Abstract
The nucleus, central to all cellular activity, relies on both direct mechanical input and its molecular transducers to sense and respond to external stimuli. While it has been shown that isolated nuclei can adapt to applied force ex vivo, the mechanisms governing nuclear mechanoadaptation in response to physiologic forces in vivo remain unclear. To investigate nuclear mechanoadaptation in cells, we developed an atomic force microscopy (AFM) based procedure to probe live nuclei isolated from mesenchymal stem cells (MSCs) following the application of low intensity vibration (LIV) to determine whether nuclear stiffness increases as a result of LIV. Results indicated that isolated nuclei were, on average, 30% softer than nuclei tested within intact MSCs prior to LIV. When the nucleus was isolated following LIV (0.7g, 90Hz, 20min) applied four times (4x) separated by 1h intervals, stiffness of isolated nuclei increased 75% compared to non-LIV controls. LIV-induced nuclear stiffening required functional Linker of Nucleoskeleton and Cytoskeleton (LINC) complex, but was not accompanied by increased levels of the nuclear envelope proteins LaminA/C or Sun-2. While depleting LaminA/C or Sun-1&2 resulted in either a 47% or 39% increased heterochromatin to nuclear area ratio in isolated nuclei, the heterochromatin to nuclear area ratio was decreased by 25% in LIV-treated nuclei compared to controls, indicating LIV-induced changes in the heterochromatin structure. Overall, our findings indicate that increased apparent cell stiffness in response to exogenous mechanical challenge of MSCs in the form of LIV is in part retained by increased nuclear stiffness and changes in heterochromatin structure.
Keywords: Mesenchymal Stem Cells, Nucleus, LINC complex, Low Intensity Vibration, Mechanical Signal
Introduction
Influence of the mechanical environment is perhaps most evident in resident stem cells which need to remodel their respective tissues in response to changing tissue mechanics (Rando and Ambrosio, 2018). For example, mesenchymal stem cells (MSCs) that replace and regenerate bone, when seeded into substrates with increasing stiffness tend to differentiate into bone lineage (Sun et al., 2018) through regulation of actin cytoskeleton dynamics and intra-nuclear Lamina (Buxboim et al., 2014; Swift et al., 2013). While cell structural changes and signaling events that take place within the focal adhesions and cytoskeletal compartments in response to environmental mechanical challenges are well studied (Lessey et al., 2012) the changes that occur inside the nucleus in response to physiological forces are less understood.
Dynamic forces applied to the exterior of the cell propagate through the cell via focal adhesions and cytoskeletal components, and reach the structural components at the outer and inner membranes of the nucleus, regulating nuclear structure (Martins et al., 2012). The nucleus is integrated within the cell structure through direct connections with cytoskeletal elements (Lombardi et al., 2011) through the LINC complexes (Linker of Nucleoskeleton and Cytoskeleton) composed of Nesprin and Sun subunits (Crisp et al., 2006). When forces are applied directly to the nucleus through LINC connections ex vivo, resulting in large strain deformations, nuclei stiffen via tyrosine phosphorylation of emerin (Guilluy et al., 2014) suggesting the nucleus is an active contributor to mechanotransduction. The nucleus also undergo structural re-organization in response to F-actin contractility by recruiting LINC complexes to apical stress fibers, leading to the accumulation of nuclear lamina element LaminA/C as well as changes in chromatin density under these stress fibers (Versaevel et al., 2014). At the level of DNA, forces applied at the cell membrane propagate through the cytoskeletal actin framework to cause deformations in chromatin (Tajik et al., 2016) and alter heterochromatin dynamics (Le et al., 2016). Nuclear stiffness is also in-part managed by the same toolset, Nesprins are anchored to the inner nuclear membrane through Sun-1&2 proteins that directly interact with structural elements such as nuclear pore complexes and LaminA/C (Hodzic et al., 2004; Padmakumar et al., 2005). Both Sun-1&2 and LaminA/C, play key roles in the structural integrity of the cell nucleus (Neelam et al., 2015). Inside the nucleus, both LaminA/C and chromatin have been shown to play independent roles in regulating nuclear mechanics (Stephens et al., 2017).
The elastic modulus of a cell nucleus can also be a marker of cell health. For example, it has been reported that the nuclei of Hepatitis C-infected cells are significantly softer than healthy controls, which was paralleled by downregulation of LaminA/C paired with upregulation of β-actin (Balakrishnan et al., 2019). Likewise, breast cancer cells exhibit large decreases in nuclear stiffness relative to healthy controls, showing downregulation of both LaminA/C and Sun-1&2 (Matsumoto et al., 2015). Furthermore, coinciding research showed heterochromatin decondensation as an additional component in highly metastatic cancer cells (Khan et al., 2018).
The nuclear envelope is subject to F-actin generated tension through LINC complex connections (Arsenovic et al., 2016). While cyto-mechanical forces can be generated in a multitude of ways, our group has focused on low intensity vibrations (LIV). LIV is a mechanical regime modeled after physiological, high frequency muscle contractions (Ozcivici et al., 2010). In MSCs, LIV promotes proliferation and osteogenic differentiation (Uzer et al., 2013) as well as increasing cell contractility by promoting GTP-bound RhoA (Ras homolog family member A)(Uzer et al., 2015). While we have further reported that daily LIV application up to 14 days increases stiffness of F-actin struts and results in increased mRNA expression of the LINC-related genes Nesprin-1&2, Sun-1&2, and LaminA/C in MSCs (Pongkitwitoon et al., 2016), the role of short term acute LIV application on the nuclear structural properties remains unknown.
Therefore, in this study we utilized AFM (atomic force microscopy) based nanoindentation measurements, immunostaining, and quantification of nuclear structural proteins to probe nuclear mechanical properties and morphology in response to acute bouts of LIV. We hypothesized that application of LIV to MSCs will increase nuclear stiffness.
Results
Cytoskeletal tension alters nuclear shape
We first investigated nuclear shape before and after our nuclear isolation protocol. Fig. 1a shows intact MSC (top) and isolated nuclei (bottom) after Hoechst 33342 staining to label DNA. As shown in Fig. 1a, the areas of isolated nuclei were visibly smaller than that of nuclei inside intact MSCs. As shown in Figures 1b & S1, DNA structure and LaminA/AC organization remained intact within the isolated nuclei. We next evaluated the relative circularity of intact MSC nuclei versus isolated nuclei by measuring their XY (top view), XZ (side view), and YZ (side view) areas and combining these three-dimensional measurements to evaluate overall sphericity (Fig. 1c). In the XY plane (i.e., the horizontal plane of the cell culture dish), intact and isolated nuclei showed no circularity differences. The isolated nuclei were 52% and 45% more circular than the nuclei of intact MSCs in the vertical XZ and YZ planes, respectively (p<.001). Combining the circularity values for each plane, the average circularity (i.e., sphericity) of the isolated nuclei was 0.809, which was significantly higher than that of the intact MSC nuclei (0.612, p<.001). Measures of nuclear height and volume (Figures 1d & 1e) showed that following nuclear isolation, height approximately doubled, increasing by 105% (p<.001) , while overall volume decreased by 44% (p<.001).
Figure 1. Nuclear geometry characteristics before and after nuclear isolation.
