Abstract
Progesterone receptor membrane component 1 (PGRMC1) has been shown to regulate some cancer hallmarks. Progesterone (P4) evokes intracellular calcium (Ca2+) changes in the triple-negative breast cancer cell lines (MDA-MB-231, MDA-MB-468, and BT-20) and in other breast cancer cell lines like the luminal MCF7 cells. PGRMC1 expression is elevated in MDA-MB-231 and MCF7 cells as compared to non-tumoral MCF10A cell line, and PGRMC1 silencing enhances P4-evoked Ca2+ mobilization. Here, we found a new P4-dependent Ca2+ mobilization pathway in MDA-MB-231 cells and other triple-negative breast cancer cells, as well as in MCF7 cells that involved Stromal interaction molecule 2 (STIM2), Calcium release-activated calcium channel protein 1 (Orai1), and Transient Receptor Potential Channel 1 (TRPC1). Stromal interaction molecule 1 (STIM1) was not involved in this novel Ca2+ pathway, as evidenced by using siRNA STIM1. PGRMC1 silencing reduced the negative effect of P4 on cell proliferation and cell death in MDA-MB-231 cells. In line with the latter observation, Nuclear Factor of Activated T-Cells 1 (NFAT1) nuclear accumulation due to P4 incubation for 48 h was enhanced in cells transfected with the small hairpin siRNA against PGRMC1 (shPGRMC1). These results provide evidence for a novel P4-evoked Ca2+ entry pathway that is downregulated by PGRMC1.
Keywords: MDA-MB-231 cells, STIM2, PGRMC1, Ca2+-homeostasis, progesterone
1. Introduction
The expression of nuclear progesterone receptor (nPR) is needed for cell proliferation and maturation of breast tissue, and further, nPR expression is found under the regulation of the estrogens [1]. Interestingly, activation of alveologenesis and, subsequently, milk production relies on the balance between progesterone receptor and prolactin/Stat5 signaling [1,2]. Reduction of circulating progesterone (P4) concentration facilitates gene expression of the prolactin receptor as well as the activation of its downstream effector, the signal transducer and activator of transcription complex 5 (STAT5) [1,2,3].
Physicians have used the lack or presence of the classical nuclear estrogen receptor (nER) or nPR, in order to histologically characterize the different breast cancer subtypes [4]. Thus, ESR1, PGR, and ERBB2 mRNAs are not detected in most of the basal tumor samples and, therefore, also known as triple-negative breast cancer subtype [5].
Regarding the role of P4 in cell proliferation, it has been reported that P4 impairs normal breast epithelial cell proliferation in vivo, as well as in some types of breast cancer cells in vitro [6]. The effect of P4 on cell proliferation has been described depending on nPR activation; thus, the triple-negative breast cancer (TNBC) cell lines should remain unaltered in the presence of P4; however, recent studies have claimed that P4 evokes alteration of the TNBC cells [7,8]. These negative effects in MDA-MB-231 cells are suggested to be mediated by membrane progesterone α (mPRα) [7], which might be modulated by progesterone receptor membrane component 1 (PGRMC1) [1]. Nonetheless, the evidence against mPRα as the P4 receptor has also been reported by others [9]. More recent studies have been performed to identify alternative P4 receptors, leading to the identification of two additional protein families that are able to transduce the P4 effects in the neuroimmune system and other tissues [10,11]: (1) Class II progestin and adipoQ receptor family members (PAQR) (such as mPRα is also known as PAQR7)); (2) B5-like heme/steroid-binding protein family that groups proteins like the PGRMC1,PGRMC2, and others [10].
PGRMC1 expression is enhanced in patients suffering from different types of tumors, resulting in poor prognosis [12,13]. In line with this observation, PGRMC1 has been linked with lipid metabolism, which would lead to enhanced breast cancer progression [14,15]. It is worth mentioning that the role of PGRMC1 is mainly evidenced in the luminal type A breast cancer cell lines (MCF7 and T47-D); meanwhile, MDA-MB-231 cell proliferation remains unaltered, even when PGRMC1 expression is artificially altered [14].
The intracellular signaling pathways activated or regulated by PGRMC1 remain barely investigated. SRC family kinase, Protein Kinase A (PKA), PKC, PI3K, and ERK-1/2 are suggested as downstream PGRMC1-dependent signaling pathways [16].
Finally, changes in the cytosolic free Ca2+ concentration ([Ca2+]c) have been reported to be relevant for cell proliferation [17]. In this sense, although P4 evokes changes in the intracellular calcium homeostasis, scientists disagree in the possible cellular Ca2+ sources. Thus, in retinal Muller glia cells, a non-permeable P4 analog evokes changes in the intracellular Ca2+ concentration, and further, authors reported a substantial calcium entry from the extracellular medium downstream of progesterone membrane receptors activation, but they did not show intracellular store calcium release [18]. Contrarily, human spermatozoa exhibit Ca2+ release from intracellular stores in response to P4 [19]. In neurons, P4 administration enhances Ca2+ release by favoring inositol trisphosphate receptor (IP3R) permeability via enhanced phosphorylation by the serine/threonine-protein kinases (AKT) [20]. Similarly, in oral squamous cancer cells (PE/CA-PJ15 cells), it has been described that P4 (5 and 10 μM) evokes both Ca2+ release and Ca2+ entry, which is inhibited by adding the PGRMC1 antagonist AG205 [21]. Contrarily, in the breast cancer luminal A type, T47-D, ATP evokes a transient increase in the [Ca2+]c that is downregulated by P4, medroxyprogesterone acetate (MPA), and also by the progesterone non-permeable analog, progesterone-CMO-BSA. The negative effect of P4 on ATP-evoked calcium mobilization is non-detected in CHO cells, and experimental data has revealed the absence of mPRs in these cells [22].
Here, we aimed to extend the knowledge regarding the possible regulation of P4 on calcium homeostasis in breast cancer cells. We provided evidence of a novel P4-activated pathway in TNBC (MDA-MB-231, BT-20, and MDA-MB-468 cells). P4 evoked Ca2+ entry in MDA-MB-231 cells and MCF-7 cells by the activation of a novel calcium pathway involving the Calcium release-activated calcium channel protein 1 (Orai1), Transient receptor potential channel 1 (TRPC1), and Stromal interaction molecule 2 (STIM2); meanwhile, STIM1 silencing had no effect in this mechanism. Interestingly, activation of PGRMC1 by P4 attenuated this Ca2+ signaling pathway, which led to a decreased Ca2+-dependent Nuclear Factor of Activated T-Cells 1 (NFAT1) nuclear accumulation and, therefore, negatively regulated MDA-MB-231 cells proliferation.
2. Results
2.1. P4 Inhibits MDA-MB-231 Cell Proliferation
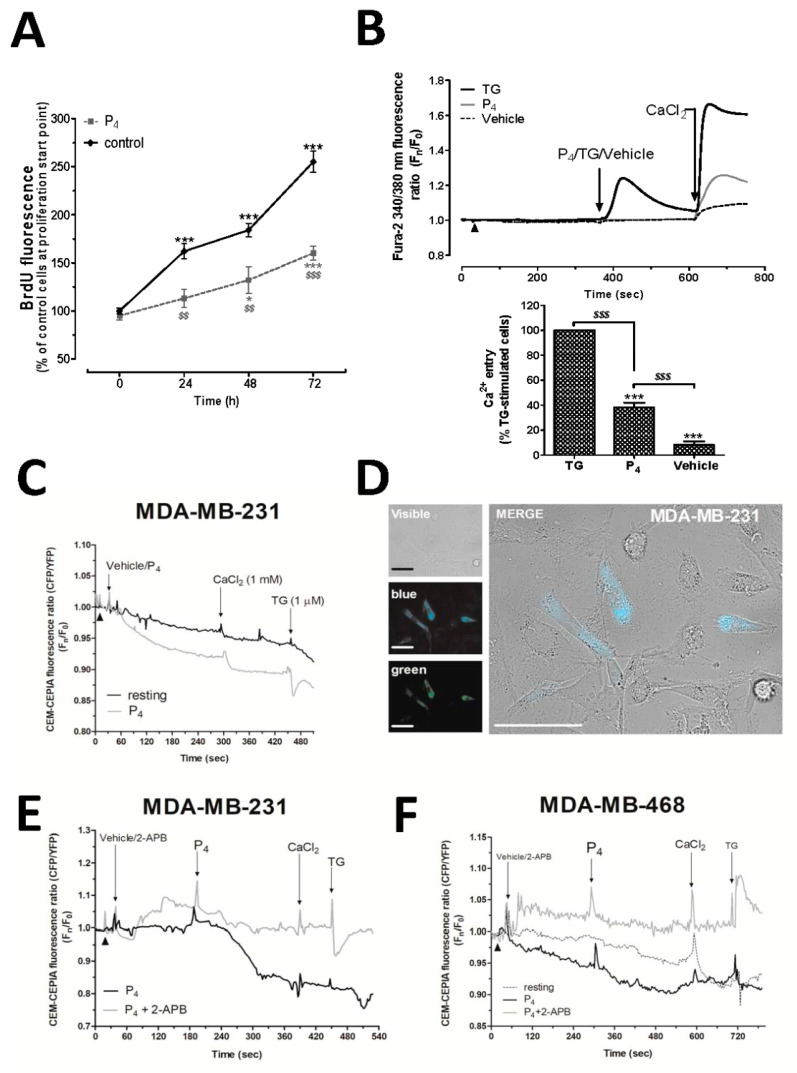
MDA-MB-231 cells lack the classical nPR [6], while these cells express a less characterized and alternatively spliced nPR, which may also bind to P4 [23]. Additionally, the effect of P4 in cell proliferation has been shown to be different according to the cell types investigated [6,8,24]. As previously reported [6], and as depicted in Figure 1A, the treatment of MDA-MB-231 cells with 1 μM of P4 resulted in a significant reduction of bromodeoxiuridin (BrdU) fluorescence at all analyzed times (24–72 h), as compared to cells treated with the vehicle. Therefore, these data revealed a negative role of P4 in the proliferation of the MDA-MB-231 cells (n = 6).
Figure 1.
Progesterone (P4) altered MDA-MB-231 cell proliferation and calcium (Ca2+) entry. (A) MDA-MB-231 cells were seeded (5 × 105 cells/mL) and allowed to proliferate for 72 h in the presence of the vehicle (control) or 1 μM of P4. At 0, 24, 48, and 72 h, cells were incubated with 5-bromo-2′-deoxyuridine (BrdU) for an additional 2 h, as described in Materials and Methods. The graph represents BrdU accumulation within the cell nucleus. *, ***: represent p < 0.05 and p < 0.001 with respect to the values of BrdU found in control cells analyzed at 0 h; $$, $$$: represent p < 0.01 and p < 0.001 with respect to the control cells at each analyzed time point. (B) Fura-2-loaded MDA-MB-231 cells were maintained in an extracellular Ca2+-free HBS medium (75 μM of ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA) was added, as indicated by arrowhead), and they were subsequently stimulated either with the vehicle or P4 (1 μM) or thapsigargin (TG; 2 μM). Ca2+ release from intracellular stores upon cell stimulation was monitored for 4 min; meanwhile, Ca2+ entry evoked by the depletion of the intracellular Ca2+ stores using the different stimuli was visualized by adding CaCl2 (1 mM) to the extracellular medium, which was monitored for additional 2 min. Graphs are representative of 6–8 independent experiments, and the histogram represents the average of the percentage ± Standard Error of the Mean (S.E.M.), resulting in the analysis of the areas under the curves corresponding to the Ca2+ entry evoked by the agonist and compared to the TG-evoked Ca2+-entry. The 60–80 cells from 6–8 independent experiments were analyzed in these experiments. ***: represents p < 0.001 with respect to the percentage of Ca2+ values found in MDA-MB-231 cells stimulated with TG; $$$: represents p < 0.001 with respect to the percentage of Ca2+ values found in non-stimulated cells (Vehicle). (C–E) MDA-MB-231 cells (C–E) and MDA-MB-468 cells (F) were transfected with the intraluminal Ca2+ dye, GEM-CEPIA1er, and at the day of the experiments upon confirmation of the positive Ca2+ dye expression by the cells (D), the cells were maintained in a Ca2+-free HBS medium (75 μM of EGTA was added, as indicated by the arrowheads), and subsequently, they were stimulated with P4 (1 μM) in the absence (C) or presence of 50 μM of 2-APB (E,F), and we alternatively monitored the changes in the fluorescence of GEM-CEPIA1er at the wavelengths of 380/510 nm (excitation: Ex/emission: Em; green) and 380/462 (Ex/Em; blue). Finally, we added TG (1 μM) at the end of each experiment, which was used as an internal experimental control. Graphs and images in (C–F) are representative of 3–4 independent cell transfections with GEM-CEPIA1er. The bar in the image (D) represents 50 μm.
Considering the relevant role of the Ca2+- homeostasis on cell proliferation, even in MDA-MB-231 cells, as previously demonstrated by our research group [17,25,26], and the controversy regarding the role of P4 in calcium homeostasis aforementioned, we analyzed the effect of P4 on the Ca2+ homeostasis in MDA-MB-231 cells [18]. MDA-MB-231 cells suspended in a Ca2+-free medium exhibited a small Ca2+ entry when it was added back to the extracellular medium (1 mM CaCl2; Figure 1B, dotted line). Incubation of MDA-MB-231 cells with P4 evoked Ca2+ entry (Figure 1B, grey line), which was significantly smaller than the one evoked by thapsigargin in these cells (2 μM; TG, black line, Figure 1B, p < 0.001). TG blocks the sarcoendoplamic reticulum Ca2+-ATPase (SERCA), which is a classic maneuver to activate store-operated Ca2+ entry (SOCE) (Figure 1B, p < 0.001, n = 6) [27,28]. Interestingly, we were unable to visualize a significant P4-induced Ca2+ release from the internal stores, contrary to the results observed using TG, and therefore, it may explain the smaller activation of the Ca2+ entry recorded under these experimental conditions if compared to the one evoked using TG.
