Summary
The Zea Mays BIG GRAIN 1 HOMOLOG 1 (ZM‐BG1H1) was ectopically expressed in maize. Elite commercial hybrid germplasm was yield tested in diverse field environment locations representing commercial models. Yield was measured in 101 tests across all 4 events, 26 locations over 2 years, for an average yield gain of 355 kg/ha (5.65 bu/ac) above control, with 83% tests broadly showing yield gains (range +2272 kg/ha to −1240 kg/ha), with seven tests gaining more than one metric ton per hectare. Plant and ear height were slightly elevated, and ear and tassel flowering time were delayed one day, but ASI was unchanged, and these traits did not correlate to yield gain. ZM‐BG1H1 overexpression is associated with increased ear kernel row number and total ear kernel number and mass, but individual kernels trended slightly smaller and less dense. The ZM‐BG1H1 protein is detected in the plasma membrane like rice OS‐BG1. Five predominant native ZM‐BG1H1 alleles exhibit little structural and expression variation compared to the large increased expression conferred by these ectopic alleles.
Keywords: kernel row number, seed size, height, plasma membrane, auxin
Introduction
Agriculture is increasingly strained to meet global demands for food, feed, fibre and fuel amid rising population, demands for economic advancement, competition for land and water, while striving to moderate environmental impacts. Genetic improvements continue to increase yield, but for many major crops including maize, the genetic gain rate has slowed, while research cost per unit gain has increased (Frey, 2000). Newer complementary approaches such as transgenics and gene editing seek to diversify and extend established breeding strategies. Hundreds of reports in recent decades describe diverse genes with positive effects on yield or yield component traits. Thousands other genes are believed to have been screened by various public and private research organizations in model or crop systems for increasing yield or improving complex agronomic traits such as drought tolerance (largely unpublished). These studies often use controlled environments and model plants, less commonly target crops and seldom elite crop germplasm in agronomically relevant field conditions measuring crop yield per unit land area. We proffer many published yield‐enhancing genes rarely validate under these restrictive conditions, or they produce effects too small or inconsistent to warrant advancement in breeding.
Biologically efficacious genes face other stiff requirements to become successfully deployed, including sufficiently large phenotype effect size, favourable germplasm interactions, minimal negative secondary traits, ease to integrate broadly into elite breeding germplasm, necessary regulatory and import–export approvals, and project continuity amid fluctuating business investment decisions (e.g. Nuccio et al., 2018). Maize receives extensive research investment, and yet while biotech alleles improving maize field yield exist, presently only one drought‐related gene has been commercialized in maize (Castiglioni et al., 2008; Nemali et al., 2015). Corteva Agriscience researchers have identified other single genes that improve maize yield and agronomic performance (Fox et al., 2017; Guo et al., 2014; Habben et al., 2014; Shi et al., 2017; Shi et al., 2015; Wu et al., 2019).
The rice BIG GRAIN 1 (herein OS‐BG1) gene was identified by gene activation tagging overexpression (OE) screens for larger grain size, and validated by transgenic OE with the rice actin promoter (Liu et al., 2015b). The OS‐BG1 OE also conferred: increased grain weight; increased yield per plot area; increased biomass; larger cell size and cell number; taller plants; larger leaves and panicles; and increased auxin basipetal transport. Additionally, depending upon the level of ectopic expression, reduced gravitropism, less erect tiller angle, grains less fully matured and grain yield per plant may increase. RNAi down‐regulation of OS‐BG1 reversed most these effects (Liu et al., 2015b). OS‐BG1 is a plasma membrane localized protein hypothesized to function in auxin transport or its regulation (Liu et al., 2015b). The OS‐BG1 belongs to a larger gene family that is vascular plant specific and has undergone extensive diversification. This family possesses uncharacterized conserved domains, and it includes at least one nuclear‐targeted family member (Mishra et al., 2017), suggesting diverse molecular functions.
This study primarily aims to determine: the likely maize BG1‐related gene family members; the likeliest maize homolog(s) to OS‐BG1; the homologs native allelic structural expression diversity in breeding germplasm; and whether ectopic expression of the maize homolog in maize increases field yield in elite commercial germplasm. We report that overexpression of the ZM‐BG1H1 homolog does increase maize grain yield across a range of field environments, demonstrating that BG1 homologs may improve yield across multiple crops.
Results
Maize BG1 gene family
Public and proprietary maize genomes and transcriptomes were extensively searched for gene family members related to rice OS‐BG1 (BIG GRAIN1) gene (Locus OS03g0175800). Mishra et al. (2017) identified 10 candidate maize family members. We determined that there are 8 members of the maize BG1‐related gene family with over 20% amino acid identity (AAID) to OS‐BG1 (Table S1). GRMZM2G027519 is a duplicate, identical to GRMZM5G843781, and the latter retained in the newer AGPv4 genome draft. We also omitted GRMZM2G117930 because of very low OS‐BG1 homology (ca. 12.5%). The closest by protein sequence (65.1% identity) to OS‐BG1 is locus GRMZM2G178852 on chromosome 1, which we name Zea Mays BIG GRAIN1 Homolog 1 (ZM‐BG1H1). A close second relative is a single or duplicated locus pair on chromosome 9 (56.3%–57.6% identity to OS‐BG1; 65% ID to ZM‐BG1H1). In the B73 genomic assemblies RefGen2.0 or AGPv4.0, this region is represented by two very closely related (97.8% AAID) and closely spaced loci GRMZM2G007134 (ZM‐BG1H2) and GRMZM2G438606 (ZM‐BG1H3). Genome drafts, RefGen2 and AGPv4 (B73), have an unresolved gap between the genes; however, a proprietary genome draft of a different line indicates these two genes are 31.5 kb apart ATG‐ATG in tandem duplication, with GRMZM2G438606 the most distal (telomeric). In some proprietary line genome drafts, this region is a single‐copy locus GRMZM2G438606. Because gene expression and genetic haplotype analyses likely conflate this closely related pair, we will refer to them together as ZM‐BG1H2(3). The other five BG1‐like family members are more distantly related and unlikely OS‐BG1 homologs.
Maize OS‐BG1 Homologs
ZM‐BG1H1 and the ZM‐BG1H2(3) locus pair are therefore candidate OS‐BG1 orthologs. Chromosomes 1 and 9 share extensive intra‐genomic synteny, and the ZM‐BG1H1 locus vicinity shares multiple gene homologs to the Chr. 9 gene vicinity around ZM‐BG1H2(3). And just as ZM‐BG1H1 and ZM‐BG1H2(3) are in forward–reverse directions on their respective chromosomes, the relative gene orders of their local syntenic neighbours are also inverted. ZM‐BG1H1 and the ZM‐BG1H2(3) locus pair are thus paralogous. Sorghum has only one OS‐BG1 close homolog, and while it has higher identity to ZM‐BG1H1 (77.5%) than to ZM‐BG1H2(3) (69.6%), the sequence is intermediate between the two. This indicates the maize–sorghum last common ancestor (ca. 11.9 m.y.a.) likely had a single BG1 homolog gene, and that the genome duplication event at ca. >4.8 m.y.a. (Swigonová et al. 2014), likely produced these paralogous maize loci, although differential retention scenarios are possible.
Gene expression analysis can help resolve the best functional homolog by determining which gene has the highest overall expression and spatial–temporal expression pattern most analogous to OS‐BG1. In plant genomes such as maize undergoing genome fractionation following ancestral whole‐genome duplications, among paralogous genes, the gene most highly expressed is likely carrying the greater functional role load (Liang and Schnable, 2018; Schnable et al., 2011). We surveyed the ZM‐BG1 family for gene expression in an atlas of 755 B73 RNAseq samples, plus the expression patterns in the Maize eFP browser (Sekhon et al., 2011). The maize gene family expression patterns are shown across these diverse tissue–treatment mRNA profiling samples organized into five major tissue categories (Figure 1). The highest average expression across all samples is ZM‐BG1H1. ZM‐BG1H2(3) expression collectively has lower expression than ZM‐BG1H1, although a few tissues of the public eFP browser indicate ZM‐BG1H2(3) has higher expression. All other BG1 family members have even lower expression levels. Gene expression correlation in using this 755 sample atlas, combined with Gene Ontological (GO) terms functional term enrichment, showed that ZM‐BG1H1 had more genes of higher correlation significance than for ZM‐BG1H2(3), which may indicate of ZM‐BG1H1 is tied into more functional biology. For ZM‐BG1H1 among the 136 correlated transcripts, the top 15 GO terms were nucleosome, nucleolus, nucleus, DNA‐binding, thylakoid, chloroplast, plasmodesma, plasma membrane, cell division and cell cycle (not shown).
Figure 1.