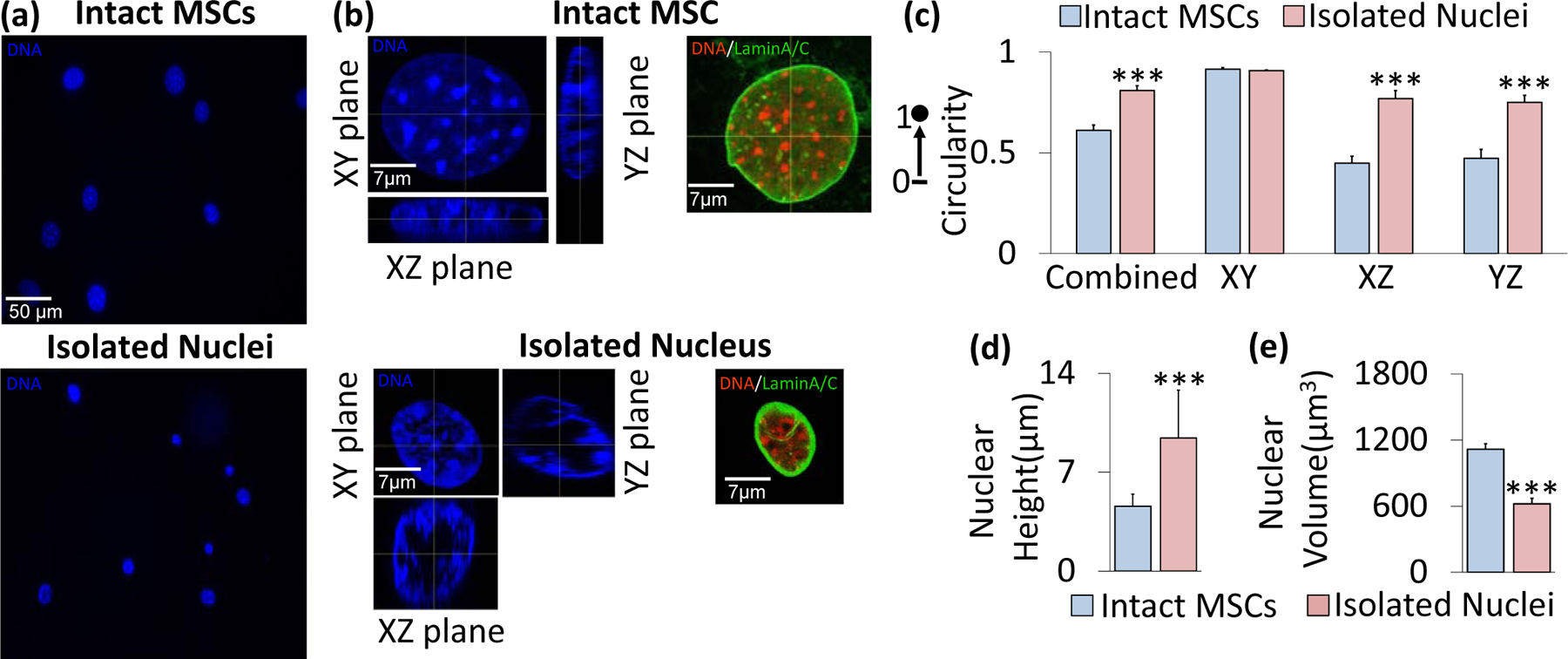
a) Mesenchymal stem cells (top) and isolated nuclei (bottom) were stained with Hoechst 33342 and subsequently imaged for DNA using a fluorescence microscope. b) Confocal imaging of an intact nucleus (top) and isolated nucleus (bottom) with Hoechst 33342 fluorescence staining under 63x focus with 16 Z-stacks for each nucleus image. Representative intact MSC (top) and isolated nucleus (bottom) shown in XY, XZ, and YZ planes of focus. Staining against LaminA/C also indicated an intact nuclear lamina post-isolation. c) Shape profiles of the intact MSC nuclei versus isolated nuclei. The isolated and intact nuclei exhibited similar circularity in the XY plane (i.e., the horizontal plane of the cell culture dish or microscope slide), but showed significant differences in shape profiles in the vertical XZ and YZ planes (p<.001, N=10 isolated nuclei, N=10 intact nuclei), with isolated nuclei showing 52% and 45% higher circularity values in those planes, respectively. The combined data, which is an average of all three planes, shows a significant difference in sphericity (p<.001) between the isolated nuclei and intact nuclei, with the two types having sphericity values of 0.809 and 0.612, respectively. d) Average isolated nuclear height (9.43 μm) is approximately twice that of intact MSC nuclei (4.59 μm, N=10/group, p<.001). e) Volume decreases from 1,116 μm3 to 621 μm3 following isolation (N=10/group, p<.001. * p<.05, ** p<.01, *** p<.001, against control.
Nucleus significantly contributes to AFM-measured MSC stiffness
To investigate the mechanical properties of the nucleus, AFM-based nanoindentation was used to measure and compare elastic moduli values of intact MSC nuclei and isolated cell nuclei (Fig.2a). Prior to testing, tipless AFM MLCT-D probes were functionalized with 10 μm glass beads (Thermo Scientific 9010, Figures 2b & S2a). Nuclei of intact MSCs and isolated nuclei were probed at the nuclear center visible in the XY plane (Fig.2a). To test whether fixation methods preserve nuclear stiffness, isolated nuclei were fixed in 2% paraformaldehyde (PFA) for 10 minutes and compared to untreated live controls. As shown in Fig.2c, the results indicated a 4-fold increase in stiffness between PFA-treated and control nuclei (p<.01).
Figure 2. Nucleus significantly contributes to AFM-measured MSC stiffness.
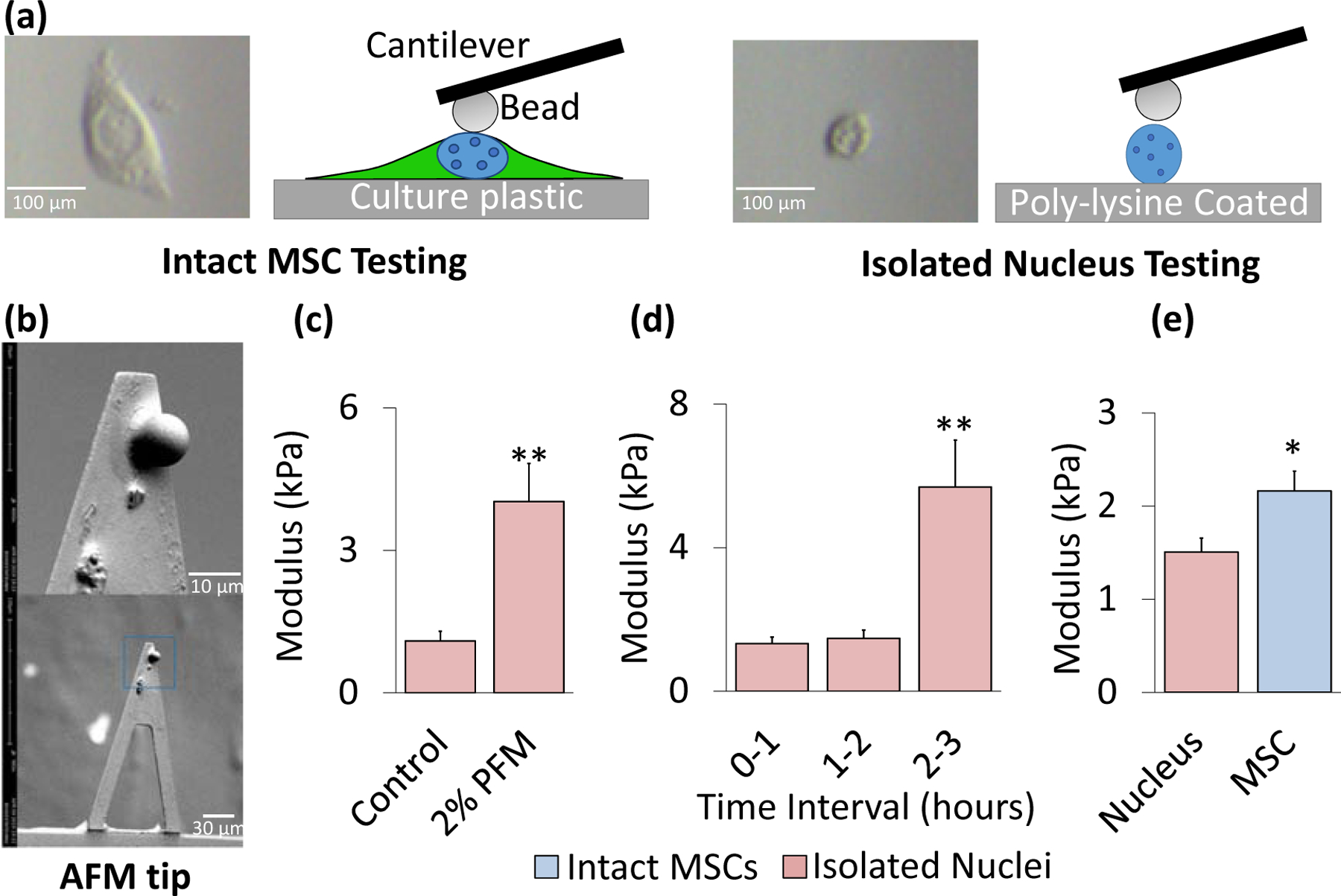
a) Nuclear isolation protocol involved plating of nuclei onto a 35 mm cell culture dish coated in poly-L-lysine. Isolated nuclei were incubated for 25 minutes at 37°C with 1 mL 1X PBS to ensure adhesion to the substrate immediately prior to AFM testing for up to 1 hour. Shown are both cartoons and actual optical images of intact MSCs and isolated nuclei as seen in the AFM camera. b) SEM image of a Bruker MLCT-D tipless AFM cantilever with a 10 μm diameter glass bead standard (Thermo Scientific 9010) attached. Scale bars were 30 μm (bottom image) and 10 μm (top image). c) Fixation of isolated nuclei in 2% paraformaldehyde (2% PFA) for 10 minutes showed an almost 4-fold increase in modulus compared to live controls (p<.01, N=10/grp). d) AFM measurements show no change in the elastic modulus of isolated nuclei over a 2-hour testing interval, after 2-hour window there was a 405% increase in the nuclear stiffness (p<.01). e) Isolated nuclei were identified via AFM and tested in the center of the nucleus to collect three individual measurements per sample. Intact MSCs and Isolated nuclei (both live) were plated on 35 mm dishes and tested for stiffness via AFM for one-hour intervals. The cell nucleus accounts for 69.7% of the total measured MSC stiffness with a modulus of 1.51 kPa (N=119). For comparison, the average stiffness of MSCs (passage 8–11) was 2.19 kPa (N=124). * p<.05, ** p<.01, *** p<.001, against control or against each other.
