AC, Fourie A, Fournier J, Furtado EL, Geering ADW, Gershenzon J, Giraldo A, Gramaje D, Hammerbacher A, He X-L, Haryadi D, Khemmuk W, Kovalenko AE, Krawczynski R, Laich F, Lechat C, Lopes UP, Madrid H, Malysheva EF, Marín-Felix Y, Martín MP, Mostert L, Nigro F, Pereira OL, Picillo B, Pinho DB, Popov ES, Peláez CA Rodas, Rooney-Latham S, Sandoval-Denis M, Shivas RG, Silva V, Stoilova-Disheva MM, Telleria MT, Ullah C, Unsicker SB, van der Merwe NA, Vizzini A, Wagner H-G, Wong PTW, Wood AR, Groenewald JZ. (2015b) Fungal Planet description sheets: 320–370. Persoonia 34: 167–266. 10.3767/003158515X688433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Schumacher RK, Summerell BA, Giraldo A, Gené J, Guarro J, Wanasinghe DN, Hyde KD, Camporesi E, Gareth Jones EB, Thambugala KM, Malysheva EF, Malysheva VF, Acharya K, Álvarez J, Alvarado P, Assefa A, Barnes CW, Bartlett JS, Blanchette RA, Burgess TI, Carlavilla JR, Coetzee MPA, Damm U, Decock CA den Breeÿen A, de Vries B, Dutta AK, Holdom DG, Latham SR, Manjón JL, Marincowitz S, Mirabolfathy M, Moreno G, Nakashima C, Papizadeh M, Fazeli SAS, Amoozegar MA, Romberg MK, Shivas RG, Stalpers JA, Stielow B, Stukely MJC, Swart WJ, Tan YP, van der Bank M, Wood AR, Zhang Y, Groenewald JZ. (2014b) Fungal Planet description sheets: 281–319. Persoonia 33: 212–289. 10.3767/003158514X685680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currey F. (1859) Synopsis of the fructification of the compound Sphaeriæ of the Hookerian herbarium. Transactions of the Linnaean Society of London 22: 257–287. 10.1111/j.1096-3642.1856.tb00098.x [DOI] [Google Scholar]
- Dai DQ, Phookamsak R, Wijayawardene NN, Li WJ, Bhat DJ, Xu JC, Taylor JE, Hyde KD, Chukeatirote E. (2016) Bambusicolous fungi. Fungal Diversity 82: 1–105. 10.1007/s13225-016-0367-8 [DOI] [Google Scholar]
- David AS, May G, Schmidt D, Seabloom EW. (2016) Beachgrass invasion in coastal dunes is mediated by soil microbes and lack of disturbance dependence. Ecosphere 11: e01527. 10.1002/ecs2.1527 [DOI]
- de Hoog GS, Gerrits van den Ende AH. (1998) Molecular diagnostics of clinical strains of filamentous Basidiomycetes. Mycoses 41: 183–189. 10.1111/j.1439-0507.1998.tb00321.x [DOI] [PubMed] [Google Scholar]
- Ellis JB. (1877) South Jersey fungi. Bulletin of the Torrey Botanical Club 6: 133–135. 10.2307/2477388 [DOI] [Google Scholar]
- Fernández FA, Huhndorf SM. (2005) New species of Chaetosphaeria, Melanopsammella and Tainosphaeria gen. nov. from the Americas. Fungal Diversity 18: 15–57. [Google Scholar]
- Fernández FA, Miller AN, Huhndorf SM, Lutzoni FM, Zoller S. (2006) Systematics of the genus Chaetosphaeria and its allied genera: morphological and phylogenetic diversity in north temperate and neotropical taxa. Mycologia 98: 121–130. 10.1080/15572536.2006.11832718 [DOI] [PubMed] [Google Scholar]
- Gams W, Holubová-Jechová V. (1976) Chloridium and some other dematiaceous hyphomycetes growing on decaying wood. Studies in Mycology 13: 1–99. [Google Scholar]