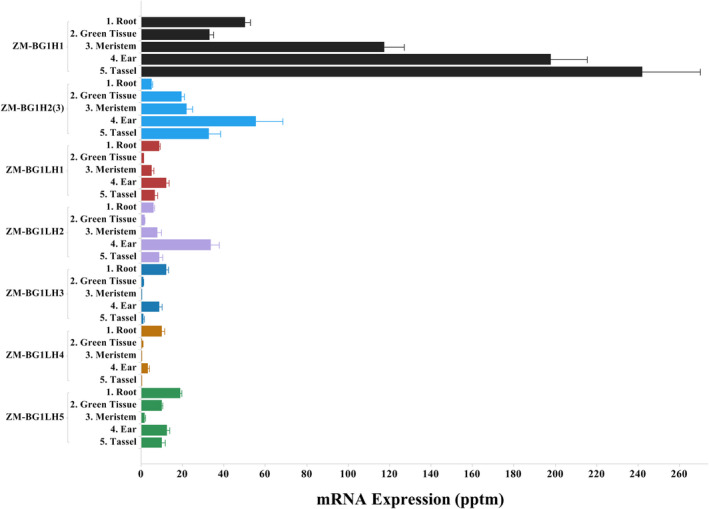
ZM‐BG1 Gene Family Tissue Expression Levels. ZM‐BG1 gene family mRNA expression (Illumina RNAseq) across five major maize tissues categories (1. Root, 2. Green Tissue, 3. Meristem, 4. Ear and 5. Tassel) from a B73‐based Gene Expression Atlas. Expression values are in mean pptm (parts per 10 million) each tissue category with SE bars.
Focusing on just ZM‐BG1H1 vs ZM‐BG1H2(3), through finer tissue–spatial resolution, ZM‐BG1H1 has highest expression in stalks, immature ear, silk and tassel, whereas ZM‐BG1H2(3) has its highest expression in husk and immature ear. In all tissues but husk, ear leaf sheath and pericarp, ZM‐BG1H1 has higher expression (Figure S1). The maize eFP browser comparison shows ZM‐BG1H1 has its highest expression in stems and shoot apical meristem, cob, tassel and silks, and for ZM‐BG1H2(3), the highest expression is in cob, Endosperm, kernels and husk. In the eFP leaf gradient expression patterns, both genes show leaf expression concentrated in the basal half of the leaf, with some extreme tip expression, especially for ZM‐BG1H2(3). ZM‐BG1H1 has low leaf or green tissue expression, but this could partly be due to most samples being day harvested, because ZM‐BG1H1 has a distinct diurnal pattern with evening dark peak expression (Figure S2). OS‐BG1 shows highest expression in the shoot apical meristem and in the developing inflorescence, but lower levels in developing seeds, lower still in leaves and roots (Rice eFP Browser at bar.utoronto.ca, querying alias LOC_0s03g07920). The ZM‐BG1H1 has especially high expression in meristematic tissues and developing inflorescences, which matches the general expression pattern for OS‐BG1. This expression analysis and protein similarity both support ZM‐BG1H1 being the closest OS‐BG1 homolog.
Transformed event evaluation
Hybrid maize breeding germplasm is commonly divided into two major heterotic groups, the Stiff Stalk (SS) commonly female lines, and Non‐Stiff Stalk (NSS) commonly male lines, their genetics differing in ways that are often broadly complementary and conducive to producing hybrid combinations with good heterotic yield gain. The ZM‐BG1H1‐A1 allele, common among SS germplasm, in the altered form ZM‐BG1H1 (MOD1), was linked to the moderate constitutive ZM‐GOS2 promoter. The transformed elite germplasm NSS inbred line PH184C possesses the ZM‐BG1H1‐A3 allele common among NSS lines. The insertion locations for events 1, 3 and 4 mapped to chromosome 2 but at separate locations in the B73 genome draft RefGen2 at positions Chr2:120.4Mbp, Chr2:1.3Mbp and Chr2:164.7Mbp, respectively. Event 2 was assigned to a distinct site absent in the B73 genome but in PH184C. T1 generation plants were top crossed to line PHW3G, a SS variety possessing the ZM‐BG1H1‐A1/2 allele.
ZM‐BG1H1 (MOD1) overexpression was assayed by qRT‐PCR in the T0 generation for event selections and in the subsequent T3 hybrid plants used for the field yield test. The relative expression of endogenous ZM‐BG1H1 gene versus ZM‐BG1H1 (MOD1) was compared in growth chamber hybrid plants at V3‐V4 leaves, and in field‐grown R1 mature ear leaves. Both indicate all 4 events have ZM‐BG1H1 (MOD1) expression, estimated at 1000‐2000 pptm by comparing to a common qRT‐PCR internal constitutive control. ZM‐BG1H1 (MOD1) exceeds expression of the ZM‐BG1H1 native locus, across all four events by an inferred relative >57‐fold in growth chamber, and in the field hybrids by >32‐fold (Table S2). We suspect these are underestimations because the native gene is expressed at very low levels, so an even modest background signal in the native gene assay could suppress the relative fold‐change for ZM‐BG1H1 (MOD1). When comparing the relative endogenous gene expression levels between the ZM‐GOS2 native gene (GRMZM2G073535) and ZM‐BG1H1 using 468 RNAseq B73 samples, the ZM‐GOS2 gene expression averaged 375‐fold higher expression than ZM‐BG1H1. When broken down by 11 major tissue types, the ratio ranged from 553‐ and 541‐fold higher in Leaf‐Shoot and Endosperm, respectively, to 21‐fold and 18‐fold in Tassel and Stem‐Stalk, respectively (Figure S3). These results also demonstrate that the native ZM‐GOS2 expression (and thus presumably the transgene) is both higher in expression than ZM‐BG1H1 and possessing a distinctive tissue–spatial–temporal expression pattern.
Field yield tests
The ZM‐BG1H1 OE events (E1–E4) were field tested as a T3 generation single hybrid background for maize grain yield in multiple field locations and environments over two years of tests. These yield tests were done across a total of 26 site locations, which across both years produced a range of yield environments with control yields from 9.4 to 17.4 t/ha. These sites were chosen to provide environment and stress variations generally, with water availability stress as a frequent driver of yield difference across sites. The lowest yielding environments below 11.2 t/ha were classified as moderate stress, those from 11.2 to 14.4 t/ha light stress, and all those above 14.4 t/ha were classified as optimal growth conditions. There were no severely stressed environments in this study. All four events increased yield per unit area relative to controls across both years, with an overall average of 355 kg/ha (5.65 bu/ac) (Figure 2). The event performance ranged from 204.7 kg/ha for event E2, and 399.1, 406.7 and 415.4 kg/ha for events E1, E4 and E3, respectively. Event‐to‐event variation was small, this classification [no difference] not rejected at alpha 0.05 using the Tukey–Kramer significance test. Events E1, E3 and E4 are indistinguishable at alpha 0.05, and together they average 407 kg/ha (6.5 bu/ac) yield advantage.
Figure 2.
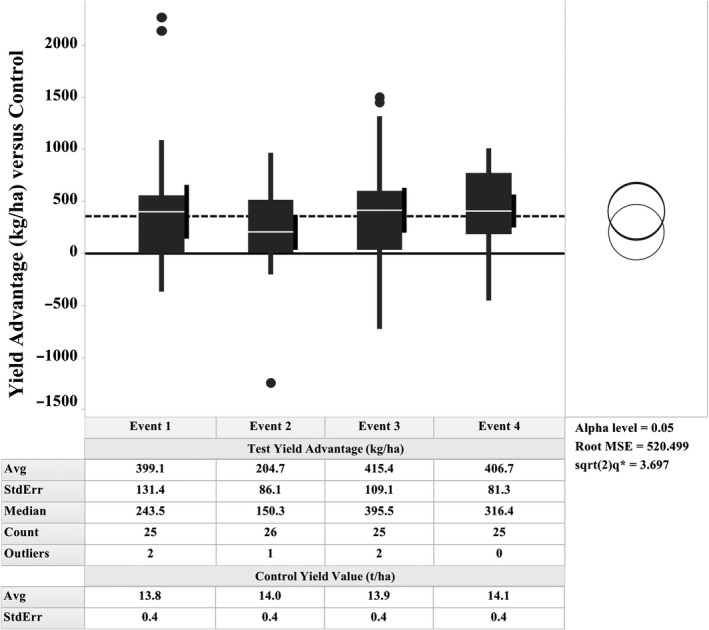
Yield Advantages of ZM‐BG1H1 OE Event versus Control. Box plot of hybrid maize yield difference (kg/ha) relative to non‐transgenic hybrid controls for each of the 4 transgenic events across two years of testing. Non‐transgenic hybrid control average yield value set to 0 on Y‐axis. The average yield advantage across all four alleles central dashed line across figure at 355 kg/ha or 5.65 bu/ac. Per each event the average (white line inside each box), 95% confidence interval (black vertical segment adhering right side each box) and outlier values above or below (dots). Significance null hypothesis test (i.e. no difference among the 4 events) not rejected at alpha level 0.05, represented by overlapping rings graph at right (Tukey–Kramer significance test). The control hybrid yields averages, and standard error values for each event in the lower table inset.
