Abstract
Interphotoreceptor retinoid‐binding protein (IRBP) is a highly expressed protein secreted by rod and cone photoreceptors that has major roles in photoreceptor homeostasis as well as retinoid and polyunsaturated fatty acid transport between the neural retina and retinal pigment epithelium. Despite two crystal structures reported on fragments of IRBP and decades of research, the overall structure of IRBP and function within the visual cycle remain unsolved. Here, we studied the structure of native bovine IRBP in complex with a monoclonal antibody (mAb5) by cryo‐electron microscopy, revealing the tertiary and quaternary structure at sufficient resolution to clearly identify the complex components. Complementary mass spectrometry experiments revealed the structure and locations of N‐linked carbohydrate post‐translational modifications. This work provides insight into the structure of IRBP, displaying an elongated, flexible three‐dimensional architecture not seen among other retinoid‐binding proteins. This work is the first step in elucidation of the function of this enigmatic protein.
Keywords: cryo‐EM, interphotoreceptor retinoid‐binding protein, monoclonal antibody, single particle analysis, visual cycle
Abbreviations
- 11c‐RAL
11‐cis‐retinal
- GlcNAc
N‐acetylglucosamine
- HexNAc
N‐acetylhexosamine
- at‐RAL
all‐trans‐retinal
- at‐ROL
all‐trans‐retinol
- CRALBP
cellular retinaldehyde‐binding protein
- ConA
concanavalin A
- CDR
complementarity determining region
- cryo‐EM
cryo‐electron microscopy
- DeoxyHex
deoxyhexose
- DDM
n‐dodecyl‐β‐D‐maltoside
- DMEM
Dulbecco's modified Eagle's medium
- ELISA
enzyme‐linked immunosorbent assay
- FBS
fetal bovine serum
- FRET
fluorescence resonance energy transfer
- FWR
framework region
- FSC
Fourier shell correlation
- GST
glutathione‐S transferase
- GO
graphene oxide
- HAT
hypoxanthine‐aminopterin‐thymidine
- Hex
hexose
- IMGT
ImMunoGeneTics information system
- Ig
immunoglobulin
- IgG1
immunoglobulin G isotype 1
- IPM
interphotoreceptor matrix
- IRBP
interphotoreceptor retinoid‐binding protein
- M1, M2, M3, M4
interphotoreceptor retinoid‐binding protein modules 1, 2, 3, 4
- IP
intraperitoneally
- LB
Luria Bertani
- Man
mannose
- mAb5
monoclonal antibody 5
- mAb5 Fab
monoclonal antibody 5 fragment antigen‐binding domain
- PMSF
phenylmethylsulfonyl fluoride
- PNGase F
peptide:N glycosidase F
- PD
protodermatan sulfate
- PKS
proteokeratan sulfate
- PUFA
polyunsaturated fatty acid
- RPE
retinal pigment epithelium
- RU
resonance units
- SPA
single particle analysis
- SPR
surface plasmon resonance
- TCEP
Tris(2‐carboxyethyl)phosphine
- VR
variable region
1. INTRODUCTION
A remarkable feature of vision lies in the ubiquitous mechanism underlying the first step of phototransduction in all vertebrates, in which the visual chromophore 11‐cis‐retinal (11c‐RAL) bound to opsin (11‐cis‐retinylidene) is isomerized to an all‐trans conformation after absorption of light. This isomerized chromophore is hydrolyzed and released as all‐trans‐retinal (at‐RAL) from the binding pocket of opsins. Another shared feature among the animal kingdom is the ability for all visual pigments to signal multiple light events. This continued transduction depends on the regeneration of the visual chromophore. 1 , 2 , 3 , 4 In the vertebrate retina, the re‐isomerization of at‐RAL to 11c‐RAL occurs in the retinal pigment epithelium (RPE), a monolayer of cells directly adjacent to the photoreceptors, in a process known as the visual (retinoid) cycle. 5 , 6 Retinoids are a class of vitamin A metabolites consisting of a β‐ionone ring, an isoprenoid tail and typically a polar head group with a varied oxidation state. Without protection, retinoids can become irreversibly oxidized or dimerize to form toxic molecules, implicated in retinal diseases. To overcome these difficulties, a number of specialized binding proteins with specific cell surface receptors have evolved to transport retinoids, which are necessary for vision transduction, embryonic development, growth, metabolism, immune function, and reproduction. 7 , 8
Interphotoreceptor retinoid‐binding protein (IRBP) is a 143‐kDa glycolipoprotein consisting of four tandem repeats, each approximately 300 amino acids in length (referred to as modules M1, M2, M3, and M4 hereafter). Previous alignments have highlighted the conservation between the modules, which indicates that RBP3, the gene encoding IRBP could have arisen from quadruplication of an ancient gene. 9 , 10 RBP3 is expressed by photoreceptor cells of the retina and secreted into the subretinal space between photoreceptors and adjacent RPE, promoting retinoid transportation and preventing degradation. The retinoid cycle is forced to accommodate multiple types of retinoids in high quantities in light and dark conditions. Previous literature suggests that IRBP can bind many of these retinoids as well as numerous polyunsaturated fatty acids (PUFAs). 11 For example, in vitro biochemical data suggest that IRBP is essential for delivery of at‐RAL to the RPE, 12 , 13 removal of 11c‐RAL from the RPE 14 , 15 , 16 , 17 , 18 and transport of retinoids to photoreceptors. 19 , 20 In addition to retinoid binding, there is evidence that IRBP plays a central role in retinal development and homeostasis because it is expressed early in retinal differentiation. 21 , 22
Mutations in the RBP3 gene can cause vision loss in humans. A conserved mutation of Asp to Asn was identified at position 1080 (D1080N) in patients with retinitis pigmentosa. 23 The Asp residue is conserved among all four modules of IRBP. Patients who inherit the D1080N mutation display progressive loss of photoreceptors with decreased function of cones and rods. Two other novel homozygous nonsense mutations (c. 1530T > A X; p.Y510 and c.3454G > T X; E1152) were identified through whole exome sequencing. 24 All four patients had similar retinal dystrophy characterized by childhood onset myopia and rod‐cone dysfunction despite an unremarkable fundus appearance. In addition, IRBP null mice also displayed delayed transfer of newly synthesized chromophore from the RPE to rod photoreceptors as well as removal of bleached chromophore from photoreceptors. 17 , 18 , 25
Currently, there is little structural evidence detailing full‐length IRBP. Two crystal structures of single modules of IRBP, IRBP module 1 (M1) from Danio rerio and IRBP module 2 (M2) from Xenopus laevis (PDB ID; 4LUR and 1J7X, respectively), provide insight into the molecular folds of the ancestral IRBP protein. 26 , 27 Both structures are similar in fold but contain a different degree of rotation along their longitudinal axis. In addition, both crystal structures display the αβα sandwich motif present in other retinoid binding proteins, adding to the evidence that modules are repeats of similar functions which possibly serve to bind hydrophobic ligands such as retinoid molecules. 28 However, the IRBP protein from Danio rerio is approximately half of the primary sequence in length as IRBP from higher ordered vertebrates, whereas bovine and human IRBP share 84.1% identity according to ExPasy SIM (ExPasy Bioinformatics Resource Portal). Other structural information was obtained from electron microscopy, detailing the flexibility, and variety of conformations that bovine IRBP can assume. 29 The primary sequence analysis of IRBP shows that 34% of residues contribute to formation of α‐helices, 20% form β‐sheets, and the remaining 45% do not form secondary structures and are likely to remain unstructured. 30 This high fraction of unstructured sequence can contribute to the difficulty in determining the crystal structure of full‐length IRBP. Despite conscientious efforts from many groups, structural insights into the full‐length protein are still lacking. Cryo‐electron microscopy (cryo‐EM) provides a unique opportunity to analyze the structure of IRBP, which has evaded crystallization for decades. Here, we present results of this effort.
2. MATERIALS AND METHODS
2.1. Purification of interphotoreceptor matrix from bovine retinas
Fresh bovine eyes obtained from a local slaughterhouse (American Beef Packers Inc, Chino, CA, USA) were dissected under dim red light and retinas were excised and kept on ice. Interphotoreceptor matrix (IPM) was isolated from retinas by slow stirring in a 1:1 ratio with HEPES buffer (50 mM HEPES HCl, pH 8.0. 300 mM NaCl, 0.1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM dithiothreitol (DTT)), for 60 minutes at 4ºC. IPM supernatant was separated from retinas by centrifugation at 10 000 g for 25 minutes at 4ºC. Retinas were resuspended in the same buffer and centrifuged again at 10 000 g for 25 minutes at 4ºC. 1 mM CaCl2, 1 mM MgCl2, and 1 mM MnCl2 were added to the combined supernatants and poured over a glass wool lined funnel and centrifuged at 100 000 g for 30 minutes at 4ºC to remove insoluble fats and tissue.
2.2. Preparation of concanavalin A sepharose
Concanavalin A (ConA) Sepharose (20 mL) was prepared from 160 mg of purified ConA (Vector Laboratories, Burlingame, CA, USA) and 5.75 g of CNBr‐activated Sepharose 4B (GE Healthcare, Chicago, IL, USA) according to the manufacturer's protocol. ConA‐Sepharose was then washed and stored in 0.01 mM Bis‐tris propane HCl pH 6.9, 0.5 M NaCl, 1 mM CaCl2, 1 mM MgCl2, 1 mM MnCl2, and 20% ethanol.