In order to investigate the mechanism underlying the P4-dependent Ca2+ entry activation, we analyzed possible changes in the intraluminal Ca2+ concentration induced by P4 using the endoplasmic reticulum Ca2+ dye, GEM-CEPIA1er, in the triple-negative breast cancer cell lines—MDA-MB-231 and MDA-MB-468 cells. As depicted in Figure 1C,D, MDA-MB-231 cells suspended in a Ca2+-free HBS medium presented changes in the decay of the blue/green fluorescence ratio of GEM-CEPIA1er after cell stimulation with P4 (1 μM) with respect to the non-stimulated resting cells (compare black and grey lines in Figure 1C). As shown in Figure 1F, similar P4-evoked Ca2+ release occurred in MDA-MB-468 cells (compare black-dotted and black-solid lines in Figure 1F). Internal control experiments consisted of the addition of TG (1 μM) at the end of each experiment, which enhanced the release of Ca2+ from the endoplasmic reticulum, as detected by the drastic change in the GEM-CEPIA1er fluorescence ratio. Following this, in an attempt to avoid the Ca2+ release evoked by P4, we used a relative specific antagonist of the inositol trisphosphate receptor (IP3R), 2-Aminoethoxydiphenyl borate (2-APB), in MDA-MB-231 cells (Figure 1E) and MDA-MB-468 cells (Figure 1F) [29]. As shown in Figure 1E,F, treatment of the triple-negative breast cancer cells with 50 μM of 2-APB for 5 min was able to almost completely block the release of Ca2+ evoked by P4. Therefore, these results allowed us to conclude that although not being reported using fura-2, P4 evokes enough intraluminal Ca2+ changes to promote the extracellular Ca2+ entry, and, further, the Ca2+ release evoked by P4 occurs through the IP3R.
2.2. Activation of PGRMC1 Downregulates P4-Dependent Ca2+ Mobilization in MDA-MB-231 Cells
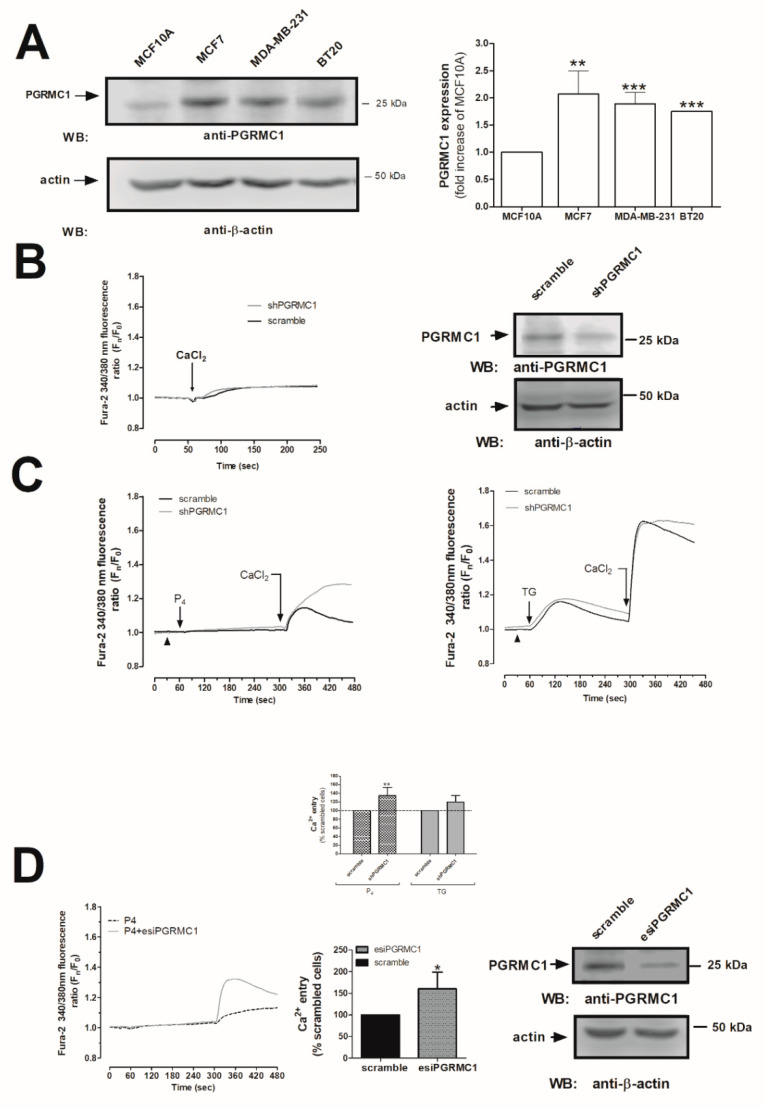
As shown in Figure 2A, MDA-MB-231 cells presented greater PGRMC1 expression values as compared to the non-tumoral cell line, MCF10A (1.9 ± 0.2-fold increase as compared to MCF10A, p < 0.001, respectively; n = 6). Similarly, enhanced PGRMC1 expression was detected in other breast cancer cell types—the luminal A type MCF7 and triple-negative BT-20 (Figure 2A, p < 0.01 and p < 0.001, respectively). In addition, considering the discrepancies mentioned above regarding the structural identity of the P4-receptor in the MDA-MB-231 cells, we silenced the expression of the PGRMC1 receptor by using the small hairpin RNA against PGRMC1 (shPGRMC1) and an endoribonuclease-prepared siRNAs against PGRMC1 (esiPGRMC1). MDA-MB-231 cell transfection with either shPGRMC1 or esiPGRMC1 efficiently reduced PGRMC1 expression in MDA-MB-231 cells after 48 h of transfection (See WB image in Figure 2B,D). Interestingly, PGRMC1 silencing did not alter Ca2+ mobilization in resting cells (Figure 2B, Ca2+ traces), but an increase in the P4-induced Ca2+ entry by a 35 ± 19% (using ShPGRMC1; Figure 2C) and 60 ± 38% (using esiPGRMC1; Figure 2D) was observed as compared to the non-genetically modified MDA-MB-231 cells (see cells transfected with scramble plasmids vs. shPGRMC1 and esiPGRMC1, respectively, and the histogram at the bottom of Figure 2C,D; p < 0.01 and p < 0.05, n = 6). On the contrary, we did not observe significant changes in the TG-evoked SOCE in MDA-MB-231 cells transfected with shPGRMC1 under these experimental conditions (Figure 2C, right-hand side graph, 20 ± 15%; p > 0.05, n = 6). According to our results, stimulation of Ca2+ entry by P4 was downregulated by PGRMC1 in MDA-MB-231 cells, and the latter results were also reproduced in MCF7 cells (See Figure 3A,B,D). These results agreed with the downregulation of (Ca2+)c concentration by P4, as previously described in spontaneously immortalized rat granulosa cells and breast cancer luminal A type, T47-D, but authors did not deeply explore this mechanism [22,30].
Figure 2.
Progesterone receptor membrane component 1 (PGRMC1) downregulated the progesterone (P4)-evoked Ca2+ entry. (A) Resting MCF10A, MCF7, MDA-MB-231, and BT20 cells were suspended at the same concentration, and they were subsequently lysed with NP40 buffer. The resulting protein samples were denaturalized by mixing with an equal volume of Laemmli’s buffer (5% sodium dodecyl sulphate, SDS). Subsequent Western blotting was performed by using the anti-PGRMC1 antibody, as described in Materials and Methods. The histogram represents the average of the fold increase ± S.E.M. of the amount of PGRMC1 found in MCF10A after they were previously normalized by considering the actin content in each gel lane. **, ***: represent p < 0.01 and p < 0.001 as compared to the PGRMC1 values found in MCF10A cells. (B,C) MDA-MB-231 cells were transfected for 48 h with 2 μg/mL of the small hairpin siRNA against PGRMC1 (shPGRMC1) plasmid (B) or esiPGRMC1 (D), and after validation of the efficiency of the silencing procedure by Western blotting, cells were incubated with 2 μM of fura-2/AM for 30 min at 37 °C. (B) In order to demonstrate that the PGRMC1 silencing did not affect Ca2+-mobilization per se in MDA-MB-231 cells, 1 mM of CaCl2 was extracellularly added to cells under resting conditions. (C) MDA-MB-231 cells transfected with the empty vector (scramble) or with shPGRMC1 and were maintained in a Ca2+-free HBS medium (75 μM of ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA) was added, arrowheads), and finally, they were stimulated either with 1 μM of P4 or 2 μM of TG. Additionally, P4 or thapsigargin (TG)-evoked Ca2+ entry was visualized by adding CaCl2 (1 mM) to the extracellular medium, which was monitored for an additional 2 min. (D) MDA-MB-231 cells were transfected with esiPGRMC1 for 48 h, and subsequently, P4-evoked Ca2+ mobilization was analyzed by using similar experimental conditions to those used in (C). Graphs are representative of 6–8 independent experiments, and the histogram represents the average of the percentage ± Standard Error of the Mean (S.E.M.), resulting in the analysis of the areas under the curves corresponding to the Ca2+ entry evoked by P4 or TG. The 60–80 cells from 6–8 independent transfections were considered during the analysis. *: represents p < 0.05; **: represents p < 0.01 with respect to the Ca2+ entry values found in scrambled MDA-MB-231 cells.
Figure 3.
Progesterone (P4) effects on Ca2+ entry in MCF7 mock cells or in cells transfected with small hairpin siRNA against progesterone receptor membrane component 1 (shPGRMC1) and endoribonuclease-prepared siRNAs against the stromal interaction molecular 2 (esiSTIM2). MCF7 cells were kept untreated (A) or were transfected for 48 h with 2 μg/mL of either shPGRMC1 plasmid (B), esiSTIM2 (C), or their respective controls, and upon confirming the efficiency of the genetic modification, cells were incubated with 2 μM of fura-2/AM for 30 min at 37 °C. Cells were finally maintained in a Ca2+-free HBS medium (75 μM of ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA) was added, arrowheads) and were stimulated with 1 μM of P4 for 4 min. P4-evoked Ca2+ entry was visualized by adding CaCl2 (1 mM) to the extracellular medium, which was further monitored for an additional 2 min. Graphs are representative of 4–6 independent experiments. (D) The histogram represents the average of the percentage ± Standard Error of the Mean (S.E.M.), resulting in the analysis of the areas under the curves corresponding to the Ca2+-entry obtained using shPGRMC1 and esiSTIM2 with respect to scrambled cells. The 20–40 cells from 4–6 independent transfections were considered during the analysis. **, ***: represent p < 0.01 and p < 0.001 with respect to the Ca2+ mobilization values found in scrambled MCF7 cells stimulated with P4.
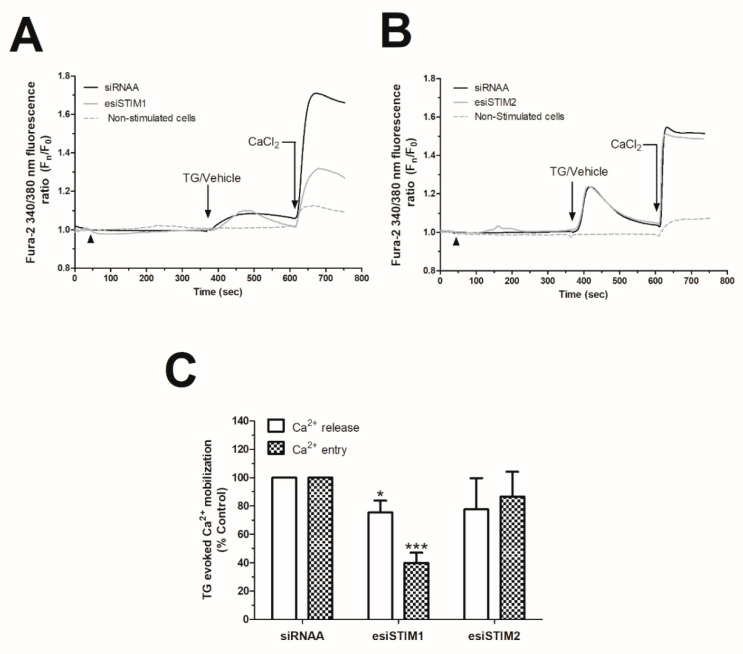
STIM1 is a very relevant component of the Ca2+ entry in non-electrical excitable cells, including breast cancer cells [31]. In order to dig into the mechanism underlying the P4-evoked changes in Ca2+ entry, we knocked-down STIM1 by using esiRNA [28,32,33,34,35]. esiRNA consists of a commercial combination of siRNAs directed against different sequences of the targeted protein that has been generated by endonuclease activity, and thus, they would present greater efficiency and specificity than siRNAs, which are generated toward a unique sequence of the targeted protein [36].