Yield differences for all 101 event‐location‐year tests (Figure 3) reveal 83% tests were positive, with 29 statistically significant at BLUP P‐value 0.1, with only two negative yielding values statistically significant. Seven tests yielded over one ton per hectare advantage. The four events were spread across the performance spectrum, with all four events having representatives in either the upper or lower 10% yield difference values. The ZM‐BG1H1 OE tests showed yield advantage across the wide range of environments encompassing light stress to optimal conditions. There was little or no observed advantage under moderate stress, but this was based upon only one location. Linear regression analysis of the ZM‐BG1H1 OE yield advantage relative to the control yield values was only r 2 = 0.05, indicating little co‐association. This indicates that ZM‐BG1H1 OE conferred yield advantages across a broad range of environments, test locations and stress levels (Figure 3).
Figure 3.
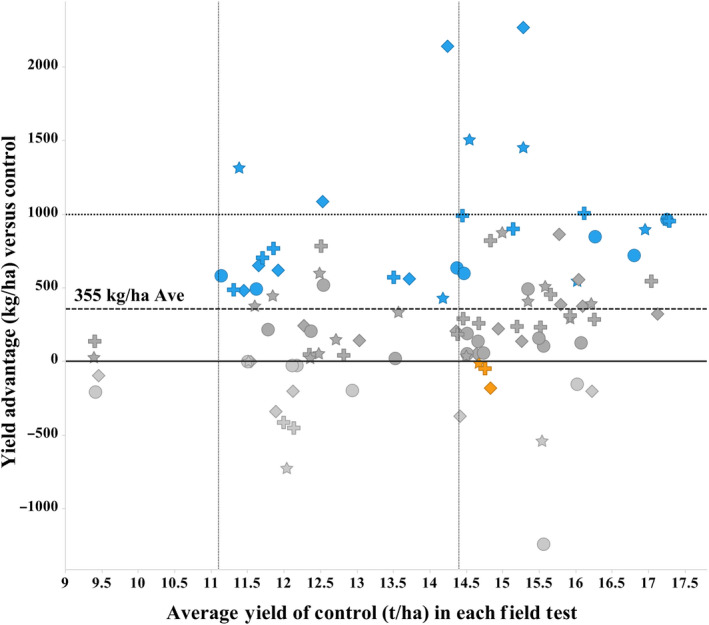
Yield versus Control Across Yield Range Environments. Hybrid maize yield difference (kg/ha) (Y‐axis) relative to non‐transgenic hybrid controls (set to 0 on Y‐axis) for each of the 101 tests, comprising the 4 independent ZM‐BG1H1 OE events for each test year and location. Non‐transgenic hybrid control yield averages (t/ha) at each test location (X‐axis). Low‐yielding sites below 11.2 t/ha are moderate stress (MS), from 11.2 to 14.4 t/ha low stress (LS), and above 14.4 t/ha optimal (OPT), these divisions separated by vertical lines and labelled at bottom of graph. The average yield advantage at 355 kg/ha is dashed line across figure, as is the 1.0 t/ha reference line. BLUP significance tests are coloured: blue, significantly positive (P < 0.1); orange, significantly negative (P < 0.1); moderate grey, positive non‐significant; light grey, negative non‐significant. Icon shapes: Event 1, diamond; Event 2, circle; Event 3, star; Event 4, cross.
The ZM‐BG1H1 OE events were assessed by aerial and ground observations in the field tests for agronomic traits relevant to maize breeding. These included traits spanning flowering, canopy and vegetative greenness, plant size and architecture, and grain moisture. All these traits including yield were converted to per cent differences from control to enable trait‐to‐trait comparisons. A linear regression analysis of each trait to the yield advantage of ZM‐BG1H1 OE was calculated (all events combined). The per cent differences from control for each trait including yield, and the correlation slope and regression for each trait to the ZM‐BG1H1 OE yield advantage are plotted together in Figure 4. Yield is the reference trait, with an average difference of 2.4%, and it necessarily self‐correlates with a slope and R 2 of 1.0. The four canopy greenness traits overall showed little difference or correlation. The two flowering time measures slightly increased (ranging 0.3% to 0.6% differences), but they had little positive slope or correlation with the yield advantage. Both plant height and ear height were above control, 2.6% and 1.5%, respectively; however, both also showed little positive slope or correlation to yield advantage. Grain moisture (MST) was slightly higher than the control (1.4%) and showed a slight positive slope and co‐association with yield (R 2 = 0.19). Grain density (TSTWT) was down an average 0.5% (R 2 = 0.31).
Figure 4.
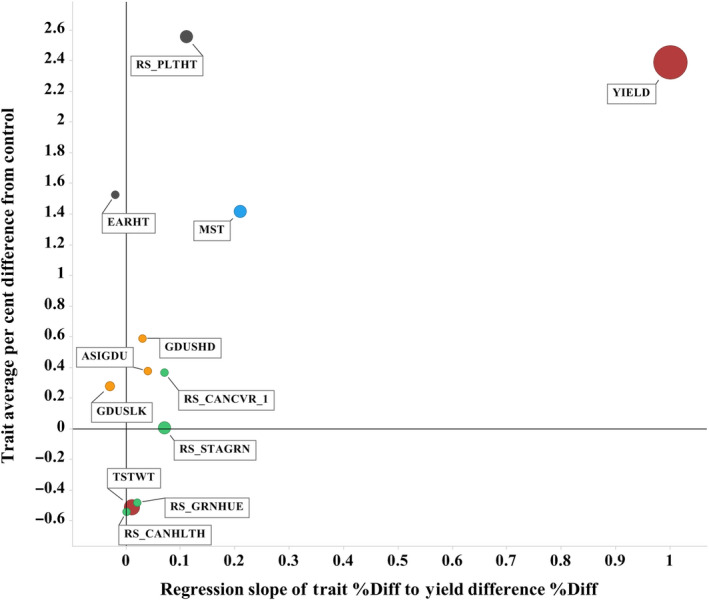
Secondary Agronomic Traits Correlations to ZM‐BG1H1 OE Yield Advantage. Eleven secondary trait associations to yield advantage in ZM‐BG1H1 overexpressing maize plants. See supplement for trait definitions. Secondary traits coloured by category groupings: canopy or greenness (green); flowering (orange); plant size (dark grey), moisture (blue), yield (maroon). All traits values are averages of all four events, and each converted to per cent difference from trait mean for the control (Y‐Axis). All trait per cent differences are linearly regressed to the yield per cent difference across available field locations and years (up to 101 measurements per trait). The slope of that correlation is projected on the X‐axis. The R 2 of the regression is the graph icon size. The overall yield difference of 2.4% therefore correlates to itself with a slope of 1.0 and icon size unit size of R 2 = 1.0 maximum.
Other traits, flowering time, plant and ear height
A targeted observation plot in year 3 (Yr3‐Obs) was used to both confirm and extend the phenotypic observations made in the yield trials. No differences from control were observed in germination, seedling stand count, canopy closure, leaf size shape or colour, tillers and plant height through V11 at 1.8 metres average plant height. We observed normal seedling gravitropism in tilt agar assays and in these field‐grown plants (not shown). Flowering measurements began at 62 days (1353 GDU growth heat units) after planting and proceeded daily through day 68 (1488 GDU). Flowering graph plots were used to interpolate the point at which pollen shed or silking reached 50%. All four events delayed pollen shed by 10–40 GDU (25 GDU average) relative to control, in order Control < E1 < E3 < E2 < E4. All four events delayed silking by 2–38 GDU (21 GDU average) relative to control, in the same relative order Control < E1 < E3 < E2 < E4. The ASI was little changed, control (31 GDU) to the 4 events ranging from 23 to 34 GDU (27 GDU average; Table S3).
Plant and ear height were measured from the ground to the first tassel branch or ear node for each plant, on days 74 and 75, respectively, by which time all plants had flowered. Average first tassel branch heights for all 4 events were taller than control by 4.9–10.1 cm (8.0 cm average, 3.2% increase, t‐test P < 1 × 10−4), in relative order E4 > E2 > E2 > E3 > Control. Average ear node heights for 3 of 4 events were taller than control, ranging from −1.3 to +7.5 cm, in relative order E4 > E2 > E1> Control > E3, and across all four events averaging 2.8 cm higher (2.1% increase, t‐test P = 0.0272). The ratio of first tassel branch height to ear node height was similar, the control at 1.94 and the events ranging from 1.92 to 1.99 (1.96 average), indicating that the plant height relative to ear height ratio was little changed. However, for this ratio the event order E3 > E1 > E2> Control > E4 did reverse relative to event order for plant or ear height, indicating a possible slight rise of ear node relative to tassel height among the taller events.
Ear and kernel morphology
We investigated the impact of ZM‐BG1H1 OE on maize grain size. The same F1 hybrid seed sources used for planting the year 1 yield trials were first evaluated for seed size and density by a combination of direct seed volume and weight measures. Across all four events measured in three replicates, the kernel volume averaged 2.5% lower than the control, and kernel weight averaged 1.5% lower, and the kernel density averaged 1.4% lower (Figure S4). The null hypothesis (no difference) is however not rejected at alpha 0.05 for each of these metrics (Tukey–Kramer test). Apart from the obvious conflict with the OS‐BG1 study where seed size increased, the smaller size indicated that the ZM‐BG1H1 OE events hybrid yield trials did not benefit at planting from larger seeds than for controls.