- Gooding GV, Lucas GB. (1959) Factors influencing sporangial formation and zoospore activity in Phytophthora parasitica var. nicotianae. Phytopathologia 49: 277–281. [Google Scholar]
- Gutell RR, Gray MW, Schnare MN. (1993) A compilation of large subunit (23S and 23S-like) ribosomal RNA structures. Nucleic Acids Research 21: 3055–3074. 10.1093/nar/21.13.3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. (1999) BioEdit 5.0.9: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- Hashimoto A, Sato G, Matsuda T, Matsumura M, Hatakeyama S, Harada Y, Ikeda H, Tanaka K. (2015) Taxonomic revision of Pseudolachnea and Pseudolachnella, and establishment of Neopseudolachnella and Pseudodinemasporium genera nova. Mycologia 107: 383–408. 10.3852/14-171 [DOI] [PubMed] [Google Scholar]
- Hawksworth DL. (2011) A new dawn for the naming of fungi: impacts of decisions made in Melbourne in July 2011 on the future publication and regulation of fungal names. IMA Fungus 2: 155–162. 10.3897/mycokeys.1.2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawksworth DL. (2012) Managing and coping with names of pleomorphic fungi in a period of transition. IMA Fungus 3: 81–86. 10.5598/imafungus.2012.03.01.03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawksworth DL, Crous PW, Redhead SA, Reynolds DR, Samson RA, Seifert KA, Taylor JW, Wingfield MJ, Abaci Ö, Aime C, Asan A, Bai FY, de Beer ZW. (2011) The Amsterdam Declaration on fungal nomenclature. IMA Fungus 2: 105–112. 10.5598/imafungus.2011.02.01.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Restrepo M, Gené J, Castañeda-Ruiz RF, Mena-Portales J, Crous PW, Guarro J. (2017) Phylogeny of saprobic microfungi from Southern Europe. Studies in Mycology 86: 53–97. 10.1016/j.simyco.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Restrepo M, Schumacher RK, Wingfield MJ, Ahmad I, Cai L, Duong TA, Edwards J, Gené J, Groenewald JZ, Jabeen S, Khalid AN, Lombard L, Madrid H, Marin-Felix Y, Marincowitz S, Miller AN, Rajeshkumar KC, Rashid A, Sarwar S, Stchigel AM, Taylor PWJ, Zhou N, Crous PW. (2016) Fungal systematics and evolution: FUSE 2. Sydowia 68: 193–230. [Google Scholar]
- Hino I. (1961) Icones fungorum bambusicolorum Japonicorum. The Fuji Bamboo Garden, Gotenba, 1–335.
- Holubová-Jechová V. (1972) Craspedodidymum, a new genus of phialosporous Hyphomycetes. Česká Mykologie 26: 70–73. [Google Scholar]
- Holubová-Jechová V. (1973) Lignicolous hyphomycetes from Czechoslovakia. 4. Menispora. Folia Geobotanica et Phytotaxonomica 8: 317–336. 10.1007/BF02852830 [DOI] [Google Scholar]
- Holubová-Jechová V. (1982) New or interesting phialidic hyphomycetes from Cuba. Mycotaxon 15: 277–292. [Google Scholar]
- Huelsenbeck JP, Ronquist F. (2001) MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Hughes SJ. (1965a) New Zealand fungi 3. Catenularia Grove. New Zealand Journal of Botany 3: 136–150. 10.1080/0028825X.1965.10876990 [DOI] [Google Scholar]
- Hughes SJ, Kendrick WB. (1965b) New Zealand fungi 4. Zanclospora gen. nov. New Zealand Journal of Botany 3: 151–158. 10.1080/0028825X.1965.10876991 [DOI] [Google Scholar]
