Abstract
Dysregulated microRNAs (miRNAs) have been implicated in the pathogenic processes of colon cancer. Epithelial mesenchymal transition (EMT) promotes metastatic progression and cancer stem cells are closely involved in colon cancer proliferation and metastasis. Functional effects of miR-377 on colon cancer stem cell phenotypes and EMT were then determined in the present study. Firstly, reduced miR-377 was found in colon cancer tissues and cell lines. Results from flow cytometry, sphere formation and western blot assays showed that miR-377 knockdown increased number of ALDH+ cells and promoted sphere formation ability. Moreover, cell migration/invasion and EMT of colon cancer cells were suppressed by miR-377 over-expression. On the contrary, miR-377 mimics caused the reversed results. ZEB2 (zinc finger E box-binding homeobox 2) was then validated as a binding target of miR-377. ZEB2 over-expression reversed the inhibitory abilities of miR-377 on cancer stem cell phenotypes, EMT, migration and invasion. In conclusion, miR-377 regulates cancer stem cell phenotypes and EMT in colon cancer cells via regulation of ZEB2, suggesting a new therapeutic strategy for colon cancer treatment.
Keywords: miR-377, cancer stem cell, EMT, ZEB2, metastasis, colon cancer
Introduction
Colon cancer ranks the second leading cause of cancer mortality.1 Nearly 50% of colon cancer patients have distance metastasis and recurrence, leading to poor prognosis, and the median survival after recurrence was only 13.3 months.2 It is well-accepted that metastasis is a complicated process and closely associated with EMT.3 Identification of agents that regulate EMT of colon cancer may be useful for the better understanding on the metastatic process in colon cancer. Furthermore, highly recurrence rate of colon cancer, related to cancer stem cells, is involved in drug resistance during treatment of colon cancer.4,5 Characterization of molecules that mediate cancer stem cell phenotypes of colon cancer may also be effective for the improvement of patient outcomes. Cancer stem cell shows mesenchymal phenotype to promote EMT for the metastasis-initiating activity of tumors.6 Moreover, EMT cells also demonstrate cancer stem cell-like features.7 Therefore, inhibition of cancer stem cell might be effective for suppression of colon cancer metastasis.8
MicroRNAs (miRNAs) negatively regulate target genes, and could be used as tumor suppressors or oncogenes.9 A large number of miRNAs were found to be abnormally expressed in different pathological states, such as cancer.10 For example, miR-21 functions as an oncomiR due to its promptive ability on the metastasis of several tumors.11,12 In colon cancer, miRNAs could be either up-regulated or down-regulated, and promoted or suppressed tumorigenesis or apoptosis.13
MiR-377, abnormally expressed in a variety of tumors, could inhibit tumor proliferation, migration and invasion.14-16 Recently, miR-377-3p was reported to be elevated in colorectal cancer, and functioned as an oncogene to promote carcinogenesis.17 However, its biological function and related mechanism o in colon cancer remains elusive. Moreover, miR-377 could target CD133 (stem cell marker),18 and is detrimental to cardiac stem cell.19 However, the role of miR-377 in colon cancer stem cell phenotypes has not been reported.
Here, the expression level of miR-377 in colon cancer was determined. The influences of miR-377 on colon cancer progression were then investigated. Moreover, impacts of miR-377 on colon cancer cell stem cell phenotype, as well as the binding target of miR-377, were also investigated.
Materials and Methods
Tumor Tissues
Sixty paired colon cancer and adjacent non-cancer tissues were collected from patients via surgical resection at Cancer Hospital of Xinjiang Medical University. Patients signed written informed consent, and diagnosed with imaging modalities. The project was approved by the Ethics Committee of Cancer Hospital of Xinjiang Medical University (Approval no. K-2020008).
Cell Culture
Colon cancer cell lines (HT29, SW480, SW620 andHCT116) and human intestinal epithelial cells (HIEC) were purchased from Chinese Academy of Sciences (Shanghai, China). Cells were cultured in RPMI 1640 medium (Lonza, Basel, Switzerland) supplemented with 10% fetal bovine serum (FBS, Gibco, MA, USA) in 37°C constant temperature incubator with 5% CO2.
Cell Transfection
MiR-377 mimics, inhibitor and the corresponding negative controls (Control, NC inh) were synthesized by GenePharma (Suzhou, China). HCT116 or HT29 cells were transfected with 20 nM miR-377 mimics/inhibitor or their negative controls (NCs) via Lipofectamine 2000. For over-expression of ZEB2, sequence of ZEB2 was constructed into pcDNA4.1 (Invitrogen, Carlsbad, CA, USA). HCT116 cells were cotransfected with miR-377 mimics and pcDNA4.1-ZEB2 via Lipofectamine 2000.
Flow Cytometry Sorting
HCT116 or HT29 cells (2 × 106/mL) were harvested by trypsin treatment and suspended in PBS. Cells were then stained with anti-CD133-APC and anti-ALDH1-FITC antibodies (BD Pharmingen, San Jose, CA, USA) at 4°C. After washing with PBS, cells were analyzed with FACSaria apparatus (BD Biosciences).
