Summary
Brassica napus is highly susceptible towards Verticillium longisporum (Vl43) with no effective genetic resistance. It is believed that the fungus reprogrammes plant physiological processes by up‐regulation of so‐called susceptibility factors to establish a compatible interaction. By transcriptome analysis, we identified genes, which were activated/up‐regulated in rapeseed after Vl43 infection. To test whether one of these genes is functionally involved in the infection process and loss of function would lead to decreased susceptibility, we firstly challenged KO lines of corresponding Arabidopsis orthologs with Vl43 and compared them with wild‐type plants. Here, we report that the KO of AtCRT1a results in drastically reduced susceptibility of plants to Vl43. To prove crt1a mutation also decreases susceptibility in B. napus, we identified 10 mutations in a TILLING population. Three T3 mutants displayed increased resistance as compared to the wild type. To validate the results, we generated CRISPR/Cas‐induced BnCRT1a mutants, challenged T2 plants with Vl43 and observed an overall reduced susceptibility in 3 out of 4 independent lines. Genotyping by allele‐specific sequencing suggests a major effect of mutations in the CRT1a A‐genome copy, while the C‐genome copy appears to have no significant impact on plant susceptibility when challenged with Vl43. As revealed by transcript analysis, the loss of function of CRT1a results in activation of the ethylene signalling pathway, which may contribute to reduced susceptibility. Furthermore, this study demonstrates a novel strategy with great potential to improve plant disease resistance.
Keywords: CRT1a, calreticulin, CRISPR/Cas9, TILLING, recessive resistance, susceptibility factor, Verticillium longisporum, Brassica napus, Arabidopsis thaliana
Introduction
Brassica napus or commonly referred to as oilseed rape (OSR) is a commercially important crop and grown to produce vegetable oil for human consumption, animal feeding and biodiesel utilization (Harloff et al., 2012). In 2018, OSR was after soya bean the second most cultivated oilseed crop with 70.91 million tons, worldwide (USDA, 2018). The advantage of OSR is the viability at low temperatures with reasonable humidity; thus, it can be cultivated in temperate zones where soya bean and sunflower are not able to be cultivated. Since the 1970s, the worldwide cultivation area has increased continuously (FAOSTAT, 2016). A consequence of the increased production area in addition to narrow crop rotation cycles and tillage operations is the enrichment of soilborne pathogens, such as Verticillium longisporum (De Coninck et al., 2015).
Verticillium longisporum is a hemibiotrophic fungal pathogen specialized to infect Brassicaceae and causes Verticillium stem striping in OSR (Depotter et al., 2016). It emerges especially in northern Europe and attacks preferentially developing plant roots (Johansson et al., 2006). The infection process starts with the germination of fungal microsclerotia that recognize root exudates and follow the nutrient gradient to reach the roots of potential host plants. The fungus can directly penetrate the epidermal cells of the root and, in addition, natural openings, for example wounds are used by the fungus to infect the host, spreading inter‐ and intracellularly, finally resulting in hyphal proliferation and the production of conidia (Depotter et al., 2016; Johansson et al., 2006). The fungus accomplishes extensive colonization of the whole plant by using the vascular system to spread its conidia. During later stages of plant colonization, the fungus resigns from the vascular elements and starts a necrotrophic life phase by feeding on senescing leaf and stem tissue (Reusche et al., 2012).
Oilseed rape is an amphidiploid species (2n = 38, AACC genome) derived from the hybridization of the two closely related species Brassica rapa (AA) and Brassica oleracea (CC) (Nagaharu, 1935). The relatively young age of this species goes along with a very limited genetic pool, which was further narrowed because the breeding focus was on double zero lines. Several problems such as inferior seed germination (Hatzig et al., 2018) or reduced pathogen resistance (Zhao and Meng, 2003) arose from these breeding strategies. For instance, genetic resources for resistance breeding to control Verticillium stem striping in OSR are hardly available, and so far, no effective resistance has been described. In practice, reducing the potential infection rate through minimizing the spore density in the soil is often a main countermeasure, but very laborious or detrimental to non‐target organisms under field conditions (Depotter et al., 2016). In addition, no fungicide treatments are available to control this soilborne disease, and due to the colonization of the host through the vascular system, a contact fungicide treatment is anyway impossible. Thus, breeding for OSR resistance to fungal diseases is also the most favoured countermeasure against V. longisporum (Dunker et al., 2008).
Worldwide, efforts have been made to identify genetic resistance resources by screening‐related wild species (Eynck et al., 2009; Happstadius et al., 2003) including the parental species B. oleracea and B. rapa and generated resynthesized lineages (Eynck et al., 2009). However, the resistance identified so far is highly polygenic (Eynck et al., 2009; Rygulla et al., 2008) and hardly suitable for breeding programmes. Thus, an alternative solution is needed. Increasing data demonstrates that a successful fungal colonization often relies on intensive plant–fungus interactions, involving negative defence regulators and susceptibility/compatibility factors (Behrens et al., 2019). The loss of such factors in plants can cause an incompatible plant–pathogen interaction, thus providing a promising alternative for resistance breeding (Langner et al., 2018). This approach was successfully applied to increase plant resistance to viral, bacterial, and fungal infections. For example, mutation of the eukaryotic translation initiation factor 4 (eIF4E/G) leads to virus resistance in a wide variety of plant species including barley, cucumber and rice (Chandrasekaran et al., 2016; Macovei et al., 2018; Stein et al., 2005), and increased resistance towards the bacterial pathogen Xanthomonas was reported by mutation of SWEET sucrose transporters in rice, cassava or citrus, respectively (Oliva et al., 2019; Zhou et al., 2015; Cohn et al., 2014; Hu et al., 2014). Furthermore, as demonstrated in barley, a nucleotide mutation in Mlo confers a durable resistance to powdery mildew since several years (Kusch and Panstruga, 2017). Loss of function in all Mlo alleles also generated powdery mildew resistance in grape, tomato and wheat (Malnoy et al., 2016; Nekrasov et al., 2017; Wang et al., 2014).
It is extremely difficult to identify natural loss‐of‐function mutations in crops with complex genomes due to functional redundancy caused by many alleles encoding the same gene. Recently, methods have been successfully developed to increase genetic variation by mutating the whole crop genome, for example using the chemical ethyl methanesulphonate (EMS). The laborious identification of such EMS‐caused point mutations can be achieved by Targeting Induced Local Lesions IN Genomes (TILLING; McCallum et al., 2000; Till et al., 2006). These mutations have to be combined by crossing, and phenotyping can be impeded by the many unwanted background mutations arising from this untargeted approach. Though it was actually possible to combine TILLING mutations for all six wheat Mlo alleles to generate a complete KO and therefore to increase resistance to powdery mildew (Acevedo‐Garcia et al., 2017), a study by Wang et al. (2014) appeared three years earlier using a targeted genome editing approach involving side‐specific nucleases (SSN).
The allotetraploid genome of Brassica napus is also very complex (Chalhoub et al., 2014). SSNs such as CRISPR/Cas have recently been successfully deployed to induce mutations in OSR to improve agronomic traits such as silique development and seed shatter resistance (Braatz et al., 2017; Yang et al., 2017; Yang et al., 2018; Zhai et al., 2019a), branching (Zheng et al., 2019), increase in the content of oleic acid (Okuzaki et al., 2018), altered oil content and fatty acid composition in seeds (Zhai et al., 2019b) or resistance to Sclerotinia sclerotiorum by WRKY70 KO (Sun et al., 2018). Thus, SSNs can efficiently mutate a target gene in less time compared to TILLING and reduce unwanted off‐target mutations in OSR.
The aim of this work was to increase the resistance of OSR towards V. longisporum infection by interrupting the compatible plant–fungus interaction via mutation of candidate host susceptibility genes identified by functional genomics. Twenty candidate genes with altered expression levels in response to infection with V. longisporum isolate 43 (Vl43) were identified from two suppression subtractive hybridization (SSH) libraries (Diatchenko et al., 1996). Here, we report that loss of function of CRT1a (calreticulin) strongly reduces plant susceptibility to V. longisporum in both A. thaliana and OSR, demonstrating thereby a strategy and working routine allowing for an efficient generation of recessive resistance against pathogens in OSR.
Results
Identification of CRT1a as putative susceptibility factor
In order to identify genes being substantially involved in the Brassica napus–Vl43 interaction, we constructed two SSH libraries. Brassica napus Express 617 plants were infected with Vl43, and mRNA was isolated at 5, 10 and 15 days postinoculation (dpi). The pooled mRNAs from infected and mock samples were used to produce the SSH libraries. For the ‘forward’ library, we used mRNA from infected material as tester and control mRNA as driver to identify up‐regulated genes and vice versa in the ‘reverse’ library to identify down‐regulated genes. In total, we obtained 1060 sequences from the forward library of which 907 were plant‐derived, representing 413 unique plant genes. 461 sequences were obtained from the reverse library with 369 being plant‐derived, representing 209 unique plant genes. The complete list of sequenced clones with homologies to plant genes is presented in Table S1. Potential susceptibility factors are expected to be up‐regulated by the pathogen to achieve colonization of its host; thus, our main interest lies on the forward library. However, it is well known that during pathogen infection genes of the plant resistance response might be also suppressed, either by the pathogen or by the host, involving, for example, negative regulators of the innate immunity. Thus, a subset of candidates from the reverse library was also selected. In order to verify the quality of the SSH libraries, we checked the expression levels of randomly selected genes by RT‐qPCR, which generally confirmed up‐ and down‐regulation of genes obtained from the forward and reverse libraries, respectively (Figure S1). The selection of candidate genes for further analysis was based on inclusion of a wide range of differentially regulated genes and genes functionally related to plant–pathogen interactions, as well as genes with so far no connection to pathogenesis to identify novel components. A GENEVESTIGATOR‐based meta‐analysis (Hruz et al., 2008) demonstrates a high functional divergence of 20 selected candidate genes (Tables S2 and S3). Since a knockout of these genes in the complex Brassica napus genome is still technically challenging and time‐consuming, we employed firstly T‐DNA insertion mutants of the corresponding Arabidopsis orthologs to investigate their functional involvement in the plant–Vl43 interaction.
For 15 of the 20 candidate genes, we obtained homozygous Arabidopsis mutants (Table S3). We challenged these mutants with Vl43 and compared the results with Col‐0 wild‐type plants with respect to disease progression and symptom development. Strikingly, as most of the Arabidopsis mutants did not show obvious effects interfering with Vl43, the crt1a knockout led to significantly reduced susceptibility and symptom development (Figure 1). While at 28 dpi, the wild‐type plants severely suffered from the infection and displayed the stunted shoot growth and senescence‐like symptoms, the crt1a mutant was less affected by Vl43 infection (Figure 1A). In accordance, the rosette diameter and the leaf area were less reduced (Figure 1B) and the fungal colonization/biomass was drastically decreased in the crt1a mutant plants as compared to the wild type (Figure 1C). In a time course experiment, we challenged the crt1a mutant and the Col‐0 plants with varying conidia concentrations as high as up to 1 × 107 mL−1. Again, we observed that the wild‐type Col‐0 plants severely suffered from the fungal infection, while the crt1a mutant plants survived and displayed impaired disease development (Figure S2). Taken together, these data strongly suggest that the knockout/loss of function of CRT1a reduces susceptibility of plants to Vl43 infection. In the next step, we analysed the expression of CRT1a in response to Vl43 infection by RT‐qPCR in both Arabidopsis and OSR and found expression of calreticulin was significantly up‐regulated in both species at 6 dpi (Figure 1D), in consistence with its identification from the SSH forward library.
Figure 1.
