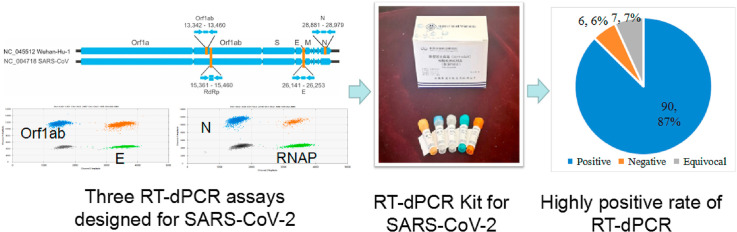
Abstract
The outbreak of COVID-19 caused by a novel Coronavirus (termed SARS-CoV-2) has spread to over 210 countries around the world. Currently, reverse transcription quantitative qPCR (RT-qPCR) is used as the gold standard for diagnosis of SARS-CoV-2. However, the sensitivity of RT-qPCR assays of pharyngeal swab samples are reported to vary from 30% to 60%. More accurate and sensitive methods are urgently needed to support the quality assurance of the RT-qPCR or as an alternative diagnostic approach. A reverse transcription digital PCR (RT-dPCR) method was established and evaluated. To explore the feasibility of RT-dPCR in diagnostic of SARS-CoV-2, a total of 196 clinical pharyngeal swab samples from 103 suspected patients, 77 close contacts and 16 supposed convalescents were analyzed by RT-qPCR and then measured by the proposed RT-dPCR. For the 103 fever suspected patients, 19 (19/25) negative and 42 (42/49) equivocal tested by RT-qPCR were positive according to RT-dPCR. The sensitivity of SARS-CoV-2 detection was significantly improved from 28.2% by RT-qPCR to 87.4% by RT-dPCR. For 29 close contacts (confirmed by additional sample and clinical follow up), 16 (16/17) equivocal and 1 negative tested by RT-qPCR were positive according to RT-dPCR, which is implying that the RT-qPCR is missing a lot of asymptomatic patients. The overall sensitivity, specificity and diagnostic accuracy of RT-dPCR were 91%, 100% and 93%, respectively. RT-dPCR is highly accurate method and suitable for detection of pharyngeal swab samples from COVID-19 suspected patients and patients under isolation and observation who may not be exhibiting clinical symptoms.
Keywords: Coronavirus disease (COVID-19), Novel coronavirus (termed SARS-CoV-2), Reverse transcription digital PCR (RT-dPCR), Reverse transcription quantitative qPCR (RT-qPCR), Sensitivity
Graphical abstract
1. Introduction
In late December 2019, a number cases of pneumonia infection were reported in Wuhan, Hubei Province, China. It was officially named Coronavirus disease (COVID-19) by the World Health Organization (WHO) and has since spread to 210 countries around the world [1,2]. The pathogen causing the outbreak of disease was identified as a novel Coronavirus (termed SARS-CoV-2), belonging to the family Coronaviridae, order Nidovirales, all of which are enveloped, non-segmented positive-sense RNA viruses [3,4]. According to the WHO and the Chinese Center for Disease Control and Prevention (CDC), the current gold standard for the diagnosis of SARS-CoV-2 infection is based on reverse transcription quantitative PCR (RT-qPCR). However, RT-qPCR is reported to have issue of false negative rates for pharyngeal swab samples [5] and there were 3% of patients who had negative RT-qPCR test results at initial presentation while chest CT checks indicated typical symptoms of viral pneumonia [6]. In order to identify and hospitalize COVID-19 patients in time, more sensitive and accurate tests are required.
Digital PCR (dPCR) is a technology which partitions nucleic acid molecules into a large number of small reactions and acquires amplification data for each partition at the endpoint based on the intensity of fluorescence [[7], [8], [9]]. Quantification is performed by applying Poisson statistics to the proportion of the negative partitions. dPCR can offer greater precision than qPCR and is far simpler to use for copy number quantification due to the binary nature in which the partitions are counted as positive or negative. Additionally, dPCR is more tolerant of PCR inhibition compared with qPCR due to partitioning and because it is an end-point PCR measurement and consequently less dependent on high PCR efficiency [10,11].
