Abstract
Prostate-specific membrane antigen (PSMA) is a cell-surface enzyme-biomarker that is actively pursued for targeted delivery of imaging and therapeutic agents for prostate cancer. Our lab has developed PSMA inhibitors based on a phosphoramidate scaffold, which has shown both high selectivity for PSMA-positive tumors and rapid clearance in vivo when radiolabeled with 18F. However, this scaffold exhibits hydrolytic instability under low pH and high temperature conditions, barring the use of other imaging or therapeutic radionuclides such as 68Ga or 177Lu. Previous studies in our lab have shown a trend in increasing acid stability as the distance between the phosphoramidate core and the α-carboxylate of the P1 residue is increased. Therefore, a new generation of phosphoramidate inhibitors was developed based on trans-4-hydroxyproline as the P1 residue to restrict the interaction of the α-carboxylate to the phosphoramidate core. These hydroxyproline inhibitors demonstrated comparable IC50 values to earlier generations as well as enhanced thermal and acid stability.
Keywords: phosphoramidate, inhibitor, prostate-specific membrane antigen, 4-hydroxyproline, prostate cancer
Prostate-specific membrane antigen (PSMA) is a cell-surface enzyme-biomarker1, 2 that continues to be actively pursued for targeted delivery of imaging3-17 and therapeutic agents18-22 for prostate cancer. PSMA has been found to be up-regulated and strongly expressed on cancer cells, including those that are metastatic.23 As a consequence, enzyme inhibitors have been developed to selectively, and in some cases, irreversibly bind to PSMA.24-27 Most recently, our lab developed, and radiolabeled with 18F, the phosphoramidate inhibitors 5 and 6 (Figure 1), which exhibits both high selectivity for PSMA-positive tumors and rapid clearance in vivo. However, this scaffold, like previous generations of this class of inhibitors, exhibits hydrolytic instability under low pH and high temperature conditions. These conditions are likely to be encountered when such scaffolds are labeled with other imaging or therapeutic radionuclides such as 68Ga or 177Lu.28-30 Hence, it is desirable to optimize the phosphoramidate scaffold for this class of PSMA inhibitors for greater acid and thermal stability.
Figure 1.
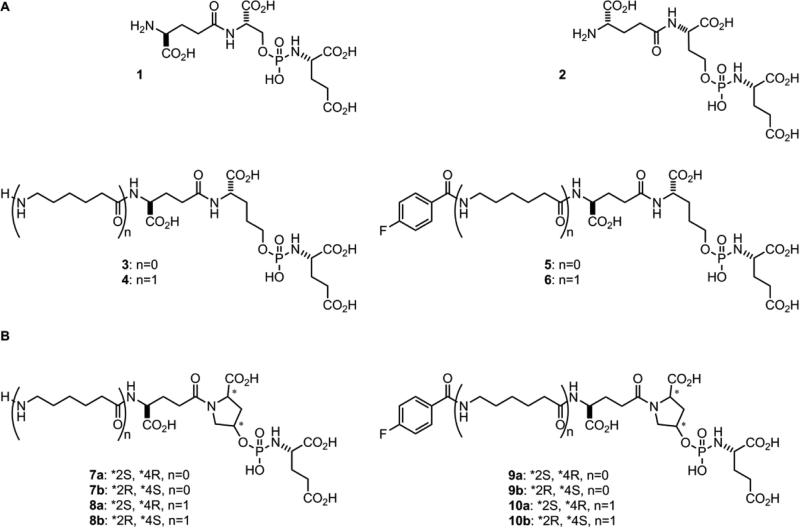
(A) Current phosphoramidate inhibitors of PSMA 1-6. (B) Phosphoramidate inhibitors of PSMA with enhanced stability 7a-10b.