2.3. Purification of glycoproteins from interphotoreceptor matrix
ConA‐Sepharose (10.0 mL) was equilibrated in the same buffer in which we isolated the interphotoreceptor matrix (50 mM HEPES HCl, pH 8.0, 300 mM NaCl, 0.1 mM PMSF, 1 mM DTT, 1mM CaCl2,1 mM MgCl2, and 1 mM MnCl2) and added to 100 mL of IPM. The sample was incubated while rocking at 4ºC for 3 hours, minimum. Using a gravity column, resin was washed with 100 mL of the same buffer at 4ºC and eluted with 4× column volumes of 50 mM HEPES HCl, pH 8.0, 0.1 M NaCl, and 400 mM methyl α‐D‐mannopyranoside. The eluate was collected and concentrated to 12.5 mL with a 100‐kDa cutoff Amicon Ultra Centrifugal Filter (EMD Millipore, Burlington, MA, USA).
2.4. Purification of IRBP
Glycoproteins obtained from the above procedures were loaded onto a HiPrep 26/10 desalting column (GE Healthcare, Chicago, IL, USA) and exchanged with buffer containing 50 mM HEPES HCl, pH 8.0, and 1.0 mM DTT. The eluate was loaded onto a Source 15Q anion exchange column (GE Healthcare, Chicago, IL, USA) and washed with 20 mL of 50 mM HEPES HCl, pH 8.0, 1.0 mM DTT. The column was then washed with an increasing “step‐wise” gradient with the same buffer plus 100 mM NaCl, 200 mM NaCl, and 300 mM NaCl, and then with an increasing “linear gradient” to 1.0 M NaCl. Many fractions contained IRBP, but only fractions eluted with 300 mM NaCl were combined and utilized for further purification and biochemical studies. The fractions were combined and concentrated to 500 µL and centrifuged at 100 000 g at 4ºC for 1 hour to remove aggregated protein. The sample was then loaded onto a Superdex‐200 column and eluted with 50 mM HEPES HCl, pH 8.0, 300 mM NaCl, 1 mM DTT, and 0.01% n‐dodecyl‐β‐D‐maltoside (DDM). IRBP‐mAb5 complex was prepared by incubating IRBP with 4 M excess of mAb5, then subjecting the material to gel filtration a with Superdex‐200 column. Every sample containing positive 280 nm absorbance was confirmed to contain IRBP and mAb5 by SDS‐PAGE and Coomassie blue staining, however, cryo‐EM samples were selected from a single 250 µL fraction with the greatest absorbance at 280 nm containing both components of the complex.
2.5. Graphene oxide coating of grids
Graphene oxide (GO) coated grids (Quanitfoil Cu 200 R1.2/1.3 grids (Electron Microscopy Sciences, Hatfield, PA, USA)) were prepared as described. 31
2.6. In‐gel digestions and mass spectrometry
Coomassie‐stained SDS‐polyacrylamide gel pieces containing intact or PNGase F‐treated bovine IRBP were cut and destained with 50% acetonitrile in 100 mM ammonium bicarbonate followed by 100% acetonitrile. Sample was reduced by incubating the sample with 20 mM DTT at room temperature for 60 minutes, followed by alkylation with 50 mM iodoacetamide for 30 minutes in the dark. The gel pieces were washed with 100 mM ammonium bicarbonate, dehydrated in acetonitrile, dried in a SpeedVac centrifuge, then rehydrated in 50 mM ammonium bicarbonate containing sequencing grade modified trypsin or chymotrypsin for overnight digestion at 37°C. Proteolytic peptides were extracted from the gel with 50% acetonitrile in 5% formic acid, dried, and reconstituted in 0.1% formic acid for MS analysis.
Liquid chromatography‐tandem MS analysis of the resulting peptides was performed using an Orbitrap Elite Hybrid Mass Spectrometer (Thermo Electron, San Jose, CA, USA) equipped with a Waters nanoAcquity UPLC system (Waters, Taunton, MA). The spectra were acquired in the positive ionization mode by two‐dependent methods consisting of a full MS scan at 120 000 resolution and MS/MS scans of the 20 most abundant precursor ions by collision‐induced dissociation at normalized collision energy of 35%. The obtained data were submitted for a database search against the IRBP primary sequence using Mascot Daemon (Matrix Science, Boston, MA, USA). Carbamidomethylation of Cys was set as a fixed modification, whereas oxidation of Met and conversion of Asn to Asp were selected as variable modifications. The mass tolerance was set as 10 ppm for precursor ions and 0.8 Da for product ions. Candidate N‐linked‐glycosylation sites were initially identified in deglycosylated peptides based on conversion of Asn residues to Asp. The corresponding glycosylated peptides were then identified in CID tandem MS. The sugar compositions of glycopeptides were determined with the assistance of the GlycoMod software and further verified by manual interpretation of their MS/MS spectra.
2.7. Fluorescence binding assays
The apparent affinity of retinoids was evaluated by monitoring changes in the fluorescence of a 25 nM solution of apo‐ and holo‐IRBP. In the retinol bound form, the protein excited at 285 nm emitted fluorescence at 350 and 480 nm due to FRET between the retinoid TRP residues and the retinoid ligand. Replacement of retinol (at‐ROL) in the binding site by tested ligands leads to disruption of FRET and consequently an increase in the fluorescence intensity at 350 nm coinciding with a diminishing signal at 480 nm. All fluorescence measurements were performed using a PerkinElmer Life Sciences LS55 spectrofluorometer. The titrations (0 nm‐2.0 µM, at‐ROL or 11c‐RAL) were performed at 25°C in 50 mM HEPES HCl buffer, pH 8.0, containing 300 mM NaCl, 0.1 mM DTT, 5% glycerol (v/v) by adding an increasing amount of compound (0‐2.0 µM) delivered in 100% ethanol. The final concentrations of ethanol never exceeded 0.5% of the sample's total volume. The K D values were calculated by fitting intensities of the protein fluorescence at the maximum emission (350 nm) or the intensity of retinoid fluorescence at 480 nm to the saturation single ligand‐binding model. The data fitting and calculations of the K D values were performed using the SigmaPlot 11 software package (Systat Software, Chicago, IL, USA).
2.8. Immunization protocol and hybridoma production
Purified bovine IRBP was diluted to 1.0 mg/mL in sterile PBS (pH 7.4). Adult female C57/B6 mice (Jackson Laboratories, Bar Harbor, ME, USA) 6‐8 weeks of age, were divided into two groups, and injected intraperitoneally (IP) with either 100 µL of a solution containing IRBP mixed at a 1:1 (v/v) ratio with QuickAntibody Adjuvant (Beijing Biodragon Immunotechnologies Co., Ltd., Beijing, China) or 100 µL of a solution containing QuickAntibody Adjuvant alone. The non‐control group was injected with 50 µg IRBP for each injection. Boosters of the same amount were given 1, 4, and 5 weeks after initial immunization. Five days after the final IP injection, serum titers were monitored by ELISA, and high‐titer antibody producing animals were sacrificed for hybridoma production by the University Hospitals Visual Sciences Research Center. 32 , 33
2.9. MAb5 purification
mAb5 was purified from serum‐free hybridoma media by thiophilic resin purification according to the manufacturer's protocol (GoldBio, St. Louis, MO, USA) (Figure S3A,B). mAb5 is of the immunoglobulin (Ig) IgG1 isotype, determined using a Pierce Rapid Isotyping Kit (Thermo Fisher Scientific, Waltham, MA, USA).
2.10. Preparation of Fab fragment from mAb5
Twenty mg of mAb5 (2.0 mL at 10 mg/mL) were digested overnight in 10 mM citrate pH 6.0, 20 mM L‐cysteine (reduced) with 0.5 mL of conjugated ficin resin according to the manufacturer's protocol (G‐Biosciences, New Delhi, India).
2.11. Paratope mapping of IRBP mAb5 variable regions
The sequence of IRBP mAb5 was determined using selective PCR amplification of the monoclonal antibody variable region (VR) derived from a hybridoma cell line. 34 The F3F5 cell line which produces mAb5 was used for generation of antibody. Total RNA was isolated from the hybridoma cell line using a RNeasy Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer's protocol. RNA concentration and purity were determined using UV absorbance and gel electrophoresis. At 1.0 µg of purified RNA was synthesized to cDNA using the Verso cDNA synthesis kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer's protocol, and confirmed by UV absorbance. PCR primers were adapted from a previous publication which examined and aligned gene nucleotide sequences from the International ImMunoGeneTics information system (IMGT, http://www.imgt.org), National Center for Biotechnology Information (NCBI) (www.ncbi.nlm.nih.gov/gene/), and the UniProt Consortium (http://www.uniprot.org/). 34 For both mAb5 heavy chain and light chain, the forward primer was located in the leader sequence, and the reverse primer was located in the hinge region (heavy chain; FWD = 5′‐ATGGGATGGAGCTGGATC‐3′, REV = 5′‐GGCTTACAACCACAATCCCTG‐3′, light chain; FWD = 5′‐ATGAAGTTGCCTGTTAGG‐3′′, REV = 5’‐ GGATACAGTTGGTGCAGCATC‐3’). PCR reactions were performed using Phusion Hot Start II High Fidelity DNA polymerase with the following reagents: 1 × Phusion HF Buffer, 200 μM of each dNTP, 0.2 μM of forward and reverse primers and 1 μL cDNA as template for each 50 μL PCR reaction (Thermo Fisher Scientific, Waltham, MA, USA). The PCR conditions for all reactions were: 98°C for 30 seconds, followed by 35 cycles of 98°C for 10 seconds, 65°C for 15 seconds for primer sets A, B, and C or 60°C for primer set D, following an extension period of 72°C for 30 seconds, and final incubation at 72 C for 10 minutes. mAb5 is of isotype immunoglobulin gamma‐1 (IgG1) with a kappa light chain. PCR products were run on 1.5% agarose gels and bands of expected sizes were extracted and DNA was purified using a QIAquick gel extraction kit (Qiagen, Hilden, Germany). Purified PCR products were then ligated into the pCRBlunt shuttle vector and transformed in One Shot Top10 chemically competent Escherichia coli using the Zero Blunt PCR cloning kit (Thermo Fisher Scientific, Waltham, MA, USA) as per manufacturer's instructions and plated onto Luria Bertani (LB) agar (Sigma‐Aldrich, St. Louis, MO, USA) with kanamycin (50 μg/mL) (Figures S4A‐C and S5A,B).