In this study, we established one step RT-dPCR method for detection of open reading frame 1 ab (ORF1ab), nucleocapsid protein (N) and envelope protein (E) gene of SARS-CoV-2. Moreover, we compared RT-qPCR and RT-dPCR on 196 clinical samples and found RT-dPCR can significantly improve the sensitivity and diagnostic accuracy of Coronavirus disease (COVID-19).
2. Experimental section
2.1. Study design
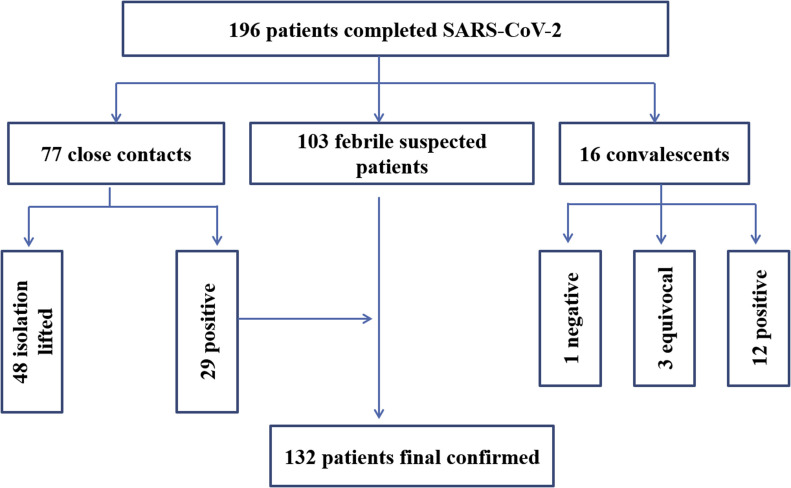
103 febrile suspected SARS-CoV-2 infected patients, 77 close contacts and 16 supposed convalescents were chosen in this study. Positive, negative and equivocal results were all included in the chosen specimens according to the RT-qPCR test, shown in Fig. 1 . RT-dPCR measurement was conducted after RT-qPCR test at the same laboratory. This study was approved by the Ethics Committee of the Wuhan CDC (WHCDCIRB-K-2020006). The analysis was performed on existing samples collected during standard diagnostic tests in the emergency state, posing no extra burden to patients.
Fig. 1.
Flow chart of the study population for SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2.
2.2. Clinical samples
Respiratory samples were obtained during February and March 2020 from hospitalized patients or close contacts of hospitalized patients tested by Beijing CDC (BJCDC), Wuhan CDC (WHCDC) and a government designated clinical test laboratory (Wuhan considerin laboratory for medical test, KXR). Samples were stored in viral transport medium (Yocon Biology) at 4 °C. RNA was extracted within 24 h from 140 μL clinical specimens and eluted into 60 μL elution buffer by using the MagMAX-96 viral RNA isolation kit (Thermo Fisher Scientific). RNA extracts containing human coronaviruses (HCoV)-229 E and (HCoV)-OC43 and avian influenza virus RNAs A/H3N2 and A/H1N1 Virus and Influenza B/Victoria Virus were provided by BJCDC. Extracted RNA was stored at −80 °C.
2.3. In vitro RNA transcription
Three in vitro transcribed RNAs were used as templates to develop RT-dPCR assays. The genome sequence of 2019-nCoV (GenBank No. NC_045512) were download from NCBI. Sequences containing ORF1ab (13201–15600), E (25381-26520), and N (28261–29820) were used to order synthetic genes from BGI (Beijing, China). All the detail information was in the supplemental information.
2.4. One step reverse transcription dPCR and RT-qPCR
Three assays for gene targets of N, ORF1ab and E of the SARS-CoV-2 were optimized on QX200 digital PCR platform [12,13]. Three different commercial RT-qPCR kits (H&R from Shanghai Huirui Biotechnology Co., Ltd, BioGerm from Shanghai BioGerm Medical Biotechnology and Daan from Daan Gene Co., Ltd) were used for the detection. The detail of RT-dPCR and RT-qPCR was in the supplemental information.
