Abstract
Long non-coding RNAs (lncRNAs) regulate diverse physiological and pathological processes via post-transcriptional, post-translational, and epigenetic mechanisms. They are also involved in tumor initiation, progression, and metastasis by functioning as key players in the tumor microenvironment. Cancer-associated fibroblasts (CAFs) promote tumor initiation, progression, metastasis, drug resistance, and immunosuppression, which can be modulated by lncRNAs. LncRNAs regulate the intrinsic properties of CAFs or cancer cells intracellularly or function extracellularly through exosomal secretion. In-depth studies on the mechanisms of lncRNA functions will enable their clinical use as diagnosis/prognosis markers and therapeutic targets in cancer treatment.
Keywords: long non-coding RNAs, cancer-associated fibroblasts, tumor microenvironment
1. Introduction
With the development of next-generation sequencing technology, tens of thousands of long non-coding RNAs (lncRNAs) have been discovered. According to LNCipedia, as of September 2020, 127,802 transcripts and 56,946 lncRNA genes have been annotated in the human genome (https://hg38.lncipedia.org/). LncRNAs are transcripts longer than 200 nucleotides in length lacking protein-coding ability that function as crucial physiological and pathological regulators via post-transcriptional, post-translational, and epigenetic mechanisms. Growing bodies of evidence indicate that numerous lncRNAs are also involved in tumor initiation, progression, and metastasis. In particular, the role of lncRNAs in the tumor microenvironment is rapidly emerging. This review focuses on the regulation, function, mechanisms of action, and clinical implications of lncRNAs in cancer-associated fibroblasts (CAFs), the major components of the tumor microenvironment.
2. Functions of lncRNAs
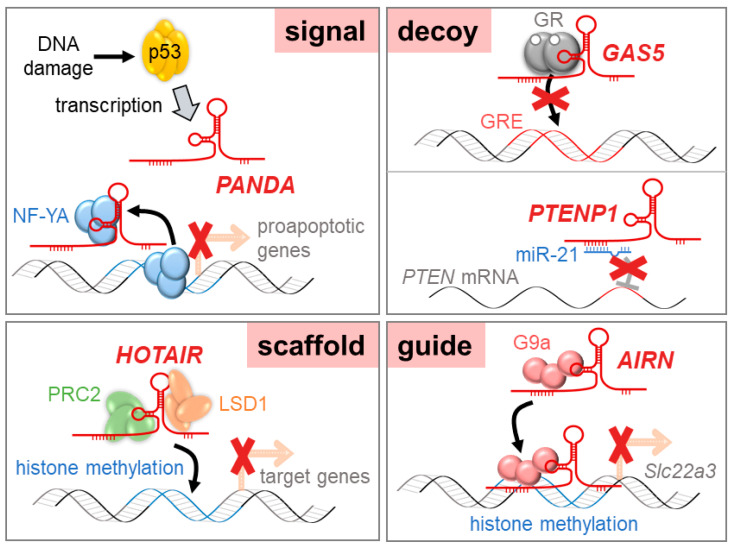
LncRNAs play crucial roles in many biological and cellular processes and are related to diseases including cancer. They function through the direct or indirect interaction with genomic DNA, mRNA, microRNAs (miRNAs), and proteins; however, the exact molecular mechanisms underlying lncRNA functions are largely unknown. Unlike protein-coding genes or miRNAs, lncRNAs have no particular sequence or conserved secondary structures to deduce their functions. However, even though lncRNAs do not share a common working mechanism, lncRNA functions can be categorized as signals, decoys, scaffolds, or guides depending on the relationship with the interacting partner.
2.1. Signal lncRNAs
Signal lncRNAs, serving as “molecular signals”, play important roles in signal regulation and response to various stimuli. These types of lncRNAs are expressed in a spatial- and temporal-specific pattern. KCNQ1 overlapping transcript 1 (KCNQ1OT1) is known to act as a signal lncRNA by interacting with histone methyltransferase G9a and polycomb repressive complex 2 (PRC2). Through the recruitment of chromatin remodeling complex, KCNQ1OT1 controls lineage-specific transcriptional silencing during embryonic liver development [1]. The p21-associated ncRNA DNA damage-activated (PANDA) lncRNA is induced in a p53-dependent manner in osteosarcoma cells. When activated by DNA damage, p53 binds to the CDKN1A locus and transactivates PANDA. PANDA then interacts with the transcription factor NF-YA to inhibit the expression of proapoptotic genes [2].
2.2. Decoy lncRNAs
Decoy lncRNAs restrict the function of regulatory factors by presenting decoy-binding sites. These lncRNAs modulate transcription by secluding regulatory factors such as transcription factors, enzymes, and miRNAs. Telomeric repeat-containing RNA (TERRA) has been shown to form an integral part of telomeric heterochromatin. TERRA interacts with both telomerase RNA hTR and telomerase reverse transcriptase protein TERT and acts as a competitive inhibitor of telomerase [3]. Growth arrest-specific 5 (GAS5), a lncRNA with hairpin structures, accumulates during nutrient- or growth factor-deprivation. GAS5 competes with glucocorticoid-response elements in binding to glucocorticoid receptors, thereby suppressing glucocorticoid-mediated transcription [4]. In addition, PTEN pseudogene 1 (PTENP1) has been reported to function as a competing endogenous RNA (ceRNA) for miRNAs. PTENP1 upregulates PTEN expression by sponging PTEN-targeting miRNAs (e.g., miR-17, miR-21, miR-214, miR-19, and miR-26) [5], leading to tumor suppression [6].
2.3. Scaffolds lncRNAs
LncRNAs can act as molecular scaffolds bringing two or more proteins together to form nucleoprotein complexes and be involved in gene activation, gene repression, and chromatin modification [7]. For example, HOX transcript antisense intergenic RNA (HOTAIR) serves as a scaffold for two distinct histone modification complexes. It was shown that the 5′-domain of HOTAIR binds to PRC2 to promote gene repression, and the 3′-domain binds to the LSD1/coREST/REST complex, which demethylates lysine 4 of histone H3 (H3K4) to repress gene activation [8]. In glioblastoma, EGFR-induced lncRNA NEAT1 interacts with EZH2 on the promoter regions of negative regulators (CTNNBIP1, GSK3B, and AXIN2) in the Wnt/β-catenin pathway, which leads to tumor progression [9]. Moreover, antisense non-coding RNA at the INK4 locus (ANRIL) is upregulated in adult T-cell leukemia and associates with EZH2 and the RelA/p65 subunit of NFκB, which enhances NFκB signaling and promotes cancer cell proliferation. An association between ANRIL and EZH2 also increases histone methylation on the promoter region of p21, which causes transcriptional silencing of p21, a cell cycle inhibitor [10].
2.4. Guide lncRNAs
Certain types of lncRNAs interact with target molecules and guide them to the proper chromosomal localization. Guide lncRNAs regulate the transcription of target genes at their genomic loci. During the mouse embryonic development of the heart and body wall, fetal-lethal non-coding developmental regulatory RNA (Fendrr) guides PRC2 to promoter regions of target genes (transcription factors for mesoderm differentiation) to increase H3K4 trimethylation, leading to attenuation of target gene expression [11]. In mouse placenta, antisense of the IGF2R non-protein coding RNA (AIRN) has been shown to silence the transcription of cis-linked genes via chromatin interaction at their promoter regions. Accumulated AIRN on the Slc22a3 promoter recruits G9a, which induces targeted histone methylation and allelic silencing [12]. LncRNA functions are shown schematically in Figure 1.
Figure 1.
Diverse functional mechanisms of long non-coding RNAs (lncRNAs).
3. Cancer-Associated Fibroblasts (CAFs)
The tumor microenvironment provides a tumor with a supportive niche to promote tumor progression and metastasis. Among the components of the tumor microenvironment are CAFs, lymphocytes, macrophages, and vascular endothelial cells, as well as the extracellular matrix. Stromal CAFs are major players in the tumor microenvironment that facilitate tumor progression and metastasis via direct or indirect interactions with neighboring cancer cells.
3.1. Origins of CAFs
Quiescent fibroblasts can differentiate into CAFs upon stimulation by diverse external cues such as cytokines/chemokines, growth factors, reactive oxygen species, hypoxia, and non-coding RNAs. TGF-β, a strong inducer of desmoplastic reaction and fibrosis, activates fibroblasts in pancreatic adenocarcinoma [13]. TGF-β stimulates the expression of myofibroblast markers and metabolic reprogramming in stromal fibroblasts of breast cancer [14]. Pro-inflammatory signals from immune cells and tumor cells educate normal fibroblasts into becoming CAFs via IL-1β and the NFκB pathway [15]. In prostate cancer, TGF-β derived from cancer cells activates fibroblasts via NADPH oxidase 4-mediated reactive oxygen species signaling [16]. Hypoxia causes the epigenetic reprogramming of breast fibroblasts, resulting in them gaining CAF-like pro-glycolytic phenotypes [17]. In addition, numerous miRNAs have been reported to be involved in the differentiation of normal fibroblasts into CAFs [18].