2.12. Sequencing and analysis of mAb5 VR
Fifteen colonies for each heavy and light chain construct were grown in 5.0 mL of LB broth supplemented with kanamycin (50 μg/mL) overnight at 37°C in 14 mL Falcon round bottom culture tubes (Corning, Corning, NY, USA). Plasmid DNA was purified using Thermo Fisher GeneJet plasmid purification (Thermo Fisher Scientific, Waltham, MA, USA). Purified samples were sequenced using GENEWIZ Sanger sequencing service (GENEWIZ, NJ, USA) and sequence files were analyzed using Serial Cloner 2.6 (http://serialbasics.free.fr/Serial_Cloner.html). Sequences were assessed for proper translation using the ExPASy translation tool (http://web.expasy.org/translate/) and the GenBank database using the BLAST program (www.blast.ncbi.nlm.nih.gov). Framework and complementarity determining regions (CDRs) were determined using the IMGT/V‐Quest program (www.imgt.org/IMGT_vquest/vquest). Contaminating transcripts are defined as VR sequences that are fully transcribed and translated into full immunoglobulin proteins with 100% homology to previously described sequences. Aberrant sequences were identified as those that did not contain a fully transcribed or translated VR sequence due to premature stop codons, missing conserved VR motifs, or are non‐specific PCR products.
2.13. Epitope mapping of IRBP mAb5
To determine which region of IRBP the mAb5 binds, we heterologously expressed the modules 1‐4 of bovine IRBP as fusion constructs with GST via pGEX2T expression vector (GE Healthcare, Chicago, IL, USA) and performed immunoblot assays of the purified protein. We constructed genes IRBPM1 (residues 1‐323), IRBPM2 (residues 320‐628), IRBPM3 (residues 625‐932), IRBPM4 (residues 930‐1286), IRBPM2_1 (residues 320‐366), IRBPM2_2 (residues 360‐488), IRBPM2_3 (residues 480‐575), IRBP M2_4 (residues 565‐630), IRBPM2_2A (residues 360‐416), and IRBP M2_2B (residues 410‐488) from the sequence of bovine IRBP and subcloned them into pGEX2t expression vector. All plasmids were transformed into the BL21 cell line (Sigma‐Aldrich, St. Louis, MO, USA) and cultured in 18.0 mL LB broth at 37°C in the presence of 2% glucose to suppress basal expression of GST from the lac promoter. When the culture OD600nm = 0.6 expression was induced with 1 mM isopropyl β‐D‐1‐thiogalactopyranoside (IPTG, Sigma‐Aldrich) for 6 hours. Cultures were then centrifuged at 10 000 g for 30 minutes. GST fusion proteins were purified using the GSTrap system (GE Healthcare, Chicago, IL, USA) with some constructs requiring SP‐200 gel filtration to remove impurities. Samples (approximately 500 ng each) were subjected to SDS‐PAGE and silver‐stained or probed by immunoblot with anti‐GST or anti‐IRBP mAb5.
2.14. Surface plasmon resonance analyses
SPR data acquisition was carried out using the Biacore S200 SPR instrument (Biacore, GE Healthcare, Boston, MA, USA) at 25°C. Purified mAb5 (30 ng mL−1) was injected at a flow rate of 10 μL min−1 in 10 mM sodium acetate pH 5.5, to achieve capture level between 700 and 1000 resonance units (RU) on a S‐CM5 sensor chip according to directions in the manufacturer's amine coupling kit (GE Healthcare, Chicago, IL, USA). After mAb5 immobilization, the surface was blocked with 1 M ethanolamine at pH 8.5, followed by regeneration using 50 mM NaOH. The interaction experiments were performed using running buffer containing 10 mM HEPES HCl pH 8.0, 300 mM NaCl, and 1 mM Tris(2‐carboxyethyl)phosphine (TCEP) and 0.075% Tween‐20, TWEEN‐20 and TCEP are suitable for detergent‐based running buffers in SPR experiments because they are less sensitive to changes in temperature than the buffer conditions of the initial IRBP purification. In addition, TWEEN‐20 is suggested by the CM5 sensorchip and Biacore S200 handbooks, which recommended avoiding high‐viscosity liquids. Binding experiments were carried out using an IRBP concentration range of 0.1‐1000 nM in running buffer at a flow rate of 20 µL min−1. The association and dissociation kinetics for the IRBP‐mAb5 complex were monitored for 120 and 320 seconds, respectively. Between each measurement, the surface was regenerated with 50 mM NaOH, a standard buffer for regeneration on the Biacore S200. SPR data processing and analysis were performed using Biacore S200 Evaluation Software (GE, version 2.0.3). For kinetic analyses, data were locally fit to a 1:1 Langmuir model to obtain on‐rate and off‐rate constants.
2.15. Cryo‐EM data collection
Purified samples of bovine IRBP or IRBP + mAb5 isolated from size‐exclusion chromatography were adsorbed to GO coated Quantifoil R1.2/1.3 Cu grids at a concentration of 0.17 mg/mL and plunge frozen in liquid ethane using an FEI Vitrobot (Thermo Fisher Scientific, Waltham, MA, USA). Sample was applied to the grids under 80% humidity and 4 ºC. Grids were plunged after incubation with sample for 5 seconds. Data were acquired at C‐CINA, University of Basel. In total, six datasets were collected on an FEI Titan Krios (Thermo Fisher Scientific, Waltham, MA, USA) operating at 300 keV and equipped with a Gatan Quantum‐LS imaging energy filter (GIF, 20 eV energy‐loss window; Gatan Inc, Pleasanton, CA, USA). Movies were acquired with a K2 Summit direct electron detector (Gatan Inc) in super resolution mode using SerialEM with a physical pixel size of either 1.058 Å or 0.831 Å and a total electron dose of 60‐81 e− Å−2 per micrograph. 35 Micrographs were motion‐corrected and dose‐weighted using MotionCor2 implemented in FOCUS. 36 , 37 Additional collection parameters can be found in Supporting Information (Table S1).
2.16. Image processing
Apo‐IRBP was initially processed with cisTEM following traditional processing methods including CTF estimation, automated ab initio free particle picking, 2D classification, ab initio 3D model generation and 3D auto‐refinement. 38 The 3D reconstruction at 13.0 Å was then low‐pass filtered to 30 Å and established as a reference for particle picking in cryoSPARC v2.14 (Structura Biotechnology Inc, Toronto, ONT, CA). In the case of IRBP‐mAb5 processing, we took advantage of the most up‐to‐date toolkit available on the FOCUS (1.1.0), MotionCor2 (1.3.1), Relion (3.1) and cryoSPARC (2.14) programs. 39 , 40 , 41 About 1377 images were selected for particle identification with the Gautomatch program (K. Zhang, MRC LMB (https://www2.mrc‐lmb.cam.ac.uk/research/locally‐developed‐software/zhang‐software/)), using low‐pass filtered projections (20 Å) of the calculated IRBP‐mAb5 structure as templates. 109 000 particles were then subjected to Relion processing. Approximately 30 000 particles entered 3D classification with the acquired IRBP‐mAb5 structure low‐pass‐filtered to 40 Å as the initial model. For structure refinement, we used the Topaz neural network picker 41 embedded in cryoSPARC 2.14 to improve particle selection. 13 000 particles were identified, subsequent 2D classification removed only 100 particles, and the non‐uniform refinement resulted in a structure at the nominal 7.4 Å resolution. However, local resolution analysis revealed that the resolution ranged from 6.5 Å in a few best resolved regions to >10Å in the major portion of the structure's density, thus clearly indicating the extents of its flexibility. Therefore, we identified three fulcrum points in the IRBP‐mAb5 structure and generated four local masks corresponding to the IRBP “N‐terminus,” IRBP “core,” IRBP “C‐terminus,” and mAb5Fab, under which we calculated four independent local non‐uniform refinements, which resulted in nominal resolutions of 6, 6.5, 6.6, and 6.8 Å, respectively, and their local resolution analyses proved consistent with the resolution across their densities. The four partial reconstructions were assembled in chimera by rigid body fitting into the 7.4 Å full‐length structure. Details of the processed dataset can be found in Supporting Information (Table S1).
2.17. Model building and refinement
The cryo‐EM map was put into an artificial crystal lattice to calculate its structure factor using the phenix.map_to_structure_factors script in the PHENIX program. 42 The 3D density map was sharpened by applying a negative B‐factor of −156 Å2 as determined with the phenix.auto_sharpen script and by manual intervention. Danio rerio IRBP module 1 (PDB: 4LUR) and Xenopus laevis IRBP module 2 (PDB: 1J7X) were used to create homology models for bovine IRBP modules M1, M2, M3, and M4 (SWISS‐MODEL). 26 , 27 , 43 mAb5 Fab homology model was created by stitching IgG1 constant regions with sequenced VR and Rosetta online server (J. Gray, ROSIE, https://rosie.graylab.jhu.edu/antibody) 44 Homology models were then filtered to 7.0 Å and rigid body fitted into the 3D density map with Chimera. 45 The quality of model fitting was utilized to finalize the directionality (N‐>C terminus). De novo modeling of IRBP M1‐M2, M2‐M3, and M3‐M4 connecting regions was performed in Coot 0.8.8 and Chimera using secondary structure predictions calculated by PSIPRED. 46 , 47 , 48 The initial de novo model was improved by multiple rounds of PHENIX real‐space refinement and REFMAC version 5.8 refinement against the overall map at a resolution of 7.0 Å. 49 Each round of refinement was followed by manual model building and adjustments with Coot 0.8.8. Simultaneous optimization of chain tracing was performed during the refinements, resulting in a final model to map correlation coefficient of 0.68.