With fixation having been shown to significantly impact measured stiffness, we next determined the time window during which stiffness of live cell nuclei remains stable. Moduli of isolated live nuclei were measured across 3 one-hour time blocks, namely 0 to 1 hour, 1 to 2 hours, and 2 to 3 hours following isolation. Fig. 2d indicates that no change was observed in the mechanical properties of isolated nuclei up to two hours post-isolation, after which a large spike in stiffness occurred 2–3 hours following isolation (+406%, p<.01). Accordingly, a one hour post-isolation window for AFM-based nanoindentation testing was deemed safe and unlikely to skew the results. With this protocol established, we compared the stiffness of intact MSC nuclei (tested in the center of the nucleus) versus isolated nuclei within 1 hr of isolation. As shown in Fig. 2e, the average modulus of the isolated nuclei (1.5 kPa) was significantly softer than that obtained for the nuclei of intact MSCs (2.19 kPa, p<.05), indicating that the nucleus itself accounted for 69.7% of the overall MSC stiffness.
Disruption of LaminA/C decreases nuclear stiffness and increases heterochromatin area in isolated nuclei
We next probed the effects of depleting known structural members in the nuclear membrane: Nuclear Lamina element, LaminA/C, and LINC complex elements Sun-1 and Sun-2 (Fig. 3a). Following siRNA depletion of LaminA/C (siLmn), co-depletion of Sun-1&2 (siSun), or control siRNA (siCntrl), groups were divided into either intact MSCs or underwent the nuclear isolation protocol. The siLmn group showed a 63.2% and 49.7% modulus decrease in intact MSCs (p<.05) and isolated nuclei (p<.01), respectively (Fig. 3b). Likewise, siSun treatment resulted in 50.7% and 27.7% decreased modulus for intact MSCs and isolated nuclei, respectively, but changes remained not significant compared to siCntrl (Fig. 3b).
Figure 3. Disruption of LaminA/C decreases nuclear stiffness and changes structure.
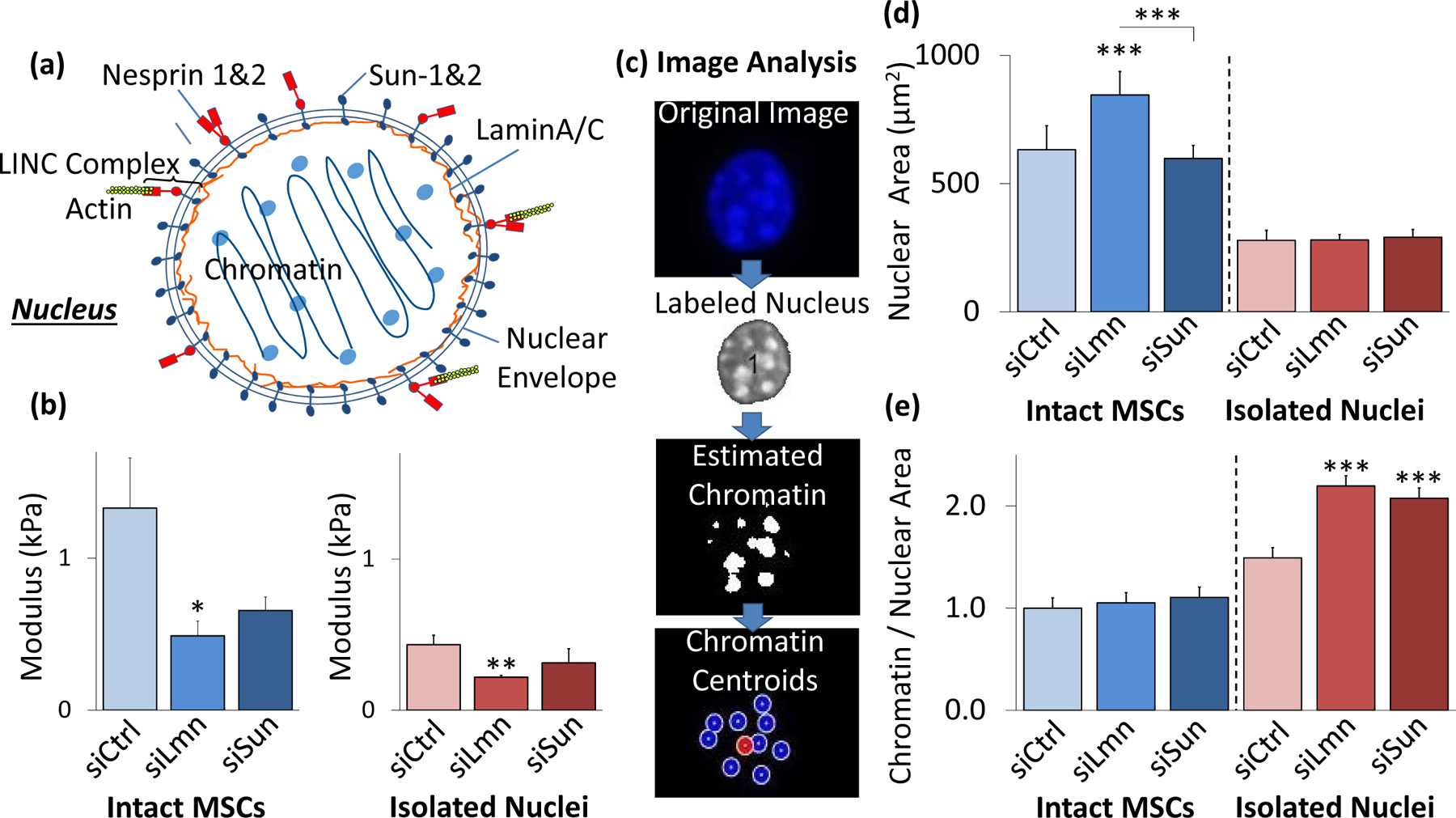
a) Schematic representation of the nucleus with nucleoskeletal and cytoskeletal connection components illustrated, including LaminA/C, Sun-1&2, Nesprin, and the KASH domain. b) SiRNA against nuclear LaminA/C (siLmn) and Sun-1&2 (siSun) decreased both isolated nuclear and intact MSC stiffness. Compared to siCtrl (N=21), siLmn and siSun treatments showed a significant decrease in the intact MSC modulus by 63.2% (p<.05, N=22) and 50.7% (p>.05, N=10), respectively. Likewise, when compared to siCtrl (N=21) the elastic modulus of isolated nuclei decreased by 49.7% (p<.01, N=21) and 27.7% (p>.05 N=10) in siLmn and siSun groups, respectively. c) A MATLAB code was constructed to evaluate differences in nuclear area and nucleoli size as determined by live Hoechst 33342 staining (see Supplementary Methods). d) Nuclear staining via Hoechst 33342 and epifluorescence imaging revealed that nuclear area increased by 33% within siLmn treated intact MSCs (p<.001, N=73), but not under Sun-1&2 depletion when compared to siCtrl (N=104). Nuclear area showed no significant changes within isolated nuclei control or experimental groups (N=245). e) Chromatin area to nuclear area ratios were calculated for all MSC and isolated nuclei for siCtrl, siLmn and siSun groups. Compared to controls (N=806), intact MSC did not show differences in chromatin to nuclear area ratio for LaminA/C (N=647) andSun-1&2 MSCs (N=489). In contrast, isolated nuclei showed significant increases in chromatin to nuclear area ratios for both LaminA/C (47.2%, p<.001, N=502) and Sun-1&2 (39.1%, p<.001, N=612) depleted nuclei compared to controls (N=416). * p<.05, ** p<.01, *** p<.001, against control or against each other.
