Abstract
Hemostasis is a complex process involving the concerted action of molecular and vascular components. Its basic understanding as well as diagnostic and therapeutic aspects have greatly benefited from the use of monoclonal antibodies. Interestingly, camelid‐derived single‐domain antibodies (sdAbs), also known as VHH or nanobodies, have become available during the previous 2 decades as alternative tools in this regard. Compared to classic antibodies, sdAbs are easier to produce and their small size facilitates their engineering and functionalization. It is not surprising, therefore, that sdAbs are increasingly used in hemostasis‐related research. In addition, they have the capacity to recognize unique epitopes unavailable to full monoclonal antibodies. This property can be used to develop novel diagnostic tests identifying conformational variants of hemostatic proteins. Examples include sdAbs that bind active but not globular von Willebrand factor or free factor VIIa but not tissue factor–bound factor VIIa. Finally, sdAbs have a high therapeutic potential, exemplified by caplacizumab, a homodimeric sdAb targeting von Willebrand factor that is approved for the treatment of thrombotic thrombocytopenic purpura. In this review, the various applications of sdAbs in thrombosis and hemostasis‐related research, diagnostics, and therapeutic strategies will be discussed.
Keywords: hemostasis, nanobodies, protein engineering, single‐domain antibodies, therapeutics
Camelid‐derived single‐domain antibodies (or nanobodies) are endowed with numerous advantages over classical antibodies. They hold the potential to represent next‐generation tools for research, diagnostic and even therapeutic purposes, also in the area of thrombosis and hemostasis.

Essentials.
Single‐domain antibodies (sdAbs) derived from camelids are increasingly used as research tools.
sdAbs against coagulation proteins have helped solve biochemical research issues.
sdAbs can be used as diagnostic tools and also for therapeutics in thrombosis and hemostasis.
Protein engineering increases the versatility of sdAbs and their potential applications.
1. INTRODUCTION
Ever since the discovery of hybridoma technology by Kohler and Milstein in 1975, 1 monoclonal antibodies have represented very powerful tools to explore biological processes. Hemostasis, a complex biological process in which a cascade of events involving the concerted action of cellular and molecular players allows for a quick response to vessel injury, is no exception in this regard. Antibodies have been invaluable biological determinants not only to better understand hemostasis but also for diagnostics of hemostasis‐related pathologies or more recently for their treatment. 2 Immunotherapy in the hemostasis field started with the use of abciximab, a humanized Fab fragment against platelet glycoprotein α2bβ3, widely used as antiplatelet therapy in patients undergoing percutaneous coronary intervention to avoid ischemic complications. 3 Abciximab has the capacity to block binding of α2bβ3 to its ligands, fibrinogen and von Willebrand factor (VWF), thereby preventing platelet aggregation. 4 Following abciximab, it has taken a number of years before new antibody‐based treatments could become available for hemorrhagic or thrombotic diseases. However, at least in hemophilia, therapeutic antibodies are now revolutionizing the treatment of patients. 5 , 6 In particular, emicizumab, a bispecific humanized IgG4 directed against coagulation factors IX and X and marketed under the name of Hemlibra has proven efficient in treating hemophilia A patients with or without inhibitors. 7 Emicizumab mimics factor VIII cofactor activity by binding simultaneously to the enzyme factor IXa and its substrate, factor X. 7 On the thrombotic side, several antibody‐based treatments are currently under development, such as ACT017, a humanized Fab fragment, directed against platelet receptor glycoprotein VI (GPVI) and intended for stroke treatment. 8 ACT017 (glenzocimab) blocks GPVI binding sites, leading to an inhibition of collagen‐induced platelet aggregation. This antibody has now demonstrated its safety profile in a clinical phase I trial. 9 Antibodies targeting coagulation factors XI/XIa and XII/XIIa also hold the promise of becoming safe antithrombotic drugs since they are not expected to increase the bleeding risk. 10 , 11
Although classical monoclonal antibodies and their Fab fragment or single‐chain variable fragments (scFvs) derivatives remain most widely used, camelid‐derived single‐domain antibodies (sdAbs) have emerged as a credible next generation of antibodies offering several advantages over classic antibodies owing to their peculiar properties, including their small size, robust behavior, high affinity and specificity, and deep tissue penetration. 12 , 13 , 14 It is also important to mention that besides camelids, sdAbs can also be found in certain sharks. 15 However, this review will focus exclusively on camelid‐derived sdAbs.
2. SINGLE‐DOMAIN ANTIBODIES: THE NEXT‐GENERATION ANTIBODIES
2.1. General features of sdAbs
The first description of functional sdAbs in camelids was reported in 1993. 16 Unlike conventional antibodies, and despite their ability to bind to their target antigen with strong affinity, a fraction of antibodies derived from camelids was found to be completely devoid of light chain. From a structural point of view, sdAbs contain the CH2 and CH3 domains but lack the first classic IgG constant domain, CH1 (Figure 1). The lack of CH1 domain is a necessity for their secretion in the absence of a light chain. 17 Consequently, their antigen‐binding regions consist of one single VH domain termed VHH. VHHs were reported to be the smallest intact antigen‐binding single‐polypeptide chains found in any natural antibody. 18
FIGURE 1.