2.5. Limit of blank (LoB) and detection (LoD) of RT-dPCR
To establish the limit of blank (LoB) [14], 60 blank measurements were obtained from 3 blank samples on three days. 70 to 76 measurements from 4 to 5 samples with low concentration (1–3 cp/reaction) were used to determine the limit of detection (LoD) according to the CLSI guideline of EP17-A [15].
3. Results and discussion
3.1. Dynamic range of the RT-dPCR assay
The linear range was investigated by varying the mean copy number per droplet, denoted as λ [16]. The precision or relative error of RT-dPCR is related to λ because RT-dPCR relies on the Poisson statistics to account for droplets with multiple molecules [17]. The upper limit of the linearity was 7.8 copies/partition tested by the N gene assay. To determine the lower limit of all three assays, serial dilutions of each RNA transcript in a human total RNA background were prepared (Table S1). The measured targets matched the anticipated values in each tested interval. A good linearity (0.93<slope<1.02, R2 ≥ 0.9997) between the measured RNA target and the prepared value was observed over the range from approximately 104 to 100 copies/reaction (Fig. 2 ). Reactions containing a mean of 60 E, 66 N or 11 ORF1ab copies fulfilled the criterion for an LoQ with a CV lower than 25%.
Fig. 2.
Validated range of the RT-dPCR assays for E, ORF1ab and N gene. Evaluation of linearity of samples containing E, ORF1ab and N gene molecules over the extended λ range (0.0002 <λ < 7.83). Data are shown in mean with standard deviation for each dilution series (3 = n ≤ 10).
3.2. Establishment of LoB and LoD for RT-dPCR assay
Sixty blank measurements obtained from 3 blank samples were analyzed to determine the LoB. As the distribution of the 60 blank measurements is skewed (Fig. S1), the LoB was estimated nonparametrically as the 95th percentile of the measurements. The 15 highest blank values for each target are displayed in Table S2. The 95th percentile corresponds to the 57.5 ordered observation (=60*(0.95/60 + 0.5)) [15]. Linear interpolation between the 57th and 58th observation yields a LoB estimate of 1.6, 1.6, and 0.8 copies/reaction for E, ORF1ab and N, respectively.
For determining the LoD of ORF1ab gene assay, 76 measurements were performed on five samples in 3 different runs on three different days to ensure the total assay variation is reflected. The distribution of the 76 measurement results from low concentration samples is not Gaussian (Fig. S2A) and so that nonparametric statistics was used according to the guideline of EP17-A [15]. Consequently, the LoD is determined to be 2 copies/reaction, the lowest level material where the β-percentile is 5%.
To determine the LoD of N and E assay, 83 measurements of E assay on 5 samples and 71 measurements of N assay on 4 samples were performed in 4 different runs. Similar to ORF1ab gene, the distribution of the 71 measurements for N gene and 83 measurements for E gene are not Gaussian (Fig. S2B and S2C), and so that nonparametric statistics was used. Consequently, the LoD is determined to be 2 copies/reaction.
The LoD of the two RT-qPCR kits (Daan gene and BioGerm) used in this study were reported in a previous study [18]. The data indicates LoD of the proposed RT-dPCR is 5 times and 10 times higher than the Daan and BioGerm kit, respectively.
3.3. Specificity testing
The Specificity of the assays for ORF1ab and E gene has been tested in a previous report [13]. To further validate the specificity of all assays, Influenza virus and other human coronavirus were collected. All assays were tested on human clinical nucleic acid samples at National institute of Metrology, China. All tests returned negative results (Table S3).
3.4. Comparison between RT-qPCR and RT-dPCR on febrile suspected patients
103 pharyngeal swabs were collected from febrile suspected SARS-CoV-2 infected patients. The relevant information, RT-qPCR result and copy number determined by the proposed RT-dPCR assay is listed in Table S4. Among the 103 specimens, 81 (P1 to P81) were tested at KXR with the H&R RT-qPCR kit, 7 (P82-88) were tested at WHCDC by the Daan qPCR kit, and 15 samples (from P89–P103) were tested at BJCDC with BioGerm RT-qPCR kit and the Chinese CDC assays.