The observation plot ear and kernel data analysis are summarized in Figure 5, with source data analysis provided in Table S4. Across all four events per ear the total kernel number increased 6.0%, total kernel volume increased 3.6%, and total kernel weight increased 2.0%. Because each plant had only one ear, this increase in kernel weight reflects a yield gain per plant. There was also a 2.6% increased ear length, a 2.3% increased ear filled length and a 2.4% increased ear diameter. The average per kernel weight on each ear however decreased 4.2%, as did per kernel volume by 2.4%, leading to a slight 1.4% decline in per kernel density (Figure 5). The ZM‐BG1H1 OE plant ears for each four events showed increased average kernel row number (KRN), collectively across all events 17.86 KRN (ZM‐BG1H1) versus 17.31 KRN (control), for a half row increase or 3.1%, with a t‐test P‐value 0.02. This upward KRN shift was observed for each of the four events, and the shift was most pronounced between 16 and 18 KRN (Figure 6).
Figure 5.
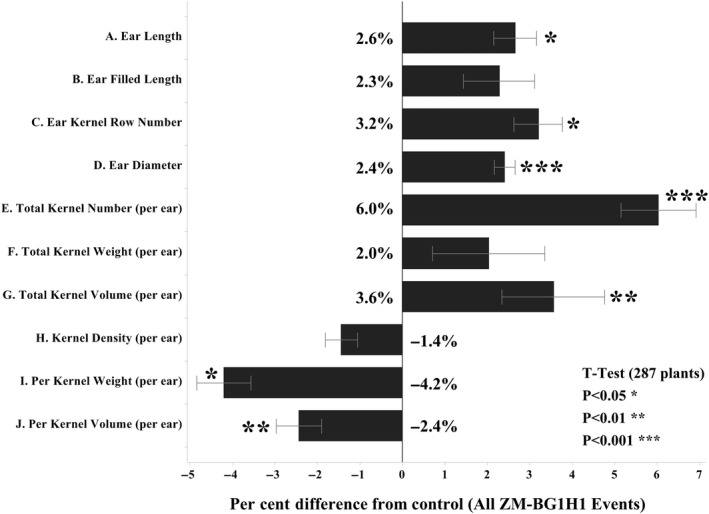
Ear and Kernel Metrics. Ear and kernel traits for ZM‐BG1H1 OE relative to control. All traits are normalized for comparison to the average per cent differences from control average for all the plants across all four events. The standard error bars are derived from the respective individual plant percentage differences from the control average. The t‐test significance was done by comparing the set of percentage differences from the control average for the all individual plants across all 4 events, to the set of per cent differences among the individual control plants to the control average value. Source data analysis provided in Table S4.
Figure 6.
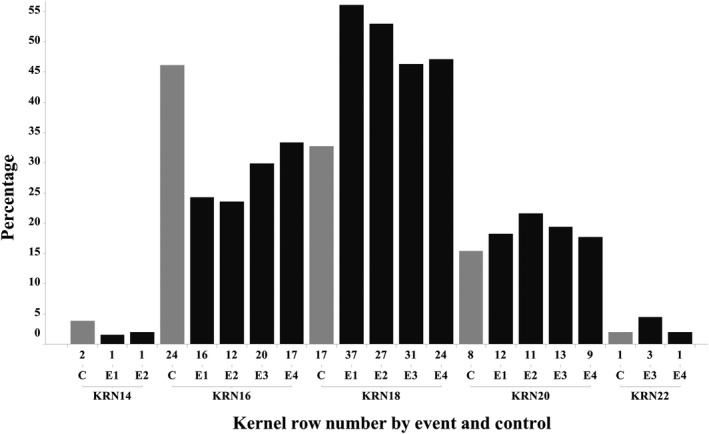
ZM‐BG1H1 OE Increases Kernel Row Number. Histogram percentage distribution of KRN among the four events (E) and Control (C) in the Yr3‐OBS plot. Note the relative shift of KRN from KRN16 to KRN18 for all four ZM‐BG1H1 OE events but a decline for the control. KRN is kernel row number followed by the specific number of rows; for example, KRN20 has 20 kernel rows. The number of plants (=ears) for each category immediately below the bars.
Pursuing the possibility that the 2.4% average increased ZM‐BG1H1 OE yield may be primarily driven by this 3.1% increased of a half kernel row, and that the decrease in average ZM‐BG1H1 OE kernel volume may relate to ear spatial constraints resulting from a proportionally increased number of higher KRN ears, we compared the ear and kernel traits again but with normalization of the KRN value (Figure S5). The results revealed that all the observed patterns of increase or decrease among ear or kernel traits observed when all the KRN values are considered together (Figure 5), persisted in roughly the same pattern and magnitude when comparing control vs ZM‐BG1H1 OE ears having the same kernel row numbers (KRN), with no statistically significant percentage differences among the various comparisons (t‐test P‐values > 0.1). Normalizing for KRN did however nominally reduce the differences for ear diameter and total kernel number, as may be expected, because kernel number and ear diameter should increase with KRN. The ear diameter does increase with KRN for both the ZM‐BG1H1 OE and control plants, yet the control ear diameters generally lagged behind ZM‐BG1H1 OE at each KRN value (Figure S6).
Subcellular localization
Subcellular localization of ZM‐BG1H1 protein was investigated to address whether ZM‐BG1H1 protein is plasma membrane (PM) like OS‐BG1, and if so does ZM‐GOS2 PRO ectopic expression overload PM localization. Maize protoplasts were transfected with two colour markers, RFP to illuminate the nucleus and normalize expression levels, and GFP, in fusion to ZM‐BG1H1 protein or not, to probe ZM‐BG1H1 cellular location. Figure 7a‐b demonstrates the control broad cellular localization of GFP and the demarcation of the nucleus when RFP is nuclear targeted by an NLS (nuclear localization signal). In Figure 7c, the GFP is fused to the N‐terminus of ZM‐BG1H1 and ectopically expressed with ZM‐GOS2 PRO. The result demonstrates GFP is localized chiefly to the cell surface consistent with the PM. In Figure 7d, a related experiment is done except that the ZM‐BG1H1 coding region is fused instead to the N‐terminus of GFP. The result is similar, indicating that the ZM‐BG1H1 protein is itself capable of directing GFP protein to the PM regardless of whether its N‐terminus or C‐terminus is occupied by the fused GFP protein. The native ZM‐BG1H1 PRO has very low expression requiring long exposure to detect the diffuse untargeted GFP expression (Figure 7e). The native ZM‐BG1H1 promoter driving GFP::ZM‐BG1H1 fusion expression produced too low of expression to detect PM localization (not shown).
Figure 7.
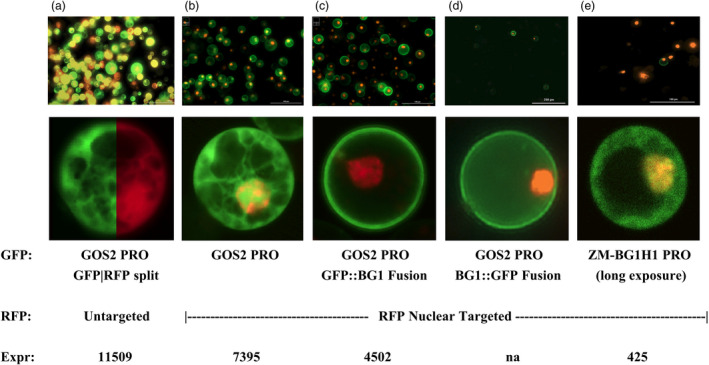
Subcellular Localization of ZM‐BG1H1 Protein and Ectopic Expression Impact. a‐e. microscopic images of protoplast transfected with various Promoter::GFP reporter gene fusions as labelled at bottom of figure. Upper set of panels show lower magnification field with multiple protoplasts in view, and lower set of panels shows single close‐up view of a representative protoplast. Most protoplast ranged 20–30 μm diameter. Green colour emanates from the GFP reporter gene, and red colour from the RFP reporter gene. Panel A lower panel, subcellular localization control. Split image of a single protoplast, with GFP (left) and RFP (right) division, revealing broadly distributed GFP and RFP, respectively, in the cell. Panels b‐e, the RFP reddish‐orange colour restricted to the nucleus using nuclear localization signals. Panels a, b and e, the GFP is broadly distributed in the cell; panels c and d, the GFP is preferentially located at the protoplast plasma membrane.