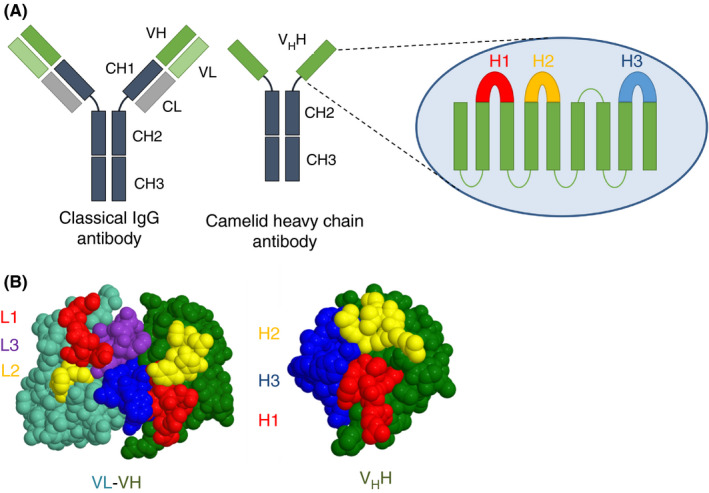
Structural features of classical and camelid heavy‐chain antibodies. (A) In a conventional IgG, pairing of a variable heavy and variable light chains are required to assemble the paratope, while in camelid heavy‐chain antibody the VHH domain does not require a light chain to bind the antigen. Inset: The secondary structure of heavy‐chain‐only antibodies showing three hypervariable loop regions known as the loop H1, H2, and H3. (B) comparison of the antigen‐binding surfaces of conventional IgG (left) and heavy‐chain‐only antibody (right). Adapted from Mitchel et al 26
Recombinant expression of VHHs allows for the generation of a soluble single‐domain antigen‐binding protein with a molecular weight of about 15 kDa. Due to their small size, these sdAbs are sometimes referred to as nanobodies. SdAbs have functional properties similar to that of conventional antibodies in terms of affinity and specificity, 19 with a binding affinity within the picomolar to nanomolar range. 20 However, distinctions also exist between sdAbs and the antigen‐binding region of IgG, which is composed of two entities in two different polypeptide chains, VL and VH. Indeed, certain residues in the VH framework regions that are part of the interface with the VL are altered at the corresponding positions in sdAbs. This endows sdAbs with a more hydrophilic character, increasing their solubility. 21 , 22 In addition, the eventual presence of an extra disulfide bond between the first and third antigen‐binding loops, H1 and H3, may rigidify the sdAb structure, contributing to an increased stability and activity at high temperatures and in high concentrations of denaturants, compared to classical IgGs. 23 The sdAb paratope is defined by three highly variable loops, H1, H2, and H3, which form an extended structural interface at one side of the protein (Figure 1). 24 Despite the smaller surface of this paratope compared to the VH‐VL paratope of IgG, sdAbs can create a higher diversity by exhibiting a greater structural variation in their H1, H2, and H3 loops and by increasing the length of the H3 loop. 24 , 25 , 26 , 27 , 28 It is noteworthy that the conserved framework regions of sdAbs show a high sequence and structural homology with human VH domains of family III (VH3) 29 . This high homology not only results in a comparable immunogenicity as human VH but also increases the chances of successful humanization of these sdAbs (see also section 5 of this review). 22 Furthermore, due to their single‐domain nature and lack of glycosylation, they can be produced in large amounts using standard microbial or yeast expression systems. 30 , 31 , 32
On the other hand, sdAbs miss some of the characteristics found in regular IgGs. For example, the absence of the Fc portion prevents these sdAbs from directly activating the complement system and immune cells, and therefore they lack cytotoxic activity. 33 , 34 Furthermore, without the Fc portion, sdAbs are incapable of entering the FcRn‐dependent recycling pathway. In combination with their small size, which is below the glomerular filtration capacity, this contributes to the short in vivo half‐life of the sdAbs. 34 , 35 Finally, with the antigen‐binding paratope being restricted in size compared to classic IgG, this implies that the number of residues within this paratope that could be used to improve antigen binding are limited as well. 36
2.2. Libraries and screening options to generate sdAbs
Three kinds of libraries—immune libraries, naïve libraries, and synthetic libraries—can be used for the selection of sdAbs. Classic library engineering and sdAb screening strategies are presented in Figure 2 and extensively reviewed by Hoogenboom. 37
FIGURE 2.
