Abstract
The use of non‐human animal models for infection experiments is important for investigating the infectious processes of human pathogenic bacteria at the molecular level. Mammals, such as mice and rabbits, are also utilized as animal infection models, but large numbers of animals are needed for these experiments, which is costly, and fraught with ethical issues. Various non‐mammalian animal infection models have been used to investigate the molecular mechanisms of various human pathogenic bacteria, including Staphylococcus aureus, Streptococcus pyogenes, and Pseudomonas aeruginosa. This review discusses the desirable characteristics of non‐mammalian infection models and describes recent non‐mammalian infection models that utilize Caenorhabditis elegans, silkworm, fruit fly, zebrafish, two‐spotted cricket, hornworm, and waxworm.
Keywords: infection model, non‐mammals, pathogenic bacteria
1. INTRODUCTION
Using mice as an animal infection model of anthrax, Koch first identified Bacillus anthracis as a human pathogen. Because B. anthracis naturally infects mammals such as cows and sheep as well as humans, it is reasonable to use a mouse infection model of B. anthracis. Koch determined three principles, referred to as Koch's postulates, for identifying infectious disease‐causing pathogens. One of these three principles is that a pure cultured microorganism causes the disease in healthy susceptible animals. Since Koch's study, several mammals that are phylogenetically close to humans have been used as healthy susceptible animals. In the dawning age of infection research around the late 19th century, the aims of animal infection experiments were to identify the causative pathogens and toxins, and evaluate antisera and vaccines. In the present day, genome science has revealed a vast number of biologic molecules that constitute pathogenic microorganisms and their host animals, prompting investigations of their functions in infectious processes. Furthermore, the biomolecules found to be involved in the infectious processes are new targets for anti‐infective drugs. Therefore, current infection research requires a large number of experiments.
Infection experiments using mammals are costly. For example, an inexpensive mouse strain costs approximately 10 US dollars per mouse. In addition, the use of mammals for research purposes is restricted internationally from an ethical point of view. 1 To perform infection experiments using mammals, research‐planning documents must adhere to international guidelines and be approved by the institutional ethics committee. The high cost and ethical issues regarding the use of mammals for research can mostly be avoided when non‐mammalian animals are used. Some review articles describe a specific non‐mammalian animal model for infection studies, but few review articles broadly describing non‐mammalian animal infection models are available to help researchers select the most appropriate non‐mammalian model for their specific experimental purposes. In this review, we summarize the advantages of non‐mammalian infection models and describe several non‐mammalian models recently used to study infection.
2. CRITERIA FOR ANIMAL INFECTION MODELS
Animals used for infection experiments should satisfy the following criteria. Researchers must select an animal species whose characteristics are most suitable for the experimental purpose.
2.1. Obtaining animals with homogeneous conditions
A large number of animals is needed for evaluating the virulence activity of a bacterial strain. Animals with different conditions, such as maturation, body weight, and health level, will exhibit different susceptibility to bacteria. Thus, ideally, researchers should obtain a large number of animals with homogeneous conditions. Animals can be raised in a laboratory or are commercially available. For insect models, however, researchers can easily obtain animals whose body weight is almost the same by selecting animals that underwent molting at about the same time.
2.2. Space for breeding and infection experiment
A large space for breeding and infection experiments requires a lot of effort to maintain. A small space is desirable to maintain animal health and to perform infection experiments. Autoclavable and disposable plastic containers are convenient for infection experiments.
2.2.1. Injection
For infection experiments, a specific amount of bacterial solution is injected into a specific part of the animal body. If the animal is large enough to use a medical syringe and needle, it is easy to conduct quantitative injection experiments. In addition, animals that can be controlled without anesthesia or a holding apparatus are easier to use for infection experiments.
2.3. Research on host factors
To investigate the host factors involved in infectious processes, it is important to perform a biochemical approach by purifying biologic molecules, as well as a genetic approach by creating genetically modified animals. In the past, genetic modification was performed only in model organisms such as Caenorhabditis elegans, Drosophila melanogaster, and mice. Recently, genome editing technology has enabled genetic modifications in many animal species. Small‐sized animals are generally not suitable for purifying biomolecules due to the small amount of biologic starting material. But when the target biomolecules are expressed in high amounts, small‐sized animals could be used for a biochemical approach.
3. NON‐MAMMALS USED AS INFECTION MODELS
The characteristics of non‐mammals used as infection models of human pathogenic bacteria are described below.