3. RESULTS
3.1. IRBP purification and retinoid binding activity
Our strategy for IRBP purification (Figure S1A,B) is similar to previous methods. 50 , 51 , 52 , 53 However, we isolated retinas in house and prepared the extract without freezing, two attributes necessary for cryo‐EM imaging of bovine IRBP. In addition, we also used increased pH and salt content while detergent concentrations were maintained below the critical micelle concentration, which yielded results suitable for imaging by cryo‐EM. Following Superdex‐200 size‐exclusion chromatography, cryo‐EM samples were selected from the 250 µL fraction corresponding to the highest relative absorbance at 280 nm, and all other positive fractions were pooled for biochemical assays. Despite being able to isolate approximately 100 µg of IRBP per eye using traditional purification techniques, our method of purification yielded ~30 µg of IRBP per eye but with minimal aggregation of the samples when imaged by cryo‐EM. To confirm that purified the IRBP maintained native binding capabilities, we utilized the intrinsic tryptophan fluorescence of IRBP to analyze its interaction with native ligands, at‐ROL and 11c‐RAL. The excitation of protein by 285 nm light leads to a significant fluorescence resonance energy transfer (FRET) between TRP residues of IRBP and the retinoid ligands with alcohol groups (Figure 1A). Two maxima at 350 and 480 nm are observable in the emission spectrum corresponding to fluorescence from Trp residues and at‐ROL, respectively. The absorbance levels were recorded after baseline was reached upon addition of ligand from 0.0 to 2.0 µM (60 seconds). The signal intensities for apo‐IRBP and IRBP/at‐ROL complex were used as the 0% and +100% change controls, respectively. Affinity was calculated by plotting the intensity of absorption against the concentration of at‐ROL and 11c‐RAL and measuring fluorescence at 50% saturation (Figure 1B). Calculation of affinity for 11c‐RAL was determined by measuring quenching of Trp residues at 350 nm maxima (Figure 1B, inset). Our results agreed with previously reported results, with a calculated Kd = 122.1 ± 13.5 nM for at‐ROL and Kd = 32.9 ± 7.4 nM for 11c‐RAL. 52 , 54 , 55
Figure 1.
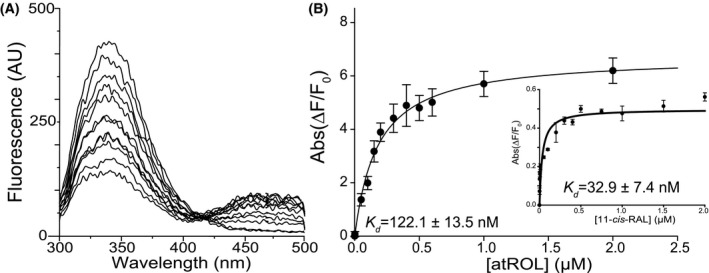
Biochemical characterization of purified IRBP. A, Fluorescence spectra of apo‐IRBP upon titration with at‐ROL. B, Changes in the emission maxima at 480 nm and 350 nm (inset) plotted vs. the concentration of at‐ROL and 11c‐RAL (inset) were fit to a one‐site saturation ligand‐binding model (R sqr = .983 for the fluorescence signal at 480 nm and R sqr = .980 for the fluorescence signal at 350 nm) and used to calculate K D values. The experiments were repeated three separate times, each time in triplicate. Data are presented as mean values ± SD
3.2. IRBP mAb5 generation, epitope mapping, and paratope mapping
We next immunized five 8‐week‐old Balb/c mice with purified bovine IRBP to generate a monoclonal antibody specific to IRBP as a means of solubilizing and stabilizing IRBP in preparation for cryo‐EM analysis. Of the five mice immunized, three produced sera capable of recognizing bovine IRBP by enzyme‐linked immunosorbent assay (ELISA) and immunoprecipitation and are referred to as mAbs3‐5 (Figure S2A,B). Of the resulting hybridomas produced, only one (hybridoma cell line F3F5) produced full‐length antibody (mAb5) (Figure S3A,B) and fragment antigen‐binding domain (mAb5 Fab) that recognized both denatured and folded forms of IRBP (Figure 2A,C, Figure S2A,B). mAb5 did not significantly alter at‐ROL binding to IRBP, which has a calculated Kd = 151.9 ± 13.5 nm (Figure S3C). To map the mAb5 epitope, we employed array‐based expression of IRBP oligopeptides. By expressing fragments of the full‐length sequence, we determined whether mAb5 binds specific regions of the sequence. Each construct was expressed as a fusion protein with glutathione‐S transferase (GST) to aid solubility, as some untagged regions aggregated due to either their hydrophobicity or their long stretches of acidic residues. Initial results demonstrated that mAb5 binds IRBP Module 2 (IRBP M2) based on immunoblot assay. We then expressed smaller fragments of IRBP M2 using an overlapping peptide strategy to delineate the epitope that mAb5 recognizes (Figure 2B). mAb5 binds IRBP in the primary sequence region of 390‐450 (Figure 2C). To examine the kinetics underlying the assembly of the IRBP‐mAb5 complex, surface plasmon resonance (SPR) spectroscopy was employed with varied concentrations of native bovine IRBP. The binding kinetics analysis revealed a rapid on‐rate constant (Kon = 4.7 × 105 M−1s−1) and a slow off‐rate constant (Koff = 2.5 × 10−2 s−1), with a high affinity interaction with a K d of 53 nM. Analysis of the protein‐protein interactions gave a similar K d of 70 nM (Figure 3A,B). To determine the paratope of mAb5, we sequenced the VR of the cDNA of the F3F5 cell line. The nucleotide and protein translation data obtained from hybridoma cell line F3F5 are provided beginning with framework region‐1 (FWR1) and ending with framework region‐4 (FWR4) for both the Hc and Lc (Figures S4A‐C and S5A,B).
Figure 2.
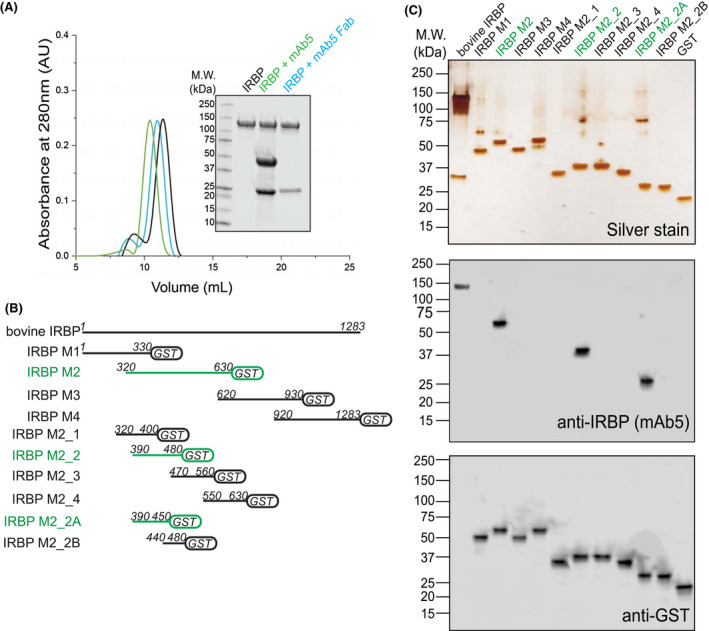
IRBP mAb5 binding characteristics and determination of epitope. A, Both full‐length mAb5 and mAb5 fragment antigen‐binding domain (mAb5 Fab) bind purified bovine IRBP. After incubating IRBP in the presence of either mAb5 or mAb5 Fab in a 1:1 molar ratio, samples were subjected to Superdex‐200 size‐exclusion chromatography. Twenty microgram of eluate peaks was subjected to SDS‐PAGE, and gels were stained with Coomassie blue (inset) showing stable association of both mAb5 and mAb5 Fab. Chromatogram corresponds to IRBP, IRBP + mAb5, and IRBP + mAb5 Fab in black, green, and blue, respectively. B, mAb5 recognizes IRBP M2_2A (residues 390‐450). IRBP Modules and smaller constructs were stably expressed as fusion proteins with glutathione S‐transferase (GST). Primary sequence lengths are represented for each construct, IRBP M1, M2, M3, M4, M2_1, M2_2, M2_3, M2_4, M2_2A, and M2_2B (residue numbers listed in methods and materials). Constructs that bound mAb5 were detected on an immunoblot and highlighted in green. C, Expressed constructs (500 ng) were subjected to SDS‐PAGE and stained by silver stain or transferred for immunoblot purposes. Transferred membranes were probed using mAb5 or an antibody targeting GST (Genscript, Piscataway, NJ, USA) as a primary probe. Purified bovine IRBP and GST were used as positive controls for the antibodies
Figure 3.