Sphere Formation
HCT116 or HT29 cells (2 × 104/well) were plated in FBS-free RPMI 1640 medium medium (PrEBM) supplemented with 20 ng/mL EGF and bFGF, 4 μg/mL insulin and B27 in ultra-low attachment plates. Two weeks later, the floating spheres were counted.
Wound Healing
HCT116 or HT29 cells (1 × 106/well) were seeded for 24 hours, and then scratched via a plastic pipette tip. After removing debris or the detached cells, cells were then cultured for 24 hours before calculation of the wound width.
Transwell
HCT116 or HT29 cells (2 × 104/mL), suspending in 200 µL FBS-free RPMI 1640 medium, were plated in the upper chamber of well with Matrigel-coated membrane (BD Biosciences). 400 µL medium with 20% FBS was added to the lower chamber. 24 hours later, cells in the lower chamber were stained with 1% crystal violet, and then counted.
Dual Luciferase Reporter Assay
Wild-type or mutant 3’-UTR of ZEB2 was constructed into pmirGLO luciferase reporter vector (Promega, Madison, Wisconsin, USA). HCT116 cells were co-transfected with miR-377 mimics or control and pmirGLO-wt-ZEB2, pmirGLO-mut-ZEB2. 48 hours later, the luciferase activities were performed via Lucifer Reporter Assay System (Promega).
qRT-PCR
RNAs or miRNAs from colon cancer tissues or cell lines were isolated by Trizol (Invitrogen) or miRcute miRNA isolation kit (Tiangen, Beijing, China), respectively. RNAs were then reverse-transcribed into cDNAs, and qRT-PCR was conducted with SYBR Green Master (Roche, Mannheim, Germany). GAPDH or U6 was used as endogenous control. Primer sequences were showed as follows:
| ID | Sequence (5’- 3’) |
|---|---|
| GAPDH-F | ACCACAGTCCATGCCATCAC |
| GAPDH-R | TCCACCACCCTGTTGCTGTA |
| miR-377-F | TGCTGATCACACAAAGG |
| miR-377-R | TGTAGTGTGTTTCCGTTGAA |
| ZEB2-F | GAGGCGCGCGAGAAAGG |
| ZEB2-R | GCCCAGCTTCCCGTAGCC |
| U6-F | CTCGCTTCGGCAGCACA |
| U6-R | AACGCTTCACGAATTTGCGT |
Western Blot
Proteins extracted from colon cancer tissues or cells (30 µg) were separated by SDS-PAGE, and then transferred onto PVDF membrane. Membranes were incubated overnight with primary antibodies: anti-ZEB2, anti-ALDH1, anti-CD133 (1:1500, Abcam, Cambridge, MA, USA), anti-E-cadherin, anti-N-cadherin anti-Vimentin (1:2000, Abcam) and anti-GAPDH (1:3000, Abcam) at 4°C after blocking with 5% BSA. After incubation with HRP labeled secondary antibody (1:5000; Abcam), the immunoreactivities were detected.
Statistical Analysis
Results were expressed as mean ± SEM, and statistical analyses were determined by GraphPad Prism software and One-way analysis of variance. Survival curves in patients with high or low expression levels of miR-377 were plotted via Kaplan–Meier method and log-rank test. P < 0.05 was considered as a significant difference.
Results
MiR-377 Was Expressed at Low Levels in Colon Cancer Tissues and Cell Lines
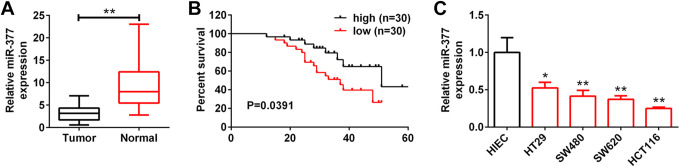
qRT-PCR revealed that miR-377 was expressed at low levels in colon cancer tissues compared with adjacent non-cancer tissues (Figure 1A). Depending on median ratio of miR-377 expression, patients with colon cancer were divided into high expression group (N = 30) and low expression group (N = 30). Moreover, patients with low expression of miR-377 showed shorter overall survival than patients with high expression of miR-377 (P = 0.0391) (Figure 1B), suggesting that miR-377 predicted poor prognosis in patients with colon cancer. Furthermore, low miR-377 expression was dramatically related to pT (P < 0.001), pN (P < 0.001), TNM stage (P < 0.001), tumor size (P = 0.011), lymph node metastasis (P = 0.007) and distant metastasis (P = 0.020) ( Table 1 ), while had no significant correlations with other clinical features such as age (P = 0.405) or gender (P = 0.190) ( Table 1 ). MiR-377 was also expressed at low levels in colon cancer cell lines compared to HIEC cells (Figure 1C). Meanwhile, HCT116 cells with lowest expression of miR-377 or HT29 with highest expression of miR-377 were selected for the following functional assays.
Figure 1.