3.1. Caenorhabditis elegans
C. elegans is an important model organism in embryology in which all cell lineages can be pursued. In the late 1990s, Ausubel et al. began utilizing this animal as an animal infection model of Pseudomonas aeruginosa. 2 This is the first case in which a non‐mammal was used as an infection model of a human pathogen. C. elegans is normally fed Escherichia coli. When C. elegans is fed pathogenic bacteria such as P. aeruginosa, the C. elegans dies. There are two known killing mechanisms: toxin secreted from the bacteria kills the animal and digesting the bacteria kills the animal. 3 When bacterial infection kills C. elegans, it is difficult to estimate the number of infected bacteria in the C. elegans because the infection inhibits feeding behavior. Genetic manipulation methods are well established in C. elegans, enabling genetic analysis of host factors. Due to its small body size, it is easy to maintain the worms in a small space, but it is difficult to inject a bacterial solution into the worm (Table 1). C. elegans can be obtained from genetic stock centers or other researchers, and can be proliferated in a laboratory. C. elegans has been used in infection experiments with various human pathogenic bacteria such as S. aureus and Klebsiella pneumoniae (Table 2).
Table 1.
Characteristics of non‐mammalian infection models and mouse infection model
| Costa | Space | Injectionb | Research on host factor | ||
|---|---|---|---|---|---|
| Model animal | Genetic mutantc | Biological material | |||
| C. elegans | Low | Small | Difficult | Available | Small |
| Silkworm (B. mori) | Low | Small | Easy | Non‐available | Large |
| Fruit fly (D. melanogaster) | Low | Small | Difficult | Available | Small |
| Zebrafish (D. rerio) | Middle | Small | Normal/Difficultd | Available | Large |
| Two‐spotted cricket (G. bimaculatus) | Low | Small | Easy | Non‐available | Large |
| Hornworm (M. sexta) | Low | Small | Normal | Non‐available | Large |
| Waxworm (G. mellonella) | Low | Small | Normal | Non‐available | Large |
| Mouse (M. musculus) | High | Large | Normal | Available | Large |
Low, less than 1 US dollar/animal; Middle, 1–5 US dollars/animal; High, more than 5 US dollars/animal.
Easy, requires no anesthesia, holding apparatus, or microscope; Normal, requires anesthesia or holding apparatus, but no microscope; Difficult, requires glass capillary and microscope.
Available means that genetically modified animals can be obtained from a genetic stock center.
Adult fish, Normal; embryo, Difficult.
Table 2.
Bacterial species evaluated in non‐mammalian infection models
| Bacterial species | C. elegans | Silkworm | Fruit fly | Zebrafish | Two‐spotted cricket | Hornworm | Waxworm |
|---|---|---|---|---|---|---|---|
| Campylobacter jejuni | − | − | − | − | − | − | Yes 36 |
| Francisella tularensis | Yes 37 | Yes 38 | Yes 9 | Yes 39 | − | − | Yes 40 |
| Legionella pneumophila | Yes 41 | − | Yes 42 | − | − | − | Yes 43 |
| Acinetobacter baumannii | Yes 44 | − | − | Yes 45 | − | − | Yes 24 |
| Pseudomonas aeruginosa | Yes 2 | Yes 4 | Yes 46 | Yes 47 | Yes 12 | Yes 48 | Yes 49 |
| Klebsiella pneumoniae | Yes 50 | − | Yes 51 | Yes 52 | − | − | Yes 53 |
| Shigella sp. | Yes 54 | − | − | Yes 55 | − | − | Yes 56 |
| Yersinia pestis | Yes 57 | − | − | − | − | − | Yes 58 |
| Mycobacterium sp. | Yes 59 | Yes 60 | Yes 61 | Yes 11 | − | − | Yes 62 |
| Enterococcus faecalis | Yes 63 | − | Yes 64 | Yes 65 | − | Yes 66 | Yes 67 |
| Staphylococcus aureus | Yes 63 | Yes 4 | Yes 7 | Yes 68 | Yes 12 | Yes 15 | Yes 69 |
| Streptococcus pyogenes | Yes 63 | Yes 26 | − | Yes 10 | − | − | Yes 70 |
| Listeria monocytogenes | Yes 71 | Yes 72 | Yes 73 | Yes 74 | Yes 12 | − | Yes 75 |
| Bacillus cereus | Yes 76 | Yes 77 | Yes 78 | − | − | − | Yes 79 |
− indicates no report available.
3.2. Silkworm ( Bombyx mori )
The silkworm is the larva of Bombyx mori, a lepidopteran insect that has been utilized in the silk industry for more than 4000 years. In 2002, we found that various human pathogenic bacteria, including S. aureus, kill silkworms (Figure 1). 4 Because the silkworm has a large body size and is slow‐moving, it is easy to inject an appropriate amount of sample solution into the silkworm hemolymph using a 27‐gauge needle and syringe without anesthesia 5 (Table 1). This accurate injection technique enables the evaluation of bacterial virulence properties by determining the median lethal dose. 5 , 6 Skilled researchers can inject 150 silkworms in 1 hr. Silkworm eggs, larvae, and artificial diets are commercially available in various regions of the world, including Europe, the United States, and Japan. Because the silkworm is a domesticated animal that cannot proliferate in nature and does not escape from its cage, the silkworm infection model is suitable for experiments using biohazardous infectious agents. The silkworm has been used as an infection model of various human pathogenic bacteria such as P. aeruginosa and S. pyogenes (Table 2).