- Hughes SJ, Kendrick WB. (1968) New Zealand fungi 12. Menispora, Codinaea, Menisporopsis. New Zealand Journal of Botany 6: 323–375. 10.1080/0028825X.1968.10428818 [DOI] [Google Scholar]
- Huhndorf SM, Fernández FA. (2005) Teleomorph-anamorph connections: Chaetosphaeria raciborskii and related species, and their Craspedodidymum-like anamorphs. Fungal Diversity 19: 23–49. [Google Scholar]
- Huhndorf SM, Miller AN, Fernández FA. (2004) Molecular systematics of the Sordariales: The order and the family Lasiosphaeriaceae redefined. Mycologia 96: 368–387. 10.1080/15572536.2005.11832982 [DOI] [PubMed] [Google Scholar]
- Hustad VP, Miller AN. (2015) Studies in the genus Glutinoglossum. Mycologia 107: 647–657. 10.3852/14-328 [DOI] [PubMed] [Google Scholar]
- Hyde KD, Tennakoon DS, Jeewon R, Bhat DJ, Maharachchikumbura SSN, Rossi W, Leonardi M, Lee HB, Mun HY, Houbraken J, Nguyen TTT, Jeon SJ, Frisvad JC, Wanasinghe DN, Lücking R, Aptroot A, Caceres MES, Karunarathna SC, Hongsanan S, Phookamsak R, Silva NI, Thambugala KM, Jayawardena RS, Senanayake IC, Boonmee S, Chen J, Luo ZL, Phukhamsakda C, Pereira OL, Abreu VP, Rosado AWC, Buyck B, Randrianjohany E, Hofstetter V, Gibertoni TB, Silva Soares AM, Plautz Jr HL, Pontes Sotaõ HM, Xavier WKS, Bezerra JDP, Oliveira TGL, Souza-Motta CM, Magalhães OMC, Bundhun D, Harishchandra D, Manawasinghe IS, Dong W, Zhang SN, Bao DF, Samarakoon MC, Pem D, Karunarathna A, Lin CG, Yang J, Perera RH, Kumar V, Huang SK, Dayarathne MC, Ekanayaka AH, Jayasiri SC, Xiao YP, Konta S, Niskanen T, Liimatainen K, Dai YC, Ji XH, Tian XM, Mešić A, Singh SK, Phutthacharoen K, Cai L, Sorvongxay T, Thiyagaraja V, Norphanphoun C, Chaiwan N, Lu YZ, Jiang HB, Zhang JF, Abeywickrama PD, Aluthmuhandiram JVS, Brahmanage RS, Zeng M, Chethana T, Wei DP, Réblová M, Fournier J, Nekvindová J, Barbosa RN, Felinto dos Santos JE, Oliveira NT, Li GJ, Ertz D, Shang QJ, Phillips AJL, Kuo CH, Camporesi E, Bulgakov TS, Lumyong S, Jones EBG, Chomnunti P, Gentekaki E, Bungartz F, Zeng XY, Fryar S, Tkalčec Z, Liang JM, Li GS, Wen TC, Singh PN, Gafforov Y, Promputtha I, Yasanthika E, Goonasekara ID, Zhao RL, Zhao Q, Kirk PM, Liu JK, Yan JY, Mortimer PE, Xu JC, Doilom MW. (2019) Fungal diversity notes 1036–1150: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 96: 1–242. 10.1007/s13225-019-00429-2 [DOI] [Google Scholar]
- Jeewon R, Yeung SYQ, Hyde KD. (2009) A novel phylogenetic group within Thozetella (Chaetosphaeriaceae): a new taxon based on morphology and DNA sequence analyses. Canadian Journal of Microbiology 55: 680–687. 10.1139/W08-148 [DOI] [PubMed] [Google Scholar]
- Johnson J, Evans C, Brown N, Skeates S, Watkinson S, Bass D. (2014) Molecular analysis shows that soil fungi from ancient semi-natural woodland exist in sites converted to non-native conifer plantations. Forestry 87: 705–717. 10.1093/forestry/cpu031 [DOI] [Google Scholar]
- Karsten PA. (1873) Mycologia Fennica. Pars secunda. Pyrenomycetes. Bidrag till Kännedom af Finlands Natur och Folk 23: I–IX, 1–252.
- Kirk PM. (2014) Nomenclatural novelties. Index Fungorum 120: 1.