Promoter analyses among native ZM‐BG1H1 homologs
OS‐BG1 and BG1 homolog promoters were reported to possess auxin response‐related motifs (Mishra et al., 2017). We initiated a de novo search for conserved motifs found in the proximal promoter, the first 1000 nts upstream from the ATG, for each of BG1 homologs across 5 species: ZM‐BG1H1, the OS‐BG1 and BG1 homologs from Sorghum, Brachypodium and Setaria (Figure S7). There exists a well‐defined TATA box context CTATATCTTC in all the genes immediately upstream from the available 5′UTRs. There is also a conserved motif GCATTG in the 5′‐UTRs amid additional 5′UTR sequence conservation. We identified five other motifs upstream from the TATA box: CGCCAC, CCCGT, CACCC, GAAAT and GGACG. Collectively, all these seven elements are conserved in relative order, and they are within 360 nts from the TATA box of ZM‐BG1H1‐A1. Apart from the TATA box, the functions of the 6 other motifs are unknown and not believed associated with auxin. The five auxin response motifs (Mishra et al., 2017) are not among these 7 conserved motifs.
ZM‐BG1H1 allelic variation
Considering the various available genome and transcriptome diversity sources for ZM‐BG1H1, we observed at least 5 major sequence variants with possibly 8–13 minor sequence variants total. The first five variants, which we here refer to as alleles A1‐A5, are represented by high‐quality gene region sequences. Other minor rare variants existed but were deemed speculative because they are not based upon quality single‐source genomic sequences, and so are not elaborated further. These five allelic sequences presented account for 93% of the germplasm lines surveyed. Alleles A1 and A2 are found almost exclusively in Stiff Stalk germplasm and account collectively for about 44% of the germplasm surveyed. Alleles A3, A4 and A5 count for 49% of the genome surveyed and are almost entirely Non‐Stiff Stalk (Table S5). There is no indication of any presence–absence variation (PAV) at this locus. A separate earlier analysis of 416 germplasm lines (63% shared with the 582‐line survey set) using an Agilent microarray and a 60‐nt hybridization probe, also did not uncover any PAV (not shown).
All five alleles presented include a complete open reading frame, with no premature truncations or obviously defective incomplete proteins. The nucleotide identities range from 94.8% to 99.3% in the CDS. The encoded proteins are all distinct, ranging from 95.4% to 99.4% ID among themselves, and to OS‐BG1 (65.1%–66.9% ID) and to sorghum SB‐BG1 (XP_021314015.1; 77.5%–80.3% AAID; Table S5). There are 7 peptide regional differences among the alleles (Figure S8). In comparison with sorghum SB‐BG1, at 3 of 7 locations, the loss of histidine in ‘MQSHQDL’ in ZM‐BG1H1‐A2(3) and losses of ‘APAP’ and ‘YGHG’ in ZM‐BG1H1‐A1, these variations appear to be maize lineage‐specific. CDS comparisons indicate additional synonymous codon variations, and that the ZM‐BG1H1‐A1 ‘APAP’ variation is likely an SSR. Each of the 7 variant peptide regions was compared to Poaceae BG1 homolog representatives. All 7 locations were also regionally variable among the cross‐species Poaceae BG1 peptides, suggesting these variants are not likely disrupting critical conserved protein function. The patterns of variation among the seven regions across the five maize alleles suggest a history of multiple intra‐genic/inter‐allelic recombination events. The five ZM‐BG1H1 alleles were also compared in the proximal 1000 nt promoter plus 5′UTR region (Figure S9). Both the 5′UTR and promoter regions show many variations, including indels and point mutations. Yet, all five alleles possess the multi‐species conserved TATA box (CTATATCTTC), and among the 6 other motifs found above to be shared across BG1 homologs from multiple species, all are also conserved across all these five alleles, suggesting these variations are not likely to disrupt conserved promoter function.
Allelic functional differences may manifest in gene expression differences. We surveyed allelic expression multiple ways. Foremost, a set of 416 inbred lines was surveyed for V6 leaf tissue expression harvested late morning between 10 and 12 AM. Marker and pedigree analyses enabled inference of the likely IBD (identity by descent) haplotypes, and through reference genomes for lines within this 416‐inbred set, the IBD could be matched to allelic IIS (identity in state) sequences for the A1–A5 alleles. However, the IBD haplotypes including IIS alleles A1 and A2 sometimes failed to separate them, likely reflecting the resolution limitations of the employed flanking genetic markers, and so these alleles were treated together for this expression analysis. Leaf expression was detected for all 5 alleles, and although leaf‐day expression is low, there was observed little RNA expression variation between the haplotypes (Figure S10). The inbred line PH184C containing the ZM‐BG1H1‐A3 allele, the line used for transformation, was subjected to a targeted RNA profiling experiment using field‐grown samples. The plants were sampled across 11 tissues at stages V10, VT/R1 and R4, and under drought and well‐watered conditions. The average expression for each tissue is shown in Figure S11. This reveals that the ZM‐BG1H1‐A3 allele in PH184C is expressed in all tissues, and in a tissue–spatial pattern consistent with the broader tissue survey done for ZM‐BG1H1‐A1 (SS, B73) (Figure 1); for example, immature ear expression is relatively high, but leaf expression is low.
We assessed whether the ZM‐BG1H1 and ZM‐BG1H2(3) loci associated with various genetic–phenotypic intervals (QTLs, GWAS, Breeding Values, etc.). Over 3000 maize public and internal genetic intervals were searched involving traits in categories classified as Yield, Kernel, Development, Architecture, Root, Fertility and Flowering. One set involved 1860 published and curated regions (licensed copy of QTLocate™, from GeneFlow Inc.) and another involved over 1180 internally computed Corteva QTL and GWAS associations. Few regions associated with either the ZM‐BG1H1 or ZM‐BG1H2(3) loci, and there appeared to be a relative deficiency of genetic regions mapped to these loci, but the statistical significance of this deficiency is difficult to ascertain because of the heterogeneous sources of information involved.
Discussion
The OS‐BG1 overexpression study (Liu et al., 2015b) highlighted a relatively unknown plant‐specific gene family with at least one member that can improve yield in rice and Arabidopsis. We sought to determine the best maize homolog, and whether this gene possesses native allelic variations that could advance plant breeding, and especially whether ectopic overexpression in maize may improve yield. The maize BG1‐like gene family was analysed to identify the best candidate OS‐BG1 homolog, which was determined by protein similarity and gene expression patterns to be ZM‐BG1H1. BG1‐like genes are found throughout higher plants and show evidence of purifying selections, suggesting important adaptive roles (Mishra et al., 2017). We found no ZM‐BG1H1 presence–absence and copy number variations in the maize germplasm, and that while the ZM‐BG1H1 locus harbours multiple (at least 5 major) allelic structural variations, there are no apparent large functional differences among these alleles in either protein function, promoter structure or gene expression. The ZM‐BG1H1 locus chromosome region is also not a common site for high‐contrasting phenotypic variations. Other ZM‐BG1H1 variants may exist, but presently there is no indication these five native endogenous alleles could drive large functional phenotypic effects to advance maize breeding.
For biotech crop improvement, and especially for hybrid maize, ectopic expression has the sought‐after and anticipated advantages of being dominant and large effect size, unlike most native variations, especially those affecting complex traits like yield. ZM‐BG1H1 ectopic OE with the ZM‐GOS2 constitutive promoter here led to an increase in hybrid maize field grain yield per unit area. This grain yield increase was observed in elite commercial hybrid maize germplasm, in heterozygous state, in four separate events and each with similar yield enhancement levels, indicating that the yield‐enhancing effect of ZM‐BG1H1 OE is dominant, expressive, penetrant and repeatable. The yield increase was observed over multiple years, in multiple environments, ranging from well‐watered optimal to stressed conditions, spanning environments exhibiting a near two‐fold range of control yield levels. This indicates the ZM‐BG1H1 OE yield‐enhancing effect is broadly achievable across varied environments. The yield advantage percentage shows an environmental stability of about 2.5% range across various environments above 11 t/ha for control yields (Figure 3). The absolute yield gains (kg/ha) trended even higher among the highest yielding environments, indicating that ZM‐BG1H1 OE can add grain yield on top of elite germplasm grown under optimal conditions.
Many plant yield‐enhancing genes exhibit secondary effects that foil their usefulness for breeding. Prior to flowering, no substantial differences for ZM‐BG1H1 OE were observed by either field trial remote sensing (RS) or by visual observations in seedling germination, gravitropism, stand establishment, canopy health or cover or greenness, and general plant habit, plant height, leaf angle or tillering. From the flowering stage onward, increases in plant height (2.6%–3.2%) and ear height (1.5%–2.1%) were observed in both the yield trials and the observation plots. Field trials indicated small delays in tassel and ear flowering, affirmed in observation plots as a one‐day delay. Interestingly, because both the male and female flowering were similarly delayed, the critically important agronomic trait maize anthesis‐to‐silking interval (ASI) was barely affected. Since ASI famously widens under stress to negatively impact maize yield, this ASI stability is consonant with the broad environment yield performance observed for ZM‐BG1H1 overexpression. None of the varied component or secondary traits investigated showed a strong correlation to yield difference (Figure 4, Table S6).
