Abstract
Favipiravir was initially developed as an antiviral drug against influenza and is currently used in clinical trials against severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) infection (COVID-19). This agent is presumably involved in RNA chain termination during influenza virus replication, although the molecular interactions underlying its potential impact on the coronaviruses including SARS-CoV-2, SARS-CoV, and Middle East respiratory syndrome coronavirus (MERS-CoV) remain unclear. We performed in silico studies to elucidate detailed molecular interactions between favipiravir and the SARS-CoV-2, SARS-CoV, MERS-CoV, and influenza virus RNA-dependent RNA polymerases (RdRp). As a result, no interactions between favipiravir ribofuranosyl-5′-triphosphate (F-RTP), the active form of favipiravir, and the active sites of RdRps (PB1 proteins) from influenza A (H1N1)pdm09 virus were found, yet the agent bound to the tunnel of the replication genome of PB1 protein leading to the inhibition of replicated RNA passage. In contrast, F-RTP bound to the active sites of coronavirus RdRp in the presence of the agent and RdRp. Further, the agent bound to the replicated RNA terminus in the presence of agent, magnesium ions, nucleotide triphosphate, and RdRp proteins. These results suggest that favipiravir exhibits distinct mechanisms of action against influenza virus and various coronaviruses.
Keywords: favipiravir, COVID-19, SARS-CoV-2, influenza, RdRp, in silico
1. Introduction
In December 2019, a new coronavirus disease (coronavirus disease 2019, COVID-19) emerged suddenly in China [1]. COVID-19 spread rapidly, resulting in a pandemic [2]. Over 34 million people were confirmed COVID-19-positive as of the end of September 2020 [2]. The agent is classified into genus Betacoronavirus, based on detailed genome analyses, and formally named severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) [3]. About 10–20% of COVID-19 cases may cause fever, fatigue, cough, and pneumonia, while the infection in some cases may result in inapparent symptoms [4]. However, some cases of COVID-19 may be complicated acute respiratory distress syndrome (ARDS) leading to death [5]. Thus, a need exists for the early development of new therapeutic drugs and applications of existing drugs for the treatment of COVID-19. To date, some antiviral agents including ciclesonide, remdesivir, and favipiravir have been tried to treat COVID-19 [6].
Inhibition of viral replication is assumed to be the mechanism for therapeutic agents for SARS-CoV-2. RNA-dependent RNA polymerase (RdRp) of SARS-CoV-2 is nonstructural protein 12 (NSP-12) and is similar to SARS-CoV RdRp [7]. NSP-7 and NSP-8 are known cofactors of SARS-CoV-2 RdRp proteins. Further, NSP-15 protein is an endonuclease from SARS-CoV-2 that plays an important role in the proofreading of viral RNA. Thus, these proteins may be targets for antiviral drugs for treating COVID-19.
Favipiravir (6-fluoro-3-hydroxypyrazine-2-carboxamide, Avigan®) was initially developed as an antiviral agent for the treatment of influenza. The mechanism of favipiravir is inhibition of viral RNA replication by inhibition of RdRp formed as a complex of PA protein, PB1 protein, and PB2 protein [8,9,10,11]. Subsequently, favipiravir was reported to have similar activity against RdRp proteins from RNA viruses other than influenza and showed efficacy for treatment of ebolavirus disease (EVD), Lassa fever, and norovirus infections [12,13,14]. Thus, favipiravir might also be effective against some RNA virus infections, and is currently in clinical evaluation as a treatment for COVID-19 [6]. However, no detailed molecular interactions between favipiravir and SARS-CoV-2 proteins, such as RdRp and NSP-15 endonuclease, are exactly known. With this background, we performed an in silico study regarding the molecular interactions among favipiravir, influenza RdRp, SARS-CoV-2 RdRp, SARS-CoV RdRp, Middle East respiratory syndrome coronavirus (MERS-CoV) RdRp, and the coronaviruses’ NSP-15 endonuclease.