- Konta S, Hongsanan S, Liu JK, Eungwanichayapant PD, Jeewon R, Hyde KD, Maharachchikumbura SSN, Boonmee S. (2017) Leptosporella (Leptosporellaceae fam. nov.) and Linocarpon and Neolinocarpon (Linocarpaceae fam. nov.) are accommodated in Chaetosphaeriales. Mycosphere 8: 1943–1974. 10.5943/mycosphere/8/10/16 [DOI] [Google Scholar]
- Landvik S. (1996) Neolecta, a fruit-body-producing genus of the basal ascomycetes, as shown by SSU and LSU DNA sequences. Mycological Research 100: 199–202. 10.1016/S0953-7562(96)80122-5 [DOI] [Google Scholar]
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. (2016) PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular biology and evolution, msw260. 10.1093/molbev/msw260 [DOI] [PubMed]
- Léveillé JH. (1846) . Descriptions des champignons de l’herbier du Muséum de Paris. Annales des Sciences Naturelles Botanique, Sér. 3, 5: 249–305.
- Lin CG, McKenzie EHC, Liu JK, Jones EBG, Hyde KD. (2019) Hyaline-spored chaetosphaeriaceous hyphomycetes from Thailand and China, with a review of the family Chaetosphaeriaceae. Mycosphere 10: 655–700. 10.5943/mycosphere/10/1/14 [DOI] [Google Scholar]
- Liu JK, Hyde KD, Jones EBG, Ariyawansa HA, Bhat DJ, Boonmee S, Maharachchikumbura SSN, McKenzie EHC, Phookamsak R, Phukhamsakda C, Shenoy BD, Abdel-Wahab MA, Buyck B, Chen J, Chethana KWT, Singtripop C, Dai DQ, Dai YC, Daranagama DA, Dissanayake AJ, Doliom M, D’souza MJ, Fan XL, Goonasekara ID, Hirayama K, Hongsanan S, Jayasiri SC, Jayawardena RS, Karunarathna SC, Li WJ, Mapook A, Norphanphoun C, Pang KL, Perera RH, Peršoh D, Pinruan U, Senanayake IC, Somrithipol S, Suetrong S, Tanaka K, Thambugala KM, Tian Q, Tibpromma S, Udayanga D, Wijayawardene NN, Wanasinghe D, Wisitrassameewong K, Zeng XY, Abdel-Aziz FA, Adamčík S, Bahkali AH, Boonyuen N, Bulgakov T, Callac P, Chomnunti P, Greiner K, Hashimoto A, Hofstetter V, Kang JC, Lewis D, Li XH, Liu ZY, Liu ZY, Matumura M, Mortimer PE, Rambold R, Randrianjohany E, Sato G, Indrasutdhi VS, Tian CM, Verbeken A, Brackel W, Wang Y, Wen TC, Xu JC, Yan JY, Zhao RL, Camporesi E. (2015) Fungal diversity notes 1–100: Taxonomic and phylogenetic contributions to fungal species. Fungal Diversity 72: 1–197. 10.1007/s13225-015-0324-y [DOI] [Google Scholar]
- Liu JK, Yang J, Maharachchikumbura SSN, McKenzie EHC, Jones EBG, Hyde KD, Liu ZY. (2016) Novel chaetosphaeriaceous hyphomycetes from aquatic habitats. Mycological Progress 15: 1157–1167. 10.1007/s11557-016-1237-1 [DOI] [Google Scholar]
- Lu YZ, Liu KJ, Hyde KD, Bhat DJ, Xiao YP, Tian Q, Wen TC, Boonmee S, Kang JC. (2016) Brunneodinemasporium jonesii and Tainosphaeria jonesii spp. nov. (Chaetosphaeriaceae, Chaetosphaeriales) from southern China. Mycosphere 7: 1322–1331. 10.5943/mycosphere/7/9/6 [DOI] [Google Scholar]
- Luo ZL, Hyde KD, Liu J-K, Maharachchikumbura SSN, Rajesh J, Bao D-F, Bhat DJ, Lin C-G, Li W-L, Yang J, Liu N-G, Lu Y-Z, Jayawardena RS, Li J-F, Su H-Y. (2019) Freshwater Sordariomycetes. Fungal Diversity 99: 451–660. 10.1007/s13225-019-00438-1 [DOI] [Google Scholar]
- Ma YR, Xia JW, Gao JM, Li Z, Zhang XG. (2016) Anacacumisporium, a new genus based on morphology and molecular analyses from Hainan, China. Cryptogamie Mycologie 37: 45–59. 10.7872/crym/v37.iss1.2016.45 [DOI] [Google Scholar]
- Magyar D, Shoemaker RA, Bobvos J, Crous PW, Groenewald JZ. (2011) Pyrigemmula, a novel hyphomycete genus on grapevine and tree bark from Hungary. Mycological Progress 10: 307–314. 10.1007/s11557-010-0703-4 [DOI] [Google Scholar]
- Malloch D. (1981) Moulds: their isolation, cultivation and identification. University of Toronto Press, Toronto, 1–97.