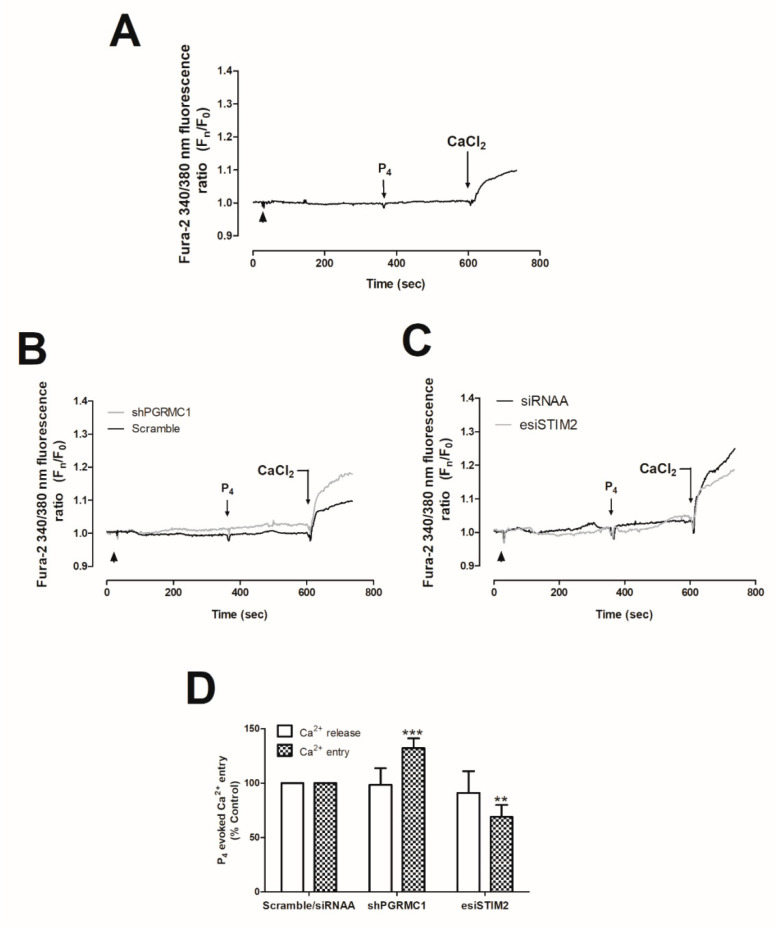
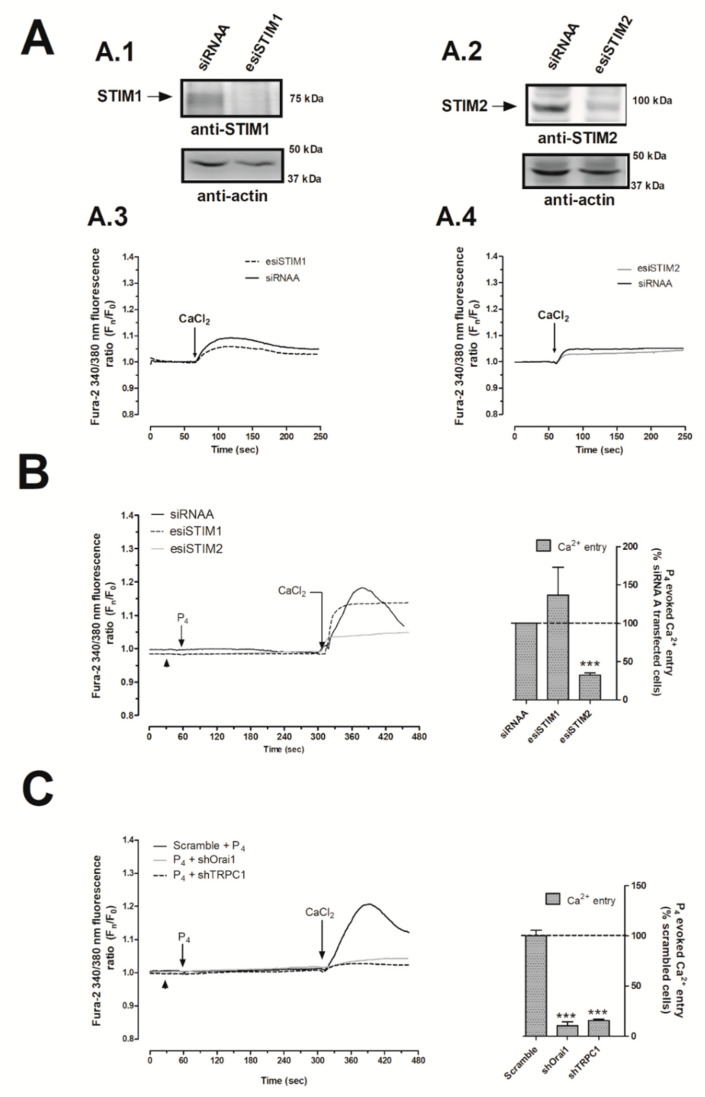
As depicted in Figure 4A.1, transfection of MDA-MB-231 cells with esiSTIM1 completely silenced STIM1 expression, which did not significantly alter the Ca2+ homeostasis of MDA-MB-231 cells maintained under normal conditions (Figure 4A.3). In our hands, esiSTIM1 did not quantitatively modify the Ca2+ entry induced by the administration of P4 (Figure 4B, black-dotted trace; p > 0.05, n = 6). Next, we explored the possible role of STIM2, a less characterized isoform of STIM1 [37,38], which is able to detect smaller changes in the endoplasmic reticulum’s Ca2+ content in response to low physiological agonist concentration, and therefore, it may detect the small P4-evoked intraluminal Ca2+ changes, previously shown in Figure 1 [39,40,41]. The transfection of MDA-MB-231 cells with the esiSTIM2 significantly reduced the expression of the targeted protein (Figure 4A.2), and as evidenced in Figure 4B, STIM2 silencing attenuated P4-evoked Ca2+ entry by 67.8 ± 3.0% (Figure 4B, grey trace; p < 0.001, n = 6) but did not significantly modify the Ca2+ homeostasis in non-stimulated cells (Figure 4A.4). Similarly, the downregulation of P4-evoked Ca2+ entry was observed in MCF7 lacking STIM2 (Figure 3C,D).
Figure 4.
Progesterone (P4) activated the stromal interaction molecule 2 (STIM2) that drives calcium (Ca2+) entry through Calcium release-activated calcium channel protein 1 (Orai1) and transient receptor potential channel 1 (TRPC1). MDA-MB-231 cells were transfected with 2 μg/mL of either endoribonuclease-generated siRNA against STIM1 (esiSTIM1) (A.1,A.3 (dotted trace) and B) or esiSTIM2 (A.2,A.4 (grey trace) and B) or with a non-specific siRNA (A.1–A.4 (black trace) and B) for 48 h. Alternatively, we used 2 μg/mL of small hairpin RNA against Orai1 (shOrai1) (C, grey solid line) or shTRPC1 (c; black dotted line) or empty vector (C, scrambled cells, black trace) for 48 h. Upon confirmation of the efficiency of the protein silencing protocols by Western blotting using the specific antibodies as indicated (A.1,A.2), Ca2+ experiments were performed according to the procedures described in Materials and Methods. Briefly, fura-2-loaded MDA-MB-231 cells were maintained in a Ca2+-rich medium (containing 1 mM of CaCl2, A.3,A.4) and were kept untreated; while other cells were maintained in a Ca2+-free HBS medium (75 μM of ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA) was added, arrowheads (B,C)) and were stimulated with 1 μM of P4 for 4 min (B,C). P4-evoked Ca2+ entry was visualized by adding CaCl2 (1 mM) to the extracellular medium, which was monitored for an additional 2 min. Graphs are representative of 6–8 independent transfections, and the histograms represent the average of the percentage ± Standar error of the Mean (S.E.M.), resulting in the analysis of the areas under the curves corresponding to the Ca2+ entry evoked by P4 in around 60–80 cells from 6–8 independent transfections. ***: represents p < 0.001 with respect to the Ca2+ entry values found in scrambled MDA-MB-231 cells.
Following this, we explored the plasma-membrane Ca2+ channels activated by P4 in MDA-MB-231 cells. Thus, the transfection of MDA-MB-231 cells with shRNAs against Orai1 and TRPC1 channels resulted in a significant reduction (89.5 ± 3.8% and 84.3 ± 1.2%) in P4-evoked Ca2+ entry as compared to the cells transfected with scrambled plasmids (Figure 4C; p < 0.001; n = 6). Therefore, our findings indicate that the P4-evoked Ca2+ entry course through a novel pathway where STIM2, Orai1, and TRPC1 are involved. Interestingly, STIM2 silencing did not alter the activation of SOCE in MDA-MB-231 cells, which is mainly conducted by STIM1 and Orai1, as depicted in Figure 5A–C, and it has been previously reported by others [31].
Figure 5.
Effect of stromal interaction molecule 1 (STIM1) and STIM2 silencing on thapsigargin (TG)-evoked store-operated Ca2+ entry (SOCE) in MDA-MB-231 cells. MDA-MB-231 cells were transfected with either endonuclease-generated siRNA against STIM1 (esiSTIM1) (A) and esiSTIM2 (B) or siRNA (A,B). Ca2+ experiments under single-cell imaging configuration were performed by loading cells with fura-2/AM. Fura-2-loaded cells were maintained in a Ca2+-free HBS medium (75 μM of ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA) was added, arrowheads) and were stimulated with 2 μM of TG or treated with the vehicle for 4 min, while we monitored the TG-evoked Ca2+ release. Subsequently, TG-evoked Ca2+ entry (SOCE) and constitutive Ca2+ entry were visualized by adding CaCl2 (1 mM) to the extracellular medium and were monitored for an additional 2 min. Graphs are representative of 6–8 independent transfections. (C) The histogram represents the average of the percentage ± Standar Erro of the Mean S.E.M., resulting in the analysis of the areas under the curves evoked by TG in around 60–80 cells from 6–8 independent transfections. *, ***: represent p < 0.05 and p < 0.001 with respect to the Ca2+ mobilization values found in scrambled MDA-MB-231 cells.
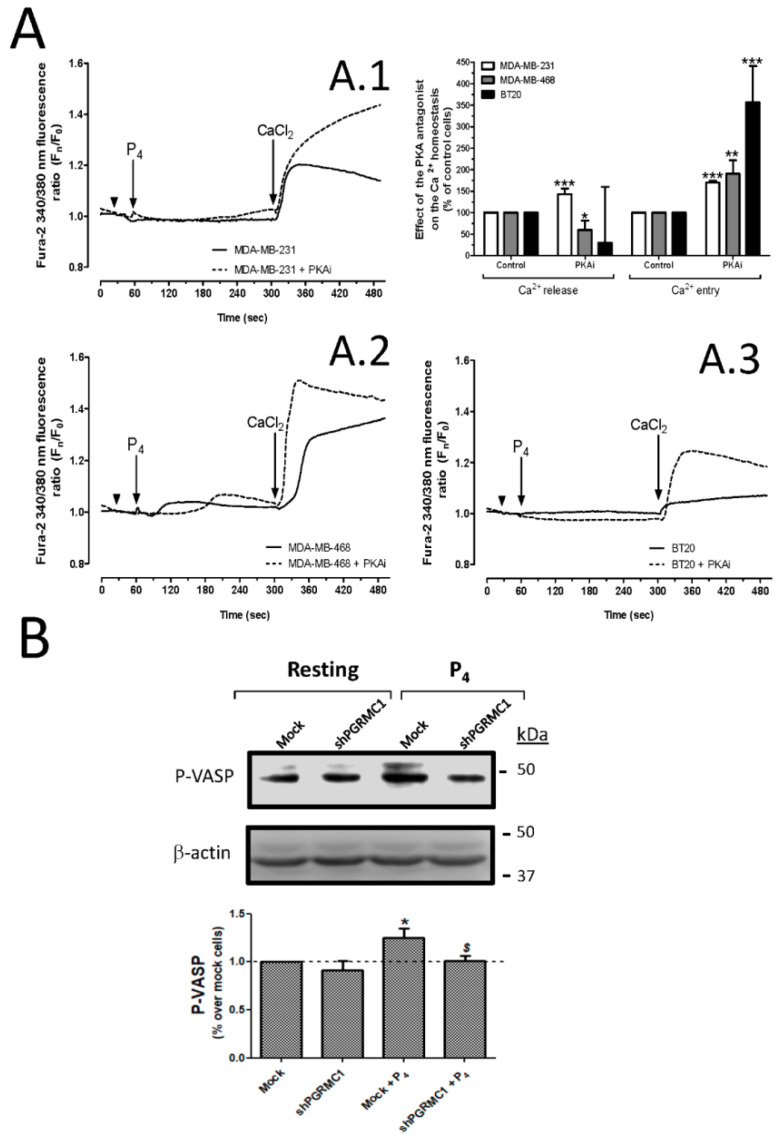
Structural analysis of the Orai1 structure revealed that Orai1 is phosphorylated at the serine-34 residue by the protein kinase A (PKA), which modulates the channel function [42]. Furthermore, it has been described that activation of PGRMC1 leads to the activation of certain protein kinases like PKA, PI3K, and SFK [16]. Therefore, we explored whether the inhibition of P4-evoked Ca2+ entry might occur through the activation of the PGRMC1/cAMP/PKA signaling pathways [43,44]. As depicted in Figure 6, MDA-MB-231 (6A.1), MDA-MB-468 (6A.2), and BT20 (6A.3) cells were preincubated with 3 μM of the specific PKA antagonist, KT5720 (PKAi), which resulted in a significant increase in the P4-evoked Ca2+ entry, as summarized in the bar graph in Figure 6A. Additionally, to further demonstrate the activation of PKA in MDA-MB-231 cells in response to P4 stimulation, we analyzed the changes in the phosphorylation at Ser239 of vasodilator stimulated phosphoprotein (VASP) that is a substrate of PKA and has commonly been used for monitoring the PKA activation. As shown in Figure 6B, P4 evoked a 1.3 ± 0.06-fold increase in P-VASP as compared to mock resting cells (p < 0.05; n = 3); meanwhile, PGRMC1 silencing reversed P-VASP to similar values than those found in mock resting cells (p < 0.05; n = 3). These data reproduced the observation noted with the shPGRMC1 (n = 6), and therefore, led us to conclude that PGRMC1 impairs P4-induced Ca2+ entry by activating PKA and, subsequently, by inducing Orai1 phosphorylation, as it has been previously demonstrated by others [42].