Hoechst staining of DNA can be used to identify heterochromatin (Imai et al., 2017). We therefore quantified average heterochromatin size following LaminA/C or Sun-1&2 depletion using Hoechst 33342 and epifluorescence imaging (Fig. 3c). As shown in Fig. 3d, nuclear area increased 33% in siLmn MSCs (p<.001), but not in siSun MSCs. While overall nuclear area was smaller in isolated nuclei, no differences were observed in nuclear area between controls and isolated nuclei subjected to siRNA treatments (Fig. 3d). Combining the results presented in Fig. 3c and 3d, the ratio of heterochromatin area to nuclear area was calculated to investigate possible changes in the heterochromatin condensation, with the results normalized relative to the intact MSC siCtrl condition. As shown in Fig. 3e, measurement of the heterochromatin area to nuclear area ratio (referred to hereinafter as chromatin-to-nuclear area ratio) of intact MSCs showed no changes between the control and siRNA-treated groups. In contrast, a comparison of the chromatin-to-nuclear area ratio of isolated nuclei showed significantly larger ratios compared to siCntrl for both the siLmn group (47.2% increase, p<.001) and the siSun group (39.1% increase p<.001).
Low Intensity Vibration (LIV) stiffens MSCs and isolated nuclei
To evaluate nuclear mechanics following LIV, we subjected intact MSCs to either 2x or 4x low intensity vibration protocols. The LIV regimen is illustrated in Fig. 4a. As shown in Figures 4b and 4c, following 2x LIV, intact MSCs showed a 71% increase in stiffness (p<.05), but there was no significant change in nuclear stiffness for isolated nuclei that underwent the same vibration protocol. In contrast, application of 4x LIV significantly increased the elastic modulus of both intact MSCs (419% increase, p<.001) and isolated nuclei (75%, p<.05).
Figure 4. LIV stiffens intact MSCs and isolated nuclei.
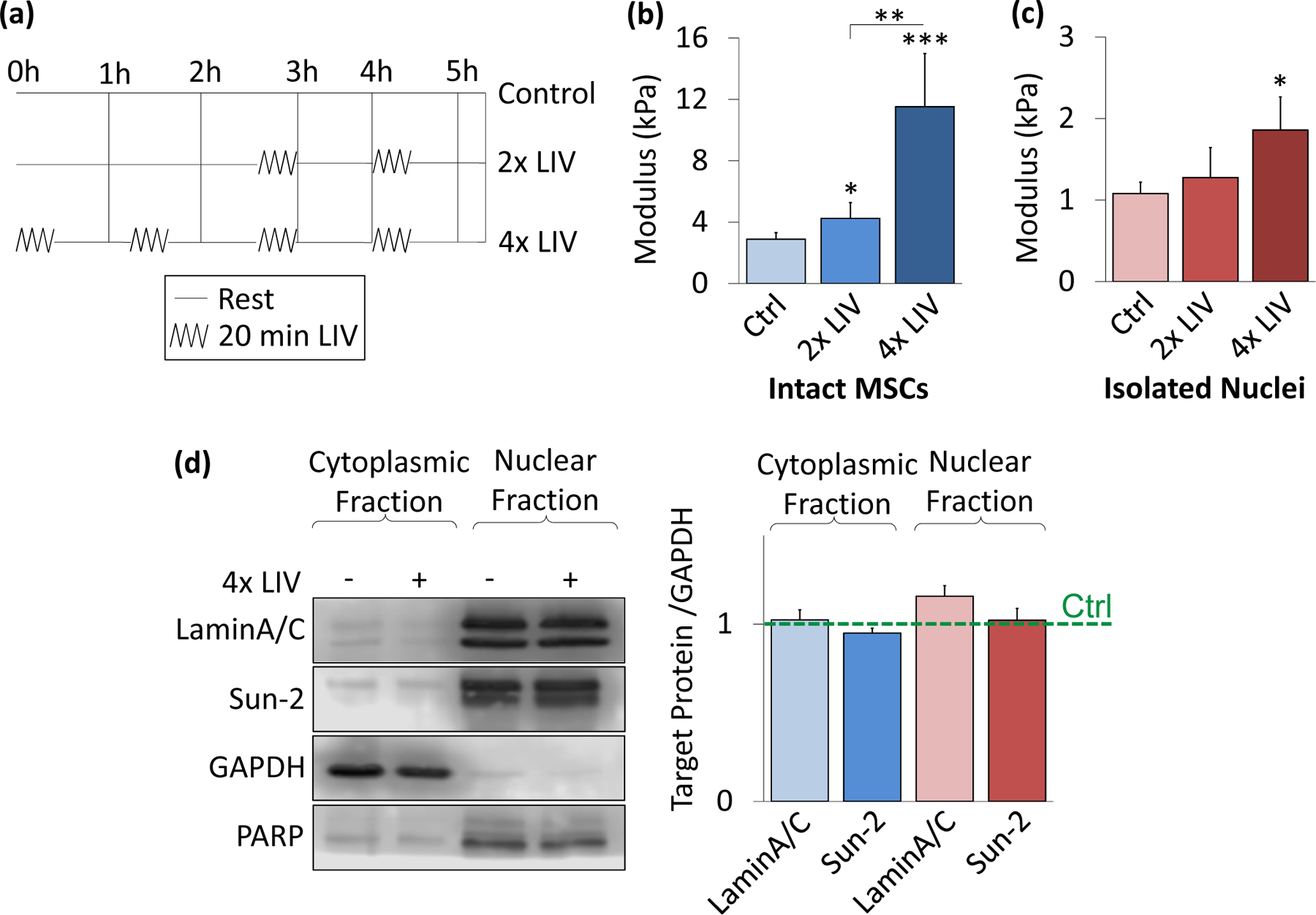
a) Low intensity vibration (0.7g, 90 Hz) was applied to MSCs at twenty-minute intervals with one-hour rest in between each vibration bout. 2x vibration included two, twenty-minute vibration periods, while 4x included four periods. b) In intact MSCs, 2x LIV increased AFM-measured elastic modulus on MSCs by 71% (N=31, p<.05). 4x LIV resulted in 419% increase in elastic modulus (N=33, p<.001) compared to control (N=32). c) Nuclear response to LIV was measured by applying the 2x LIV protocol to intact MSCs and then isolating nuclei to test stiffness. 2x LIV showed no significant increase in stiffness in comparison to control (N=20). Nuclei responded to 4x LIV by showing a 75% increase in modulus (N=24, p<0.05) when compared to control nuclei (N=27) post-LIV isolation. d) Western blotting for LaminA/C and Sun-2 show no changes in protein levels for 4x LIV groups compared to their respective controls (N=3/grp). * p<.05, ** p<.01, *** p<.001, against control or against each other.
To investigate the source of the observed changes in mechanical properties following LIV, the cytoplasmic and nuclear compartments were probed via western blotting. LaminA/C and Sun-2 protein levels following the 4x LIV protocol was quantified to test whether LIV results in upregulation of either protein. These protein levels were then compared to PARP and GAPDH as control markers. Fig. 4d shows no significant differences were observed in LaminA/C or Sun-2 protein levels, either in the cytoplasmic or nuclear fractions, following a 4x LIV protocol.
Isolated nuclei maintain a smaller chromatin-to-nuclear area ratio after vibration
As LIV-induced stiffness increase was not accompanied by changes in LaminA/C or Sun-2 proteins, we probed possible changes in heterochromatin by comparing the chromatin-to-nuclear area ratio between 4x LIV and control samples for intact and isolated nuclei (Fig 5a). As shown in Fig.5b, intact MSCs subjected to 4x LIV showed no difference in nuclear area compared to controls. Likewise, there was no difference in nuclear area between isolated nuclei subjected to 4x LIV and the corresponding controls. As shown in Fig.5c, intact MSCs subject to 4x LIV showed no difference in chromatin-to-nuclear area ratio compared to controls. In contrast to LaminA/C and Sun-1&2 depleted nuclei (Fig.3e) which showed higher chromatin-to-nuclear area ratio compared to control siRNA, isolated nuclei subjected to 4x LIV, showed a 25.4% decrease in chromatin-to-nuclear area ratio compared to non-LIV controls (p<.05).