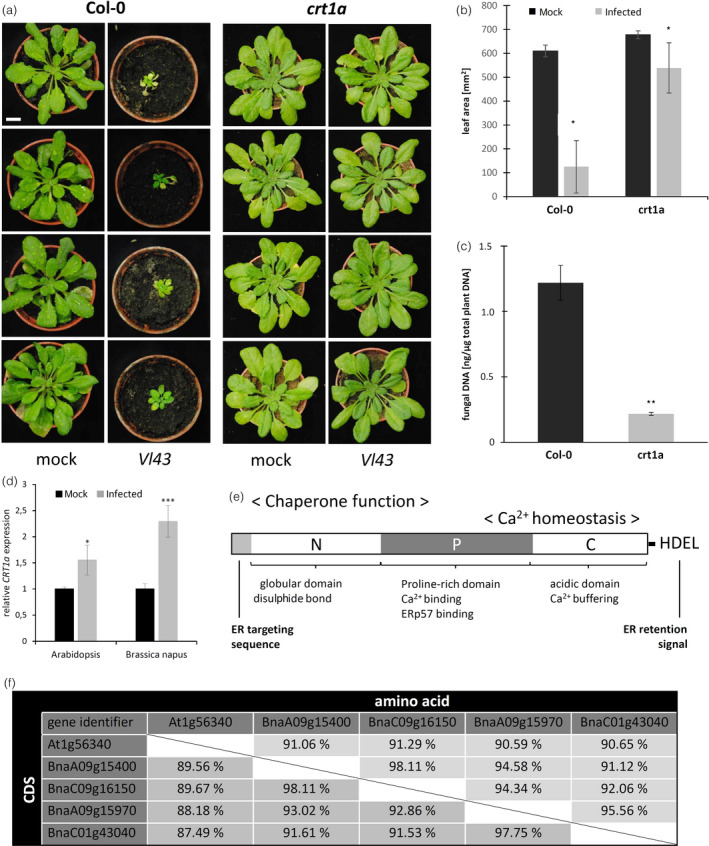
The Arabidopsis CRT1a mutant displays reduced susceptibility to VL43 infection. (a) Disease symptom development of crt1a as compared to Col‐0 wild‐type plants infected with 1 × 106 conidia/mL. Photographs were taken at 28 dpi. The scale bar equals 1 cm. (b) Leaf area analysis as measured at 35 dpi. (c) Fungal proliferation within infected Arabidopsis roots as measured by quantification of fungal DNA via qPCR at 28 dpi. (d) CRT1a is up‐regulated in response to Vl43 infection at 6 dpi in both in Arabidopsis thaliana (left) seedlings and Brassica napus (right) roots. (e) Typical calreticulin structure, according to Christensen et al., 2008. (f) Comparison of Arabidopsis and Brassica napus CRT1a sequence homology on CDS and amino acid level. Numbers indicate the % identity. Statistical significance was determined on the basis of three independent biological replicates by Student's t‐test and was evident for differences between wild‐type and the respective mutants (*P ≤ 0.05; **P ≤ 0.01).
In Arabidopsis, calreticulin (CRT) has three paralogs (CRT1a, CRT1b and CRT3), being an ER‐localized protein with three major domains (Figure 1E). Calreticulins function in protein folding and Ca2+ homeostasis (TAIR; Berardini et al., 2015). When searching for the corresponding Brassica napus orthologous copies in the GENOSCOPE database (Chalhoub et al., 2014), four sequences with high homology on cDNA and amino acid level were identified (Figure 1F). However, the sequences of BnaA09g15970 and BnaC01g43040 contained duplicated regions, probably due to wrong NGS annotation (Figure S3A). After removal of these duplicated regions, both sequences resemble very well CRT1a and all loci appear to be expressed (Figure S3B). Thus, we believe there are four functional CRT1a loci within the Brassica napus genome. In addition, a GENEVESTIGATOR meta‐analysis revealed that Arabidopsis CRT1a is up‐regulated in response to several other pathogens, such as Blumeria graminis, Golovinomyces orontii, Phytophthora infestans, Sclerotinia sclerotiorum and Xanthomonas campestris, and the PAMP elf18 and salicylic acid (Table S2). Furthermore, the impact of the AtCRT1a KO on agronomic traits, such as flowering time, plant height and number of siliques, was determined (Table S4). Collectively, the data suggest that CRT1a loss of function reduced the susceptibility of Arabidopsis to Vl43 without negatively affecting the agronomic traits analysed. Thus, this gene was chosen for further investigation in Brassica napus.
TILLING of CRT1a in Brassica napus
To generate allele‐specific primer for TILLING at the CRT1a locus, we did bioinformatic analysis of the 4 CRT1a loci in the Brassica napus genome (Figures S3 and S4). From these, the CRT1a locus BnaA09g15970D is matching 100% to the EST 6F11‐F8 from the SSH library; thus, we focused on identification of TILLING mutations in this gene, which we believe is the ‘active’ locus (Figures 6A and S4). We developed allele‐specific primers for BnaA09g15970D covering the CRT1a genomic region containing exons 4–9 (Table S5; Figure 2A) and used them for TILLING at the CRT1a locus of an EMS‐mutagenized OSR population established at the CAU (Harloff et al., 2012). This TILLING fragment of 1082 bps represents 41.7% of the genomic region (Figure 2B). Screening of 3840 individual plants resulted in 131 potential mutation signals. With the aid of a specific pooling strategy (Harloff et al., 2012), 64 signals could be attributed to single plants (Table S5). Due to degradation of some storage plant DNAs and failure of PCRs, only 27 single plants were available and subsequently subjected to sequencing in order to validate the mutations (Table S5). In total, 16 of 27 sequenced candidates showed an expected single nucleotide mutation triggered by EMS. Considering only mutations in the exonic region leading to an alternation in the amino acid sequence (mis‐sense mutation), we focused on five cases with a nucleotide transition from G to A and on four cases with a C‐to‐T transition or SNP. Three of these mutants showed a change from tryptophan to a pre‐mature stop codon, thus leading to a potential knockout of CRT1a (Figure 2C).
Figure 6.
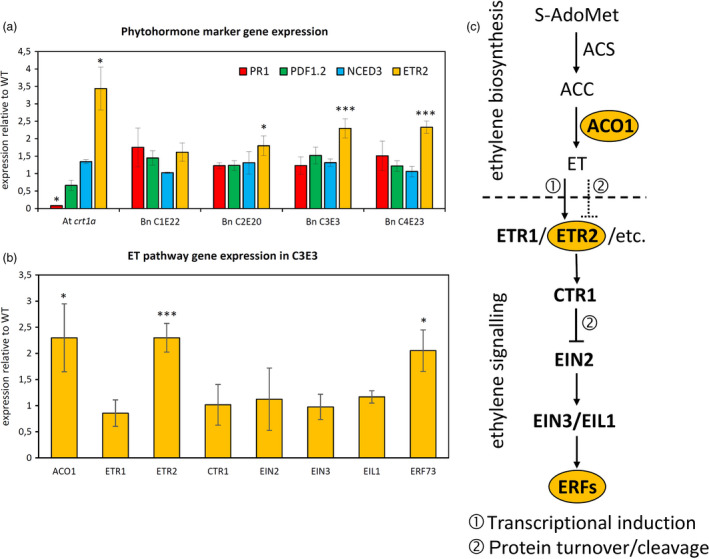
Up‐regulation of the ethylene pathway in crt1a mutants. (a) Marker gene expression analysis by qPCR relative to wild type. Expression of PR1 for the salicylic acid pathway, PDF1.2 for the jasmonic acid pathway, NCED3 for abscisic acid and ETR2 for the ethylene pathway was investigated in both Arabidopsis crt1a (SALK_055452) and several OSR crt1a mutant backgrounds. (b) ET gene expression analysis in C3E3 plants by qPCR relative to wild type. Root material was harvested from 4‐week‐old Arabidopsis plants and 2‐week‐old Brassica napus plants. Statistical significance was checked on the basis of three independent biological replicates by Student's t‐test (*P ≤ 0.05; **P ≤ 0.01). (c) Simplified overview of the ET signalling pathway in plants (modified from Ju and Chang, 2015).
Figure 2.
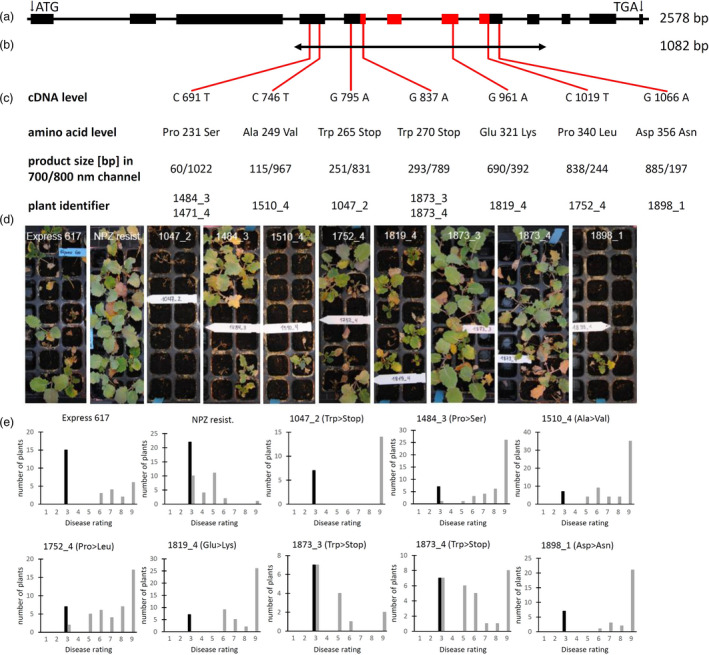
TILLING analysis of the CRT1a locus BnaA09g15400D. (a) Genomic structure consisting of 12 exons (boxes) with the EST region identified in the SSH library indicated in red (see also Figure S4A). (b) Region selected for TILLING containing exons 4–9. (c) Location of mutations identified by TILLING according to the cDNA and amino acid level. (d) CRT1a oilseed rape TILLING mutants were infected with Verticillium longisporum in comparison with the susceptible line Express 617 and a less susceptible NPZ‐resistant reference. Photographs were taken at 35 dpi. Squares are 4 × 4 cm. (e) Rating of disease symptoms. The disease rating was conducted according to Zeise and von Tiedemann (2002) and used to monitor disease severity in infected and non‐infected oilseed rape plants. The rating is distributed from 1 (no symptoms) to 9 (dead plant).
To investigate possible effects of these changes on OSR in regard to Vl43 infection, we challenged the mutant plants with Vl43 under greenhouse conditions in two independent experiments in which the susceptible line Express 617, the donor of the EMS population, and a resistant breeding line of NPZ served as controls. The plant growth and the development of disease symptoms were photographically documented weekly. As shown in Figure 2D, typical disease symptoms such as yellowing of leaves, necrosis and dieback of plants appeared as expected in the control plants, while the OSR mutants showed varied responses to the V. longisporum infection.
A disease rating was conducted at 35 dpi (Zeise and von Tiedemann, 2002) showing the resistant line has a constant rating of about 3 similar to non‐infected plants, while the susceptible control Express 617 exhibited the typical disease symptoms capable of ratings up to 9 for dead plants. Plants from mutants 1510_4, 1819_4 and 1898_1 showed comparable disease symptoms to Express 617. Mutant 1047_2, from which all plants died at 35 dpi, appeared to be hyper‐susceptible to Vl43 infection. Mutant 1484_3 showed a similar rating as Express 617. Nevertheless, one plant from 1484_3 and two plants from 1752_4 could be scored 3 for resistance. However, seven individual plants from mutants 1873_3 and 1873_4 were scored similar to non‐infected control plants (Figure 2E).
In addition, stunting symptoms were measured at 35 dpi. As shown in Figure 3A, the susceptible control Express 617 displays clear reduction in plant height to 60.5% of non‐infected control plants. The hyper‐susceptible mutant 1047_2 showed 100% stunting since all plants were dead. Mutants 1898_1, 1819_4 and 1752_4 revealed a comparable stunting as observed for Express 617 (60.5%), while the mutants 1484_3 and 1510_4 displayed a slightly reduced stunting effect. The knockout mutant 1873_4 showed a reduced stunting degree of only 37.2% compared to non‐infected control plants and the mutant 1873_3 even less stunting (17.9%). These data suggest that the OSR crta1a mutants 1873_3 and 1873_4 represent the most promising mutant plants with decreased susceptibility to V. longisporum infection. In support for this, the fungal biomass, quantified by qPCR in root material of mutants 1873_3 and 1873_4, was much less than in the susceptible control Express 617 (Figure 3B), suggesting an impeded infection process of Vl43 in both mutants. When comparing the disease phenotype of control and infected WT and mutant (1873_4) plants, we observed no growth reduction by infection in the mutant (Figure 3C).
Figure 3.
