Abstract
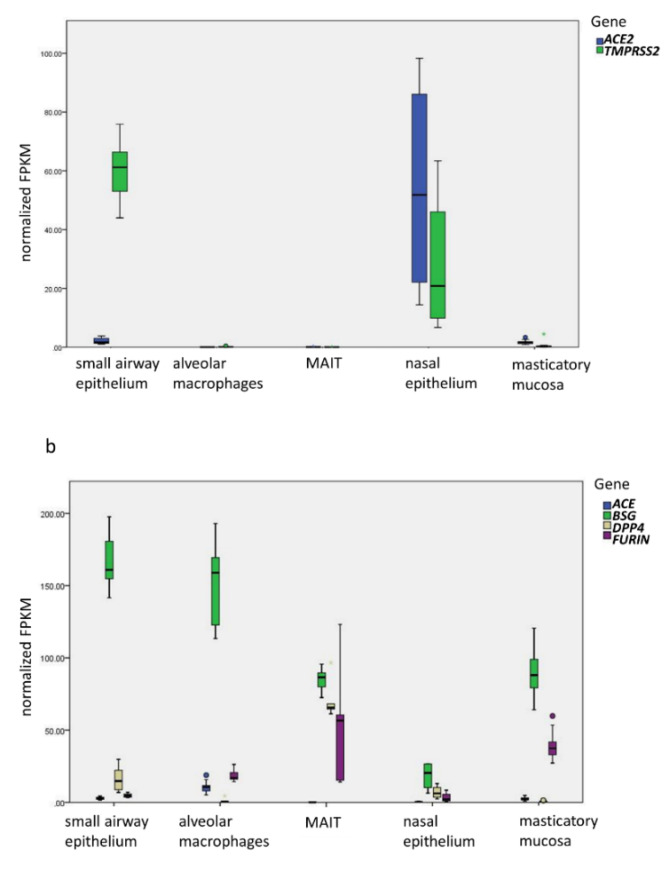
To address the expression pattern of the SARS-CoV-2 receptor ACE2 and the viral priming protease TMPRSS2 in the respiratory tract, this study investigated RNA sequencing transcriptome profiling of samples of airway and oral mucosa. As shown, ACE2 has medium levels of expression in both small airway epithelium and masticatory mucosa, and high levels of expression in nasal epithelium. The expression of ACE2 is low in mucosal-associated invariant T (MAIT) cells and cannot be detected in alveolar macrophages. TMPRSS2 is highly expressed in small airway epithelium and nasal epithelium and has lower expression in masticatory mucosa. Our results provide the molecular basis that the nasal mucosa is the most susceptible locus in the respiratory tract for SARS-CoV-2 infection and consequently for subsequent droplet transmission and should be the focus for protection against SARS-CoV-2 infection.
Keywords: ACE2, COVID-19, SARS-CoV-2, TMPRSS2
1. Introduction
Concerning the pandemic of coronavirus disease 2019 (COVID-19), on 26 September 2020, it had been diagnosed in 32.6 million people globally, causing 990,000 deaths. COVID-19 is caused by infection with the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). For this highly infectious and deadly disease, there is no effective antiviral treatment [1]. The human angiotensin-I-converting enzyme 2 (ACE2) has been suggested to serve as the receptor for the cell entry of SARS-CoV-2 to cause infection [2]. ACE2 is a member of the renin–angiotensin system (RAS), with the function of converting angiotensin II to angiotensin-(1-7) (with seven amino acids), and converting angiotensin I to angiotensin-(1-9) [3], thereby negatively regulating the effects of angiotensin-I-converting enzyme (ACE) and the RAS system. In addition to its critical roles in RAS, ACE2 binds the S1 domain of the SARS-CoV Spike (S) protein as the viral receptor, and accounts for the infection of SARS-CoV and syncytia formation [4]. The genome sequence of SARS-CoV-2 shows significant similarity (79%) to that of SARS-CoV, while its receptor-binding domain shows even higher similarity to that of SARS-CoV [2], further supporting ACE2 as the receptor of SARS-CoV-2. COVID-19 is a highly infectious respiratory disease with a basic reproduction number, R0 (95% CI), of 3.28 (1.4, 6.49) [5]. After binding with ACE2, SARS-CoV-2 priming by the serine protease encoded by the transmembrane serine protease 2 gene (TMPRSS2) is also required for the viral entry into host cells [6,7]. In addition, three other genes that may be involved in the SARS-CoV-2 infection were highlighted by a recent study, i.e., furin, the paired basic amino acid cleaving enzyme gene (FURIN); the gene encoding the proprotein convertase subtilisin/kexin 3 (PCSK3); the dipeptidyl peptidase 4 gene (DPP4); and the basigin (Ok blood group) gene (BSG) [8]. PCSK3 may activate the SARS-CoV-2 S protein when TMPRSS2 has low expression [9]. DPP4 and BSG have the possibility of serving as the alternative receptor of SARS-CoV-2 invasion [8].