Figure 6.
Protein kinase A (PKA) activation by progesterone (P4) impaired calcium (Ca2+) entry in triple-negative breast cancer cells. MDA-MB-231 cells (A.1), MDA-MB-468 (A.2), and BT20 cells (A.3) were loaded with fura-2/AM. Single-cell imaging experiments were done using fura-2 loaded triple-negative breast cancer cells that were previously incubated for 30 min with the vehicle (black solid traces) or with 3 μM of the PKA antagonist, KT5720 (PKAi, black dotted traces). Following this, the triple-negative breast cancer cells were stimulated for 4 min with P4 (1 μM) in a Ca2+-free HBS medium (75 μM of ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA) was added, as indicated by arrowheads), and subsequently, P4-evoked Ca2+ entry was visualized by adding 1 mM of CaCl2 to the extracellular medium. Areas under the curves evoked by P4 and CaCl2 administration to the cells were determined, and they were used for comparison between both experimental conditions. The histogram represents the average of the percentage ± Standard error of the Mean (S.E.M.), resulting in the analysis of the areas under the curves corresponding to the P4-evoked Ca2+ release and Ca2+ entry evoked in the absence or presence of the PKA antagonist. The 20-30 cells from 6 independent experiments were considered during the analysis. *, **, ***: represent p < 0.05, p < 0.01, and p < 0.001 with respect to the control PKAi untreated cells, respectively. (B) The activation of PKA was analyzed by determining the phosphorylation rates of vasodilator stimulated phosphoprotein (VASP; at the Ser239) in MDA-MB-231 cells lacking or expressing the progesterone receptor membrane component 1 (PGRMC1), which were subsequently stimulated for 1 min with the vehicle or 1 μM of P4. The image is representative of 3 independent transfections, and the bar graph represents the increase in the phosphorylation rate of VASP with respect to the control mock cells under resting conditions. *: represents p < 0.05; meanwhile, $: represents p < 0.05 with respect to the cell stimulated with P4 but non-transfected with the small hairpin siRNA against PGMRC1 (shPGRMC1).
2.3. PGRMC1 Silencing Reduces the Negative Effect of P4 in MDA-MB-231 Cell Proliferation
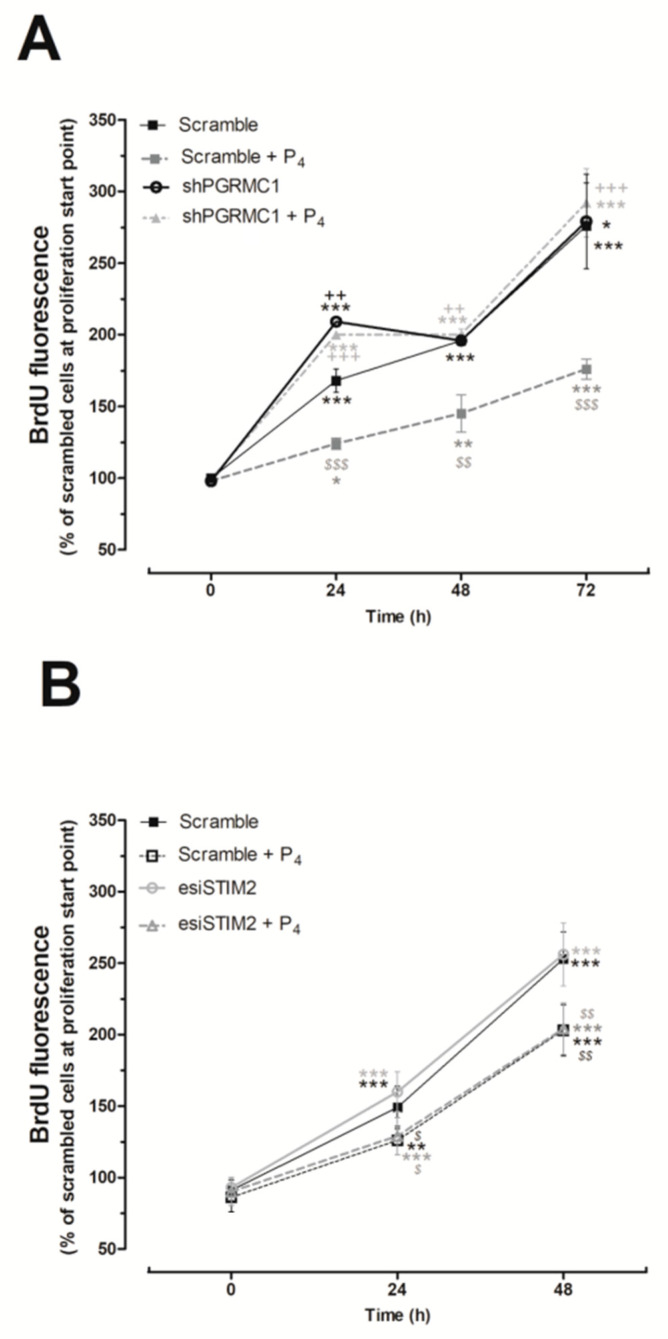
A positive effect of P4 in proliferation has been reported in cells expressing the classical nPR, such as MCF7 or glioblastoma cells [45,46,47]; meanwhile, the negative effect of P4 on cell proliferation was observed in cells lacking the nPR like the human BxPC3 pancreatic adenocarcinoma cells [48]. In TNBC cells, we explored the role of PGRMC1 on the P4-mediated inhibition of cell proliferation, previously described in Figure 1. As shown in Figure 7A, the silencing of PGRMC1 reversed the inhibitory effect of P4 on MDA-MB-231 cell proliferation, thus suggesting that this negative effect is mediated by the P4 binding and activation of PGRMC1. Furthermore, our results indicated that the silencing of STIM2 did not alter the proliferation pattern found in MDA-MB-231 cultured in the presence of P4 for 24 h (See Figure 7B); then, STIM2 would not be involved in the negative role of P4 on MDA-MB-231 cells proliferation.
Figure 7.
Effect of silencing the progesterone receptor membrane component 1 (PGRMC1) and the stromal interaction molecule 2 (STIM2) in the proliferation of MDA-MB-231 cells. MDA-MB-231 cells were transfected for 48 h with either small hairpin siRNA against PGRMC1 (shPGRMC1) (A) or the respective empty vector (A; scramble), and alternatively, cells were transfected with either endoribonuclease-generated siRNA against STIM2 (esiSTIM2) (B) or siRNA for 48 h (B; scramble). Upon confirming the efficiency of the silencing protocol by Western blotting, the cells were seeded and allowed to proliferate in the absence or presence of progesterone (P4; 1 μM). The proliferation rate was determined by analyzing the bromodeoxyuridine (BrdU) uptake by the cells for 2 h at 0, 24, 48, and 72 h from the beginning of the proliferation experiments. Graphs represent the average of the percentage ± Standard of the Mean (S.E.M.) of the BrdU fluorescence detected in the scrambled cells at the beginning of the cell culture (time 0 h). *, **, ***: represent p < 0.05, p < 0.01, and p < 0.001 with respect to the scrambled cells at time 0 h; meanwhile, $,$$, $$$: represent p < 0.05, p < 0.01, and p < 0.001 with respect to the BrdU values found in scrambled cells grown in the absence of P4 at the different analyzed time points, and ++,+++: represent p < 0.01 and p < 0.001 with respect to the values found in cells expressing the targeted protein.
2.4. PGRMC1 Is Involved in the P4-Evoked Cell Death in Breast Cancer Cells
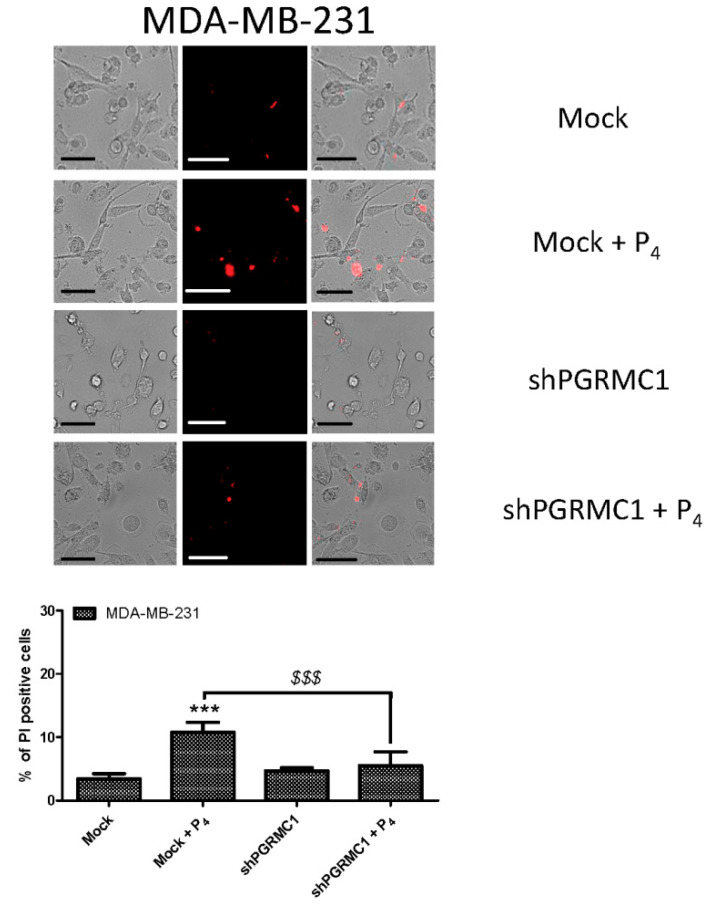
In order to confirm that the negative effect of P4 on breast cancer cells course by activation of PGRMC1, we analyzed the number of dead cells by using the PI uptake protocol in MDA-MB-231 cells that were previously transfected with the scramble or shPGRMC1 plasmids. As shown in Figure 8, MDA-MB-231 cells exhibited a significant increase in the percentage of positive PI cells after incubation with P4 (1 μM) for 24 h. Interestingly, MDA-MB-231 cells transfected with shPGRMC1 presented a reduction in the percentage of dead cells after the incubation with P4, as compared to the scrambled control cells (Mock cells, p < 0.001; n = 3–4). These results might explain the reduction in the cell count observed at similar times that are shown in Figure 1 and Figure 4.
Figure 8.
Protesterone (P4) induced cell death of the MDA-MB-231 cells. MDA-MB-231 cells were transfected with the scrambled (Mock) or the small hairpin siRNA against the progesterone receptor membrane component 1 (shPGRMC1) plasmids for 48 h. After confirmation of the efficiency of the silencing protocol by Western blotting, cells were cultured for an additional 24 h in the absence or presence of P4 (1 μM). Following this, cells were incubated on the day of the experiment with 4 μM of propidium iodide (PI) for 45 min at 37 °C inside the incubator. Cell images were acquired at 555/624 nm of wavelength (excitation/emission, respectively), by using a 40× objective mounted in a fluorescence microscope. The percentage of positive PI loaded cells with respect to the total counted cells in the fields was estimated, considering the mean ± Standard error of the mean (S.E.M.) of 3–4 independent experiments, and was represented in the bar chart. ***: represents p < 0.001 with respect to the mock resting cells; meanwhile, $$$: represents p < 0.001 with respect to the cells expressing PGRMC1 and treated with P4. The bar in the images represents 50 μm.
2.5. PGRMC1 Activation by P4 Impairs Nuclear Factor of Activated T-Cells 1 (NFAT1) Nuclear Translocation
Finally, we analyzed the effect of P4 on the activation of NFAT1, an important calcium-dependent and pro-proliferative transcription factor that acts as checkpoints in the cell cycle [49,50,51,52,53].
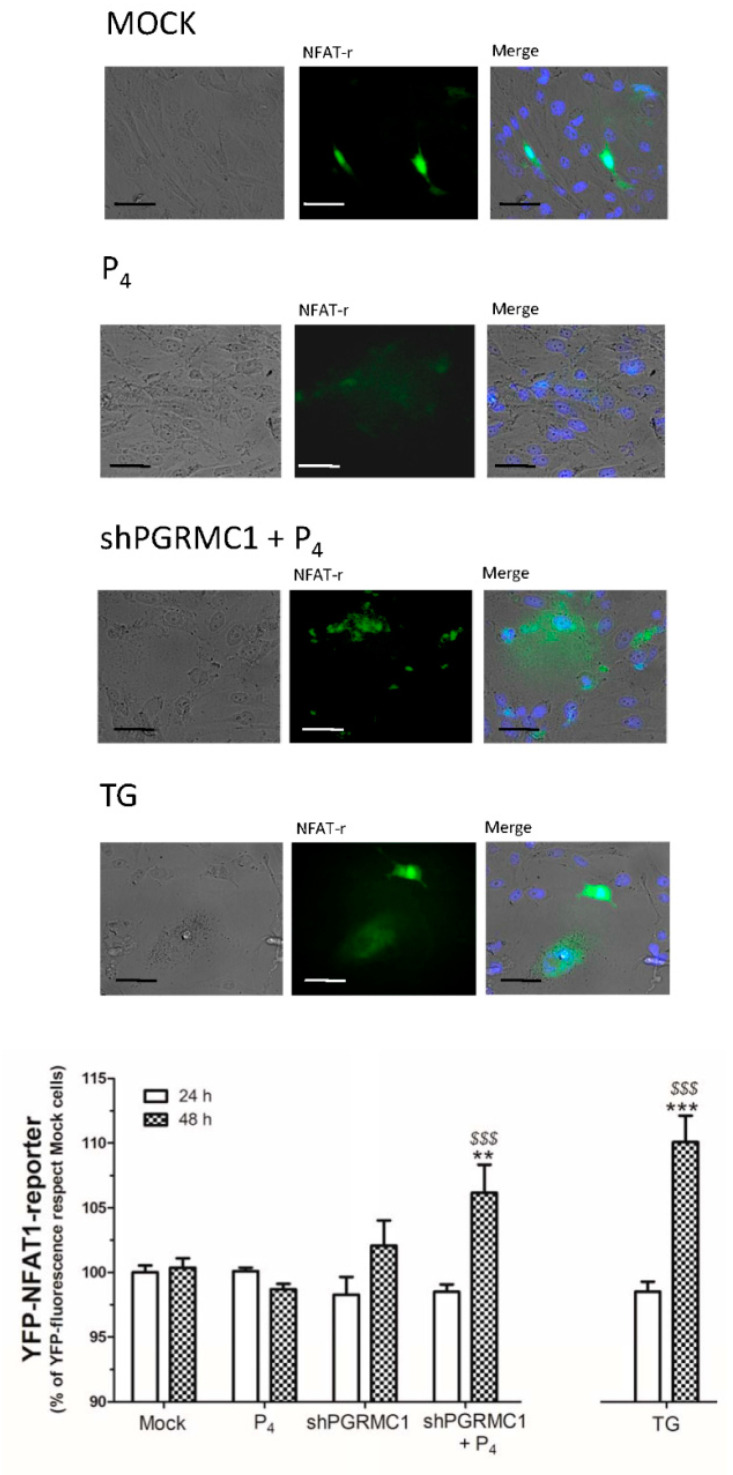
As depicted in Figure 9, NFAT1-reporter fluorescence was significantly enhanced in cells where PGRMC1 was silenced and, subsequently, stimulated for 48 h with 1 μM of P4, perhaps, enhanced NFAT1 reporter visualization results of the biggest Ca2+ entry previously shown in these cells lacking PGRMC1 under similar experimental conditions (Figure 2), and it has been also reported by others.