Figure 1.
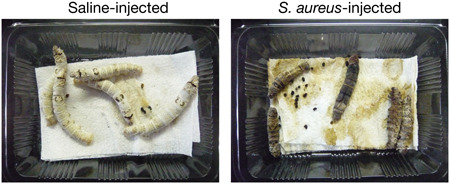
Silkworm Staphylococcus aureus infection model. Silkworms are injected into the hemolymph with S. aureus (107 CFU/larva) or saline. Images obtained 3 days after infection are shown. All the larvae injected with S. aureus died. [Color figure can be viewed at wileyonlinelibrary.com]
3.3. Fruit fly ( Drosophila melanogaster )
The fruit fly is used as a model organism for genetic analysis. The Toll‐like receptor, an important receptor in the innate immune system, was discovered in a fruit fly infection model with E. coli. The fruit fly has been used as an infection model for many human pathogenic bacteria, including S. aureus. 7 To perform infection experiments using the fruit fly, anesthesia is induced by carbon dioxide gas and a needle dipped in bacterial solution is used to cause injury under a microscope, or the fly can be injected with bacterial solution using a glass capillary under a microscope (Table 1). 8 , 9 Biohazard prevention requires considerable attention, because flies can escape by flying away. Fruit flies can be obtained from stock centers or other researchers, and can be proliferated in a laboratory.
3.4. Zebrafish ( Danio rerio )
The zebrafish is a well‐known aquarium fish and model organism. The embryo is transparent and thus suitable for observing organ development. Both adult fish and embryos are used for infection experiments (Table 1). To infect an adult fish, the fish is anesthetized with tricaine and injected with a bacterial solution using a syringe and a 29‐gauge needle. 10 To infect an embryo, the bacterial solution is injected using a glass capillary under a microscope. 11 The transparency of the embryo enables real‐time imaging of the infectious process by Mycobacterium 11 (Table 2). Zebrafish can be purchased from ornamental fish shops, or the fish strain used for genetic analysis can be obtained from genetic stock centers or other researchers. It should be noted that fish experiments now require approval from research ethics committees.
3.5. Two‐spotted crickets
Crickets have long been used as model animals of calling behavior and aggressive behavior. Two‐spotted crickets and house crickets are easy to purchase and maintain, because these crickets are captive‐bred as food for reptiles. The two‐spotted cricket is a tropical insect that can be kept at 37°C, human body temperature. We examined two‐spotted crickets as an animal infection model of human pathogens and found that human pathogenic bacteria and fungi kill two‐spotted crickets 12 , 13 (Figure 2). Furthermore, we revealed that the sensitivity of two‐spotted crickets to S. aureus and P. aeruginosa is not different between 27°C and 37°C, whereas the sensitivity of two‐spotted crickets to Listeria monocytogenes is greater at 37°C than at 27°C. 12 These findings suggest that two‐spotted crickets do not have increased sensitivity to all pathogens at a high temperature, but rather that specific pathogens exhibit increased virulence properties in high temperature conditions. Utilization of the two‐spotted cricket model allows for comparisons of the virulence of pathogenic bacteria at low and high temperatures, and identification of temperature‐dependent virulence mechanisms of bacteria. In infection experiments, two‐spotted crickets are injected with a bacterial solution using a syringe with a 30‐gauge needle without anesthesia 12 (Table 1).
Figure 2.

Infection model using the two‐spotted cricket. (a) A disposable plastic cage (diameter 130 mm × height 100 mm) used for infection experiments with two‐spotted crickets is shown, in which five crickets are grouped together and provided cricket food, water, and shelter. (b) Two‐spotted crickets were injected with Staphylococcus aureus (108 CFU/cricket) or saline. Images obtained 1 day after infection are shown. All the crickets injected with S. aureus died. [Color figure can be viewed at wileyonlinelibrary.com]
3.6. Hornworm ( Manduca sexta )
The hornworm is a larva of M. sexta, a lepidopteran insect that damages tobacco leaves, and has been used for research on hormones and neurons. Hornworms are captive‐bred as food for reptiles or fish, and fertilized eggs and artificial foods are commercially available (Table 1). The hornworm was first examined as an animal infection model of Bacillus cereus, a human pathogenic bacterium 14 and subsequently used for evaluating the virulence factors of other pathogens, such as S. aureus 15 and Streptococcus pneumoniae 16 (Table 2). After anesthetization by ice, hornworms are injected with bacterial solution using a Hamilton syringe with a 30‐gauge needle. 17
3.7. Waxworm ( Galleria mellonella )
The waxworm is a larva of G. mellonella, which feeds on honeybee nests. Waxworms are captive‐bred worldwide as food for amphibians, reptiles, and fish, and are easy to purchase and maintain (Table 1). For injection of waxworms, the worms are anesthetized on ice and injected using a syringe with a 30‐gauge needle, 18 , 19 or held in a special apparatus and injected. 20 Because waxworms can grow at 37°C, 21 infection experiments can be performed at 37°C. All human pathogens that have been examined in the waxworm infection model at 37°C, including S. aureus, 22 Cryptococcus neoformans, 23 and Acinetobacter baumannii, 24 exhibited increased killing activity against waxworms at 37°C compared with a low temperature, raising the possibility that the waxworm immune system is damaged at 37°C and evaluation of the temperature effects on the virulence properties of pathogens should be evaluated cautiously in this model. A number of human pathogenic bacteria have been examined in the waxworm infection model (Table 2).