Figure 5. Isolated nuclei maintain a smaller chromatin-to-nuclear area ratio after vibration.
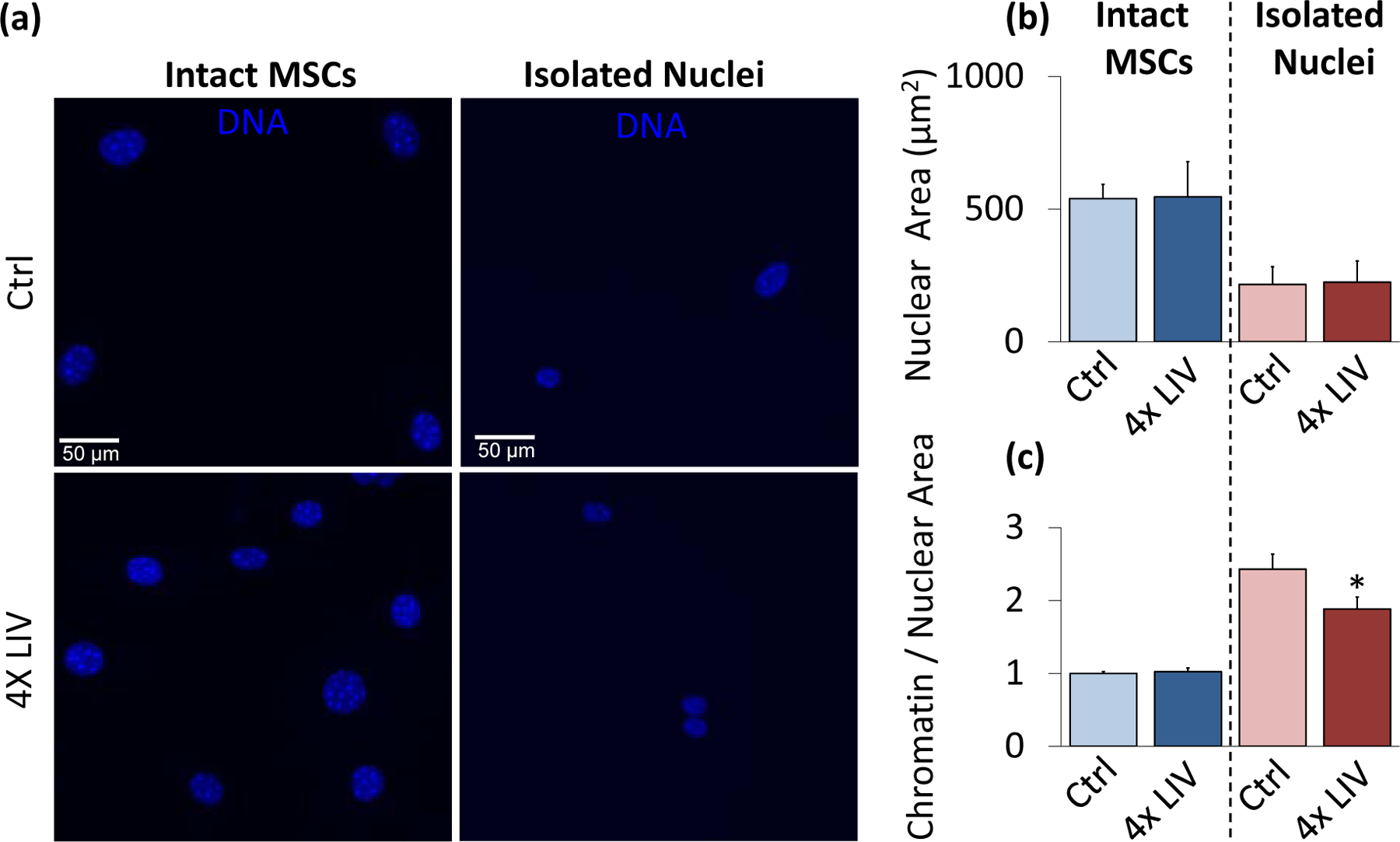
a) Epi-fluorescence images of intact control MSC nuclei (upper left), intact 4x LIV MSC nuclei (lower left), isolated control nuclei (upper right), and isolated 4x LIV nuclei (lower right). Cells were treated with Hoechst 33342 to stain heterochromatin, fixed in 2% paraformaldehyde, and kept in PBS for imaging. b) Nuclear area was measured using MATLAB code to identify nuclear bounds under Hoechst 33342 staining. Intact MSCs subject to 4x LIV (N=216) showed no difference in nuclear area compared to controls (N=214). Likewise, there was no difference in nuclear area between control (N=90) and 4x LIV isolated nuclei (N=102). c) Chromatin measurements were analyzed via MATLAB analysis. Individual chromatin were averaged and compared to respective nuclear area. Intact MSCs subject to 4x LIV (N=2136) showed no difference in chromatin to nuclear area ratio compared to controls (N=2082). Isolated nuclei subject to 4x LIV showed a 25.4% lower chromatin to nuclear area ratio (N=480) than isolated controls (p<.05, N=360). * p<.05, ** p<.01, *** p<.001, against control.
LINC function is required for LIV-induced nuclear stiffening
To test the requirement of cytoskeletal connections to the nucleus in the LIV response, we used LINC-compromised MSCs (referred to as +KASH). As depicted in Fig.6a, using previously established methods (Uzer et al., 2015), LINC complex function was disrupted by overexpressing a dominant negative form of the Nesprin KASH domain (pCDH-EF1-MSC1-puro-mCherry-Nesprin-1αKASH), which serves to disrupt the connection between Nesprin and Sun proteins via competitive binding to Sun proteins. Compared to empty control plasmid (pCDH-EF1-MSC1-puro-mCherry, referred to as mCherry), LINC-disrupted intact cells and isolated nuclei did not show any changes in stiffness (Fig.S3). As shown in Figures 6b and 6c, comparing intact MSC and isolated nuclear stiffness with or without 4x LIV showed no significant changes in elastic modulus between non-vibrated and vibrated MSCs with +KASH insertion. Further, neither mCherry nor KASH-expressing MSCs showed significant differences between intact MSC and isolated nuclei stiffness.
Figure 6. LINC function is required for LIV-induced nuclear stiffening.
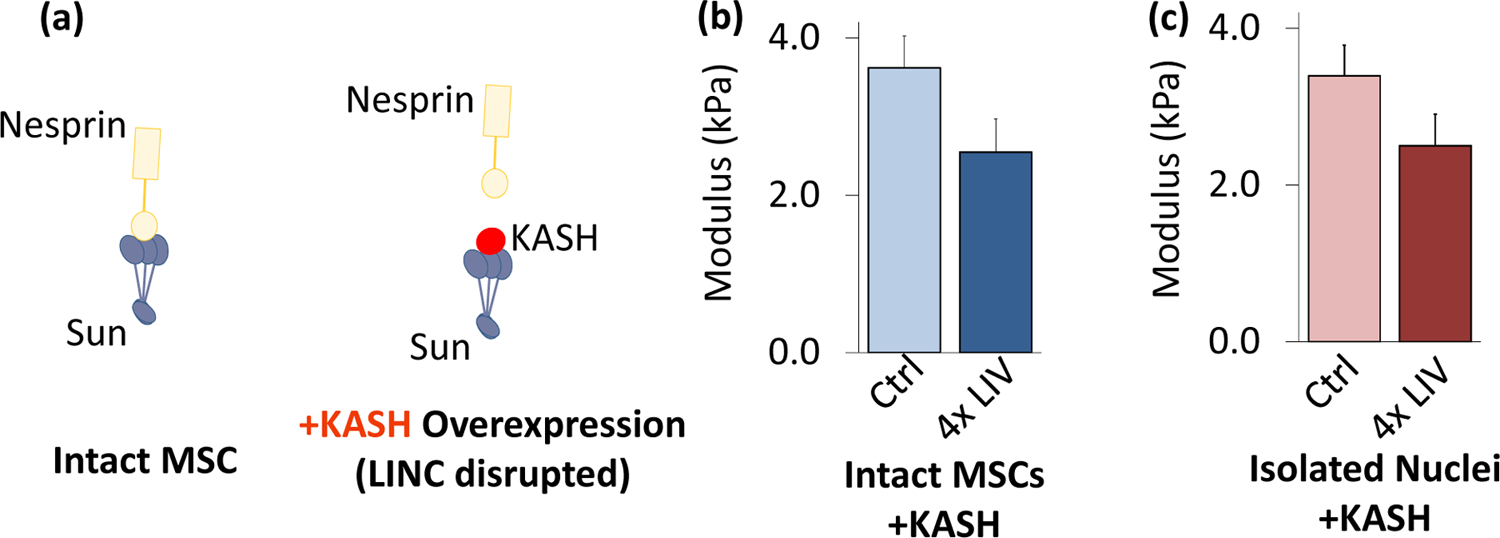
a) Overexpressing dominant negative form of Nesprin KASH domain was used to disrupt LINC function in MSCs. b) Intact MSCs and c) isolated nuclei did not show any response to 4x LIV (N=20/grp). * p<.05, ** p<.01, *** p<.001, against control.