Besides simply differentiating from normal fibroblasts, CAFs are able to originate from different types of cells residing in the tumor microenvironment (e.g., epithelial cells, endothelial cells, pericytes, and adipocytes) or migrating from distant sites (e.g., mesenchymal stem cells) [19,20,21]. Through the epithelial-to-mesenchymal transition (EMT) process, epithelial cells adjacent to cancer cells can differentiate into CAFs [22,23]. TGF-β-induced endothelial-to-mesenchymal transition in endothelial cells also contributes to the accumulation of CAFs in the tumor microenvironment [24]. Vascular pericytes stimulated by PDGF-BB acquire stromal fibroblast features [25]. Even adipose tissue-derived stem cells around breast cancer cells differentiate toward CAFs in response to TGF-β and Smad3 signaling [26].
Cells recruited from outside the tumor microenvironment can be converted to CAFs. In gastric cancer, it was shown that quite a significant population of CAFs originated from bone marrow-derived mesenchymal stem cells [27]. SDF-1α/CXCR4 and TGF-β signaling promote the differentiation of bone marrow-derived mesenchymal stem cells into myofibroblasts, which are then recruited to the tumor microenvironment. In prostate cancer, mesenchymal stem cells are recruited from bone marrow contact with prostate cancer cells in the tumor microenvironment and then trans-differentiated into CAF-like cells via TGF-β signaling [28]. CAFs can also derive from hematopoietic stem cells [29], which create a pro-tumorigenic microenvironment [30].
3.2. Effects of CAFs on Cancer Development
CAFs are involved in the entire cancer developmental process, from tumorigenesis to progression and metastasis. Most CAFs exert pro-tumorigenic effects, while certain subpopulations of CAFs have anti-tumorigenic effects [19].
CAFs promote tumor initiation and proliferation by secreting growth factors and cytokines. CAFs isolated from ovarian cancer patients promote cancer cell proliferation through the paracrine secretion of FGF-1, which is mediated by FGFR4 [31]. CAF-derived HGF also activates the proliferation of ovarian cancer cells thorough c-Met/PI3K/Akt signaling [32]. In endometrial cancer, CAFs promote cancer cell proliferation via the secretion of CXCL12 or IL-6 [33,34]. CAF-derived CCL2 stimulates the sphere-forming activity and self-renewal of breast cancer stem cells through NOTCH signaling [35]. In non-small cell lung cancer, CAFs promote the growth and stemness of lung cancer stem cells via IGF-II secretion and Nanog expression [36].
CAFs in the tumor microenvironment promote the progression of tumors in various ways. CAFs secrete IL-6 and activate STAT3 to induce the EMT, migration, and invasion of bladder cancer cells [37]. CAF-derived IL-6 also induces EMT, migration, and clonogenicity in esophageal adenocarcinoma [38]. CAFs promote the EMT of recipient lung cancer cells through the exosomal delivery of SNAIL, an EMT-inducing transcription factor [39]. The transfer of miR-181d-5p from CAFs to breast cancer cells via exosomes then induces EMT by the direct targeting of CDX2, a transcription factor for HOXA5 [40]. TGF-β from CAFs promotes EMT and invasion in bladder cancer cells through the regulation of lncRNA ZEB2NAT [41], which will be described in more detail below. Consequently, CAFs can induce cancer cell invasion through direct physical interaction [42].
Galectin-1 originating from CAFs in gastric cancer promotes proliferation, migration, and tube formation of vascular endothelial cells and enhanced tumor angiogenesis in vivo [43]. The upregulation of WNT2 in colorectal CAFs stimulates the migration and invasion of endothelial cells and promotes angiogenesis through the production of pro-angiogenic factors [44]. VEGF-bound small extracellular vesicles are secreted by CAFs to enhance tumor angiogenesis [45]. In addition, CAFs induce biomechanical deformation of the extracellular matrix and promote tumor-supportive vascularization [46].
CAFs also modulate glycolytic metabolism in cancer cells. Downregulation of focal adhesion kinase in CAFs increases CCL6 and CCL12 expression, leading to elevated glycolysis in cancer cells [47]. CAFs educated by neighboring cancer cells reversely secrete cytokines to stimulate glycogen metabolism in cancer cells, which accelerates energy production and tumor progression [48]. Epigenetic changes in prostate CAFs drive the synthesis of glutamine, which is then transported to adjacent cancer cells to promote energy production, differentiation, and the aggressiveness of prostate cancer cells [49]. Interestingly, CAFs have been reported to transfer their own mitochondrial DNA via exosomes or mitochondria via intercellular bridges to adjacent cancer cells, resulting in metabolic reprogramming and enhanced malignancy [50,51].
The tumor microenvironment provides cancer cells with shelter from the attack of anticancer drugs [52]. The interplay between CAFs and cancer cells contributes significantly to drug resistance. CAFs reduce cisplatin-induced apoptosis in bladder cancer cells by activating signaling cascades, leading to Bcl-2 upregulation [53]. CAFs secrete IL-6, which activates the JAK/STAT3 pathway and reduces the susceptibility of gastric cancer cells to 5-fluorouracil [54]. In lung and ovarian cancers, CAF-derived HGF activates PI3K/Akt signaling and suppresses the paclitaxel-induced apoptosis of cancer cells [32,55]. Neuregulin 1 from CAFs induces HER3 activation in prostate cancer cells, which is responsible for reduced sensitivity to androgen deprivation therapy [56]. Moreover, CAFs release extracellular vesicles containing Annexin A6, which induces FAK-YAP activation and drug resistance in gastric cancer cells [57].
In addition, CAFs are involved in immunosuppression and tumor-promoting inflammation through interactions with other stromal cells. In breast cancer, the glycoprotein Chitinase-3-like-1 secreted from CAFs enhances the recruitment and M2-type polarization of macrophages, leading to immunosuppression and the promotion of tumor growth [58]. In lung squamous cell carcinoma, CAFs recruit monocytes to the tumor microenvironment by secreting CCL2 and facilitate their differentiation into immunosuppressive myeloid-derived suppressor cells [59]. Furthermore, CAF-driven antigen presentation leads to the depletion of CD8+ T cells via PD-L2 and FasL within the tumor microenvironment [60].
4. LncRNAs that Regulate the Interaction between CAFs and Cancer Cells
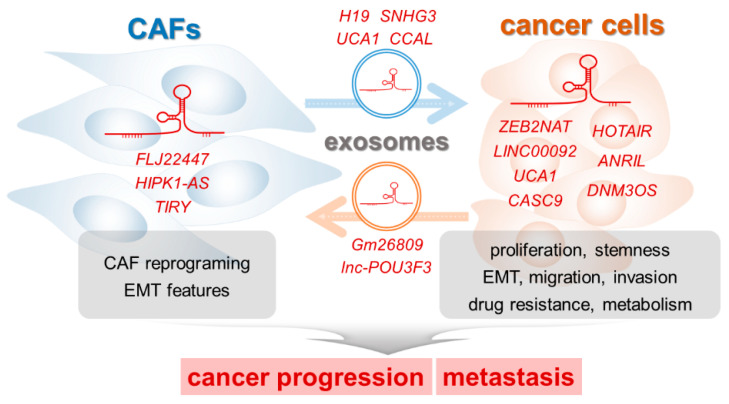
As described above, CAFs and cancer cells interact together directly or indirectly, and they influence each other to promote tumor progression and metastasis. Reviewing the recently accumulated literature reveals that lncRNAs are also important mediators between cancer cells and CAFs. The lncRNAs function within cells (CAFs and cancer cells) or are transported to other cells via exosomes (from CAFs to cancer cells and vice versa).
4.1. LncRNAs Working within CAFs
The lncRNA profiling of CAFs has been attempted in various cancers to examine overall expression changes and to select novel candidates working in the tumor microenvironment. High-throughput methods to profile lncRNA expression levels include quantitative reverse transcription (qRT)-PCR-based arrays, microarrays, and RNA sequencing, and each of these methods has advantages and disadvantages [61]. Compared with RNA sequencing, PCR arrays and microarrays are limited to known transcripts. However, microarrays generally do not involve PCR-based amplifications, which can cause unwanted bias.
