Abstract
Among the natural bases, guanine is the most oxidizable base. The damage caused by oxidation of guanine, commonly referred to as oxidative guanine damage, results in the formation of several products, including 2,5-diamino-4H-imidazol-4-one (Iz), 2,2,4-triamino-5(2H)-oxazolone (Oz), guanidinoformimine (Gf), guanidinohydantoin/iminoallantoin (Gh/Ia), spiroiminodihydantoin (Sp), 5-carboxamido-5-formamido-2-iminohydantoin (2Ih), urea (Ua), 5-guanidino-4-nitroimidazole (NI), spirodi(iminohydantoin) (5-Si and 8-Si), triazine, the M+7 product, other products by peroxynitrite, alkylated guanines, and 8,5′-cyclo-2′-deoxyguanosine (cG). Herein, we summarize the present knowledge about base pairs containing the products of oxidative guanine damage and guanine. Of these products, Iz is involved in G-C transversions. Oz, Gh/Ia, and Sp form preferably Oz:G, Gh/Ia:G, and Sp:G base pairs in some cases. An involvement of Gf, 2Ih, Ua, 5-Si, 8-Si, triazine, the M+7 product, and 4-hydroxy-2,5-dioxo-imidazolidine-4-carboxylic acid (HICA) in G-C transversions requires further experiments. In addition, we describe base pairs that target the RNA-dependent RNA polymerase (RdRp) of RNA viruses and describe implications for the 2019 novel coronavirus (SARS-CoV-2): When products of oxidative guanine damage are adapted for the ribonucleoside analogs, mimics of oxidative guanine damages, which can form base pairs, may become antiviral agents for SARS-CoV-2.
Keywords: oxidative guanine damage, base incorporation, base pair, G-C transversions, 2019 novel coronavirus, SARS-CoV-2
1. Introduction
The DNA bases guanine, adenine, thymine, and cytosine have oxidation potentials (vs. NHE) of 1.29, 1.42, 1.6, and 1.7 V, respectively, and among them, guanine is most susceptible to oxidation [1]. Oxidation of guanine triggers mutations. Indeed, mutations resulting from a change from guanine to other bases have been detected in the oncogene K-ras and the tumor suppressor gene p53 [2,3,4]. G-T transversions and G-C transversions are preferentially caused by sunlight, ultraviolet light in the presence of riboflavin or menadione, visible light with methylene blue, Fe2+, hydrogen peroxide, peroxy radicals, endoperoxide, dioxetane, γ irradiation, and smoking [5], and all these processes are induced by oxidative stress. The products of oxidative guanine damage that can pair with adenine cause G-T transversions. Alternatively, those that can pair with guanine cause G-C transversions.
8-Oxoguanine (8-oxoG), which is known as oxidative guanine damage, causes G-T transversions by pairing with adenine [6,7]. Many products of oxidative guanine damage cause G-T transversions, but few cause G-C transversions [8,9,10]. Actually, adenine is predominantly incorporated opposite an apurinic/apyrimidinic site; this “A rule” suggests that the insertion of adenine may not depend on the formation of hydrogen bonds with the DNA damage [11]. Conversely, when guanine is preferentially incorporated, hydrogen bonds certainly form between the DNA damage and guanine [12]. Therefore, base pairs containing guanine damages with guanine are highlighted in this review, and these base pairs can lead to G-C transversions. In addition, we pick up base pairs containing antiviral drugs, which target the RNA-dependent RNA polymerase of RNA viruses and describe implications involving oxidative guanine damage for the 2019 novel coronavirus (SARS-CoV-2).
2. 2,5-Diamino-4H-imidazol-4-one (Iz) and 2,2,4-triamino-5(2H)-oxazolone (Oz)
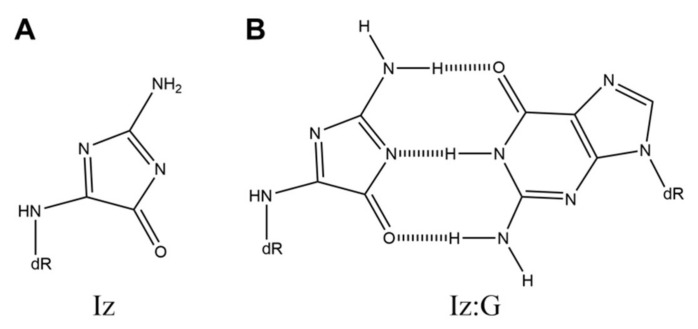
2,5-Diamino-4H-imidazol-4-one (Iz) (Figure 1A) is a product of oxidative guanine damage. Iz can be produced by the oxidation of guanine with Mn-TMPyP/KHSO5, photooxidation in the presence of riboflavin or anthraquinone, or oxidation by γ radiation [13,14,15,16]. Iz can also be produced by the oxidation of guanine and 8-oxoG with peroxynitrite [17] or by photooxidation of 8-oxoG in the presence of riboflavin and methylene blue under basic conditions [14,18,19].
Figure 1.
The structures of (A) Iz and (B) Iz:G.
The structure of Iz can form a base pair with guanine (Figure 1B). In 1998, their ab initio molecular orbital calculations of the base pair containing Iz indicated that the Iz:G base pair is predicted to be stable [15]. Other calculations for the Iz:G base pair have also been confirmed [20]. In addition, Iz:G base pairs are the most thermally stable because the Tm values of base pairs are as follows: Iz:G, 37.1 °C; Iz:T, 27.3 °C; Iz:A, 24.9 °C; Iz:C, 23.7 °C [5]. In 2001, base incorporation studies were performed using Escherichia coli (E. coli) DNA polymerase I. The results of these studies show that guanine is dominantly incorporated opposite Iz, and that the Iz triphosphate is only incorporated opposite guanine [14]. It is also suggested that Iz forms a base pair with guanine [21]. In addition, guanine is incorporated opposite Iz when human DNA polymerase α or rat DNA polymerase β is used [22]. Furthermore, human DNA polymerase η incorporates guanine in addition to cytosine, adenine, and thymine [23]. Thus, all of the DNA polymerases incorporate guanine opposite Iz. Furthermore, when replicated in vivo in E. coli, Iz induces mainly G-C transversions [24].
Based on these experimental results, Iz can be considered to be a product of oxidative guanine damage that can cause G-C transversions. However, Iz has low thermal stability (half-life of 2.5 h at 37 °C [16]) and decomposes into 2,2,4-triamino-5(2H)-oxazolone (Oz) (Figure 2A). Oz has been detected in rat liver DNA [25], and this report shows that Iz is degraded under the near-neutral condition in rat liver.
Figure 2.
The structures of (A) Oz (the closed-ring structure), (B) Oz (the open-ring structure), and (C) Oz:G.
The large fragment of E. coli DNA polymerase I exonuclease minus (Klenow fragment exo-) incorporates adenine opposite Oz, and Taq DNA polymerase mostly incorporates adenine opposite Oz [26]. However, when base incorporation opposite Oz using human DNA polymerase α was first analyzed, their findings reveal that guanine, rather than adenine, is predominantly incorporated [22].
Oz has been reported to have a closed ring structure [26], which does not appear to be able to form base pairs with guanine. However, since the closed-ring structure of Oz has an ester structure, the ring can be opened by hydrolysis, forming an open-ring structure. It has been noticed that the open-ring structure of Oz (Figure 2B) could form two hydrogen bonds with guanine. Ab initio calculation suggests the existence of the base pair structure shown in Figure 2C [22]. Moreover, unlike the closed-ring structure of Oz, the open-ring structure of Oz can be planar, contributing to the stabilization of stacking and conferring an advantage in the DNA elongation reaction because a planar structure has less steric hindrance. In addition, we note that the Tm values of base pairs are as follows: G:C, 55.1 °C; T:A, 48.9 °C; Oz:G, 45.7 °C [27]. A G:C base pair has three hydrogen bonds, and a T:A base pair has two. Since the Tm value of an Oz:G base pair is close to that of T:A, Oz:G base pairs are thought to have two hydrogen bonds (Figure 2C) [27]. Other calculations also show that there are two hydrogen bonds in Oz:G base pairs [20].
In addition to human DNA polymerase α, guanine is incorporated opposite Oz by calf thymus DNA polymerase α, human DNA polymerase β, rat DNA polymerase β, human DNA polymerase δ, and yeast DNA polymerase ε [22,28]. Sulfolobus solfataricus DNA polymerase IV, human DNA polymerase γ, and human DNA polymerase κ incorporate guanine and adenine opposite Oz [22,28]. Human DNA polymerase ι and yeast DNA polymerase ζ incorporate guanine, cytosine, adenine, and thymine opposite Oz [28]. Human DNA polymerase η incorporates guanine, adenine, and cytosine opposite Oz [23]. Thus, for all of the polymerases mentioned above, guanine is either the base or is among the bases that could be incorporated opposite Oz.
In E. coli, G-T transversions are predominant in the mutation spectrum analysis, whereas G-C transversions are barely detected [29]. As mentioned earlier, adenine is predominantly incorporated opposite Oz [26]. The difference between these results [26] and the subsequent results [22,27] may be due to sequence-dependent effects. The possibility of sequence-dependent effects on base incorporation by polymerases should be examined in the future.
Based on these findings, we propose that in addition to Iz, Oz might also cause G-C transversions.
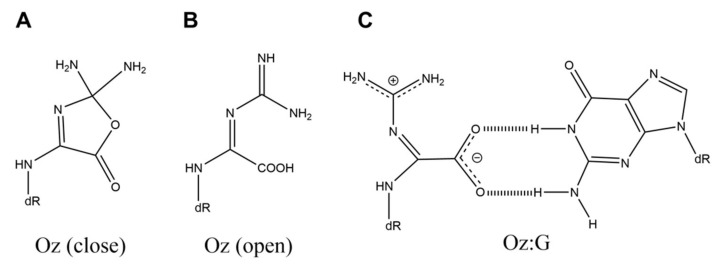
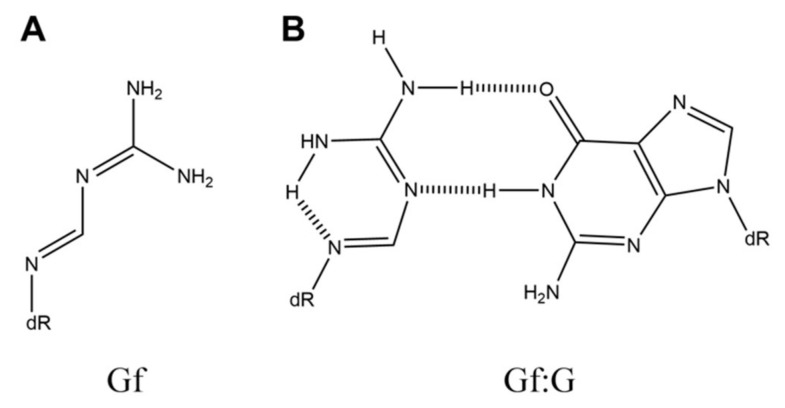
3. Guanidinoformimine (Gf)
Guanidinoformimine (Gf) (Figure 3A) can be produced by decarboxylation of Oz [30]. Base incorporation opposite Gf has been analyzed using the pyrosequencing method with Klenow fragment exo-, human DNA polymerase κ, and yeast DNA polymerase η [30]. In the case of Klenow fragment exo-, cytosine, adenine, and guanine are incorporated opposite Gf. Human DNA polymerase κ incorporates guanine, cytosine, and adenine opposite Gf. When yeast DNA polymerase η is used, guanine and cytosine are incorporated opposite Gf. As a result of base incorporation, Gf is predicted to be able to form a Gf:G base pair with two hydrogen bonds (Figure 3B) [30].