ZM‐BG1H1 OE plants in observation plots had multiple ear–kernel phenotypic shifts, with KRN and per ear kernel number and mass all increased, with ear length and ear filled length also trending longer. In sharp contrast to the OS‐BG1 rice study, individual maize kernel average volume, weight and density all declined slightly. Increased KRN itself is expected to accommodate more kernels per ear, but kernel size normally declines with increasing KRN from ear–kernel packing constraints. KRN increase alone is not however the only factor affecting these ear and kernel changes, because even when comparing ZM‐BG1H1 OE to control for the same KRN values, all these ear–kernel patterns persisted. This indicates ZM‐BG1H1 OE plants have increased capacity to increase kernel set and overall kernel weight, but not simply due to KRN increases. These kernel number and mass increases are partially offset by the reduced individual kernel volume, weight and density, yet nonetheless leaving the net positive gain (average 2.0%) in grain weight per plant. With uniform planting densities and nearly all plants having one ear, this individual plant difference in the observation plots presumably translates to the (average 2.4%) gain in field yield per area.
ZM‐BG1H1 and OS‐BG1 both increase yield; however, the signature OS‐BG1 rice trait ‘bigger grain’ was not observed, and instead maize kernels were smaller. How can two related orthologous genes from closely related plants both produce yield gains by overexpression, yet by opposite effects on grain size? Unlike loose panicle head bearing grains such as rice, maize has a uniquely structured ear that governs seed size through ear fill packing spatial dynamics, and this ear sits mid‐plant level in the canopy. Rice is multi‐tillered with comparatively loose panicles heads at the plant top. Maize selection has strongly channelled into a single‐culm and single‐ear architecture, with all the kernels subordinate to the pollination dynamics, grain fill and packing constraints on that ear. These rice vs. maize architectural differences can dramatically affect source–sink dynamics and may explain why ZM‐BG1H1 OE manifests yield gain differently. The lower individual maize kernel weight may be a compensatory divisional response of the ZM‐BG1H1 OE plant's increased capacity to set and fill more kernels overall, and yet this is not merely a zero‐sum response, as the net harvested kernel weight per plant or area is still increased. This suggests ZM‐BG1H1 OE may have elevated the sink capacity and through which yield gain comes from the ability to capture more of an otherwise excess source capacity, thus shifting these plants a portion from sink‐limited towards source‐limited. The OS‐BG1 OE rice may have increased grain size itself as the direct manifestation of increased sink capacity. For maize, sink size may be the whole ear unit, not individual kernels. Importantly, specifically looking for maize kernel size increase would have marked this homolog overexpression test a failure to confirm. Establishing maize field yield gain first, and then working backward through maize‐specific kernel‐ear morphology, has revealed a different no‐less interesting result.
Kernel row number (KRN) is a major determinant of maize yield and has been actively selected in maize breeding, likely since emerging from its teosinte(‐like) common ancestor having presumed 2‐row KRN. Despite many genetic regions and some specific genes and their functions known to control KRN (e.g. Brommert et al., 2013; Doebley and Stec, 1991; Liu et al., 2015a; Liu et al., 2016; Lu et al., 2011), none of these yet appear related to ZM‐BG1H1 or its chromosome region. The low structure–expression functional variation and strong heterotic group segregation at the ZM‐BG1H1 locus region suggests a history of selective sweep, but it is unknown whether this relates to KRN. Weak FEA3 alleles that moderately increase KRN without distorting ear structure can increase maize yield (Je et al., 2016). Perhaps ZM‐BG1H1 OE acts similarly, although as noted its effect is not primarily due to KRN.
The physiological mechanisms leading to BG1 yield gains in rice and maize are unresolved. The observed physiological changes in ZM‐BG1H1 OE plants are considered. Flowering time delay is unlikely related however because a delay typically jeopardizes yield (Baum et al., 2019). However, as floral transition occurs in the shoot apical meristem (SAM), and SAM is also implicated in determining KRN, perhaps subtle changes in SAM development may both (independently) underlie the increased KRN and delayed flowering. Auxin is long known to play a critical role in SAM development and differentiation. Regardless whether auxin mediates these yield increases, auxin has been extensively associated with crop productivity (see Rasmussen, 2001). This study did not focus on the basipetal auxin transport hypothesis nor on benchmarking the ZM‐BG1H1 OE plants against all the rice OS‐BG1 OE observations. Some OS‐BG1 OE rice effects were not observed, such as reduced gravitropism or stem/leaf angle change; however, both the rice and maize BG1 OE plants were taller. Maize height and ear height have slight downward trends in maize breeding (Duvick, 2005); however, height can be associated with increased yield (e.g. Yin et al., 2011), although breeders typically manage plant and ear height downward to control lodging and harvestability issues. Nonetheless, other reported maize transgenes with diverse molecular–physiological functions have also increased yield and been associated with plant or ear height increases (Habben et al., 2014; Shi et al., 2015; Wu et al., 2019). As ZM‐BG1H1 OE height increases occur after canopy closure, a quickening of the normally suppressed shade avoidance response may be possible, but SAR typically counteracts crop yield. Apical dominance, governed partly by basipetal auxin transport (reviewed in Tamas, 1995), is an important process breeding has altered to give rise to the characteristic high‐yielding modern maize architecture, particularly reduced tassel over ear dominance (Duvick, 2005), increased upper over lower ear dominance (Wills et al., 2013) and single culm over multi‐tiller dominance (Doebley et al., 1997). There is no indication any of these factors are altered by ZM‐BG1H1 OE.
OS‐BG1 is a plasma membrane‐localized protein believed involved in auxin transport or its regulation (Liu et al., 2015b). In Mishra et al., 2017, the BG1 family is instead presented as nuclear located, with Ath‐BG2 experimentally shown to be. Nonetheless, in this study ZM‐BG1H1 appears to be plasma membrane located like OS‐BG1. The ZM‐BG1H1‐GFP fusion was able to direct protein to the PM whether the GFP fusion was located at the N‐ or C‐terminus of ZM‐BG1H1, supporting non‐transmembrane affiliation, and not likely dependent upon translation‐coupled translocation into the membrane (Cymer et al., 2015). There is also no indication that the (ZM‐GOS2 promoter) ectopic expression causes ‘spillover’ redirection of ZM‐BG1H1 protein into other cellular compartments that might then lead to neomorphic side‐effect behaviours. The sites that the ZM‐BG1H1 protein interacts in the PM are unknown, but it appears they are not readily saturated.
These ZM‐BG1H1 overexpression alleles represent a large step change relative to the known native maize allelic structure–expression functional diversity. We do not yet know whether it is the level of expression and/or specific spatial–temporal expression conferred by the ZM‐GOS2 promoter that contributes to the positive yield gain effect; however, the rice OS‐BG1 study demonstrates that two different promoters can boost yield. Regulatory and plant hormone‐related genes commonly have a constrained ideal zone for expression level to produce a healthy high‐performing plant, especially so in elite germplasm. The fact that ZM‐BG1H1 ectopic expression preserves plant health and increases yield indicates the ZM‐BG1H1 gene product's beneficial expression range is quite broad. Given the considerable peptide diversity space spanning between ZM‐BG1H1 and OS‐BG1, we predict that the other maize gene family members, especially the ZM‐BG1H2(3) gene pair, along with numerous other species BG1 homologs, may function to produce similar yield promoting effects in maize.
Methods
Native gene and expression analyses
The BG1‐related genes described herein were derived from public sources, and the loci are identified in the manuscript. For maize, a chief source was the B73 RefGen v2, its associated gene models and AGP B73v4 genome drafts. ZM‐BG1H1 allelic sequences derived from public and proprietary sources, including high‐quality genome drafts for key reference lines containing alleles A1‐A5. Corteva Agriscience's heritage organization DuPont Pioneer had generated extensive genomic, genetic marker, haplotype diversity and gene expression data over many years using a variety of technology platforms. These were used for the gene structural and tissue–spatial gene expression analyses of the BG1 family and ZM‐BG1H1 allelic variants.
Gene expression cassette and plant transformation
The gene expression cassette possessed the following features: ZM‐GOS1 PRO; ZM‐GOS2 5UTR1; ZM‐GOS2 Intron1; ZM‐GOS1 5UTR2; nt C to create a NCO1 site at the start codon; ZM‐BG1H1 (MOD1) ORF; and OS‐T28 TERM (MOD1) (derived from rice locus LOC_Os03g60090.1). This cassette sequence (3672 bp) is provided (Figure S12). The ZM‐BG1H1 ORF sequence contained two purposeful amino acid changes and (R119S and D261N) amid 24.5% nt changes from the original ORF. The isolated ZM‐GOS2 promoter is described (Barbour et al., 2003; Shi et al., 2017). The vector backbone and helper vectors for Agrobacterium transformation follow closely those described (Anand et al., 2018), and the final vector was assembled by Gateway® (Invitrogen) technology. The elite proprietary maize germplasm NSS inbred line PH184C was transformed with techniques described using maize immature embryos and a CHV (cohabitating vector) (Cho et al., 2014; Zhi et al., 2015). Transformants were selected with the PMI marker gene and identified by PCR fragment amplification (Zhi et al., 2015). Single‐copy quality events were propagated in the greenhouse, transgene expression verified by qRT‐PCR and transgene chromosomal locations identified by Southern‐by‐Sequencing (Zastrow‐Hayes et al., 2015). After self‐fertilization, events were selected for genetic nursery, where they were top crossed to PHW3G inbred for T3 generation hybrid seed production. Transgene ZM‐BG1H1 (MOD1) and endogenous ZM‐BG1H1 genes relative expression analyses were assayed by qRT‐PCR analysis.