MiR-377 was expressed at low levels in colon cancer tissues and cell lines. (A) Expression level of miR-377 in colon cancer tissues and adjacent noncancer tissues detected by qRT-PCR. ** represent Normal vs. Tumor, P < 0.01. (B) Overall survival analysis of colon cancer patients with high miR-377 expression and low miR-377 expression. (C) Expression level of miR-377 in colon cancer cell lines (HT29, SW480, SW620, HCT116) and HIEC was detected by qRT-PCR. *, ** represent colon cancer cell lines vs. HIEC, P < 0.05, P < 0.01.
Table 1.
Relationship Between miR-377 Expression and Clinicopathological Characteristics in Patients With Colon Cancer.
| Characteristics | Number of patients | miR-377 Low expression (≤ medin) | miR-377 High expression (> medin) | P value |
|---|---|---|---|---|
| Number | 60 | 30 | 30 | |
| Ages(years) | 0.405 | |||
| <65 | 19 (31.67%) | 11 (36.67%) | 8 (26.67%) | |
| ≥65 | 41 (68.33%) | 19 (63.33%) | 22 (73.33%) | |
| Gender | 0.190 | |||
| Male | 35 (58.33%) | 15 (50.00%) | 20 (66.67%) | |
| Female | 25 (41.67%) | 15 (50.00%) | 10 (33.33%) | |
| Location | 0.795 | |||
| Right | 27 (45.00%) | 13 (43.33%) | 14 (46.67%) | |
| Others | 33 (55.00%) | 17 (56.67%) | 16 (53.33%) | |
| pT stage | <0.001 | |||
| T1 | 25 (41.67%) | 5 (16.67%) | 20 (66.67%) | |
| T2 | 24 (40.00%) | 15 (50.00%) | 9 (30.00%) | |
| T3 | 9 (15.00%) | 8 (26.67%) | 1 (3.33%) | |
| T4 | 2 (3.33%) | 2 (6.67%) | 0 (0.00%) | |
| pN stage | ||||
| N0 | 27 (45.00%) | 4 (13.33%) | 23 (76.67%) | <0.001 |
| N1 | 25 (41.67%) | 19 (63.33%) | 6 (20.00%) | |
| N2 | 8 (13.33%) | 7 (23.33%) | 1 (3.33%) | |
| TNM stage | <0.001 | |||
| I/II | 35 (58.33%) | 10 (33.33%) | 25 (83.33%) | |
| III | 25 (41.67%) | 20 (66.67%) | 5 (16.67%) | |
| Differentiation | 0.275 | |||
| Well | 27 (45.00%) | 12 (40.00%) | 15 (50.00%) | |
| Moderate | 30 (50.00%) | 15 (50.00%) | 15 (50.00%) | |
| Poor | 3 (5.00%) | 3 (10.00%) | 0 (36.67%) | |
| Vessel invasion | 0.347 | |||
| No | 47 (78.33%) | 22 (73.33%) | 25 (83.33%) | |
| Yes | 13 (21.67%) | 8 (26.67%) | 5 (16.67%) | |
| Tumor size (cm) | 0.011* | |||
| <5 | 35 (58.33%) | 13 (43.33%) | 22 (73.33%) | |
| ≥5 | 25 (41.67%) | 17 (56.67%) | 8 (26.67%) | |
| Lymph node metastasis | 0.007* | |||
| No | 49 (81.67%) | 18 (60.00%) | 27 (90.00%) | |
| Yes | 11 (18.33%) | 12 (40.00%) | 3 (10.00%) | |
| Distant metastasis | 0.020* | |||
| No | 57 (95.00%) | 25 (83.33%) | 30 (100.00%) | |
| Yes | 3 (5.00%) | 5 (16.67%) | 0 (0.0047%) |
* p < 0.05,**p < 0.001*.
Over-Expression of miR-377 Suppressed Colon Cancer Stem Cell Phenotype
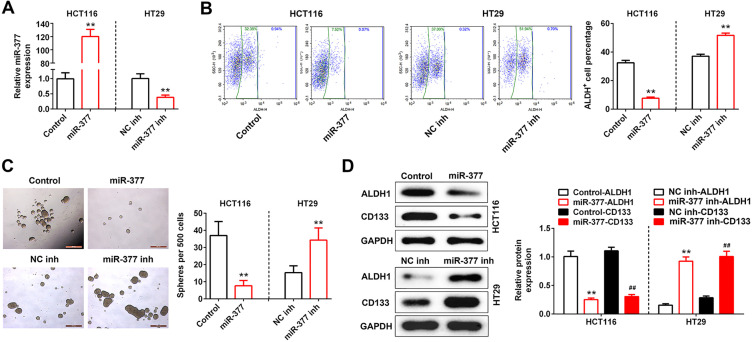
The biological roles of miR-377 in colon cancer stem cell phenotypes were investigated by in vitro gain- and loss-of functional analysis. Firstly, HCT116 cells were transfected with miR-377 mimics or HT29 cells were transfected with miR-377 inhibitor (Figure 2A). Secondly, ALDH, as a cancer stem cell marker, was enriched in HT29 cells caused by miR-377 inhibitor, as shown by the increased percentage of ALDH+ cells (Figure 2B). However, miR-377 mimics decreased the percentage of ALDH+ cells in HCT116 cells (Figure 2B). Notably, tumor sphere formation assay showed that miR-377 over-expression could inhibit self-renewal of spherogenic HCT116 cells, while miR-377 knockdown promoted self-renewal of spherogenic HT29 cells (Figure 2C). MiR-377 mimics also decreased the protein expression levels of ALDH1 and CD133 in HCT116 cells, while miR-377 inhibitor increased the protein expression levels of ALDH1 and CD133 in HT29 cells (Figure 2D).