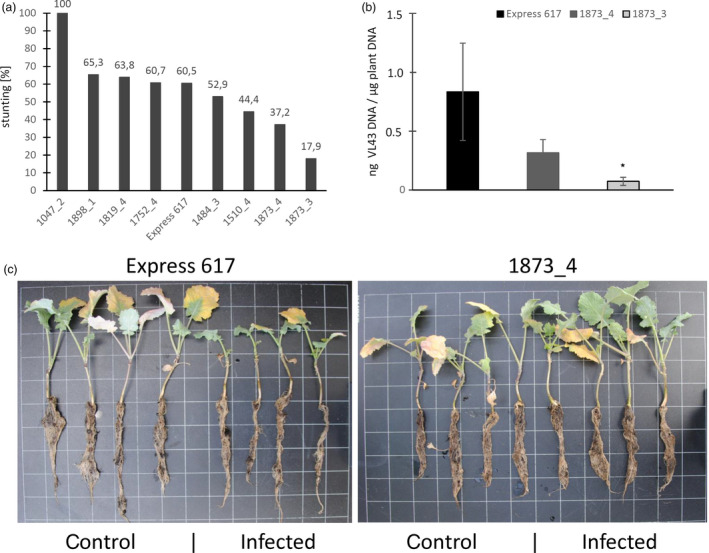
Resistance phenotype of TILLING mutants. (a) Plant size reduction for infected oilseed rape candidates compared to the susceptible cultivar Express 617 at 35 dpi. With a 60.5% stunting degree, Express 617 is in the middle field among the investigated plants. 1484_3 and 1510_4, and above all 1873_4 and 1873_3 show clear reduction in stunting, demonstrating an enhanced resistance against Vl43. (b) Results of qPCR to quantify Verticillium longisporum DNA in roots of selected resistant oilseed rape mutants 1873_3 and 1873_4 at 42 dpi in comparison with the susceptible Express 617 cultivar. Significances were calculated using Student's t‐test (*P ≤ 0.05). (c) Disease phenotype of infected wild‐type (Express 617) and mutant (1873_4) plants at 42 dpi. Squares are 3 × 3 cm.
Unexpectedly, mutant 1047_2 also carrying a stop codon in BnaA09g15400D was rather hyper‐susceptible and not able to survive the infection. This might be attributed to background mutations caused by EMS mutagenesis in the plant genome. To address the impact of background mutations on the decreased susceptibility observed in mutants 1873_3 and 1873_4, a successive backcross is in progress and genome editing using CRISPR/Cas was applied for a targeted CRT1a KO.
Allele‐specific mutations of BnCRT1 by CRISPR/Cas
To exclude potential side effects from unwanted off‐target background mutations frequently occurring by the EMS treatment, we employed the CRISPR/Cas technology for a target‐specific mutation of the BnCRT1a loci. Thus, we constructed a vector, referred to as MP_23, by utilizing gene synthesis to obtain a codon‐optimized Cas9 for Brassica napus (Figure S5). We designed a single sgRNA allowing to discriminate between ‘on‐target’ (BnaA09g15400D and BnaC09g16150D) and ‘off‐target’ (BnaA09g15970D and BnaC01g43040D, as well as all CRT1b copies) alleles. The BnaA09g15400D locus is the same as in the TILLING approach corresponding to the SSH library EST (6F11‐F8). This is visualized in Figure 4A by a phylogenetic analysis in combination with the aligned target region (Figure 4B). In total, 20 independent transgenic oilseed rape plants were obtained, from which four T0 transformation events (C1 to C4) were identified as mutants carrying expected mutations in the target region as revealed by locus‐specific sequencing (Figure 4C).
Figure 4.
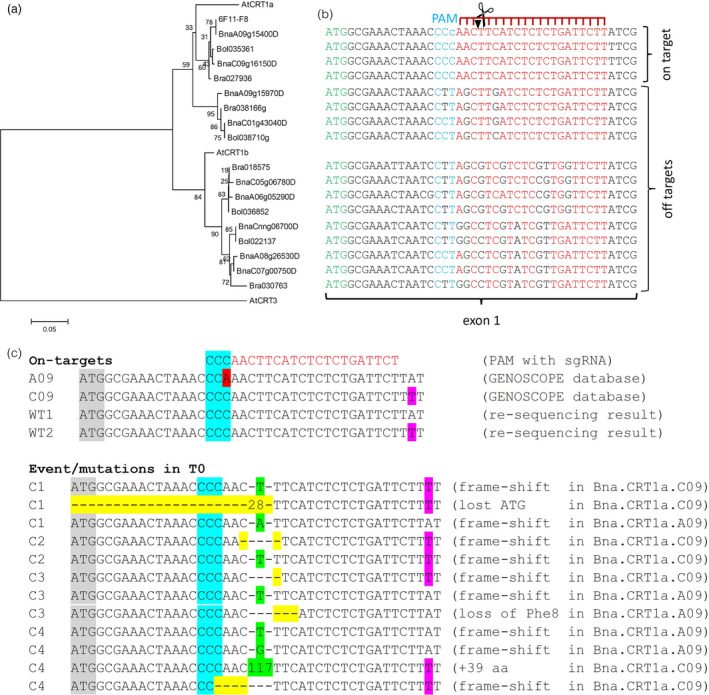
Identification of CRISPR/Cas‐induced mutations. (a) The neighbour‐joining tree was based on corrected cDNA sequences and calculated with MEGA6 applying 1000 bootstraps, maximum composite Likelihood, d; uniform rates, same (homogenous); and complete settings and AtCRT3 as an outlier to root the tree. (b) CRT1a target region analysis. Shown is the beginning of exon 1, starting with the ATG (green letters). Only the two closely related CRT1a copies BnaA09g15400D and BnaC09g16150D are expected to be cleaved by the sgRNA guided Cas9, since other copies contain SNPs either in the PAM (blue letters) or the seed region of the sgRNA (red letters). (c) Detection of CRISPR/Cas‐induced mutations in four independent T0 events (C1 to C4). Grey marks the start codon of the CRT1a ORF. Blue marks the PAM sequence of the Cas9‐sgRNA. Insertions are coloured green and deletions in yellow. Purple indicates SNPs distinguishing copies from the AA and CC genomes and the red SNP a difference between database and re‐sequencing results, but this has no negative impact on CRISPR/Cas specificity or functionality of the PAM (NGG).
For confirmation, genomic DNA of the four T0 plants was subsequently subjected to a ‘derived cleaved amplified polymorphic sequence’ (dCAPS) assay (Figure S6A/D). As expected, PCR fragments from wild‐type plants were almost completely digested by PdmI, while the T0 plants all showed partial digestions with varying intensities (Figure S6A), indicating varied mutation rates in the BnCRT1a alleles. Thus, this screening with the dCAPS assay is an efficient and a reliable tool to quickly identify CRISPR/Cas‐derived mutations, especially for crops with complex genomes also proving that our CRISPR/Cas system is functional.
Next, the T1 and T2 generations for all four independent mutant events were analysed. To simplify the screening procedure, three T1 and T2 plants descending from each event were propagated and characterized at first by dCAPS to identify progenies with complete mutation of all BnCRT1a alleles. Figure S6B,C show examples of the progenies selected by dCAPS assay of the T1 and T2 individuals, demonstrating that mutations were stably transmitted to the T2 generation. The large insertion of 117 bp detected in the T0 event C4 was successfully deselected from progenies for further analysis since this mutation was in frame causing no potential KO. A complete mutation of the BnCRT1a locus was indicated only in the C1‐ and C4‐derived T2 plants, while the C2 and C3 individuals showed still varied intensities of digestion patterns in the dCAPS assay (Figure S6C).
First, we challenged T2 plants with Vl43 (1 × 106 conidia) under greenhouse conditions and compared them with wild‐type plants as control. Without fungal infection, most mutant plants showed no obvious difference in plant growth and development as compared to the control (Figure 5A). While at 28 dpi typical disease symptoms such as yellowing of leaves and stunting of plant growth appeared in the wild type, the mutant plants with the exception of plants derived from event C2 showed overall decreased disease symptoms as calculated by the area under the disease progress curve (AUDPC; Figure 5B), less stunting as measured by plant height (Figure 5C) when compared to the control. In particular, the T2 progenies derived from the T1 parents C3E3 and C4E23 showed a significantly reduced susceptibility as indicated by impaired development of disease symptoms and less stunting (Figure 5D), as well as reduced fungal growth (Figure 5E). Even under a higher infection pressure with 1x107 conidia, the mutants of C3E3 and C4E23 survived the infection until 28 dpi, while the control plants were completely deceased (Figures 5F left and S8).
Figure 5.
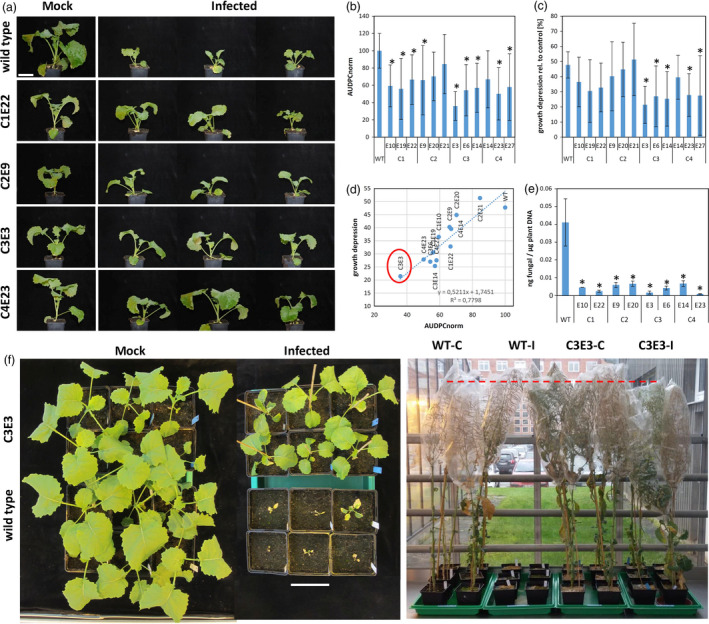
CRT1a CRISPR mutants display reduced susceptibility. (a) T2 progenies of four independent events were photographed at 28 dpi with Verticillium longisporum (after infection with 1 × 106 spores/mL). Some CRT1a mutants display better growth under infected conditions when compared to the wild type shown on top. The scale bar equals 9 cm. (b) Disease rating according to Eynck et al. (2007). The area under the disease progress curve (AUDPC) was determined starting at 7 dpi weekly until 28 dpi, and growth depression (c) was determined at 28 dpi and calculated from 10 individual plants. (d) Correlation analysis between (a) and (b) shows the best value for C3E3. (e) Fungal DNA was quantified via qPCR using the petioles of the first two true leaves harvested at 28 dpi. Statistical analysis was done by Dunnett's t‐test, and significant changes are marked by asterisk (*P ≤ 0.05). (f) The mutant C3E3 was infected a second time, now with 1 × 107 spores/mL and photographed at 28 dpi (left) and after 21 weeks (right). The scale bar equals 13 cm.
Furthermore, there was no statistically significant difference in the 1000 corn weight between the T2 progenies from all four independent T0 events (Figure S7), indicating that the crt1a mutation has no negative effect on this agronomic trait. Though we observe a slight growth suppression after infection (Figure 5F right), the mock‐treated C3E3 mutant plants grew similar to wild‐type plants. Additionally, randomly chosen five individual plants were checked from each tested T2 population for the transgene construct by PCR and three plants appeared to be transgene‐free (Figure S9).
In a next step, we genotyped T2 individuals by BnCRT1a locus‐specific Sanger sequencing, including the off‐target CRT1a alleles. On average, 40 clones per individual were analysed. No mutations were found in the off‐target alleles (data not shown), while the detected mutations in BnaA09g15400D were mostly corresponding to those observed in the T0 generation (Figure 4C) with exception for C2E9‐3 (Table 1). This indicates a stable inheritance of several CRISPR/Cas‐induced mutations and confirms our observations from the dCAPS analysis (Figure S6C). Interestingly, we observed that the C2E9 plant harbours an intact allele of the BnaA09g15400D copy displaying the weakest phenotype with respect to the reduced susceptibility, while the C3E3 plant has two intact alleles of the closely related BnaC09g16150D copy but being most resistant, which is in agreement with our TILLING results. This suggests BnaA09g15400D is the ‘active’ CRT1a copy in Brassica napus being involved in the plant–fungus interaction.