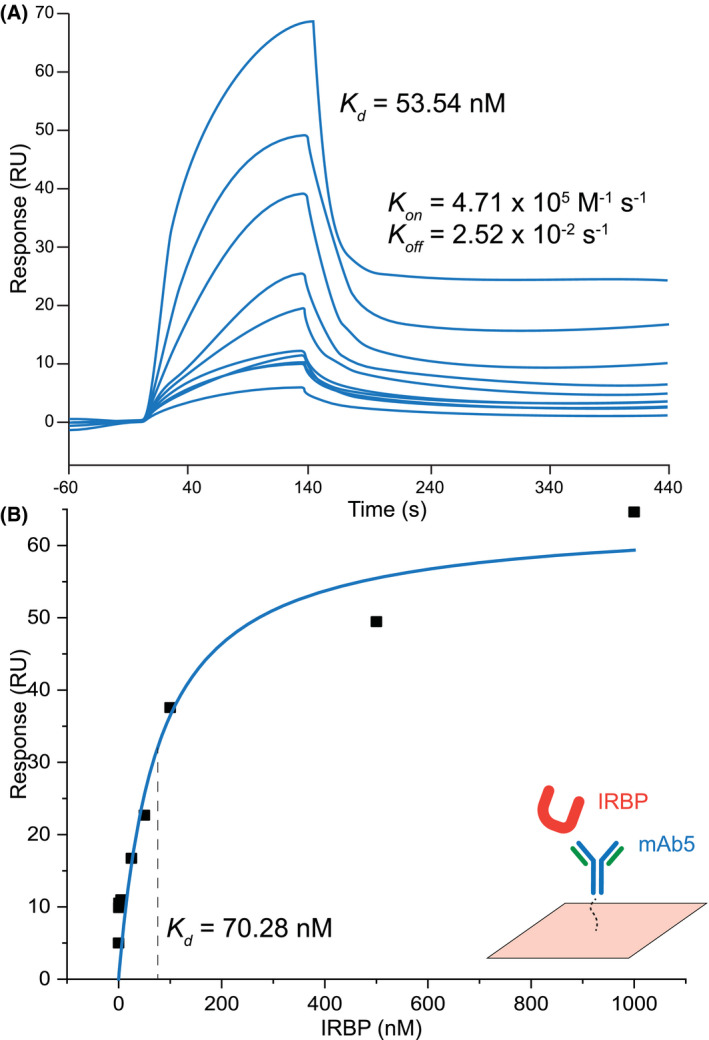
Kinetic profiling and mAb5‐IRBP complex. Binding of IRBP to immobilized mAb5 in SPR equilibrium binding experiments. Association of the IRBP protein with mAb5 was investigated by single‐cycle kinetics (A) and affinity‐based analysis (B). Resonance signals are indicated in response units (RU). The determined equilibrium dissociation constants (K d) are shown as insets with values of 53.34 and 70.28 nM, respectively. Kinetic parameters (Kon, Koff) are also shown as inset of (A) with values of 5.71 × 105 M‐1 s‐1, and 2.52 × 10‐2 s‐1
3.3. IRBP N‐linked glycosylation sites and sequencing
We next identified five N‐linked glycosylation sites with a characteristic 1 Da mass increase resulting from transformation of Asn to Asp residues by peptide:N glycosidase F (PNGase F) during cleavage of β‐aspartylglycosylamine linkages (Figure 4A,B). We determined the composition of the five IRBP oligosaccharides based on patterns of their collision‐induced fragmentation observed in tandem mass spectra (MS/MS) of the corresponding glycopeptides (Figure 4B and Figure S6‐S10, Table 1). Modified Asn residues include N107, N161, N205, N513, and N1114 (Figure 4C). These experimentally determined sites coincided with five sites predicted on the basis of the Asn‐X‐(Ser/Thr) sequence motif of N‐linked glycosylation (NetNGlyc). 56 Notably, peptides carrying glycosylation sites at positions 107, 161, 205, 513, and 1114 were not detected in a free form in glycosylated IRBP digests, which indicates nearly 100% glycosylation. The sugar chains attached to the protein were relatively short and homogeneous in composition, creating the sharp IRBP band observed in SDS‐polyacrylamide gels and a shift in mobility detected after protein deglycosylation. The mild heterogeneity observed in MS experiments could be partially induced by in‐source glycan fragmentation. 57
Figure 4.
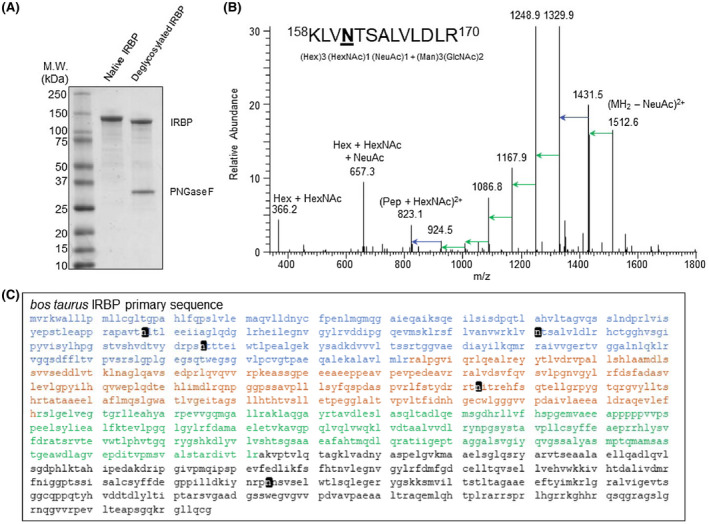
Determination of glycan composition and N‐glycosylation types of native bovine IRBP. A, SDS‐PAGE of bovine IRBP peptide:N‐glycosidase F (PNGase‐F) treated, deglycosylated IRBP showing shift in MW. B, A MS/MS spectrum of a glycosylated, native IRBP peptide (KLVNTSALVLDLR, residues 158‐170) illustrates the collision‐induced dissociation of N‐linked polysaccharide. Inset shows a list of glycans detected by MS/MS for each site determined. C, Location of 5 novel glycosylation sites (black) on the bovine IRBP primary sequence. IRBP modules 1‐4 are displayed in blue, orange, green and black, respectively. Abbreviations: DeoxyHex; deoxyhexose, GlcNAc; N‐acetylglucosamine, Hex; hexose, HexNAc; N‐acetylhexosamine, Man; mannose
Table 1.
Glycopeptides and respected glycan compositions of bovine IRBP
| Glycopeptides | Sites | Glycan compositions | Obs. mass | Error (ppm) |
|---|---|---|---|---|
| APAVTNLTLEEIIAGLQDGLR | N107 | (HexNAc)1 + (Man)3(GlcNAc)2 | 3289.5946 | −2.9 |
| (Hex)2 (HexNAc)1 + (Man)3(GlcNAc)2 | 3613.7014 | −2.2 | ||
| (Hex)3 (HexNAc)1 + (Man)3(GlcNAc)2 | 3775.7530 | −2.4 | ||
| (HexNAc)1 (Deoxyhexose)1 + (Man)3(GlcNAc)2 | 3435.649 | −1.8 | ||
| (Hex)2 (HexNAc)1 (Deoxyhexose)1 + (Man)3(GlcNAc)2 | 3759.7552 | −3.8 | ||
| (Hex)2 (HexNAc)1 (NeuAc)1 + (Man)3(GlcNAc)2 | 3904.7959 | −3.2 | ||
| (Hex)3 (HexNAc)1 (NeuAc)1 + (Man)3(GlcNAc)2 | 4066.8499 | −2.6 | ||
| KLVNTSALVLDLR | N161 | (Hex)2 (HexNAc)1 + (Man)3(GlcNAc)2 | 2861.3677 | −2.6 |
| (Hex)3 (HexNAc)1 + (Man)3(GlcNAc)2 | 3023.4220 | −2.1 | ||
| (Hex)2 (HexNAc)1 (NeuAc)1 + (Man)3(GlcNAc)2 | 3152.4625 | −2.7 | ||
| (Hex)3 (HexNAc)1 (NeuAc)1 + (Man)3(GlcNAc)2 | 3314.5219 | −0.4 | ||
| (Hex)2 (HexNAc)2 (NeuAc)1 + (Man)3(GlcNAc)2 | 3335.5424 | −2.3 | ||
| (Hex)2 (HexNAc)2 (NeuAc)2 + (Man)3(GlcNAc)2 | 3646.6351 | −2.7 | ||
| (Hex)3 (HexNAc)1 (Deoxyhexose)1 + (Man)3(GlcNAc)2 | 3169.4776 | −2.7 | ||
| (Hex)2 (HexNAc)1 (Deoxyhexose)1 (NeuAc)1 + (Man)3(GlcNAc)2 | 3298.5265 | −1.1 | ||
| (Hex)3 (HexNAc)1 (Deoxyhexose)1 (NeuAc)1 + (Man)3(GlcNAc)2 | 3460.5763 | −1.4 | ||
| DRPSNTTTEIW | N205 | (Hex)2 (HexNAc)2 (NeuAc)1 + (Man)3(GlcNAc)2 | 3233.2993 | 0 |
| (Hex)3 (HexNAc)1 (NeuAc)1 + (Man)3(GlcNAc)2 | 3192.2650 | −2.3 | ||
| (Hex)2 (HexNAc)1 (NeuAc)2 + (Man)3(GlcNAc)2 | 3321.3088 | −1.9 | ||
| (Hex)3 (HexNAc)1 (Deoxyhexose)1 (NeuAc)1 + (Man)3(GlcNAc)2 | 3338.3227 | −2.6 | ||
| (Hex)2 (HexNAc)2 (Deoxyhexose)1 (NeuAc)1 + (Man)3(GlcNAc)2 | 3379.3591 | −0.5 | ||
| TNITR | N513 | (HexNAc)1 + (Man)3(GlcNAc)2 | 1699.7342 | −2.4 |
| (Hex)1 (HexNAc)1 (NeuAc)1 + (Man)3(GlcNAc)2 | 2152.8796 | −3.1 | ||
| (Hex)2 (HexNAc)1 (NeuAc)1 + (Man)3(GlcNAc)2 | 2314.9312 | −3.5 | ||
| (Hex)3 (HexNAc)1 (NeuAc)1 + (Man)3(GlcNAc)2 | 2476.9831 | −3.3 | ||
| (Hex)1 (HexNAc)2 (NeuAc)1 + (Man)3(GlcNAc)2 | 2355.9604 | −2.1 | ||
| (Hex)2 (HexNAc)2 (NeuAc)1 + (Man)3(GlcNAc)2 | 2518.0114 | −2.7 | ||
| (Hex)2 (HexNAc)1 (NeuAc)2 + (Man)3(GlcNAc)2 | 2606.0269 | −2.8 | ||
| (Hex)2 (HexNAc)2 (NeuAc)2 + (Man)3(GlcNAc)2 | 2809.1068 | −2.2 | ||
| (Hex)3 (HexNAc)2 (NeuAc)2 + (Man)3(GlcNAc)2 | 2971.1566 | −3.2 | ||
| (Hex)2 (HexNAc)2 (NeuAc)3 + (Man)3(GlcNAc)2 | 3100.2001 | −2.9 | ||
| IYNRPNNSVSELWTLSQLEGER | N1114 | (Hex)2 (HexNAc)1 (NeuAc)1 + (Man)3(GlcNAc)2 | 4315.8874 | −2.5 |
| (Hex)3 (HexNAc)1 (NeuAc)1 + (Man)3(GlcNAc)2 | 4477.9422 | −1.1 | ||
| (Hex)3 (HexNAc)1 (Deoxyhexose)1 (NeuAc)1 + (Man)3(GlcNAc)2 | 4624.0014 | −1.5 |
3.4. Cryo‐EM single particle analysis of IRBP‐mAb5
To assess the 3D architecture of the IRBP, we used cryo‐EM single particle analysis (SPA). Detection of apo‐IRBP particles proved challenging. Despite the high purity and quality of the gel filtration elution peak, particles were extremely heterogeneous in shape (Figure S11A). IRBP is elongated, glycosylated, and has multiple degrees of rotations. To reduce the denaturation and aggregation of the protein at the air‐water interface, we coated grids with graphene oxide (GO) as a support layer to improve monodispersity of IRBP, and thus particle detection. We took advantage of cisTEM template‐free particle picking and 2D classification to maximize the extraction of IRBP particles from the cryo‐EM images. 38 The low‐pass filtered 2D class averages (30 Å) were used for 3D reconstruction. Processing of the initial dataset yielded 2D class averages in numerous orientations (Figure S11B). From this data it is possible to establish an ab initio 3D reconstruction volume. Using this as an input for cryoSPARC leads to a reconstruction with a global resolution of 8.1 Å as determined by Fourier shell correlation (FSC) = 0.143.