- Maire R. (1937) Fungi Catalaunici: Series altera. Contributions a l’étude de la flore mycologique de la Catalogne. Publicacions del Instituto Botánico Barcelona 3: 1–128. [Google Scholar]
- Marvanová L. (1980) New or noteworthy aquatic hyphomycetes. Transactions of the British Mycological Society 75: 221–231. 10.1016/S0007-1536(80)80083-0 [DOI] [Google Scholar]
- Matsushima T. (1985) Matsushima mycological memoirs 4: 1–68.
- Matsushima T. (2003) [2001]. Matsushima mycological memoirs 10: 1–214. [Google Scholar]
- Miller AN, Huhndorf SM. (2004) A natural classification of Lasiosphaeria based on nuclear LSU rDNA sequences. Mycological Research 108: 26–34. 10.1017/S0953756203008864 [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE), 14 Nov. 2010, New Orleans, 1–8. 10.1109/GCE.2010.5676129 [DOI]
- Müller E, Harr J, Sulmont P. (1969) Deux ascomycètes dont le stade conidien présente des conidies phaeophragmiées endogènes. Revue de Mycologie 33: 369–378. [Google Scholar]
- Penzig AJO, Saccardo PA. (1897) Diagnoses fungorum novorum in insula Java collectorum. Ser. I. Malpighia 11: 387–409. 10.5962/bhl.title.4921 [DOI] [Google Scholar]
- Perera RH, Maharachchikumbura SSN, Bhat JD, Al-Sadi AM, Liu JK, Hyde KD, Liu ZY. (2016) New species of Thozetella and Chaetosphaeria and new records of Chaetosphaeria and Tainosphaeria from Thailand. Mycosphere 7: 1301–1321. 10.5943/mycosphere/7/9/5 [DOI] [Google Scholar]
- Plaingam N, Somrithipol S, Jones EBG. (2003) Infundibulomyces: a new genus of coelomycetes from Thailand. Canadian Journal of Botany 81: 732–737. 10.1139/b03-069 [DOI] [Google Scholar]
- Réblová M. (2004) Four new species of Chaetosphaeria from New Zealand and redescription of Dictyochaeta fuegiana. Studies in Mycology 50: 171–186. [Google Scholar]
- Réblová M, Barr ME, Samuels GJ. (1999) Chaetosphaeriaceae, a new family for Chaetosphaeria and its allies. Sydowia 51: 49–70. 10.33585/cmy.51101 [DOI] [Google Scholar]
- Réblová M, Gams W. (1999) Teleomorph-anamorph connections in Ascomycetes. 1. Cylindrotrichum and Cacumisporium anamorphs of Chaetosphaeria. Czech Mycology 51: 1–41. [Google Scholar]
- Réblová M, Miller AN, Rossman AY, Seifert KA, Crous PW, Hawksworth DL, Abdel-Wahab MA, Cannon PF, Daranagama DA, De Beer ZW, Huang SK, Hyde KD, Jayawardena R, Jaklitsch W, Jones EBG, Ju YM, Judith C, Maharachchikumbura SSN, Pang KL, Petrini LE, Raja HA, Romero AI, Shearer C, Senanayake IC, Voglmayr H, Weir BS, Wijayawarden NN. (2016) Recommendations for competing sexual-asexually typified generic names in Sordariomycetes (except Diaporthales, Hypocreales, and Magnaporthales). IMA Fungus 7: 131–153. 10.5598/imafungus.2016.07.01.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Réblová M, Nekvindová J, Kolařík M, Hernández-Restrepo M. (2020) Delimitation and phylogeny of Dictyochaeta, and introduction of Achrochaeta and Tubulicolla genera nova. Mycologia (Accepted). [DOI] [PubMed]
- Réblová M, Seifert KA. (2003) Six new species of Chaetosphaeria from tropical rain forests in Thailand and redescription of Chaetosphaeria hiugensis. Sydowia 55: 313–347. [Google Scholar]