Firstly, the criteria claimed by the H&R kit manufacturer are: Ct value ≤ 35 are positive, Ct value > 39.2 are negative, and 35 < Ct < 39.2 are equivocal. The criteria of the Daan qPCR kit are: Ct > 40, negative, Ct ≤ 40, positive, and equivocal if only one gene with Ct ≤ 40 and no amplification for another gene. According to such criteria, 14 positive, 25 negative and 49 suspected SARS-CoV-2 infections were reported by qPCR (P1 to P88).
For RT-dPCR, three targets are tested in parallel in the same laboratory and the determination of a positive result should meet the following criteria: quantification of any one of the three gene targets is ≥ 2 copies/reaction. If no positive droplet was detected in FAM channel but positive droplets were detected in VIC indicating RNAseP positive for human reference control15, the sample can be judged negative. If 0<result<2, it should be attributed to equivocal and needs further verification. According to such criteria, 44 out of 49 equivocal and 17 out of 25 negatives were corrected to be positive by RT-dPCR and the positive rate significantly increased. No positive droplet was detected for the 6 negatives and copy numbers were quantified under the established LoD for 7 equivocal, due to either no virus sampled or ultra-low virus load in these specimens.
For the 15 samples tested at Beijing CDC (P89 to P103), Ct values were not available and only negative or positive information were reported. Single gene target positive was determined to be SARS-CoV-2 positive based on parallel test with a commercial kit and the Chinese CDC assays. Therefore, these 15 samples were reported positive by BJCDC. 8 RT-qPCR negatives for ORF1ab were positive tested by RT-dPCR, showing high sensitivity for ORF1ab by RT-dPCR. Only 3 negatives for ORF1ab can be complemented by E gene targets.
Among the 103 specimens, 29 positive, 25 negative and 49 equivocal were reported by RT-qPCR. However, 61 samples including 19 negative and 42 equivocal tested by RT-qPCR were confirmed to be positive by RT-dPCR, thus 90 patients in total could be reported SARS-CoV-2 nucleic acid positive and diagnosed with COVID-19. According to a follow-up survey, all the 103 patients were clinically diagnosed with SARS-CoV-2 infection through later test of re-sampling by RT-qPCR and clinical symptoms. Thus, the true positive rate of SARS-CoV-2 detection was significantly improved from 28.2% to 87.4% for the 103 patients (Fig. 3 A and B).
Fig. 3.
Diagnosis of SARS-CoV-2 by RT-qPCR (A,C) and RT-dPCR (B,D) for 103 febrile suspected patients and 77 close contacts. (A) 25 positive, 29 negative and 49 suspected were reported by RT-qPCR for 103 suspected patients. (B) 90 positive, 6 negative and 7 equivocal were determined by RT-dPCR for 103 suspected patients. (C) 12 positive, 49 negative and 16 suspected were reported by RT-qPCR for 77close contacts. (D) 28 positive, 48 negative and 1 equivocal were determined by RT-dPCR for 77 close contacts.
Furthermore, the 61 samples (either negative or equivocal tested by RT-qPCR but positive by RT-dPCR, Table S4) were reported with averaged viral load of 31, 25 and 26 copies/reaction for ORF1ab, N and E, respectively. Those 29 positive samples by RT-qPCR showed a relatively high viral load with an average of 998, 1099 and 2594 copies/reaction for ORF1ab, N and E, respectively.
3.5. Detection of SARS-CoV-2 for close contacts
77 specimens were collected from contacts and close contacts. 48 specimens from contacts were reported negative based RT-qPCR test by BJCDC on Feb 6 and were confirmed by RT-dPCR on Feb 7 in Table S5. According to a follow-up survey, all of them were in good health and isolation was lifted after 14 days.