Table 1.
Summary of detected mutations in on‐target loci of T2 individuals
| Plant | BnCTR1a | Allele a | Allele b | Resistance level |
|---|---|---|---|---|
| C1E22 |
BnaA09g15400D BnaC09g16150D |
+1 bp (A) −28 bp |
+1 bp (A) −4 bp |
++ |
| C2E9 |
BnaA09g15400D BnaC09g16150D |
WT −4 bp |
+1 bp (T) +1 bp (T) |
+ |
| C3E3 |
BnaA09g15400D BnaC09g16150D |
−3 bp (Phe8) WT |
+1 bp (T) WT |
+++ |
| C4E23 |
BnaA09g15400D BnaC09g16150D |
+1 bp (T) −4 bp |
+1 bp (T) −4 bp |
++ |
A bioinformatic analysis (Blom et al., 2004) suggests that two potential phosphorylation sites might be responsible for this difference between BnaA09g15400D and BnaC09g16150D, being present in the former and missing in the latter copy (Figure S10A). It appears that the first site was lost in BnaC09g16150D, while the second was gained in BnaA09g15400D (Figure S10B). Furthermore, the complete KO by frame‐shift mutations as observed in C1E22 and C4E23 affecting all four BnCRT1a on‐target alleles results in a less strong phenotype, suggesting the type of mutation might also play a role. The 3‐bp deletion observed in BnaA09g15400D of C3E3 causes loss of Phe8 within the N‐terminal ER targeting sequence (Figure S10B) and might cause aberrant localization of CRT1a. Additionally, we still detected WT alleles in some T2 individuals (Table 1). Thus, it appears CRISPR/Cas becomes less active once plants were regenerated from callus culture, though we still observed Cas9 expression in T2 plants (data not shown).
The ethylene pathway appears to be induced in the crt1a mutants
In order to get a first clue about the underlying molecular mechanism causing reduced susceptibility in the crt1a Arabidopsis and OSR mutant plants, we analysed the expression of several phytohormone marker genes involved in plant defence towards pathogens, such as PR1 (salicylic acid), PDF1.2 (jasmonic acid) and ETR2 (ethylene), as well as NCED3 for abscisic acid often involved in abiotic stress defence. Comparing wild‐type and mutant backgrounds of Arabidopsis and OSR, we found no statistically significant differences for PR1, PDF1.2 and NCED3. However, for ETR2 we could observe an overall increase in OSR mutants, as well as in the Arabidopsis KO line when compared to the respective wild type (Figure 6A). Following this, we investigated several ET pathway‐related genes in the highly resistant OSR mutant C3E3 and found that in addition to ETR2, also the expression of ACO1 and ERF73 was significantly elevated (Figure 6B/C).
Discussion
In this study, we demonstrate a strategy and working routine allowing for an efficient generation of recessive resistance against pathogens in oilseed rape. By differential gene expression deploying a SSH library, we identified several putative candidates for susceptibility factors in the Brassica napus–Verticillium longisporum interaction. The related model plant Arabidopsis thaliana can be infected with Vl43 as well. Thus, we obtained for 20 corresponding Arabidopsis orthologous gene loci T‐DNA knockout lines and infected 15 homozygous mutants with Vl43. One loss‐of‐function mutant displaying increased resistance compared to the wild‐type Col‐0 was identified as crt1a. The T‐DNA insertion into the Arabidopsis CRT1a gene resulted in decreased symptom development and fungal DNA accumulation, suggesting this might be a novel factor that plays an important role in the establishment of compatibility/successful host colonization by V. longisporum. For Col‐0 wild‐type plants, we observed a reduction in plant height of 41.9%, which was only 31.4% in crt1a, while the decrease in silique numbers reached 87.8% in wild‐type plants compared to only 65.9% in crt1a. Also, the leaf area reduction was significantly less pronounced in crt1a when compared to infected Col‐0 plants, and in consistence with these observations, we measured significantly less Vl43 DNA in crt1a roots as compared to WT plants, suggesting that altered responses are mainly attributed to lower susceptibility towards V. longisporum.
The characterization of Arabidopsis knockout mutants was followed by the identification of corresponding OSR mutants in an EMS‐mutagenized population by TILLING. A database search at GENOSCOPE revealed four distinct CRT1a copies in the Brassica napus genome, all being expressed in cDNA preparations from infected and mock‐treated roots (Figure S4B). We identified BnaA09g15400D as the BnCRT1a copy matching 100% to the EST identified from the SSH library. Thus, we focused on this CRT1a copy when designing specific TILLING primer. Nine OSR mutant candidates with single nucleotide mutations in the target locus (BnaA09g15400D) were identified. These mutations mostly led to amino acid changes in the protein (mis‐sense mutation). Three lines with stop codons in the CRT1a ORF (potential knockouts) displayed converse phenotypes: the line 1047_2 was hyper‐sensitive to Vl43 infection, while the lines 1873_3 and 1873_4 appeared more resistant, suggesting strong side effects from the EMS mutational background. To clarify whether mutation of BnCRT1a really results in a decreased susceptibility to Vl43 infection, a successive, time‐consuming backcross is required. But in the light of legal issues concerning GMO regulations, the TILLING approach is still attractive since EMS‐mutated plants are not regulated as transgenic GMOs (Henikoff et al., 2004), while in Europe, SSN‐mutated plants currently fall under GMO regulations (Urnov et al., 2018).
Nevertheless, to verify the observed reduced susceptibility to Vl43 in OSR, we applied CRISPR/Cas to specifically target the ‘active’ CRT1a locus BnaA09g15400D. We developed a CRISPR/Cas system optimized for Brassica napus and designed a sgRNA targeting BnaA09g15400D and also BnaC09g16150D, but not in the off‐target CRT1a copies BnaA09g15970D and BnaC01g43040D. It is interesting to note that several of the identified mutations were identical between the T0 and T2 plants (Figure 4C and Table 1) and that even though CRISPR/Cas is assumed not to rest unless all target loci are mutated, still some wild‐type alleles could be found in two sequenced individuals. This indicates that CRISPR/Cas‐induced mutations did not generally increase over two generations, albeit Cas9 was still expressed in T2 plants (data not shown). One explanation might be that the construct is mainly active during tissue culturing, an observation also made by others (Mikami et al., 2015). Alternatively, such incomplete mutation might be due to certain epigenetic modification at the target loci (Kallimasioti‐Paz et al., 2018; Verkuijl and Rots, 2019).
It is to note that plants of C3E3 and C4E23 displayed a stronger mutation effect, even under a higher infection pressure with 10−7 spores/mL as Vl43 inoculum (Figures 5F and S9). The mutations in the C3E3 plants affected only the ‘active’ TILLING copy BnaA09g15400D, while in C4E23 plants, both on‐target copies were mutated. Though C2E9 plants are completely mutated in the BnaC09g16150D copy, they showed the weakest resistance phenotype, suggesting this copy is less important (Table 1). This might be explained by differences at the amino acid level between both on‐target copies. An in silico analysis reveals that BnaA09g15400D contains two potential phosphorylation sites, which are missing in BnaC09g16150D (Figure S10A). Furthermore, it appears that the first site was lost in BnaC09g16150D, while the second was gained in BnaA09g15400D (Figure S10B). But in how far this affects CRT1a function still remains to be elucidated. Thus, we conclude that the BnaA09g15400D copy is a dominant functional homeolog being important for the plant–fungus interaction and that its loss of function impedes the compatible interaction and reduces plant susceptibility to Vl43. In addition, five T2 progenies for each selected T1 plant (Figure S9A) were tested by PCR for the presence of the CRISPR/Cas construct. From them, three plants appeared to be transgene‐free (Figure S9B). Thus, we believe that it is possible to generate transgene‐free plants carrying only the mutation in the target locus already in T2 generation. Analysis of the 1000 seed weight showed additionally that the CRISPR/Cas‐mutated crt1a lines were not negatively affected in comparison with the wild‐type Mozart (Figure S7).
The finding that CRT1a knockout results in decreased susceptibility to Vl43 in two Brassicaceae species, Arabidopsis thaliana and OSR, strongly supports the importance of CRT1a in a compatible plant–fungus interaction. It is to note that the C3E3 mutant lacks only Phe8 in one BnaA09g15400D allele but confers higher resistance than the other mutants (Figure 5F). We speculate that this mutation might impair the ER targeting of CRT1a, resulting in an atypical allocation of the protein in plant cells, and that the still‐intact BnaC09g16150D alleles compensate for some other CRT1a function in plant resistance. Calreticulin (CRT) was initially described from rabbit skeletal muscle sarcoplasmic reticulum serving as a Ca2+ binding protein (Ostwald and MacLennan, 1974). This highly conserved protein is found in higher eukaryotes and locates to the endoplasmic reticulum (ER), where it is involved in the regulation of many cellular processes (Krause and Michalak, 1997). Plant CRTs were first isolated from spinach and tobacco (Denecke et al., 1995; Menegazzi et al., 1993) and in Arabidopsis thaliana exist three CRT family members (CRT1a, 1b and CRT3), which share protein sequence similarities from of 57%–86%. The predicted amino acid sequences of plant calreticulins have structural similarities to those found in animals (Coppolino and Dedhar, 1998), and it was shown that AtCRT1a could substitute the chaperone and Ca2+‐ homeostasis function of animal CRTs, indicating that basic functions are conserved between the two kingdoms (Christensen et al., 2008). Since CRTs are ubiquitously expressed, it is supposed that they play a general role in plant growth and developmental processes (Jia et al., 2009). Structurally, calreticulins contain three different domains, which are the N‐terminal globular domain with chaperone function, a central proline‐rich domain for high‐affinity Ca2+ binding adding to the chaperone function and a C‐terminal acidic domain responsible for Ca2+ buffering with a C‐terminal ER retention signal (Sarwat and Tuteja, 2017). It appears that isoform CRTs have different functions, for example CRT3 being distinct from CRT1a and 1b, which are in contrast co‐expressed with a multitude of ER genes (Christensen et al., 2010; Jia et al., 2009).
Besides their role in calcium regulation, CRTs are involved in the ER quality control, functioning as chaperones to ensure proper protein folding (Danilczyk et al., 2000) and this has also an impact on abiotic and biotic stress responses (Garg et al., 2015, Kang et al., 2010). For example, the Arabidopsis triple mutant of CRT1a, CRT1b and CRT3 showed growth defects in response to water stress, suggesting that CRTs are involved in drought stress responses. The absence of defects in single isoform mutants could be explained by compensation of certain functions because of functional redundancy (Kim et al., 2013). Other potential roles of plant CRTs in abiotic and biotic stress responses have been recently reviewed (Joshi et al., 2019). For biotic stress, tobacco calreticulin was shown to be involved in cell‐to‐cell movement of TMV (Chen et al., 2005) and it has been shown that CRT3 plays a role in folding of EFR (but not FLS2), establishing also a link to induction of PAMP‐triggered immunity (Christensen et al., 2010; Li et al., 2009). A knockdown of CRTs led to a disturbance of the Ve1‐mediated resistance against Verticillium dahliae in tomato, indicating a function in proper folding of Ve1 receptor‐like protein (Liebrand et al., 2014). Thus, it is reasonable to speculate that the loss of CRT1a function could also result in an indirect effect leading to misfolding and degradation of other susceptibility factors, which are then absent and therefore impede pathogen development.