2. Materials and Methods
2.1. Structural Modeling
Sequences of the RdRp proteins of influenza A/Northern Territory/60/1968/H3N2 (PDBID: 6qnw), bat influenza A polymerase (PDBID: 6szu), and SARS-CoV-2 (PDBID: 6 m71, 7bv2) were downloaded from Protein Data Bank Japan (https://pdbj.org/). We also downloaded NSP-15 of SARS-CoV-2 (PDBID: 6x1b), SARS-CoV (PDBID: 2h85), and MERS-CoV (PDBID: 5yvd) from Protein Data Bank Japan. Sequences of the RdRp proteins of influenza A/California/07/2009(H1N1) (Protein ID: YP_009118630.1, YP_009118628.1, YP_009118631.1), RdRp proteins of MERS-CoV (Protein ID:YP_009047223.1, YP_009047220.1, YP_009047219.1) were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/protein/). The three-dimensional (3D) structure of influenza H3N2 PB1 protein was constructed based on data available on the structure of the B chain of influenza H3N2 RdRp protein.
The homology model of influenza A/California/07/2009(H1N1) RdRp protein was built with the template structure of bat influenza A polymerase (PDBID: 6szu) using MODELLER 9.23 software on the Windows operating system [15]. The query sequences from A/California/07/2009(H1N1) strain were searched to find related protein structures for use as templates for the BLAST (Basic Local Alignment Search Tool) program (https://blast.ncbi.nlm.nih.gov/Blast.cgi) against PDB [16]. The 3D model obtained was evaluated by Ramachandran’s map using RNA Annotation and Mapping of Promoters for the Analysis of Gene Expression (RAMPAGE) and by the Swiss Protein Databank Viewer (SPDBV) 4.10 software [17,18]. Nucleotide triphosphates (NTPs) ions were manually inserted to the RdRp proteins referring to bat influenza A polymerase (PDBID:6szu). We used coronavirus RdRp protein that contained NSP-12, NSP-7, and NSP-8. The model of SARS-CoV-2 RdRp protein with NTP and ions was manually constructed from SARS-CoV-2 RdRp (PDBID: 7bv2). The homology model of SARS-CoV RdRp protein and MERS-CoV RdRp protein was also built with the template structure of SARS-CoV-2 RdRp protein (PDBID: 7bv2). NTPs ions were manually inserted into SARS-CoV RdRp proteins and MERS-CoV RdRp proteins referring to SARS-CoV-2 RdRp proteins. The homology model for NSP-15 of SARS-CoV-2 was built as previously described [19]. NSP-15 of SARS-CoV-2 (PDBID: 6x1b) were downloaded from Protein Data Bank Japan.
The 3D structure of favipiravir (Accession Number: DB12466) used for the docking simulation analysis was obtained from the DRUGBANK database (https://www.drugbank.ca/), including the structural data. The putative structure of favipiravir ribofuranosyl-5′-triphosphate (F-RTP) was manually reconstructed using Molview v2.4 [20]. Favipiravir ribofuranosyl-5′-monophosphate (F-RMP) was manually reconstructed as previously described [10].
2.2. Docking Simulation
Computational simulation of the molecular recognition process was performed using AutoDock Vina 1.1.2 software according to software instructions [21]. Before docking compounds on the target, the protein was edited using AutoDockTools 1.5.6. Polar hydrogen atoms were added to amino acid residues, and Gasteiger charges were assigned to all atoms of the protein. The protein in PDBQT format was then used as an input to AutoDock Vina. The grid box for analysis was set to a size that included the entire protein region. Detailed procedures used for docking simulations have been previously reported [19,21,22]. Top 20 score poses were evaluated by using PyMOL 2.3.4 to visualize protein–ligand interactions. We evaluated binding energy, which was simply provided by programs. The active sites in influenza PB1 protein were added manually; these included Ser443, Asp444, and Asp445 based on findings in a previous report [23]. Similarly, the active site of SARS-CoV-2 RdRp protein was added manually and included residues Ser 759, Asp760, and Asp761 [24]. The active site of SARS-CoV RdRp and MERS-CoV RdRp was added manually including a similar motif in SARS-CoV-2 RdRp. Among the ligands bound to the active site, those associated with a root mean square deviation of 2 or more compared to values obtained prior to binding analysis were excluded from further consideration.
3. Results
3.1. Molecular Interactions between Favipiravir and F-RTP with Influenza PB1 Proteins
Neither favipiravir nor F-RTP bound to the active site of influenza virus subtype H1N1 or H3N2 PB1 proteins. Interestingly, F-RTP bound to sites in both the H1N1 and H3N2 PB1 proteins that were implicated in double-strand RNAs synthesis pathways. The chemical binding energy between F-RTP and the two PB1 proteins was estimated at −6.1 and −6.0 kcal/mol, respectively (Figure 1a,b). The F-RTP binding site on the H3N2 PB1 protein included Gln124, Arg249, Glu256, and Met411; the F-RTP binding site on the H1N1 PB1 protein included Asp743, Glu934, Arg940, Thr1009, Lys1010, and Lys1182.