To overcome the limited solubility of IRBP, we took advantage of mAb5 to increase the protein concentration, thereby increasing IRBP‐mAb5 particle density on GO coated grids. In addition, the increased particle mass facilitated more efficient particle detection and improves amplitude measurements, allowing for better motion correction. 2D classes (Figure 5A) show a distinct complex of IRBP bound to mAb5, yielding details which were unattainable in the apo‐IRBP class averages (Figure S11). IRBP‐mAb5 3D reconstruction was performed in cryoSPARC 3.1. The structure was refined after applying local masks corresponding to each “hinge” region (Figures 5B and 6A). The 3D reconstruction of IRBP‐mAb5 revealed an open box or “pi” architecture as seen in the apo‐IRBP 2D classes (Figure S11B), but with an additional density corresponding to mAb5. Antibodies possess a high degree of flexibility, which explains why only the Fab portion of mAb5 bound to IRBP is mainly visualized in the reconstruction. As expected, we observed two additional densities protruding out of the Fab portion of IRBP‐mAb5 complex. These partial cryo‐EM densities likely belong to the other Fab portion and the Fc domain of mAb5. Local masks used to improve resolution corresponded to densities of mAb5 Fab, IRBP N‐terminus, core region, and C‐terminus. The dimensions of IRBP were determined directly from the reconstructed map of IRBP (Figure 5B). The largest diameter of IRBP is 14.1 nm, and the entire length of the molecule from the N‐C termini is approximately 31.9 nm, with a width of 2.2‐4.0 nm. In addition, we observed few particles and hence no 2D class averages in the “straight, or linear” conformation presented by Adler et al. 29 FSC calculation revealed consistency in change of resolution across the 3D reconstruction (Figure 6A). The reconstruction had preferred orientation (Figure 6B), and although the FSC at 0.143 (Figure 6C) determines the resolution of the reconstructed map to be at 7.4 Å, the reconstruction only partly resolved alpha‐helical densities, suggesting a resolution around 8 Å for most parts of the complex. To interpret the density, we docked bovine modules M1, M2, M3, and M4 modeled from the previously reported crystal structures of IRBP M1 and M2 (PDB ID’s: 4LUR, 1J7X) and the homology modeled VR of mAb5 (PIGSpro, Rosetta) at 7.0 Å using an unbiased, bidirectional docking approach to gain more insight into the IRBP‐mAb5 interaction seen in the EM reconstruction (Figure 7). 26 , 27 , 58 The N to C directionality was confirmed in the automated docking because the epitope determined was placed in close proximity to the complementarity determining regions of mAb5 density (Figure 7, inset). Confirmation of directionality allowed manual construction of a partial structure of IRBP‐mAb5. After refinement, it was possible to map connecting regions between M1‐M2, M2‐M3, and M3‐M4, which defines how these tandem modules interact with each other with high confidence (CC = 0.7) (Figure 8). Despite this new knowledge we are still limited by the intermediate resolution of IRBP, evidenced by low occurrence of secondary structure elements and large degree of flexibility as seen in the 2D class averages.
Figure 5.
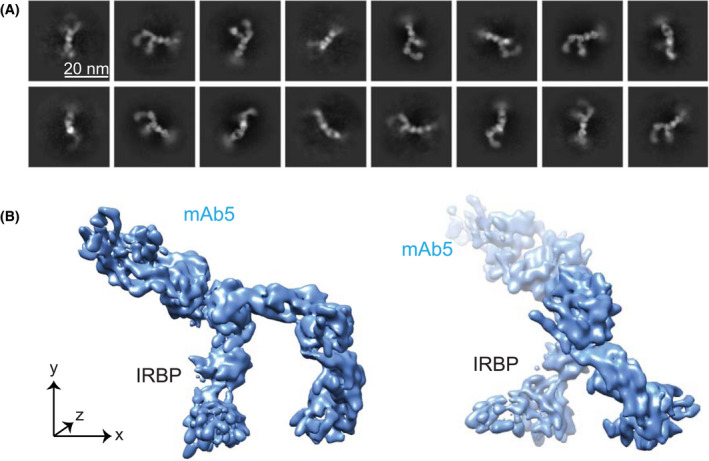
2D classes and 3D reconstruction of IRBP with a bound mAb5. A, 2D classes of IRBP and mAb5 complex, with scale bar shown in inlay (20 nm). The variable and constant regions of the Fab portion of mAb5 are visible. B, Reconstructed 3D model at viewing threshold 0.277. IRBP and mAb5 are labeled in black and blue, respectively. The left 3D model represents the front view, whereas the right one is turned 90° clockwise around the y‐axis
Figure 6.

Local resolution, refinement, and model statistics of IRBP‐mAb5 cryo‐EM density map. A, Cryo‐EM density map filtered and colored according to local resolution as estimated by REFMAC with Fourier Shell Correlation (FSC) plots for the refined 7.4 Å IRBP‐mAb5 map. Densities attributed to local masks are labeled accordingly as mAb5, IRBP N‐terminus, core, and C‐terminus, and separated for clarity. B, Azimuth plot of angular distribution based on particle distribution displays preferred orientation by IRBP‐mAb5 complex in cryo‐EM. C, FSC plot generated in cryoSPARC illustrating the 7.4 Å resolution of the reconstructed model with no ultimately corrected masks corresponding to masks mAb5, IRBP N‐terminus, core, and C‐terminus colored in gold, gray, blue, and orange, respectively
Figure 7.
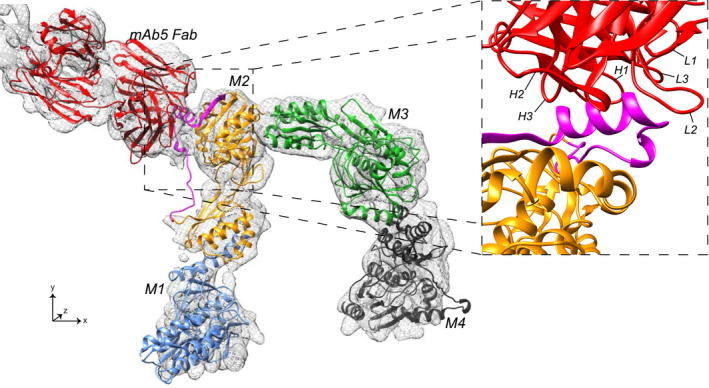
Fitting of homology modeled mAb5 Fab, IRBP modules M1, M2, M3 and M4 into cryo‐EM density of IRBP‐mAb5 as ribbon diagrams displayed in red, blue, orange, green, and black, respectively. The enlarged inset helps show detailed mAb5 and M2 interactions with a 90° clockwise rotation around the x‐ and y‐axes. Heavy chain CDR's (H1, H2, and H3) and light chain CDR’s (L1, L2, and L3) are labeled. IRBP sequence determined to bind mAb5 (residues 390‐450) is colored in magenta to show interactions with CDR's
Figure 8.