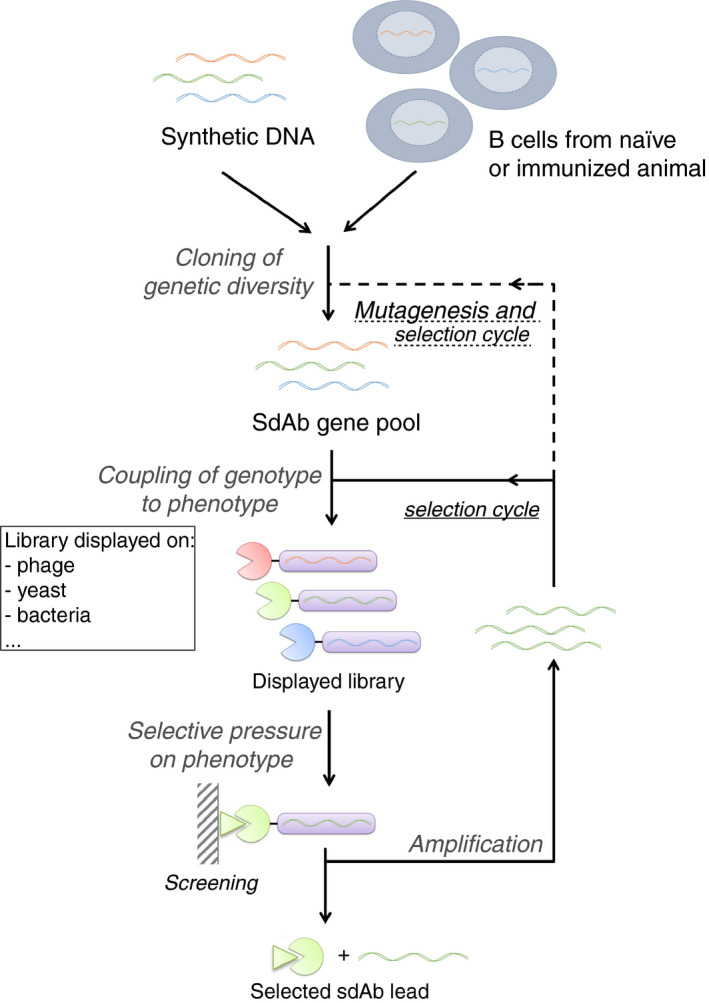
Strategies for creation and selection of sdAbs libraries. sdAb diversity is obtained from B cells of naïve or immunized animal, or from synthetic DNA library. Expression of single sequences results in the presence of a unique sdAb variant at the surface of each individual host (typically phages, bacteria, or yeast cells). Multiple rounds of selective pressure are applied to the host, resulting in the identification of strong antigen binders. Adapted from Hoogenboom 37 . sdAbs, single‐domain antibodies
Traditionally, immune libraries of sdAbs are obtained upon immunization of an animal (llama, alpaca, or dromedary) with the target antigen. The immunization process takes up to 6 weeks and requires weekly injections of variable amounts of the target antigen supplemented with an adjuvant to boost its immunogenicity. Animals can be injected with multiple antigens (5‐10), improving the economics of producing sdAbs and reducing the number of animals. Total mRNA from peripheral blood monocytic cells is then isolated, and the V‐gene repertoire is amplified to generate cDNAs encoding sdAbs, which will make up the library.
sdAbs libraries can also be generated from nonimmunized animals and are therefore called naïve libraries. They come with the advantage of not being selective for a specific antigen and represent the natural diversity of the immune repertoire of the animal. This can be useful when the structure that needs to be targeted by the sdAb is poorly antigenic. It should be noted that naïve libraries often lead to generation of low‐affinity sdAbs, which then require subsequent maturation steps. 37
Synthetic libraries are increasingly used and consist of in vitro randomized libraries. 38 Starting from a unique sdAb scaffold, random primers are used to generate various complementarity‐determining region (CDR) lengths and sequences by polymerase chain reaction, creating a novel library that does not rely on the immune system of an animal. This allows the generation of unique CDR3 sequences that may have never existed in vivo, leading to the identification of antigen binders that can target poorly antigenic structures. One pitfall could be that such unnatural CDR3 sequences may increase immunogenicity, a potential issue if the sdAb is selected for therapeutic purposes.
The immune libraries are generally superior due to the higher affinities of the recovered sdAbs. This is likely due to the deletions/insertions seen in the VHH genes, which occurs as clones are selected by the immune response. Such recombination events do not occur in the naïve libraries and are not naturally selected for using synthetic libraries.
One important characteristic of using cDNA‐based libraries (whether they may be immune, naïve or synthetic), is that they allow for a reproducible expression of the sdAbs during each round of the selection process. This is an important advantage over the use of hybridomas, in which loss of IgG expression may occur during selection.
Independent of the approach underlying library engineering, numerous selection techniques including phage display, yeast display, bacterial display, mRNA/cDNA display, intracellular two‐hybrid system, and ribosome display can be applied to facilitate screening and panning for a given antigen‐specific sdAb. 39 The generation of a sdAb library is advantageous in the sense that it can be screened multiple times, allowing for the use of several setups for antigen presentation. This is of particular interest when the target antigen presents with complicated features, as is often the case with coagulation proteins. Indeed, coagulation factors are found in a zymogen state in the circulation and needs to be activated to exert their functions. 40 Hence, activated coagulation factors are often extremely flexible in their structure, making it sometimes difficult to identify specific binders.
Following the selection protocol, sequence optimization may also be necessary to improve sdAb production efficiency, solubility, or any other desired property.
2.3. Versatility of sdAbs
The combined properties of sdAbs (eg, selection of conformation‐specific binders, ease of engineering, production via routine recombinant techniques) have led to a plethora of applications for sdAbs as research tools. For instance, sdAbs have been used as crystallization chaperones for the structural investigation of diverse conformational states of flexible proteins. 39 , 41 Green fluorescent protein–targeting sdAbs have been applied to optimize superresolution microscopy. 42 Since sdAbs are easily expressed in a variety of cell types, they also have been used to study intracellular processes (for which they are referred to as intrabodies). In this respect, they can be used to specifically inhibit target proteins, signal the appearance of a specific conformational state, or direct the target protein to the intracellular degradation pathway. 38 , 43 , 44 In addition, numerous fusion variants of sdAbs have been described. Apart from the classic fusion candidates (fluorescent proteins or IgG Fc domain), these also include horseradish peroxidase and the phospholipid‐binding C1C2 domains of lactadherin. 45 , 46 Besides these general applications showing (in a nonexhaustive fashion) how powerful the sdAb technology can be, more specific use of sdAbs in the thrombosis and hemostasis field will be the focus of the next few paragraphs.