Such impaired ER quality control by CRT1a loss of function could furthermore prevent correct folding of negative regulators in ethylene (ET) signalling, since we observed up‐regulation of the ET‐responsive gene ETR2 in crt1a roots (Figure 6A) and a constitutively activated ET signalling pathway might contribute to VL43 resistance, as has been shown also for other pathogenic fungi infecting roots (Okubara and Paulitz, 2005). Interestingly, ETR2 is one of five ET receptors located in the ER and regulated by MAPK (mitogen‐activated protein kinase) cascades (Das et al., 2015; Shakeel et al., 2015) and itself involved in negative regulation of the ET‐activated signalling pathway via ET perception (Ju and Chang, 2015). ETR2 is transcriptionally activated in response to ET, but the protein is also degraded after ET perception (Chen et al., 2007; Shakeel et al., 2015). Concerning ETR2 expression, we could observe a dose effect in OSR roots, being highest in the more resistant lines C3E3 and C4E23 (Figure 6A). We investigated transcriptional activation of several ET pathway genes in the C3E3 mutant, and besides, ETR2 only ACO1 and ERF73 were significantly up‐regulated (Figure 6B). This is not surprising since ET signalling is largely regulated post‐transcriptionally by protein modification and degradation (Gallie, 2015; Merchante et al., 2013). An at least partially induced ET pathway would imply a fundamental change in the plants alert status, which might also explain to some extend why crt1a mutants become less susceptible. That ET can contribute to the plant defence towards Verticillium ssp. is further corroborated by observations that V. dahliae degrades the ET precursor 1‐aminocyclopropane‐1‐carboxylic (ACC) to increase its virulence, while ACC pretreatment reduced symptoms in tomato (Tsolakidou et al., 2019). Very recently, Xiong et al. (2020) reported that a knockdown of GhWRKY70D13 improved cotton resistance to V. dahliae going along with up‐regulation of genes involved in ET and jasmonic acid (JA) biosynthesis as well as increased ACC, JA and JA‐isoleucine levels. Also, Arabidopsis ERF TFs are induced in response to V. longisporum and their over‐expression caused a reduction of fungal DNA within the mutants, while RNAi against ERFs had the opposite effect (Fröschel et al., 2019). But also the opposite effect has been observed with Arabidopsis etr1 mutants being more resistant to V. dahliae (Pantelides et al., 2010). However, the constitutively expressed ETR1 is not responsive to ET and has a higher affinity for CTR1 as compared to ETR2, pointing to different functional roles (Chen et al., 2007; Shakeel et al., 2015). For example, ETR1 and ETR2 act reversely in abscisic acid‐mediated seed germination under salt stress (Wilson et al., 2014). In summary, we demonstrate a strategy and working routine to increase natural variation for improving plant disease resistance in OSR. Following this, we identified CRT1a as a novel susceptibility factor, which loss of function reduces OSR and also Arabidopsis susceptibility to V. longisporum without negatively affecting overall plant performance under greenhouse conditions, being therefore a potentially interesting target for breeding programmes.
Material and Methods
Fungal cultivation
Verticillium longisporum isolate Vl43 provided by Dr. Elke Diederichsen (FU Berlin, Germany) was stored as 22% glycerol stock culture and conidia were produced by transferring mycelium plugs from one‐week‐old PDA (Duchefa, the Netherlands) plates into liquid Czapek‐Dox (Duchefa, the Netherlands) medium for subculturing in the dark at 25 °C on a rotating shaker. After 1 week, conidia were harvested with the help of gaze filter (mesh 200 µm; Hydro‐Bios Kiel, Germany), centrifuged for 8 min at 8000 rpm and resuspended in sterile Czapek‐Dox medium to increase the concentration. Stocks for infection were supplemented with 86% glycerol to a final concentration of 22% and kept at −80°C till the day of inoculation. At the day of inoculation, stocks were thawed, centrifuged for 8 min at 8000 rpm and resuspended in tap water (for ‘in vivo’ infection on soil) or sterile Czapek‐Dox medium (for ‘in vitro’ infection) to a final concentration of 1 × 106 conidia/mL if not otherwise indicated. Tap water and Czapek‐Dox medium were used as mock treatments, respectively.
Plant material and growth
Arabidopsis and B. napus seeds were surface‐sterilized with a 6% sodium hypochlorite/Tween‐20 solution and 70% ethanol followed by three times washing with sterile dist. water. The Arabidopsis ecotype Col‐0 (wild type) was purchased from Lehle Seeds, Round Rock, TX, USA, while T‐DNA insertion mutants were obtained from NASC, Nottingham, UK. Homozygous plants were selected using the T‐DNA‐specific primer LBb1.3 (http://signal.salk.edu/tdnaprimers.2.html) in combination with locus‐specific primers (Table S6). Surface‐sterilized seeds were placed in B5 agar medium and grown under short‐day conditions (8‐h light/16‐h dark) at 22 °C in a growth chamber for four weeks. Seeds of the susceptible OSR cultivar ‘Express 617’, a NPZ‐resistant reference line and the TILLING mutants were provided by the Norddeutsche Pflanzenzucht Hans‐Georg Lembke KG (NPZ). Surface‐sterilized seeds were placed on B5 agar medium and grown under sterile long‐day conditions (16‐h light/ 8‐h dark) at 22 °C in a growth chamber for 10 days.
In vivo infection of Brassica napus
Seeds of Brassica napus were surface‐sterilized with 70% ethanol and a 5% sodium hypochlorite solution with a drop Tween‐20 followed by three times washing with sterile dist. water. They were sown out on sterile 1‐mm‐thick filter paper saturated with MS media without sucrose and placed into a climate cabinet (Vötsch VB 0714‐A, Reiskirchen, Germany) with short‐day conditions at room temperature. When the cotyledons were fully expanded after 7 days’ incubation, V. longisporum stock culture was thawed on ice and then centrifuged at 6000 g for 10 min. The supernatant was discarded, and the pellet was dissolved in with the same amount of tap water. Roots of the Brassica napus seedling were cut with a razor blade to a length of 2 cm, and inoculation was performed according to Eynck et al. (2007) by the root‐dip method using 1 × 106 conidia for mild infection or 1 × 107 conidia for strong infection. After inoculation for 5 min, plants were transferred to soil with an initial application of WUXAL® universal fertilizer (Bayer CropScience, Leverkusen, Germany) and grown under long‐day conditions in the greenhouse until the day of harvest.
In vitro Infection assay of Brassica napus
Seeds of Brassica napus were surface‐sterilized with 70% ethanol and a 5% sodium hypochlorite solution with a drop Tween‐20 followed by three times washing with sterile dist. water. The seeds were placed in plastic salad boxes with solid MS media and grown under short‐day conditions until the cotyledons were fully expanded. Seedlings were transferred to 120 mm square Petri dishes containing ½ MS (0.5% sucrose) as described by Behrens et al. (2019). Roots were inoculated by brushing them with V. longisporum conidia suspension, and plants were grown under short‐day conditions for six days in a climate cabinet (Vötsch VB 0714‐A) with short‐day conditions at room temperature. The root material was harvested, frozen in liquid nitrogen and stored at −80 °C until used for RNA isolation for gene expression studies.
In vivo infection of Arabidopsis thaliana
Inoculation was performed according to Eynck et al. (2007) by the root‐dip method for plants grown on soil. Arabidopsis plants grown under short‐day conditions for 3 weeks in a climate cabinet (Vötsch VB 0714‐A) were dipped for 5 min into a conidia suspension with different concentrations (1 × 103 to 1 × 107 conidia/mL). Control plants were dipped for the same time into filtrated and autoclaved tap water. Immediately after the inoculation, plants were transferred into pots filled with a sand:vermiculite:soil mixture in a ratio 3:1:1, and additionally, 5 mL from the conidia suspension was given directly to each plant. The plants were photographed at the indicated time points, and roots were harvested at 28 dpi, flash‐frozen in liquid nitrogen and stored at −80 °C.
DNA and RNA isolation
Leaf and root tissue of at least three independent biological replicates from each infected and mock‐treated plants was flash‐frozen in liquid nitrogen and stored at −80 °C. DNA was isolated using the cetyltrimethylammonium bromide (CTAB) method (Rogers and Bendich, 1985), and the resulting DNA pellet was resolved in 50 µg ddH2O, treated with RNaseI (Fermentas) for 1 h by 37 °C and stored at −20 °C until further use.
Total RNA was isolated using the TRIzol® Reagent (Invitrogen, Karlsruhe, Germany) according to the manufacturer´s recommendation including a DNaseI (Fermentas, Vilnius, Lithuania) treatment. The resulting RNA pellet was resolved in 20–50 µL DEPC‐treated ddH2O and stored at −20 °C until further use.
Suppression subtractive hybridization (SSH library)
For the construction of the SSH libraries, mRNA was isolated with the ‘PolyATract® mRNA Isolation System III’ (Promega, Madison, Wisconsin) according to the manufacturer's instructions. After Vl43 infection B. napus root material was collected at 5 dpi, 10 dpi and 15 dpi. Total RNA from the three harvesting time points was pooled into infected and mock‐treated samples. The RNA quality and quantity was measured with the NanoVue Spectrophotometer (GE Healthcare, Chicago, Illinois) and by electrophoresis on agarose denaturing gel.
A forward (up‐regulated genes after infection) and reverse (down‐regulated genes after infection) SSH library was constructed using the PCR‐select cDNA subtractive kit (ClonTech, TaKaRa Biotech, Saint‐Germain‐en‐Laye, France) according to the manufacturer´s manual from infected and non‐infected B. napus root mRNA at the three harvesting time points 5 dpi, 10 dpi and 15 dpi. The pooled mRNA of infected and non‐infected B. napus root was used. For the forward library, mRNA of non‐infected roots was used as the driver, and the mRNA of infected roots was used as the tester. For the reverse library, tester and driver were changed. Double‐stranded cDNAs were obtained according to the manufacturer's instructions and then used to initiate SSH procedure. Both, tester and driver were incubated for 1 h at 37 °C with RsaI. Adapter 1 was ligated with half of the digested tester cDNA, and adapter 2R was ligated with the other half. Adequate driver cDNA was mixed with both of them in separate tubes for the first subtractive hybridization. The resulted products were mixed together, freshly denaturated driver cDNA was added, and the second subtractive hybridization took place. To enrich the differential expression fragment, PCR amplification was conducted.
To construct the SSH‐cDNA library, the purified differentially expressed fragments were cloned into pGEM®‐T Easy Vector (Promega) and transformed into electrocompetent Escherichia coli strain DH5α (Invitrogen), cultured on LB medium containing Amp/IPTG/X‐Gal for blue‐white screening at 37 °C. Positive clones were selected on new LB medium, grown overnight at 37 °C and analysed by PCR with vector‐specific standard primer pairs SP6/T7 or M13F/M13R (both Invitrogen). Positive clones were sent for sequencing. PCRs were carried out in a total volume of 20 µL with 10 pmol for each primer, 10 mm PCR buffer (pH 8.3), 1 U of Taq DNA polymerase (Invitrogen), 2 mm MgCl2 and each dNTP at 5 mm in a standard thermocycler (Biometra, Göttingen, Germany). PCR was performed using a PCR programme of 94 °C for 1 min, 56 °C for 30 s and 72 °C for 30 s for 35 cycles, followed by 5 min at 72 °C.
Sequences from the SSH clones were determined by the Department for Clinical Molecular biology at the University of Kiel, and sequence analysis was carried out using the software CHROMAS Lite (Technelysium Pty Ltd, South Brisbane, Australia). The obtained EST sequences from the suppressive subtractive hybridization were blasted at the NCBI database (http://blast.ncbi.nlm.nih.gov/Blast.cgi) using the BLASTX algorithm (Altschul et al., 1990) and TAIR10 database for Arabidopsis (Berardini et al., 2015).
Phylogenetic analysis
A neighbour‐joining phylogenetic tree based on corrected cDNA sequences of CRT1a and CRT1b was created using MEGA6 software (Tamura et al., 2013). The tree was calculated by applying 1000 bootstraps with the following settings: maximum composite likelihood, d; uniform rates, same (homogenous). AtCRT3 was included as an outlier to root the tree.
TILLING
The sequence of CRT1a (AT1G56340) was retrieved from the Arabidopsis database TAIR (The Arabidopsis Information Resource) and submitted to BLAST analysis at the GENOSCOPE B. napus database (http://www.genoscope.cns.fr/brassicanapus). Additionally, homologous genomic sequences from the Brassica database (BRAD) for B. rapa and B. oleracea were retrieved as well. A sequence alignment was performed comparing all sequences with the CRT1a EST, which was found in our comparative transcriptome analysis, matching 100% to the ‘active copy’ (BnaA09g15400D). Gene copy‐specific primers were designed to amplify 1082 bp, covering 41.7% of the CRT1a ORF (Table S5). Gene‐specific primers, as well as fluorescence‐labelled primers, were tested according to the protocol of Till et al. (2006), before the actual TILLING procedure started.