Figure 2.
Over-expression of miR-377 suppressed colon cancer stem cell phenotype. (A) Transfection efficiency of miR-377 mimics in HCT116 cells and miR-377 inhibitor in HT29 cells was determined by qRT-PCR. ** represent miR-377 mimics vs. Control or miR-377 inhibitor vs. NC inh, P < 0.01. (B) Effect of miR-377 on the percentage of ALDH+ cells was detected by flow cytometry sorting. ** represent miR-377 mimics vs. Control or miR-377 inhibitor vs. NC inh, P < 0.01. (C) Effect of miR-377 on self-renewal of spherogenic HCT116 and HT29 cells was detected by sphere formation assay. ** represent miR-377 mimics vs. Control or miR-377 inhibitor vs. NC inh, P < 0.01. (D) Effect of miR-377 on the protein expression of ALDH1 and CD133 in HCT116 and HT29 cells was detected by western blot. ** or ## represent miR-377 mimics vs. Control or miR-377 inhibitor vs.NC inh, P < 0.01.
Over-Expression of miR-377 Suppressed the Migration, Invasion and EMT of Colon Cancer Cells
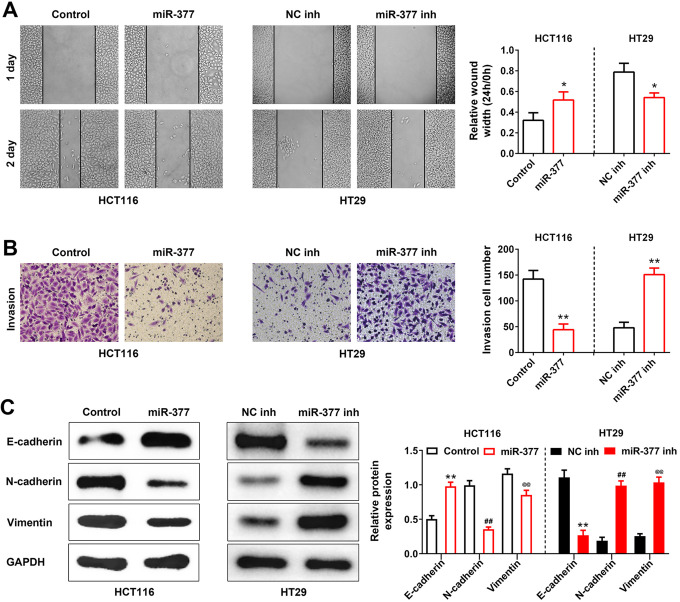
The impacts of miR-377 on growth of colon cancer cells were also evaluated. Wound healing (Figure 3A) showed that miR-377 over-expression inhibited the migration of HCT116 cells, and the invasion of HCT116 cells was also suppressed by miR-377mimics (Figure 3B). However, the migratory and invasive ability of HT29 cells were enhanced by miR-377 knockdown, but the migratory and invasive ability of HCT116 cells were reduced by miR-377mimics (Figure 3A and 3B). Moreover, as deciphered in Figure 3C, miR-377 mimics increased the protein expression of E-cadherin, while decreased the protein expressions of N-cadherin and Vimentin in HCT116 cells. However, knockdown of miR-377 increased the protein expressions of N-cadherin, E-cadherin and Vimentin in HT29 cells (Figure 3C).
Figure 3.
Over-expression of miR-377 suppressed EMT of colon cancer cells. (A) Effect of miR-377 on HCT116 and HT29 cells migration was detected by wound healing assay. * represent miR-377 mimics vs. Control or miR-377 inhibitor vs.NC inh, P < 0.05. (B) Effect of miR-377 on HCT116 and HT29 cells invasion was detected by transwell assay. ** represent miR-377 mimics vs. Control or miR-377 inhibitor vs. NC inh, P < 0.01. (C) Effect of miR-377 on protein expression of E-cadherin, N-cadherin and vimentin in HCT116 and HT29 cells was detected by sphere formation assay. **, ## or @@ represent miR-377 mimics vs. Control or miR-377 inhibitor vs. NC inh, P < 0.01.