3. SINGLE‐DOMAIN ANTIBODIES AS RESEARCH TOOLS IN HEMOSTASIS
In the context of hemostasis, a certain number of sdAbs have been developed in the past 15‐20 years, and many of those have been extremely useful, especially for biochemical studies. Indeed, many proteins relevant to maintain the hemostatic balance can exist in different forms. Best described are enzymes and cofactors that require proteolytic activation. sdAbs have the potential to differentiate small conformational differences and have therefore contributed to answer important research questions. Table 1 and Figure 3 summarize sdAbs targeting hemostasis proteins.
TABLE 1.
Overview of sdAbs used in the thrombosis and hemostasis field
| Target protein | Camelid species/Synthetic library | sdAb nomenclature | Binding site | Activity | Application | References |
|---|---|---|---|---|---|---|
| Antithrombin | Llama | KB‐AT‐01 ‐ 07; KB‐AT‐23 | None defined | Neutralization of antithrombin function | Research/Therapy | 79 |
| Factor VIIa | Llama | αFVIIa‐VHH | FVIIa protease domain | Detection of FVIIa but not FVII | Diagnostics | 75 |
| Factor VIIa | Llama | KB‐FVIIa‐004 | FVIIa protease domain | Inhibits free FVIIa but not TF‐bound FVIIa activity | Research | 49 |
| Factor XIIa | Llama | A10 | Catalytic domain | Binds selectively to α‐FXIIa | Research/Diagnostics | 52 |
| Factor XIIa | Llama | B7 | Catalytic domain | Binds both α‐FXIIa and β‐FXIIa | Research/Diagnostics | 52 |
| Fibrinogen | Dromedary | VFIB1, ‐2, ‐3 | None defined | High‐affinity binding to fibrinogen | Diagnostics | 76, 77 |
| PAI‐1 | Alpaca | VHH‐s‐a98 | Surface loops around central β‐sheet A | Switches PAI‐1/PA to substrate pathway | Research | 105 |
| PAI‐1 | Alpaca | VHH‐2w‐64 (Nb64) | Surface loops around central β‐sheet A | Switches PAI‐1/PA to substrate pathway | Research/Therapy | 60, 105 |
| PAI‐1 | Alpaca | VHH‐s‐a27 | None defined | Neutralization of PAI‐1 function | Research | 105 |
| PAI‐1 | Alpaca | VHH‐s‐a93 | Vicinity of reactive center loop | Interferes with initial PAI‐1/PA complex formation | Research | 105 |
| PAI‐1 | Alpaca | VHH‐2g‐42 (Nb42) | Vicinity of reactive center loop | Interferes with initial PAI‐1/PA complex formation | Research/Therapy | 60, 105 |
| PN‐1 | Synthetic library | KB‐PN1‐A08, ‐B11, ‐F06, ‐H12 | Reactive center loop | Neutralization of human and murine PN‐1 function | Research | 106 |
| hTAFI | Alpaca | VHH‐TAFI‐i and VHH‐TAFI‐a series | None defined | Interfere with TAFI activation and/or activity | Research | 54, 55, 57 |
| hTAFI | Alpaca |
VHH‐TAFI‐a51 VHH‐TAFI‐i103 |
None defined | Stimulate activity of TAFI zymogen | Research | 56 |
| mTAFI | Alpaca | VHH‐TAFI‐i49 | Lys212 & Arg227 | Stimulates activity of TAFI zymogen | Research | 107 |
| u‐PA | Alpaca | Nb4 | Active site | Inhibits u‐PA activity | Research | 59 |
| u‐PA | Alpaca | Nb7 | 37s and 70s loops of the N‐terminal β‐barrel | Allosteric inhibitor of u‐PA activity | Research | 58 |
| u‐PA | Alpaca | Nb22 | Active site | Inhibits u‐PA activity | Research | 58 |
| VWF | Llama | ALX‐0081 (caplacizumab) | Human A1 domain | Inhibition of VWF/glycoprotein Ibα interaction | Therapy | 80, 82, 83 |
| VWF | Llama | KB‐VWF‐006bv | Human/murine A1 domain | Inhibition of VWF/glycoprotein Ibα interaction | Research | 63, 64 |
| VWF | Llama | AU‐VWFa‐11 | Human/murine A1 domain | Specifically detects platelet‐binding VWF conformation | Diagnostics | 66, 67, 69, 72, 73, 74 |
Abbreviations: hTAFI, thrombin‐activatable fibrinolysis inhibitor; mTAFI, murine thrombin‐activatable fibrinolysis inhibitor; PA, plasminogen activator; PAI‐1, plasminogen activator inhibitor‐1; PN‐1, protease nexin‐1; u‐PA, urokinase plasminogen activator; TAFI, thrombin‐activatable fibrinolysis inhibitor; VWF, von Willebrand factor.