DNA samples from an EMS treated OSR M2 population were provided by AG Prof. Dr. Jung, Institute of Plant Breeding, Christian‐Albrechts‐University of Kiel. In total, 3840 M2 individual plant DNAs, pooled in ten microtitre plates, were employed in the TILLING screening as described in Harloff et al. (2012). Obtained DNA bands were extracted from the gel after electrophoresis and purified with the NucleoSpin® Gel and PCR Clean‐up Kit (Macherey‐Nagel, Düren, Germany). The locus specificity was confirmed by Sanger sequencing. The TILLING PCR of DNA pools was performed using 96‐well plates (Bio‐Rad) and CFX96 TouchTM Real‐Time Detection System (Bio‐Rad, Hercules, California). A mixture of unlabelled and 5′‐end labelled (DY‐681 and DY‐781) forward and reverse primers was used for amplification, utilizing the proofreading Phusion High‐Fidelity DNA Polymerase (Thermo Fisher Scientific, Waltham, MA, USA) and the following PCR programme: 95 °C 5 min, 40 cycles of 95 °C 30 s, 62 °C 30 s, 72 °C 90 s, 72 °C 10 min. Portions of the amplicon were checked by gel electrophoresis to confirm product specificity.
According to Till et al. (2006), the PCR products of mutated (EMS) plants and non‐mutated (WT) plants were hybridized to form heteroduplex structures and celery juice extract was prepared from commercial celery. This juice extract contains CEL1, a heteroduplex‐specific restriction endonuclease, which cuts DNA exactly at base‐pair mismatch sites. The digestion was stopped with 50 mm EDTA. Samples were cleaned from salts and buffer residues by Sephadex gel purification. The sample volume was reduced by evaporation on a heat block at 90 °C. The IRD fluorescence‐labelled PCR products were separated on polyacrylamide gels by using a LI‐COR 4300 DNA analyser (LI‐COR Bioscience) tool (4 h at 1.500 V, 40 mA and 40 W). Gel images were studied by the freely available software GelBuddy. After the identification of positive mutation signals in a DNA pool, corresponding single plant DNAs were used to confirm the single nucleotide mutation by Sanger sequencing (IKMB, Kiel, Germany). For sequencing, two independent PCRs were performed. The products were gel‐purified with the NucleoSpin® Gel and PCR Clean‐up Kit (Macherey‐Nagel) and ligated into pGEM®‐T Easy Vector System (Promega) according to the manufacturer's instructions. The T‐Vector was transformed into heat shock‐competent E. coli DH5α cells, and bacteria were selected on LB plates containing IPTG/X‐gal and ampicillin. Positive clones were identified by blue‐white screening. Plasmid DNA was isolated and sequenced using standard vector primers M13/T7. The obtained sequences were aligned with the specific B. napus gene copy by using CLUSTALW to identify nucleotide exchanges, allowing also the exact localization of the single nucleotide mutation in CRT1a. Visible double spikes in the electropherograms indicate that the plant is heterozygous for the corresponding point mutation. Unambiguous single peaks indicate homozygosity. Plants with homozygous genetic background were kept for seed production.
dCAPS assay
Derived cleaved amplified polymorphic sequences (dCAPS) is a simple technique to genetically analyse nucleotide polymorphisms (Neff et al., 1998). dCAPS is an alternative if the target region of sgRNA‐directed CRISPR/Cas does not contain a natural restriction site to quickly screen for mutational events. In case of the CRT1a loci, primers have been designed (Table S6) to generate an artificial PdmI restriction site, which would not be cleaved if the locus was successfully mutated, while wild‐type sequences are cleaved into 29‐ and 63‐bp fragments. The PCR products were digested with FastDigest PdmI (Thermo Fisher Scientific) for 1 h before heat inactivation at 80 °C for 20 min and analysis by gel electrophoresis on a 2% agarose gel.
Vl43 gDNA detection in plant tissue
Arabidopsis roots and B. napus petioles from infected and mock‐treated plants were sampled at the indicated time points and further processed as described in Behrens et al. (2019).
Assessment of projected leaf area
The leaf area analysis was used to compare the leaf rosettes of control and Vl43‐inoculated Arabidopsis mutant and wild‐type plants. The plants were grown and infected in a 60‐mm pot. After infection, the plants were photographed once a week in a precisely defined distance with a digital camera (Canon EOS 450D). The program WinCAM 2004a (Regent Instruments) was used to analyse the leaf area. A 1 cm × 1 cm white square served as scale to calculate the pixel area of the visible surface.
Disease rating and AUDPC calculation
The scoring of disease symptoms was conducted according to Eynck et al. (2007) with a rating scale from 1 to 9 as shown in Table S7.
Symptoms of the Vl43 infection of Brassica napus CRISPR/Cas mutants were assessed every 7 days until 28 dpi. From disease severity values, the area under the disease progress curve (AUDPC) values were calculated according to Campbell and Madden (1990):
where yi = disease score for observation number i; ti = days postinoculation, n = number of observations.
Adjusted AUDPC values were calculated to not overestimate the disease severity of each inoculated variant through natural occurring senescence. Therefore, AUDPC values from the mock control were subtracted from the AUDPC values from inoculated plants resulting in the AUDPCnet values:
The AUDPCnet values of each accession were further normalized against the WT resulting in AUDPCnorm values. In addition to the AUDPC values, the plant height was measured at each scoring date as well. The growth depression (GD) compared to the mock control was calculated by dividing the plant height of the infected variant through the mock variant of each accession:
cDNA synthesis and gene expression analysis by qRT‐PCR
One µg of total RNA was deployed in first‐strand cDNA synthesis, diluted in a ratio 1:10 and subjected to qPCR analysis as described in Behrens et al. (2019). For Brassica napus, the reference genes Bn‐ß‐tubulin 4 (224 bp) and BnActin 7 (216 bp) served as control, while in Arabidopsis, AtActin2 (298 bp) and AtPP2A (208 bp) were used as housekeeping genes. The primer sequences for all genes analysed can be found in Table S6.
CRISPR/Cas
The Brassica napus reference genome used for all in silico analysis and sgRNA design was accessed via the GENOSCOPE Darmor‐bzh v4.1 database (Chalhoub et al., 2014). Annotated genes were obtained from the respective database, and the following genes served as template for sgRNA design conducted using the web tool Cas‐Designer (Bae et al., 2014; Park et al., 2015). The PAM type was set to 5′‐NGG‐3′ (Cas9) and guide sequences with a hit in both B. napus target alleles, with localization at the beginning of the ORF within exons, were chosen as good candidates for sgRNA design. For B. napus a separate off‐target analysis was conducted using the sgRNA target sequence and a second reference genome. Only hits present in both genomes were considered potential off‐targets. The sgRNA was created according to Li et al. (2013) involving three rounds of PCR to replace the guide sequence within the sgRNA template (Figure S5) and expressed under the Arabidopsis U6‐26 promoter described by Schiml et al. (2014).
To efficiently apply CRISPR/Cas to Brassica napus we established a vector system involving a codon‐optimized Cas9 (http://www.kazusa.or.jp/codon/cgi‐bin/showcodon.cgi?species=3708) with a C‐terminal Mammalian Importin alpha type nucleus localization sequence (NLS). The entry vector for gateway cloning (BS_01) was synthesized based on a pUC57 backbone containing a sgRNA template driven by an AtU6 promoter within a multiple cloning site and the Cas9‐NLS under the control of the CaMV 35S promoter. The recombination sites allow a gateway LR reaction to clone the CRISPR/Cas components into the binary vector pGWB401 (Nakagawa et al., 2007), adding the NOS terminator to the Cas9‐NLS and the nptII plant selection marker (kanamycin resistance). This resulted in the plant transformation vector MP_23, ready for Agrobacterium tumefaciens‐mediated gene transfer (Figure S5). Stable transgenic lines were produced by the Saaten‐Union Biotec GmbH (Leopoldshöhe, Germany).
Statistical analyses
Graphs and plots were created with Microsoft® Office Excel® 2016 and statistical calculations were carried out in R. Simple comparisons of means for only two groups were done using the Student's or Dunnett t‐test (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001).
Conflicts of interest
There is no conflict of interest.
Author contributions
Dirk Schenke: Bioinformatic analysis, gene expression analysis, design of CRISPR vector system, sequencing of target loci, data interpretation, arrangement of data, confirmation of CRISPR results, drafting and writing of manuscript. Michael Pröbsting: OSR CRISPR: sgRNA design to target BnCRT1a, cloning, transformation, dCAPS analysis, phenotyping of CRISPR mutants, sequencing of target loci, gene expression analysis. Roxana Hossain: Arabidopsis phenotype confirmation, gene expression analysis, OSR TILLING: Identification of mutations, sequencing of target loci. Claudia Häder: Establishment of pathosystem, SSH library analysis, selection of candidate genes, Arabidopsis mutant genotyping and pre‐screening, gene expression analysis. Tim Thurau: Establishment of SSH library, selection of candidate genes, gene expression analysis. Lisa Krapoth: Arabidopsis mutant phenotyping. Andrea Schuster: TILLING mutant phenotyping. Zheng Zhou/Wanzhi Ye: gene expression analysis. Steffen Rietz/Gunhild Leckband: Project industry partner, responsible for plant transformation and seed propagation. Daguang Cai: Project strategies and experimental design, finalizing the manuscript.
Supporting information
Figure S1 Verification of the SSH libraries.
Figure S2 Time course of infection with Vl43 and determination of optimal conidia concentration.
Figure S3 There are 4 CRT1a loci in the Brassica napus genome.
Figure S4 The SSH‐EST 6F11‐F8 corresponds to BnaA09g15400D (SNPs highlighted in red).
Figure S5 Flow chart for CRISPR/Cas vector construction.
Figure S6 dCAPS‐analysis of positive CRISPR events.
Figure S7 Effect of CRT1a mutations on 1000 seed weight.
Figure S8 Phenotype of C4E23 at 28 dpi with Verticillium longisporum using 1 × 107 spores/mL.
Figure S9 Detection of transgene‐free plants.
Figure S10 Analysis of CRT1a on amino acid level.
Table S1 SSH library (EXCEL).
Table S2 Genevestigator Meta‐Analysis of SSH candidate genes (EXCEL).
Table S3 Candidate genes selected from the SSH libraries.
Table S4 Agronomic traits investigated under control and infection conditions.
Table S5 TILLING statistics of CRT1a screen within an EMS mutagenized population.
Table S6 Primer‐Information (EXCEL).
Table S7 Disease rating scale for Verticillium infection of Brassica napus plants (Enyck et al. 2007).
Acknowledgements
We thank Dr. Hans Harloff and Prof. Dr. Christian Jung from the Plant Breeding Institute at the Christian‐Albrechts‐University Kiel and our industry partner Norddeutsche Pflanzenzucht Hans‐Georg Lembke KG and NPZ Innovation GmbH, Germany, for support with the TILLING material and equipment. For technical assistance in the greenhouse, we thank Christin Belz and Martina Wittke. Furthermore, we thank the Bundesministerium für Ernährung und Landwirtschaft (BMEL, Grant no. 22006516), the Bundesanstalt für Landwirtschaft and Ernährung (BLE, Grant No. 2814IP004), and the Bundesministerium für Bildung und Forschung (BMBF, Grant no. 031B0033C), Germany, for financial support.
Pröbsting, M. , Schenke, D. , Hossain, R. , Häder, C. , Thurau, T. , Wighardt, L. , Schuster, A. , Zhou, Z. , Ye, W. , Rietz, S. , Leckband, G. and Cai, D. (2020) Loss of function of CRT1a (calreticulin) reduces plant susceptibility to Verticillium longisporum in both Arabidopsis thaliana and oilseed rape (Brassica napus). Plant Biotechnol J, 10.1111/pbi.13394
Contributor Information
Dirk Schenke, Email: d.schenke@phytomed.uni-kiel.de.
Daguang Cai, Email: dcai@phytomed.uni-kiel.de.