4. COVERAGE OF NON‐MAMMALIAN INFECTION MODEL IN INFECTION RESEARCH
In infection experiments, animals must exhibit sensitivity against the pathogen. Whether a non‐mammal is susceptible to a human‐pathogenic bacterium depends on the conservation of the organ, tissue, cell, and cell signaling involved in the infection process between non‐mammals and mammals. The innate immune system constitutes the first defense mechanism against invading pathogens and is similar in many aspects between mammals and insects. For example, antimicrobial peptides that act as humoral antimicrobials, Toll‐like receptors that recognize pathogens by pattern recognition, and intracellular signaling pathways that are triggered by Toll‐like receptors are conserved between mammals and insects. 25 In addition, although insect appearance is distinct from that of mammals, insects have compartments that function as the blood, gut, liver, kidney, and heart. Therefore, interactions between the pathogen and host that depend on conserved biologic systems can be analyzed in insect infection models. Various human pathogenic bacteria are analyzed in many insect infection models (Table 2). We previously reported that S. aureus, P. aeruginosa, S. pyogenes, and enterohemorrhagic E. coli kill silkworms 4 , 26 (Table 2), identified the bacterial genes required for killing silkworms, and revealed that the identified genes contribute to virulence against mammals. 6 , 26 , 27 , 28 , 29 , 30 Some of the bacterial genetic mutants with attenuated killing activity against silkworms are sensitive to oxidative stress in macrophages, antimicrobial peptides, and complements. 30 , 31 Because bacterial resistance to the host innate immune system is essential for bacterial infection in both mammals and insects, the silkworm infection model can be used to evaluate the virulence properties of human pathogenic bacteria. In contrast, there are differences between mammals and silkworms in the host–pathogen interaction. For example, neurotoxins for mammals, including botulinum toxin and morphine, are less toxic to silkworms, although both alpha and beta hemolysin of S. aureus are toxic to silkworms. 32 , 33 This difference could be due to the presence or absence of toxin receptors in silkworms. When pathogens infect animals via mammalian‐specific routes, non‐mammalian infection models cannot be used to evaluate bacterial virulence. Recent progress in genome editing technologies may solve such problems of non‐mammalian infection models by constructing humanized non‐mammalian models in future.
Chronic infection is a unique aspect of some human pathogenic bacteria such as Mycobacterium tuberculosis. At present, most non‐mammalian infection models are used for acute infection by human pathogenic bacteria, in which bacteria proliferate in hemolymph or blood and kill animals within 10 days. Chronic infection experiments using non‐mammalian models are not well established and challenging. In the zebrafish model, chronic infection by Mycobacterium marinum has been studied as a model of M. tuberculosis infection in humans. M. marinum infection in adult zebrafish is prolonged for more than 8 weeks and forms granulomas, a characteristic of chronic tuberculosis infection. 34 , 35
5. CONCLUDING REMARKS
This review outlines non‐mammalian infection models used over the past two decades. In the absence of an appropriate established infection model among current infection models, a new animal infection model must be established to proceed with the infection research. Most of the non‐mammals mentioned in this review are utilized as foods for animals such as reptiles and fish. Food animals are easily maintained and bred, grow rapidly, have large bodies, and are inexpensive, which are desirable characteristics for animal infection models. Various non‐mammalian species that are used as food animals are good resources for establishing new animal infection models and will be powerful tools for infection research.
CONFLICT OF INTEREST
All authors declare there are no conflicts of interest.
ACKNOWLEDGMENTS
JSPS Grants‐in‐Aid for Scientific Research, Grant/Award Number: 19H03466, 19K22523, Takeda Science Foundation, and The Ichiro Kanehara Foundation.