Figure 3.
The structures of (A) Gf and (B) Gf:G.
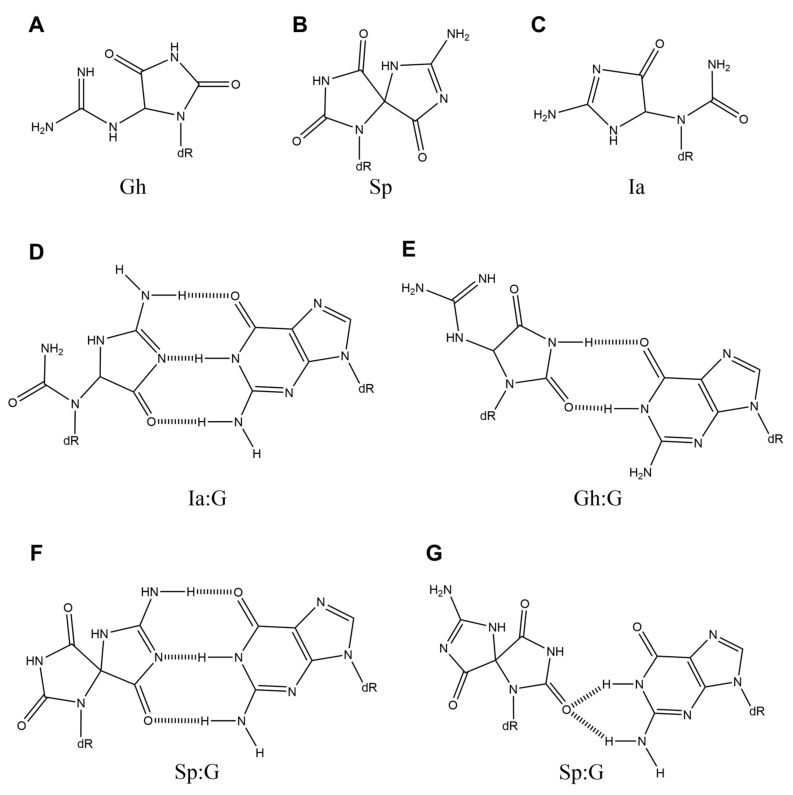
4. Guanidinohydantoin/Iminoallantoin (Gh/Ia) and Spiroiminodihydantoin (Sp)
Using reaction mixtures of 8-oxoG with Na2IrCl6 without isolating each lesion as templates, guanine is incorporated in addition to adenine opposite the products by Klenow fragment exo- [31]. No base is incorporated by calf thymus DNA polymerase α or human DNA polymerase β [31]. Two products are found: One product has a mass corresponded to 10 amu (M-10) below that of 8-oxoG. The other product has a mass corresponded to 16 amu (M+16) above that of 8-oxoG. The authors noted that the M-10 product is guanidinohydantoin (Gh) (Figure 4A), and the M+16 product is 5-OH-8-oxoG [31]. However, a year later, these authors reidentified 5-OH-8-oxoG as spiroiminodihydantoin (Sp) (Figure 4B) [32]. Furthermore, 4-OH-8-oxoG, as reported by Ravanat et al. [33], is actually Sp [34]. After that, Gh and Sp are separated [35].
Figure 4.
The structures of (A) Gh, (B) Sp, (C) Ia, (D) Ia:G, (E) Gh:G, and (F and G) Sp:G.
In this paragraph, we discuss Gh. Klenow fragment exo- incorporates guanine and adenine opposite Gh, and the kcat/Km for the incorporation of adenine is less than that for guanine [36]. However, Gh:A base pairs are more thermally stable than Gh:G base pairs because the Tm values of Gh:A base pairs are higher than those of Gh:G base pairs [36]. The efficiency of incorporation of nucleotides opposite Gh is influenced by adjacent bases [36]. Moreover, the stabilities of Gh:G and Gh:A seem to depend on their positions in the sequence [37].
Gh is in equilibrium with iminoallantoin (Ia) (Figure 4C) [38], and, in 2005, it was described that Ia could form a base pair with guanine as Ia has the same structural moiety as Iz (Figure 4D) [5]. Subsequently, a Gh:G base pair (Figure 4E), which is based on the G:T wobble base pair, was proposed in 2007 [39]. Ab initio calculations for the base pairs shown in Figure 4D,E have indicated that the energy of the base pair shown in Figure 4D is more stable [40]. This result was confirmed by Jena et al. [41,42]. Indeed, in further experiments, human DNA polymerase α [22], calf thymus DNA polymerase α [22], human DNA polymerase β [22], rat DNA polymerase β [22], human DNA polymerase γ [22], yeast DNA polymerase ε [22], human DNA polymerase η [23], Klenow fragment exo- [22,36], Sulfolobus solfataricus DNA polymerase IV [22], and bacteriophage DNA polymerase RB69 exo- [43,44] commonly incorporated guanine opposite Gh/Ia. During transcription by yeast RNA polymerase II, although adenine is predominantly incorporated opposite Gh/Ia, guanine is also incorporated [45]. In reverse transcription by SuperScript III, guanine is incorporated opposite Gh/Ia, in addition to adenine [46]. Furthermore, mutational spectrum analysis in E. coli [47,48,49] yields results that suggest the presence of Gh/Ia:G base pairs.
In 2016, it was reported that Gh dominates at pH < 10.1 and Ia at pH > 10.1 [50]. Since the amount of guanine insertion by Klenow fragment exo- increases at higher pH [50], Ia, but not Gh, can form base pairs with guanine and causes G-C transversions.
Finally, we discuss Sp. After Gh/Ia and Sp are separated [35], the incorporation reaction of Klenow fragment exo- with Sp has been analyzed [36]. As a result, guanine is incorporated in addition to adenine. The Tm values of Sp:A and Sp:G base pairs differ depending on the surrounding sequence [36]. Similarly, the result of experiments of T4 DNA ligation suggests that the stabilities of Sp:G and Sp:A depend on their position in the sequence [37].
Like Ia, Sp has the same structural moiety as Iz, and Sp can form a base pair with guanine that has three hydrogen bonds (Figure 4F) [5]. Base pairs of Sp and guanine have been calculated, and the optimized structures have been obtained [40]. Similar calculations are also reported [51]. In addition, another Sp:G base pair (Figure 4G) has been proposed [52], and it is different from the base pair shown in Figure 4F.
Sp has a chiral carbon that can be separated by HPLC as two diastereomers, Sp1 and Sp2. Although the absolute stereochemistry remains to be determined, analyses of the differences in thermal stability and base incorporation of Sp1 and Sp2 have been performed, and the results are as follows. When the Tm values of both Sp1 and Sp2 are measured, the thermal stability of a base pair with guanine is found to be higher than that with adenine [53]. Mutation spectrum analysis in E. coli [47,48,49] suggests that guanine forms base pairs with both Sp1 and Sp2. DNA polymerase V in E. coli increases G-C transversions in the Sp2 site [54] and then is thought to incorporate guanine opposite Sp2.
In 2009, the absolute configurations of two stereoisomers, Sp1 and Sp2, were determined: Sp1 is in the (−)-S configuration, and Sp2 is in the (+)-R configuration [55]. Regarding S-Sp and R-Sp, Klenow fragment exo-, Sulfolobus solfataricus DNA polymerase IV, and hemo KlenTaq DNA polymerase incorporate guanine opposite S-Sp and R-Sp in addition to adenine [56]. However, S-Sp and R-Sp triphosphates are incorporated only opposite cytosine, indicating that Sp:G base pairs are not formed when S-Sp and R-Sp triphosphates are used [57]. In a transcription reaction with yeast RNA polymerase II, adenine is incorporated opposite S-Sp and R-Sp more often than guanine [45]. Besides, SuperScript III incorporates guanine and adenine opposite both S-Sp and R-Sp [46].
Based on these findings, Ia and Sp are oxidative guanine damages, which are involved in the generation of G-C transversions.
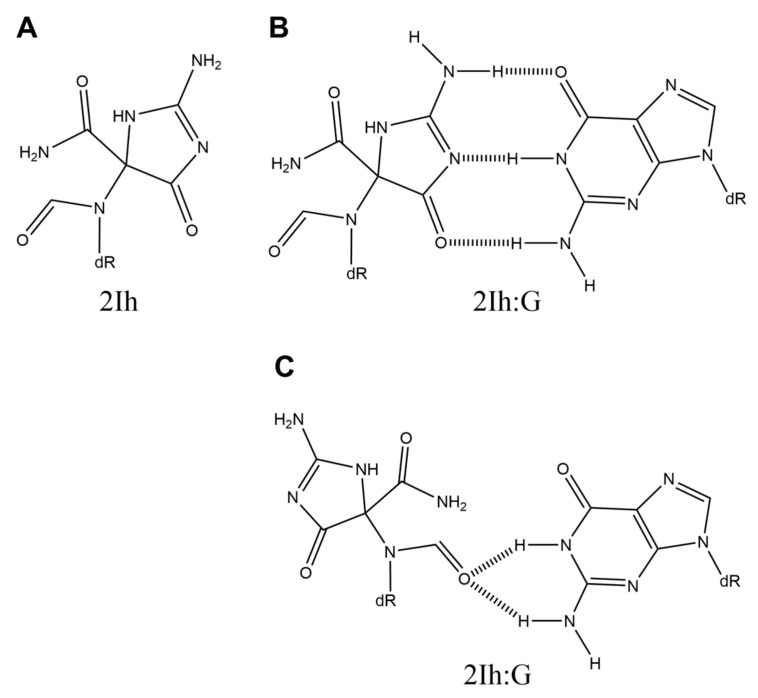
5. 5-Carboxamido-5-formamido-2-iminohydantoin (2Ih)
5-Carboxamido-5-formamido-2-iminohydantoin (2Ih) (Figure 5A) is produced from guanine by Fe(II) or Cu(II) under reducing conditions, X-ray irradiation, or by manganese or nickel complex with KHSO5 [58,59,60,61,62].