29 specimens (Table S6 and Fig. 3C and D) were tested at WHCDC by RT-qPCR with a kit from Daan gene on March 2, 4 and 6. According to RT-qPCR result, 12 positive, one negative and 16 equivocal were reported. It is very difficult to detect the SARS-CoV-2 nucleic acids due the low virus load at the early stage for the close contacts. However, 15 out of 16 equivocal and one negative were positive by RT-dPCR. The equivocal rate was significantly decreased from 21% down to 1% according to the detection of RT-dPCR. Subsequently, the 16 R T-dPCR positive were confirmed as SARS-CoV-2 infected patients by both re-sampling test and a clinical follow up. This indicates that RT-qPCR can miss asymptomatic patients at first time.
16 samples (1 negative or 15 equivocal tested by RT-qPCR but positive by RT-dPCR) were reported with averaged viral load 66, 0.6 and 13 copies/reaction for ORF1ab, N and E, respectively, by RT-dPCR. Those 12 positive samples by RT-qPCR showed a relatively high viral load with an average of 666, 807 and 773 copies/reaction for ORF1ab, N and E, respectively.
Furthermore, among the 16 specimens corrected by RT-dPCR, 6 persons (P14,18-21and P23 in supplemental information: Table S7) were directed for secondary testing following an initial negative test 2–10 days before. Based on RT-qPCR results, further isolation and observation was still needed to be conducted as the testing result is equivocal or negative and no clinical symptoms were observed for them. However, if based on RT-dPCR, all the six patients can be diagnosed with COVID-19 infected by SARS-CoV-2 and treatment could be conducted earlier. This indicates RT-dPCR is more sensitive and suitable for low virus load specimens from the patients under isolation and observation without clinical symptoms, which is in agreement with the very recent online report [19].
3.6. Detection of SARS-CoV-2 for convalescent
16 pharyngeal swabs were collected from convalescent patients (Table 1 ). 12 positive, 3 equivocal and 1 negative were reported by qPCR. However, all of these 16 patients are diagnosed to be positive by RT-dPCR, indicating that all of them still need to be observed in hospital.
Table 1.
Comparison of SARS-CoV-2 RNA measurement on convalescent patients by RT-qPCR and RT-dPCR.
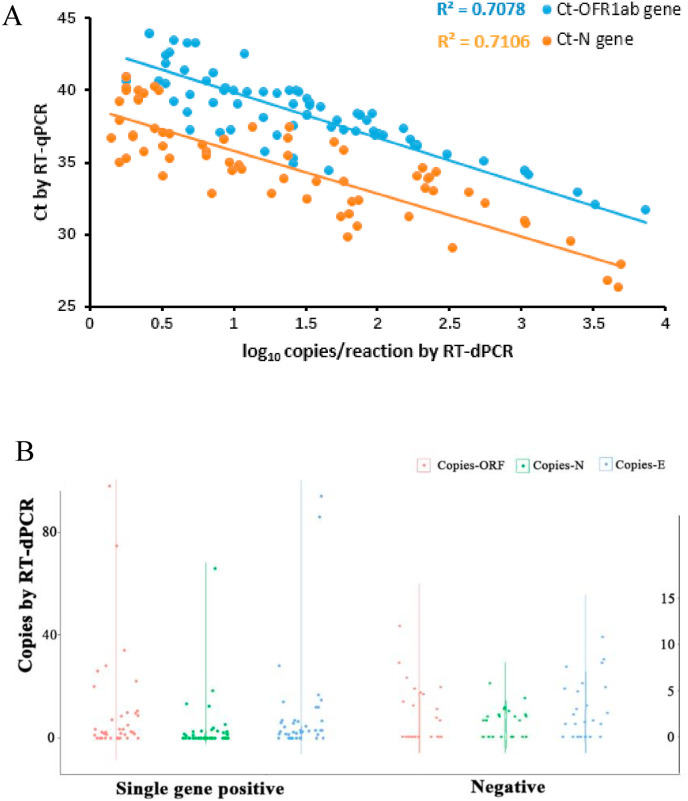
3.7. Correlation between Ct value and copy number
The correlation between the Ct value and copy number/reaction for both ORF1ab and N of all specimens was analyzed in Fig. 4 A. Ct value of RT-PCR was highly correlated with the log copy number determined by RT-dPCR (ORF1ab, R2 = 0.7078; N, R2 = 0.7106). The RT-dPCR assay for ORF1ab was correlated well with N assay. The viral load was distributed in the range of 2–100 and 2–15 copy number/reaction for the specimens with the single gene positive and negative, respectively, detected by RT-qPCR (Fig. 4B). This very low viral load could explain why these specimens were single gene positive or negative reported by RT-qPCR.