FIGURE 3.
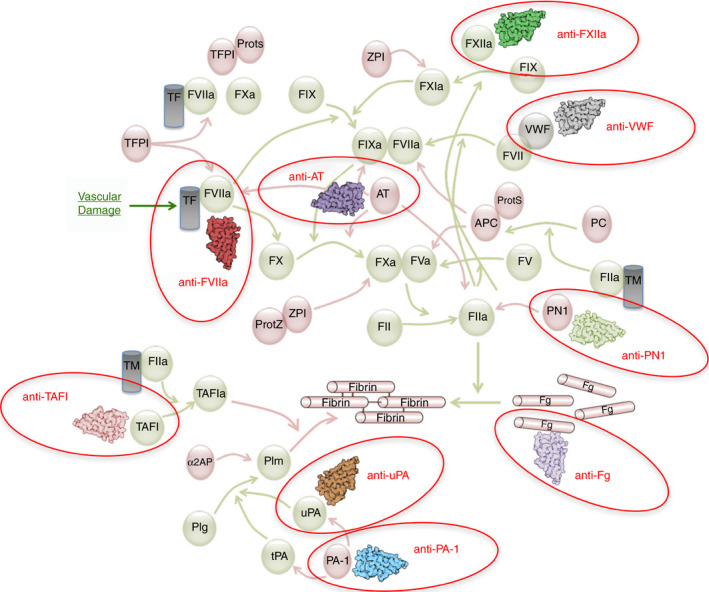
Targets for sdAbs in the coagulation and fibrinolysis pathways. An overview of the major activation and inactivation reactions in the coagulation and fibrinolysis pathway are shown. Red circles indicate sdAbs and their targets that have been mentioned in this review. APC, activated Protein C; α2AP, α2‐antiplasmin; AT, antithrombin; F, factor; Plm, plasmin; Plg, plasminogen; PAI‐1, plasminogen activator inhibitor‐1; PC, protein C; PN1, Protease nexin‐1; ProtS, protein S; ProtZ, protein Z; TAFI, thrombin‐activatable fibrinolysis inhibitor; TM, thrombomodulin; TF, tissue factor; TFPI, tissue factor pathway inhibitor; t‐PA, tissue‐type plasminogen activator; u‐PA, urokinase plasminogen activator; VWF, von Willebrand factor; ZPI, protein Z–dependent protease inhibitor
3.1. sdAbs against factor VIIa
One example of sdAb that allowed to resolve a scientific issue relates to factor VII (FVII) which can exist in a precursor (FVII) and an activated (FVIIa) state. 47 In the 1990s, FVIIa has become available for the treatment of hemophilia inhibitor patients. Its efficacy is based on the capacity of FVIIa to activate coagulation factors IX and X, thereby leading to thrombin activation. One unanswered question remained about the mechanism of action of FVIIa, based on the apparent discrepancy between the high therapeutic doses needed compared to the low physiological concentration of FVIIa. One hypothesis was that FVIIa had to compete with endogenous FVII for binding to its cofactor tissue factor (TF). 48 To assess this hypothesis, our group has recently developed a sdAb (KB‐FVIIa‐004) capable of selectively detecting and inhibiting proteolytic activities of FVIIa but not TF‐bound FVIIa. 49 Using this unique sdAb, we have been able to demonstrate the TF‐independent mode of action of FVIIa in vivo in hemophilia A mice.
3.2. sdAbs against factor XIIa
sdAbs have also helped decipher the sequential activation steps of factor XII (FXII), a key protein of the contact system that has recently gained a lot of attention for its potential as an antithrombotic target. 50 , 51 Upon surface binding, small amounts of FXII are converted into an activated derivative, α‐FXIIa. α‐FXIIa is able to convert plasma prekallikrein (PPK) into plasma kallikrein (PK), and in turn PK will further amplify α‐FXIIa formation. However, PK will also generate β‐FXIIa, a smaller derivative of α‐FXIIa that lacks the surface binding heavy chain. α‐FXIIa and β‐FXIIa differ in that only α‐FXIIa is able to activate factor XI (FXI) into FXIa. In contrast, both variants are able to convert PPK into PK. Since PK is responsible for the release of bradykinin, a small peptide that evokes vascular leakage and tissue swelling, it is thus important to know how much α‐FXIIa and β‐FXIIa is present under various physiological and pathological conditions. To improve the analysis of α‐FXIIa and β‐FXIIa formation, de Maat et al 52 have developed sdAbs that are able to distinguish between both forms. One sdAb (VHH‐B7) recognizes both variants, whereas VHH‐A10 selectively binds to α‐FXIIa. These sdAbs thus represent an essential biochemical research tool to capture these molecules at different time points of the contact system and reveal previously unidentified agonist‐dependent FXII activation pathways, which might or might not lead to activation of the coagulation cascade. 52
3.3. sdAbs against thrombin‐activatable fibrinolysis inhibitor
Another hemostatic protein which has significantly benefited from sdAbs technology for its structural and mechanistic studies is thrombin‐activatable fibrinolysis inhibitor (TAFI), known for its modulatory function of the fibrinolytic process. TAFI circulates as an inactive carboxypeptidase precursor, which becomes active via proteolytic activation predominantly by the thrombin/thrombomodulin complex. This proteolytic step releases the N‐terminal activation peptide and opens the active site for its substrates. The main function of TAFIa is to downregulate fibrinolysis by removing C‐terminal lysines from fibrin. 53 Its physiological relevance is illustrated by the notion that elevated TAFI levels are associated with an increased thrombotic risk.