Figure 5.
The structures of (A) 2Ih and (B and C) 2Ih:G.
2Ih has a chiral carbon, which can be separated by HPLC as two diastereomers. The thermal stability and base incorporation of S-2Ih and R-2Ih have been analyzed. The results of Tm analyses reveal that both S-2Ih and R-2Ih have higher thermal stability when base-paired with guanine than they do when base-paired with adenine [56]. Because the structure of 2Ih is similar to that of Sp, the 2Ih:G base pairs appear to be the structures shown in Figure 5B,C. In addition, analyses of base incorporation using Klenow fragment exo-, Sulfolobus solfataricus DNA polymerase IV, and hemo KlenTaq DNA polymerase show that guanine is predominantly incorporated opposite both S-2Ih and R-2Ih [56].
Although there has thus far been only one report of these findings, 2Ih is a candidate for a product of oxidative guanine damage that can cause G-C transversions.
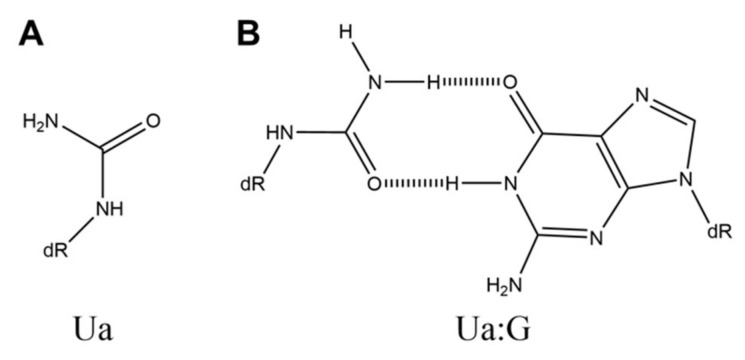
6. Urea (Ua)
Urea (Ua) is generated from thymine by osmium tetroxide (Figure 6A) [63] and has been reported to be a product of oxidative guanine damage [64].
Figure 6.
The structures of (A) Ua and (B) Ua:G.
Molecular mechanics calculations have proposed that the Ua:G base pair has one hydrogen bond [65]. However, the Ua:G base pair was previously proposed to have two hydrogen bonds (Figure 6B) [66]. Analysis of the mutation spectrum in E. coli [48,64,65] also suggests that Ua:G base pairs are formed. DNA polymerase V in E. coli increases G-C transversions in the Ua site [54] and then is thought to incorporate guanine opposite Ua.
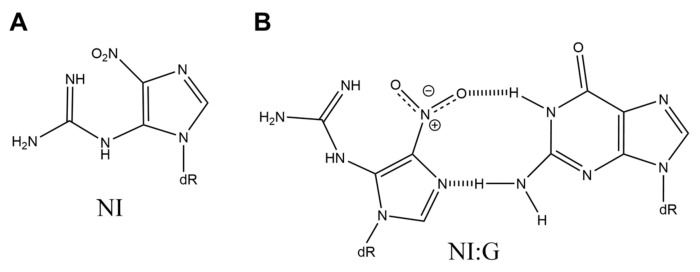
7. 5-Guanidino-4-nitroimidazole (NI)
In 2001, it was reported that 5-guanidino-4-nitroimidazole (NI) (Figure 7A) is produced by the oxidation of guanine by peroxynitrite [17]. NI is also generated by photooxidation of guanine with NaHCO3, NaNO2, and Na2S2O8 [67]. The Tm values of NI:G base pairs are higher than those of NI:A, NI:C, and NI:T base pairs [68], and the ab initio calculations for the base pairs have indicated that NI:G base pairs, shown in Figure 7B, are stable [69]. Calf thymus DNA polymerase α incorporates guanine and adenine opposite NI [70]. However, cytosine is predominantly incorporated opposite NI by human DNA polymerase β [70], Klenow fragment exo- [70], human RNA polymerase II [71], and bacteriophage T7 RNA polymerase [71]. Furthermore, in E. coli, G-C transversions are the minor point mutation, and cytosine is preferentially inserted opposite NI, which leads to no mutation [24,54].
Figure 7.
The structures of (A) NI and (B) NI:G.
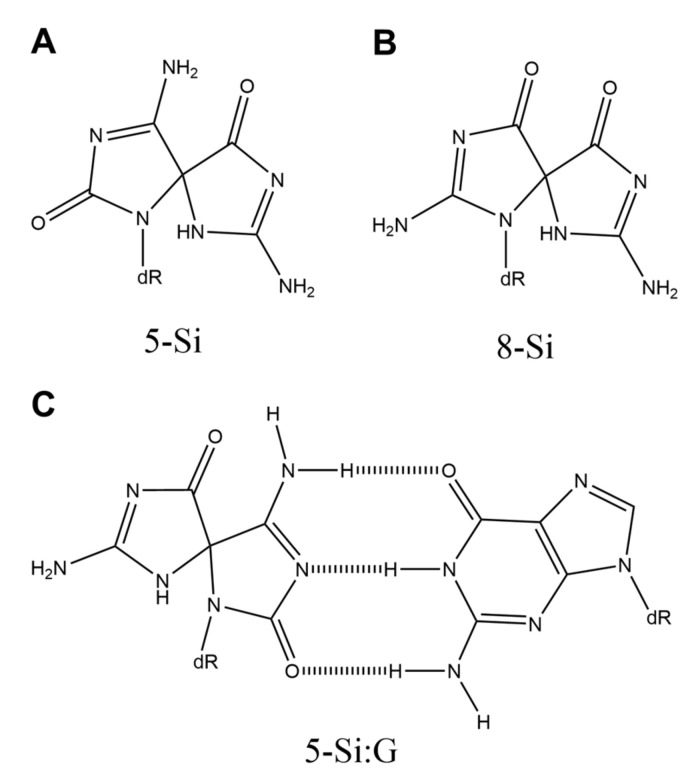
8. Spirodi(iminohydantoin) (5-Si and 8-Si)
Two diastereomers of spirodi(iminohydantoin) (5-Si and 8-Si) (Figure 8A,B) were first reported in 2015 and are produced by oxidation of guanine in the presence of NH4Cl [72]. Though base incorporation opposite 8-Si has not been analyzed, Klenow fragment exo- incorporates adenine and guanine opposite 5-Si [72]. Since 5-Si mimics the hydrogen-bonding pattern of cytosine [72], a 5-Si:G base pair with three hydrogen bonds appears to be the structure shown in Figure 8C. On the other hand, in 2007, it was reported that the product having the same molecular weight as 5-Si and 8-Si is produced by oxidation of 8-oxoG with peroxynitrite [73]. Moreover, this unidentified product has caused G-C and G-T transversions in E. coli, and G-T transversions are the major point mutation [73].
Figure 8.
The structures of (A) 5-Si, (B) 8-Si, and (C) 5-Si:G.
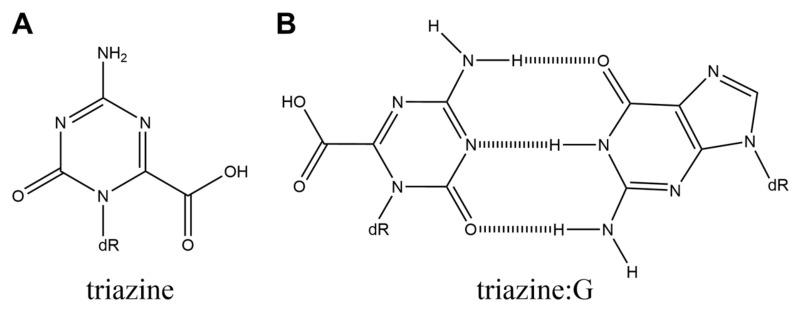
9. Triazine and Unknown M+7 Product
Triazine (Figure 9A) is an oxidation product of guanine and 8-oxoG [74,75]. The product having the same molecular weight as triazine causes G-C and G-T transversions in E. coli, although the product has not been identified as triazine [73]. However, the thermal stability of DNA duplexes, base incorporation with polymerases, and calculations of base pairs have not been analyzed for triazine. Based on the structure of triazine, it can form a base pair with guanine (Figure 9B).
Figure 9.
The structures of (A) triazine and (B) triazine:G.
Moreover, the unidentified product having a mass corresponded to 7 amu (M+7) above that of guanine has been reported, and the M+7 product causes the greatest amounts of G-C transversions in addition to G-A transitions and G-T transversions [73].
10. Other Products by Peroxynitrite
In addition to NI, 8-nitroguanine is a nitration product of guanine [76]. Parabanic acid and N-nitro-dehydroguanidinohydantoin are produced by oxidation of 8-oxoG with peroxynitrite [77,78]. However, since these three products are unstable, researches on these products are unrealistic.
On the other hand, 4-hydroxy-2,5-dioxo-imidazolidine-4-carboxylic acid (HICA) is a stable product of oxidative guanine damage [79,80,81], and the thermal stability of DNA duplexes containing HICA and base incorporation with polymerases opposite HICA have not been analyzed.
11. Alkylated Guanines
In the mutation spectrum for N7-methylguanine, N1-methylguanine, 8-methylguanine, and 1,N2-ethenoguanine, G-C transversions are observed [82]. However, opposite these alkylated guanines, cytosine is predominantly incorporated, and G-C transversions at these alkylated guanines are the minor point mutation [82].
12. 8,5′-Cyclo-2′-deoxyguanosine (cG)
8,5′-cyclo-2′-deoxyguanosine (cG) is the smallest tandem lesion generated by hydroxyl radical [83]. Guanine is the least incorporated base opposite cG by Klenow fragment exo- [84] and Sulfolobus solfolobus P2 DNA polymerase B1 [85]. In addition, cytosine is preferentially incorporated by Sulfolobus solfolobus DNA polymerase IV [84,85], DNA polymerase IV [84], human and Saccharomyces cerevisiae DNA polymerase η [86], human DNA polymerase ι [87]. Human DNA polymerase κ [87] and Saccharomyces cerevisiae DNA polymerase ζ [87] incorporate cytosine and guanine. In E. coli, cG causes no mutations and G-A mutations [88,89]. From the above, cG is not a product of oxidative guanine damage that can cause G-C transversions.
13. Base Pairs Related to New Medicines Against Novel Coronavirus
The spread of the 2019 novel coronavirus (SARS-CoV-2) has caused a global pandemic. Many researchers are considering whether several existing antiviral agents against the influenza virus and others are effective in SARS-CoV-2 [90,91].