Fig. 4.
Correlation analysis between the Ct value of RT-qPCR and the viral load determined by RT-dPCR (A) and copy number distribution for the single gene positive and negative specimens (B).
RT-qPCR, as the standard method of diagnostics of SARS-CoV-2, plays an important role in this outbreak, though a low positive rate has been reported [5]. A number of factors could affect RT-PCR testing results including sample collection and transportation, RNA extraction and storage, and proper performance of the kit [20]. More recently, more than 145 RT-qPCR kits have been developed by the in vitro diagnostic manufactures (IVDs) in China [21]. Among the RT-qPCR kits, those with low sensitivity would cause high false negative rate or high equivocal rate. For the equivocal results it is necessary to conduct a retest and this would improve the positive rate of RT-qPCR. However, in the clinical practice under the current situation, it is impossible to do a same day retest due to the daily burden of thousands of incoming samples. The testing laboratory should initially report a result based on a single test, while secondary sampling for a later retest does not need to be sent to the same laboratory. Therefore, availability of a highly sensitive and accurate confirmatory method is of particular importance for the diagnosis of SARS-CoV-2 in this outbreak.
Currently, besides RT-qPCR, other methods such as next generation sequencing (NGS) and immunological detection of IgM and IgG could be used as confirmatory methods for diagnosis of COVID-19 according to the latest guideline of Diagnosis and Treatment of Pneumonitis Caused by SARS-CoV-2 (trial seventh version) published by National Health Commission [22]. This would decrease the false negative rate by applying multiple methods. However, nucleic acid testing is still considered the gold standard as this is the most direct way to detect the presence of the virus. Thus, digital PCR method could be a powerful method because it can significantly improve the sensitivity for pharyngeal swab samples of the suspect patients. The overall sensitivity and diagnostic accuracy of RT-qPCR in our study were 36% and 52% (Table S8), respectively, according to our follow-up survey on the 196 pharyngeal swab samples: identified 48 negatives and 148 positives based on either repeated RT-qPCR tests or clinical evidences tracked on National Infectious Disease Information System (NIDIS). The RT-qPCR sensitivity is in agreement with the previous report for the pharyngeal swab samples [5]. However, both sensitivity and diagnostic accuracy of RT-dPCR dramatically increased to 91% and 93%, respectively. This is very meaningful as pharyngeal swab is much easier to sampling. Thus RT-dPCR is very sensitive for the very low viral load in suspected patients and the asymptomatic close contacts.
Furthermore, it is suitable for monitoring the change of the virus load in the convalescent patients. An additional advantage of quantification of SARS-CoV-2 copy number by RT-dPCR is that comparisons can be conducted between different dates and different laboratories as absolute quantitation of targets by RT-dPCR provides high concordance between sites, runs and operators [14,23,24]. However, it is not possible to compare Ct values on different runs or different machines. Thus, RT-dPCR is an ideal method to for measuring the change of virus load in the convalescent patients.
4. Conclusions
This work demonstrates that RT-dPCR significantly improves accuracy and reduces the false negative rate of diagnostics of SARS-CoV-2 in pharyngeal swab specimens, which is more convenient and simpler to sampling. Furthermore, dPCR is more sensitive and suitable for low virus load specimens from the patients under isolation and observation who may not be exhibiting clinical symptoms. Finally, RT-dPCR could be used to quantitative monitoring the convalescents to evaluate disease progression.