In our studies with earlier generations of phosphoramidate-based PSMA inhibitors, we observed a trend in increasing acid stability as the distance between the phosphoramidate centers and the α-carboxylate of the P1 residue was increased. This is evidenced by the observed rates of decomposition at pH 6.0 and 4.5 for scaffolds containing a P1 serine (1), homoserine (2), and hydroxypropylglycine residue (3).31 The mechanism for the cleavage of the P-N bond of the phosphoramidate core under these conditions remains conjectural, however, the proximity of the P1 α-carboxylate to the phosphoramidate center does appear to contribute to its lability. While this mechanism is a topic for a future study, this observed trend inspired the design of a new scaffold based on trans-4-hydroxyproline as the P1 residue. Due to the conformational restrictions provided by the proline ring, the trans-orientation between the α-carboxylate and hydroxyl group was expected to afford enhanced acid stability. Therefore, the focus of this study was aimed at preparing and evaluating a limited series of phosphoramidate-based PSMA inhibitors comprised of a trans-4-hydroxyproline in the P1 position for both acid and thermal stability as well as inhibitory potency against PSMA. Because of the restricted conformational freedom imposed by the pyrrolidine ring, it was also expected that a degree of stereoselective inhibition of PSMA would be observed. In terms of assessing the impact of introducing a relatively inflexible pyrrolidine ring in the P1 position (e.g., 7a-b, 8a-b, 9a-b, and 10a-b), we chose to compare the inhibitory potencies and mode of inhibition to the more conventional acyclic analog 3 (Figure 1).31
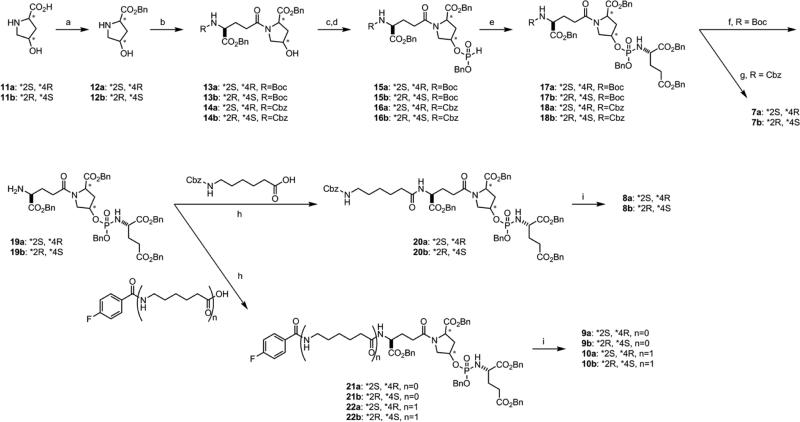
A common method was employed to prepare the trans-4-hydroxyproline-based phosphoramidate inhibitors (Scheme 1). The starting amino acids 11a-b were protected as the benzyl esters 12a-b, which were subsequently coupled to N-protected glutamic acid to provide the corresponding alcohols 13a-b and 14a-b. The reaction of these alcohols with diphenyl phosphite followed by the addition of benzyl alcohol provided H-phosphonates 15a-b and 16a-b, which were then subjected to standard Atherton-Todd conditions to generate the protected phosphoramidate intermediates 17a-b and 18a-b. Deprotection of 18a-b under hydrogenolysis conditions yielded the stereoisomeric inhibitors 7a-b. The Boc-protected intermediates 17a-b were N-deprotected and functionalized to provide globally protected precursors 20a-b, 21a-b, and 22a-b. Global deprotection of these precursors under hydrogenolysis conditions yielded 8a-b, 9a-b, and 10a-b.
Scheme 1.
(a) BnOH, p-toluene-SO3H, Benzene, 125 °C, 20 h reflux; (b) R-Glu-OBzl (R=Cbz or Boc), HBTU, Et3N, DMF; (c) (PhO)2P(O)H, pyridine, −5 °C to rt, 2 h; (d) BnOH, rt, 3 h; (e) H-Glu(OBzl)-OBzl HCl, CCl4, Et3N, CH3CN; (f) 30% TFA/CH2Cl2, rt, 1.5 h; (g) H2, 10% Pd/C, K2CO3, ddH2O, 1,4-dioxanes; (h) HBTU, Et3N, DMF; (i) H2, 10% Pd/C, K2CO3, ddH2O, 1,4-dioxanes
To assess the stability of the trans-4-hydroxyproline scaffold compared to an acyclic conventional type of phosphoramidate PSMA inhibitor scaffold under mildly acidic conditions, the decomposition of 10a and 631, were monitored by 31P NMR at pH 4.5. The compounds were analyzed at both 50 and 70 °C (see Supporting Information for procedures). The respective rates of decomposition (Table 1) of 10a and 6 confirmed that the trans-4-hydroxyproline scaffold exhibited enhanced the stability of the phosphoramidate P-N bond over the acyclic analog 6. These results further support the contribution of the α-carboxylate of the P1 amino acid on P-N hydrolysis of this class of phosphoramidate inhibitors of PSMA.