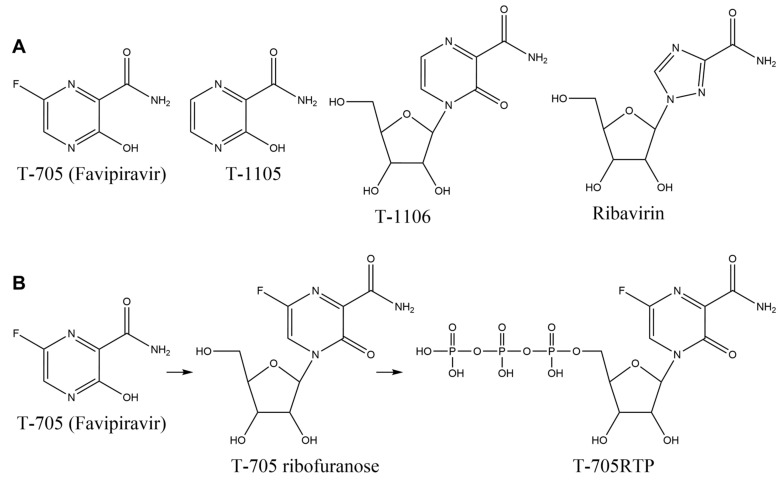
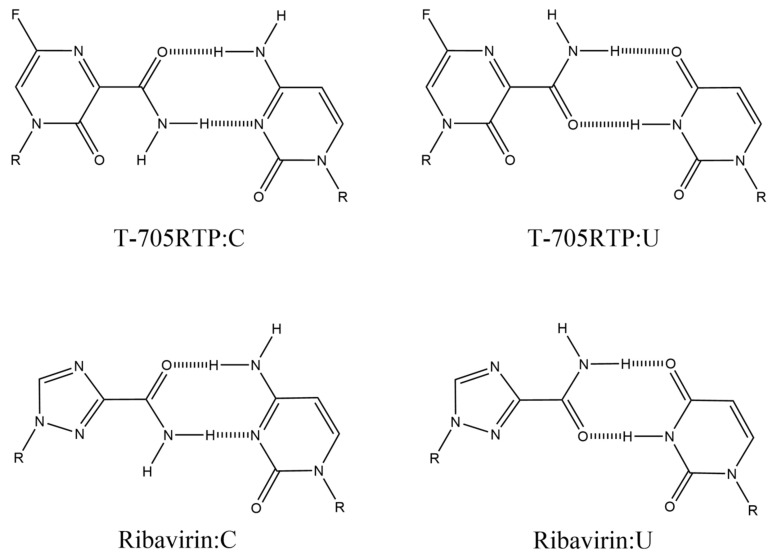
T-705 (favipiravir) (Figure 10A) is an antiviral drug that inhibits the RNA-dependent RNA polymerase (RdRp) of the influenza virus. T-705 eventually leads to its active form, T-705RTP (Figure 10B) [92,93]. T-705 has a broad-spectrum activity against other RNA viruses, such as the Ebola virus and others [94,95]. Since the catalytic region of the RdRp is widely conserved among the RNA viruses, T-705 is said to be effective against a wide range of RNA viruses. The inhibition of RdRp activity affects viral genomic replication, and forming base pairs is important for the viral suppression mechanism: T-705RTP has been proposed to form the base pairs with cytidine and uridine (Figure 11) [93].
Figure 10.
The existing agents against RdRp. (A) The structures of T-705, T-1105, T-1106, and ribavirin. (B) Converting T-705 to T-705RTP.
Figure 11.
The structures of base pairs containing T-705 ribofuranose and ribavirin.
The structure of T-1105 (Figure 10A) is similar to T-705, and T-1105 is also effective in the influenza virus [96]. T-1105 and T-1106 (Figure 10A) have potent antiviral activities against the dengue virus [97]. The base moiety has potential base pairs with cytidine and uridine like T-705 [97]. Via similar considerations, C-nucleoside analogs having amide show anti-influenza activity, and base pairs with cytidine and uridine have been proposed [98].
Ribavirin (Figure 10A) also shows antiviral activity against various RNA viruses, especially in the hepatitis C virus (HCV), and the structure having amide leads to suggested base pairs with cytidine and uridine (Figure 11) [99]. On the other hand, it has been reported that N4−hydroxycytidine, which acts as an analog of cytosine or uridine [100], is effective against several viruses [101].
Remdesivir leads to its active triphosphate and inhibits the RdRp of the Ebola virus [102] and others [103,104]. This drug, which is effective against a wide range of RNA viruses, also has antiviral activity against SARS-CoV-2 [105]. Because remdesivir mimics the structure of adenosine as a substrate, delayed chain termination of RNA synthesis using RdRps of SARS-CoV-2 [106], Nipah virus [107], and Ebola virus [108] show that remdesivir forms the base pair with uridine [105].
Thus, T-705 ribofuranose, T-1106, ribavirin, N4−hydroxycytidine, and remdesivir can act as natural nucleosides in the RNA replication, and the fact that drugs with different structures have effects on RdRps of several viruses suggests that the nucleic acid derivatives capable of forming base pairs have broad-spectrum antiviral activities. Therefore, some ribonucleoside analogs, which can form base pairs, are possible to be effective in SARS-CoV-2.
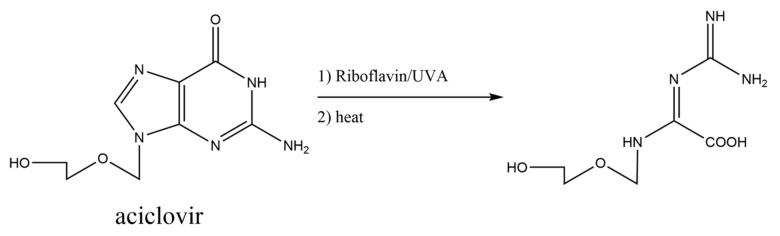
In the above sections, the base pairs containing oxidative guanine damages have been described. The reactions that produce DNA damages can also be adapted for the ribonucleoside analogs. In Figure 12, an example using aciclovir is shown. Taking into consideration that forming base pairs are important in the antiviral mechanisms against RNA viruses, some reaction products capable of forming hydrogen bonds with natural ribonucleotides may be drug candidates for SARS-CoV-2.
Figure 12.
Proposed photosensitization adapted for aciclovir.
14. Conclusions and Future Studies
In this review, we have described the findings so far concerning products of oxidative guanine damage that can form base pairs with guanine. Iz forms mainly Iz:G base pairs. Oz, Gh/Ia, and Sp form base pairs, preferably with guanine in some cases. Gf, 2Ih, Ua, 5-Si, 8-Si, triazine, the M+7 product, and HICA have a few or no experiments on thermal stability, base incorporations, or mutations in E.coli. NI, alkylated guanines, and cG are not involved in G-C transversions. Recently, it has been reported that DNA base pairing is not controlled by DNA properties alone but by appropriateness for substrates in the polymerase active site [109]. Especially this discussion is important in consideration of a wide variety of bases incorporated by translesion synthesis polymerases. Conversely, when the preferentially incorporated base is independent of polymerases, forming base pairs is certainly important: We have collected information on typical generations of oligomer containing the damage, the Tm data, bases incorporated by polymerases except for translesion synthesis polymerases, and mutations in E. coli in Table 1. We note, however, that some lesions have not been analyzed with some polymerases, and that mutation spectrum analysis in human cells has not yet been performed. Further analyses are likely to reveal more detailed information about the mechanisms of G-C transversion.
Table 1.
Oxidative guanine damages that mainly cause G-C transversions.
| Damage | Typical Generations of Oligomer Containing the Damage 1 | The Ease of Formation of Base Pairs 3,4 |
|---|---|---|
| Iz | G/riboflavin, 366 nm [14,15] | Tm: Iz:G > Iz:T > Iz:A > Iz:C [5] |
| Pol I: Iz:G > Iz:A [14] | ||
| Pol α: Iz:G ~ Iz:C > Iz:A [22] | ||
| Pol β: Iz:G ~ Iz:C > Iz:A [22] | ||
| E. coli: Iz:G > Iz:C [24] | ||
| Oz | Iz/heat [26] | Tm: only Oz:G 2 [27] |
| Kf exo-: only Oz:A [26] | ||
| Kf exo-: Oz:A ~ Oz:G [22] | ||
| Taq: Oz:A > Oz:G ~ Oz:C [26] | ||
| Pol α: Oz:G > Oz:A [22] | ||
| Pol β: Oz:G > Oz:A [22] | ||
| Pol γ: Oz:G > Oz:A [22] | ||
| Pol δ: only Oz:G [28] | ||
| Pol ε: only Oz:G [22] | ||
| E. coli: Oz:A > Oz:C [29] | ||
| Gf | Oz/heat [30] | Kf exo-: Gf:A > Gf:G > Gf:C [30] |
| Gh/Ia | 8-oxoG/Na2IrCl6, 4 °C [35] | Tm: Gh/Ia:A > Gh/Ia:C > Gh/Ia:G > Gh/Ia:T [36] |
| 8-oxoG/I2, KI, pH 5.7 [81] | Kf exo-: Gh/Ia:A > Gh/Ia:G [22,36] | |
| Kf exo-: Gh/Ia:G > Gh/Ia:A [36] | ||
| RB69 exo-: Gh/Ia:A > Gh/Ia:G [43,44] | ||
| Pol α: Gh/Ia:G ~ Gh/Ia:A [22] | ||
| Pol β: Gh/Ia:G > Gh/Ia:A [22] | ||
| Pol γ: Gh/Ia:A > Gh/Ia:G [22] | ||
| Pol ε: Gh/Ia:A ~ Gh/Ia:G [22] | ||
| RNA Pol II: Gh/Ia:A > Gh/Ia:G [45] | ||
| SuperScript III: Gh/Ia:G > Gh/Ia:A [46] | ||
| E. coli: Gh:G > Gh:A [47,48,54] | ||
| Sp | 8-oxoG/Na2IrCl6, 50 °C [35] | Tm: Sp:G > Sp:A > Sp:C > Sp:T [36] |
| 8-oxoG/I2, KI, pH 7.7 [81] | Tm: Sp:C > Sp:G > Sp:A > Sp:T [36] | |
| Tm: Sp1:G > Sp1:T > Sp1:A > Sp1:C [53] | ||
| Tm: Sp2:G ~ Sp2:T > Sp2:A > Sp2:C [53] | ||
| Tm: Sp2:G > Sp2:A > Sp2:T > Sp2:C [53] | ||
| Kf exo-: Sp:A > Sp:G [36] | ||
| Kf exo-: Sp:G > Sp:A [36] | ||
| Klen Taq: Sp:G > Sp:A [56] | ||
| RNA Pol II: Sp:A > Sp:G [45] | ||
| SuperScript III: Sp:G > Sp:A [46] | ||
| E. coli: Sp:G > Sp:A [47] | ||
| E. coli: Sp1:A > Sp1:G [48,54] | ||
| E. coli: Sp2:A ~ Sp2:G [48] | ||
| E. coli: Sp2:G > Sp2:A [54] | ||
| SOS-induced E. coli: Sp2:A > Sp2:G [54] | ||
| 2Ih | G/Mn-TMPyP, KHSO5 [59] | Tm: 2Ih:G > 2Ih:A > 2Ih:C > 2Ih:T [56] |
| G/X-ray, ascorbate [62] | Kf exo-: 2Ih:G > 2Ih:A [56] | |
| G/Fe(II)-EDTA, H2O2, ascorbate [62] | Klen Taq: 2Ih:G > 2Ih:A [56] | |
| G/NiCR, KHSO5 [60] | ||
| G/(AcO)2Cu, ascorbate, H2O2 [61] | ||
| Ua | oxalurate/ NaHCO3 [64] | E. coli: Ua:A > Ua:G > Ua:T [48] |
| E. coli: Ua:A > Ua:G > Ua:T > Ua:C [54] | ||
| E.coli: only Ua:A [64] | ||
| SOS-induced E. coli: Ua:G > Ua:A > Ua:T [64] | ||
| NI | G/peroxynitrate [17] | Tm: NI:G > NI:A > NI:C > NI:T [68] |
| G/308 nm, NaHCO3, NaNO2, Na2S2O8 [67] | Pol α: NI:A > NI:G > NI:C [70] | |
| Pol β: NI:C > NI:A > NI:G [70] | ||
| Kf exo-: NI:C > NI:A > NI:G [70] | ||
| T7 RNA Pol: NI:C > NI:A > NI:G > NI:U [71] | ||
| RNA Pol II: only NI:C [71] | ||
| E. coli: NI:C > NI:A > NI:T > NI:G [24] | ||
| E. coli: NI:C > NI:A > NI:G > NI:T [54] | ||
| SOS-induced E. coli: NI:C > NI:A > NI:T > NI:G [54] | ||
| 5-Si | G/riboflavin, NH4Cl, 350 nm [72] | Kf exo-: 5-Si:A > 5-Si:G > 5-Si:C ~ 5-Si:T [72] |
| G/rose bengal, NH4Cl, 350 nm [72] | (E. coli: ?:A > ?:T > ?:G > ?:C)5 [73] | |
| G/Na2IrCl6, NH4Cl [72] | (SOS-induced E. coli: ?:A > ?:G > ?:T > ?:C)5 [73] | |
| (8-oxoG/peroxynitrate) [73] | ||
| triazine | (8-oxoG/peroxynitrate) [73] | (E. coli: ?:G > ?:A > ?:T > ?:C)5 [73] |
| (SOS-induced E. coli: ?:A > ?:G > ?:T > ?:C)5 [73] | ||
| M+7 | (8-oxoG/peroxynitrate) [73] | (E. coli: ?:G > ?:T > ?:A > ?:C)5 [73] |
| cG | Kf exo-: cG:C > cG:T > cG:A ~ cG:G [84] | |
| Pol B1: cG:C > cG:A > cG:T > cG:G [85] | ||
| E. coli: cG:C > cG:T [88,89] |