Credit author statement
Lianhua Dong, Junbo Zhou and Chunyan Niu wrote the manuscript and analyzed the data. Lianhua Dong, Xinhua Dai and Xiang Fang conceived the project and designed the study. Chunyan Niu, Xia Wang, Yongzhuo Zhang and Jiayi Yang validated the dPCR assay. Junbo Zhou, Quanyi Wang, Yang Pan, Sitong Sheng, Manqing Liu, Xiaoying Zhang and Tao Zhu analyzed the clinical samples by RT-qPCR and dPCR. Yang Zhao, Tao Peng, Jie Xie, Yunhua Gao, Di Wang did the LoD measurements. And all authors have reviewed the manuscript.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgments
We would like to thank Academy of Military Medical Sciences for providing the purified virus RNA for method validation, Dr. Huggett from the National Measurement Laboratory, NML for his valuable comments and Bio-Rad Laboratories, China for donating the one step RT-dPCR mastermix. Fundamental Research Funds for Central Public welfare Scientific research Institutes sponsored by National Institute of Metrology, P.R. China (31-ZYZJ2001/AKYYJ2009).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.talanta.2020.121726.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Simpson D., Feeney S., Boyle C., Stitt A.W. Retinal VEGF mRNA measured by SYBR green I fluorescence: a versatile approach to quantitative PCR. Mol. Vis. 2000;6:178–183. [PubMed] [Google Scholar]
- 2.Coronavirus disease (COVID-19) outbreak. Available at: https://www.Who.Int/emergencies/diseases/novel-coronavirus-2019..
- 3.Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han Q., Lin Q., Jin S., You L. Coronavirus 2019-nCoV: a brief perspective from the front line. J. Infect. 2020 doi: 10.1016/j.jinf.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Y. Yang, M. Yang, C. Shen, F. Wang, J. Yuan, J. Li, M. Zhang, Z. Wang, L. Xing, J. Wei, L. Peng, G. Wong, H. Zheng, M. Liao, K. Feng, J.Li, Q. Yang, J. Zhao, Z. Zhang, L. Liu, Y. Liu, Evaluating the accuracy of different respiratory specimens in the laboratory diagnosis andmonitoring the viral shedding of 2019-ncov infections, MedRxiv 20021493 [Preprint]..
- 6.Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest CT for typical 2019-ncov pneumonia: relationship to negative RT-PCR testing. Radiology. 2020 doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hindson B.J., Ness K.D., Masquelier D.A., Belgrader P., Heredia N.J., Makarewicz A.J., Bright I.J., Lucero M.Y., Hiddessen A.L., Legler T.C., Kitano T.K., Hodel M.R., Petersen J.F., Wyatt P.W., Steenblock E.R., Shah P.H., Bousse L.J., Troup C.B., Mellen J.C., Wittmann D.K., Erndt N.G., Cauley T.H., Koehler R.T., So A.P., Dube S., Rose K.A., Montesclaros L., Wang S., Stumbo D.P., Hodges S.P., Romine S., Milanovich F.P., White H.E., Regan J.F., Karlin-Neumann G.A., Hindson C.M., Saxonov S., Colstonet B.W. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011;83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hindson C.M., Chevillet J.R., ., Briggs H.A., Gallichotte E.N., Ruf I.K., Hindson B.J., Vessella R.L., Tewari M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods. 2013;10:1003–1005. doi: 10.1038/nmeth.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vogelstein B., Kinzler K.W. Digital PCR. Proc. Natl. Acad. Sci. U.S.A. 1999;96:9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dingle T.C., Sedlak R.H., Cook L., Jerome K.R. Tolerance of droplet-digital PCR vs real-time quantitative PCR to inhibitory substances. Clin. Chem. 2013;59:1668–1670. doi: 10.1373/clinchem.2013.211045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strain M.C., Lada S.M., Luong T., Rought S.E., Gianella S., Terry V.H., Spina C.A., Woelk C.H., Richman D.D. Highly precise measurement of HIV DNA by droplet digital PCR. PloS One. 2013;8 doi: 10.1371/journal.pone.0055943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.China-prevention and control plan of new coronavirus pneumonia. http://www.chinacdc.cn/jkzt/crb/xcrxjb/202002/W020200207737109070291.pdf Available at:
- 13.Diagnostic detection of 2019-ncov by real-time RT-PCR. 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/laboratory-guidance Available at: [DOI] [PMC free article] [PubMed]
- 14.Dong L., Wang X., Wang S., Du M., Niu C., Yang J., Li L., Zhang G., Fu B., Gao Y., Wang J. Interlaboratory assessment of droplet digital pcr for quantification of braf v600e mutation using a novel DNA reference material. Talanta. 2020;207:120293. doi: 10.1016/j.talanta.2019.120293. [DOI] [PubMed] [Google Scholar]
- 15.W.T. Daniel, L. Kristian, K. Marina, A.A. David, E.G. Patricia, L.J. Robert, H.K.Martin, M.L. Rudolf, J.P. Thomas, G.A. Scassellati, S. Heinz, T. Jane, Protocols for Determination of Limits of Detection and Limits of Quantitation; Approved Guideline, EP17-A;24.