Both activation and inactivation pathways of TAFI are complex and involve multiple steps. Particularly, inactivation of TAFIa is autoregulated by a self‐destruction mechanism that involves sequential conformational changes within the protein. To better understand this process, the laboratory of Dr Declerck has developed numerous sdAbs specific to the different conformations of the TAFI zymogen and its activated derivatives. 54 , 55 , 56 , 57 These sdAbs have been used in activity assays as well as in structural studies to obtain specific crystal structures, which together led to the discovery of previously unidentified conformations of this intriguing enzyme. In addition, they were useful to identify the activation peptide as an anchor for the thrombin/thrombomodulin complex, necessary to achieve physiological relevant activation rates. Together, these studies provide an excellent example of how the unique properties of sdAbs can generate novel structural insights with regard to the highly flexible structures of hemostatic proteins.
3.4. sdAbs against urokinase plasminogen activator
sdAbs have also been used for the analysis of other fibrinolytic proteins, such as Plasminogen Activator Inhibitor‐1 and urokinase plasminogen activator (u‐PA). 58 , 59 , 60 u‐PA in particular is mainly known as a plasminogen activator, but is also involved in processes that do not require plasmin generation. 61 In search for inhibitors of u‐PA function, it has been observed that some of the peptide bond–based inhibitors also function as substrates. It is unclear if this dual behavior originates (i) from conformation changes within the peptide segment of the inhibitor/substrate that inserts into the active site pocket of the enzyme, (ii) from conformational changes within the enzyme structure or (iii) both. To get insight into this important question, Kromann‐Hansen and coworkers developed sdAbs that penetrate into the u‐PA active site. 58 , 59 Via crystallographic analysis, they observed that the presence of such sdAbs changed the structure of u‐PA, favoring option ii. Interestingly, they noticed that despite the strong inhibitory effect of the sdAb, it also was slowly proteolyzed. Following mutagenesis aiming to introduce more flexibility into the rigid H3 loop, the sdAb was proteolyzed more efficiently. This suggests that conformational changes within the peptide segment that protrudes from the active site can also contribute to whether this segment is acting as inhibitor, substrate, or both. It would be interesting to investigate whether sdAbs that penetrate the active site of other serine proteases display similar features, as this may help to develop agents that display partial or transient inhibitory properties toward their targets.
3.5. sdAbs against VWF
Although most of the above‐mentioned studies have used sdAbs as biochemical tools, they can also be essential for functional studies. One such example relates to sdAbs raised against VWF, the main molecular player in platelet adhesion to the injured vessel during primary hemostasis. 62 As many other hemostatic proteins, VWF displays important roles beyond hemostasis. In this context, our group has recently generated a sdAb binding to the VWF A1 domain (KB‐VWF‐006) with the unique capacity to recognize human and murine VWF. 63 This sdAb interferes with ristocetin‐induced human and murine platelet aggregation by inhibiting VWF‐platelet glycoprotein Ibα (GPIbα) interaction. Because our sdAb can also recognize murine VWF, it holds the potential to be used in vivo to test the implication of the VWF‐GPIbα axis in different pathological settings. When applied to inflammatory in vivo models, ie immune‐complex‐mediated vasculitis and irritant contact dermatitis, KB‐VWF‐006bi (the bivalent version of KB‐VWF‐006) was able to reduce hemorrhages and leukocyte recruitment, supporting the previously hypothesized direct role of the VWF‐GPIbα pathway in inflammatory circumstances and during vascular permeability. 63 Similarly, in a stroke model, KB‐VWF‐006bi reduced thromboinflammation, highlighting the important role of the VWF A1 domain in this process. 64
4. SINGLE‐DOMAIN ANTIBODIES AS DIAGNOSTIC TOOLS IN HEMOSTASIS
Although there is not any sdAb‐based routine diagnostic assay available yet in clinical hematology laboratories, several sdAbs have been used for clinical research purposes, proving their usefulness in detecting specific disease‐associated protein distinctions.