1 All detection methods are HPLC. 2 The Tm values of Oz:C, Oz:T, or Oz:A are not determined and below 40 °C. 3 This table does not contain the data of base incorporations by translesion synthesis polymerases. 4 “Damage:Base 1 > Damage:Base 2” means that the Tm value of Damage:Base 1 is higher than that of Damage:Base 2, or that Base 1 is incorporated more preferentially than Base 2 by polymerases or in E. coli. “Damage:Base 1 ~ Damage:Base 2” means that the Tm value of Damage:Base 1 is almost the same as that of Damage:Base 2, or that Base 1 and Base 2 are incorporated to the same degree by polymerases or in E. coli. “Pol” is an abbreviation of “Polymerase”. “Kf” is an abbreviation of “Klenow fragment”. 5 The unidentified product “?” having the same mass as each damage is used.
In addition, to prevent the occurrence of mutations, DNA damages must be repaired [110]. Oxidative damages are usually repaired by base excision repair enzymes. For example, human NEIL1 and human NTH1 are active against Oz, but similar activities against Oz:C, Oz:G, and Oz:A are shown [111]. Considering the facts, at least human NEIL1 and human NTH1 do not depend on the stability of base pairs containing Oz. Therefore, in addition to the previously known results, it is necessary to newly measure the activity of various repair enzymes to determine whether the enzyme depends on the stability of the base pair or not.
On the other hand, for nucleotide excision repair, it is important for XPC-RAD23B to detect the bulge of the structure of DNA duplex. Previously, the stability of base pairs containing 5-formyluracil, which is oxidative damage to thymine, was correlated with nucleotide excision repair activity [112]. Therefore, in the future, it is necessary to study the correlation between the stability of base pairs containing oxidative guanine damage and nucleotide excision repair activity.
Lastly, in the Section 13 of this review, several antiviral drugs are described and forming base pairs inhibit the RdRp. Since some oxidative guanine damages can form base pairs, researchers may find drug candidates for SARS-CoV-2 when reactions producing oxidative guanine damage are adapted for the ribonucleoside analogs.
Acknowledgments
The authors would like to thank T. Ohshima (Tokushima Bunri University).
Abbreviations
| Iz | 2,5-diamino-4H-imidazol-4-one |
| Oz | 2,2,4-triamino-5(2H)-oxazolone |
| Gf | guanidinoformimine |
| Gh | guanidinohydantoin |
| Ia | iminoallantoin |
| Sp | spiroiminodihydantoin |
| 2Ih | 5-carboxamido-5-formamido-2-iminohydantoin |
| Ua | urea |
| 8-oxoG | 8-oxoguanine |
| E. coli | Escherichia coli |
| Klenow fragment exo- | the large fragment of Escherichia coli DNA polymerase I exonuclease minus |
| dR | deoxyribose |
| NI | 5-guanidino-4-nitroimidazole |
| Si | spirodi(iminohydantoin) |
| HICA | 4-hydroxy-2,5-dioxo-imidazolidine-4-carboxylic acid |
| cG | 8,5′-cyclo-2′-deoxyguanosine |
| SARS-CoV-2 | 2019 novel coronavirus |
| RdRp | RNA-dependent RNA polymerase |
| HCV | hepatitis C virus |
| R | ribose |
Author Contributions
Conceptualization, K.K.; writing—original draft preparation, K.K., T.K., and M.H.-S.; writing—review and editing, K.K., T.K., M.M., M.H.-S., and H.M.; visualization, K.K. and T.K.; supervision, K.K. and H.M.; project administration, K.K.; funding acquisition, K.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by research grants from Tokushima Bunri University, from LNest Grant L-RAD Award, from Radiation Effects Association, from the Nakatomi Foundation, from some JSPS KAKENHIs, and from the Japan Prize Foundation. The APC was funded by JSPS KAKENHI 17K00558.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Steenken S., Jovanovic S.V. How easily oxidizable is DNA? One-electron reduction potentials of adenosine and guanosine radicals in aqueous solution. J. Am. Chem. Soc. 1997;119:617–618. doi: 10.1021/ja962255b. [DOI] [Google Scholar]
- 2.Maehira F., Miyagi I., Asato T., Eguchi Y., Takei H., Nakatsuki K., Fukuoka M., Zaha F. Alterations of protein kinase C, 8-hydroxydeoxyguanosine, and K-ras oncogene in rat lungs exposed to passive smoking. Clin. Chim. Acta. 1999;289:133–144. doi: 10.1016/S0009-8981(99)00163-1. [DOI] [PubMed] [Google Scholar]
- 3.Pfeifer G.P., Besaratinia A. Mutational spectra of human cancer. Hum. Genet. 2009;125:493–506. doi: 10.1007/s00439-009-0657-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oliva M.R., Ripoll F., Muñiz P., Iradi A., Trullenque R., Valls V., Drehmer E., Sáez G.T. Genetic alterations and oxidative metabolism in sporadic colorectal tumors from a Spanish community. Mol. Carcinog. 1997;18:232–243. doi: 10.1002/(SICI)1098-2744(199704)18:4<232::AID-MC7>3.0.CO;2-F. Erratum in 1997, 19, 280. [DOI] [PubMed] [Google Scholar]
- 5.Kino K., Sugiyama H. UVR-induced G-C to C-G transversions from oxidative DNA damage. Mutat. Res. 2005;571:33–42. doi: 10.1016/j.mrfmmm.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 6.Klein J.C., Bleeker M.J., Saris C.P., Roelen H.C.P.F., Brugghe H.F., van den Elst H., van der Marel G.A., van Boom J.H., Westra J.G., Kriek E., et al. Repair and replication of plasmids with site-specific 8-oxodG and 8-AAFdG residues in normal and repair-deficient human cells. Nucleic Acids Res. 1992;20:4437–4443. doi: 10.1093/nar/20.17.4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moriya M. Single-stranded shuttle phagemid for mutagenesis studies in mammalian cells: 8-oxoguanine in DNA induces targeted G•C->T•A transversions in simian kidney cells. Proc. Natl. Acad. Sci. USA. 1993;90:1122–1126. doi: 10.1073/pnas.90.3.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bjelland S., Seeberg E. Mutagenicity toxicity and repair of DNA base damage induced by oxidation. Mutat. Res. 2003;531:37–80. doi: 10.1016/j.mrfmmm.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Turesky R.J. Heterocyclic aromatic amine metabolism, DNA adduct formation, mutagenesis, and carcinogenesis. Drug Metab. Rev. 2002;34:625–650. doi: 10.1081/DMR-120005665. [DOI] [PubMed] [Google Scholar]
- 10.Schut H.A.J., Snyderwine E.G. DNA adduct of heterocyclic amine food mutagens: Implications for mutagenesis and carcinogenesis. Carcinogenesis. 1999;20:353–368. doi: 10.1093/carcin/20.3.353. [DOI] [PubMed] [Google Scholar]
- 11.Kool E.T. Hydrogen bonding, base stacking, and steric effects in DNA replication. Annu. Rev. Biophys. Biomol. Struct. 2001;30:1–22. doi: 10.1146/annurev.biophys.30.1.1. [DOI] [PubMed] [Google Scholar]
- 12.Kino K., Sugiyama H., Miyazawa H. Molecular basis of guanine oxidation under UV-A/VIS radiation and its biological effects. In: Miura S., Nakano S., editors. Progress in DNA Damage Research. Nova Science Publishers; Hauppauge, NY, USA: 2008. pp. 271–276. [Google Scholar]
- 13.Vialas C., Claparols C., Pratviel G., Meunier B. Guanine oxidation in double-stranded DNA by Mn-TMPyP/KHSO5: 5,8-dihydroxy-7,8-dihydroguanine residue as a key precursor of imidazolone and parabanic acid derivatives. J. Am. Chem. Soc. 2000;122:2157–2167. doi: 10.1021/ja992860p. [DOI] [Google Scholar]
- 14.Kino K., Sugiyama H. Possible cause of G•C->C•G transversion mutation by guanine oxidation product, imidazolone. Chem. Biol. 2001;8:369–378. doi: 10.1016/S1074-5521(01)00019-9. [DOI] [PubMed] [Google Scholar]
- 15.Kino K., Saito I., Sugiyama H. Product analysis of GG-specific photooxidation of DNA via electron transfer: 2-aminoimidazolone as a major guanine oxidation product. J. Am. Chem. Soc. 1998;120:7373–7374. doi: 10.1021/ja980763a. [DOI] [Google Scholar]
- 16.Cadet J., Berger M., Buchko G.W., Joshi P.C., Raoul S., Ravanat J.-L. 2,2-Diamino-4-[(3,5-di-O-acetyl-2-deoxy-β-D-erythro-pentofuranosyl)amino]-5-(2H)-oxazolone: A novel and predominant radical oxidation product of 3′,5′-di-O-acetyl-2′-deoxyguanosine. J. Am. Chem. Soc. 1994;116:7403–7404. doi: 10.1021/ja00095a052. [DOI] [Google Scholar]
- 17.Niles J.C., Wishnok J.S., Tannenbaum S.R. A novel nitroimidazole compound formed during the reaction of peroxynitrite with 2′,3′,5′-tri-O-acetyl-guanosine. J. Am. Chem. Soc. 2001;123:12147–12151. doi: 10.1021/ja004296k. [DOI] [PubMed] [Google Scholar]
- 18.Luo W., Muller J.G., Burrows C.J. The pH-dependent role of superoxide in riboflavin-catalyzed photooxidation of 8-oxo-7,8-dihydroguanosine. Org. Lett. 2001;3:2801–2804. doi: 10.1021/ol0161763. [DOI] [PubMed] [Google Scholar]
- 19.Raoul S., Cadet J. Photosensitized reaction of 8-oxo-7,8-dihydro-2′-deoxyguanosine: Identification of 1-(2-deoxy-β-D-erythro-pentofuranosyl)cyanuric acid as the major singlet oxygen oxidation product. J. Am. Chem. Soc. 1996;118:1892–1898. doi: 10.1021/ja952347l. [DOI] [Google Scholar]