Kaito C, Murakami K, Imai L, Furuta K. Animal infection models using non‐mammals. Microbiology and Immunology. 2020;64:585–592. 10.1111/1348-0421.12834
DATA AVAILABILITY STATEMENT
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
REFERENCES
- 1. Cheluvappa R, Scowen P, Eri R. Ethics of animal research in human disease remediation, its institutional teaching: and alternatives to animal experimentation. Pharmacol Res Perspect. 2017;5:e00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tan MW, Mahajan‐Miklos S, Ausubel FM. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci USA. 1999;96:715‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aballay A, Ausubel FM. Caenorhabditis elegans as a host for the study of host‐pathogen interactions. Curr Opin Microbiol. 2002;5:97‐101. [DOI] [PubMed] [Google Scholar]
- 4. Kaito C, Akimitsu N, Watanabe H, Sekimizu K. Silkworm larvae as an animal model of bacterial infection pathogenic to humans. Microb Pathog. 2002;32:183‐90. [DOI] [PubMed] [Google Scholar]
- 5. Kurokawa K, Kaito C, Sekimizu K. Two‐component signaling in the virulence of Staphylococcus aureus: a silkworm larvae‐pathogenic agent infection model of virulence. Methods Enzymol. 2007;422:233‐44. [DOI] [PubMed] [Google Scholar]
- 6. Miyazaki S, Matsumoto Y, Sekimizu K, Kaito C. Evaluation of Staphylococcus aureus virulence factors using a silkworm model. FEMS Microbiol Lett. 2012;326:116‐24. [DOI] [PubMed] [Google Scholar]
- 7. Needham AJ, Kibart M, Crossley H, Ingham PW, Foster SJ. Drosophila melanogaster as a model host for Staphylococcus aureus infection. Microbiology. 2004;150:2347‐55. [DOI] [PubMed] [Google Scholar]
- 8. Vodovar N, Acosta C, Lemaitre B, Boccard F. Drosophila: a polyvalent model to decipher host‐pathogen interactions. Trends Microbiol. 2004;12:235‐42. [DOI] [PubMed] [Google Scholar]
- 9. Vonkavaara M, Telepnev MV, Ryden P, Sjostedt A, Stoven S. Drosophila melanogaster as a model for elucidating the pathogenicity of Francisella tularensis . Cell Microbiol. 2008;10:1327‐38. [DOI] [PubMed] [Google Scholar]
- 10. Neely MN, Pfeifer JD, Caparon M. Streptococcus‐zebrafish model of bacterial pathogenesis. Infect Immun. 2002;70:3904‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Davis JM, Clay H, Lewis JL, Ghori N, Herbomel P, Ramakrishnan L. Real‐time visualization of mycobacterium‐macrophage interactions leading to initiation of granuloma formation in zebrafish embryos. Immunity. 2002;17:693‐702. [DOI] [PubMed] [Google Scholar]
- 12. Kochi Y, Miyashita A, Tsuchiya K, Mitsuyama M, Sekimizu K, Kaito C. A human pathogenic bacterial infection model using the two‐spotted cricket, Gryllus bimaculatus . FEMS Microbiol Lett. 2016;363:fnw163. [DOI] [PubMed] [Google Scholar]
- 13. Kochi Y, Matsumoto Y, Sekimizu K, Kaito C. Two‐spotted cricket as an animal infection model of human pathogenic fungi. Drug Discov Ther. 2017;11:259‐66. [DOI] [PubMed] [Google Scholar]
- 14. Harvie DR, Vilchez S, Steggles JR, Ellar DJ. Bacillus cereus Fur regulates iron metabolism and is required for full virulence. Microbiology. 2005;151:569‐77. [DOI] [PubMed] [Google Scholar]
- 15. Fleming V, Feil E, Sewell AK, Day N, Buckling A, Massey RC. Agr interference between clinical Staphylococcus aureus strains in an insect model of virulence. J Bacteriol. 2006;188:7686‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Roth A, Reichmann P, Hakenbeck R. The capsule of Streptococcus pneumoniae contributes to virulence in the insect model Manduca sexta . J Mol Microbiol Biotechnol. 2012;22:326‐34. [DOI] [PubMed] [Google Scholar]
- 17. Hussa E. Rearing and injection of Manduca sexta larvae to assess bacterial virulence. J Vis Exp. 2012;70:e4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bokhari H, Ali A, Noreen Z, Thomson N, Wren BW. Galleria mellonella is low cost and suitable surrogate host for studying virulence of human pathogenic Vibrio cholerae . Gene. 2017;628:1‐7. [DOI] [PubMed] [Google Scholar]
- 19. Rossoni RD, Ribeiro FC, Dos Santos HFS, et al. Galleria mellonella as an experimental model to study human oral pathogens. Arch Oral Biol. 2019;101:13‐22. [DOI] [PubMed] [Google Scholar]
- 20. Dalton JP, Uy B, Swift S, Wiles S. A novel restraint device for injection of Galleria mellonella larvae that minimizes the risk of accidental operator needle stick injury. Front Cell Infect Microbiol. 2017;7:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Warren LO, Huddleston P. Life history of the greater wax moth, Galleria mellonella L., in Arkansas. J Kans Entomol Soc. 1962;35:212‐16. [Google Scholar]