Figure 1.
Detailed interactions between favipiravir ribofuranosyl-5′-triphosphate (F-RTP) and the active sites (red-colored regions) of (a) influenza H3N2 PB1 protein, (b) influenza H1N1 PB1 protein, (c) severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) RNA-dependent RNA polymerases (RdRp), (d) SARS-CoV RdRp, and (e) Middle East respiratory syndrome coronavirus (MERS-CoV) RdRp. Three-dimensional configurations of F-RTP and proteins were constructed with space-filling or stick models.
3.2. Molecular Interactions between Favipiravir and F-RTP with SARS-CoV-2/SARS-CoV/MERS-CoV Replication-Associated Proteins RdRp and NSP-15 Endonuclease Alone
First, we examined molecular interactions using favipiravir/F-RTP and various proteins alone. As shown in Figure 1c, F-RTP bound directly to the active site of the SARS-CoV-2 RdRp; Asp760 was the critical amino acid residue facilitating this interaction. The binding energy between F-RTP and the SARS-CoV-2 RdRp was −6.6 kcal/mol. Uncharged favipiravir also bound to the SARS-CoV-2 RdRp at Asp760 and Asp761 with a binding energy of −4.0 kcal/mol. F-RTP bound to Ser679 in the active site of the SARS-CoV RdRp protein with a binding energy of −6.4 kcal/mol (Figure 1d). Similarly, F-RTP bound to the active site of MERS-CoV RdRp at Ser 678 and Asp680 with a binding energy of −7.3kcal/mol (Figure 1e). Both favipiravir and F-RTP have the potential to inhibit RNA polymerization catalyzed by SARS-CoV-2 RdRp, SARS-CoV RdRp, and MERS-CoV RdRp. In contrast, F-RMP did not interact with SARS-CoV-2 NSP-15, SARS-CoV NSP-15, and MERS-CoV NSP-15 (data not shown).
3.3. Molecular Interactions among Various Polymerases, Nucleotide Triphosphate (NTP), Magnesium Ion, and Favipiravir/F-RTP
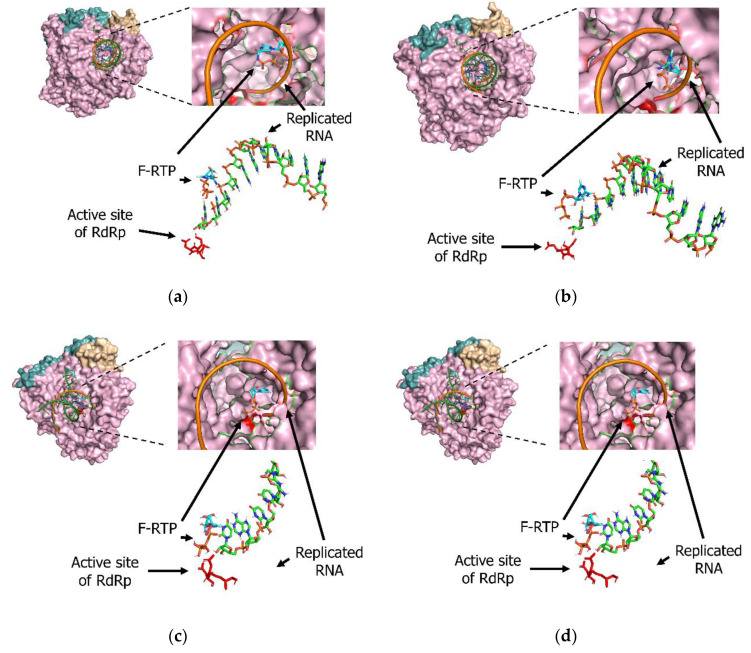
We also analyzed molecular interactions using various viral polymerases, NTP, magnesium ions (Mg2+), and favipiravir/F-RTP. F-RTP bound to termini of replicated RNA of influenza RdRp protein with a binding energy of −9.8kcal/mol (Figure 2a). Similarly, F-RTP bound to termini of replicated RNA of all coronavirus RdRp proteins. Binding energy among the termini of the replicated RNA by SARS-CoV-2, SARS-CoV, and MERS-CoV were estimated as −8.4, −7.5, and −8.9 kcal/mol, respectively (Figure 2b–d).
Figure 2.