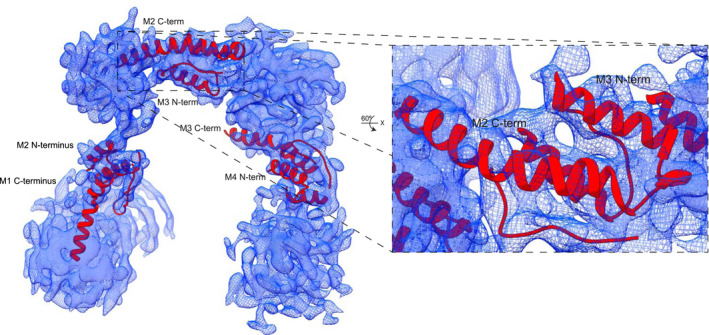
Connecting regions between IRBP M1, M2, M3, and M4. Modeled N‐termini of M2, M3, and M4, as well as modeled C‐ termini of IRBP M1, M2, and M3 in 3D reconstruction of IRBP. The enlarged section highlights the accuracy of alpha helix modeling into EM density, rotated 60° about the x‐axis
4. DISCUSSION
In this study, we report the production of a specific IRBP mAb5 and map its specificity to the module 2 of IRBP. We also determined that N‐linked glycosylation sites in IRBP (Bos taurus) exist in modules M1, M2, and M4 (N107, N161, N205, N513, and N1114), identical to the 5x predicted consensus sequences according to the sequence motif Asn‐X‐(Ser/Thr). The abundance of N‐linked glycosylation sites in IRBP could underlie its ability to interact with the extracellular matrix of photoreceptors. Additionally, glycosylation sites rarely mediate protein‐protein interaction, and thus M3 is most likely interacting with an unknown partner located within RPE or photoreceptor membranes. 59 , 60 , 61
mAb5 enabled cryo‐EM single particle analysis to determine the native 3D structure of IRBP isolated from bovine retinas to a resolution of 6‐7 Å. Our results display an atypical architecture for IRBP exhibiting an open ended box or “pi” shape with a diameter of 14.1 nm, formed by a bent filament of 39.1 nm length and 2.2‐4 nm width. The excellent electron microscopic analysis by Adler et al of IRBP prepared by glycerol spraying and metal shadowing describes a similar “pi” architecture with a width of 3‐4 nm but a shorter length of approximately 24 nm. In contrast to this pioneering work, we observe only few particles and hence no 2D class averages in the “straight, or linear” conformation presented in Adler et al. 29 The 143‐kDa protein is extremely flexible owing to multiple degrees of rotational freedom in the hinge regions. Utilizing a monoclonal antibody for IRBP helped increase solubility, thereby increasing particle count. More importantly, an increase in MW from addition of mAb5 promoted better motion correction of the micrographs and improved measurements of structure factor amplitudes of high spatial frequencies.
Imaging of the IRBP‐mAb5 complex enabled us to fit bovine IRBP modules M1, M2, M3, and M4 into the density map in addition to the modeled Fab of mAb5. Crystal structures of M1 and M2 from X laevis and D rerio IRBP provide direct structural information of the molecular folds of portions of the full‐length protein, but the continuous structure provides the ability to interpret how all modules interact with each other. We report the overall, unique architecture of full‐length IRBP. Importantly, an unbiased bidirectional rigid‐body refinement and automated docking place our resulting epitope directly in the CDR binding region of mAb5.
The box‐like architecture of IRBP, determined at 7.4 Å resolution is unlike any other retinoid binding protein structure to date. 62 The shape could facilitate binding to photoreceptor or RPE cell receptors and promote the transport of retinoids. It is likely that the inherent flexibility of IRBP contributes to the broad specificity of binding hydrophobic molecules necessary for the health and maintenance of photoreceptors. Flexibility in structure might be responsible for the protein's numerous implied functions. It is also possible that the quadruple, tandem module arrangement allows a single protomer of IRBP to bind different ligands and act as a buffer for the interphotoreceptor matrix, which facilitates high turnover of retinoids and shedding of photoreceptor outer segments every day. Distinct cavities (~14.0 Å) observed in the 3D structure could fit a variety of retinoid moieties and completely sequester them from bulk solvent. Considering that the crystal structures of D rerio IRBP M1 and X laevis IRBP M2 as well as the homology modeled bovine M1‐M4 all contain the canonical αβα sandwich motif present in cellular retinaldehyde‐binding protein (CRALBP), it is possible that each module can bind a retinoid molecule. 28 In conclusion, we have defined a reproducible protocol for isolating pure, native IRBP from bovine retinas. We identified multiple N‐linked glycosylation sites in 3 of the 4 IRBP modules and generated a monoclonal antibody specific to IRBP M2. This antibody enabled a cryo‐EM structure of native IRBP with resolution of 7.4 Å, revealing the interconnections between modules that form a box‐like structure not seen among other retinoid‐binding proteins.
CONFLICT OF INTEREST
None.
AUTHOR CONTRIBUTIONS
A.E. Sears, S. Gulati, and K. Palczewski designed the research; A.E. Sears, S. Gulati, P. Kiser, and K. Palczewski wrote the paper; A.E. Sears, S. Albiez, S. Gulati, L. Kovacik, and H. Stahlberg performed the cryo‐EM data acquisition and analyses; A.E. Sears and B. Wang performed mass spectrometry analyses; AES performed the biochemical analyses (protein purification, FRET, TRP quenching, ELSA, and SPR); S. Albiez, B. Wang, L. Kovacik, A. Engel, and H. Stahlberg contributed to the written paper; A.E. Sears, S. Albiez, S. Gulati, B. Wang, P. Kiser, L. Kovacik, A. Engel, H. Stahlberg, and K. Palczewski, contributed to methodology; H. Stahlberg and K. Palczewski contributed to resources and funding acquisition; A. Engel, H. Stahlberg, and K. Palczewski provided supervision.
Supporting information
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Palczewski laboratory for their helpful comments regarding this project. We also thank the members of the University Hospitals Visual Sciences Research Center, supported by the National Eye Institute (NEI) P30 EY011373. This research was supported in part by grants from the National Institutes of Health (NIH) (EY009339) and the MSTP training grant (T32 GM002750), the US Department of Veterans Affairs (IK2BX002683), and the Swiss National Science Foundation (NCCR TransCure). K.P is the Irving H. Leopold Chair of Ophthalmology. The authors also acknowledge support from an RPB unrestricted grant to the Department of Ophthalmology, University of California, Irvine.
Sears AE, Albiez S, Gulati S, et al. Single particle cryo‐EM of the complex between interphotoreceptor retinoid‐binding protein and a monoclonal antibody. The FASEB Journal. 2020;34:13918–13934. 10.1096/fj.202000796RR
Avery E. Sears and Stefan Albiez contributed equally to this work.
Contributor Information
Avery E. Sears, Email: aes31@case.edu.
Henning Stahlberg, Email: henning.stahlberg@unibas.ch.
Krzysztof Palczewski, Email: kpalczew@uci.edu.
DATA AVAILABILITY STATEMENT
All data supporting the findings of this study are available within this paper. Additional data supporting the findings of this manuscript are available from the corresponding authors upon reasonable request. The cryo‐EM map of IRBP mAb5 complex and atomic coordinates of docked IRBP modules and mAb5 Fab fragment have been deposited in the Electron Microscopy Data Bank under accession codes EMD‐22474 and PDB ID 7JTI, respectively. The cryo‐EM movie files have been deposited at the EMPIAR under accession code EMPIAR‐10388.
REFERENCES
- 1. Wald G, Durell J, St George CC. The light reaction in the bleaching of rhodopsin. Science. 1950;111:179‐181. [DOI] [PubMed] [Google Scholar]
- 2. Wald G. Vitamin a in eye tissues. J Gen Physiol. 1935;18:905‐915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Marmor MF, Martin LJ. 100 years of the visual cycle. Surv Ophthalmol. 1978;22:279‐285. [DOI] [PubMed] [Google Scholar]
- 4. Palczewski K. G protein‐coupled receptor rhodopsin. Annu Rev Biochem. 2006;75:743‐767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kiser PD, Golczak M, Palczewski K. Chemistry of the retinoid (visual) cycle. Chem Rev. 2014;114:194‐232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kiser PD, Golczak M, Maeda A, Palczewski K. Key enzymes of the retinoid (visual) cycle in vertebrate retina. Biochem Biophys Acta. 2011;1821:137‐151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blomhoff R, Blomhoff HK. Overview of retinoid metabolism and function. J Neurobiol. 2006;66:606‐630. [DOI] [PubMed] [Google Scholar]
- 8. O'Byrne SM, Blaner WS. Retinol and retinyl esters: biochemistry and physiology. J Lipid Res. 2013;54:1731‐1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Borst DE, Redmond TM, Elser JE, et al. Interphotoreceptor retinoid‐binding protein. Gene characterization, protein repeat structure, and its evolution. J Biol Chem. 1989;264:1115‐1123. [PubMed] [Google Scholar]
- 10. Dettai A, Lecointre G. New insights into the organization and evolution of vertebrate IRBP genes and utility of IRBP gene sequences for the phylogenetic study of the Acanthomorpha (Actinopterygii: Teleostei). Mol Phylogenet Evol. 2008;48:258‐269. [DOI] [PubMed] [Google Scholar]
- 11. Chen Y, Houghton LA, Brenna JT, Noy N. Docosahexaenoic acid modulates the interactions of the interphotoreceptor retinoid‐binding protein with 11‐cis‐retinal. J Biol Chem. 1996;271:20507‐20515. [DOI] [PubMed] [Google Scholar]
- 12. Okajima TI, Pepperberg DR, Ripps H, Wiggert B, Chader GJ. Interphotoreceptor retinoid‐binding protein: role in delivery of retinol to the pigment epithelium. Exp Eye Res. 1989;49:629‐644. [DOI] [PubMed] [Google Scholar]
- 13. Cook JD, Ng SY, Lloyd M, et al. Peropsin modulates transit of vitamin A from retina to retinal pigment epithelium. J Biol Chem. 2017;292:21407‐21416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Flannery JG, O'Day W, Pfeffer BA, Horwitz J, Bok D. Uptake, processing and release of retinoids by cultured human retinal pigment epithelium. Exp Eye Res. 1990;51:717‐728. [DOI] [PubMed] [Google Scholar]