Teng et al. performed lncRNA profiling to identify differentially expressed lncRNAs between CAFs and adjacent normal fibroblasts obtained from patients with non-small cell lung cancer [62]. They performed lncRNA microarrays using three matched pairs of CAFs and normal fibroblasts. A total of 766 lncRNAs showed differential expression patterns in CAFs: 322 lncRNAs were upregulated and 444 lncRNAs were downregulated. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses revealed that upregulated lncRNAs in CAFs are involved in immune network pathways such as type I interferon signaling, defense response to viruses, NOD-like receptor signaling, Wnt signaling, and toll-like receptor signaling. These results suggest that CAF-specific lncRNAs mediate an immune response during lung cancer progression.
In ovarian cancer, Colvin et al. profiled CAF-specific lncRNAs associated with patient overall survival [63]. Using microarrays and Kaplan–Meier survival analysis, they identified ten CAF-specific lncRNAs: nine lncRNAs (CRNDE, DANCR, LOC642852, MALAT1, MEG3, MGC2752, NEAT1, TP73-AS1, and XIST) were associated with shorter survival, and one (MIR155HG) was associated with longer survival. A co-expressed interactome with protein-coding genes and pathway (GO and KEGG) analyses showed that MIR155HG-related genes are associated with the immune system, with involvement in processes such as T-cell activation, antigen presentation, and leukocyte migration. Gene clusters involving DANCR, LOC642852, MALAT1, MEG3, MGC2752, TP73-AS1, and XIST are associated with metabolic processes and autophagy. The study showed that CAF-specific lncRNAs are linked to the survival of patients with ovarian cancer, and these lncRNAs regulate the tumor microenvironment to boost tumor growth and immune evasion.
Beyond global lncRNA profilings, in-depth studies on the roles and working mechanisms of individual lncRNAs in cancer development have also been conducted. In oral squamous cell carcinoma, RNA sequencing was performed to profile the changes of stromal lncRNAs during CAF transformation from normal fibroblasts [64]. Among those lncRNAs upregulated in CAFs, an intergenic lncRNA FLJ22447 (referred to as Lnc-CAF in the paper) proved to be required to maintain the stromal features of CAFs. The working mechanism of FLJ22447 was investigated through siRNA-mediated knockdown or overexpression. FLJ22447 increased the stability of IL-33 by preventing autophagic degradation, which promotes CAF reprogramming and tumor growth. In terms of clinical relevance, high FLJ22447 expression was associated with a high TNM stage and poor patient survival. Based on these findings, FLJ22447 promotes CAF activation and tumor progression in oral squamous cell carcinoma.
CAF-specific lncRNAs can be molecular targets of anticancer drugs. Sipi soup, a traditional Chinese medicine against infection and inflammation, prevents cancer cell-guided CAF activation and cancer progression in cervical cancer [65]. Through lncRNA microarray profiling, HIPK1 antisense RNA (HIPK1-AS) was shown to be induced in CAFs after treatment with conditioned medium from HeLa cells, and this was attenuated by Sipi soup. The shRNA-mediated knockdown of HIPK1-AS blocked the activation of CAFs by HeLa conditioned medium. In addition, HIPK1-AS was expressed at higher levels in cervicitis and cervical squamous cell carcinoma than in normal cervical cells. Upregulated HIPK1-AS in tumor stroma may cause inflammation and progression of cervical cancer; therefore, HIPK1-AS could be a good therapeutic target.
Another study in oral squamous cell carcinoma showed a more detailed working mechanism of a lncRNA in CAFs [66]. LncRNAs with significantly differential expression in CAFs from oral squamous cell carcinoma tissues compared with fibroblasts from normal tissues were identified using RNA sequencing: 29 lncRNAs were upregulated and 17 were downregulated. Among those, TIRY was highly expressed in CAFs. Bioinformatic and consequent in vitro approaches (RNA pull-down assays) found that TIRY can function as an endogenous sponge against miR-14. In oral squamous cell carcinoma tissues, TIRY expression was inversely correlated with miR-14 expression. Patients with high TIRY expression had poorer overall and progression-free survival than those with low TIRY expression. CAFs with higher TIRY expression present strong EMT features and promote the invasion and metastasis of co-cultured cancer cells. Interestingly, TIRY in CAFs suppressed exosomal delivery of miR-14 to neighboring cancer cells, which caused the upregulation of WNT3A, a miR-14 target, and activation of Wnt/β-catenin signaling in cancer cells. TIRY induces EMT in CAFs and promotes the progression and metastasis of oral squamous cell carcinoma by blocking miR-14 delivery; therefore, TIRY could be a prognostic and therapeutic target.
4.2. LncRNAs Working within Cancer Cells
A vast amount of research has been reported on the roles and mechanisms of lncRNAs in various types of cancer cells [67]. In this section, we focus on the studies of lncRNAs within cancer cells interacting with CAFs in the tumor microenvironment.
In urinary bladder cancer, conditioned medium from CAFs induced the EMT, migration, and invasion of cancer cells [41]. The main player within the conditioned medium was TGF-β, a strong EMT inducer [68]. To discover lncRNA mediators, a PCR array with 72 probes detecting cancer-related lncRNAs was performed in bladder cancer cells treated with conditioned medium from CAFs: three upregulated and six downregulated lncRNAs were identified. ZEB2NAT, a natural antisense transcript to the ZEB2 gene (also known as ZEB2-AS1), was among the upregulated lncRNAs. TGF-β induced ZEB2NAT transcription, and ZEB2NAT induced the EMT, migration, and invasion of bladder cancer cells. Inversely, ZEB2NAT depletion by siRNAs restored the enhanced invasion by conditioned medium from CAFs or TGF-β. ZEB2NAT is known to positively regulate ZEB2 expression via a post-transcriptional mechanism [69]. In bladder cancer samples, TGF-β, ZEB2NAT, and ZEB2 expression levels correlate positively and increase significantly in invasive tumors compared with non-invasive tumors. Therefore, ZEB2NAT responds to EMT and invasion signals from CAFs and mediates EMT and invasion in bladder cancer cells.
CXCL14 is known to mediate CAF-induced pro-tumorigenic effects (migration, invasion, and metastasis) in ovarian cancer [70]. CXCL14 is highly expressed in tumor stroma compared with normal stroma and is negatively correlated with the overall survival of patients with ovarian cancer. Using lncRNA microarrays, it was found that LINC00092 was one of the lncRNAs upregulated in ovarian cancer cells after CXCL14 treatment. LINC00092 knockdown by siRNAs reduced invasion and induced anoikis in ovarian cancer cells, and suppressed metastasis and extended mouse survival duration in a xenograft model. LINC00092 knockdown also causes a reduction in glycolysis metabolites, such as lactate. RNA pull-down assays proved that LINC00092 binds directly to a glycolytic enzyme, PFKFB2. In ovarian cancer patients, a positive correlation between LINC00092 and PFKFB2 protein expression was observed, which suggests that LINC00092 directly interacts with PFKFB2, maintains PFKFB2 enzyme stability, and modulates glycolysis during ovarian cancer progression and metastasis. Reciprocally, the glycolytic phenotype augmented by LINC00092 and PFKFB2 in ovarian cancer cells helps CAFs to remain activated and facilitates metastasis in the tumor microenvironment.
Another lncRNA, urothelial carcinoma-associated 1 (UCA1), was also reported to be induced in glioma cells by paracrine CXCL14 secretion from glioblastoma-associated stromal cells [71]. UCA1 positively regulates PFKFB2 expression in glioma cells by functioning as a sponge against miR-182, which directly targets PFKFB2. Modulating the interaction between glioma cells and glioblastoma-associated stromal cells, the CXCL14-UCA1-miR-182-PFKFB2 axis promotes glycolysis and the invasion of glioma. In colorectal cancer cells, UCA1 expression is induced by conditioned medium from CAFs [72]. UCA1 is involved in the promotion of the proliferation, migration, invasion, and EMT of colorectal cancer cells, which is mediated by blocking miR-143 and relieving KRAS inhibition [73].
Functioning as a ceRNA or a miRNA sponge is one of the major mechanisms of lncRNAs, as mentioned above. Cancer susceptibility candidate 9 (CASC9) is another example of a ceRNA in cervical cancer [74]. TGF-β released from CAFs elevates CASC9 expression in cervical cancer cells. CASC9 is increased in cancer tissues compared to normal tissues, and cervical cancer patients with low CASC9 expression have better overall survival rates than those with high CASC9 expression. CASC9 expression levels were also associated with cancer pathological stages. Through RNA immunoprecipitation (IP), miRNA pull-down, and luciferase assays, it was found that CASC9 competitively binds to miR-215 and releases its target TWIST2 from suppression. Thus, signaling from CAFs (TGF-β) contributes to cervical cancer progression via the regulation of CASC9/miR-215/TWIST2 signaling in cancer cells.