Discussion
Here we investigated the effects of a low magnitude, low intensity vibration (LIV) protocol on mesenchymal stem cell nuclei mechanical properties. Our results show that nucleus retains about 70% of its mechanical properties when isolated. Reduced nuclear stiffness was also reported in fibroblastic cells (Liu et al., 2014). Further, the nucleus responds to low intensity vibration by increasing its apparent stiffness, not due to an increase in LaminA/C or Sun-2 protein amounts, but instead perhaps through changes in heterochromatin organization. When cyto-mechanical forces that keep nucleus elongated was removed during nuclear isolation, isolated nuclei changed shape from 60% to 80% sphericity (Fig. 1c). We also observed a reduction in nuclear volume post isolation. While cytoskeletal force may, in part, account for the observed volume changes, nuclear volume depends on other factors including changes in pH, salt conditions, and temperature (Chan et al., 2017). Additionally, loss of protein structures during nuclear isolation has been reported (Paine et al., 1983). Although a potential limitation exist in the isolation technique, we confirmed changes in isolated nuclei stiffness via AFM-based measurements of elastic modulus (Fig. 4c).
When MSCs treated with siRNA protocols, both intact MSC and isolated nuclei modulus were consistently lower than non-siRNA counterparts (Fig. 3b), similar batch effects are also seen in KASH inserted MSCs (Fig.6). Increased stiffness of KASH inserted MSCs is probably due to extended culturing period during puromycin mediated selection process (see supplementary methods) as later passage MSCs were stiffer (Fig. S4). Our results in Fig. 6 show that there was no significant difference between intact MSCs and isolated nuclei stiffness. While we do not know the exact reason why these findings do not agree with the data presented in Fig 2, we can make two observations. First, nuclei were softer at early passages when compared to intact cells in three separate data sets (Figs. 2e, 3b, and 4b–c); when plasmid expressions (i.e., later passages) were used, this difference between intact MSC and isolated nuclei stiffness was less and non-significant (Figs. 6b–c and S3). Second, overexpressed KASH fragments localize to the nuclear envelope (Uzer et al., 2015), and our data shown in Fig. S3 indicates that overexpressing KASH or empty mCherry plasmid does not make a difference in either intact cell or isolated nuclei stiffness. Therefore, the lack of stiffness difference between intact cells and isolated nuclei cannot simply be explained by the presence of the KASH fragment. Accordingly, while the observations in each figure were made across groups at the same cell passage to ensure self-consistency, due to the observed global shift in stiffness in later passages (Fig. S4), we cannot rule out the possibility that the lack of stiffness difference between intact cells and isolated nuclei as well as the lack of LIV-induced stiffening in KASH-expressing MSCs were simply due to increased cell passage. While this global shift in stiffness under siRNA treatment and plasmid overexpression remains as a limitation, in agreement with literature, depletion of LaminA/C showed significantly lower modulus compared to control siRNA (Lammerding et al., 2006; Schäpe et al., 2009). Similar to Lamin depletion studies in mouse embryonic fibroblasts (Kim et al., 2017), softer nuclei, due to LaminA/C depletion, also resulted in increased nuclear area in MSCs (Fig.3d). This softening and concomitant increased nuclear area points to a weakening of nuclear structure. As expected, when Sun-1&2 were co-depleted, we did not observe any changes in the nuclear area of intact MSCs (Fig.3d), owing to an absence of cytoskeletal connections (Uzer et al., 2018). Following nuclear isolation (i.e. in the absence of cytoskeletal connections), both LaminA/C and Sun-1&2 depleted nuclei showed large increases in heterochromatin area. This observation suggests that in the absence of cytoskeletal constraints, nuclear Lamina and Sun proteins can have secondary effects on heterochromatin structure.
Acute application of LIV caused a significant increase in stiffness for both intact MSC nuclei and isolated nuclei (Fig.4b), suggesting that the changes in cellular stiffness and cytoskeletal tension (Pagnotti et al., 2019) in response to LIV were in-part retained by nucleus as increased stiffness even after nuclear isolation protocol. Though the stiffness was increased, there were no short-term changes in the levels LaminA/C and Sun-2 (Fig.4c). As the LIV-induced stiffening of isolated nuclei were lower than that of intact MSCs, our findings suggests other mechanisms responsible for MSC stiffening in response to LIV, such as the cytoskeletal connections to the nucleus. Additionally, disrupting the LINC complex connections and applying the same 4x LIV protocol showed no changes in elastic modulus. This further suggests that cytoskeletal components contribute to the overall stiffening response of both intact MSCs and isolated nuclei.
Image analysis indicated a retention of chromatin-to-nuclear area ratio in LIV treated MSC nuclei post nuclear isolation (Fig. 5c). When cytoskeletal constraints were removed during nuclear isolation, chromatin-to-nuclear area ratio was increased in both control and LIV treated MSC nuclei, although nuclei subjected to 4x LIV showed significantly smaller, or more condensed heterochromatin compared to controls. These findings show that the effects of LIV were maintained by heterochromatin after the nucleus was mechanically separated from the cytoskeleton. While the mechanism by which this happens is again outside the scope of this current work, differences in DNA organization between isolated nuclei subject to LIV vs non-LIV counterparts may contribute to the increased stiffening of the nucleus (Stephens et al., 2018), suggesting that DNA organization plays a role in LIV-induced mechanoadaptation. Consequently LIV-induced decrease in the chromatin-to-nuclear area ratio post-isolation can potentially be a marker of altered gene access in response to environmental factors (Miroshnikova et al., 2017; Rubin et al., 2018).
Until now, it was previously unknown whether the nucleus responds to low intensity vibration in living cells. Our findings show that the nucleus is a mechanoresponsive element and responds to low intensity vibration by increasing its stiffness as confirmed by elastic modulus measurements. The mechanism(s) responsible for this stiffness regulation are not fully known, but our findings suggest that heterochromatin structure and cytoskeletal connections likely play a role in nuclear stiffening in response to LIV. Thus future studies aimed at understanding how LIV and other forms of mechanical signals regulate the mechanics of nucleoskeletal and heterochromatin structures may uncover new mechanisms by which forces regulate gene expression and cellular decisions.
Materials and Methods
Low Intensity Vibration (LIV) Protocol
Vibrations were applied with a peak magnitude of 0.7g, where g was defined as the acceleration due to earth’s gravitational field (9.81 m/s2). These dynamic accelerations were induced in the vertical direction via a sine wave oscillating at 90 Hz. Application of this sine wave excitation to the entire plate results in three types of forces: dynamic acceleration, vibration-induced fluid shear, and substrate deformation. In vivo, application of LIV results in bone deformations of 5–10 με, fluid shear stress of 0.5–1 Pa at the bone/bone marrow interface, and peak acceleration of 0.7 g (Chan et al., 2013). In vitro, we have shown that LIV response generates <10 με substrate deformation (Sen et al., 2011). Further, by modulating cell culture viscosity, we have reported that the cell response to LIV is independent of fluid shear as two conditions that generate 2 Pa and 0.015 Pa fluid shear, respectively, generated the same amount of osteogenic differentiation in MSCs (Uzer et al., 2013). In the current situation, where LIV was applied in the vertical direction, we have estimated fluid shear to be as small as 0.0008 Pa (Uzer et al., 2015). In light of the extremely small substrate strains and fluid shear, we report acceleration magnitude and frequency in the vertical direction as the primary mechanical signal. Controls were handled the same way, but the LIV device was not turned on. LIV was applied as either, two (2x) or four (4x), twenty-minute vibration periods with one-hour rest in-between.