Detailed interactions among favipiravir ribofuranosyl-5′-triphosphate (F-RTP), nucleotide triphosphate (NTP), ions, and the active sites (red-colored regions) of (a) influenza H1N1, (b) severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), (c) SARS-CoV, and (d) Middle East respiratory syndrome coronavirus (MERS-CoV) RNA-dependent RNA polymerases (RdRp). Three-dimensional configurations of F-RTP, NTP, ions, and RdRp protein were constructed with space-filling or stick models.
4. Discussion
We showed in the present in silico study that F-RTP (the active form of favipiravir) could bind to RdRp active sites of SARS-CoV-2, SARS-CoV, and MERS-CoV in the presence of the agent and protein. Moreover, the F-RTP bound to the replicated RNA termini in the presence of the agent, magnesium ions, nucleotide triphosphate, and RdRp proteins. Conversely, F-RTP did not bind to PB1 (RdRp) active sites of influenza virus H1N1 in the presence of agent and protein. Further, F-RTP may bind to the tunnel of the PB1 protein leading to the inhibition of the replicated RNA passage. Thus, F-RTP displays distinct pharmacological effects on various coronaviruses and influenza virus subtype AH1N1.
Favipiravir is a nucleic acid analog and was developed as a therapeutic drug to be used to treat influenza [25]. The drug was approved in Japan in 2014 and was stockpiled for use in the event of a new influenza epidemic. Favipiravir is activated by intracellular conversion to F-RTP by hypoxanthine-guanine phosphoribosyltransferase [8]. Previous reports revealed that the actions of favipiravir/F-RTP against influenza involves the termination of genome replication, although the detailed molecular interactions between the drug and the PB1 polymerase had not been fully elucidated. Favipiravir is currently undergoing clinical evaluation for use in treating COVID-19 in some countries, including Japan. Some reports suggest that favipiravir is effective in this setting [26,27], although the molecular mechanisms underlying drug efficacy against SARS-CoV-2 have not been fully explored. To date, a few antiviral agents against coronaviruses have been approved. However, our results suggest favipiravir may show antiviral activity against SARS-CoV-2, SARS-CoV, and MERS-CoV, though the present study was purely in silico. Thus, further clinical studies may be needed to demonstrate efficacy.
We examined the presence of the agent and coronavirus RdRp proteins and also the presence of the agent, magnesium ions, nucleotide triphosphate, and the viral RdRp proteins. The agent could bind to RdRp proteins and could inhibit genome replication. A previous report showed that the agent inhibits SARS-CoV-2 in vitro [28]. This study suggests that inhibition of genome replication is termination [28]. However, detailed molecular interactions between the agent and the viral replication systems may not currently be known. Thus, the present molecular pharmacological results may be the first observations.
Further, previous reports suggest that the antiviral effect of the agent was premature termination of genome replication [8,29,30]. In our study, F-RTP could bind to the replicated RNA termini of influenza RdRp proteins, suggesting that the inhibition of the genome replication mechanism of influenza virus is termination. This may be compatible with earlier reports [8,29,30]. The present in silico study suggests that F-RTP does not bind to PB1 protein active sites of influenza virus subtype AH1N1 in the presence of agent and protein, but does suggest that F-RTP may bind to the tunnel of the protein. This binding may result in inhibition of replicated RNA passage. Based on the results and speculation, we suggest that favipiravir may exhibit two mechanisms of antiviral activity. This observation may also be a first.
The half-maximal effective concentration (EC50) of favipiravir against SARS-CoV-2 is 61.88 μΜ (9.4 μg/mL), which is comparable to Ebola virus (10.8–63 μg/mL) but higher than EC50′s of 0.030–0.46 μg/mL for influenza virus [25,31,32,33]. Thus, the clinical dose of favipiravir for COVID-19 will be higher than the dose for influenza but comparable to the dose for Ebola hemorrhagic fever [34]. However, the underlying cause of differences in EC50 for influenza and SARS-CoV-2 is unknown. The difference in mechanisms of action of favipiravir in influenza and SARS-CoV-2 may explain this difference in EC50.
Favipiravir binding to proteins other than SARS-CoV-2 RdRp has been investigated [35,36]. NSP-15 protein is an endonuclease from SARS-CoV-2 which plays an important role in the replication of viral RNA. We previously reported that ciclesonide inhibits viral replication in SARS-CoV-2 by binding to active sites of NSP-15 [19]. Hence, we also examined a docking simulation for interactions between favipiravir/F-RTP and NSP-15. However, favipiravir/F-RMP did not bind to these active sites. Thus, NSP-15 is not involved in differences in EC50 of favipiravir between influenza virus and SARS-CoV-2.