Maize hybrid yield testing
Two years of field trials was conducted in North America. These trials are like other published transgene or gene edit yield trials by DuPont Pioneer (e.g. Shi et al., 2017; Shi et al., 2015; Wu et al., 2019; Zastrow‐Hayes et al., 2015). All field test comparisons were made between the heterozygous F1 transgenic single hybrid (PH184C × PHW3G) for each of four transgenic events and a control in that same hybrid genetic background. Yield was calculated using 56 lb/bu (25.4 kg/bu). These early discovery evaluation yield trails involved numerous small plot studies for each event and control, with replicates at the same location, and across locations, environments and years. The total area involved was 0.234 hectares (0.579 acres). See supplement for more detailed descriptions of the field locations, including the managed stress sites, plot sizes and layout and statistics treatments, and definitions of yield and other field assayed agronomic traits.
Plant morphometric observations
Hybrid seed for the four events was grown in the field at Johnston Iowa in year 3 (Yr3‐Obs). Plant photography, canopy appearance, architecture, plant height, ear height, silking and pollen shed flowering times were recorded. The Yr3‐Obs plots were also used for ear and kernel morphometric traits, including KRN, ear length, ear fill length, ear diameter and kernel number, size, weight and density. Other assays included kernel size and density metrics on ‘parent’ field trial remnant hybrid seed, and seedling gravitropism assays.
Subcellular localization
Maize protoplasts were used for ZM‐BG1H1 peptide subcellular location. This involved TagRFP (Evrogen) red fluorescence protein colour, when in conjunction with the VIRD2 NLS demarcating nuclei. The plasma membrane vs cytoplasm test colour marker gene was AcGFP1 (Takara Bio USA, Inc.) monomeric green fluorescent protein (GFP), used in various fusion arrangements with the ZM‐BG1H1 ORF, and the ZM‐BG1H1 or ZM‐GOS2 promoters. Maize protoplasts were isolated using a protocol adapted from publications by the Jen Sheen laboratory (e.g. see Yoo et al., 2007), transfection used the PEG method, with gene expression detected with microscopic spectrophotometric analyses. [For more details of all methods, see the Data S1].
Conflict of interest
The authors are scientists employed at Corteva Agriscience, a world leader in developing agricultural products, with the goal to ‘enrich the lives of those who produce and those who consume, ensuring progress for generations to come’. Patent applications covering this work have been filed that include one or more of the listed authors. Materials reported in this paper may be subject to third‐party ownership and/or to governmental regulations. Availability of materials reported in this paper to academic investigators for non‐commercial research purposes under an applicable material transfer agreement will be subject to the requisite permission from any third‐party owners of all or parts of the materials and to governmental regulation considerations. Obtaining the applicable permission from such third‐party owners will be the responsibility of the requestor. Transgenic materials reported in this paper would only be made available if in full accordance with all applicable governmental regulations.
Author contributions
C.R.S. performed manuscript writing and coordination, gene family–orthology, genomics and transcriptomics analyses, targeted field and phenotypic analyses; B.P.W. performed field testing design and plant performance analyses; K.S.R. performed transgene expression analyses and plant and kernel morphometrics; S.E.A. and M.J.F. performed protoplast expression and subcellular localization; B.S. performed project concept and construct testing initiation, J.W. and W.W. involved in the project management, and J.E.H. project supervisor.
Supporting information
Figure S1 ZM‐BG1H1 and ZM‐BG1H2(3) Gene Expression in 19 Tissues.
Figure S2 ZM‐BG1H1 and ZM‐BG1H2(3) Leaf diurnal gene expression.
Figure S3 Relative ZM‐GOS2 vs ZM‐BG1H1 native gene expression across tissues.
Figure S4 Hybrid parent seed volume, weight, and density.
Figure S5 Ear and kernel differences at same KRN value.
Figure S6 Ear diameter per KRN with BG1 exceeding control.
Figure S7 Promoter motif analyses in ZM‐BG1H1 alleles and among four species homologs.
Figure S8 ZM‐BG1H1 Alleles A1‐A5 Peptide Alignments.
Figure S9 Maize ZM‐BG1H1 five alleles promoter regions with conserved motifs.
Figure S10 ZM‐BG1H1 Alleles A1‐A5 leaf gene expression.
Figure S11 ZM‐BG1H1‐A3 mRNA expression across tissues in field‐grown transformation line PH184C.
Figure S12 Gene expression cassette used for ZM‐BG1H1 (MOD1) Overexpression.
Table S1 Maize BG1 and BG1‐like family members.
Table S2 Endogenous and transgene ZM‐BG1H1 gene expression levels compared.
Table S3 Flowering time differences for ZM‐BG1H1 OE plants.
Table S4 Ear and kernel metrics data for Figures 5 and S5.
Table S5 Maize allelic diversity and heterotic group relationships at ZM‐BG1H1 Locus.
Table S6 Summary of all trait observations.
Data S1 Supplemental experimental procedures.
Table S4, S6
Acknowledgments
Certain aspects of his work were part of a collaboration between Corteva Agriscience and Institute of Genetics and Developmental Biology (IGDB). We thank Professor Chengcai Chu at IGDB for his input. We thank many Corteva researchers supporting this project including: Ashley Burns, David Habier, Deb Wetterberg, Dennis O'Neill, Hua Mo, Mary Rupe, Jesse Ourada, Jing Wang, Julie Vogel, Katherine Thilges, Elizabeth Stephenson, Mary Trimnell, Bill Van Zante, Cory Robinson, Anthony Blake, Michelle Paulsen, Norbert Brugiere, Olga Danilevskaya, Renee Lafitte, Salim Hakimi, Terry Hu and Tom Greene.
Simmons, C. R. , Weers, B. P. , Reimann, K. S. , Abbitt, S. E. , Frank, M. J. , Wang, W. , Wu, J. , Shen, B. and Habben, J. E. (2020) Maize BIG GRAIN1 homolog overexpression increases maize grain yield. Plant Biotechnol. J., 10.1111/pbi.13392
References
- Anand, A. , Bass, S.H. , Wu, E. , Wang, N. , McBride, K.E. , Annaluru, N. , Miller, M. et al. (2018) An improved ternary vector system for Agrobacterium‐mediated rapid maize transformation. Plant Mol. Biol. 97, 187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour, E. , Meyer, T.E. and Saad, M.E. (2003) Maize GOS‐2 Promoters. United States Patent US 6,504,083, B1.