Table 1.
Comparative stability of representative phosphoramidates at pH 4.5
| Entry | Temp (°C) | t½ (min) |
|---|---|---|
| 6 | 50 | 75 |
| 6 | 70 | 12 |
| 10a | 50 | 105 |
| 10a | 70 | 32 |
While greater acid stability of the trans-4-hydroxyproline scaffold was observed, it was important to determine if this structural change resulted in a loss of inhibitory potency toward PSMA and/or a change in the mode of inhibition when compared to acyclic analogs. It was noted that the trans-4-hydroxyproline-based inhibitors of PSMA with unmodified N-terminal amines generally exhibited lower inhibitory potency than their respective 4-fluorobenzamide derivatives; a trend that is consistent with small molecule PSMA inhibitors based on phosphoramidate and urea scaffolds.31 With respect to the impact of stereochemistry of the P1 4-trans-hydroxyproline residue on PSMA inhibition, it was observed that the natural L-isomer of this amino acid was preferred (e.g., 9a and 10a), which is consistent with our previous findings.32 Of the 4-fluorobenzamide derivatives of the inhibitors based on the trans-4-hydroxyproline scaffold (9 and 10), the one that was most comparable to the analogs based on the hydroxypropylglycine scaffold (5 and 6)31 was 10a (Table 2) based on IC50 values. Interestingly, the mode of inhibition studies demonstrated that all the analogs based on the trans-4-hydroxyproline scaffold (7-10) exhibited slowly reversible binding, in contrast to the irreversible mode of inhibition for the compounds based on the acyclic scaffold 1-631 suggesting that rigidity or increased molecular volume introduced by the trans-4-hydroxyproline residue does not allow the same interactions that the acyclic analogs experience with the binding site of PSMA that lead to irreversible binding.
Table 2.
Inhibition potency of phosphoramidate inhibitors of PSMA
| Entry | IC50 (nM)a |
|---|---|
| 3 | 27 (3) |
| 4 | 19 (1) |
| 5 | 1.3 (0.08) |
| 6 | 0.4 (0.05) |
| 7a | 60 (11) |
| 7b | 357 (29) |
| 8a | 79 (6) |
| 8b | 112 (8) |
| 9a | 2.6 (0.2) |
| 9b | 3.0 (0.3) |
| 10a | 1.3 (0.2) |
| 10b | 11 (0.8) |
Standard deviation in parentheses.
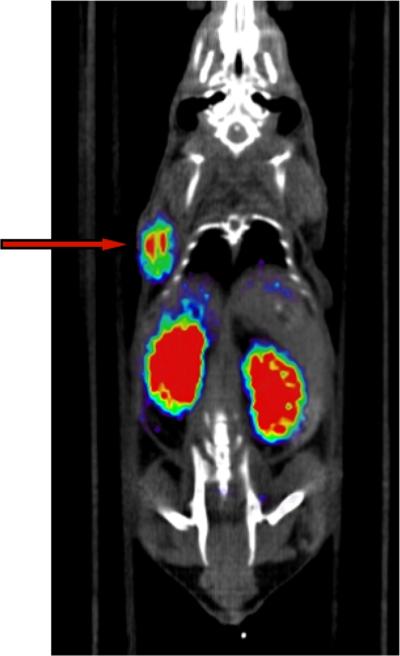
With considerable inhibitory potency against PSMA and a slowly-reversible mode of inhibition, it was of interest to confirm that the 4-trans-hydroxyproline-based phosphoramidate inhibitor scaffold could also perform sufficiently in vivo as the targeting motif of a PET imaging agent for PSMA-positive tumors. For a preliminary assessment of this new scaffold's in vivo performance, the 18F-radiolabled analog [18F]10a was prepared as described previously using N-succinimidyl 4-[18F]fluorobenzoate ([18F]SFB).15 As shown in the PET/CT image (Figure 2), there was significant and selective uptake of [18F]10a tracer in CWR22Rv1 (PSMA+) tumor xenografts at 2 h post-injection. In addition, there was the expected uptake in the kidneys due to the expression of PSMA in mouse kidneys23, 33, 34 but minimal uptake in all other organs.
Figure 2.
PET/CT image (20 minute static scan) of a male nude mouse bearing a CWR22Rv1 tumor xenograft at 2 h post-injection of [18F]10a. Arrow indicates tumor placement.