ZEB2 Was Directly Repressed by miR-377
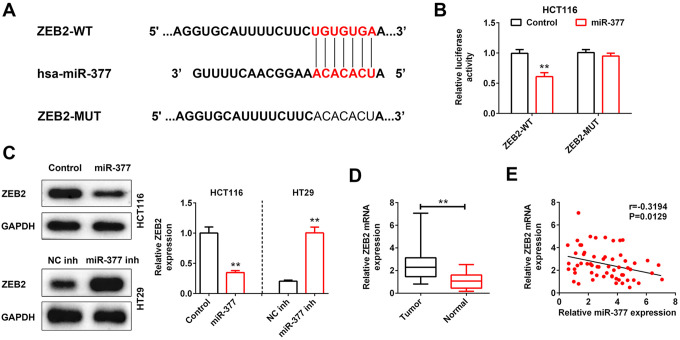
By bioinformatics tools, we confirmed that ZEB2 had conserved binding sites for miR-377 (Figure 4A). Luciferase reporter assay showed the decreased luciferase activity in HCT116 cells that co-transfected with miR-377 mimics and pmirGLO-wt-ZEB2 as compared to the Control. However, mutations in the binding site of ZEB2 reversed the suppressive influence of miR-377 mimics on the luciferase activity (Figure 4B). Importantly, miR-377 mimics decreased ZEB2 expression in HCT116 cells, while miR-377 knockdown increased ZEB2 expression in HT29 cells (Figure 4C). Furthermore, ZEB2 was elevated in colon cancer tissues as compared to adjacent non-cancer tissues (Figure 4D). A negatively correlation between miR-377 and ZEB2 was validated in patients with colon cancer (Figure 4E).
Figure 4.
ZEB2 was directly repressed by miR-377. (A) The predicted wildtype (WT) binding site of miR-377 in ZEB2. Mutant (MUT) with disrupted binding site of miR-377 in ZEB2 was also shown. (B) The effect of miR-377 mimics on the luciferase activity of reporter gene with wild-type or mutant ZEB2 was detected by luciferase reporter assay. ** represent miR-377 mimics vs. Control, P < 0.01. (C) Effect of miR-377 on protein expression of ZEB2 in HCT116 and HT29 cells invasion was detected by western blot. ** represent miR-377 mimics vs. Control or miR-377 inhibitor vs. NC inh, P < 0.01. (D) Expression level of ZEB2 in colon cancer tissues and adjacent non-cancer tissues was detected by qRT-PCR. ** represent Normal vs. Tumor, P < 0.01. (E) Negative correlation between miR-377 and ZEB2 in colon cancer patients.
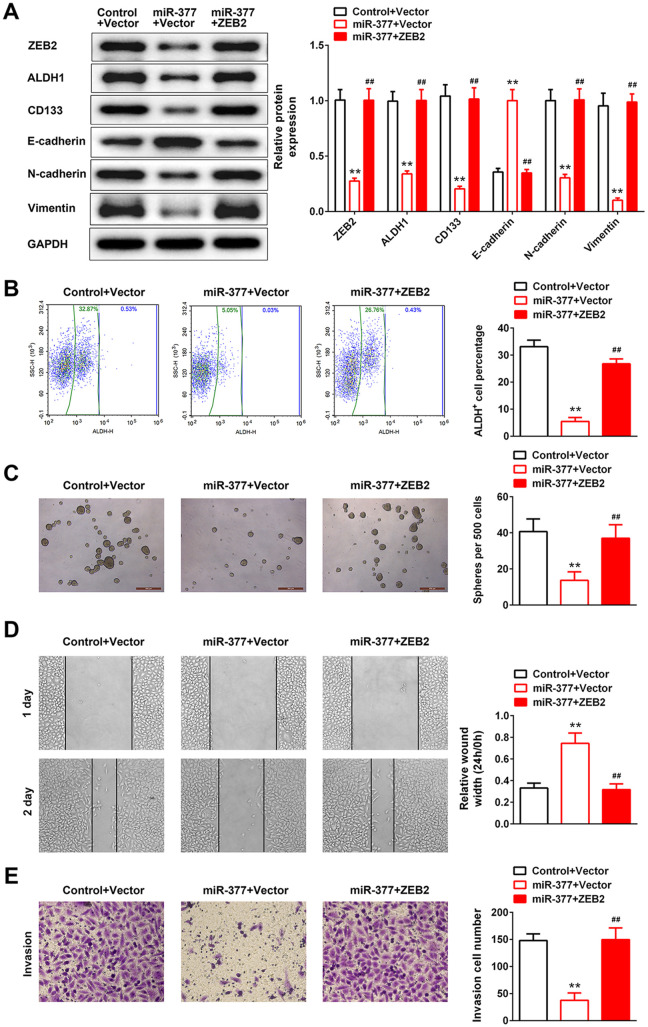
Over-Expression of miR-377 Suppressed Colon Cancer Stem Cell Phenotype and EMT Via Repression of ZEB2
To investigate the functional significance of ZEB2 in miR-377-mediated colon cancer stem cell phenotype and EMT, HCT116 cells were co-transfected with miR-377 mimics and pcDNA4.1-ZEB2. As shown in Figure 5A, over-expression of ZEB2 weakened the induction in E-cadherin expression and reductions in the expressions of ZEB2, ALDH1, CD133, N-cadherin and Vimentin caused by miR-337 over-expression in HCT116 cells. Moreover, over-expression of ZEB2 reversed miR-377-induced suppression of percentage of ALDH+ cells (Figure 5B) and self-renewal of spherogenic HCT116 cells (Figure 5C). Notably, over-expression of ZEB2 attenuated miR-377-suppressed cell migration (Figure 5D) and invasion (Figure 5E) in HCT116 cells.