- 20.Jena N.R., Mishra P.C. Normal and reverse base pairing of Iz and Oz lesions in DNA: Structural implications for mutagenesis. RSC Adv. 2016;6:64019–64027. doi: 10.1039/C6RA14031A. [DOI] [Google Scholar]
- 21.Mourgues S., Trzcionka J., Vasseur J.-J., Pratviel G., Meunier B. Incorporation of oxidized guanine nucleoside 5′-triphosphates in DNA with DNA polymerases and preparation of single-lesion carrying DNA. Biochemistry. 2008;47:4788–4799. doi: 10.1021/bi7022199. [DOI] [PubMed] [Google Scholar]
- 22.Kino K., Sugasawa K., Mizuno T., Bando T., Sugiyama H., Akita M., Miyazawa H., Hanaoka F. Eukaryotic DNA polymerases α β and ε incorporate guanine opposite 2,2,4-triamino-5(2H)-oxazolone. ChemBioChem. 2009;10:2613–2616. doi: 10.1002/cbic.200900492. [DOI] [PubMed] [Google Scholar]
- 23.Kino K., Ito N., Sugasawa K., Sugiyama H., Hanaoka F. Translesion synthesis by human DNA polymerase η across oxidative products of guanine. Nucleic Acids Symp. Ser. 2004;48:171–172. doi: 10.1093/nass/48.1.171. [DOI] [PubMed] [Google Scholar]
- 24.Neeley W.L., Delaney J.C., Henderson P.T., Essigmann J.M. In vivo bypass efficiencies and mutational signatures of the guanine oxidation products 2-aminoimidazolone and 5-guanidino-4-nitroimidazole. J. Biol. Chem. 2004;279:43568–43573. doi: 10.1074/jbc.M407117200. [DOI] [PubMed] [Google Scholar]
- 25.Matter B., Malejka-Giganti D., Csallany A.S., Tretyakova N. Quantitative analysis of the oxidative DNA lesion, 2,2-diamino-4-(2-deoxy-β-D-erythro-pentofuranosyl)amino]-5(2H)-oxazolone (oxazolone), in vitro and in vivo by isotope dilution-capillary HPLC-ESI-MS/MS. Nucleic Acids Res. 2006;34:5449–5460. doi: 10.1093/nar/gkl596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duarte V., Gasparutto D., Jaquinod M., Cadet J. In vitro DNA synthesis opposite oxazolone and repair of this DNA damage using modified oligonucleotides. Nucleic Acids Res. 2000;28:1555–1563. doi: 10.1093/nar/28.7.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki M., Ohtsuki K., Kino K., Kobayashi T., Morikawa M., Kobayashi T., Miyazawa H. Effects of stability of base pairs containing an oxazolone on DNA elongation. J. Nucleic Acids. 2014;2014:178350. doi: 10.1155/2014/178350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki M., Kino K., Kawada T., Morikawa M., Kobayashi T., Miyazawa H. Analysis of nucleotide insertion opposite 2,2,4-triamino-5(2H)-oxazolone by eukaryotic B- and Y-family DNA polymerases. Chem. Res. Toxicol. 2015;28:1307–1316. doi: 10.1021/acs.chemrestox.5b00114. [DOI] [PubMed] [Google Scholar]
- 29.Henderson P.T., Delaney J.C., Gu F., Tannenbaum S.R., Essigmann J.M. Oxidation of 7,8-dihydro-8-oxoguanine affords lesions that are potent sources of replication errors in vivo. Biochemistry. 2002;41:914–921. doi: 10.1021/bi0156355. [DOI] [PubMed] [Google Scholar]
- 30.Stathis D., Lischke U., Koch S.C., Deiml C.A., Carell T. Discovery and mutagenicity of a guanidinoformimine lesion as a new intermediate of the oxidative deoxyguanosine degradation pathway. J. Am. Chem. Soc. 2012;134:4925–4930. doi: 10.1021/ja211435d. [DOI] [PubMed] [Google Scholar]
- 31.Duarte V., Muller J.G., Burrows C.J. Insertion of dGMP and dAMP during in vitro DNA synthesis opposite an oxidized form of 7,8-dihydro-8-oxoguanine. Nucleic Acids Res. 1999;27:496–502. doi: 10.1093/nar/27.2.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo W., Muller J.G., Rachlin E.M., Burrows C.J. Characterization of spiroiminodihydantoin as a product of one-electron oxidation of 8-oxo-7,8-dihydroguanosine. Org. Lett. 2000;2:613–616. doi: 10.1021/ol9913643. [DOI] [PubMed] [Google Scholar]
- 33.Ravanat J.-L., Cadet J. Reaction of singlet oxygen with 2-deoxyguanosine and DNA. Isolation and characterization of the main oxidation products. Chem. Res. Toxicol. 1995;8:379–388. doi: 10.1021/tx00045a009. [DOI] [PubMed] [Google Scholar]
- 34.Niles J.C., Wishnok J.S., Tannenbaum S.R. Spiroiminodihydantoin is the major product of the 8-oxo-7,8-dihydroguanosine reaction with peroxynitrite in the presence of thiols and guanosine photooxidation by methylene blue. Org. Lett. 2001;3:963–966. doi: 10.1021/ol006993n. [DOI] [PubMed] [Google Scholar]
- 35.Leipold M.D., Muller J.G., Burrows C.J., David S.S. Removal of hydantoin products of 8-oxoguanine oxidation by the Escherichia coli. Biochemistry. 2000;39:14984–14992. doi: 10.1021/bi0017982. [DOI] [PubMed] [Google Scholar]
- 36.Kornyushyna O., Berges A.M., Muller J.G., Burrows C.J. In vitro nucleotide misinsertion opposite the oxidized guanosine lesions spiroiminodihydantoin and guanidinohydantoin and DNA synthesis past the lesions using Escherichia coli DNA polymerase I (Klenow Fragment) Biochemistry. 2002;41:15304–15314. doi: 10.1021/bi0264925. [DOI] [PubMed] [Google Scholar]
- 37.Zhao X., Muller J.G., Halasyam M., David S.S., Burrows J.C. In vitro ligation of oligodeoxynucleotides containing C8-oxidized purine lesions using bacteriophage T4 DNA ligase. Biochemistry. 2007;46:3734–3744. doi: 10.1021/bi062214k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo W., Muller J.G., Rachlin E.M., Burrows C.J. Characterization of hydantoin products from one-electron oxidation of 8-oxo-7,8-dihydroguanosine in a nucleoside model. Chem. Res. Toxicol. 2001;14:927–938. doi: 10.1021/tx010072j. [DOI] [PubMed] [Google Scholar]
- 39.Krishnamurthy N., Muller J.G., Burrows C.J., David S.S. Unusual structural features of hydantoin lesions translate into efficient recognition by Escherichia coli Fpg. Biochemistry. 2007;46:9355–9365. doi: 10.1021/bi602459v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki M., Kino K., Morikawa M., Kobayashi T., Komori R., Miyazawa H. Calculation of the stabilization energies of oxidatively damaged guanine base pairs with guanine. Molecules. 2012;17:6705–6715. doi: 10.3390/molecules17066705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jena N.R., Gaur V., Mishrac P.C. The R- and S-diastereoisomeric effects on the guanidinohydantoin-induced mutations in DNA. Phys. Chem. Chem. Phys. 2015;17:18111–18120. doi: 10.1039/C5CP02636A. [DOI] [PubMed] [Google Scholar]
- 42.Jena N.R., Bansal M., Mishrac P.C. Conformational stabilities of iminoallantoin and its base pairs in DNA: Implications for mutagenicity. Phys. Chem. Chem. Phys. 2016;18:12774–12783. doi: 10.1039/C6CP02212J. [DOI] [PubMed] [Google Scholar]
- 43.Aller P., Ye Y., Wallace S.S., Burrows C.J., Doublié S. Crystal structure of a replicative DNA polymerase bound to the oxidized guanine lesion guanidinohydantoin. Biochemistry. 2010;49:2502–2509. doi: 10.1021/bi902195p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beckman J., Wang M., Blaha G., Wang J., Konigsberg W.H. Substitution of Ala for Tyr567 in RB69 DNA polymerase allows dAMP and dGMP to be inserted opposite guanidinohydantoin. Biochemistry. 2010;49:8554–8563. doi: 10.1021/bi100913v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oh J., Fleming A.M., Xu J., Chong J., Burrows C.J., Wang D. RNA polymerase II stalls on oxidative DNA damage via a torsion-latch mechanism involving lone pair–π and CH–π interactions. Proc. Natl. Acad. Sci. USA. 2020;117:9338–9348. doi: 10.1073/pnas.1919904117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alenko A., Fleming A.M., Burrows C.J. Reverse transcription past products of guanine oxidation in RNA leads to insertion of A and C opposite 8-oxo-7,8-dihydroguanine and A and G opposite 5-guanidinohydantoin and spiroiminodihydantoin diastereomers. Biochemistry. 2017;56:5053–5064. doi: 10.1021/acs.biochem.7b00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Henderson P.T., Delaney J.C., Muller J.G., Neeley W.L., Tannenbaum S.R., Burrows C.J., Essigmann J.M. The hydantoin lesions formed from oxidation of 7,8-dihydro-8-oxoguanine are potent sources of replication errors in vivo. Biochemistry. 2003;42:9257–9262. doi: 10.1021/bi0347252. [DOI] [PubMed] [Google Scholar]
- 48.Delaney S., Neeley W.L., Delaney J.C., Essigmann J.M. The substrate specificity of MutY for hyperoxidized guanine lesions in vivo. Biochemistry. 2007;46:1448–1455. doi: 10.1021/bi061174h. [DOI] [PubMed] [Google Scholar]
- 49.Hori M., Suzuki T., Minakawa N., Matsuda A., Harashima H., Kamiya H. Mutagenicity of secondary oxidation products of 8-oxo-7,8-dihydro-2′-deoxyguanosine 5′-triphosphate (8-hydroxy-2′-deoxyguanosine 5′-triphosphate) Mutat. Res. 2011;714:11–16. doi: 10.1016/j.mrfmmm.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 50.Zhu J., Fleming A.M., Orendt A.M., Burrows J. CpH-Dependent equilibrium between 5-guanidinohydantoin and iminoallantoin affects nucleotide insertion opposite the DNA lesion. J. Org. Chem. 2016;81:351–359. doi: 10.1021/acs.joc.5b02180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shukla P.K., Mishra P.C. Base pairing patterns of DNA base lesion spiroiminodihydantoin: A DFT study. Int. J. Quantum Chem. 2013;113:2600–2604. doi: 10.1002/qua.24512. [DOI] [Google Scholar]