Figure 9.
Progesterone receptor membrane component 1 (PGRMC1) silencing enhanced nuclear factor of activated T-cells 1 (NFAT1) translocation to the cell nuclei in MDA-MB-231 cells. MDA-MB-231 cells were transfected for 48 h with yellow fluorescent protein (YFP)-NFAT1-reporter plasmid alone or in combination with the small hairpin siRNA against PGRMC1 (shPGRMC1). Fluorescence of NFAT1-reporter (NFAT-r) was analyzed using cell images of the cultures kept under regular condition or after being incubated with 1 μM of progesterone (P4) for 24 and 48 h. Images are representative of 3 independent transfections after 48 h of P4 administration to the respective cell batches. The histogram represents the percentage ± Standard Error of the Mean (S.E.M.) of the NFAT1-reporter fluorescence found in 4–8 images obtained from 3 independent transfections and after normalizing using the fluorescence found in scrambled and P4-untreated cells (mock). Internal experimental control consisted of the quantification of NFAT1 translocation due to store-operated Calcium entry (SOCE) evoked by thapsigargin (TG) administration. **, ***: represent p < 0.01 and 0.001 with respect to the scrambled and P4-untreated cells; meanwhile, $$$: represents p < 0.001 with respect to the scrambled cells and cells treated with P4, respectively. The bar represents 50 μm.
3. Discussion
The role of P4 on cell proliferation has been widely debated due to the presence of different receptors that may be targeted by this hormone in the different tissues [8,9,35,49,54]. In our hands, stimulation of MDA-MB-231 cells with P4 (1 μM) resulted in a PGRMC1-dependent reduction of cell proliferation, as demonstrated by the observation that PGRMC1 silencing impairs the inhibitory effect of P4 in cell proliferation. Furthermore, P4-evoked Ca2+ entry was strongly dependent on the activation of STIM2, Orai1, and TRPC1. Furthermore, P4-evoked Ca2+ entry was bigger in cells lacking PGRMC1 and in those cells pre-incubated with the PKA antagonist KT5720, thus, suggesting that the PGRMC1 plays a negative role in the P4-induced Ca2+ signaling. PGRMC1 silencing reversed the P4 negative effect on the Ca2+ entry reported in the MDA-MB-231 cells, which would lead to an enhanced NFAT1 translocation to the nucleus, as evidenced by using the NFAT1-reporter.
MDA-MB-231 cells express at least three different types of P4 receptors: PAQR family, PGRMC family, and sigma-2R. The latter was initially considered to be the PGRMC1; in fact, current evidence supports this idea [55,56]. Nonetheless, sigma-2R has been recently claimed to be different from PGRMC1 based on genetic and biochemical evidence [57,58,59,60]. In MDA-MB-231 cells, we demonstrated that inhibition of sigma-2R with NO1 reported a different effect on TG-evoked SOCE to the one presented here by silencing PGRMC1 [26]. The different physiological roles of both proteins would confirm the previous observations done by other groups [55]; however, we could not discard that both proteins would be part of a macromolecular complex targeted by P4. On the other hand, regarding the possible relationship between the members of the PAQR and PGRMC1 families, controversy still remains. In this sense, it has been reported that mPRα expressed in seatrout oocyte membranes would be activated by P4, which would subsequently downregulate cAMP production by impairing the activation of the adenylate cyclase [54,61], but according to the results obtained with the PKA antagonist and further demonstrated with the analysis of P-VASP, we believed that the latter did not take place in our cell models. Additionally, these authors reported that PGRMC1 might be involved in this pathway since PGRMC1 supports the access of mPRα to the plasma membrane [11,60]. Contrarily, artificial human mPRα expressed in HEK293 cells and MDA-MB-231 cells has failed to evoke Ca2+ mobilization in response to P4, and it is not able to interact with G proteins, and therefore, it is not able to activate cAMP or MAPK (ERK1/2 or p38). In fact, these authors concluded that mPRα was not expressed in the plasma membrane [30]. By silencing PGRMC1, we were able to completely abolish the negative effect of P4 on the proliferation of the MDA-MB-231 cells, which denotes a relevant role of this protein in the physiology of the MDA-MB-231 cells, as previously indicated by others [8,10]. Interestingly, PGRMC1 knockdown in the human colon cancer cell line, HCT116, has reported the contrary effect since authors evidenced a decrease in proliferation and metastasis to the liver in cells lacking this protein [62].
Regarding the role of P4 in the regulation of the intracellular Ca2+ homeostasis, it has been reported that the hormone reduces the expression of Ca2+ channels and Ca2+-dependent proteins involved in cell contraction in mouse embryonic stem cells (such as Ryr2, Calm2, Trpv2, and Mylk3) [63]. In line with this observation, spontaneously immortalized granulosa cells exhibit a dramatic reduction in the (Ca2+)c in response to prolonged P4 administration [30]. In the breast cancer cell line MCF7, P4 elicits changes in the expression of S100A11, S100A10, calreticulin, VDAC1, SERCA3, and SERCA1, resulting in a barely Ca2+ efflux from the endoplasmic reticulum as well as impaired membrane potential in the mitochondria [64]. Here, we reported for the very first time that P4-evoked Ca2+ entry requires STIM2 activation in MDA-MB-231 and MCF7 cell lines (Figure 3 and Figure 4), which operates Ca2+ entry in response to small changes in the amount of Ca2+ accumulated in the endoplasmic reticulum [28,36]. The latter was evidenced here by using GEM-CEPIA1er, a Ca2+ dye that was specifically designed to monitor the intraluminal Ca2+ changes in the transfected cells [65]. Interestingly, the combined use of this Ca2+ dye and 2-APB allowed us to demonstrate that in the triple-negative breast cancer cells (MDA-MB-231 and MDA-MB-468), the release of Ca2+ evoked by P4 occurred through the IP3R (Figure 1). 2-APB was previously reported to be a relatively high specific IP3R type 1 antagonist, but it may also affect IP3R type 3 when used at very elevated concentrations [29]. P4-evoked Ca2+ entry was also found to be dependent on Orai1 and TRPC1 activation. Therefore, PGRMC1 silencing enhanced P4-evoked Ca2+ entry and reversed the P4 inhibitory role in cell proliferation. In line with this observation, the antagonist of PKA, KT5720, enhanced the P4-evoked Ca2+ entry in all the triple breast cancer cell types evaluated (Figure 6A), so it did the silencing of PGRMC1 in MDA-MB-231 cells and MCF7 cells. Additional evidence of the existence of a negative regulatory pathway downstream of PGRMC1, which depends on PKA activation results of the evaluation of the level of P-VASP, being the phosphorylation of VASP, impaired in MDA-MB-231 cells lacking PGRMC1 (See Figure 6B), might support the existence of a P4/PGRMC1/cAMP/PKA signaling pathway in breast cancer cells, modulating the P4-evoked Ca2+ entry via phosphorylation of Orai1 by PKA, as previously reported [43,44].
The regulation of intracellular Ca2+ homeostasis is crucial for the activation of several key checkpoint proteins involved in the cell cycle; hence, alteration of this Ca2+ fluxes might have negative effects on cell proliferation [66]. It has been demonstrated that transient Ca2+ currents like those conducted by voltage-operated Ca2+ channels activation are very effective, activating transcription factors to maintain the cells in a quiescent status [67,68]. Ca2+ currents through the plasma membrane, such as SOCE, have been reported to evoke the activation of the pro-proliferative factor NFAT1 and its translocation to the nucleus [52,69]. Stimulation of cardiac myocytes with P4 leads to NFAT1 activation that is reversed in the presence of cyclosporine, an antagonist of the Ca2+-dependent phosphatase calcineurin, which is the main protein responsible for NFAT1 dephosphorylation, and subsequently, it is required for NFAT1 migration to the cell nucleus [70]. Activation of this pathway has been widely described downstream of the canonical SOCE Ca2+ channel, Orai1 [70]. Here, in MDA-MB-231 cells, P4-STIM2 dependent Ca2+ entry conducted the activation of Orai1 and TRPC1, and it led to NFAT1 nuclear translocation, as evidenced by the increase in the fluorescence of the NFAT1 reporter. In line with this, PGRMC1 silencing favored an enhanced NFAT1 accumulation in cells treated with P4 (Figure 9), thus favoring cell proliferation similar to that in prostate cancer cells [71,72]. In addition, NFAT1 has been reported to be overexpressed in several breast types like pancreatic, lung, and other cancer types [73,74]. Interestingly, Ca2+ entry through SOCE, a mechanism that relies on Orai1/STIM1 interaction, leads to NFAT1 activation due to a calcineurin-dependent dephosphorylation mechanism, as described above. Lack of STIM2 role in proliferation has been recently described in other cancer cell lines. Contrarily, a recent paper has claimed that STIM2 is responsible for the epithelial-mesenchymal transition (EMT), while others have pointed out that it may be involved in migration and promoting metastasis [75,76,77]. So, during cancer onset, PGRMC1 might be temporally downregulated, and thus, cancer cells might avoid the negative effect of P4 in proliferation (shown in Figure 7 and Figure 8), and in parallel, these cells might get the benefit of the activation of NFAT1 that would allow the MDA-MB-231 cells to migrate and initiate other cancer sprouts (shown in Figure 9).
Figure 10 summarizes the main ideas supported by our experimental data, and according to the experimental evidence shown in the present manuscript, patients with TNBC might get the benefit of P4 administration if an elevated PGRMC1 expression is previously confirmed in their breast tissues samples. Although future investigation will be required to confirm this hypothesis, this idea is reinforced by the observations done in mice where P4 reduces tumor size and brain metastasis evoked by exogenously injected MDA-MB-231 cells [5,8,54]. Although MDA-MB-231 cells present a truncated nPR with the ability of binding P4, the reported effects of this hormone should be limited to the presence of alternative receptors to the classic nPR [23]. In fact, P4 enhances proliferation in T47-D, HER2-overexpressing cells, MCF7, and MCF10A cell lines, exhibiting normal to high nPR expression [78,79]. As previously mentioned, this discrepancy in the P4 effects among these cell types and the TNBCs can be explained by the different expression of the classic progesterone receptors, and particularly, by the absence or presence of the nPR, which should be considered in future investigations.
Figure 10.
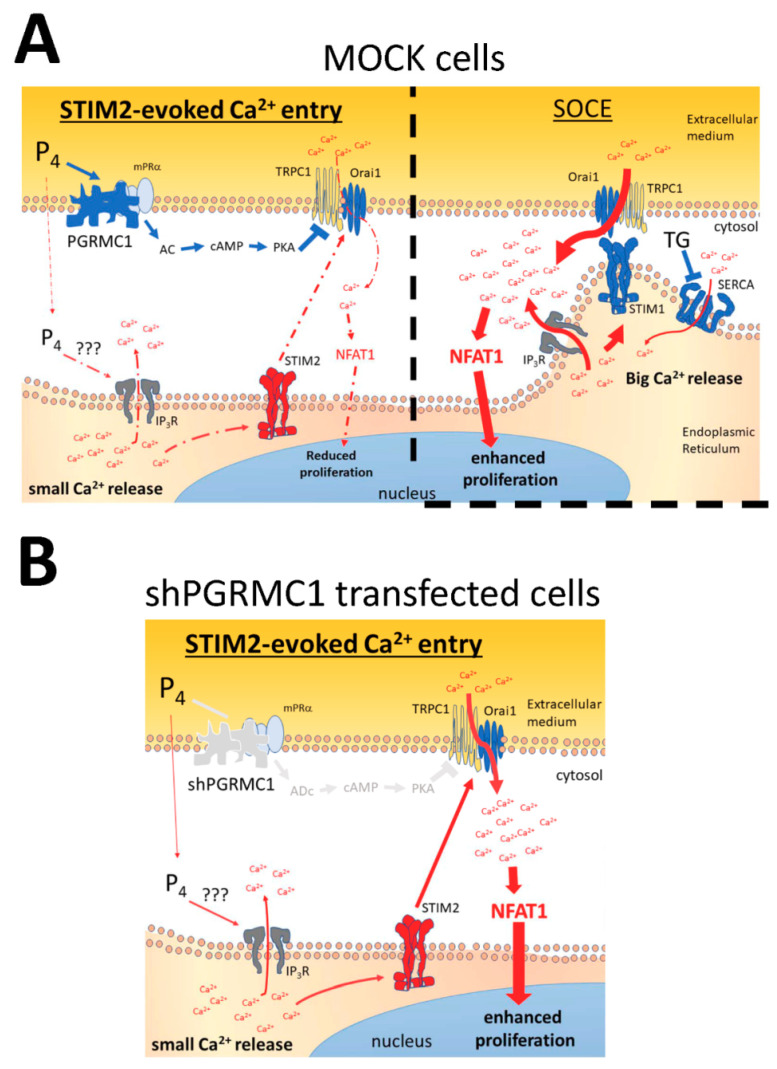
Schematic representation of the proposed mechanism of the P4 effects on Ca2+ homeostasis. (A) In MDA-MB-231 cells, STIM2 detects the small intraluminal Ca2+ changes evoked by P4 and activates a Ca2+ entry through Orai1 and TRPC1, which is limited due to P4-dependent PGRMC1 activation. PGRMC1 downstream mechanisms involve the activation of PKA that phosphorylates Orai1 and, thus, downregulates Ca2+ entry and cell proliferation. (B) Silencing of PGRMC1 results in an increase in the P4-evoked STIM2-dependent Ca2+ entry that leads to NFAT1 translocation to the cell nucleus and, therefore, reverses the negative effect of P4 in the proliferation of MDA-MB-231 cells.