4.1. sdAbs against active VWF
Despite its nonenzymatic characteristic, VWF can transition from an inactive‐ to an active conformation in specific pathophysiologic circumstances. Indeed, procoagulant VWF properties largely rely on conformational changes that switch the molecule from a globular, inactive form to an open, GPIbα‐binding state, also referred as active VWF. 65 Physical relaxation and/or mechanical activation of VWF‐A1 domain in response to mechanical supraphysiologic forces allow active VWF to interact with its partners, among which the most important are subendothelial collagen and platelets. Several years ago, a sdAb (AU/VWFa‐11) was identified to selectively detect GPIbα‐binding VWF state but not globular VWF. 66 Importantly, AU/VWFa‐11 has no inhibitory function. AU/VWFa‐11 binding assays have been developed to directly reveal circulating active VWF and provided an alternative easy method as compared to indirect measurement of platelet aggregation in the presence of various ristocetin concentrations. Major advantage of the AU/VWFa‐11 binding assay is the ability to detect active VWF even under conditions of low VWF antigen levels. It is important to mention that under physiological conditions, extremely low levels of active VWF are present in the circulation since this platelet‐binding form of VWF is considered prothrombotic. AU/VWFa‐11 assays have therefore been used to detect excessive VWF activation in several pathologic conditions. 67 Best described are a bleeding‐associated rare subset of von Willebrand disease (VWD) classified as VWD‐type 2B and thrombotic thrombocytopenic purpura (TTP), which is related to disseminated thrombotic events in the microcirculation. VWD‐type 2B is caused by gain‐of‐function genetic variants within or close to the VWF‐A1 domain. These mutations destabilize VWF toward its platelet‐binding conformation. 68 Using the AU/VWFa‐11 binding assay, it was found that active VWF was raised 2‐ to 15‐fold. 66 Moreover, an inverse correlation was observed between the amount of active VWF and platelet count, which in turn is associated with the severity of the bleeding phenotypes. 69
Similar rises in active VWF have also been measured in TTP patients, with a 2‐ to 12‐fold increase over resting, despite a very different underlying molecular mechanism. Active VWF in TTP stems from a lack of proteolytic inactivation of VWF by its specific enzyme, ADAMTS13. 70 Interestingly, higher levels of active VWF were consistently measured in patients with congenital forms of the disease, while samples from patients suffering from the acquired forms showed smaller, albeit supraphysiologic levels of active VWF. Therefore, AU/VWFa‐11 is able to discriminate between congenital and acquired TTP forms, which is relevant to apply therapeutic decisions. 66
A number of pathologic conditions characterized by VWF‐dependent platelet aggregates have also been associated with elevated levels of active VWF, as quantified by AU/VWFa‐11–based assays. 67 This was the case in patients suffering from HELLP syndrome (hemolysis, elevated liver enzymes, low platelets) a severe complication of preeclampsia, 71 parasitic malaria, which is associated with thrombocytopenia and cerebral thrombotic events, 72 antiphospholipid syndrome, again characterized by low platelet counts and microthrombopathy 73 and sickle cell disease, associated with chronic hemolytic anemia and sporadic vaso‐occlusion. 74
4.2. sdAbs against FVIIa
Besides the sdAb against FVIIa developed by our group and used for mechanistic studies (see section 3.1), another group has generated a bivalent clone (αFVIIa‐VHH), which was elegantly employed in different pathologic settings. Indeed, specific levels of FVII versus FVIIa have been evaluated in cases of systemic inflammatory response syndrome. In this context, quantification of FVII versus FVIIa appeared useful to stratify patients with higher mortality risk. Indeed, elevated FVIIa and low FVII levels were found to associate with high mortality rate. This study highlights how constant implementation of highly selective research tools is not only beneficial for diagnosis but allows to test unexplored aspects of complex pathophysiological pathways, for example, coagulation and inflammation. 75
4.3. sdAbs against fibrinogen
Another advantage of sdAbs is that the C‐terminus is opposite to the antigen‐binding surface, allowing it to be used for functionalization of the antibody. This feature has recently been used for the development of a novel diagnostic assay to measure fibrinogen levels. 76 , 77 Purified sdAbs targeting fibrinogen were used in an immunobased amperometric test, allowing the rapid detection of as little as 44 μg fibrinogen/L in plasma (normal range, 1.5‐4.5 g/L). In follow‐up studies, the same group is now developing an agglutination‐based assay, that relies on the expression of sdAbs at the Escherichia coli surface. Incubation of plasma samples containing fibrinogen with these sdAb‐expressing bacteria results in a dose‐dependent agglutination. 78 Given the high affinity and specificity, this type of approaches could be used for the detection of other hemostatic proteins as well.
5. SINGLE‐DOMAIN ANTIBODIES AS THERAPEUTIC TOOLS IN THROMBOSIS AND HEMOSTASIS
Probably owing to their relatively recent discovery, sdAbs have not yet widely found their way to the clinic. There is no doubt that this situation is likely to evolve in the next few years since given their unique properties, sdAbs have a high potential in this regard. First, despite their nonhuman origin, sdAbs are less immunogenic compared to murine‐ or rat‐derived monoclonal IgG antibodies because of their high degree of homology to the human VH3 domain. 29 In addition, humanization of sdAbs is relatively straightforward compared to the humanization of IgG molecules. The main issue is to substitute the amino acids that correspond to those in human IgG that are involved in the VH/VL interaction, which are located in framework region 2. 22 Since such substitutions may affect solubility or affinity, it is not uncommon that additional mutagenesis is performed to restore or even improve these parameters. A second advantage relates to the half‐life of the sdAbs, which can be tailored dependent on its purpose. Due to its small size, a single unbound sdAb is rapidly eliminated from the circulation via renal clearance. This could be advantageous when used for in vivo imaging as explained in part 6.2. Alternatively, sdAbs can form a complex with their target in the circulation, and in this complex sdAbs adopt the circulatory half‐life of their target. Examples hereof are caplacizumab (discussed in part 5.1), or sdAbs targeting antithrombin, which are eliminated while bound to their respective targets VWF and antithrombin. 79 , 80 Finally, sdAbs can be engineered to have a prolonged half‐life, for instance via fusion to proteins with a long half‐life (such as the Fc region of IgG molecules or albumin), via fusion to a second sdAb that binds for example albumin, via multimerization to a size above the renal clearance limit, or via the formation of sdAb‐containing nanoparticles. 81