- 15. Okajima TI, Pepperberg DR, Ripps H, Wiggert B, Chader GJ. Interphotoreceptor retinoid‐binding protein promotes rhodopsin regeneration in toad photoreceptors. Proc Natl Acad Sci USA. 1990;87:6907‐6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Carlson A, Bok D. Promotion of the release of 11‐cis‐retinal from cultured retinal pigment epithelium by interphotoreceptor retinoid‐binding protein. Biochemistry. 1992;31:9056‐9062. [DOI] [PubMed] [Google Scholar]
- 17. Chen C, Adler LT, Goletz P, Gonzalez‐Fernandez F, Thompson DA, Koutalos Y. Interphotoreceptor retinoid‐binding protein removes all‐trans‐retinol and retinal from rod outer segments, preventing lipofuscin precursor formation. J Biol Chem. 2017;292:19356‐19365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jin M, Li S, Nusinowitz S, et al. The role of interphotoreceptor retinoid‐binding protein on the translocation of visual retinoids and function of cone photoreceptors. J Neurosci. 2009;29:1486‐1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Duffy M, Sun Y, Wiggert B, Duncan T, Chader GJ, Ripps H. Interphotoreceptor retinoid binding protein (IRBP) enhances rhodopsin regeneration in the experimentally detached retina. Exp Eye Res. 1993;57:771‐782. [DOI] [PubMed] [Google Scholar]
- 20. Edwards RB, Adler AJ. IRBP enhances removal of 11‐ cis ‐retinaldehyde from isolated RPE membranes. Exp Eye Res. 2000;70:235‐245. [DOI] [PubMed] [Google Scholar]
- 21. Gonzalez‐Fernandez F, Healy JI. Early expression of the gene for interphotoreceptor retinol‐binding protein during photoreceptor differentiation suggests a critical role for the interphotoreceptor matrix in retinal development. J Cell Biol. 1990;111:2775‐2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liou GI, Wang M, Matragoon S. Timing of interphotoreceptor retinoid‐binding protein (IRBP) gene expression and hypomethylation in developing mouse retina. Dev Biol. 1994;161:345‐356. [DOI] [PubMed] [Google Scholar]
- 23. den Hollander AI, McGee TL, Ziviello C, et al. A homozygous missense mutation in the IRBP gene (RBP3) associated with autosomal recessive retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2009;50:1864‐1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Arno G, Hull S, Robson AG, et al. Lack of interphotoreceptor retinoid binding protein caused by homozygous mutation of RBP3 is associated with high myopia and retinal dystrophy. Invest Ophthalmol Vis Sci. 2015;56:2358‐2365. [DOI] [PubMed] [Google Scholar]
- 25. Kolesnikov AV, Tang PH, Parker RO, Crouch RK, Kefalov VJ. The mammalian cone visual cycle promotes rapid M/L‐cone pigment regeneration independently of the interphotoreceptor retinoid‐binding protein. J Neurosci. 2011;31:7900‐7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ghosh D, Haswell KM, Sprada M, Gonzalez‐Fernandez F. Structure of zebrafish IRBP reveals fatty acid binding. Exp Eye Res. 2015;140:149‐158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Loew A, Gonzalez‐Fernandez F. Crystal structure of the functional unit of interphotoreceptor retinoid binding protein. Structure. 2002;10:43‐49. [DOI] [PubMed] [Google Scholar]
- 28. Crabb JW, Nie Z, Chen Y, et al. Cellular retinaldehyde‐binding protein ligand interactions. Gln‐210 and Lys‐221 are in the retinoid binding pocket. J Biol Chem. 1998;273:20712‐20720. [DOI] [PubMed] [Google Scholar]
- 29. Adler AJ, Stafford WF 3rd, Slayter HS. Size and shape of bovine interphotoreceptor retinoid‐binding protein by electron microscopy and hydrodynamic analysis. J Biol Chem. 1987;262:13198‐13203. [PubMed] [Google Scholar]
- 30. Drozdetskiy A, Cole C, Procter J, Barton GJ. JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 2015;43:W389‐W394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Martin TGB, Andreas WPF, Anthony S, Sjors HW. Graphene oxide grid preparation. Figshare. 2016. 10.6084/m9.figshare.3178669.v1. [DOI] [Google Scholar]
- 32. Kohler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495‐497. [DOI] [PubMed] [Google Scholar]
- 33. Holzlöhner P, Hanack K. Generation of Murine Monoclonal Antibodies by Hybridoma Technology. J Vis Exp. 2017. 10.3791/54832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Babrak L, McGarvey JA, Stanker LH, Hnasko R. Identification and verification of hybridoma‐derived monoclonal antibody variable region sequences using recombinant DNA technology and mass spectrometry. Mol Immunol. 2017;90:287‐294. [DOI] [PubMed] [Google Scholar]
- 35. Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36‐51. [DOI] [PubMed] [Google Scholar]
- 36. Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA. MotionCor2: anisotropic correction of beam‐induced motion for improved cryo‐electron microscopy. Nat Methods. 2017;14:331‐332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Biyani N, Righetto RD, McLeod R, et al. Focus: The interface between data collection and data processing in cryo‐EM. J Struct Biol. 2017;198:124‐133. [DOI] [PubMed] [Google Scholar]
- 38. Grant T, Rohou A, Grigorieff N. cisTEM, user‐friendly software for single‐particle image processing. eLife. 2018;7:e35383 10.7554/elife.35383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. cryoSPARC: algorithms for rapid unsupervised cryo‐EM structure determination. Nat Methods. 2017;14:290‐296. [DOI] [PubMed] [Google Scholar]
- 40. Zivanov J, Nakane T, Forsberg B, Kimanius D, Hagen W, Lindahl E, Scheres S. New tools for automated high‐resolution cryo‐EM structure determination in RELION‐3. eLife. 2018;7:e42166 10.7554/elife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bepler T, Morin A, Rapp M, et al. Positive‐unlabeled convolutional neural networks for particle picking in cryo‐electron micrographs. Nat Methods. 2019;16:1153‐1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Adams PD, Afonine PV, Bunkoczi G, et al. PHENIX: a comprehensive Python‐based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213‐221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Waterhouse A, Bertoni M, Bienert S, et al. SWISS‐MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296‐W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Moretti R, Lyskov S, Das R, Meiler J, Gray JJ. Web‐accessible molecular modeling with Rosetta: the Rosetta Online Server that Includes Everyone (ROSIE). Protein Sci. 2018;27:259‐268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605‐1612. [DOI] [PubMed] [Google Scholar]
- 46. Emsley P, Cowtan K. Coot: model‐building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126‐2132. [DOI] [PubMed] [Google Scholar]
- 47. Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT. Scalable web services for the PSIPRED protein analysis workbench. Nucleic Acids Res. 2013;41:W349‐W357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486‐501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Collaborative Computational Project, N . The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760‐763. [DOI] [PubMed] [Google Scholar]
- 50. Pfeffer B, Wiggert B, Lee L, Zonnenberg B, Newsome D, Chader G. The presence of a soluble interphotoreceptor retinol‐binding protein (IRBP) in the retinal interphotoreceptor space. J Cell Physiol. 1983;117:333‐341. [DOI] [PubMed] [Google Scholar]
- 51. Fong SL, Irimura T, Landers RA, Bridges CD. The carbohydrate of bovine interstitial retinol‐binding protein. Prog Clin Biol Res. 1985;190:111‐128. [PubMed] [Google Scholar]
- 52. Chen Y, Saari JC, Noy N. Interactions of all‐trans‐retinol and long‐chain fatty acids with interphotoreceptor retinoid‐binding protein. Biochemistry. 1993;32:11311‐11318. [DOI] [PubMed] [Google Scholar]
- 53. Gonzalez‐Fernandez F, Sung D, Haswell KM, Tsin A, Ghosh D. Thiol‐dependent antioxidant activity of interphotoreceptor retinoid‐binding protein. Exp Eye Res. 2014;120:167‐174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen Y, Noy N. Retinoid specificity of interphotoreceptor retinoid‐binding protein. Biochemistry. 1994;33:10658‐10665. [DOI] [PubMed] [Google Scholar]
- 55. Noy N. Measurement of rates of dissociation of retinoids from the interphotoreceptor retinoid‐binding protein. Methods Mol Biol. 1998;89:177‐189. [DOI] [PubMed] [Google Scholar]
- 56. Blom N, Sicheritz‐Pontén T, Gupta R, Gammeltoft S, Brunak S. Prediction of post‐translational glycosylation and phosphorylation of proteins from the amino acid sequence. PROTEOMICS. 2004;4:1633‐1649. 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- 57. Peterman SM, Mulholland JJ. A novel approach for identification and characterization of glycoproteins using a hybrid linear ion trap/FT‐ICR mass spectrometer. J Am Soc Mass Spectrom. 2006;17:168‐179. [DOI] [PubMed] [Google Scholar]
- 58. Lepore R, Olimpieri PP, Messih MA, Tramontano A. PIGSPro: prediction of immunoglobulin structures v2. Nucleic Acids Res. 2017;45:W17‐W23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Varki A. Biological roles of glycans. Glycobiology. 2017;27:3‐49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hubmacher D, Apte SS. The biology of the extracellular matrix: novel insights. Curr Opin Rheumatol. 2013;25:65‐70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kiessling LL, Grim JC. Glycopolymer probes of signal transduction. Chem Soc Rev. 2013;42:4476‐4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhang YR, Zhao YQ, Huang JF. Retinoid‐binding proteins: similar protein architectures bind similar ligands via completely different ways. PLoS ONE. 2012;7:e36772. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Data Availability Statement
All data supporting the findings of this study are available within this paper. Additional data supporting the findings of this manuscript are available from the corresponding authors upon reasonable request. The cryo‐EM map of IRBP mAb5 complex and atomic coordinates of docked IRBP modules and mAb5 Fab fragment have been deposited in the Electron Microscopy Data Bank under accession codes EMD‐22474 and PDB ID 7JTI, respectively. The cryo‐EM movie files have been deposited at the EMPIAR under accession code EMPIAR‐10388.