In breast cancer, TGF-β has been reported to be a key cytokine mediating the crosstalk between CAFs and cancer cells [75]. TGF-β is enriched in conditioned medium from CAFs and is required for CAFs to induce EMT in breast cancer cells. TGF-β stimulation encourages cancer cells to express more HOTAIR, which is a well-known epigenetic silencer that promotes tumor progression and metastasis [76]. Downstream of TGF-β, Smad2/3/4 transactivates HOTAIR by directly binding to its promoter region. HOTAIR, in turn, activates the EMT, migration, and invasion of breast cancer cells by the epigenetic suppression of EGR1 and CDK5RAP1 promoters. In addition, HOTAIR expression is higher in metastatic breast carcinoma than in ductal carcinoma in situ. Collectively, the TGF-β/HOTAIR axis controls breast cancer development and metastasis by facilitating communication between CAFs and breast cancer cells in the tumor microenvironment.
CAFs in the tumor microenvironment endow cancer cells with therapy resistance [77]. In oral squamous cell carcinoma, midkine, a member of the heparin-binding growth factor family, was shown to mediate CAF-induced cisplatin resistance in cancer cells [78]. Midkine released from CAFs can block cisplatin-induced cancer cell death. CAF-derived midkine enhances ANRIL expression in cancer cells, which promotes cell proliferation and cisplatin resistance. ANRIL upregulates the expression of ATP-binding cassette transporter proteins and thus increases the efflux of cancer drugs. Moreover, high ANRIL expression is associated with high TNM stage and lymph node metastasis in patients with oral squamous cell carcinoma.
In esophageal squamous cell carcinoma, CAFs confer cancer cells with resistance to radiotherapy [79]. The expression of DNM3 opposite strand/antisense RNA (DNM3OS), one of the highly expressed lncRNAs screened by PCR array, increases in radioresistant esophageal cancer cells compared with parental cells. DNM3OS expression was also higher in tumor tissues than in normal tissues and was associated with tumor stage. Conditioned medium derived from CAFs stimulated DNM3OS expression in cancer cells via PDGFβ/PDGFRβ signaling, which modulates the irradiation-induced DNA damage response. Transcriptionally, FOXO1, a downstream transcription factor of PDGFβ/PDGFRβ signaling, binds to the promoter region of DNM3OS and upregulates its expression. CAF-induced lncRNA DNM3OS can be a therapeutic target to restore radio-resistance in esophageal squamous cell carcinoma.
4.3. Exosomal lncRNAs
Exosomes or extracellular vesicles function as essential mediators of cell-to-cell communication in the tumor microenvironment [80,81]. They transport specific cargo molecules including proteins, mRNAs, miRNAs, and lncRNAs. Recent findings have demonstrated that various exosomal lncRNAs are involved in the interaction between CAFs and cancer cells and contribute to cancer progression, drug resistance, cancer stemness, and metastasis.
In a mouse model of colorectal cancer, RNA sequencing was performed to identify differentially expressed transcripts in tumor tissues compared with normal colon tissues [82]. LncRNA H19 was highly expressed in tumor tissues and increased according to cancer development stages and metastasis status. H19 overexpression in colorectal cancer cells promotes cancer stem cell-like phenotypes and oxaliplatin resistance. RNA fluorescence in situ hybridization revealed that H19 was predominantly expressed in tumor stroma, and CAFs expressed higher levels of H19 than normal fibroblasts. Interestingly, H19 is enriched in CAF-derived exosomes and is transported to cancer cells. In colon cancer cells, transported H19 functions as a ceRNA for miR-141, which targets β-catenin. Through the exosomal transport of H19, CAFs activate the Wnt/β-catenin signaling in colorectal cancer cells, facilitating cancer stemness and drug resistance.
Colorectal cancer-associated lncRNA (CCAL), which was found to be upregulated in colorectal cancer through PCR arrays [83], was also shown to be involved in drug resistance. CCAL is highly expressed in oxaliplatin-resistant colorectal cancer cells and promotes drug resistance [84]. Just like H19, CCAL is highly enriched in CAF-derived exosomes, and exosomal CCAL has been observed to be transferred from CAFs to cancer cells, promoting drug resistance. CCAL in cancer cells activates Wnt/β-catenin signaling by modulating β-catenin mRNA stability. Through RNA IP and RNA pull-down assays, CCAL was shown to bind directly to HuR, an RNA-binding protein that increases β-catenin mRNA stability. Thus, exosomal CCAL secreted from CAFs promotes the drug resistance of colorectal cancer cells by the regulation of HuR and β-catenin.
In vulvar squamous cell carcinoma tissues, UCA1 was highly expressed compared with normal tissues [85]. UCA1 expression is associated with tumor stages and lymph node metastasis in vulvar squamous cell carcinoma. Again, high UCA1 expression was detected in exosomes secreted from CAFs, and exosomal UCA1 was transported to cancer cells. UCA1 endowed cancer cells with resistance to cisplatin by functioning as a miRNA sponge against miR-103a which targets WEE1, a cell-cycle kinase regulating G2-M checkpoint. This study shows that CAFs promote the cisplatin resistance of cancer cells via exosomal UCA1 transport.
Exosomes released from CAFs derived from breast cancer patients were found to promote the growth and change the metabolic pathways of cancer cells. This was mediated by small nucleolar RNA host gene 3 (SNHG3) enclosed in the exosomes [86]. SNHG3 serves as a miRNA sponge against miR-330-5p and releases PKM from direct targeting by miR-330-5p. PKM is a glycolytic enzyme that converts phosphoenolpyruvate to pyruvate and is involved in cancer cell proliferation and metabolism [87]. Therefore, the exosomal transport of SNHG3 from CAFs to neighboring cancer cells modulates the miR-330-5p/PKM axis and then stimulates cell proliferation and metabolism reprogramming in breast cancer cells.
Conversely, cancer cells secrete exosomes containing lncRNAs to induce reprogramming and differentiation of CAFs in the tumor microenvironment. Exosomes secreted from mouse melanoma cells were found to stimulate the expression of CAF-specific markers (α-SMA and FAP) and the migratory activity of fibroblasts (NIH/3T3) [88]. In melanoma-derived exosomes, the lncRNA Gm26809 was significantly enriched. Gm26809 is a key player in mediating the effect of melanoma-derived exosomes on the reprogramming of normal fibroblasts into CAFs. Reprogrammed fibroblasts, in turn, promote melanoma cell proliferation and migration. Supplemented with further investigation on the exact working mechanisms, exosomal Gm26809 could be a good target for melanoma treatment.
Esophageal squamous cell carcinoma cancer cell-secreted exosomes upregulate the expression of CAF-specific markers in normal fibroblasts when compared with normal cell-secreted exosomes [89]. Among 14 esophageal squamous cell carcinoma-related lncRNAs, lncRNA POU3F3 (lnc-POU3F3) in exosomes facilitates the reprogramming of normal fibroblasts to CAFs. The lnc-POU3F3 lncRNA is transferred from cancer cells to fibroblasts via exosomes and activated fibroblasts. Fibroblasts activated by exosomal lnc-POU3F3, in turn, promote the proliferation and cisplatin resistance of cancer cells through the paracrine secretion of an inflammatory cytokine, IL-6. In esophageal squamous cell carcinoma patients, high plasma lnc-POU3F3 levels are associated with resistance to chemoradiotherapy, suggesting that plasma exosomal lnc-POU3F3 expression can be used as a liquid biopsy marker predicting the prognosis of patients with esophageal squamous cell carcinoma cancer. LncRNAs regulating the interaction between CAFs and cancer cells are summarized in Table 1 and Figure 2.
Table 1.