4. Materials and Methods
4.1. Material
Fura-2/AM and anti-STIM2 antibody were from ThermoFisher Scientific (Molecular Probes®, Madrid, Spain). PKA antagonist, KT5720, was from Abcam® (Cambridge, UK). DharmaFECT Kb transfection reagent was from Dharmacon Inc. (Lafayette, CO, USA). The anti-STIM1 antibody was from BD-bioscience® (Madrid, Spain). The anti-PGRMC1 antibody was from Santa Cruz Biotechnology Inc. (Dallas, USA). Polyclonal anti-VASP (Ser239) antibody was from Cell Signaling Technology® (Danvers, MA, USA). HRP-conjugated secondary antibodies were from Jackson Immunoresearch Europe Ltd. (Cambridge, UK). YFP-NFAT1-reporter overexpression plasmid was kindly provided by Christoph Romanin (Johannes Kepler Institute of Biophysic, University of Linz, Linz, Austria). pCMV GEM-CEPIA1er was a gift from Masamitsu Iino (Addgene plasmid #58215; http://n2t.net/addgene:58215; RRID:Addgene_58215) [65]. BrdU cell proliferation assay kit was from BioVision Inc. (Milpitas, CA, USA). shPGRMC1 was from OriGene Technologies Inc. (Rockville, MD, USA). shOrai1 was kindly provided by Mohamed Trebak (Department of Cellular and Molecular Physiology, The Pennsylvania State University College of Medicine, Hershey, PA 17033, USA). shTRPC1 plasmid was kindly provided by Geoffrey E. Woodard (Department of Surgery, Uniformed Services University of the Health Sciences, Bethesda, MD, USA). siRNA was from Santacruz biotechnology Inc. (Heidelberg, Germany). esiSTIM1, esiPGRMC1, and esiSTIM2, thapsigargin (TG), progesterone (P4), 2-Aminoethoxydiphenyl borate (2-APB), anti-β-actin antibody, and other reagents of analytical grade were ordered from Sigma-Aldrich® (Madrid, Spain).
4.2. Cell Lines and Genetic Modification
MCF10A cell line was provided by Potier-Cartereau (Université François Rabelais, Tours, France). MCF7 and the triple-negative breast cancer cell lines—MDA-MB-231, MDA-MB-468, and BT20, were collected from ATCC® (Manassas, VA, USA). Cells were cultured following the manufacturer’s instructions or the protocols described elsewhere [25]. MCF7 and the TNBC cells were cultured at 37 °C with 5% CO2 in DMEM medium supplemented with FBS and penicillin/streptomycin; meanwhile, MCF10A cells were cultured in DMEM F-12 supplemented with horse serum, insulin, hydrocortisone, EGF, cholera toxin, and penicillin/streptomycin. Cell transfection was achieved by cell permeabilization with DharmaFECT transfection reagent, and subsequently, cells were incubated for 48 h either with 1–3 μg/mL of esiRNAs against STIM1 and STIM2 or siRNA. Alternatively, we used 2 μg/mL of scrambled plasmid, shPGRMC1, shOrai1, and shTRPC1 plasmids. In order to ascertain the efficiency of silencing protocols, Western blotting using the specific antibodies was done, as described below. Finally, cell transfection was done using the YFP-NFAT1-reporter overexpression plasmid (2 μg/mL), which was evaluated upon 72 h of transfection in order to ascertain NFAT1 translocation to the nucleus due to Ca2+ entry activation [80].
4.3. Cell Proliferation Assay
Non-genetically modified MDA-MB-231 cells (scrambled cells) or MDA-MB-231 cells transfected with the respective genetic agents were seeded at 5 × 105 cells/mL in 96-well plates. Following this, cells were cultured for 72 h either in the absence (control) or presence of P4 (1 μM). Proliferation cell rate at time 0, 24, 48, and 72 h was analyzed by using a commercial BrdU-based proliferation kit from Abcam [25]. Briefly, cells were incubated at the time indicated with BrdU for 2 h and subsequently fixed. Upon cell permeabilization, samples were incubated with an anti-BrdU antibody and with the reporter dye for 30 min. Immediately after the addition of the stop solution, the light emitted at 650 nm by the reporter dye was monitored by using a Plate-reader (Epoch, Bioteck®; Swindon, UK).
4.4. Analysis of the Changes in the Cytosolic Free Ca2+ Concentration (Ca2+)c
MDA-MB-231 cells were either transfected for 48 h with the indicated genetic material or were kept untreated. The day before the experiments, cells were detached, the medium was replaced, and finally, they were seeded again at 1 × 105 cells/mL in a 6-well plate containing a sterile coverslip that was deposited at the bottom of the wells. Cells were allowed to spread onto the coverslips for an additional 24 h. The next day, cells were incubated for 30 min with 2 μM of fura-2/AM at 37 °C in the incubation chamber.
Coverslips were mounted in a perfusion chamber and were maintained in a Ca2+-free HBS medium (in mM: 125 NaCl, 25 HEPES, 5 KCl, 1 MgCl2, pH 7.4, and supplemented with 0.1% (w/v) bovine serum albumin and 5 mM of D-glucose). Fluorescence emitted by the samples at 505 nm, as a result of the alternative excitation at 340/380 nm wavelengths, was recorded using a cooled sCMOS camera (Zyla 4.2, Andor, Belfast, UK). Fura-2 fluorescence was represented as the changes in the ratio of fluorescence (Fn/F0) upon normalizing the ratio by considering the basal values on each experiment, which result from removing the extracellular free Ca2+ by supplementation of the medium with 75 μM of EGTA (F0). To compare among different experimental conditions, the areas under the curves were calculated by using the integral of the rise in the ratio of fura-2 fluorescence for 2–4 min after the addition of the stimuli to the cells. Ca2+ entry evoked by emptying the intracellular Ca2+ stores was visualized upon the addition of 1 mM of CaCl2 to the extracellular medium, as widely reported elsewhere [25].
Additionally, Ca2+ mobilization from the endoplasmic reticulum was determined by using the endoplasmic reticulum Ca2+ dye, GEM-CEPIA1er, which can be excited at 380 nm and presents two maximum emission wavelengths: at 510 nm in the absence of Ca2+ (green) and at 462 nm in the presence of Ca2+ (blue). Therefore, the ratio between both emission wavelength (blue/green) can be used to determine the intraluminal Ca2+ changes within the endoplasmic reticulum, as previously reported by others [65].
4.5. Quantification of Cell Death
MDA-MB-231 cells were transfected with shPGRMC1 or the scrambled plasmids, as aforementioned. Cells were cultured for 24 h in the absence or presence of P4 (1 μM) and, subsequently, they were loaded with propidium iodide (4 μM) for 45 min at 37 °C inside the incubator. Cell middle plane images were acquired at 555/624 of wavelength (Ex/Em, respectively) using a cooled digital CMOS camera (Zyla 4.2, Andor, Belfast, UK) and a fluorescence 40x objective mounted in an inverted microscope (Nikon Eclipse Ti-2, Amsterdam, The Netherlands) that is controlled by the NIS-Elements AR software (Nikon, Amsterdam, The Netherlands). Cell death was expressed as a percentage of positive PI cells with respect to the total cell account in each field.
4.6. Visualization of NFAT1 Translocation to the Nucleus
MDA-MB-231 cells (1 × 105 cells/mL) were transfected for 48 h with the YFP-NFAT1-reporter overexpression plasmid and shPGRMC1 or with the NFAT1-reporter plasmid alone. Upon confirmation of the positive expression of NFAT1, which was done by visualizing the YFP fluorescence derived from the NFAT1-reporter in control proliferating cells, they were seeded at the appropriate concentration (1 × 105 cells/mL). The following day, cells were stimulated for 48 h with P4 (1 μM) and were maintained growing under regular culture conditions (Mock). Alternatively, as an internal control, we artificially forced the internalization of the YFP-NFAT1-reporter into the nuclei by stimulating the cells for 24 h with 1 μM of thapsigargin (TG). This maneuver should activate SOCE and, subsequently, facilitate downstream NFAT1 constant translocation to the nucleus [69]. The analysis of the fluorescence emitted by the NFAT1-reporter was done by using an inverted fluorescence microscope at 24 h and 48 h after the beginning of the cell stimulation with P4. Data were normalized by comparing with the NFAT1 fluorescence with respect to the one emitted by the mock cells growing under regular conditions.
4.7. Western Blotting
MCF10A, MCF7, and MDA-MB-231 cells (1 × 106 cells/mL) were lysed by mixing with an equal volume of NP40 buffer. The protein quantification was done using the BCA-based protocol, and all samples were normalized to 1 mg/mL, before proteins were denatured by mixing with an equal volume of Laemmli’s buffer (supplemented with SDS, 5%), following which, they were heated at 95 °C for 5 min. Proteins were separated by SDS-PAGE and were subsequently transferred to nitrocellulose membranes. Non-specific binding sites were blocked by incubating the membranes with the blocking buffer (10 % of BSA). Western blotting was done by incubating the membranes overnight either with the specific monoclonal anti-STIM1, anti-STIM2, anti-PGRMC1, and anti-VASP antibodies diluted in TBST (supplemented with BSA at 1:1000). The excess of the primary antibody was removed by washing for 30 min with TBST. Following this, the membranes were incubated for an additional hour with the appropriate HRP-conjugated secondary antibodies (diluted 1:10,000 in TBST). Finally, the membranes were exposed to the dura-ECL solution for 5 min. Chemiluminescence images were acquired using a CD-digit chemiluminescence device (Licor®, Bonsai Lab, Madrid, Spain), and optic densitometry of the images was analyzed by using the Image J (Fiji plugging) (free-software from NIH). Membranes were reprobed with anti-β-actin antibody as a loading control.
4.8. Statistical Analysis
A student’s t-test was used for establishing a comparison between two groups. Meanwhile, for multiple comparisons, the analysis of the variance (ANOVA), combined with post hoc Tukey’s test and Dunnett’s tests, was used. p < 0.05 was considered a significant difference between groups.
5. Conclusions
P4 activates a STIM2-dependent Ca2+ entry that occurs through Orai1 and TRPC1 in MDA-MB-231 cells.
PGRMC1 activates PKA, which phosphorylates Orai1 to downregulate the P4-evoked Ca2+ entry.
P4-evoked PGRMC1 activation downregulates cell proliferation in MDA-MB-231 cells.
The silencing of PGRMC1 impairs P4-evoked cell death in MDA-MB-231 cells.
The silencing of PGRMC1 results in an elevated Ca2+ entry and, subsequently, increases NFAT1 translocation to the cell nucleus.
PGRMC1 could be targeted to prevent uncontrolled cell proliferation in TNBC.
Acknowledgments
We thank Mercedes Gomez and Sandra Alvarado for their technical assistance.
Abbreviations
| 2-APB | 2-aminoethoxydiphenyl borate |
| cAMP | 3′,5′-cyclic adenosine monophosphate |
| GEM-CEPIA1er | Endoplasmic reticulum intraluminal Ca2+ dye |
| IP3R | Inositol 1,4,5-trisphosphate receptor |
| mPRα | Progesterone receptor membrane α |
| NFAT1 | Nuclear factor of activated T-cells 1 |
| nPR | Nuclear progesterone receptor |
| P4 | Progesterone |
| PGRMC1 | Progesterone receptor membrane component 1 |
| PKAi | PKA antagonist, KT5720 |
| SERCA | Sarcoendoplamic reticulum Ca2+-ATPase |
| SOCE | Store-operated calcium entry |
| STIM1 & 2 | Stromal interaction molecule 1 & 2 |
| TG | Thapsigargin |
| TNBC | Triple negative breast cancer cells |
| EGTA | Ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid |
Author Contributions
C.C. had done most of the experiments. G.M.S. and J.A.R. discussed the results, designed the experiments, and edited the manuscript. P.C.R. designed the experiments, analyzed, presented, and discussed the results, and finally, wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work has been supported by MICIN (BFU-2016-74932-C2-1-P) and Junta de Extremadura-FEDER (IB16046, IB18020, and GR18061). C.C. has been awarded a Junta de Extremadura Predoctoral Fellowship (PD16072).