In conclusion, we found, in silico, that favipiravir/F-RTP could bind to active sites of coronavirus RdRp proteins and replicated RNA termini. We also showed that F-RTP binds near the tunnel of influenza RdRp protein. Distinct mechanisms underlying favipiravir-mediated interactions with influenza RdRp and coronavirus RdRp may help explain the need for different doses of the drug for effective clinical responses for treating influenza vs. SARS-CoV-2 infections.
Author Contributions
Conceptualization, M.S. and H.K.; writing—original draft preparation, M.S.; writing—review and editing, H.K. K.O. and Y.H. supervised the data collection; T.S., H.I., T.T., A.N., K.M., M.K., and A.R. provided expertise in infectious disease and revised it carefully from a professional point of view; funding acquisition, M.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by AMED (https://www.amed.go.jp/) under Grant Number JP20fk0108103. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zhu N.A., Zhang D., Wang W., Li X., Yang B.O., Song J., Zhao X., Huang B., Shi W., Lu R., et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization Coronavirus Disease (COVID-19) [(accessed on 30 September 2020)]; Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
- 3.The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahumud R.A., Kamara J.K., Renzaho A.M.N. The epidemiological burden and overall distribution of chronic comorbidities in coronavirus disease-2019 among 202,005 infected patients: Evidence from a systematic review and meta-analysis. Infection. 2020 doi: 10.1007/s15010-020-01502-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang D., Hu B.O., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., et al. Clinical Characteristics of 138 Hospitalized Patients with 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esposito S., Noviello S., Pagliano P. Update on treatment of COVID-19: Ongoing studies between promising and disappointing results. Le Infez. Med. 2020;28:198–211. [PubMed] [Google Scholar]
- 7.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sangawa H., Komeno T., Nishikawa H., Yoshida A., Takahashi K., Nomura N., Furuta Y. Mechanism of action of T-705 ribosyl triphosphate against influenza virus RNA polymerase. Antimicrob. Agents Chemother. 2013;57:5202–5208. doi: 10.1128/AAC.00649-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takashita E. Influenza Polymerase Inhibitors: Mechanisms of Action and Resistance. Cold Spring Harb. Perspect. Med. 2020:a038687. doi: 10.1101/cshperspect.a038687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldhill D.H., Te Velthuis A.J.W., Fletcher R.A., Langat P., Zambon M., Lackenby A., Barclay W.S. The mechanism of resistance to favipiravir in influenza. Proc. Natl. Acad. Sci. USA. 2018;115:11613–11618. doi: 10.1073/pnas.1811345115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Te Velthuis A.J.W., Fodor E. Influenza virus RNA polymerase: Insights into the mechanisms of viral RNA synthesis. Nat. Rev. Microbiol. 2016;14:479–493. doi: 10.1038/nrmicro.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bai C.-Q., Mu J.-S., Kargbo D., Song Y.-B., Niu W.-K., Nie W.-M., Kanu A., Liu W.-W., Wang Y.-P., Dafae F., et al. Clinical and Virological Characteristics of Ebola Virus Disease Patients Treated with Favipiravir (T-705)-Sierra Leone, 2014. Clin. Infect. Dis. 2016;63:1288–1294. doi: 10.1093/cid/ciw571. [DOI] [PubMed] [Google Scholar]
- 13.Raabe V.N., Kann G., Ribner B.S., Morales A., Varkey J.B., Mehta A.K., Lyon G.M., Vanairsdale S., Faber K., Becker S., et al. Favipiravir and Ribavirin Treatment of Epidemiologically Linked Cases of Lassa Fever. Clin. Infect. Dis. 2017;65:855–859. doi: 10.1093/cid/cix406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arias A., Thorne L., Goodfellow I. Favipiravir elicits antiviral mutagenesis during virus replication in vivo. eLife. 2014;3:e03679. doi: 10.7554/eLife.03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Webb B., Sali A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinform. 2014;47:5.6.1–5.6.32. doi: 10.1002/0471250953.bi0506s47. [DOI] [PubMed] [Google Scholar]
- 16.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 17.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 18.Raborn R.T., Brendel V.P. Using RAMPAGE to Identify and Annotate Promoters in Insect Genomes. Methods Mol. Biol. 2019;1858:99–116. doi: 10.1007/978-1-4939-8775-7_9. [DOI] [PubMed] [Google Scholar]