LncRNAs regulating the interaction between cancer-associated fibroblasts (CAFs) and cancer cells. EMT: epithelial-to-mesenchymal transition.
| LncRNA | Function | Working Target | Reference |
|---|---|---|---|
| LncRNAs within CAFs | |||
| FLJ22447 | Promotes CAF activation and the progression of oral squamous cell carcinoma | IL-33 | [64] |
| HIPK1-AS | Promotes CAF activation, inflammation, and the progression of cervical cancer | - | [65] |
| TIRY | Promotes EMT in CAFs and the progression and metastasis of oral squamous cell carcinoma | miR-14/WNT3 | [66] |
| LncRNAs within Cancer Cells | |||
| ZEB2NAT | Promotes the EMT, migration, and invasion of bladder cancer cells | ZEB2 | [41] |
| LINC00092 | Promotes the EMT, glycolytic phenotype, and metastasis of ovarian cancer cells | PFKFB2 | [70] |
| UCA1 | Promotes the glycolysis and invasion of glioma cells | miR-182/PFKFB2 | [71] |
| UCA1 | Promotes the proliferation, migration, invasion, and EMT of colorectal cancer cells | miR-143/KRAS | [72] |
| CASC9 | Promotes the progression of cervical cancer | miR-215/TWIST2 | [74] |
| HOTAIR | Promotes the EMT, migration, invasion, and metastasis of breast cancer cells | EGR1, CDK5RAP1 | [75] |
| ANRIL | Promotes the proliferation, cisplatin resistance, and progression of oral squamous cell carcinoma | ABC transporters | [78] |
| DNM3OS | Promotes radiotherapy resistance in esophageal squamous cell carcinoma | - | [79] |
| LncRNAs within Exosomes | |||
| H19 | Promotes cancer stem cell phenotypes and oxaliplatin resistance in colorectal cancer cells | miR-141/β-catenin | [82] |
| CCAL | Promotes oxaliplatin resistance in colorectal cancer cells | HuR/β-catenin | [84] |
| UCA1 | Promotes cisplatin resistance in vulvar squamous cell carcinoma cells | miR-103a/WEE1 | [85] |
| SNHG3 | Promotes proliferation and glycolytic metabolism in breast cancer cells | miR-330/PKM | [86] |
| Gm26809 | Promotes CAF reprogramming and activation | - | [88] |
| lnc-POU3F3 | Promotes CAF reprogramming and activation | IL-6 | [89] |
Figure 2.
LncRNAs influence the interaction between CAFs and cancer cells in the tumor microenvironment.
5. Conclusions and Perspectives
In the tumor microenvironment, cancer cells interact with various types of cells, such as lymphocytes, macrophages, endothelial cells, and CAFs. CAFs occupy an important position in the tumor microenvironment: they modulate the EMT, proliferation, migration, invasiveness, drug resistance, and metastasis of neighboring cancer cells. Among the regulatory molecules in the interaction between CAFs and cancer cells, lncRNAs are relatively novel biomolecules and are receiving more and more attention from cancer researchers. However, because they have lower expression levels, lower stability, less accurate annotation, and more complicated functional mechanisms compared with protein-coding mRNAs and miRNAs, more difficult and higher barriers lie ahead of lncRNA research [90]. Recently, the development of novel methodologies, such as RNA IP followed by sequencing, high-throughput sequencing cross-linking IP, and chromatin isolation by RNA purification sequencing, has enabled broader and more detailed studies to elucidate the functional molecular mechanisms of lncRNAs [91].
In the clinical aspect, lncRNAs in the tumor microenvironment can be used as potential markers for diagnosis and prognosis and therapeutic targets [92]. LncRNAs, such as FLJ22447, HIPK1-AS, and TIRY, are highly expressed in tumor stroma and are associated with tumor stages or metastatic status. Working within cancer cells, the lncRNAs described above also promote cancer progression and metastasis. ANRIL and DNM3OS are responsible for resistance to cancer therapy and could be used as predictive markers for therapeutic response. Exosomal lncRNAs, such as H19, CCAL, and UCA1, are also involved in drug resistance, suggesting that these are potential candidates for liquid biopsy. Given that lncRNAs that mediate the interaction between CAFs and cancer cells can promote cancer progression and metastasis, therapeutic targeting of these lncRNAs by RNA interference-mediated lncRNA silencing or inhibition of lncRNA-protein/mRNA/miRNA interactions will be applicable in the clinical treatment of cancer [93].
Abbreviations
| lncRNAs | Long non-coding RNAs |
| miRNAs | microRNAs |
| KCNQ10T1 | KCNQ1 overlapping transcript 1 |
| PRC2 | Polycomb repressive complex 2 |
| PANDA | p21-associated ncRNA DNA damage-activated |
| TERRA | Telomeric repeat-containing RNA |
| GAS5 | Growth arrest-specific 5 |
| PTENP1 | PTEN pseudogene 1 |
| ceRNAs | Competing endogenous RNAs |
| HOTAIR | HOX transcript antisense intergenic RNA |
| H3K4 | Lysine 4 of histone 3 |
| ANRIL | Antisense non-coding RNA at the INK4 locus |
| Fendrr | Fetal-lethal non-coding developmental regulatory RNA |
| AIRN | Antisense of IGF2R non-protein coding RNA |
| CAFs | Cancer-associated fibroblasts |
| qRT-PCR | Quantitative reverse transcription-PCR |
| GO | Gene ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| HIPK1-AS | HIPK1 antisense RNA |
| EMT | Epithelial-to-mesenchymal transition |
| ZEB2NAT | ZEB2 natural antisense transcript |
| UCA1 | Urothelial carcinoma-associated 1 |
| CASC9 | Cancer susceptibility candidate 9 |
| IP | Immunoprecipitation |
| DNM3OS | DNM3 opposite strand/antisense RNA |
| CCAL | Colorectal cancer-associated lncRNA |
| SNHG3 | Small nucleolar RNA host gene 3 |
Author Contributions
Y.-H.A. and J.S.K.; writing—original draft preparation; Y.-H.A. and J.S.K.; writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grants funded by the government of Korea (MSIT) (NRF-2019R1F1A1057968 and NRF-2020R1A5A2019210).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Pandey R.R., Mondal T., Mohammad F., Enroth S., Redrup L., Komorowski J., Nagano T., Mancini-Dinardo D., Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 2.Hung T., Wang Y., Lin M.F., Koegel A.K., Kotake Y., Grant G.D., Horlings H.M., Shah N., Umbricht C., Wang P., et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Redon S., Reichenbach P., Lingner J. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. 2010;38:5797–5806. doi: 10.1093/nar/gkq296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kino T., Hurt D.E., Ichijo T., Nader N., Chrousos G.P. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poliseno L., Salmena L., Zhang J., Carver B., Haveman W.J., Pandolfi P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marsit C.J., Zheng S., Aldape K., Hinds P.W., Nelson H.H., Wiencke J.K., Kelsey K.T. PTEN expression in non-small-cell lung cancer: Evaluating its relation to tumor characteristics, allelic loss, and epigenetic alteration. Hum. Pathol. 2005;36:768–776. doi: 10.1016/j.humpath.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 7.Spitale R.C., Tsai M.C., Chang H.Y. RNA templating the epigenome: Long noncoding RNAs as molecular scaffolds. Epigenetics. 2011;6:539–543. doi: 10.4161/epi.6.5.15221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Q., Cai J., Wang Q., Wang Y., Liu M., Yang J., Zhou J., Kang C., Li M., Jiang C. Long Noncoding RNA NEAT1, Regulated by the EGFR Pathway, Contributes to Glioblastoma Progression Through the WNT/β-Catenin Pathway by Scaffolding EZH2. Clin. Cancer Res. 2018;24:684–695. doi: 10.1158/1078-0432.CCR-17-0605. [DOI] [PubMed] [Google Scholar]