- 52.Jia L., Shafirovich V., Shapiro R., Geacintov N.E., Broyde S. Structural and thermodynamic features of spiroiminodihydantoin damaged DNA duplexes. Biochemistry. 2005;44:13342–13353. doi: 10.1021/bi050790v. [DOI] [PubMed] [Google Scholar]
- 53.Gruessner B., Dwarakanath M., Stewart E., Bae Y., Jamieson E.R. Effect of base-pairing partner on the thermodynamic stability of the diastereomeric spiroiminodihydantoin lesion. Chem. Res. Toxicol. 2016;29:279–284. doi: 10.1021/acs.chemrestox.5b00453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neeley W.L., Delaney S., Alekseyev Y.O., Jarosz D.F., Delaney J.C., Walker G.C., Essigmann J.M. DNA polymerase V allows bypass of toxic guanine oxidation products in vivo. J. Biol. Chem. 2007;282:12741–12748. doi: 10.1074/jbc.M700575200. [DOI] [PubMed] [Google Scholar]
- 55.Ding S., Jia L., Durandin A., Crean C., Kolbanovskiy A. Absolute configurations of spiroiminodihydantoin and allantoin stereoisomers: Comparison of computed and measured electronic circular dichroism spectra. Chem. Res. Toxicol. 2009;22:1189–1193. doi: 10.1021/tx900107q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alshykhly O.R., Fleming A.M., Burrows C.J. Guanine oxidation product 5-carboxamido-5-formamido-2-iminohydantoin induces mutations when bypassed by DNA polymerases and is a substrate for base excision repair. Chem. Res. Toxicol. 2015;28:1861–1871. doi: 10.1021/acs.chemrestox.5b00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang J., Yennie C.J., Delaney S. Klenow fragment discriminates against the incorporation of the hyperoxidized dGTP lesion spiroiminodihydantoin into DNA. Chem. Res. Toxicol. 2015;28:2325–2333. doi: 10.1021/acs.chemrestox.5b00330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lapi A., Pratviel G., Meunier B. Guanine oxidation in double-stranded DNA by MnTMPyP/KHSO5: At least three independent reaction pathways. Met. Based Drugs. 2001;8:47–56. doi: 10.1155/MBD.2001.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mourgues S., Kupan A., Pratviel G., Meunier B. Use of short duplexes for the analysis of the sequence-dependent cleavage of DNA by a chemical nuclease, a manganese porphyrin. Chembiochem. 2005;6:2326–2335. doi: 10.1002/cbic.200500254. [DOI] [PubMed] [Google Scholar]
- 60.Ghude P., Schallenbeiger M.A., Fleming A.M., Muller J.G., Burrows C.J. Comparison of transition metal-mediated oxidation reactions of guanine in nucleoside and single-stranded oligodeoxynucleotide contexts. Inorg. Chim. Acta. 2011;369:240–246. doi: 10.1016/j.ica.2010.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fleming A.M., Muller J.G., Ji I., Burrows C.J. Characterization of 2′-deoxyguanosine oxidation products observed in the Fenton-like system Cu(Il)/H2O2/reductant in nucleoside and oligodeoxynucleotide contexts. Org. Biomol. Chem. 2011;9:3338–3348. doi: 10.1039/c1ob05112a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alshykhly O.R., Fleming A.M., Burrows C.J. 5-Carboxamido-5-formamido-2-iminohydantoin, in addition to 8-oxo-7,8-dihydroguanine, is the major product of the iron-Fenton or X-ray radiation-induced oxidation of guanine under aerobic reducing conditions in nucleoside and DNA contexts. J. Org. Chem. 2015;80:6996–7007. doi: 10.1021/acs.joc.5b00689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ide H., Kow Y.W., Wallace S.S. Thymine glycols and urea residues in M13 DNA constitute replicative blocks in vitro. Nucleic Acids Res. 1985;13:8035–8052. doi: 10.1093/nar/13.22.8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Henderson P.T., Neeley W.L., Delaney J.C., Gu F., Niles J.C., Hah S.S., Tannenbaum S.R., Essigmann J.M. Urea lesion formation in DNA as a consequence of 7,8-dihydro-8-oxoguanine oxidation and hydrolysis provides a potent source of point mutations. Chem. Res. Toxicol. 2005;18:12–18. doi: 10.1021/tx049757k. [DOI] [PubMed] [Google Scholar]
- 65.Maccabee M., Evans J.S., Glackin M.P., Hatahet Z., Wallace S.S. Pyrimidine ring fragmentation products. Effects of lesion structure and sequence context on mutagenesis. J. Mol. Biol. 1994;236:514–530. doi: 10.1006/jmbi.1994.1162. [DOI] [PubMed] [Google Scholar]
- 66.Suzuki M., Kino K., Miyazawa H. Selectivity of bases incorporated opposite oxidative guanine damages by DNA polymerases. Rad. Biol. Res. Comm. 2012;47:137–164. [Google Scholar]
- 67.Joffe A., Mock S., Yun B.H., Kolbanovskiy A., Geacintov N.E., Shafirovich V. Oxidative generation of guanine radicals by carbonate radicals and their reactions with nitrogen dioxide to form site specific 5-guanidino-4-nitroimidazole lesions in oligodeoxynucleotides. Chem. Res. Toxicol. 2003;16:966–973. doi: 10.1021/tx025578w. [DOI] [PubMed] [Google Scholar]
- 68.Neeley W.L., Henderson P.T., Essigmann J.M. Efficient synthesis of DNA containing the guanine oxidation-nitration product 5-guanidino-4-nitroimidazole: Generation by a postsynthetic substitution reaction. Org. Lett. 2004;6:245–248. doi: 10.1021/ol036188j. [DOI] [PubMed] [Google Scholar]
- 69.Jia L., Shafirovich V., Shapiro R., Geacintov N.E., Broyde S. Flexible 5-guanidino-4-nitroimidazole DNA lesions: Structures and thermodynamics. Biochemistry. 2006;45:6644–6655. doi: 10.1021/bi0601757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gu F., Stillwell W.G., Wishnok J.S., Shallop A.J., Jones R.A., Tannenbaum S.R. Peroxynitrite-induced reactions of synthetic oligo 2′-deoxynucleotides and DNA containing guanine: Formation and stability of a 5-guanidino-4-nitroimidazole lesion. Biochemistry. 2002;41:7508–7518. doi: 10.1021/bi020148q. [DOI] [PubMed] [Google Scholar]
- 71.Dimitri A., Jia L., Shafirovich V., Geacintov N.E., Broyde S., Scicchitano D.A. Transcription of DNA containing the 5-guanidino-4-nitroimidazole lesion by human RNA polymerase II and bacteriophage T7 RNA polymerase. DNA Repair. 2008;7:1276–1288. doi: 10.1016/j.dnarep.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fleming A.M., Armentrout E.I., Zhu J., Muller J.G., Burrows C.J. Spirodi(iminohydantoin) products from oxidation of 2′-deoxyguanosine in the presence of NH4Cl in nucleoside and oligodeoxynucleotide contexts. J. Org. Chem. 2015;80:711–721. doi: 10.1021/jo502665p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Delaney S., Delaney J.C., Essigmann J.M. Chemical-biological fingerprinting: Probing the properties of DNA lesions formed by peroxynitrite. Chem. Res. Toxicol. 2007;20:1718–1729. doi: 10.1021/tx700273u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moschel R.C., Behrman E.J. Oxidation of nucleic acid bases by potassium peroxodisulfate in alkaline aqueous solution. J. Org. Chem. 1974;39:1983–1989. doi: 10.1021/jo00928a001. [DOI] [PubMed] [Google Scholar]
- 75.Torres M.C., Varaprasad C.V., Johnson F., Iden C.R. Formation of s-triazines during aerial oxidation of 8-oxo-7,8-dihydro-2′-deoxyguanosine in concentrated ammonia. Carcinogenesis. 1999;20:167–172. doi: 10.1093/carcin/20.1.167. [DOI] [PubMed] [Google Scholar]
- 76.Yermilov V., Rubio J., Becchi M., Friesen M.D., Pignatelli B., Ohshima H. Formation of 8-nitroguanine by the reaction of guanine with peroxynitrite in vitro. Carcinogenesis. 1995;16:2045–2050. doi: 10.1093/carcin/16.9.2045. [DOI] [PubMed] [Google Scholar]
- 77.Niles J.C., Burney S., Singh S.P., Wishnok J.S., Tannenbaum S.R. Peroxynitrite reaction products of 3′,5′-di-O-acetyl-8-oxo-7,8-dihydro-2′-deoxyguanosine. Proc. Natl. Acad. Sci. USA. 1999;96:11729–11734. doi: 10.1073/pnas.96.21.11729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Niles J.C., Wishnok J.S., Tannenbaum S.R. A novel nitration product formed during the reaction of peroxynitrite with 2′,3′,5′-tri-O-acetyl-7,8-dihydro-8-oxoguanosine: N-nitro-N′-[1-(2,3,5-tri-O-acetyl-β-D-erythro-pentofuranosyl)-2,4-dioxoimidazolidin-5-ylidene]guanidine. Chem. Res. Toxicol. 2000;13:390–396. doi: 10.1021/tx0000318. [DOI] [PubMed] [Google Scholar]
- 79.Niles J.C., Wishnok J.S., Tannenbaum S.R. Mass spectrometric identification of 4-hydroxy-2,5-dioxo-imidazolidine-4-carboxylic acid during oxidation of 8-oxoguanosine by peroxynitrite and KHSO5/CoCl2. Chem. Res. Toxicol. 2004;17:1501–1509. doi: 10.1021/tx040003f. [DOI] [PubMed] [Google Scholar]