Knowledge about the expression of ACE2 and TMPRSS2, as well as other potential entry genes of SARS-CoV-2, namely FURIN, DPP4, and BSG, is extremely important to understand the infection of SARS-CoV-2 and to find ways to prevent the infection. For this purpose, we investigated RNA sequencing transcriptome profiling of samples of airway and oral mucosa, including small airway epithelium, alveolar macrophages, nasal epithelium, and masticatory mucosa. In addition, considering the critical role of mucosal-associated invariant T (MAIT) cells in mucosal immune defense against viral infection [10], transcriptome profiling of MAIT was also examined in this study.
2. Materials and Methods
Five datasets of transcriptome profiling by RNA sequencing (RNAseq) were acquired from the NCBI Gene Expression Omnibus (GEO) database (Table 1). We mapped and quantified the trimmed RNAseq reads using HISAT2 (https://ccb.jhu.edu/software/hisat2/index.shtml) to hg19 refSeq for each sample at default thresholds. The expression matrix was generated based on Cuffnorm functions in Cufflink package Version 2.2.1 [11]. Library sizes (i.e., sequencing depths) were normalized by the classic-fpkm method. Comparisons of the levels of ACE, ACE2, and TMPRSS2 across different samples were based on the control or pre-exposure samples in each dataset, i.e., small airway epithelium of 10 healthy never-smokers before smoking E-cigarettes; alveolar macrophages of 10 healthy never-smokers before smoking E-cigarettes; nasal epithelium of 4 nonsmoker females before exposure to third-hand smoke; masticatory mucosa of 21 never-smokers; and MAIT of 5 healthy-bodyweight donors. The relative levels of the target genes were presented as the fragments per kilobase of transcript per million mapped reads (FPKM). Different types of human tissues were compared in this study. The expression of a single housekeeping gene (HKG) may not be constant in different cell types or different stages of the cell cycle. Instead, we chose 6 most stable HKGs with different essential functions for cell survival, to correct the expression levels of target genes. All values of the target genes were corrected by the average of relative levels of 6 HKGs, i.e., ACTB, GAPDH, HMBS, HPRT1, RPL13A, and TBP.
Table 1.
Five datasets of RNAseq profiling analyzed in this study.
| GEO Accession | Sample * | Data Description | Library Prep | Sequencing | Spots (M) | Bases | Size | GC Content | Reference |
|---|---|---|---|---|---|---|---|---|---|
| GSE85121 | small airway epithelium [12] | 10 healthy never-smokers before and after smoking E-cigarettes | TruSeq v2 | Illumina HiSeq2500 | 39.1 | 9.8 Gbp | 3.7 G | 44.27% | hg19 |
| GSE85121 | alveolar macrophages [12] | 10 healthy never-smokers before and after smoking E-cigarettes | TruSeq v2 | Illumina HiSeq2500 | 37.5 | 9.4 Gbp | 3.6 G | 45.62% | hg19 |
| GSE129959 | nasal epithelium | 4 nonsmoker females before and after exposure to third-hand smoke | Nextera XT | Illumina NextSeq500 | 51.1 | 4.2 Gbp | 1.6 G | 43.39% | hg19 |
| GSE136262 | masticatory mucosa [13] | 21 never-smokers; 17 current smokers | TruSeq | Illumina HiSeq3000 | 16.5 | 840.4 Mbp | 305.7 M | 51.43% | hg19 |
| GSE126169 | MAIT [10] | 5 healthy-bodyweight donors; 4 morbidly obese donors | SMART-Seq v4 | Illumina NextSeq 500 | 25.2 | 2.1 Gbp | 794.4 M | 47.29% | hg19 |
* Includes the references showing the original source. MAIT: mucosal-associated invariant T (cells).
3. Results and Discussion
Gene expression patterns in five types of normal tissues are shown in Figure 1. As the major counterpart of ACE2 expression, the expression of ACE is also investigated (Figure 1b). ACE and ACE2 have medium and comparable levels of expression in both small airway epithelium and masticatory mucosa. These findings suggest that SARS-CoV-2 can infect both small airway epithelium and oral mucosa. The ACE2 expression with the highest level of TMPRSS2 expression in small airway epithelium provides explanation for the vulnerability infected individuals have for the characteristic pneumonia of COVID-19. The ACE2 expression in masticatory mucosa helps explain the high level of infectivity via droplet transmission from SARS-CoV-2 infection residing in the oral mucous membrane. In addition to this study, the analysis on single-cell RNAseq (scRNAseq) data of human tissues showed high expression of ACE2 in pulmonary type II alveolar cells (AT2) and respiratory epithelial cells [14]. Liao et al. further demonstrated expression of ACE2 and its binding with the SARS-CoV-2 S protein in human bronchial epithelial cells [15]. In addition, a number of studies demonstrated the expression of ACE2 in airway epithelial cells and its correlation with risk factors of severe COVID-19 [16,17].