In summary, the stability and inhibition studies demonstrated that limiting the interaction of the α-carboxylate of the P1 amino acid to the phosphoramidate core improved the acid and thermal stability of the P-N bond without completely sacrificing inhibitory potency against PSMA. As a result, it is expect that the trans-4-hydroxyproline scaffold could be radiolabeled with imaging and therapeutic radionuclides such as 68Ga and 177Lu that might require harsher conditions for the installation of the isotopes. Such findings would extend the versatility of the phosphoramidate scaffold in the targeting molecule motif for targeted imaging and therapeutic agents for prostate cancer.
Supplementary Material
Acknowledgments
This work was supported in part by the National Institutes of Health (R01CA140617).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Material
For detailed experimental procedures, refer to the Supplemental Material.
References
- 1.Perner S, Hofer MD, Kim R, Shah RB, Li H, Moller P, Hautmann RE, Gschwend JE, Kuefer R, Rubin MA. Hum Pathol. 2007;38:696. doi: 10.1016/j.humpath.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 2.Bander NH, Milowsky MI, Nanus DM, Kostakoglu L, Vallabhajosula S, Goldsmith SJ. J Clin Oncol. 2005;23:4591. doi: 10.1200/JCO.2005.05.160. [DOI] [PubMed] [Google Scholar]
- 3.Rosenthal SA, Haseman MK, Polascik TJ, Division of Radiation Oncology, R. A. o. S. M. G. I. C. U. S. A. Techniques in urology. 2001;7(1):27. [PubMed] [Google Scholar]
- 4.Chu TC, Shieh F, Lavery LA, Levy M, Richards-Kortum R, Korgel BA, Ellington AD. Biosens Bioelectron. 2006;21:1859. doi: 10.1016/j.bios.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 5.Farokhzad OC, Khademhosseini A, Jon S, Hermmann A, Cheng J, Chin C, Kiselyuk A, Teply B, Eng G, Langer R. Analytical chemistry. 2005;77:5453. doi: 10.1021/ac050312q. [DOI] [PubMed] [Google Scholar]
- 6.Foss CA, Mease RC, Fan H, Wang Y, Ravert HT, Dannals RF, Olszewski RT, Heston WD, Kozikowski AP, Pomper MG. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11:4022. doi: 10.1158/1078-0432.CCR-04-2690. [DOI] [PubMed] [Google Scholar]
- 7.Gao X, Cui Y, Levenson RM, Chung LW, Nie S. Nature biotechnology. 2004;22:969. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 8.Guilarte TR, McGlothan JL, Foss CA, Zhou J, Heston WD, Kozikowski AP, Pomper MG. Neurosci Lett. 2005;387:141. doi: 10.1016/j.neulet.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 9.Humblet V, Lapidus R, Williams LR, Tsukamoto T, Rojas C, Majer P, Hin B, Ohnishi S, De Grand AM, Zaheer A, Renze JT, Nakayama A, Slusher BS, Frangioni JV. Mol Imaging. 2005;4:448. doi: 10.2310/7290.2005.05163. [DOI] [PubMed] [Google Scholar]
- 10.Milowsky MI, Nanus DM, Kostakoglu L, Sheehan CE, Vallabhajosula S, Goldsmith SJ, Ross JS, Bander NH. J Clin Oncol. 2007;25:540. doi: 10.1200/JCO.2006.07.8097. [DOI] [PubMed] [Google Scholar]
- 11.Pomper MG, Musachio JL, Zhang J, Scheffel U, Zhou Y, Hilton J, Maini A, Dannals RF, Wong DF, Kozikowski AP. Mol Imaging. 2002;1:96. doi: 10.1162/15353500200202109. [DOI] [PubMed] [Google Scholar]
- 12.Smith KA, Sheppard SD, Johnson DW, Johnson RT. Journal of Engineering Education. 2005;94:87. [Google Scholar]
- 13.Smith-Jones PM, Vallabhajosula S, Navarro V, Bastidas D, Goldsmith SJ, Bander NH. Journal of nuclear medicine : official publication, Society of Nuclear Medicine. 2003;44:610. [PubMed] [Google Scholar]