- 22. Desbois AP, Coote PJ. Wax moth larva (Galleria mellonella): an in vivo model for assessing the efficacy of antistaphylococcal agents. J Antimicrob Chemother. 2011;66:1785‐90. [DOI] [PubMed] [Google Scholar]
- 23. Mylonakis E, Moreno R, El Khoury JB, et al. Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect Immun. 2005;73:3842‐50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Peleg AY, Jara S, Monga D, Eliopoulos GM, Moellering RC Jr, Mylonakis E. Galleria mellonella as a model system to study Acinetobacter baumannii pathogenesis and therapeutics. Antimicrob Agents Chemother. 2009;53:2605‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sheehan G, Garvey A, Croke M, Kavanagh K. Innate humoral immune defences in mammals and insects: the same, with differences? Virulence. 2018;9:1625‐39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kaito C, Kurokawa K, Matsumoto Y, et al. Silkworm pathogenic bacteria infection model for identification of novel virulence genes. Mol Microbiol. 2005;56:934‐44. [DOI] [PubMed] [Google Scholar]
- 27. Matsumoto Y, Kaito C, Morishita D, Kurokawa K, Sekimizu K. Regulation of exoprotein gene expression by the Staphylococcus aureus cvfB gene. Infect Immun. 2007;75:1964‐72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kyuma T, Kimura S, Hanada Y, Suzuki T, Sekimizu K, Kaito C. Ribosomal RNA methyltransferases contribute to Staphylococcus aureus virulence. FEBS J. 2015;282:2570‐84. [DOI] [PubMed] [Google Scholar]
- 29. Kaito C, Yoshikai H, Wakamatsu A, et al. Non‐pathogenic Escherichia coli acquires virulence by mutating a growth‐essential LPS transporter. PLoS Pathog. 2020;16:e1008469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miyashita A, Iyoda S, Ishii K, Hamamoto H, Sekimizu K, Kaito C. Lipopolysaccharide O‐antigen of enterohemorrhagic Escherichia coli O157: H7 is required for killing both insects and mammals. FEMS Microbiol Lett. 2012;333:59‐68. [DOI] [PubMed] [Google Scholar]
- 31. Kyuma T, Kizaki H, Ryuno H, Sekimizu K, Kaito C. 16S rRNA methyltransferase KsgA contributes to oxidative stress resistance and virulence in Staphylococcus aureus . Biochimie. 2015;119:166‐74. [DOI] [PubMed] [Google Scholar]
- 32. Hamamoto H, Tonoike A, Narushima K, Horie R, Sekimizu K. Silkworm as a model animal to evaluate drug candidate toxicity and metabolism. Comp Biochem Physiol C Toxicol Pharmacol. 2009;149:334‐9. [DOI] [PubMed] [Google Scholar]
- 33. Hossain MS, Hamamoto H, Matsumoto Y, et al. Use of silkworm larvae to study pathogenic bacterial toxins. J Biochem. 2006;140:439‐44. [DOI] [PubMed] [Google Scholar]
- 34. Swaim LE, Connolly LE, Volkman HE, Humbert O, Born DE, Ramakrishnan L. Mycobacterium marinum infection of adult zebrafish causes caseating granulomatous tuberculosis and is moderated by adaptive immunity. Infect Immun. 2006;74:6108‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Van Der Sar AM, Spaink HP, Zakrzewska A, Bitter W, Meijer AH. Specificity of the zebrafish host transcriptome response to acute and chronic mycobacterial infection and the role of innate and adaptive immune components. Mol Immunol. 2009;46:2317‐32. [DOI] [PubMed] [Google Scholar]
- 36. Champion OL, Karlyshev AV, Senior NJ, et al. Insect infection model for Campylobacter jejuni reveals that O‐methyl phosphoramidate has insecticidal activity. J Infect Dis. 2010;201:776‐82. [DOI] [PubMed] [Google Scholar]
- 37. Jayamani E, Tharmalingam N, Rajamuthiah R, et al. Characterization of a Francisella tularensis‐Caenorhabditis elegans pathosystem for the evaluation of therapeutic compounds. Antimicrob Agents Chemother. 2017;61:e00310‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Suzuki J, Uda A, Watanabe K, Shimizu T, Watarai M. Symbiosis with Francisella tularensis provides resistance to pathogens in the silkworm. Sci Rep. 2016;6:31476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brudal E, Ulanova LS, E OL, Rishovd AL, Griffiths G, Winther‐Larsen HC. Establishment of three Francisella infections in zebrafish embryos at different temperatures. Infect Immun. 2014;82:2180‐94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Aperis G, Fuchs BB, Anderson CA, Warner JE, Calderwood SB, Mylonakis E. Galleria mellonella as a model host to study infection by the Francisella tularensis live vaccine strain. Microbes Infect. 