Figure 5.
Over-expression of miR-377 suppressed colon cancer stem cell phenotype and EMT via repression of ZEB2. (A) Effect of miR-377 and ZEB2 on the protein expression of ZEB2, ALDH1, CD133, E-cadherin, N-cadherin and vimentin in HCT116 cells was detected by western blot. ** represents miR-377 + Vector vs. Control + Vector, P < 0.01. ## represents miR-377 + Vector vs. miR-377 + ZEB2, P < 0.01. (B) Effect of miR-377 and ZEB2 on the percentage of ALDH+ cells detected by flow cytometry sorting. ** represents miR-377 + Vector vs. Control + Vector, P < 0.01. ## represents miR-377 + Vector vs. miR-377 + ZEB2, P < 0.01. (C) Effect of miR-377 and ZEB2 on the self-renewal of spherogenic HCT116 cells was detected by sphere formation assay. ** represents miR-377 + Vector vs. Control + Vector, P < 0.01. ## represents miR-377 + Vector vs. miR-377 + ZEB2, P < 0.01. (D) Effect of miR-377 mimics and ZEB2 overexpression on HCT116 cell migration was detected by wound healing assay. ** represents miR-377 + Vector vs. Control + Vector, P < 0.01. ## represents miR-377 + Vector vs. miR-377 + ZEB2, P < 0.01. (E) Effect of miR-377 mimics and ZEB2 overexpression on HCT116 cell invasion was detected by transwell assay. ** represents miR-377 + Vector vs. Control + Vector, P < 0.01. ## represents miR-377 + Vector vs. miR-377 + ZEB2, P < 0.01.
Discussion
Metastasis could attribute to the dysregulated survival ability of tumors, especially in colon cancer.20 Colon stem cells have properties including self-renewal and multipotency,21 are implicated in metastasis.22 Therefore, therapies targeting colon stem cells have become a critical strategy to treat the metastasis of colon cancer.23 For example, inhibition of extracellular signal-regulated kinase 5 could eliminate cancer stem cells, thus improving colon cancer treatment.24 Recently, through regulation of pathways involved in colon cancer stem cell, miRNAs have been reported to provide perspective on metastasis and recurrence of colon cancer.25 Here, our results demonstrated that miR-377 could regulate cancer stem cell phenotypes, and inhibited EMT of colon cancer, suggesting that miR-377 had tumor suppressive ability against colon cancer via suppression of colon cancer stem cells and metastasis.
Previous study has shown that miR-377-3p was up-regulated in colorectal cancer tissues.17 However, our results were in line with Huang et al (插入文献MiR-377-3p Suppresses Colorectal Cancer Through Negative Regulation on Wnt/β-catenin Signaling by Targeting XIAP and ZEB2), showing that miR-377 was decreased in colon cancer tissues. Moreover, Huang et al (插入文献MiR-377-3p Suppresses Colorectal Cancer Through Negative Regulation on Wnt/β-catenin Signaling by Targeting XIAP and ZEB2) indicated the low miR-377 expression was not only related to the overall survival of colon cancer patients, but also tightly related to pT, pN and TNM stage of the patients, predicting a poor prognosis. The different miR-377 expression in colorectal or colon cancer may be caused by the small sample sizes of patients. Therefore, a larger patient cohort is needed to indicate clinical significance of miR-377 in colon cancer.
Cancer stem cell with surface markers (CD133, ALDH1 and CD44) has been identified in various tumors.26 FACS analysis showed that colon cancer stem cells was characterized by ALDH+, while non-colon cancer stem cells was ALDH-.27 Increase of ALDH1 and CD133 promoted tumorigenic capacity of colon cancer stem cells.28 This study revealed that miR-377 reduced the enrichment of ALDH+ HCT116 cells, inhibited self-renewal of spherogenic HCT116 cells and decreased protein expression of ALDH1 and CD133, suggesting its inhibitory effect on colon cancer stem cell phenotype. Moreover, miR-377 has been shown to target CD133 to suppress esophageal cancer progression,18 and activation of cancer stem cell is tightly associated with recurrence, metastasis and poor prognosis of colon cancer.29 Influences of miR-377 on colon cancer metastasis were then evaluated.
Consistently, a previous report showed that miR-377 exerted inhibitory effect on colon cancer progression through reduction of E2F3 (插入文献CircPRMT5 circular RNA promotes proliferation of colorectal cancer through sponging miR-377 to induce E2F3 expression), and function assays revealed that miR-377 suppressed colon cancer cell migration and invasion. Moreover, miR-377 was shown to enhance epithelial cell marker (E-cadherin), while reduce mesenchymal markers (N-cadherin and vimentin), thus repressing EMT of colon cancer. Previous research showed that miR-377-3p promoted EMT of colorectal cancer through targeting glycogen synthase kinase-3β.17 However, miR-377 was shown to suppress EMT through targeting ZEB2 in colorectal cancer (插入文献MiR-377-3p Suppresses Colorectal Cancer Through Negative Regulation on Wnt/β-catenin Signaling by Targeting XIAP and ZEB2). The functional target of miR-337 involved in colon cancer progression in this study was then determined.