- 14.Tsukamoto T, Wozniak KM, Slusher BS. Drug Discov Today. 2007;12:767. doi: 10.1016/j.drudis.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 15.Lapi SE, Wahnishe H, Pham D, Wu LY, Nedrow-Byers JR, Liu T, Vejdani K, VanBrocklin HF, Berkman CE, Jones EF. Journal of nuclear medicine : official publication, Society of Nuclear Medicine. 2009;50:2042. doi: 10.2967/jnumed.109.066589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nedrow-Byers JR, Jabbes M, Jewett C, Ganguly T, He H, Liu T, Benny P, Bryan JN, Berkman CE. The Prostate. 2012;72:904. doi: 10.1002/pros.21493. [DOI] [PubMed] [Google Scholar]
- 17.Nedrow-Byers JR, Moore AL, Ganguly T, Hopkins MR, Fulton MD, Benny PD, Berkman CE. The Prostate. 2013;73:355. doi: 10.1002/pros.22575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tasch J, Gong M, Sadelain M, Heston WD. Crit Rev Immunol. 2001;21:249. [PubMed] [Google Scholar]
- 19.Lu J, Celis E. Cancer research. 2002;62(20):5807. [PubMed] [Google Scholar]
- 20.Liu T, Wu LY, Berkman CE. Cancer letters. 2010;296:106. doi: 10.1016/j.canlet.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu T, Wu LY, Choi JK, Berkman CE. International journal of oncology. 2010;36:777. doi: 10.3892/ijo_00000553. [DOI] [PubMed] [Google Scholar]
- 22.Liu T, Nedrow-Byers JR, Hopkins MR, Wu LY, Lee J, Reilly PT, Berkman CE. Bioorganic & medicinal chemistry letters. 2012;22:3931. doi: 10.1016/j.bmcl.2012.04.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bacich DJ, Pinto JT, Tong WP, Heston WD. Mammalian genome : official journal of the International Mammalian Genome Society. 2001;12(2):117. doi: 10.1007/s003350010240. [DOI] [PubMed] [Google Scholar]
- 24.Liu T, Toriyabe Y, Kazak M, Berkman CE. Biochemistry. 2008;47:12658. doi: 10.1021/bi801883v. [DOI] [PubMed] [Google Scholar]
- 25.Liu T, Wu LY, Hopkins MR, Choi JK, Berkman CE. Bioorganic & medicinal chemistry letters. 2010;20:7124. doi: 10.1016/j.bmcl.2010.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Majer P, Jackson PF, Delahanty G, Grella BS, Ko YS, Li W, Liu Q, Maclin KM, Polakova J, Shaffer KA, Stoermer D, Vitharana D, Wang EY, Zakrzewski A, Rojas C, Slusher BS, Wozniak KM, Burak E, Limsakun T, Tsukamoto T. J Med Chem. 2003;46:1989. doi: 10.1021/jm020515w. [DOI] [PubMed] [Google Scholar]
- 27.Thomas AG, Wozniak KM, Tsukamoto T, Calvin D, Wu Y, Rojas C, Vornov J, Slusher BS. Adv Exp Med Biol. 2006;576:327. doi: 10.1007/0-387-30172-0_24. [DOI] [PubMed] [Google Scholar]
- 28.Wunderlich G, Schiller E, Bergmann R, Pietzsch H-J. Nuclear Medicine and Biology. 2010;37:861. doi: 10.1016/j.nucmedbio.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 29.Liu S, Cheung E, Ziegler MC, Rajopadhye M, Edwards DS. Bioconjugate Chemistry. 2001;12:559. doi: 10.1021/bc000146n. [DOI] [PubMed] [Google Scholar]
- 30.Sosabowski JK, Mather SJ. Nat. Protocols. 2006;1:972. doi: 10.1038/nprot.2006.175. [DOI] [PubMed] [Google Scholar]
- 31.Ganguly T. Ph.D. Thesis. Washington State University; 2013. [Google Scholar]
- 32.Wu LY, Anderson MO, Toriyabe Y, Maung J, Campbell TY, Tajon C, Kazak M, Moser J, Berkman CE. Bioorganic & medicinal chemistry. 2007;15:7434. doi: 10.1016/j.bmc.2007.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Troyer JK, Beckett ML, Wright GL., Jr. Int J Cancer. 1995;62:552. doi: 10.1002/ijc.2910620511. [DOI] [PubMed] [Google Scholar]
- 34.Israeli RS, Powell CT, Corr JG, Fair WR, Heston WD. Cancer research. 1994;54:1807. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.