2007;9:729‐34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Komura T, Yasui C, Miyamoto H, Nishikawa Y. Caenorhabditis elegans as an alternative model host for legionella pneumophila, and protective effects of Bifidobacterium infantis . Appl Environ Microbiol. 2010;76:4105‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kubori T, Shinzawa N, Kanuka H, Nagai H. Legionella metaeffector exploits host proteasome to temporally regulate cognate effector. PLoS Pathog. 2010;6:e1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Harding CR, Schroeder GN, Reynolds S, et al. Legionella pneumophila pathogenesis in the Galleria mellonella infection model. Infect Immun. 2012;80:2780‐90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Smith MG, Des Etages SG, Snyder M. Microbial synergy via an ethanol‐triggered pathway. Mol Cell Biol. 2004;24:3874‐84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bhuiyan MS, Ellett F, Murray GL, et al. Acinetobacter baumannii phenylacetic acid metabolism influences infection outcome through a direct effect on neutrophil chemotaxis. Proc Natl Acad Sci USA. 2016;113:9599‐604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. D'Argenio DA, Gallagher LA, Berg CA, Manoil C. Drosophila as a model host for Pseudomonas aeruginosa infection. J Bacteriol. 2001;183:1466‐71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Clatworthy AE, Lee JS, Leibman M, Kostun Z, Davidson AJ, Hung DT. Pseudomonas aeruginosa infection of zebrafish involves both host and pathogen determinants. Infect Immun. 2009;77:1293‐303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Del Campo ML, Halitschke R, Short SM, Lazzaro BP, Kessler A. Dietary plant phenolic improves survival of bacterial infection in Manduca sexta caterpillars. Entomol Exp Appl. 2013;146:321‐31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hendrickson EL, Plotnikova J, Mahajan‐Miklos S, Rahme LG, Ausubel FM. Differential roles of the Pseudomonas aeruginosa PA14 rpoN gene in pathogenicity in plants, nematodes, insects, and mice. J Bacteriol. 2001;183:7126‐34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fuursted K, Scholer L, Hansen F, et al. Virulence of a Klebsiella pneumoniae strain carrying the New Delhi metallo‐beta‐lactamase‐1 (NDM‐1). Microbes Infect. 2012;14:155‐8. [DOI] [PubMed] [Google Scholar]
- 51. Lee H, Baek JY, Kim SY, et al. Comparison of virulence between matt and mucoid colonies of Klebsiella pneumoniae coproducing NDM‐1 and OXA‐232 isolated from a single patient. J Microbiol. 2018;56:665‐72. [DOI] [PubMed] [Google Scholar]
- 52. Cheepurupalli L, Raman T, Rathore SS, Ramakrishnan J. Bioactive molecule from Streptomyces sp. mitigates MDR Klebsiella pneumoniae in Zebrafish infection model. Front Microbiol. 2017;8:614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Insua JL, Llobet E, Moranta D, et al. Modeling Klebsiella pneumoniae pathogenesis by infection of the wax moth Galleria mellonella . Infect Immun. 2013;81:3552‐65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Burton EA, Pendergast AM, Aballay A. The Caenorhabditis elegans ABL‐1 tyrosine kinase is required for Shigella flexneri pathogenesis. Appl Environ Microbiol. 2006;72:5043‐51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mostowy S, Boucontet L, Mazon Moya MJ, et al. The zebrafish as a new model for the in vivo study of Shigella flexneri interaction with phagocytes and bacterial autophagy. PLoS Pathog. 2013;9:e1003588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mahmoud RY, Li W, Eldomany RA, Emara M, Yu J. The Shigella ProU system is required for osmotic tolerance and virulence. Virulence. 2017;8:362‐74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Joshua GWP, Karlyshev AV, Smith MP, Isherwood KE, Titball RW, Wren BW. A Caenorhabditis elegans model of Yersinia infection: biofilm formation on a biotic surface. Microbiology. 2003;149:3221‐29. [DOI] [PubMed] [Google Scholar]
- 58. Erickson DL, Russell CW, Johnson KL, Hileman T, Stewart RM. PhoP and OxyR transcriptional regulators contribute to Yersinia pestis virulence and survival within Galleria mellonella . Microb Pathog. 2011;51:389‐95. [DOI] [PubMed] [Google Scholar]
- 59. Everman JL, Ziaie NR, Bechler J, Bermudez LE. Establishing Caenorhabditis elegans as a model for Mycobacterium avium subspecies hominissuis infection and intestinal colonization. Biol Open. 2015;4:1330‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yagi A, Uchida R, Hamamoto H, Sekimizu K, Kimura KI, Tomoda H. Anti‐Mycobacterium activity of microbial peptides in a silkworm infection model with Mycobacterium smegmatis . J Antibiot. 2017;70:685‐90. [DOI] [PubMed] [Google Scholar]