References
- Acevedo‐Garcia, J. , Spencer, D. , Thieron, H. , Reinstädler, A. , Hammond‐Kosack, K. , Phillips, A.L. and Panstruga, R. (2017) mlo‐based powdery mildew resistance in hexaploid bread wheat generated by a non‐transgenic TILLING approach. Plant Biotechnol. J. 15, 367–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F. , Gish, W. , Miller, W. , Myers, E.W. and Lipman, D.J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Bae, S. , Park, J. and Kim, J.S. (2014) Cas‐OFFinder: a fast and versatile algorithm that searches for potential off‐target sites of Cas9 RNA‐guided endonucleases. Bioinformatics, 30, 1473–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens, F.H. , Schenke, D. , Hossain, R. , Häder, C. , Zhao, Y. , Zhu, W. and Cai, D. (2019) Suppression of abscisic acid (ABA) biosynthesis at the early infection stage of Verticillium longisporum in oilseed rape (Brassica napus). Mol. Plant Pathol. 20,1645–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini, T.Z. , Reiser, L. , Li, D. , Mezheritsky, Y. , Muller, R. , Strait, E. and Huala, E. (2015) The Arabidopsis information resource: making and mining the “gold standard” annotated reference plant genome. Genesis, 53, 474–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom, N. , Sicheritz‐Ponten, T. , Gupta, R. , Gammeltoft, S. and Brunak, S. (2004) Prediction of post‐translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics, 4, 1633–1649. [DOI] [PubMed] [Google Scholar]
- Braatz, J. , Harloff, H.J. , Mascher, M. , Stein, N. , Himmelbach, A. and Jung, C. (2017) CRISPR‐Cas9 targeted mutagenesis leads to simultaneous modification of different homoeologous gene copies in polyploid oilseed rape (Brassica napus). Plant Physiol. 174, 935–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, C.L. and Madden, L.V. (1990) Introduction to plant disease epidemiology. New York: Wiley. http://www.loc.gov/catdir/enhancements/fy0706/89034349‐b.html. [Google Scholar]
- Chalhoub, B. , Denoeud, F. , Liu, S. , Parkin, I.A.P. , Tang, H. et al. (2014) Early allopolyploid evolution in the post‐Neolithic Brassica napus oilseed genome. Science, 354, 950–953. [DOI] [PubMed] [Google Scholar]
- Chandrasekaran, J. , Brumin, M. , Wolf, D. , Leibman, D. , Klap, C. , Pearlsman, M. , Sherman, A. et al. (2016) Development of broad virus resistance in non‐transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 17, 1140–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M.‐H. , Tian, G.‐W. , Gafni, Y. and Citovsky, V. (2005) Effects of Calreticulin on viral cell‐to‐cell movement. Plant Physiol. 138, 1866–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y.F. , Shakeel, S.N. , Bowers, J. , Zhao, X.C. , Etheridge, N. and Schaller, G.E. (2007) Ligand‐induced degradation of the ethylene receptor ETR2 through a proteasome‐dependent pathway in Arabidopsis. J. Biol. Chem. 282, 24752–8. [DOI] [PubMed] [Google Scholar]
- Christensen, A. , Svensson, K. , Persson, S. , Jung, J. , Michalak, M. , Widell, S. and Sommarin, M. (2008) Functional characterization of Arabidopsis Calreticulin1a: a key alleviator of endoplasmic reticulum stress. Plant Cell Physiol. 49, 912–924. [DOI] [PubMed] [Google Scholar]
- Christensen, A. , Svensson, K. , Thelin, L. , Zhang, W. , Tintor, N. , Prins, D. , Funke, N. et al. (2010) Higher plant Calreticulins have acquired specialized functions in Arabidopsis. PLoS ONE, 5, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppolino, M.G. and Dedhar, S. (1998) Molecules in focus: calreticulin. Int. J. Biochem. Cell Biol. 30, 553–558. [DOI] [PubMed] [Google Scholar]
- Cohn, M. , Bart, R.S. , Shybut, M. , Dahlbeck, D. , Gomez, M. , Morbitzer, R. , Hou, B.H. et al. (2014) Xanthomonas axonopodis virulence is promoted by a transcription activator‐like effector‐mediated induction of a SWEET sugar transporter in cassava. Mol. Plant Microbe Interact. 27, 1186–98. [DOI] [PubMed] [Google Scholar]
- Depotter, J.R. , Deketelaere, S. , Inderbitzin, P. , von Tiedemann, A. , Höfte, M. , Subbarao, K.V. , Wood, T.A. et al. (2016) Verticillium longisporum, the invisible threat to oilseed rape and other Brassicaceous plant hosts. Mol. Plant Pathol. 17, 1004–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Coninck, B. , Timmermans, P. , Vos, C. , Cammue, B.P.A. and Kemal, K. (2015) What lies beneath: belowground defense strategies in plants. Trends Plant Sci. 20, 91–101. [DOI] [PubMed] [Google Scholar]
- Diatchenko, L. , Lau, Y.‐F.C. , Campbell, A.P. , Chenchik, A. , Moqadam, F. , Huang, B. , Lukyanov, S. et al. (1996) Suppression subtractive hybridization: a method for generating differentially regulated or tissue‐specific cDNA probes and libraries. Proc. Natl. Acad. Sci. USA, 93, 6025–6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilczyk, U.G. , Cohen‐Doyle, M.F. and Williams, D.B. (2000) Functional relationship between Calreticulin, Calnexin, and the endoplasmic reticulum luminal domain of Calnexin. J. Biol. Chem. 275, 13089–13097. [DOI] [PubMed] [Google Scholar]
- Das, S. , Dutta, S.S. , Chowdhury, S. and Das, K. (2015) Ethylene signal transduction and signaling roles – a review. Agri. Rev. 36, 133–139. [Google Scholar]
- Denecke, J. , Carlsson, L.E. , Vidal, S. , Höglund, A.‐S. , Ek, B. , van Zeijl, M.J. , Sinjorgo, K.M.C. et al. (1995) The tobacco homolog of mammalian Calreticulin is present in protein complexes in vivo . Plant Cell, 7, 391–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker, S. , Keunecke, H. , Steinbach, P. and von Tiedemann, A. (2008) Impact of Verticillium longisporum on yield and morphology of winter oilseed rape (Brassica napus) in relation to systemic spread in the plant. J. Phytopathol. 156, 698–707. [Google Scholar]
- Eynck, C. , Koopmann, B. , Grunewald‐Stoecker, G. , Karlovsky, P. and von Tiedemann, A. (2007) Differential interactions of Verticillium longisporum and V. dahliae with Brassica napus detected with molecular and histological techniques. Eur. J. Plant Pathol. 118, 259–274. [Google Scholar]
- Eynck, C. , Koopmann, B. , Karlovsky, P. and von Tiedemann, A. (2009) Internal resistance in winter oilseed rape inhibits systemic spread of the vascular pathogen Verticillium longisporum . Phytopathology, 99, 802–811. [DOI] [PubMed] [Google Scholar]
- FAOSTAT . (2016) Statistical database. Food and Agriculture Organization of the United Nations: Statistic Division. Available at: http://www.fao.org/faostat/en/#data/QC/visualize. Accessed: 09/12/2016. [Google Scholar]
- Fröschel, C. , Iven, T. , Walper, E. , Bachmann, V. , Weiste, C. and Dröge‐Laser, W. (2019) A gain‐of‐function screen reveals redundant ERF transcription factors providing opportunities for resistance breeding toward the vascular fungal pathogen Verticillium longisporum . Mol. Plant Microbe Interact. 32, 1095–1109. [DOI] [PubMed] [Google Scholar]
- Gallie, D.R. (2015) Ethylene receptors in plants – why so much complexity? F1000Prime Rep. 7, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg, G. , Yadav, S. and Ruchi, Y.G. (2015) Key roles of Calreticulin and Calnexin proteins in plant perception under stress conditions: a review. Adv. Life Sci. 5, 18–26. [Google Scholar]
- Happstadius, I. , Ljungberg, A. , Kristiansson, B. and Dixelius, C. (2003) Identification of Brassica oleracea germplasm with improved resistance to Verticillium wilt. Plant Breed. 122, 30–34. [Google Scholar]
- Harloff, H.‐J. , Lemcke, S. , Mittasch, J. , Frolov, A. , Wu, J.G. , Dreyer, F. , Leckband, G. et al. (2012) A mutation screening platform for rapeseed (Brassica napus L.) and the detection of sinapine biosynthesis mutants. Theor. Appl. Genet. 124, 957–969. [DOI] [PubMed] [Google Scholar]
- Hatzig, S. , Breuer, F. , Nesi, N. , Ducournau, S. , Wagner, M.H. , Leckband, G. , Abbadi, A. et al. (2018) Hidden effects of seed quality breeding on germination in oilseed rape (Brassica napus L.). Front. Plant Sci. 9, 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S. , Till, B.J. and Comai, L. (2004) TILLING. Traditional mutagenesis meets functional genomics. Plant Physiol. 135, 630–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz, T. , Laule, O. , Szabo, G. , Wessendorp, F. , Bleuler, S. , Oertle, L. , Widmayer, P. et al. (2008) Genevestigator v3: a reference expression database for the meta‐analysis of transcriptomes. Adv. Bioinformatics, 420747. 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y. , Zhang, J. , Jia, H. , Sosso, D. , Li, T. , Frommer, W.B. , Yang, B. et al. (2014) Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc. Natl. Acad. Sci. USA, 111, E521–E529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson, A. , Staal, J. and Dixelius, C. (2006) Early responses in the Arabidopsis‐Verticillium longisporum pathosystem are dependent on NDR1, JA‐ and ET‐associated signals via cytosolic NPR1 and RFO1 . Mol. Plant Microbe Interact. 19, 958–969. [DOI] [PubMed] [Google Scholar]
- Joshi, R. , Paul, M. , Kumar, A. and Pandey, D. (2019) Role of calreticulin in biotic and abiotic stress signalling and tolerance mechanisms in plants. Gene, 714, 144004. [DOI] [PubMed] [Google Scholar]
- Jia, X.‐J. , He, L.‐H. , Jing, R.‐L. and Li, R.‐Z. (2009) Calreticulin: conserved protein and diverse functions in plants. Physiol. Plant. 136, 127–138. [DOI] [PubMed] [Google Scholar]
- Ju, C. and Chang, C. (2015) Mechanistic insights in ethylene perception and signal transduction. Plant Physiol. 169, 85–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallimasioti‐Pazi, E.M. , Thelakkad, C.K. , Taylor, G.C. , Meynert, A. , Ballinger, T. , Kelder, M.J.E. , Lalevée, S. et al. (2018) Heterochromatin delays CRISPR‐Cas9 mutagenesis but does not influence the outcome of mutagenic DNA repair. PLoS Biol. 16, e2005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, H.‐G. , Oh, C.‐S. , Sato, M. , Katagiri, F. , Glazebrook, J.‐ , Takahashi, H. et al. (2010) Endosome‐associated CRT1 functions early in resistance gene‐mediated defense signaling in Arabidopsis and tobacco. Plant Cell, 22, 918–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.H. , Nguyen, N.H. , Nguyen, N.T. , Hong, S.‐H. and Lee, H. (2013) Loss of all three calreticulins, CRT1, CRT2 and CRT3, causes enhanced sensitivity to water stress in Arabidopsis. Plant Cell Rep. 32, 1843–1853. [DOI] [PubMed] [Google Scholar]
- Krause, K.‐H. and Michalak, M. (1997) Calreticulin. Cell, 88, 439–443. [DOI] [PubMed] [Google Scholar]
- Kusch, S. and Panstruga, R. (2017) mlo‐based resistance: an apparently universal “weapon” to defeat powdery mildew disease. Mol. Plant Microbe Interact. 30, 179–189. [DOI] [PubMed] [Google Scholar]
- Langner, T. , Kamoun, S. and Belhaj, K. (2018) CRISPR Crops: plant genome editing toward disease resistance. Annu. Rev. Phytopathol. 56, 479–512. [DOI] [PubMed] [Google Scholar]