- Réblová M, Seifert KA. (2007) A new fungal genus, Teracosphaeria, with a phialophora-like anamorph (Sordariomycetes, Ascomycota). Mycological Research 111: 287–298. 10.1016/j.mycres.2006.12.005 [DOI] [PubMed] [Google Scholar]
- Réblová M, Seifert KA. (2008) A new species of Chaetosphaeria with Menispora ciliata and phialophora-like anamorphs. Fungal Diversity 29: 99–105. [Google Scholar]
- Réblová M, Winka K. (2000) Phylogeny of Chaetosphaeria and its anamorphs based on morphological and molecular data. Mycologia 92: 939–954. 10.1080/00275514.2000.12061238 [DOI] [Google Scholar]
- Saccardo PA. (1879) Fungi Gallici lecti a cl. viris P. Brunaud, C.C. Gillet et Abb. Letendre. Michelia 1: 500–538. [Google Scholar]
- Saccardo PA. (1880) Conspectus generum fungorum Italiae inferiorum nempe ad Sphaeropsideas, Melanconieas et Hyphomyceteas pertinentium systemate sporologico dispositorum. Michelia 2: 1–38. [Google Scholar]
- Saccardo PA. (1883) Sylloge Pyrenomycetum. Sylloge Fungorum 2: 1–813. [Google Scholar]
- Samuels GJ, Müller E. (1978) Life-history studies of Brazilian Ascomycetes. 1. Two new genera of Sphaeriaceae having, respectively, Sporoschisma-like and Codinaea anamorphs. Sydowia 31: 126–136. [Google Scholar]
- Sayers EW, Cavanaugh M, Clark K, Ostell J, Pruitt KD, Karsch-Mizrachi I. (2019) GenBank. Nucleic Acids Research 47: D94–D99. 10.1093/nar/gky989 [DOI] [PMC free article] [PubMed]
- Schoch CL, Crous PW, Groenewald JZ, Boehm EWA, Burgess TI, De Gruyter J, de Hoog GS, Dixon LJ, Grube M, Gueidan C, Harada Y, Hatakeyama S, Hirayama K, Hosoya T, Hyde KD, Jones EBG, Kohlmeyer J, Kruys Å, Lücking R, Lumbsch HT, Lutzoni F, Marvanová L, McVay AH, Mbatchou JS, Miller AN, Mugambi GK, Muggia L, Nelsen MP, Nelson P, Owensby CA, Li YM, Phillips AJL, Phongpaichit S, Pointing SB, Pujade-Renaud V, Raja HA, Rivas Plata E, Robbertse B, Ruibal C, Sakayaroj J, Sano T, Selbmann L, Shearer CA, Shirouzu T, Slippers B, Suetrong S, Tanaka K, Volkmann-Kohlmeyer B, Wingfield MJ, Wood AR, Woudenberg JHC, Yonezawa H, Zhang Y, Spatafora JW. (2009) A class-wide phylogenetic assessment of Dothideomycetes. Studies in Mycology 64: 1–15. 10.3114/sim.2009.64.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert K, Morgan-Jones G, Gams W, Kendrick B. (2011) The genera of hyphomycetes. CBS-KNAW Fungal Biodiversity Centre, Utrecht, CBS Biodiversity Series no. 9: 1–997. 10.3767/003158511X617435 [DOI] [Google Scholar]
- Shenoy BD, Jeewon R, Wang H, Amandeep K, Ho HW, Bhat DJ, Crous PW, Hyde KD. (2010) Sequence data reveals phylogenetic affinities of fungal anamorphs Bahusutrabeeja, Diplococcium, Natarajania, Paliphora, Polyschema, Rattania and Spadicoides. Fungal Diversity 44: 161–169. 10.1007/s13225-010-0059-8 [DOI] [Google Scholar]
- Somrithipol S, Sakayaroj J, Rungjindamai N, Plaingam N, Jones EBG. (2008) Phylogenetic relationship of the coelomycete genus Infundibulomyces based on nuclear rDNA data. Mycologia 100: 735–741. 10.3852/07-040 [DOI] [PubMed] [Google Scholar]