- 61. Dionne MS, Ghori N, Schneider DS. Drosophila melanogaster is a genetically tractable model host for Mycobacterium marinum . Infect Immun. 2003;71:3540‐50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rwegasila E, Mubofu EB, Nyandoro SS, Erasto P, Munissi JJ. Preparation, characterization and in vivo antimycobacterial studies of panchovillin‐chitosan nanocomposites. Int J Mol Sci. 2016;17:1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Garsin DA, Sifri CD, Mylonakis E, et al. A simple model host for identifying Gram‐positive virulence factors. Proc Natl Acad Sci USA. 2001;98:10892‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cox CR, Gilmore MS. Native microbial colonization of Drosophila melanogaster and its use as a model of Enterococcus faecalis pathogenesis. Infect Immun. 2007;75:1565‐76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Prajsnar TK, Renshaw SA, Ogryzko NV, Foster SJ, Serror P, Mesnage S. Zebrafish as a novel vertebrate model to dissect enterococcal pathogenesis. Infect Immun. 2013;81:4271‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mason KL, Stepien TA, Blum JE, et al. From commensal to pathogen: translocation of Enterococcus faecalis from the midgut to the hemocoel of Manduca sexta . mBio. 2011;2:e00065‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Park SY, Kim KM, Lee JH, Seo SJ, Lee IH. Extracellular gelatinase of Enterococcus faecalis destroys a defense system in insect hemolymph and human serum. Infect Immun. 2007;75:1861‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Prajsnar TK, Cunliffe VT, Foster SJ, Renshaw SA. A novel vertebrate model of Staphylococcus aureus infection reveals phagocyte‐dependent resistance of zebrafish to non‐host specialized pathogens. Cell Microbiol. 2008;10:2312‐25. [DOI] [PubMed] [Google Scholar]
- 69. Peleg AY, Monga D, Pillai S, Mylonakis E, Moellering RC Jr, Eliopoulos GM. Reduced susceptibility to vancomycin influences pathogenicity in Staphylococcus aureus infection. J Infect Dis. 2009;199:532‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Olsen RJ, Watkins ME, Cantu CC, Beres SB, Musser JM. Virulence of serotype M3 Group A Streptococcus strains in wax worms (Galleria mellonella larvae). Virulence. 2011;2:111‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Thomsen LE, Slutz SS, Tan MW, Ingmer H. Caenorhabditis elegans is a model host for Listeria monocytogenes . Appl Environ Microbiol. 2006;72:1700‐1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Castillo Y, Suzuki J, Watanabe K, Shimizu T, Watarai M. Effect of vitamin A on Listeria monocytogenes infection in a silkworm model. PLoS One. 2016;11:e0163747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mansfield BE, Dionne MS, Schneider DS, Freitag NE. Exploration of host‐pathogen interactions using Listeria monocytogenes and Drosophila melanogaster . Cell Microbiol. 2003;5:901‐11. [DOI] [PubMed] [Google Scholar]
- 74. Levraud JP, Disson O, Kissa K, et al. Real‐time observation of listeria monocytogenes‐phagocyte interactions in living zebrafish larvae. Infect Immun. 2009;77:3651‐60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mukherjee K, Altincicek B, Hain T, Domann E, Vilcinskas A, Chakraborty T. Galleria mellonella as a model system for studying Listeria pathogenesis. Appl Environ Microbiol. 2010;76:310‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Rae R, Iatsenko I, Witte H, Sommer RJ. A subset of naturally isolated Bacillus strains show extreme virulence to the free‐living nematodes Caenorhabditis elegans and Pristionchus pacificus . Environ Microbiol. 2010;12:3007‐21. [DOI] [PubMed] [Google Scholar]
- 77. Usui K, Miyazaki S, Kaito C, Sekimizu K. Purification of a soil bacteria exotoxin using silkworm toxicity to measure specific activity. Microb Pathog. 2009;46:59‐62. [DOI] [PubMed] [Google Scholar]
- 78. Ma J, Benson AK, Kachman SD, Hu Z, Harshman LG. Drosophila melanogaster selection for survival of Bacillus cereus infection: life history trait indirect responses. Int J Evol Biol. 2012;2012:935970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Salamitou S, Ramisse F, Brehelin M, et al. The plcR regulon is involved in the opportunistic properties of Bacillus thuringiensis and Bacillus cereus in mice and insects. Microbiology. 2000;146:2825‐32. 2000. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.