- 19.Kimura H., Kurusu H., Sada M., Kurai D., Murakami K., Kamitani W., Tomita H., Katayama K., Ryo A. Molecular pharmacology of ciclesonide against SARS-CoV-2. J. Allergy Clin. Immunol. 2020;146:330–331. doi: 10.1016/j.jaci.2020.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith T.J. MolView: A program for analyzing and displaying atomic structures on the Macintosh personal computer. J. Mol. Graph. 1995;13:122–125. doi: 10.1016/0263-7855(94)00019-O. [DOI] [PubMed] [Google Scholar]
- 21.Trott O., Olson A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forli S., Huey R., Pique M.E., Sanner M.F., Goodsell D.S., Olson A.J. Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 2016;11:905–919. doi: 10.1038/nprot.2016.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li C., Wu A., Peng Y., Wang J., Guo Y., Chen Z., Zhang H., Wang Y., Dong J., Wang L., et al. Integrating computational modeling and functional assays to decipher the structure-function relationship of influenza virus PB1 protein. Sci. Rep. 2014;4:7192. doi: 10.1038/srep07192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hillen H.S., Kokic G., Farnung L., Dienemann C., Tegunov D., Cramer P. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020;584:154–156. doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- 25.Furuta Y., Takahashi K., Fukuda Y., Kuno M., Kamiyama T., Kozaki K., Nomura N., Egawa H., Minami S., Watanabe Y., et al. In vitro and in vivo activities of anti-influenza virus compound T-705. Antimicrob. Agents Chemother. 2002;46:977–981. doi: 10.1128/AAC.46.4.977-981.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen C., Zhang Y., Huang J., Yin P., Cheng Z., Wu J., Chen S., Zhang Y., Chen B., Lu M., et al. Favipiravir versus Arbidol for COVID-19: A Randomized Clinical Trial. medRxiv. 2020 doi: 10.1101/2020.03.17.20037432. [DOI] [Google Scholar]
- 27.Cai Q., Yang M., Liu D., Chen J., Shu D., Xia J., Liao X., Gu Y., Yang Y., Shen C., et al. Experimental Treatment with Favipiravir for COVID-19: An Open-Label Control Study. Engineering. 2020 doi: 10.1016/j.eng.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shannon A., Selisko B., Le N., Huchting J., Touret F., Piorkowski G., Fattorini V., Ferron F., Decroly E., Meier C., et al. Favipiravir strikes the SARS-CoV-2 at its Achilles heel, the RNA polymerase. BioRxiv. 2020 doi: 10.1101/2020.05.15.098731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shiraki K., Daikoku T. Favipiravir, an anti-influenza drug against life-threatening RNA virus infections. Pharmacol. Ther. 2020;209:107512. doi: 10.1016/j.pharmthera.2020.107512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gordon C.J., Tchesnokov E.P., Woolner E., Perry J.K., Feng J.Y., Porter D.P., Götte M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020;295:6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W.U., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smither S.J., Eastaugh L.S., Steward J.A., Nelson M., Lenk R.P., Lever M.S. Post-exposure efficacy of oral T-705 (Favipiravir) against inhalational Ebola virus infection in a mouse model. Antivir. Res. 2014;104:153–155. doi: 10.1016/j.antiviral.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 33.Oestereich L., Lüdtke A., Wurr S., Rieger T., Muñoz Fontela C., Günther S. Successful treatment of advanced Ebola virus infection with T-705 (favipiravir) in a small animal model. Antivir. Res. 2014;105:17–21. doi: 10.1016/j.antiviral.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 34.Eloy P., Solas C., Touret F., Mentré F., Malvy D., De Lamballerie X., Guedj J. Dose Rationale for Favipiravir Use in Patients Infected with SARS-CoV-2. Clin. Pharmacol. Ther. 2020;108:188. doi: 10.1002/cpt.1877. [DOI] [PubMed] [Google Scholar]
- 35.Hagar M., Ahmed H.A., Aljohani G., Alhaddad O.A. Investigation of Some Antiviral N-Heterocycles as COVID 19 Drug: Molecular Docking and DFT Calculations. Int. J. Mol. Sci. 2020;21:3922. doi: 10.3390/ijms21113922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harismah K., Mirzaei M. Favipiravir: Structural Analysis and Activity against COVID-19. Adv. J. Chem. Sect. B. 2020;2:55–60. [Google Scholar]