Our result revealed that ZEB2, belonging to ZEB families, was also validated as a target of miR-377, in line with a previous report showing that miR-377/ZEB2 axis inhibited cervical cancer progression.15 ZEB has been shown to be potent inducers of EMT.30 ZEB2 could promote EMT in various tumors.31,32 Moreover, ZEB2 was elevated in colorectal cancer tissues and functioned as a biomarker for colorectal cancer.33 ZEB2 was involved in miRNAs-regulated metastasis and EMT of colon cancer.34,35 Over-expression of ZEB2 promoted colorectal cancer invasion,36 and EMT for the activation of colorectal cancer stem cells.37 Here, ZEB2 over-expression reversed the inhibitory effects of miR-377 on colon cancer stem cell phenotype and EMT. However, in vivo animal experiments should be conducted to further investigate the role of miR-377/ZEB2 axis in colon cancer.
In conclusion, this study firstly demonstrated that miR-377 suppressed colon cancer stem cell phenotype and EMT, thus repressing metastasis of colon cancer. Additionally, ZEB2 was closely associated with the anticancer role of miR-377 in colon cancer. These findings might contribute to the development of a promising anticancer agent for colon cancer.
Abbreviation
- EMT
Epithelial mesenchymal transition
- miRNAs
microRNAs
Footnotes
Authors’ Contributions: Paerhati shayimu and Aikeremu yusufu conceived and designed the experiments,Aizimaiti rehemutula and Darebai redati analyzed and interpreted the results of the experiments, Rexida jiapaer and Rousidan tuerdi performed the experiments
Availability of data and materials: All data generated or analyzed during this study are included in this published article.
Consent to participate: All the patients signed written informed consent.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethics approval: The project was approved by the Ethics Committee of Cancer Hospital of Xinjiang Medical University.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by Natural Science Foundation of Xinjiang Uygur Autonomous Region (Grant No. 2019D01C261).
ORCID iD: Aikeremu Yusufu  https://orcid.org/0000-0002-7839-6928
https://orcid.org/0000-0002-7839-6928
References
- 1. Haggar F, Boushey R. Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clin Colon Rectal Surg. 2009;22(4):191–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. O’Connell M, Campbell M, Goldberg R, et al. Survival following recurrence in stage II and III colon cancer: Findings from the ACCENT data set. J Clin Onco. 200;26(14):2336–2341. [DOI] [PubMed] [Google Scholar]
- 3. Mehlen P, Puisieux A. Metastasis: a question of life or death. Nature Rev Cancer. 2006;6(6):449–458. [DOI] [PubMed] [Google Scholar]
- 4. Sanders M, Majumdar A. Colon cancer stem cells: implications in carcinogenesis. Front Biosci. 2011;16:1651–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nature Rev Cancer. 2005;5(4):275–284. [DOI] [PubMed] [Google Scholar]
- 6. Al-Hajj M, Becker MW, Wicha M, Weissman I, Clarke MF. Therapeutic implications of cancer stem cells. Curr Opin Genet Dev. 2004;14(1):43–47. [DOI] [PubMed] [Google Scholar]
- 7. Wang Z, Li Y, Ahmad A, et al. Targeting miRNAs involved in cancer stem cell and EMT regulation: an emerging concept in overcoming drug resistance. Drug Resist Updat. 2010;13(4):109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ordóñez-Morán P, Dafflon C, Imajo M, Nishida E, Huelsken J. HOXA5 counteracts stem cell traits by inhibiting Wnt signaling in colorectal cancer. Cancer Cell. 2015;28(6):815–829. [DOI] [PubMed] [Google Scholar]
- 9. Hutvagner G, Zamore P. A MicroRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297(5589):2056–2060. [DOI] [PubMed] [Google Scholar]
- 10. Di Leva G, Croce CM. miRNA profiling of cancer. Curr Opin Genet Dev. 2013;23(1):3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Medina P, Nolde M, Slack F. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467(7311):86–90. [DOI] [PubMed] [Google Scholar]
- 12. Ziyan W, Shuhua Y, Xiufang W, Xiaoyun L. MicroRNA-21 is involved in osteosarcoma cell invasion and migration. Med Oncol. 2011;28(4):1469–1474. [DOI] [PubMed] [Google Scholar]
- 13. Yang L, Belaguli N, Berger DH. MicroRNA and Colorectal Cancer. World J Surg. 2009;33(4):638–646. [DOI] [PubMed] [Google Scholar]
- 14. Wang R, Ma Y, Yu D, Zhao J, Ma P. miR-377 functions as a tumor suppressor in human clear cell renal cell carcinoma by targeting ETS1. Biomed Pharmacother. 2015;70:64–71. [DOI] [PubMed] [Google Scholar]