- Baum, M.E. , Archontoulis, S.V. and Licht, M.A. (2019) Planting date, hybrid maturity, and weather effects on maize yield and crop stage. Agronomy J. 3, 303–313. [Google Scholar]
- Bommert, P. , Nagasawa, N.S. and Jackson, D. (2013) Quantitative variation in maize kernel row number is controlled by the FASCIATED EAR2 locus. Nat. Genet. 45, 334–337. [DOI] [PubMed] [Google Scholar]
- Castiglioni, P. , Warner, D. , Bensen, R.J. , Anstrom, D.C. , Harrison, J. , Stoecker, M. , Abad, M. et al. (2008) Bacterial RNA chaperones confer abiotic stress tolerance in plants and improved grain yield in maize under water‐limited conditions. Plant Physiol. 147, 446–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, M.‐J. , Wu, E. , Kwan, J. , Yu, M. , Banh, J. , Linn, W. , Anand, A. et al. (2014) Agrobacterium mediated high‐frequency transformation of an elite commercial maize (Zea mays L.) inbred line. Plant Cell Rep. 33, 1767–1777. [DOI] [PubMed] [Google Scholar]
- Cymer, F. , von Heijne, G. and White, S.H. (2015) Mechanisms of integral membrane protein insertion and folding. J. Mol. Biol. 427, 999–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J. and Stec, A. (1991) Genetic analysis of the morphological differences between maize and teosinte. Genetics, 129, 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J. , Stec, A. and Hubbard, L. (1997) The evolution of apical dominance in maize. Nature, 386, 485–488. [DOI] [PubMed] [Google Scholar]
- Duvick, D.N. (2005) The contribution of breeding to yield advances in maize (Zea mays L). Adv. Agronomy, 86, 83–145. [Google Scholar]
- Fox, T. , DeBruin, J. , Haug Collet, K. , Trimnell, M. , Clapp, J. , Leonard, A. , Li, B. et al. (2017) A single point mutation in Ms44 results in dominant male sterility and improves nitrogen use efficiency in maize. Plant Biotechnol. J. 15, 942–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey, K.J. (2000) A National Strategy for Plant Breeding in the United States. In: Public‐Private Collaboration in Agricultural Research: New Institutional Arrangements and Economic Implications ( Fuglie, K.O. and Schimmelpfennig, D.E. eds), pp 77–98. Ames: Iowa State University Press. pp. 356. ISBN 0‐8138‐2789‐2. [Google Scholar]
- Guo, M. , Rupe, M.A. , Wei, J. , Winkler, C. , Goncalves‐Butruille, M. , Weers, B.P. , Cerwick, S.F. et al. (2014) Maize ARGOS1 (ZAR1) transgenic alleles increase hybrid maize yield. J. Exp. Bot. 65, 249–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habben, J.E. , Bao, X. , Bate, N.J. , DeBruin, J.L. , Dolan, D. , Hasegawa, D. , Helentjaris, T.G. et al. (2014) Transgenic alteration of ethylene biosynthesis increases grain yield in maize under field drought‐stress conditions. Plant Biotechnol. J. 12, 685–693. [DOI] [PubMed] [Google Scholar]
- Je, B.I. , Gruel, J. , Lee, Y.K. , Bommert, P. , Arevalo, E.D. , Eveland, A.L. , Wu, Q. et al. (2016) Signaling from maize organ primordia via FASCIATED EAR3 regulates stem cell proliferation and yield traits. Nat. Genet. 48, 785–791. [DOI] [PubMed] [Google Scholar]
- Liang, Z. and Schnable, J.C. (2018) Functional divergence between subgenomes and gene pairs after whole genome duplications. Mol. Plant, 11, 388–397. [DOI] [PubMed] [Google Scholar]
- Liu, L. , Du, Y. , Shen, X. , Li, M. , Sun, W. , Huang, J. , Liu, Z. et al. (2015a) KRN4 controls quantitative variation in maize kernel row number. PLOS Genet. 11, e1005670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L. , Tong, H. , Xiao, Y. , Che, R. , Xu, F. , Hu, B. , Liang, C. et al. (2015b) Activation of Big Grain1 significantly improves grain size by regulating auxin transport in rice. Proc. Natl. Acad. Sci USA, 112, 11102–11107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C. , Zhou, Q. , Dong, L. , Wang, H. , Liu, F. , Weng, J. , Li, X. and et al. (2016) Genetic architecture of the maize kernel row number revealed by combining QTL mapping using a high‐density genetic map and bulked segregant RNA sequencing. BMC Genom. 17(1), 915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, M. , Xie, C.‐X. , Li, X.‐H. , Hao, Z.‐F. , Li, M.‐S. , Weng, J. , Zhang, D.‐G. et al. (2011) Mapping of quantitative trait loci for kernel row number in maize across seven environments. Mol. Breed. 28, 143–152. [Google Scholar]
- Mishra, B.S. , Jamsheer, K.M. , Singh, D. , Sharma, M. and Laxmi, A. (2017) Genome‐wide identification and expression, protein‐protein interaction and evolutionary analysis of the seed plant‐specific BIG GRAIN and BIG GRAIN LIKE gene family. Front. Plant Sci. 8, 1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemali, K.S. , Bonin, C. , Dohleman, F.G. , Stephens, M. , Reeves, W.R. , Nelson, D.E. , Castiglioni, P. et al. (2015) Physiological responses related to increased grain yield under drought in the first biotechnology‐derived drought‐tolerant maize. Plant Cell Environ. 38, 1866–1880. [DOI] [PubMed] [Google Scholar]
- Nuccio, M.L. , Paul, M. , Bate, N.J. , Cohn, J. and Cutler, S.R. (2018) Where are the drought tolerant crops? An assessment of more than two decades of plant biotechnology effort in crop improvement. Plant Sci. 273, 110–119. [DOI] [PubMed] [Google Scholar]
- Rasmussen, N. (2001) Plant hormones in war and peace: science, industry, and government in the development of herbicides in 1940s America. Isis, 92, 291–316. [DOI] [PubMed] [Google Scholar]
- Schnable, J.C. , Springer, N.M. and Freeling, M. (2011) Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc. Natl. Acad. Sci. USA, 108, 4069–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekhon, R.S. , Lin, H. , Childs, K.L. , Hansey, C.N. , Buell, C.R. , De Leon, N. and Kaeppler, S.M. (2011) Genome‐wide atlas of transcription during maize development. Plant J. 66, 553–563. [DOI] [PubMed] [Google Scholar]
- Shi, J. , Habben, J.E. , Archibald, R.L. , Drummond, B.J. , Chamberlin, M.A. , Williams, R.W. , Lafitte, H.R. and et al. (2015) Overexpression of ARGOS genes modifies plant sensitivity to ethylene, leading to improved drought tolerance in both Arabidopsis and maize. Plant Physiol. 169, 266–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J. , Gao, H. , Wang, H. , Lafitte, H.R. , Archibald, R.L. , Yang, M. , Hakimi, S.M. et al. (2017) ARGOS8 variants generated by CRISPR‐Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 15, 207–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigonová, Z. , Lai, J. , Ma, J. , Ramakrishna, W. , Llaca, V. , Bennetzen, J.L. and Messing, J. (2004) Close split of sorghum and maize genome progenitors. Genome Res. 14, 1916–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamas, I.A. (1995) Hormonal regulation of apical dominance. In: Plant Hormones ( Davies, P.J. ed). Dordrecht: Springer. Print ISBN 978‐0‐7923‐2985‐5; Online ISBN 978‐94‐011‐0473‐9; doi: 10.1007/978-94-011-0473-9_27. [DOI] [Google Scholar]
- Wills, D.M. , Whipple, C.J. , Takuno, S. , Kursel, L.E. , Shannon, L.M. , Ross‐Ibarra, J. and Doebley, J.F. (2013) From many, one: genetic control of prolificacy during maize domestication. PLoS Genet. 9(6), e1003604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J. , Lawit, S.J. , Weers, B. , Sun, J. , Mongar, N. , Van Hemert, J. , Melo, R. et al. (2019) Overexpression of zmm28 increases maize grain yield in the field. Proc. Natl. Acad. Sci. USA, 116, 23850–13858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, X. , Mcclue, A. , Jaja, N. , Tyler, D.D. and Hayes, R.M. (2011) In‐season prediction of corn yield using plant height under major production systems. Agronomy J. 103, 923–929. [Google Scholar]
- Yoo, S.D. , Cho, Y.H. and Sheen, J. (2007) Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protocols, 2, 1565–1575. [DOI] [PubMed] [Google Scholar]
- Zastrow‐Hayes, G.M. , Lin, H. , Sigmund, A.L. , Hoffman, J.L. , Alarcon, C.M. , Hayes, K.R. , Richmond, T.A. et al. (2015) Southern‐by‐sequencing: a robust screening approach for molecular characterization of genetically modified crops. Plant Genome, 8, 1–15. [DOI] [PubMed] [Google Scholar]
- Zhi, L. , TeRonde, S. , Meyer, S. , Arling, M.L. , Register, J.C. III , Zhao, Z.‐Y. , Jones, T.J. and et al. (2015) Effect of Agrobacterium strain and plasmid copy number on transformation frequency, event quality and usable event quality in an elite maize cultivar. Plant Cell Rep. 34, 745–754. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 ZM‐BG1H1 and ZM‐BG1H2(3) Gene Expression in 19 Tissues.
Figure S2 ZM‐BG1H1 and ZM‐BG1H2(3) Leaf diurnal gene expression.
Figure S3 Relative ZM‐GOS2 vs ZM‐BG1H1 native gene expression across tissues.
Figure S4 Hybrid parent seed volume, weight, and density.
Figure S5 Ear and kernel differences at same KRN value.
Figure S6 Ear diameter per KRN with BG1 exceeding control.
Figure S7 Promoter motif analyses in ZM‐BG1H1 alleles and among four species homologs.
Figure S8 ZM‐BG1H1 Alleles A1‐A5 Peptide Alignments.
Figure S9 Maize ZM‐BG1H1 five alleles promoter regions with conserved motifs.
Figure S10 ZM‐BG1H1 Alleles A1‐A5 leaf gene expression.
Figure S11 ZM‐BG1H1‐A3 mRNA expression across tissues in field‐grown transformation line PH184C.
Figure S12 Gene expression cassette used for ZM‐BG1H1 (MOD1) Overexpression.
Table S1 Maize BG1 and BG1‐like family members.
Table S2 Endogenous and transgene ZM‐BG1H1 gene expression levels compared.
Table S3 Flowering time differences for ZM‐BG1H1 OE plants.
Table S4 Ear and kernel metrics data for Figures 5 and S5.
Table S5 Maize allelic diversity and heterotic group relationships at ZM‐BG1H1 Locus.
Table S6 Summary of all trait observations.
Data S1 Supplemental experimental procedures.
Table S4, S6