Conflicts of Interest
All authors have read and given their consent for publishing the present results. The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kastner P., Krust A., Turcotte B., Stropp U., Tora L., Gronemeyer H., Chambon P. Two distinct estrogen-regulated promoters generate transcripts encoding the two functionally different human progesterone receptor forms A and B. EMBO J. 1990;9:1603–1614. doi: 10.1002/j.1460-2075.1990.tb08280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Obr A.E., Grimm S.L., Bishop K.A., Pike J.W., Lydon J.P., Edwards D.P. Progesterone receptor and Stat5 signaling cross talk through RANKL in mammary epithelial cells. Mol. Endocrinol. 2013;27:1808–1824. doi: 10.1210/me.2013-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohammad M.A., Hadsell D.L., Haymond M.W. Gene regulation of UDP-galactose synthesis and transport: Potential rate-limiting processes in initiation of milk production in humans. Am. J. Physiol. Endocrinol. Metab. 2012;303:E365–E376. doi: 10.1152/ajpendo.00175.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin Y.H., Hua Q.F., Zheng J.J., Ma X.H., Chen T.X., Zhang S., Chen B., Dai Q., Zhang X.H. Diagnostic Value of ER, PR, FR and HER-2-Targeted Molecular Probes for Magnetic Resonance Imaging in Patients with Breast Cancer. Cell Physiol. Biochem. 2018;49:271–281. doi: 10.1159/000492877. [DOI] [PubMed] [Google Scholar]
- 5.Bertucci F., Finetti P., Birnbaum D. Basal breast cancer: A complex and deadly molecular subtype. Curr. Mol. Med. 2012;12:96–110. doi: 10.2174/156652412798376134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Formby B., Wiley T.S. Progesterone inhibits growth and induces apoptosis in breast cancer cells: Inverse effects on Bcl-2 and p53. Ann. Clin. Lab. Sci. 1998;28:360–369. [PubMed] [Google Scholar]
- 7.Zhou L., Zhou W., Zhang H., Hu Y., Yu L., Zhang Y., Wang S., Wang P., Xia W. Progesterone suppresses triple-negative breast cancer growth and metastasis to the brain via membrane progesterone receptor alpha. Int. J. Mol. Med. 2017;40:755–761. doi: 10.3892/ijmm.2017.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark N.C., Friel A.M., Pru C.A., Zhang L., Shioda T., Rueda B.R., Peluso J.J., Pru J.K. Progesterone receptor membrane component 1 promotes survival of human breast cancer cells and the growth of xenograft tumors. Cancer Biol. Ther. 2016;17:262–271. doi: 10.1080/15384047.2016.1139240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krietsch T., Fernandes M.S., Kero J., Losel R., Heyens M., Lam E.W., Huhtaniemi I., Brosens J.J., Gellersen B. Human homologs of the putative G protein-coupled membrane progestin receptors (mPRalpha, beta, and gamma) localize to the endoplasmic reticulum and are not activated by progesterone. Mol. Endocrinol. 2006;20:3146–3164. doi: 10.1210/me.2006-0129. [DOI] [PubMed] [Google Scholar]
- 10.Petersen S.L., Intlekofer K.A., Moura-Conlon P.J., Brewer D.N., Del Pino Sans J., Lopez J.A. Nonclassical progesterone signalling molecules in the nervous system. J. Neuroendocr. 2013;25:991–1001. doi: 10.1111/jne.12060. [DOI] [PubMed] [Google Scholar]
- 11.Thomas P., Pang Y., Dong J. Enhancement of cell surface expression and receptor functions of membrane progestin receptor alpha (mPRalpha) by progesterone receptor membrane component 1 (PGRMC1): Evidence for a role of PGRMC1 as an adaptor protein for steroid receptors. Endocrinology. 2014;155:1107–1119. doi: 10.1210/en.2013-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cahill M.A., Jazayeri J.A., Catalano S.M., Toyokuni S., Kovacevic Z., Richardson D.R. The emerging role of progesterone receptor membrane component 1 (PGRMC1) in cancer biology. Biochim. Biophys. Acta. 2016;1866:339–349. doi: 10.1016/j.bbcan.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 13.Ruan X., Zhang Y., Mueck A.O., Willibald M., Seeger H., Fehm T., Brucker S., Neubauer H. Increased expression of progesterone receptor membrane component 1 is associated with aggressive phenotype and poor prognosis in ER-positive and negative breast cancer. Menopause. 2017;24:203–209. doi: 10.1097/GME.0000000000000739. [DOI] [PubMed] [Google Scholar]
- 14.Asperger H., Stamm N., Gierke B., Pawlak M., Hofmann U., Zanger U.M., Marton A., Katona R.L., Buhala A., Vizler C., et al. Progesterone receptor membrane component 1 regulates lipid homeostasis and drives oncogenic signaling resulting in breast cancer progression. Breast Cancer Res. 2020;22:75. doi: 10.1186/s13058-020-01312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riad A., Zeng C., Weng C.C., Winters H., Xu K., Makvandi M., Metz T., Carlin S., Mach R.H. Sigma-2 Receptor/TMEM97 and PGRMC-1 Increase the Rate of Internalization of LDL by LDL Receptor through the Formation of a Ternary Complex. Sci. Rep. 2018;8:16845. doi: 10.1038/s41598-018-35430-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X., Rhee D.K., Malhotra R., Mayeur C., Hurst L.A., Ager E., Shelton G., Kramer Y., McCulloh D., Keefe D., et al. Progesterone receptor membrane component-1 regulates hepcidin biosynthesis. J. Clin. Investig. 2016;126:389–401. doi: 10.1172/JCI83831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jardin I., Lopez J.J., Salido G.M., Rosado J.A. Store-Operated Ca2+ Entry in Breast Cancer Cells: Remodeling and Functional Role. Int. J. Mol. Sci. 2018;19:4053. doi: 10.3390/ijms19124053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swiatek-De Lange M., Stampfl A., Hauck S.M., Zischka H., Gloeckner C.J., Deeg C.A., Ueffing M. Membrane-initiated effects of progesterone on calcium dependent signaling and activation of VEGF gene expression in retinal glial cells. Glia. 2007;55:1061–1073. doi: 10.1002/glia.20523. [DOI] [PubMed] [Google Scholar]
- 19.Espino J., Mediero M., Lozano G.M., Bejarano I., Ortiz A., Garcia J.F., Pariente J.A., Rodriguez A.B. Reduced levels of intracellular calcium releasing in spermatozoa from asthenozoospermic patients. Reprod. Biol. Endocrinol. 2009;7:11. doi: 10.1186/1477-7827-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwang J.Y., Duncan R.S., Madry C., Singh M., Koulen P. Progesterone potentiates calcium release through IP3 receptors by an Akt-mediated mechanism in hippocampal neurons. Cell Calcium. 2009;45:233–242. doi: 10.1016/j.ceca.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palmerini C.A., Mazzoni M., Radicioni G., Marzano V., Granieri L., Iavarone F., Longhi R., Messana I., Cabras T., Sanna M.T., et al. Antagonistic Effect of a Salivary Proline-Rich Peptide on the Cytosolic Ca2+ Mobilization Induced by Progesterone in Oral Squamous Cancer Cells. PLoS ONE. 2016;11:e0147925. doi: 10.1371/journal.pone.0147925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee K.L., Dai Q., Hansen E.L., Saner C.N., Price T.M. Modulation of ATP-induced calcium signaling by progesterone in T47D-Y breast cancer cells. Mol. Cell Endocrinol. 2010;319:109–115. doi: 10.1016/j.mce.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cork D.M., Lennard T.W., Tyson-Capper A.J. Progesterone receptor (PR) variants exist in breast cancer cells characterised as PR negative. Tumour Biol. 2012;33:2329–2340. doi: 10.1007/s13277-012-0495-z. [DOI] [PubMed] [Google Scholar]
- 24.Salazar M., Lerma-Ortiz A., Hooks G.M., Ashley A.K., Ashley R.L. Progestin-mediated activation of MAPK and AKT in nuclear progesterone receptor negative breast epithelial cells: The role of membrane progesterone receptors. Gene. 2016;591:6–13. doi: 10.1016/j.gene.2016.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jardin I., Diez-Bello R., Lopez J.J., Redondo P.C., Salido G.M., Smani T., Rosado J.A. TRPC6 Channels Are Required for Proliferation, Migration and Invasion of Breast Cancer Cell Lines by Modulation of Orai1 and Orai3 Surface Exposure. Cancers. 2018;10:331. doi: 10.3390/cancers10090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cantonero C., Camello P.J., Abate C., Berardi F., Salido G.M., Rosado J.A., Redondo P.C. NO1, a New Sigma 2 Receptor/TMEM97 Fluorescent Ligand, Downregulates SOCE and Promotes Apoptosis in the Triple Negative Breast Cancer Cell Lines. Cancers. 2020;12:257. doi: 10.3390/cancers12020257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bird G.S., Bian X., Putney J.W., Jr. Calcium entry signal? Nature. 1995;373:481–482. doi: 10.1038/373481b0. [DOI] [PubMed] [Google Scholar]
- 28.Redondo P.C., Rosado J.A. Store-operated calcium entry: Unveiling the calcium handling signalplex. Int. Rev. Cell Mol. Biol. 2015;316:183–226. doi: 10.1016/bs.ircmb.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 29.Iagaru A., Minamimoto R., Jamali M., Barkodhodari A., Gambhir S.S., Vasanawala S. Imaging patients with breast and prostate cancers using combined 18F NaF/18F FDG and TOF simultaneous PET/MRI. EJNMMI Phys. 2015;2:A65. doi: 10.1186/2197-7364-2-S1-A65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peluso J.J., Fernandez G., Pappalardo A., White B.A. Membrane-initiated events account for progesterone’s ability to regulate intracellular free calcium levels and inhibit rat granulosa cell mitosis. Biol. Reprod. 2002;67:379–385. doi: 10.1095/biolreprod67.2.379. [DOI] [PubMed] [Google Scholar]
- 31.Motiani R.K., Abdullaev I.F., Trebak M. A novel native store-operated calcium channel encoded by Orai3: Selective requirement of Orai3 versus Orai1 in estrogen receptor-positive versus estrogen receptor-negative breast cancer cells. J. Biol. Chem. 2010;285:19173–19183. doi: 10.1074/jbc.M110.102582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopez J.J., Salido G.M., Pariente J.A., Rosado J.A. Interaction of STIM1 with endogenously expressed human canonical TRP1 upon depletion of intracellular Ca2+ stores. J. Biol. Chem. 2006;281:28254–28264. doi: 10.1074/jbc.M604272200. [DOI] [PubMed] [Google Scholar]
- 33.Williams R.T., Manji S.S., Parker N.J., Hancock M.S., Van Stekelenburg L., Eid J.P., Senior P.V., Kazenwadel J.S., Shandala T., Saint R., et al. Identification and characterization of the STIM (stromal interaction molecule) gene family: Coding for a novel class of transmembrane proteins. Biochem. J. 2001;357:673–685. doi: 10.1042/bj3570673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roos J., DiGregorio P.J., Yeromin A.V., Ohlsen K., Lioudyno M., Zhang S., Safrina O., Kozak J.A., Wagner S.L., Cahalan M.D., et al. STIM1, an essential and conserved component of store-operated Ca2+ channel function. J. Cell. Biol. 2005;169:435–445. doi: 10.1083/jcb.200502019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liou J., Kim M.L., Heo W.D., Jones J.T., Myers J.W., Ferrell J.E., Jr., Meyer T. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr. Biol. 2005;15:1235–1241. doi: 10.1016/j.cub.2005.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Theis M., Buchholz F. MISSION esiRNA for RNAi screening in mammalian cells. J. Vis. Exp. 2010;39:e2008. doi: 10.3791/2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nelson H.A., Leech C.A., Kopp R.F., Roe M.W. Interplay between ER Ca2+ Binding Proteins, STIM1 and STIM2, Is Required for Store-Operated Ca2+ Entry. Int. J. Mol. Sci. 2018;19:1522. doi: 10.3390/ijms19051522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subedi K.P., Ong H.L., Son G.Y., Liu X., Ambudkar I.S. STIM2 Induces Activated Conformation of STIM1 to Control Orai1 Function in ER-PM Junctions. Cell Rep. 2018;23:522–534. doi: 10.1016/j.celrep.2018.03.065. [DOI] [PubMed] [Google Scholar]
- 39.Stathopulos P.B., Zheng L., Ikura M. Stromal interaction molecule (STIM) 1 and STIM2 calcium sensing regions exhibit distinct unfolding and oligomerization kinetics. J. Biol. Chem. 2009;284:728–732. doi: 10.1074/jbc.C800178200. [DOI] [PubMed] [Google Scholar]
- 40.Zheng L., Stathopulos P.B., Li G.Y., Ikura M. Biophysical characterization of the EF-hand and SAM domain containing Ca2+ sensory region of STIM1 and STIM2. Biochem. Biophys. Res. Commun. 2008;369:240–246. doi: 10.1016/j.bbrc.2007.12.129. [DOI] [PubMed] [Google Scholar]