- 10.Song Z., Wu W., Chen M., Cheng W., Yu J., Fang J., Xu L., Yasunaga J.I., Matsuoka M., Zhao T. Long Noncoding RNA ANRIL Supports Proliferation of Adult T-Cell Leukemia Cells through Cooperation with EZH2. J. Virol. 2018;92:e00909–e00918. doi: 10.1128/JVI.00909-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grote P., Wittler L., Hendrix D., Koch F., Währisch S., Beisaw A., Macura K., Bläss G., Kellis M., Werber M., et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell. 2013;24:206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagano T., Mitchell J.A., Sanz L.A., Pauler F.M., Ferguson-Smith A.C., Feil R., Fraser P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 13.Löhr M., Schmidt C., Ringel J., Kluth M., Müller P., Nizze H., Jesnowski R. Transforming growth factor-beta1 induces desmoplasia in an experimental model of human pancreatic carcinoma. Cancer Res. 2001;61:550–555. [PubMed] [Google Scholar]
- 14.Guido C., Whitaker-Menezes D., Capparelli C., Balliet R., Lin Z., Pestell R.G., Howell A., Aquila S., Andò S., Martinez-Outschoorn U., et al. Metabolic reprogramming of cancer-associated fibroblasts by TGF-β drives tumor growth: Connecting TGF-β signaling with “Warburg-like” cancer metabolism and L-lactate production. Cell Cycle. 2012;11:3019–3035. doi: 10.4161/cc.21384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erez N., Truitt M., Olson P., Arron S.T., Hanahan D. Cancer-Associated Fibroblasts Are Activated in Incipient Neoplasia to Orchestrate Tumor-Promoting Inflammation in an NF-kappaB-Dependent Manner. Cancer Cell. 2010;17:135–147. doi: 10.1016/j.ccr.2009.12.041. [DOI] [PubMed] [Google Scholar]
- 16.Sampson N., Brunner E., Weber A., Puhr M., Schäfer G., Szyndralewiez C., Klocker H. Inhibition of Nox4-dependent ROS signaling attenuates prostate fibroblast activation and abrogates stromal-mediated protumorigenic interactions. Int. J. Cancer. 2018;143:383–395. doi: 10.1002/ijc.31316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Becker L.M., O’Connell J.T., Vo A.P., Cain M.P., Tampe D., Bizarro L., Sugimoto H., McGow A.K., Asara J.M., Lovisa S., et al. Epigenetic Reprogramming of Cancer-Associated Fibroblasts Deregulates Glucose Metabolism and Facilitates Progression of Breast Cancer. Cell Rep. 2020;31:107701. doi: 10.1016/j.celrep.2020.107701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schoepp M., Ströse A.J., Haier J. Dysregulation of miRNA Expression in Cancer Associated Fibroblasts (CAFs) and Its Consequences on the Tumor Microenvironment. Cancers (Basel) 2017;9:54. doi: 10.3390/cancers9060054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LeBleu V.S., Kalluri R. A peek into cancer-associated fibroblasts: Origins, functions and translational impact. Dis. Model. Mech. 2018;11:dmm029447. doi: 10.1242/dmm.029447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ganguly D., Chandra R., Karalis J., Teke M., Aguilera T., Maddipati R., Wachsmann M.B., Ghersi D., Siravegna G., Zeh H.J., 3rd, et al. Cancer-Associated Fibroblasts: Versatile Players in the Tumor Microenvironment. Cancers (Basel) 2020;12:2652. doi: 10.3390/cancers12092652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shoucair I., Weber Mello F., Jabalee J., Maleki S., Garnis C. The Role of Cancer-Associated Fibroblasts and Extracellular Vesicles in Tumorigenesis. Int. J. Mol. Sci. 2020;21:6837. doi: 10.3390/ijms21186837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalluri R., Zeisberg M. Fibroblasts in cancer. Nat. Rev. Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 23.Kalluri R., Neilson E.G. Epithelial-mesenchymal transition and its implications for fibrosis. J. Clin. Investig. 2003;112:1776–1784. doi: 10.1172/JCI200320530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeisberg E.M., Potenta S., Xie L., Zeisberg M., Kalluri R. Discovery of endothelial to mesenchymal transition as a source for carcinoma-associated fibroblasts. Cancer Res. 2007;67:10123–10128. doi: 10.1158/0008-5472.CAN-07-3127. [DOI] [PubMed] [Google Scholar]
- 25.Hosaka K., Yang Y., Seki T., Fischer C., Dubey O., Fredlund E., Hartman J., Religa P., Morikawa H., Ishii Y., et al. Pericyte-fibroblast transition promotes tumor growth and metastasis. Proc. Natl. Acad. Sci. USA. 2016;113:E5618–E5627. doi: 10.1073/pnas.1608384113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jotzu C., Alt E., Welte G., Li J., Hennessy B.T., Devarajan E., Krishnappa S., Pinilla S., Droll L., Song Y.H. Adipose tissue derived stem cells differentiate into carcinoma-associated fibroblast-like cells under the influence of tumor derived factors. Cell. Oncol. (Dordr.) 2011;34:55–67. doi: 10.1007/s13402-011-0012-1. [DOI] [PubMed] [Google Scholar]
- 27.Quante M., Tu S.P., Tomita H., Gonda T., Wang S.S., Takashi S., Baik G.H., Shibata W., Diprete B., Betz K.S., et al. Bone marrow-derived myofibroblasts contribute to the mesenchymal stem cell niche and promote tumor growth. Cancer Cell. 2011;19:257–272. doi: 10.1016/j.ccr.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barcellos-de-Souza P., Comito G., Pons-Segura C., Taddei M.L., Gori V., Becherucci V., Bambi F., Margheri F., Laurenzana A., Del Rosso M., et al. Mesenchymal Stem Cells are Recruited and Activated into Carcinoma-Associated Fibroblasts by Prostate Cancer Microenvironment-Derived TGF-β1. Stem Cell. 2016;34:2536–2547. doi: 10.1002/stem.2412. [DOI] [PubMed] [Google Scholar]
- 29.LaRue A.C., Masuya M., Ebihara Y., Fleming P.A., Visconti R.P., Minamiguchi H., Ogawa M., Drake C.J. Hematopoietic origins of fibroblasts: I. In vivo studies of fibroblasts associated with solid tumors. Exp. Hematol. 2006;34:208–218. doi: 10.1016/j.exphem.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 30.McDonald L.T., Russell D.L., Kelly R.R., Xiong Y., Motamarry A., Patel R.K., Jones J.A., Watson P.M., Turner D.P., Watson D.K., et al. Hematopoietic stem cell-derived cancer-associated fibroblasts are novel contributors to the pro-tumorigenic microenvironment. Neoplasia. 2015;17:434–448. doi: 10.1016/j.neo.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun Y., Fan X., Zhang Q., Shi X., Xu G., Zou C. Cancer-associated fibroblasts secrete FGF-1 to promote ovarian proliferation, migration, and invasion through the activation of FGF-1/FGFR4 signaling. Tumour Biol. 2017;39:1010428317712592. doi: 10.1177/1010428317712592. [DOI] [PubMed] [Google Scholar]
- 32.Deying W., Feng G., Shumei L., Hui Z., Ming L., Hongqing W. CAF-derived HGF promotes cell proliferation and drug resistance by up-regulating the c-Met/PI3K/Akt and GRP78 signalling in ovarian cancer cells. Biosci. Rep. 2017;37:BSR20160470. doi: 10.1042/BSR20160470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teng F., Tian W.Y., Wang Y.M., Zhang Y.F., Guo F., Zhao J., Gao C., Xue F.X. Cancer-associated fibroblasts promote the progression of endometrial cancer via the SDF-1/CXCR4 axis. J. Hematol. Oncol. 2016;9:8. doi: 10.1186/s13045-015-0231-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subramaniam K.S., Omar I.S., Kwong S.C., Mohamed Z., Woo Y.L., Mat Adenan N.A., Chung I. Cancer-associated fibroblasts promote endometrial cancer growth via activation of interleukin-6/STAT-3/c-Myc pathway. Am. J. Cancer Res. 2016;6:200–213. [PMC free article] [PubMed] [Google Scholar]
- 35.Tsuyada A., Chow A., Wu J., Somlo G., Chu P., Loera S., Luu T., Li A.X., Wu X., Ye W., et al. CCL2 mediates cross-talk between cancer cells and stromal fibroblasts that regulates breast cancer stem cells. Cancer Res. 2012;72:2768–2779. doi: 10.1158/0008-5472.CAN-11-3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen W.J., Ho C.C., Chang Y.L., Chen H.Y., Lin C.A., Ling T.Y., Yu S.L., Yuan S.S., Chen Y.J., Lin C.Y., et al. Cancer-associated fibroblasts regulate the plasticity of lung cancer stemness via paracrine signalling. Nat. Commun. 2014;5:3472. doi: 10.1038/ncomms4472. [DOI] [PubMed] [Google Scholar]