Atomic Force Microscopy (AFM)
Force-displacement curves for both intact MSC nuclei and isolated nuclei were acquired using a Bruker Dimension FastScan Bio AFM. Tipless MLCT-D probes (0.03 N/m spring constant) were functionalized with 10 μm diameter borosilicate glass beads (Thermo Scientific 9010, 10.0 ± 1.0 μm NIST-traceable 9000 Series Glass Particle Standards) prior to AFM experiments using UV-curable Norland Optical Adhesive 61 (Fig. S2a). To ensure accurate force measurements, each probe was individually calibrated. Accordingly, a thermal tune was conducted on each probe immediately prior to use to determine its spring constant and deflection sensitivity (Fig. S2b). MSCs and nuclei were located using the AFM’s optical microscope and engaged on using a minimal force setpoint (2–3 nN) to ensure contact while minimizing applied force and resultant deformation prior to testing. Ramps were performed over the approximate center of each nucleus for all samples. After engaging on a selected nucleus to ensure probe/nucleus contact as described above, force curve ramping was performed at a rate of 2 μm/sec over 2 μm total travel (1 μm approach, 1 μm retract; Fig. S2c). Three replicate force-displacement curves were acquired and saved for each nucleus tested, with at least 3 seconds of rest between conducting each test. Measurements that showed less than 600 nm contact with the nucleus were discarded.
Measured force-displacement curves were analyzed assuming Hertzian (spherical contact) mechanics (Guo et al., 2012), employing Bruker’s Nanoscope Analysis software package v1.8 to obtain the elastic modulus of the samples. Accordingly, MSC and nuclear stiffness were quantified in Nanoscope Analysis using a best-fit curve to a Hertzian model, with the point of initial contact was visually selected. While visual selection of the initial contact point can potentially lead to variability in the resultant calculated elastic modulus, averaging of three consecutive measurements was employed to minimize introduction of error due to selection uncertainties. The fitted curves were analyzed until the R2 value was greater than 0.95 (p<0.05). To examine the effect of Poisson’s ratio (which can vary and is not precisely known for MSC nuclei) on the calculated modulus, values of both 0.3 (a common default for unknown materials that assumes some degree of compressibility) and 0.5 (corresponding to purely elastic deformation of an incompressible isotropic solid) were tested for the sample Poisson’s ratio. We found that the group differences were unaltered by the choice of Poisson’s ratio (data not shown). Therefore, due to limitations of the Hertzian contact model and the inability to measure experimental deformations, we did not employ a variable Poisson’s ratio, instead utilizing a value of 0.5 throughout the study as a best estimate for the effective Poisson’s ratio of the cells. While these simplifications may not be completely realistic, we additionally kept our deformation small (i.e., <1 μm) in order to avoid non-linear effects and changes in Poisson’s ratio. The modulus of each individual sample (i.e., intact MSC or isolated nucleus) was reported as the average of three consecutive force curve measurements. Group averages were then obtained from the average values for the individual samples.
Statistical Analysis
Unless indicated otherwise in figure legends, results are expressed as mean ± standard error of the mean throughout. All experiments were replicated at least three times to ensure reproducibility. Densitometry and other analyses were performed on at least three independent experiments. As we previously reported (Uzer et. al 2018, Touchstone et al. 2019), for western blot data, differences between treatments within each biological replicate were assumed to follow a normal distribution due to large mean sample; thus for these comparisons, we have used two-tailed un-paired T-tests (Figure 4d). For other comparisons, we used a non-parametric two-tailed Mann-Whitney U-test (Figures 1, 2c, 2e, 5, 6, S3 and S4) or Kruskal-Wallis test followed by Tukey multiple comparison (Figures 2d, 3 and 4b). P-values of less than 0.05 were considered significant.
Supplementary Material
Figure S1. Nuclei were either left intact inside MSCs or isolated (isolation protocol, methods), stained with Hoechst 33342, and fixed in 2% paraformaldehyde. As shown in these videos, a Leica 6500 confocal microscope was then used to acquire vertical stacked images of both nuclei inside intact MSC samples (left) and isolated nuclei (right).
Fig. S2 a) SEM image of a Bruker MLCT-D AFM probe functionalized with a 10 µm diameter glassbead (Thermo Scientific 9010 NIST-traceable 9000 Series Glass Particle Standard, 10.0 ± 1.0 µm). b) Representative thermal tune of an MLCT-D probe functionalized with such a 10 µm glass bead. Nominal resonance frequency of the MLCT-D cantilever prior to bead functionalization is 10–20 kHz, with the added weight of the bead causing the resonance to shift to lower frequency. A simple harmonic oscillator fit was used (red) to model the resonant response of the cantilever in a fluid environment. c) Representative ramp data. Ramping to a maximum excursion/extension of 1 µm was performed at a rate of 1 Hz (i.e., 2 µm/sec load/unload velocity).
Fig S3. Compared to empty control plasmid (pCDH-EF1-MSC1-puro-mCherry), LINC-disrupted intact cells (N=10/grp) and isolated nuclei (N=20/grp) did not show any changes in stiffness. * p<.05, ** p<.01, *** p<.001, against control.
Figure S4. MSCs measured via AFM-based nanoindentation showed 132% higher elastic modulus in passage 15 samples (P15, N=10, p<.001) compared to samples between passages 8 and 11 (P8–11, N=45). * p<.05, ** p<.01, *** p<.001, against control or against each other.
Figure S5. Unprocessed blots as obtained by LiCor C-DiGit blot scanner. Red lines outline the representative blots used.
Acknowledgments
Funding support: This study was supported by NIH R01AG059923, 5P2CHD086843–03, P20GM109095, and P20GM103408 and NSF 1929188.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The author(s) declare no competing financial interests
Conflict of Interest
The author(s) declare no conflict of interest, financial or otherwise.
References
- Arsenovic PT, Ramachandran I, Bathula K, Zhu R, Narang JD, Noll NA, Lemmon CA, Gundersen GG, Conway DE, 2016. Nesprin-2G, a Component of the Nuclear LINC Complex, Is Subject to Myosin-Dependent Tension. Biophysical journal 110, 34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan S, Mathad SS, Sharma G, Raju SR, Reddy UB, Das S, Ananthasuresh GK, 2019. A Nondimensional Model Reveals Alterations in Nuclear Mechanics upon Hepatitis C Virus Replication. Biophysical journal 116, 1328–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxboim A, Swift J, Irianto J, Spinler KR, Dingal PC, Athirasala A, Kao YR, Cho S, Harada T, Shin JW, Discher DE, 2014. Matrix elasticity regulates lamin-A,C phosphorylation and turnover with feedback to actomyosin. Current biology : CB 24, 1909–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case N, Xie Z, Sen B, Styner M, Zou M, O’Conor C, Horowitz M, Rubin J, 2010. Mechanical activation of β-catenin regulates phenotype in adult murine marrow-derived mesenchymal stem cells. Journal of Orthopaedic Research 28, 1531–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CJ, Li W, Cojoc G, Guck J, 2017. Volume Transitions of Isolated Cell Nuclei Induced by Rapid Temperature Increase. Biophysical journal 112, 1063–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan ME, Uzer G, Rubin C, 2013. The Potential Benefits and Inherent Risks of Vibration as a Non-Drug Therapy for the Prevention and Treatment of Osteoporosis. Current osteoporosis reports, 1–9. [DOI] [PMC free article] [PubMed]
- Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D, 2006. Coupling of the nucleus and cytoplasm: role of the LINC complex. The Journal of Cell Biology 172, 41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilluy C, Osborne LD, Van Landeghem L, Sharek L, Superfine R, Garcia-Mata R, Burridge K, 2014. Isolated nuclei adapt to force and reveal a mechanotransduction pathway in the nucleus. Nature cell biology 16, 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q, Xia Y, Sandig M, Yang J, 2012. Characterization of cell elasticity correlated with cell morphology by atomic force microscope. Journal of biomechanics 45, 304–309. [DOI] [PubMed] [Google Scholar]
- Hodzic DM, Yeater DB, Bengtsson L, Otto H, Stahl PD, 2004. Sun2 Is a Novel Mammalian Inner Nuclear Membrane Protein. Journal of Biological Chemistry 279, 25805–25812. [DOI] [PubMed] [Google Scholar]