- 80.Niles J.C., Wishnok J.S., Tannenbaum S.R. Spiroiminodihydantoin and guanidinohydantoin are the dominant products of 8-oxoguanosine oxidation at low fluxes of peroxynitrite: Mechanistic studies with 18O. Chem. Res. Toxicol. 2004;17:1510–1519. doi: 10.1021/tx0400048. [DOI] [PubMed] [Google Scholar]
- 81.Kino K., Morikawa M., Kobayashi T., Kobayashi T., Komori R., Sei Y., Miyazawa H. The oxidation of 8-oxo-7,8-dihydroguanine by iodine. Bioorg. Med. Chem. Lett. 2010;20:3818–3820. doi: 10.1016/j.bmcl.2010.04.032. [DOI] [PubMed] [Google Scholar]
- 82.Shrivastav N., Li D., Essigmann J.M. Chemical biology of mutagenesis and DNA repair: Cellular responses to DNA alkylation. Carcinogenesis. 2010;31:59–70. doi: 10.1093/carcin/bgp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fleming A.M., Burrows C.J. Iron Fenton oxidation of 2′-deoxyguanosine in physiological bicarbonate buffer yields products consistent with the reactive oxygen species carbonate radical anion not the hydroxyl radical. Chem. Commun. 2020;56:9779–9782. doi: 10.1039/D0CC04138F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pednekar V., Weerasooriya S., Jasti V.P., Basu A.K. Mutagenicity and genotoxicity of (5′S)-8,5′-cyclo-2′-deoxyadenosine in Escherichia coli and replication of (5′S)-8,5′-cyclopurine-2′-deoxynucleosides in vitro by DNA polymerase IV, exo-free Klenow fragment, and Dpo4. Chem. Res. Toxicol. 2014;27:200–210. doi: 10.1021/tx4002786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu W., Ouellette A.M., Wawrzak Z., Shriver S.J., Anderson S.M., Zhao L. Kinetic and structural mechanisms of (5′S)-8,5′-cyclo-2′-deoxyguanosine-induced dna replication stalling. Biochemistry. 2015;54:639–651. doi: 10.1021/bi5014936. [DOI] [PubMed] [Google Scholar]
- 86.Swanson A.L., Wang J., Wang Y. Accurate and efficient bypass of 8,5′-cyclopurine-2′-deoxynucleosides by human and yeast DNA polymerase η. Chem. Res. Toxicol. 2012;25:1682–1691. doi: 10.1021/tx3001576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.You C., Swanson A.L., Dai X., Yuan B., Wang J., Wang Y. Translesion synthesis of 8,5′-cyclopurine-2′-deoxynucleosides by DNA polymerases η, ι, and ζ. J. Biol. Chem. 2013;288:28548–28556. doi: 10.1074/jbc.M113.480459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jasti V.P., Das R.S., Hilton B.A., Weerasooriya S., Zou Y., Basu A.K. (5′S)-8,5′-cyclo-2′-deoxyguanosine is a strong block to replication, a potent pol V-dependent mutagenic lesion, and is inefficiently repaired in Escherichia coli. Biochemistry. 2011;50:3862–3865. doi: 10.1021/bi2004944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yuan B., Wang J., Cao H., Sun R., Wang Y. High-throughput analysis of the mutagenic and cytotoxic properties of DNA lesions by next-generation sequencing. Nucleic Acids Res. 2011;39:5945–5954. doi: 10.1093/nar/gkr159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 91.Liu C., Zhou Q., Li Y., Garner L.V., Watkins S.P., Carter L.J., Smoot J., Gregg A.C., Daniels A.D., Jervey S., et al. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent. Sci. 2020;6:315–331. doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Furuta Y., Takahashi K., Kuno-Maekawa M., Sangawa S., Uehara K., Kozaki N., Nomura N., Egawa H., Shiraki K. Mechanism of action of T-705 against influenza virus. Antimicrob. Agents Chemother. 2005;49:981–986. doi: 10.1128/AAC.49.3.981-986.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jin Z., Smith L.K., Rajwanshi V.K., Kim B., Deval J. The ambiguous base-pairing and high substrate efficiency of T-705 (Favipiravir) ribofuranosyl 5′-triphosphate towards influenza a virus polymerase. PLoS ONE. 2013;8:e68347. doi: 10.1371/journal.pone.0068347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rocha-Pereira J., Jochmans D., Dallmeier K., Leyssen P., Nascimento M.S., Neyts J. Favipiravir (T-705) inhibits in vitro norovirus replication. Biochem. Biophys. Res. Commun. 2012;424:777–780. doi: 10.1016/j.bbrc.2012.07.034. [DOI] [PubMed] [Google Scholar]
- 95.Smither S.J., Eastaugh L.S., Steward J.A., Nelson M., Lenk R.P., Lever M.S. Post-exposure efficacy of oral T-705 (Favipiravir) against inhalational Ebola virus infection in a mouse model. Antivir. Res. 2014;104:153–155. doi: 10.1016/j.antiviral.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 96.Furuta Y., Egawa H. Nitrogenous Heterocyclic Carboxamide Derivatives or Salts Thereof and Antiviral Agents Containing Both. PCT/JP1999/004429. International Patent Application No. 1999 Aug 18;
- 97.Qiu L., Patterson S.E., Bonnac L.F., Geraghty R.J. Nucleobases and corresponding nucleosides display potent antiviral activities against dengue virus possibly through viral lethal mutagenesis. PLoS Negl. Trop. Dis. 2018;12:e0006421. doi: 10.1371/journal.pntd.0006421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang G., Wan J., Hu Y., Wu X., Prhavc M., Dyatkina N., Rajwanshi V.K., Smith D.B., Jekle A., Kinkade A., et al. Synthesis and anti-influenza activity of pyridine, pyridazine, and pyrimidine C-nucleosides as favipiravir (T-705) analogues. J. Med. Chem. 2016;59:4611–4624. doi: 10.1021/acs.jmedchem.5b01933. [DOI] [PubMed] [Google Scholar]
- 99.Crotty S., Maag D., Arnold J.J., Zhong W., Lau J.Y., Hong Z., Andino R., Cameron C.E. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 2000;6:1375–1379. doi: 10.1038/82191. [DOI] [PubMed] [Google Scholar]
- 100.Moriyama K., Negishi K., Briggs M.S., Smith C.L., Hill F., Churcher M.J., Brown D.M., Loakes D. Synthesis and RNA polymerase incorporation of the degenerate ribonucleotide analogue rPTP. Nucleic Acids Res. 1998;26:2105–2111. doi: 10.1093/nar/26.9.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sheahan T.P., Sims A.C., Zhou S., Graham R.L., Pruijssers A.J., Agostini M.L., Leist S.R., Schäfer A., Dinnon K.H., 3rd, Stevens L.J., et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12:eabb5883. doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Warren T.K., Jordan R., Lo M.K., Ray A.S., Mackman R.L., Soloveva V., Siegel D., Perron M., Bannister R., Hui H.C., et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lo M.K., Jordan R., Arvey A., Sudhamsu J., Shrivastava-Ranjan P., Hotard A.L., Flint M., McMullan L.K., Siegel D., Clarke M.O., et al. GS-5734 and its parent nucleoside analog inhibit Filo-, Pneumo-, and Paramyxoviruses. Sci. Rep. 2017;7:43395. doi: 10.1038/srep43395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sheahan T.P., Sims A.C., Graham R.L., Menachery V.D., Gralinski L.E., Case J.B., Leist S.R., Pyrc K., Feng J.Y., Trantcheva I., et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017;9:eaal3653. doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yin W., Mao C., Luan X., Shen D.D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gordon C.J., Tchesnokov E.P., Woolner E., Perry J.K., Feng J.Y., Porter D.P., Götte M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020;295:6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jordan P.C., Liu C., Raynaud P., Lo M.K., Spiropoulou C.F., Symons J.A., Beigelman L., Deval J. Initiation, extension, and termination of RNA synthesis by a paramyxovirus polymerase. PLoS Pathog. 2018;14:e1006889. doi: 10.1371/journal.ppat.1006889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tchesnokov E.P., Feng J.Y., Porter D.P., Götte M. Mechanism of Inhibition of Ebola Virus RNA-Dependent RNA Polymerase by Remdesivir. Viruses. 2019;11:326. doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Oertell K., Harcourt E.M., Mohsen M.G., Petruska J., Kool E.T., Goodman M.F. Kinetic selection vs. free energy of DNA base pairing in control of polymerase fidelity. Proc. Natl. Acad. Sci. USA. 2016;113:E2277–E2285. doi: 10.1073/pnas.1600279113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dizdaroglu M., Coskun E., Jaruga P. Repair of oxidatively induced DNA damage by DNA glycosylases: Mechanisms of action, substrate specificities and excision kinetics. Mutat. Res. 2017;771:99–127. doi: 10.1016/j.mrrev.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kino K., Takao M., Miyazawa H., Hanaoka F. A DNA oligomer containing 2,2,4-triamino-5(2H)-oxazolone is incised by human NEIL1 and NTH1. Mutat. Res. 2012;734:73–77. doi: 10.1016/j.mrfmmm.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 112.Kino K., Shimizu Y., Sugasawa K., Sugiyama H., Hanaoka F. Nucleotide excision repair of 5-formyluracil in vitro is enhanced by the presence of mismatched bases. Biochemistry. 2004;43:2682–2687. doi: 10.1021/bi0361416. [DOI] [PubMed] [Google Scholar]