- 15. Tang L, Yang B, Cao X, Li Q, Jiang L, Wang D. MicroRNA-377-3p inhibits growth and invasion through sponging JAG1 in ovarian cancer. Genes Genomics. 2019;41(8):919–926. [DOI] [PubMed] [Google Scholar]
- 16. Chen G, Lu L, Liu C, Shan L, Yuan D. MicroRNA-377 suppresses cell proliferation and invasion by inhibiting tiam1 expression in hepatocellular carcinoma. PloS One. 2015;10(3):e0117714. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17. Liu W-Y, Yang Z, Sun Q, et al. miR-377-3p drives malignancy characteristics via upregulating GSK-3β expression and activating NF-κB pathway in hCRC cells. J Cell Biochem. 2018;119(2):2124–2134. [DOI] [PubMed] [Google Scholar]
- 18. Li B, Xu WW, Han L, et al. MicroRNA-377 suppresses initiation and progression of esophageal cancer by inhibiting CD133 and VEGF. Oncogene. 2017;36(28):3986–4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Joladarashi D, Garikipati VNS, Thandavarayan RA, et al. Enhanced cardiac regenerative ability of stem cells after ischemia-reperfusion injury: role of human CD34+ cells deficient in MicroRNA-377. J Am Coll Cardiol. 2015;66(20):2214–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Riihimäki M, Hemminki A, Sundquist J, et al. Patterns of metastasis in colon and rectal cancer. Scientific reports. 2016;6:29765 10.1038/srep29765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Munro MJ, Wickremesekera SK, Peng L, Tan ST, Itinteang T. Cancer stem cells in colorectal cancer: a review. J Clin Pathol. 2018;71(2):110–116. [DOI] [PubMed] [Google Scholar]
- 22. Shiozawa Y, Nie B, Pienta K, Morgan T, Taichman R. cancer stem cells and their role in metastasis. Pharmacol Therapeutics. 2013;138(2):285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Medema JP. Targeting the colorectal cancer stem Cell. N Engl J Med. 2017;377(9):888–890. [DOI] [PubMed] [Google Scholar]
- 24. Pereira DM, Gomes SE, Borralho PM, Rodrigues CMP. MEK5/ERK5 activation regulates colon cancer stem-like cell properties. Cell Death Discov. 2019;5(1):68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fang Y, Xiang J, Chen Z, et al. miRNA expression profile of colon cancer stem cells compared to non-stem cells using the SW1116 cell line. Oncol Rep. 2012;28(6):2115–2124. [DOI] [PubMed] [Google Scholar]
- 26. Visvader J. Cells of origin in cancer nature. Nature. 2011;469(7330):314–322. [DOI] [PubMed] [Google Scholar]
- 27. Huang EH, Hynes MJ, Zhang T, et al. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 2009;69(8):3382–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bu P, Chen K-Y, Joyce H, et al. A microRNA miR-34a-regulated bimodal switch targets notch in colon cancer stem cells. Cell Stem Cell. 2013;12(5):602–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Peitzsch C, Tyutyunnykova A, Pantel K, Dubrovska A. Cancer stem cells: the root of tumor recurrence and metastases. Semin Cancer Biol. 2017;44:10–24. [DOI] [PubMed] [Google Scholar]
- 30. Browne G, Sayan AE, Tulchinsky E. ZEB proteins link cell motility with cell cycle control and cell survival in cancer. Cell Cycle. 2014;9(5):886–891. [DOI] [PubMed] [Google Scholar]
- 31. Wang T, Chen X, Qiao W, Kong L, Sun D, Li Z. Transcription factor E2F1 promotes EMT by regulating ZEB2 in small cell lung cancer. BMC Cancer. 2017;17(1):719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Beltran M, Puig I, Pena C, et al. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Deve. 2008;22(6):756–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sreekumar R, Harris S, Moutasim K, et al. Assessment of nuclear ZEB2 as a biomarker for colorectal cancer outcome and TNM risk stratification. JAMA Netw Open. 2018;1(6):e183115–e183115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zheng Y-B, Luo H-P, Shi Q, et al. miR-132 inhibits colorectal cancer invasion and metastasis via directly targeting ZEB2. World J Gastroenterol. 2014;20(21):6515–6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhu W, Luo X, Fu H, Liu L, Sun P, Wang Z. MiR-3653 inhibits the metastasis and epithelial-mesenchymal transition of colon cancer by targeting Zeb2. Pathol Res Pract. 2019;215(10):152577. [DOI] [PubMed] [Google Scholar]
- 36. Kahlert C, Lahes S, Radhakrishnan P, et al. Overexpression of ZEB2 at the invasion front of colorectal cancer is an independent prognostic marker and regulates tumor invasion in vitro. Clin Cancer Res. 2011;17(24):7654. [DOI] [PubMed] [Google Scholar]
- 37. Li N, Jadidi R, Lorenzi F, et al. Oncogenesis An FBXW7-ZEB2 axis links EMT and tumour microenvironment to promote colorectal cancer stem cells and chemoresistance. Oncogenesis. 2019;8(3):13. [DOI] [PMC free article] [PubMed] [Google Scholar]