- Li, J. , Zhao‐Hui, C. , Batoux, M. , Nekrasov, V. , Roux, M. , Chinchilla, D. , Zipfel, C. et al. (2009) Specific ER quality control components required for biogenesis of the plant innate immune receptor EFR. Proc. Natl. Acad. Sci. USA, 106, 15973–15978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J.F. , Norville, J.E. , Aach, J. , McCormack, M. , Zhang, D. , Bush, J. , Church, G.M. et al. (2013) Multiplex and homologous recombination‐mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31, 688–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebrand, T.W.H. , Kombrink, A. , Zhang, Z. , Sklenar, J. , Jones, A.M.E. , Robatzek, S. , Thomma, B.P.H.J. et al. (2014) Chaperones of the endoplasmic reticulum are required for Ve1‐mediated resistance to Verticillium . Mol. Plant Pathol. 15, 109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macovei, A. , Sevilla, N.R. , Cantos, C. , Jonson, G.B. , Slamet‐Loedin, I. , Cermak, T. , Voytas, D.F. et al. (2018) Novel alleles of rice eIF4G generated by CRISPR/Cas9‐targeted mutagenesis confer resistance to rice tungro spherical virus. Plant Biotechnol. J. 16, 1918–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnoy, M. , Viola, R. , Jung, M.‐H. , Koo, O.‐J. , Kim, S. , Kim, J.‐S. , Velasco, R. et al. (2016) DNA‐free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front. Plant Sci. 7, 1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum, C.M. , Comai, L. , Greene, E.A. and Henikoff, S. (2000) Targeting Induced Local Lesions IN Genomes (TILLING) for plant functional genomics. Plant Physiol. 123, 439–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menegazzi, P. , Guzzo, F. , Baldan, B. , Mariani, P. and Treves, S. (1993) Purification of Calreticulin‐like protein(s) from spinach leaves. Biochem. Biophys. Res. Comm. 190, 1130–1135. [DOI] [PubMed] [Google Scholar]
- Merchante, C. , Alonso, J.M. and Stepanova, A.N. (2013) Ethylene signaling: simple ligand, complex regulation. Curr. Opin. Plant Biol. 16, 554–560. [DOI] [PubMed] [Google Scholar]
- Mikami, M. , Toki, S. and Endo, M. (2015) Parameters affecting frequency of CRISPR/Cas9 mediated targeted mutagenesis in rice. Plant Cell Rep. 34, 1807–1815. [DOI] [PubMed] [Google Scholar]
- Nagaharu, U. (1935) Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Japan. J. Bot. 7, 389–452. [Google Scholar]
- Nakagawa, T. , Suzuki, T. , Murata, S. , Nakamura, S. , Hino, T. , Maeo, K. , Tabata, R. et al. (2007) Improved Gateway binary vectors: high‐performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71, 2095–2100. [DOI] [PubMed] [Google Scholar]
- Neff, M.M. , Neff, J.D. , Chory, J. and Pepper, A.E. (1998) dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J. 14, 387–92. [DOI] [PubMed] [Google Scholar]
- Nekrasov, V. , Wang, C. , Win, J. , Lanz, C. , Weigel, D. and Kamoun, S. (2017) Rapid generation of a transgene‐free powdery mildew resistant tomato by genome deletion. Sci. Rep. 7, 482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okubara, P.A. and Paulitz, T.C. (2005) Root defense responses to fungal pathogens: a molecular perspective. Plant Soil. 274, 215–226. [Google Scholar]
- Oliva, R. , Ji, C. , Atienza‐Grande, G. , Huguet‐Tapia, J.C. , Perez‐Quintero, A. , Li, T. , Eom, J.‐S. et al. (2019) Broad‐spectrum resistance to bacterial blight in rice using genome editing. Nat. Biotechnol. 37, 1344–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuzaki, A. , Ogawa, T. , Koizuka, C. , Kaneko, K. , Inaba, M. , Imamura, J. and Koizuka, N. (2018) CRISPR/Cas9‐mediated genome editing of the fatty acid desaturase 2 gene in Brassica napus. Plant Physiol Biochem. 131, 63–69. 10.1016/j.plaphy.2018.04.025 [DOI] [PubMed] [Google Scholar]
- Ostwald, T.J. and MacLennan, D.H. (1974) Isolation of a high affinity calcium‐binding protein from sarcoplasmic reticulum. J. Biol. Chem. 249, 974–979. [PubMed] [Google Scholar]
- Pantelides, I.S. , Tjamos, S.E. and Paplomatas, E.J. (2010) Ethylene perception via ETR1 is required in Arabidopsis infection by Verticillium dahliae. Mol Plant Pathol. 11(2), 191–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J. , Bae, S. and Kim, J.S. (2015) Cas‐Designer: a web‐based tool for choice of CRISPR‐Cas9 target sites. Bioinformatics, 31, 4014–4016. [DOI] [PubMed] [Google Scholar]
- Reusche, M. , Thole, K. , Janz, D. , Truskina, J. , Rindfleisch, S. , Drübert, C. , Polle, A. et al. (2012) Verticillium infection triggers VASCULAR‐RELATED NAC DOMAIN7‐dependent de novo xylem formation and enhances drought tolerance in Arabidopsis . Plant Cell, 24, 3823–3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, S.O. and Bendich, A.J. (1985) Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol. Biol. 5, 69–76. [DOI] [PubMed] [Google Scholar]
- Rygulla, W. , Snowden, R.J. , Friedt, W. , Happstadius, I. , Cheung, W.Y. and Chen, D. (2008) Identification of quantitative trait loci for resistance against Verticillium longisporum in oilseed rape (Brassica napus). Phytopathology, 98, 215–221. [DOI] [PubMed] [Google Scholar]
- Sarwat, M. and Tuteja, N. (2017) How the ER Stress Protein Calreticulins Differ from Each Other in Plants?. In Plant Bioinformatics, pp. 403–415. Cham: Springer. 10.1007/978-3-319-67156-7_17 [DOI] [Google Scholar]
- Schiml, S. , Fauser, F. and Puchta, H. (2014) The CRISPR/Cas system can be used as nuclease for in planta gene targeting and as paired nickases for directed mutagenesis in Arabidopsis resulting in heritable progeny. Plant J. 80, 1139–50. [DOI] [PubMed] [Google Scholar]
- Shakeel, S.N. , Gao, Z. , Amir, M. , Chen, Y.F. , Rai, M.I. , Haq, N.U. and Schaller, G.E. (2015) Ethylene regulates levels of ethylene receptor/CTR1 signaling complexes in Arabidopsis thaliana . J. Biol. Chem. 290, 12415–12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, N. , Perovic, D. , Kumlehn, J. , Pellio, B. , Stracke, S. , Streng, S. , Ordon, F. et al. (2005) The eukaryotic translation initiation factor 4E confers multiallelic recessive Bymovirus resistance in Hordeum vulgare (L.). Plant J. 42, 912–22. [DOI] [PubMed] [Google Scholar]
- Sun, Q. , Lin, L. , Liu, D. , Wu, D. , Fang, Y. , Wu, J. and Wang, Y. (2018) CRISPR/Cas9‐mediated multiplex genome editing of the BnWRKY11 and BnWRKY70 genes in Brassica napus L. Int. J. Mol. Sci. 19(9), pii: E2716. 10.3390/ijms19092716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K. , Stecher, G. , Peterson, D. , Filipski, A. and Kumar, S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till, B.J. , Zerr, T. , Comai, L. and Henikoff, S. (2006) A protocol for TILLING and Ecotilling in plants and animals. Nat. Protoc. 5, 2465–2477. [DOI] [PubMed] [Google Scholar]
- Tsolakidou, M.D. , Pantelides, I. , Tzima, A.K. , Kang, S. , Paplomatas, E.J. and Tsaltas, D. (2019) Disruption and overexpression of the gene encoding ACC (1‐Aminocyclopropane‐1‐Carboxylic Acid) deaminase in soil‐borne fungal pathogen Verticillium dahliae revealed the role of ACC as a potential regulator of virulence and plant defense. Mol. Plant Microbe Interact. 32, 639–653. [DOI] [PubMed] [Google Scholar]
- Urnov, F.D. , Ronald, P.C. and Carroll, D. (2018) A call for science‐based review of the European court's decision on gene‐edited crops. Nat. Biotechnol. 36, 800–802. [DOI] [PubMed] [Google Scholar]
- USDA: Crop Production . (2018) Summary. February 2019. ISSN: 1057‐7823. [Google Scholar]
- Verkuijl, S.A. and Rots, M.G. (2019) The influence of eukaryotic chromatin state on CRISPR‐Cas9 editing efficiencies. Curr. Opin. Biotechnol. 55, 68–73. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Cheng, X. , Shan, Q. , Zhang, Y. , Liu, J. , Gao, C. and Qiu, J.‐L. (2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. [DOI] [PubMed] [Google Scholar]
- Wilson, R.L. , Kim, H. , Bakshi, A. and Binder, B.M. (2014) The ethylene receptors ETHYLENE RESPONSE1 and ETHYLENE RESPONSE2 have contrasting roles in seed germination of Arabidopsis during salt stress. Plant Physiol. 165, 1353–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, X. , Sun, S. , Zhang, X. , Li, Y. , Liu, F. , Zhu, Q. , Xeu, F. et al. (2020) GhWRKY70D13 regulates resistance to Verticillium dahliae in cotton through the ethylene and jasmonic acid signaling pathways. Front. Plant Sci., 11, 69. 10.3389/fpls.2020.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H. , Wu, J.J. , Tang, T. , Liu, K.D. and Dai, C. (2017) CRISPR/Cas9‐mediated genome editing efficiently creates specific mutations at multiple loci using one sgRNA in Brassica napus . Sci. Rep. 7, 7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y. , Zhu, K. , Li, H. , Han, S. , Meng, Q. , Khan, S.U. , Fan, C. et al. (2018) Precise editing of CLAVATA genes in Brassica napus L. regulates multilocular silique development. Plant Biotechnol. J. 16, 1322–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeise, K. and von Tiedemann, A. (2002) Host specialization among vegetative compatibility groups of Verticillium dahliae in relation to Verticillium longisporum . J. Phytopathol. 150, 112–119. [Google Scholar]
- Zhai, Y. , Cai, S. , Hu, L. , Yang, Y. , Amoo, O. , Fan, C. and Zhou, Y. (2019a) CRISPR/Cas9‐mediated genome editing reveals differences in the contribution of INDEHISCENT homologues to pod shatter resistance in Brassica napus L. Theor. Appl. Genet. 132, 2111–2123. [DOI] [PubMed] [Google Scholar]
- Zhai, Y. , Yu, K. , Cai, S. , Hu, L. , Amoo, O. , Xu, L. , Yang, Y. et al. (2019b) Targeted mutagenesis of BnTT8 homologs controls yellow seed coat development for effective oil production in Brassica napus L. Plant Biotechnol. J. 18, 1153–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J. and Meng, J. (2003) Detection of loci controlling seed glucosinolate content and their association with Sclerotinia resistance in Brassica napus . Plant Breed. 122, 19–23. [Google Scholar]
- Zheng, M. , Zhang, L. , Tang, M. , Liu, J. , Liu, H. , Yang, H. , Fan, S. et al. (2019) Knockout of two BnaMAX1 homologs by CRISPR/Cas9‐targeted mutagenesis improves plant architecture and increases yield in rapeseed (Brassica napus L.). Plant Biotechnol. J. 18, 644–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J. , Peng, Z. , Long, J. , Yang, B. (2015) Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J. 82, 632–643. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Verification of the SSH libraries.
Figure S2 Time course of infection with Vl43 and determination of optimal conidia concentration.
Figure S3 There are 4 CRT1a loci in the Brassica napus genome.
Figure S4 The SSH‐EST 6F11‐F8 corresponds to BnaA09g15400D (SNPs highlighted in red).
Figure S5 Flow chart for CRISPR/Cas vector construction.
Figure S6 dCAPS‐analysis of positive CRISPR events.
Figure S7 Effect of CRT1a mutations on 1000 seed weight.
Figure S8 Phenotype of C4E23 at 28 dpi with Verticillium longisporum using 1 × 107 spores/mL.
Figure S9 Detection of transgene‐free plants.
Figure S10 Analysis of CRT1a on amino acid level.
Table S1 SSH library (EXCEL).
Table S2 Genevestigator Meta‐Analysis of SSH candidate genes (EXCEL).
Table S3 Candidate genes selected from the SSH libraries.
Table S4 Agronomic traits investigated under control and infection conditions.
Table S5 TILLING statistics of CRT1a screen within an EMS mutagenized population.
Table S6 Primer‐Information (EXCEL).
Table S7 Disease rating scale for Verticillium infection of Brassica napus plants (Enyck et al. 2007).