- Imai R, Nozaki T, Tani T, Kaizu K, Hibino K, Ide S, Tamura S, Takahashi K, Shribak M, Maeshima K, 2017. Density imaging of heterochromatin in live cells using orientation-independent-DIC microscopy. Molecular biology of the cell 28, 3349–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan ZS, Santos JM, Hussain F, 2018. Aggressive prostate cancer cell nuclei have reduced stiffness. Biomicrofluidics 12, 014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J-K, Louhghalam A, Lee G, Schafer BW, Wirtz D, Kim D-H, 2017. Nuclear lamin A/C harnesses the perinuclear apical actin cables to protect nuclear morphology. Nature communications 8, 2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammerding J, Fong LG, Ji JY, Reue K, Stewart CL, Young SG, Lee RT, 2006. Lamins A and C but Not Lamin B1 Regulate Nuclear Mechanics. Journal of Biological Chemistry 281, 25768–25780. [DOI] [PubMed] [Google Scholar]
- Le HQ, Ghatak S, Yeung CY, Tellkamp F, Gunschmann C, Dieterich C, Yeroslaviz A, Habermann B, Pombo A, Niessen CM, Wickstrom SA, 2016. Mechanical regulation of transcription controls Polycomb-mediated gene silencing during lineage commitment. Nature cell biology 18, 864–875. [DOI] [PubMed] [Google Scholar]
- Lessey EC, Guilluy C, Burridge K, 2012. From mechanical force to RhoA activation. Biochemistry 51, 7420–7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wen J, Xiao Y, Liu J, Hopyan S, Radisic M, Simmons CA, Sun Y, 2014. In situ mechanical characterization of the cell nucleus by atomic force microscopy. ACS nano 8, 3821–3828. [DOI] [PubMed] [Google Scholar]
- Lombardi ML, Jaalouk DE, Shanahan CM, Burke B, Roux KJ, Lammerding J, 2011. The Interaction between Nesprins and Sun Proteins at the Nuclear Envelope Is Critical for Force Transmission between the Nucleus and Cytoskeleton. Journal of Biological Chemistry 286, 26743–26753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins RP, Finan JD, Farshid G, Lee DA, 2012. Mechanical Regulation of Nuclear Structure and Function. Annual review of biomedical engineering 14, 431–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A, Hieda M, Yokoyama Y, Nishioka Y, Yoshidome K, Tsujimoto M, Matsuura N, 2015. Global loss of a nuclear lamina component, lamin A/C, and LINC complex components SUN1, SUN2, and nesprin-2 in breast cancer. Cancer Med. [DOI] [PMC free article] [PubMed]
- Miroshnikova YA, Nava MM, Wickstrom SA, 2017. Emerging roles of mechanical forces in chromatin regulation. Journal of cell science 130, 2243–2250. [DOI] [PubMed] [Google Scholar]
- Neelam S, Chancellor TJ, Li Y, Nickerson JA, Roux KJ, Dickinson RB, Lele TP, 2015. Direct force probe reveals the mechanics of nuclear homeostasis in the mammalian cell. Proceedings of the National Academy of Sciences of the United States of America 112, 5720–5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcivici E, Luu YK, Adler B, Qin YX, Rubin J, Judex S, Rubin CT, 2010. Mechanical signals as anabolic agents in bone. Nature reviews. Rheumatology 6, 50–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmakumar VC, Libotte T, Lu W, Zaim H, Abraham S, Noegel AA, Gotzmann J, Foisner R, Karakesisoglou I, 2005. The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. Journal of cell science 118, 3419–3430. [DOI] [PubMed] [Google Scholar]
- Pagnotti GM, Styner M, Uzer G, Patel VS, Wright LE, Ness KK, Guise TA, Rubin J, Rubin CT, 2019. Combating osteoporosis and obesity with exercise: leveraging cell mechanosensitivity. Nature Reviews Endocrinology [DOI] [PMC free article] [PubMed]
- Paine PL, Austerberry CF, Desjarlais LJ, Horowitz SB, 1983. Protein loss during nuclear isolation. J Cell Biol 97, 1240–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peister A, Mellad JA, Larson BL, Hall BM, Gibson LF, Prockop DJ, 2004. Adult stem cells from bone marrow (MSCs) isolated from different strains of inbred mice vary in surface epitopes, rates of proliferation, and differentiation potential. Blood 103, 1662–1668. [DOI] [PubMed] [Google Scholar]
- Pongkitwitoon S, Uzer G, Rubin J, Judex S, 2016. Cytoskeletal Configuration Modulates Mechanically Induced Changes in Mesenchymal Stem Cell Osteogenesis, Morphology, and Stiffness. Scientific reports 6, 34791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando TA, Ambrosio F, 2018. Regenerative Rehabilitation: Applied Biophysics Meets Stem Cell Therapeutics. Cell Stem Cell 22, 306–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin J, Styner M, Uzer G, 2018. Physical Signals May Affect Mesenchymal Stem Cell Differentiation via Epigenetic Controls. Exercise and sport sciences reviews 46, 42–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäpe J, Prausse S, Radmacher M, Stick R, 2009. Influence of lamin A on the mechanical properties of amphibian oocyte nuclei measured by atomic force microscopy. Biophysical journal 96, 4319–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen B, Xie Z, Case N, Styner M, Rubin CT, Rubin J, 2011. Mechanical signal influence on mesenchymal stem cell fate is enhanced by incorporation of refractory periods into the loading regimen. Journal of biomechanics 44, 593–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, Banigan EJ, Adam SA, Goldman RD, Marko JF, 2017. Chromatin and lamin A determine two different mechanical response regimes of the cell nucleus. Molecular biology of the cell 28, 1984–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, Liu PZ, Banigan EJ, Almassalha LM, Backman V, Adam SA, Goldman RD, Marko JF, 2018. Chromatin histone modifications and rigidity affect nuclear morphology independent of lamins. Molecular biology of the cell 29, 220–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PCDP, Pinter J, Pajerowski JD, Spinler KR, Shin J-W, Tewari M, Rehfeldt F, Speicher DW, Discher DE, 2013. Nuclear Lamin-A Scales with Tissue Stiffness and Enhances Matrix-Directed Differentiation. Science (New York, N.Y.) 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzer G, Bas G, Sen B, Xie Z, Birks S, Olcum M, McGrath C, Styner M, Rubin J, 2018. Sun-mediated mechanical LINC between nucleus and cytoskeleton regulates betacatenin nuclear access. Journal of biomechanics 74, 32–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzer G, Pongkitwitoon S, Ete Chan M, Judex S, 2013. Vibration induced osteogenic commitment of mesenchymal stem cells is enhanced by cytoskeletal remodeling but not fluid shear. Journal of biomechanics 46, 2296–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzer G, Thompson WR, Sen B, Xie Z, Yen SS, Miller S, Bas G, Styner M, Rubin CT, Judex S, Burridge K, Rubin J, 2015. Cell Mechanosensitivity to Extremely Low-Magnitude Signals Is Enabled by a LINCed Nucleus. STEM CELLS 33, 2063–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versaevel M, Braquenier J-B, Riaz M, Grevesse T, Lantoine J, Gabriele S, 2014. Super-resolution microscopy reveals LINC complex recruitment at nuclear indentation sites. Sci. Rep 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Nuclei were either left intact inside MSCs or isolated (isolation protocol, methods), stained with Hoechst 33342, and fixed in 2% paraformaldehyde. As shown in these videos, a Leica 6500 confocal microscope was then used to acquire vertical stacked images of both nuclei inside intact MSC samples (left) and isolated nuclei (right).
Fig. S2 a) SEM image of a Bruker MLCT-D AFM probe functionalized with a 10 µm diameter glassbead (Thermo Scientific 9010 NIST-traceable 9000 Series Glass Particle Standard, 10.0 ± 1.0 µm). b) Representative thermal tune of an MLCT-D probe functionalized with such a 10 µm glass bead. Nominal resonance frequency of the MLCT-D cantilever prior to bead functionalization is 10–20 kHz, with the added weight of the bead causing the resonance to shift to lower frequency. A simple harmonic oscillator fit was used (red) to model the resonant response of the cantilever in a fluid environment. c) Representative ramp data. Ramping to a maximum excursion/extension of 1 µm was performed at a rate of 1 Hz (i.e., 2 µm/sec load/unload velocity).
Fig S3. Compared to empty control plasmid (pCDH-EF1-MSC1-puro-mCherry), LINC-disrupted intact cells (N=10/grp) and isolated nuclei (N=20/grp) did not show any changes in stiffness. * p<.05, ** p<.01, *** p<.001, against control.
Figure S4. MSCs measured via AFM-based nanoindentation showed 132% higher elastic modulus in passage 15 samples (P15, N=10, p<.001) compared to samples between passages 8 and 11 (P8–11, N=45). * p<.05, ** p<.01, *** p<.001, against control or against each other.
Figure S5. Unprocessed blots as obtained by LiCor C-DiGit blot scanner. Red lines outline the representative blots used.