- Spegazzini C. (1923) Algunos hongos de Tierra del Fuego. Physis Revista de la Sociedad Argentina de Ciencias Naturales 7: 9–23. [Google Scholar]
- Stamatakis A. (2014) RAxML Version 8: A tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian CV, Bhat DJ. (1977) Bahusutrabeeja, a new genus of the hyphomycetes. Canadian Journal of Botany 55: 2202–2206. 10.1139/b77-249 [DOI] [Google Scholar]
- Sukosd Z, Knudsen B, Kjems J, Pedersen CN. (2012) PPfold 3.0: Fast RNA secondary structure prediction using phylogeny and auxiliary data. Bioinformatics 28: 2691–2692. 10.1093/bioinformatics/bts488 [DOI] [PubMed] [Google Scholar]
- Sutton BC. (1965) Typification of Dendrophoma and a reassessment of D. obscurans. Transactions of the British Mycological Society 48: 611–616. 10.1016/S0007-1536(65)80038-9 [DOI] [Google Scholar]
- Swofford DL. (2003) PAUP*. Phylogenetic analysis using parsimony (* and other methods). Version 4. Sinauer Associates, Sunderland.
- Tibpromma S, Hyde KD, McKenzie EHC, Bhat DJ, Phillips AJL, Wanasinghe DN, Samarakoon MC, Jayawardena RS, Dissanayake AJ, Tennakoon DS, Doilom M, Phookamsak R, Tang AMC, Xu JC, Mortimer PE, Promputtha I, Maharachchikumbura SSN, Khan S, Karunarathna SC. (2018) Fungal diversity notes 840–928: micro-fungi associated with Pandanaceae. Fungal Diversity 93: 1–160. 10.1007/s13225-018-0408-6 [DOI] [Google Scholar]
- Tulasne ELR, Tulasne C. (1863) Selecta Fungorum Carpologia, Tomus Secundus. Xylariei - Valsei - Sphaeriei. Paris, Imperial, Typograph, I–XIX, 1–319.
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. 10.1128/JB.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu D, Groenewald M, de Vries M, Gehrmann T, Stielow B, Eberhardt U, Al-Hatmi A, Groenewald JZ, Cardinali G, Houbraken J, Boekhout T, Crous PW, Robert V, Verkley GJM. (2019) Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Studies in Mycology 92: 135–154. 10.1016/j.simyco.2018.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns TD, Lee SB, Taylor JW. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (Eds) PCR protocols: A guide to the methods and applications.Academic Press, New York, 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI]
- Yang J, Liu JK, Hyde KD, Bhat DJ, Jones EBG, Liu ZY. (2016) New species of Sporoschisma (Chaetosphaeriaceae) from aquatic habitats in Thailand. Phytotaxa 289: 147–157. 10.11646/phytotaxa.289.2.4 [DOI] [Google Scholar]
- Yang J, Liu NG, Liu JK, Hyde KD, Jones EBG, Liu ZY. (2018) Phylogenetic placement of Cryptophiale, Cryptophialoidea, Nawawia, Neonawawia gen. nov. and Phialosporostilbe. Mycosphere 9: 1132–1150. 10.5943/mycosphere/9/6/5 [DOI] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. (2000) A greedy algorithm for aligning DNA sequences. Journal of Computational Biology 7: 203–214. 10.1089/10665270050081478 [DOI] [PubMed] [Google Scholar]
- Zuker M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Research 31: 3406–3415. 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.