- 37.Goulet C.R., Champagne A., Bernard G., Vandal D., Chabaud S., Pouliot F., Bolduc S. Cancer-associated fibroblasts induce epithelial-mesenchymal transition of bladder cancer cells through paracrine IL-6 signalling. BMC Cancer. 2019;19:137. doi: 10.1186/s12885-019-5353-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ebbing E.A., van der Zalm A.P., Steins A., Creemers A., Hermsen S., Rentenaar R., Klein M., Waasdorp C., Hooijer G.K.J., Meijer S.L., et al. Stromal-derived interleukin 6 drives epithelial-to-mesenchymal transition and therapy resistance in esophageal adenocarcinoma. Proc. Natl. Acad. Sci. USA. 2019;116:2237–2242. doi: 10.1073/pnas.1820459116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.You J., Li M., Cao L.M., Gu Q.H., Deng P.B., Tan Y., Hu C.P. Snail1-dependent cancer-associated fibroblasts induce epithelial-mesenchymal transition in lung cancer cells via exosomes. QJM. 2019;112:581–590. doi: 10.1093/qjmed/hcz093. [DOI] [PubMed] [Google Scholar]
- 40.Wang H., Wei H., Wang J., Li L., Chen A., Li Z. MicroRNA-181d-5p-Containing Exosomes Derived from CAFs Promote EMT by Regulating CDX2/HOXA5 in Breast Cancer. Mol. Nucl. Acid. 2020;19:654–667. doi: 10.1016/j.omtn.2019.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhuang J., Lu Q., Shen B., Huang X., Shen L., Zheng X., Huang R., Yan J., Guo H. TGFbeta1 secreted by cancer-associated fibroblasts induces epithelial-mesenchymal transition of bladder cancer cells through lncRNA-ZEB2NAT. Sci. Rep. 2015;5:11924. doi: 10.1038/srep11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamaguchi H., Sakai R. Direct Interaction between Carcinoma Cells and Cancer Associated Fibroblasts for the Regulation of Cancer Invasion. Cancers (Basel) 2015;7:2054–2062. doi: 10.3390/cancers7040876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang D., Gao J., Wang S., Ye N., Chong Y., Huang Y., Wang J., Li B., Yin W., Wang D. Cancer-associated fibroblasts promote angiogenesis in gastric cancer through galectin-1 expression. Tumour Biol. 2016;37:1889–1899. doi: 10.1007/s13277-015-3942-9. [DOI] [PubMed] [Google Scholar]
- 44.Unterleuthner D., Neuhold P., Schwarz K., Janker L., Neuditschko B., Nivarthi H., Crncec I., Kramer N., Unger C., Hengstschläger M., et al. Cancer-associated fibroblast-derived WNT2 increases tumor angiogenesis in colon cancer. Angiogenesis. 2020;23:159–177. doi: 10.1007/s10456-019-09688-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J., Liu X., Zang S., Zhou J., Zhang F., Sun B., Qi D., Li X., Kong J., Jin D., et al. Small extracellular vesicle-bound vascular endothelial growth factor secreted by carcinoma-associated fibroblasts promotes angiogenesis in a bevacizumab-resistant manner. Cancer Lett. 2020;492:71–83. doi: 10.1016/j.canlet.2020.08.030. [DOI] [PubMed] [Google Scholar]
- 46.Sewell-Loftin M.K., Bayer S.V.H., Crist E., Hughes T., Joison S.M., Longmore G.D., George S.C. Cancer-associated fibroblasts support vascular growth through mechanical force. Sci. Rep. 2017;7:12574. doi: 10.1038/s41598-017-13006-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Demircioglu F., Wang J., Candido J., Costa A.S.H., Casado P., de Luxan Delgado B., Reynolds L.E., Gomez-Escudero J., Newport E., Rajeeve V., et al. Cancer associated fibroblast FAK regulates malignant cell metabolism. Nat. Commun. 2020;11:1290. doi: 10.1038/s41467-020-15104-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Curtis M., Kenny H.A., Ashcroft B., Mukherjee A., Johnson A., Zhang Y., Helou Y., Batlle R., Liu X., Gutierrez N., et al. Fibroblasts Mobilize Tumor Cell Glycogen to Promote Proliferation and Metastasis. Cell Metab. 2019;29:141–155.e9. doi: 10.1016/j.cmet.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mishra R., Haldar S., Placencio V., Madhav A., Rohena-Rivera K., Agarwal P., Duong F., Angara B., Tripathi M., Liu Z., et al. Stromal epigenetic alterations drive metabolic and neuroendocrine prostate cancer reprogramming. J. Clin. Investig. 2018;128:4472–4484. doi: 10.1172/JCI99397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ippolito L., Morandi A., Taddei M.L., Parri M., Comito G., Iscaro A., Raspollini M.R., Magherini F., Rapizzi E., Masquelier J., et al. Cancer-associated fibroblasts promote prostate cancer malignancy via metabolic rewiring and mitochondrial transfer. Oncogene. 2019;38:5339–5355. doi: 10.1038/s41388-019-0805-7. [DOI] [PubMed] [Google Scholar]
- 51.Sansone P., Savini C., Kurelac I., Chang Q., Amato L.B., Strillacci A., Stepanova A., Iommarini L., Mastroleo C., Daly L., et al. Packaging and transfer of mitochondrial DNA via exosomes regulate escape from dormancy in hormonal therapy-resistant breast cancer. Proc. Natl. Acad. Sci. USA. 2017;114:E9066–E9075. doi: 10.1073/pnas.1704862114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li X.Y., Hu S.Q., Xiao L. The cancer-associated fibroblasts and drug resistance. Eur. Rev. Med. Pharm. Sci. 2015;19:2112–2119. [PubMed] [Google Scholar]
- 53.Long X., Xiong W., Zeng X., Qi L., Cai Y., Mo M., Jiang H., Zhu B., Chen Z., Li Y. Cancer-associated fibroblasts promote cisplatin resistance in bladder cancer cells by increasing IGF-1/ERβ/Bcl-2 signalling. Cell Death Dis. 2019;10:375. doi: 10.1038/s41419-019-1581-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ham I.H., Oh H.J., Jin H., Bae C.A., Jeon S.M., Choi K.S., Son S.Y., Han S.U., Brekken R.A., Lee D., et al. Targeting interleukin-6 as a strategy to overcome stroma-induced resistance to chemotherapy in gastric cancer. Mol. Cancer. 2019;18:68. doi: 10.1186/s12943-019-0972-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ying L., Zhu Z., Xu Z., He T., Li E., Guo Z., Liu F., Jiang C., Wang Q. Cancer Associated Fibroblast-Derived Hepatocyte Growth Factor Inhibits the Paclitaxel-Induced Apoptosis of Lung Cancer A549 Cells by Up-Regulating the PI3K/Akt and GRP78 Signaling on a Microfluidic Platform. PLoS ONE. 2015;10:e0129593. doi: 10.1371/journal.pone.0129593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Z., Karthaus W.R., Lee Y.S., Gao V.R., Wu C., Russo J.W., Liu M., Mota J.M., Abida W., Linton E., et al. Tumor Microenvironment-Derived NRG1 Promotes Antiandrogen Resistance in Prostate Cancer. Cancer Cell. 2020;38:279–296. doi: 10.1016/j.ccell.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Uchihara T., Miyake K., Yonemura A., Komohara Y., Itoyama R., Koiwa M., Yasuda T., Arima K., Harada K., Eto K., et al. Extracellular Vesicles from Cancer-Associated Fibroblasts Containing Annexin A6 Induces FAK-YAP Activation by Stabilizing β1 Integrin, Enhancing Drug Resistance. Cancer Res. 2020;80:3222–3235. doi: 10.1158/0008-5472.CAN-19-3803. [DOI] [PubMed] [Google Scholar]
- 58.Cohen N., Shani O., Raz Y., Sharon Y., Hoffman D., Abramovitz L., Erez N. Fibroblasts drive an immunosuppressive and growth-promoting microenvironment in breast cancer via secretion of Chitinase 3-like 1. Oncogene. 2017;36:4457–4468. doi: 10.1038/onc.2017.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xiang H., Ramil C.P., Hai J., Zhang C., Wang H., Watkins A.A., Afshar R., Georgiev P., Sze M.A., Song X.S., et al. Cancer-Associated Fibroblasts Promote Immunosuppression by Inducing ROS-Generating Monocytic MDSCs in Lung Squamous Cell Carcinoma. Cancer Immunol. Res. 2020;8:436–450. doi: 10.1158/2326-6066.CIR-19-0507. [DOI] [PubMed] [Google Scholar]
- 60.Lakins M.A., Ghorani E., Munir H., Martins C.P., Shields J.D. Cancer-associated fibroblasts induce antigen-specific deletion of CD8 (+) T Cells to protect tumour cells. Nat. Commun. 2018;9:948. doi: 10.1038/s41467-018-03347-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Uchida S. High-Throughput Methods to Detect Long Non-Coding RNAs. High Throughput. 2017;6:12. doi: 10.3390/ht6030012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teng C., Huang G., Luo Y., Pan Y., Wang H., Liao X., Li Y., Yang J. Differential long noncoding RNAs expression in cancer-associated fibroblasts of non-small-cell lung cancer. Pharmacogenomics. 2019;20:143–153. doi: 10.2217/pgs-2018-0102. [DOI] [PubMed] [Google Scholar]