Figure 1.
The expression of SARS-CoV-2 entry genes. (a) Established SARS-CoV-2 entry genes, ACE2 and TMPRSS2, in five different types of samples. The difference between ACE2 and TMPRSS2 in the small airway epithelium is highly significant (p = 1.00 × 10-12). In the nasal epithelium, the expression of ACE2 is significantly higher than that in small airway epithelium (p = 6.92 × 10−4), alveolar macrophages (p = 5.02 × 10−4), MAIT (p = 0.016), and masticatory mucosa (p = 5.48 × 10−7); the expression of TMPRSS2 is lower than that in small airway epithelium (p = 5.20 × 10−3), but higher than in alveolar macrophages (p = 2.98 × 10−3), MAIT (p = 0.040), and masticatory mucosa (p = 1.40 × 10−5). (b) The expression of ACE and possible SARS-CoV-2 alternative entry genes, BSG, DPP4, and FURIN. Y-axis represents normalized FPKM values by the average of relative levels of 6 HKGs. The boxplot produced by the IBM SPSS Statistics Version 23 shows the mean, the first quartile and the third quartile, and the 95% confidence interval (CI). FPKM: fragments per kilobase of transcript per million mapped reads.
Interestingly, the expression level of both ACE2 and TMPRSS2 in nasal epithelium is much higher than the levels of ACE expression, which is consistent with the studies by Sungnak et al. [18] and Bunyavanich et al. [19]. These results provide mechanistic evidence that the SARS-CoV-2 virus resides in both the oral and nasal mucosa of the upper respiratory tract, where it is able to bind to the ACE2 receptor, serving as the principal locus of infections. These results also provide explanation for the high level of viral load in the oral and nasal mucosa and resulting high level of droplet transmission, with the lower airways being responsible for the severe form of pneumonia as well as the aerosol transmission of the virus. With ACE2 and TMPRSS2 as the major entry genes, however, other potential entry genes of SARS-CoV-2, namely FURIN, DPP4, and BSG, in the respiratory tract should not be neglected for the possibility of serving as the alternative pathway of SARS-CoV-2 infection.
The expressions of ACE, ACE2, and TMPRSS2 are low in MAIT cells. The expression of ACE is high in alveolar macrophages, but the expression of ACE2 cannot be detected with a low level of TMPRSS2 in alveolar macrophages. These patterns of gene expression in the two types of innate immune cells suggest that SARS-CoV-2 has no or little direct impact on these two components of the innate immune system. In addition, these findings suggest that the infection of SARS-CoV-2 can be limited to the respiratory tract, which explains the absent of viremia in many COVID-19 patients [20,21].
In parallel, we examined whether the exposure factor in each RNAseq dataset affected the expression levels of ACE, ACE2, and TMPRSS2, to assess whether those common factors (i.e., smoking E-cigarettes, third-hand smoke, smoking, and obesity) are associated with the susceptibility of SARS-CoV-2 infection. FPKM values of ACE, ACE2, and TMPRSS2 within each dataset were compared by paired T test (for small airway epithelium and nasal epithelium) or independent T test (for masticatory mucosa and MAIT). However, there was no difference observed (p > 0.05) between the dataset for ACE, ACE2, or TMPRSS2. Accordingly, while the SARS-CoV-2 virus is highly infectious, our results do not suggest a significant change in susceptibility to SARS-CoV-2 infection due to these factors. However, because of the modest sample size for each dataset, we must acknowledge that we are underpowered to identify minor effects.
In summary, this study highlights that the nasal mucosa is the most susceptible locus in the respiratory tract for SARS-CoV-2 infection and replication, is responsible for the subsequent high level of droplet transmission, and should be the focus for protection against SARS-CoV-2 infection, in line with a recent virological analysis [22]. Accordingly, local interventions with ACE2 inhibitors [23] or TMPRSS2 inhibitors (e.g., camostat mesylate) [7] may represent novel interventions to block SARS-CoV-2 cell entry and treat COVID-19.
Author Contributions
Y.L., H.-Q.Q., and J.Q. analyzed and interpreted the RNAseq data. Y.L., H.-Q.Q., L.T., and H.H. wrote the manuscript. All authors read and approved the final manuscript.