- 16.Huggett J.F., Foy C.A., Benes V., Emslie K., Garson J.A., Haynes R., Hellemans J., Kubista M., Mueller R.D., Nolan T., Pfaffl M.W., Shipley G.L., Vandesompele J., Wittwer C.T., Bustin S.A. The digital miqe guidelines: minimum information for publication of quantitative digital PCR experiments. Clin. Chem. 2013;59:892–902. doi: 10.1373/clinchem.2013.206375. [DOI] [PubMed] [Google Scholar]
- 17.Weaver S., Dube S., Mir A., Qin J., Sun G., Ramakrishnan R. Taking qPCR to a higher level: analysis of cnv reveals the power of high throughput qPCR to enhance quantitative resolution. Methods. 2010;50:271–276. doi: 10.1016/j.ymeth.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 18.Wang X., Yao H., Xu X., Zhang Pe, Zhang M., Shao J., Xiao Y., Wang H. Limits of detection of 6 approved RT–PCR kits for the novel SARS-coronavirus-2 (SARS-CoV-2) Clin. Chem. 2020;66:977–979. doi: 10.1093/clinchem/hvaa099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suo T., Liu X., Guo M., Feng J., Hu W., Guo D., UIIah H., Yang Y., Zhang Q., Wang X., Sajiad M., Huang Z., Deng L., Chen T., Liu F., Xu K., Liu Y., Zhang Q., Liu Y., Xiong Y., Chen G., Lan K., Chen Y. ddPCR: a more sensitive and accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg. Microb. Infect. 2020;9:1259–1268. doi: 10.1080/22221751.2020.1772678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest ct and rt-pcr testing in coronavirus disease 2019 (covid-19) in China: a report of 1014 cases. Radiology. 2020 doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.More than 100 new coronavirus detection reagent market research report! https://www.Sohu.Com/a/377833693_120051606 Available at:
- 22.National Health Commission . 2020. Diagnosis and Treatment protocol for COVID-19 (Trial version 7)http://www.nhc.gov.cn/yzygj/s7652m/202003/a31191442e29474b98bfed5579d5af95.shtml [Google Scholar]
- 23.Whale A.S., Jones G.M., Jernej P., Dreo T., Redshaw N., Akyürek S., Akgöz M., Divieto C., Sassi M.P., He H., Cole K.D., Bae Y.-K., Park S.-R., Deprez L., Corbisier P., Garrigou S., Taly V., Larios R., Cowen S., O'Sullivan D.M., Bushell C.A., Goenaga-Infante H., Foy C.A., Woolford A.J., Parkes H., Huggett J.F., Devonshire A.S. Assessment of digital PCR as a primary reference measurement procedure to support advances in precision medicine. Clin. Chem. 2018;64:1296–1307. doi: 10.1373/clinchem.2017.285478. [DOI] [PubMed] [Google Scholar]
- 24.Yoo H.-B., Park S.-R., Dong L., Wang J., Sui Z., Pavšič J., Milavec M., Akgoz M., Mozioğlu E., Corbisier P., Janka M., Cosme B., Cavalcante J.J.V., Flatshart R.B., Burke D., Forbes-Smith M., McLaughlin J., Emslie K., Whale Jf Huggett A.S., Parkes H., Kline M.C., Harenza J.L., Vallone P.M. International comparison of enumeration-based quantification of DNA copy-concentration using flow cytometric counting and digital polymerase chain reaction. Anal. Chem. 2016;88:12169–12176. doi: 10.1021/acs.analchem.6b03076. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.