- 63.Colvin E.K., Howell V.M., Mok S.C., Samimi G., Vafaee F. Expression of long noncoding RNAs in cancer-associated fibroblasts linked to patient survival in ovarian cancer. Cancer Sci. 2020;111:1805–1817. doi: 10.1111/cas.14350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ding L., Ren J., Zhang D., Li Y., Huang X., Hu Q., Wang H., Song Y., Ni Y., Hou Y. A novel stromal lncRNA signature reprograms fibroblasts to promote the growth of oral squamous cell carcinoma via LncRNA-CAF/interleukin-33. Carcinogenesis. 2018;39:397–406. doi: 10.1093/carcin/bgy006. [DOI] [PubMed] [Google Scholar]
- 65.Zhou B., Yu Y., Yu L., Que B., Qiu R. Sipi soup inhibits cancer-associated fibroblast activation and the inflammatory process by downregulating long non-coding RNA HIPK1-AS. Mol. Med. Rep. 2018;18:1361–1368. doi: 10.3892/mmr.2018.9144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jin N., Jin N., Bu W., Li X., Liu L., Wang Z., Tong J., Li D. Long non-coding RNA TIRY promotes tumor metastasis by enhancing epithelial-to-mesenchymal transition in oral cancer. Exp. Biol. Med. 2020;245:585–596. doi: 10.1177/1535370220903673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schmitt A.M., Chang H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu J., Lamouille S., Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009;19:156–172. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Beltran M., Puig I., Peña C., García J.M., Alvarez A.B., Peña R., Bonilla F., de Herreros A.G. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22:756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao L., Ji G., Le X., Wang C., Xu L., Feng M., Zhang Y., Yang H., Xuan Y., Yang Y., et al. Long Noncoding RNA LINC00092 Acts in Cancer-Associated Fibroblasts to Drive Glycolysis and Progression of Ovarian Cancer. Cancer Res. 2017;77:1369–1382. doi: 10.1158/0008-5472.CAN-16-1615. [DOI] [PubMed] [Google Scholar]
- 71.He Z., You C., Zhao D. Long non-coding RNA UCA1/miR-182/PFKFB2 axis modulates glioblastoma-associated stromal cells-mediated glycolysis and invasion of glioma cells. Biochem. Biophys. Res. Commun. 2018;500:569–576. doi: 10.1016/j.bbrc.2018.04.091. [DOI] [PubMed] [Google Scholar]
- 72.Jahangiri B., Khalaj-Kondori M., Asadollahi E., Sadeghizadeh M. Cancer-associated fibroblasts enhance cell proliferation and metastasis of colorectal cancer SW480 cells by provoking long noncoding RNA UCA1. J. Cell Commun. Signal. 2019;13:53–64. doi: 10.1007/s12079-018-0471-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tuo Y.L., Li X.M., Luo J. Long noncoding RNA UCA1 modulates breast cancer cell growth and apoptosis through decreasing tumor suppressive miR-143. Eur. Rev. Med. Pharm. Sci. 2015;19:3403–3411. [PubMed] [Google Scholar]
- 74.Zhang J., Wang Q., Quan Z. Long non-coding RNA CASC9 enhances breast cancer progression by promoting metastasis through the meditation of miR-215/TWIST2 signaling associated with TGF-beta expression. Biochem. Biophys. Res. Commun. 2019;515:644–650. doi: 10.1016/j.bbrc.2019.05.080. [DOI] [PubMed] [Google Scholar]
- 75.Ren Y., Jia H.H., Xu Y.Q., Zhou X., Zhao X.H., Wang Y.F., Song X., Zhu Z.Y., Sun T., Dou Y., et al. Paracrine and epigenetic control of CAF-induced metastasis: The role of HOTAIR stimulated by TGF-ss1 secretion. Mol. Cancer. 2018;17:5. doi: 10.1186/s12943-018-0758-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J., Tsai M.C., Hung T., Argani P., Rinn J.L., et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fiori M.E., Di Franco S., Villanova L., Bianca P., Stassi G., De Maria R. Cancer-associated fibroblasts as abettors of tumor progression at the crossroads of EMT and therapy resistance. Mol. Cancer. 2019;18:70. doi: 10.1186/s12943-019-0994-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang D., Ding L., Li Y., Ren J., Shi G., Wang Y., Zhao S., Ni Y., Hou Y. Midkine derived from cancer-associated fibroblasts promotes cisplatin-resistance via up-regulation of the expression of lncRNA ANRIL in tumour cells. Sci. Rep. 2017;7:16231. doi: 10.1038/s41598-017-13431-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang H., Hua Y., Jiang Z., Yue J., Shi M., Zhen X., Zhang X., Yang L., Zhou R., Wu S. Cancer-associated Fibroblast-promoted LncRNA DNM3OS Confers Radioresistance by Regulating DNA Damage Response in Esophageal Squamous Cell Carcinoma. Clin. Cancer Res. 2019;25:1989–2000. doi: 10.1158/1078-0432.CCR-18-0773. [DOI] [PubMed] [Google Scholar]
- 80.Fan Q., Yang L., Zhang X., Peng X., Wei S., Su D., Zhai Z., Hua X., Li H. The emerging role of exosome-derived non-coding RNAs in cancer biology. Cancer Lett. 2018;414:107–115. doi: 10.1016/j.canlet.2017.10.040. [DOI] [PubMed] [Google Scholar]
- 81.Yang X., Li Y., Zou L., Zhu Z. Role of Exosomes in Crosstalk Between Cancer-Associated Fibroblasts and Cancer Cells. Front. Oncol. 2019;9:356. doi: 10.3389/fonc.2019.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ren J., Ding L., Zhang D., Shi G., Xu Q., Shen S., Wang Y., Wang T., Hou Y. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics. 2018;8:3932–3948. doi: 10.7150/thno.25541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ma Y., Yang Y., Wang F., Moyer M.P., Wei Q., Zhang P., Yang Z., Liu W., Zhang H., Chen N., et al. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/β-catenin signalling pathway via suppression of activator protein 2α. Gut. 2016;65:1494–1504. doi: 10.1136/gutjnl-2014-308392. [DOI] [PubMed] [Google Scholar]
- 84.Deng X., Ruan H., Zhang X., Xu X., Zhu Y., Peng H., Zhang X., Kong F., Guan M. Long noncoding RNA CCAL transferred from fibroblasts by exosomes promotes chemoresistance of colorectal cancer cells. Int. J. Cancer. 2020;146:1700–1716. doi: 10.1002/ijc.32608. [DOI] [PubMed] [Google Scholar]
- 85.Gao Q., Fang X., Chen Y., Li Z., Wang M. Exosomal lncRNA UCA1 from cancer-associated fibroblasts enhances chemoresistance in vulvar squamous cell carcinoma cells. J. Obs. Gynaecol. Res. 2020 doi: 10.1111/jog.14418. [DOI] [PubMed] [Google Scholar]
- 86.Li Y., Zhao Z., Liu W., Li X. SNHG3 Functions as miRNA Sponge to Promote Breast Cancer Cells Growth Through the Metabolic Reprogramming. Appl. Biochem. Biotechnol. 2020;191:1084–1099. doi: 10.1007/s12010-020-03244-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dayton T.L., Jacks T., Vander Heiden M.G. PKM2, cancer metabolism, and the road ahead. EMBO Rep. 2016;17:1721–1730. doi: 10.15252/embr.201643300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hu T., Hu J. Melanoma-derived exosomes induce reprogramming fibroblasts into cancer-associated fibroblasts via Gm26809 delivery. Cell Cycle. 2019;18:3085–3094. doi: 10.1080/15384101.2019.1669380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tong Y., Yang L., Yu C., Zhu W., Zhou X., Xiong Y., Wang W., Ji F., He D., Cao X. Tumor-Secreted Exosomal lncRNA POU3F3 Promotes Cisplatin Resistance in ESCC by Inducing Fibroblast Differentiation into CAFs. Mol. Oncolytics. 2020;18:1–13. doi: 10.1016/j.omto.2020.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhao Y., Teng H., Yao F., Yap S., Sun Y., Ma L. Challenges and Strategies in Ascribing Functions to Long Noncoding RNAs. Cancers (Basel) 2020;12:1458. doi: 10.3390/cancers12061458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang H.V., Chekanova J.A. An Overview of Methodologies in Studying lncRNAs in the High-Throughput Era: When Acronyms ATTACK! Methods Mol. Biol. 2019;1933:1–30. doi: 10.1007/978-1-4939-9045-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hauptman N., Glavač D. Long non-coding RNA in cancer. Int. J. Mol. Sci. 2013;14:4655–4669. doi: 10.3390/ijms14034655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lu T., Wang Y., Chen D., Liu J., Jiao W. Potential clinical application of lncRNAs in non-small cell lung cancer. Onco. Target. Ther. 2018;11:8045–8052. doi: 10.2147/OTT.S178431. [DOI] [PMC free article] [PubMed] [Google Scholar]