Funding
The study was supported by Institutional Development Funds from the Children’s Hospital of Philadelphia to the Center for Applied Genomics, and The Children’s Hospital of Philadelphia Endowed Chair in Genomic Research to H.H.
Conflicts of Interest
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wu Z., McGoogan J.M. Characteristics of and Important Lessons from the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72 314 Cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 2.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferrario C.M., Ahmad S., Nagata S., Simington S.W., Varagic J., Kon N., Dell’italia L.J. An evolving story of angiotensin-II-forming pathways in rodents and humans. Clin. Sci. 2014;126:461–469. doi: 10.1042/CS20130400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y., Gayle A.A., Wilder-Smith A., Rocklöv J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J. Travel Med. 2020;27:taaa021. doi: 10.1093/jtm/taaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lukassen S., Chua R.L., Trefzer T., Kahn N.C., Schneider M.A., Muley T., Winter H., Meister M., Veith C., Boots A.W. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020;39:e105114. doi: 10.15252/embj.2020105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Latini A., Agolini E., Novelli A., Borgiani P., Giannini R., Gravina P., Smarrazzo A., Dauri M., Andreoni M., Rogliani P., et al. COVID-19 and Genetic Variants of Protein Involved in the SARS-CoV-2 Entry into the Host Cells. Genes. 2020;11:1010. doi: 10.3390/genes11091010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Millet J.K., Whittaker G.R. Host cell proteases: Critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Brien A., Loftus R.M., Pisarska M.M., Tobin L.M., Bergin R., Wood N.A.W., Foley C., Mat A., Tinley F.C., Bannan C., et al. Obesity Reduces mTORC1 Activity in Mucosal-Associated Invariant T Cells, Driving Defective Metabolic and Functional Responses. J. Immunol. 2019;202:3404–3411. doi: 10.4049/jimmunol.1801600. [DOI] [PubMed] [Google Scholar]
- 11.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Staudt M.R., Salit J., Kaner R.J., Hollmann C., Crystal R.G. Altered lung biology of healthy never smokers following acute inhalation of E-cigarettes. Respir. Res. 2018;19:78. doi: 10.1186/s12931-018-0778-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richter G.M., Kruppa J., Munz M., Wiehe R., Hasler R., Franke A., Martins O., Jockel-Schneider Y., Bruckmann C., Dommisch H., et al. A combined epigenome- and transcriptome-wide association study of the oral masticatory mucosa assigns CYP1B1 a central role for epithelial health in smokers. Clin. Epigenetics. 2019;11:105. doi: 10.1186/s13148-019-0697-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zou X., Chen K., Zou J., Han P., Hao J., Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020;14:1–8. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liao Y., Li X., Mou T., Zhou X., Li D., Wang L., Zhang Y., Dong X., Zheng H., Guo L., et al. Distinct infection process of SARS-CoV-2 in human bronchial epithelial cells line. J. Med. Virol. 2020 doi: 10.1002/jmv.26200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ziegler C.G., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035.e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai G., Bossé Y., Xiao F., Kheradmand F., Amos C.I. Tobacco smoking increases the lung gene expression of ACE2, the receptor of SARS-CoV-2. Am. J. Respir. Crit. Care Med. 2020;201:1557–1559. doi: 10.1164/rccm.202003-0693LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bunyavanich S., Do A., Vicencio A. Nasal gene expression of angiotensin-converting enzyme 2 in children and adults. JAMA. 2020;323:2427–2429. doi: 10.1001/jama.2020.8707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lescure F.-X., Bouadma L., Nguyen D., Parisey M., Wicky P.-H., Behillil S., Gaymard A., Bouscambert-Duchamp M., Donati F., Le Hingrat Q. Clinical and virological data of the first cases of COVID-19 in Europe: A case series. Lancet Infect. Dis. 2020;20:697–706. doi: 10.1016/S1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang L., Yan Y., Wang L. Coronavirus Disease 2019: Coronaviruses and Blood Safety. Transfus. Med. Rev. 2020;34:75–80. doi: 10.1016/j.tmrv.2020.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ehmann K.Z., Drosten C., Wendtner C., Zange M., Vollmar P., Rosina Ehmann D., Zwirglmaier K., Guggemos M., Seilmaier M., Niemeyer D. Virological assessment of hospitalized cases of coronavirus disease 2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 23.Huentelman M.J., Zubcevic J., Hernandez Prada J.A., Xiao X., Dimitrov D.S., Raizada M.K., Ostrov D.A. Structure-based discovery of a novel angiotensin-converting enzyme 2 inhibitor. Hypertension. 2004;44:903–906. doi: 10.1161/01.HYP.0000146120.29648.36. [DOI] [PubMed] [Google Scholar]