- 41.Ong H.L., de Souza L.B., Zheng C., Cheng K.T., Liu X., Goldsmith C.M., Feske S., Ambudkar I.S. STIM2 enhances receptor-stimulated Ca2+ signaling by promoting recruitment of STIM1 to the endoplasmic reticulum-plasma membrane junctions. Sci. Signal. 2015;8:ra3. doi: 10.1126/scisignal.2005748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang X., Pathak T., Yoast R., Emrich S., Xin P., Nwokonko R.M., Johnson M., Wu S., Delierneux C., Gueguinou M., et al. A calcium/cAMP signaling loop at the ORAI1 mouth drives channel inactivation to shape NFAT induction. Nat. Commun. 2019;10:1971. doi: 10.1038/s41467-019-09593-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang M., Robitaille M., Showalter A.D., Huang X., Liu Y., Bhattacharjee A., Willard F.S., Han J., Froese S., Wei L., et al. Progesterone receptor membrane component 1 is a functional part of the glucagon-like peptide-1 (GLP-1) receptor complex in pancreatic beta cells. Mol. Cell. Proteom. 2014;13:3049–3062. doi: 10.1074/mcp.M114.040196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Machelon V., Nome F., Grosse B., Lieberherr M. Progesterone triggers rapid transmembrane calcium influx and/or calcium mobilization from endoplasmic reticulum, via a pertussis-insensitive G-protein in granulosa cells in relation to luteinization process. J. Cell. Biochem. 1996;61:619–628. doi: 10.1002/(SICI)1097-4644(19960616)61:4<619::AID-JCB16>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 45.Gonzalez-Orozco J.C., Hansberg-Pastor V., Valadez-Cosmes P., Nicolas-Ortega W., Bastida-Beristain Y., Fuente-Granada M., Gonzalez-Arenas A., Camacho-Arroyo I. Activation of membrane progesterone receptor-alpha increases proliferation, migration, and invasion of human glioblastoma cells. Mol. Cell. Endocrinol. 2018;477:81–89. doi: 10.1016/j.mce.2018.06.004. [DOI] [PubMed] [Google Scholar]
- 46.Hegde S.M., Kumar M.N., Kavya K., Kumar K.M., Nagesh R., Patil R.H., Babu R.L., Ramesh G.T., Sharma S.C. Interplay of nuclear receptors (ER, PR, and GR) and their steroid hormones in MCF7 cells. Mol. Cell. Biochem. 2016;422:109–120. doi: 10.1007/s11010-016-2810-2. [DOI] [PubMed] [Google Scholar]
- 47.Neubauer H., Adam G., Seeger H., Mueck A.O., Solomayer E., Wallwiener D., Cahill M.A., Fehm T. Membrane-initiated effects of progesterone on proliferation and activation of VEGF in breast cancer cells. Climacteric. 2009;12:230–239. doi: 10.1080/13697130802635637. [DOI] [PubMed] [Google Scholar]
- 48.Goncharov A.I., Maslakova A.A., Polikarpova A.V., Bulanova E.A., Guseva A.A., Morozov I.A., Rubtsov P.M., Smirnova O.V., Shchelkunova T.A. Progesterone inhibits proliferation and modulates expression of proliferation-Related genes in classical progesterone receptor-negative human BxPC3 pancreatic adenocarcinoma cells. J. Steroid Biochem. Mol. Biol. 2017;165:293–304. doi: 10.1016/j.jsbmb.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 49.Rashmi K.C., Harsha Raj M., Paul M., Girish K.S., Salimath B.P., Aparna H.S. A new pyrrole based small molecule from Tinospora cordifolia induces apoptosis in MDA-MB-231 breast cancer cells via ROS mediated mitochondrial damage and restoration of p53 activity. Chem. Biol. Interact. 2019;299:120–130. doi: 10.1016/j.cbi.2018.12.005. [DOI] [PubMed] [Google Scholar]
- 50.van Ginkel P.R., Yan M.B., Bhattacharya S., Polans A.S., Kenealey J.D. Natural products induce a G protein-mediated calcium pathway activating p53 in cancer cells. Toxicol. Appl. Pharm. 2015;288:453–462. doi: 10.1016/j.taap.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chung E., Yeung F., Leinwand L.A. Calcineurin activity is required for cardiac remodelling in pregnancy. Cardiovasc. Res. 2013;100:402–410. doi: 10.1093/cvr/cvt208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Albarran L., Lopez J.J., Jardin I., Sanchez-Collado J., Berna-Erro A., Smani T., Camello P.J., Salido G.M., Rosado J.A. EFHB is a Novel Cytosolic Ca2+ Sensor That Modulates STIM1-SARAF Interaction. Cell. Physiol. Biochem. 2018;51:1164–1178. doi: 10.1159/000495494. [DOI] [PubMed] [Google Scholar]
- 53.Lee S.H., Rigas N.K., Lee C.R., Bang A., Srikanth S., Gwack Y., Kang M.K., Kim R.H., Park N.H., Shin K.H. Orai1 promotes tumor progression by enhancing cancer stemness via NFAT signaling in oral/oropharyngeal squamous cell carcinoma. Oncotarget. 2016;7:43239–43255. doi: 10.18632/oncotarget.9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wiebe J.P., Zhang G., Welch I., Cadieux-Pitre H.A. Progesterone metabolites regulate induction, growth, and suppression of estrogen- and progesterone receptor-negative human breast cell tumors. Breast Cancer Res. 2013;15:R38. doi: 10.1186/bcr3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu J., Zeng C., Chu W., Pan F., Rothfuss J.M., Zhang F., Tu Z., Zhou D., Zeng D., Vangveravong S., et al. Identification of the PGRMC1 protein complex as the putative sigma-2 receptor binding site. Nat. Commun. 2011;2:380. doi: 10.1038/ncomms1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bali N., Arimoto J.M., Morgan T.E., Finch C.E. Progesterone antagonism of neurite outgrowth depends on microglial activation via Pgrmc1/S2R. Endocrinology. 2013;154:2468–2480. doi: 10.1210/en.2012-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chu U.B., Mavlyutov T.A., Chu M.L., Yang H., Schulman A., Mesangeau C., McCurdy C.R., Guo L.W., Ruoho A.E. The Sigma-2 Receptor and Progesterone Receptor Membrane Component 1 are Different Binding Sites Derived From Independent Genes. EBioMedicine. 2015;2:1806–1813. doi: 10.1016/j.ebiom.2015.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pati M.L., Groza D., Riganti C., Kopecka J., Niso M., Berardi F., Hager S., Heffeter P., Hirai M., Tsugawa H., et al. Sigma-2 receptor and progesterone receptor membrane component 1 (PGRMC1) are two different proteins: Proofs by fluorescent labeling and binding of sigma-2 receptor ligands to PGRMC1. Pharm. Res. 2017;117:67–74. doi: 10.1016/j.phrs.2016.12.023. [DOI] [PubMed] [Google Scholar]
- 59.Abate C., Niso M., Infantino V., Menga A., Berardi F. Elements in support of the ‘non-identity’ of the PGRMC1 protein with the sigma2 receptor. Eur. J. Pharm. 2015;758:16–23. doi: 10.1016/j.ejphar.2015.03.067. [DOI] [PubMed] [Google Scholar]
- 60.Hiranita T. Identification of the Sigma-2 Receptor: Distinct from the Progesterone Receptor Membrane Component 1 (PGRMC1) J. Alcohol. Drug Depend. 2016;4 doi: 10.4172/2329-6488.1000e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thomas P. Characteristics of membrane progestin receptor alpha (mPRalpha) and progesterone membrane receptor component 1 (PGMRC1) and their roles in mediating rapid progestin actions. Front. Neuroendocr. 2008;29:292–312. doi: 10.1016/j.yfrne.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kabe Y., Nakane T., Koike I., Yamamoto T., Sugiura Y., Harada E., Sugase K., Shimamura T., Ohmura M., Muraoka K., et al. Haem-dependent dimerization of PGRMC1/Sigma-2 receptor facilitates cancer proliferation and chemoresistance. Nat. Commun. 2016;7:11030. doi: 10.1038/ncomms11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kang H.Y., Choi Y.K., Jeung E.B. Inhibitory effect of progesterone during early embryonic development: Suppression of myocardial differentiation and calcium-related transcriptome by progesterone in mESCs: Progesterone disturb cardiac differentiation of mESCs through lower cytosolic Ca2+ Reprod. Toxicol. 2016;64:169–179. doi: 10.1016/j.reprotox.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 64.Azeez J.M., Vini R., Remadevi V., Surendran A., Jaleel A., Santhosh Kumar T.R., Sreeja S. VDAC1 and SERCA3 Mediate Progesterone-Triggered Ca2+ Signaling in Breast Cancer Cells. J. Proteome Res. 2018;17:698–709. doi: 10.1021/acs.jproteome.7b00754. [DOI] [PubMed] [Google Scholar]
- 65.Boillot M., Morin-Brureau M., Picard F., Weckhuysen S., Lambrecq V., Minetti C., Striano P., Zara F., Iacomino M., Ishida S., et al. Novel GABRG2 mutations cause familial febrile seizures. Neurol. Genet. 2015;1:e35. doi: 10.1212/NXG.0000000000000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu H.G., McLaughlin S., Newman M., Brundage K., Ammer A., Martin K., Coad J. Altering calcium influx for selective destruction of breast tumor. BMC Cancer. 2017;17:169. doi: 10.1186/s12885-017-3168-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cartin L., Lounsbury K.M., Nelson M.T. Coupling of Ca2+ to CREB activation and gene expression in intact cerebral arteries from mouse: Roles of ryanodine receptors and voltage-dependent Ca2+ channels. Circ. Res. 2000;86:760–767. doi: 10.1161/01.RES.86.7.760. [DOI] [PubMed] [Google Scholar]
- 68.Feske S., Giltnane J., Dolmetsch R., Staudt L.M., Rao A. Gene regulation mediated by calcium signals in T lymphocytes. Nat. Immunol. 2001;2:316–324. doi: 10.1038/86318. [DOI] [PubMed] [Google Scholar]
- 69.Kar P., Parekh A.B. Distinct spatial Ca2+ signatures selectively activate different NFAT transcription factor isoforms. Mol. Cell. 2015;58:232–243. doi: 10.1016/j.molcel.2015.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu B.X., Yuan J.N., Zhang F.R., Liu Y.Y., Zhang T.T., Li K., Lv X.F., Zhou J.G., Huang L.Y., Shang J.Y., et al. Inhibition of Orai1-mediated Ca2+ entry limits endothelial cell inflammation by suppressing calcineurin-NFATc4 signaling pathway. Biochem. Biophys. Res. Commun. 2018;495:1864–1870. doi: 10.1016/j.bbrc.2017.12.034. [DOI] [PubMed] [Google Scholar]
- 71.Thebault S., Flourakis M., Vanoverberghe K., Vandermoere F., Roudbaraki M., Lehen’kyi V., Slomianny C., Beck B., Mariot P., Bonnal J.L., et al. Differential role of transient receptor potential channels in Ca2+ entry and proliferation of prostate cancer epithelial cells. Cancer Res. 2006;66:2038–2047. doi: 10.1158/0008-5472.CAN-05-0376. [DOI] [PubMed] [Google Scholar]
- 72.Manda K.R., Tripathi P., Hsi A.C., Ning J., Ruzinova M.B., Liapis H., Bailey M., Zhang H., Maher C.A., Humphrey P.A., et al. NFATc1 promotes prostate tumorigenesis and overcomes PTEN loss-induced senescence. Oncogene. 2016;35:3282–3292. doi: 10.1038/onc.2015.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baumgart S., Chen N.M., Zhang J.S., Billadeau D.D., Gaisina I.N., Kozikowski A.P., Singh S.K., Fink D., Strobel P., Klindt C., et al. GSK-3beta Governs Inflammation-Induced NFATc2 Signaling Hubs to Promote Pancreatic Cancer Progression. Mol. Cancer Ther. 2016;15:491–502. doi: 10.1158/1535-7163.MCT-15-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu J.F., Zhao S.H., Wu S.S. Depleting NFAT1 expression inhibits the ability of invasion and migration of human lung cancer cells. Cancer Cell. Int. 2013;13:41. doi: 10.1186/1475-2867-13-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Oskay Halacli S. FOXP1 enhances tumor cell migration by repression of NFAT1 transcriptional activity in MDA-MB-231 cells. Cell. Biol. Int. 2017;41:102–110. doi: 10.1002/cbin.10702. [DOI] [PubMed] [Google Scholar]
- 76.Shadboorestan A., Tarfiei G.A., Montazeri H., Sepand M.R., Zangooei M., Khedri A., Ostad S.N., Ghahremani M.H. Invasion and migration of MDA-MB-231 cells are inhibited by block of AhR and NFAT: Role of AhR/NFAT1/beta4 integrin signaling. J. Appl. Toxicol. 2019;39:375–384. doi: 10.1002/jat.3728. [DOI] [PubMed] [Google Scholar]
- 77.Zhang S., Miao Y., Zheng X., Gong Y., Zhang J., Zou F., Cai C. STIM1 and STIM2 differently regulate endogenous Ca2+ entry and promote TGF-beta-induced EMT in breast cancer cells. Biochem. Biophys. Res. Commun. 2017;488:74–80. doi: 10.1016/j.bbrc.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 78.Wang H.C., Lee W.S. Molecular mechanisms underlying progesterone-induced cytoplasmic retention of p27 in breast cancer cells. J. Steroid Biochem. Mol. Biol. 2018;183:202–209. doi: 10.1016/j.jsbmb.2018.06.015. [DOI] [PubMed] [Google Scholar]
- 79.Tian J.M., Ran B., Zhang C.L., Yan D.M., Li X.H. Estrogen and progesterone promote breast cancer cell proliferation by inducing cyclin G1 expression. Braz. J. Med. Biol. Res. 2018;51:1–7. doi: 10.1590/1414-431x20175612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Frischauf I., Zayats V., Deix M., Hochreiter A., Jardin I., Muik M., Lackner B., Svobodova B., Pammer T., Litvinukova M., et al. A calcium-accumulating region, CAR, in the channel Orai1 enhances Ca2+ permeation and SOCE-induced gene transcription. Sci. Signal. 2015;8:ra131. doi: 10.1126/scisignal.aab1901. [DOI] [PMC free article] [PubMed] [Google Scholar]