Together, these properties make sdAbs attractive candidates for therapeutic applications whether in the area of thrombosis and hemostasis or other pathologies. So far, only one sdAb has been approved for treatment, but a number of other sdAbs are currently in clinical trials for treatment of rheumatoid arthritis, breast cancer, or other pathologies. 13
5.1. sdAbs in thrombosis
As previously mentioned, only one sdAb, caplacizumab, a homodimeric sdAb targeting the A1‐domain of VWF, has been approved for therapeutic purposes. Caplacizumab interferes with the interaction between VWF and GPIbα, preventing the adhesion of platelets to VWF. 80 Originally designed to prevent thrombotic events during percutaneous coronary intervention, caplacizumab is now approved for the management of TTP. TTP is characterized by the functional deficiency of ADAMTS13, a protease important for the regulation of VWF multimer size. The absence of ADAMTS13 activity allows the secretion of ultra‐large platelet‐binding VWF multimers, which provokes the formation of VWF/platelet aggregates that occlude the microvasculature. 70 Caplacizumab has been evaluated in two clinical trials, the TITAN (Study to assess efficacy and safety of anti‐von willebrand factor [vWF] nanobody in patients with acquired thrombotic thrombocytopenic purpura [aTTP]) and HERCULES (Phase III trial with caplacizumab in patients with acquired thrombotic thrombocytopenic purpura) trials. 82 , 83 In both trials, caplacizumab treatment was characterized by a faster resolution of thrombocytopenia, a reduction in TTP‐related death, relapse, and major thrombotic events as well as shorter hospitalization times. Importantly, only 3% of the patients in the HERCULES trial developed anti‐caplacizumab antibodies, with none of them affecting clinical efficacy or caplacizumab function. It is of further interest to mention that caplacizumab has been shown to reduce infarct size in an animal model for cerebral artery thrombosis, 84 while studies using a caplacizumab mimetic (sdAb KB‐VWF‐006bi described in part 3.5) revealed that blocking VWF/platelet interactions reduces the inflammatory response following vascular injury. 63 , 64 It would be interesting therefore to evaluate caplacizumab in other thrombotic conditions, such as ischemic stroke.
Although not yet in clinical development, there are also a number of other sdAbs that could be of interest for the management of thrombotic complications. In particular, sdAbs that target natural inhibitors of the fibrinolytic pathway could be useful in this regard. Inhibitory sdAbs targeting TAFI (described in section 3.3) and plasminogen activator inhibitor‐1 (PAI‐1), developed by the laboratory of Dr Declerck could be used to enhance thrombolysis. 54 , 57 In particular, these sdAbs could be applied to enhance thrombolysis during exaggerated pulmonary thrombosis. Interestingly, a clinical trial is under consideration to improve thrombolysis via nebulization of tissue‐type plasminogen activator. 85 Since sdAbs have excellent bioavailability following nebulization, 86 , 87 it would be interesting to add sdAbs targeting TAFI or PAI‐1 as stand‐alone or adjuvant therapy.
5.2. sdAbs in hemorrhagic disorders
Also in hemorrhagic conditions, sdAbs could be useful, and our laboratory has recently reported two potential examples in which sdAbs have been tested in preclinical settings. First, we used engineered sdAbs neutralizing the anticoagulant antithrombin. 79 By neutralizing antithrombin, thrombin generation in the absence of factor VIII (FVIII) or factor IX was normalized (irrespective of the presence of inhibitors). In subsequent mouse models for hemophilia A or B with inhibitors, the presence of anti‐antithrombin sdAbs normalized the bleeding tendency of these mice. Interestingly, correction of bleeding was achieved not only via a single intravenous infusion of the purified protein but also in a gene therapy setting. Apparently, adeno‐associated virus (AAV)‐mediated expression of sdAbs is sufficiently strong to neutralize plasma proteins present in high concentrations (2.5 μmol/L for antithrombin). It would be of interest to also investigate sdAbs targeting other natural anticoagulants, including protease nexin‐1, tissue factor pathway inhibitor, protein Z–dependent protease inhibitor, activated protein C and protein S, for their efficacy in correcting the bleeding tendency in hemophilia.
In a second approach, we replaced the B domain of FVIII with sdAbs targeting VWF, the carrier protein of FVIII. 88 This increased the affinity of FVIII for VWF about 25‐fold (13 pmol/L instead of 330 pmol/L), while leaving in vitro and in vivo FVIII activity unaffected. The increased affinity resulted in a twofold prolonged half‐life. Unexpectedly, increased affinity for VWF resulted in a strongly reduced immune‐response, with sevenfold fewer mice developing anti‐FVIII antibodies compared to wild‐type FVIII. Although in the early stages of development, this FVIII variant could be an attractive alternative in the treatment of patients having increased risks of inhibitor development.
6. SINGLE‐DOMAIN ANTIBODIES AND CHALLENGES IN HEMOSTASIS
Because sdAbs are so versatile and can be used in so many different ways, as shown in the example of the FVIII‐sdAb fusion molecule, there are (almost) unlimited options in their potential applications. A few examples will be discussed below and are summarized on Figure 4.
FIGURE 4.
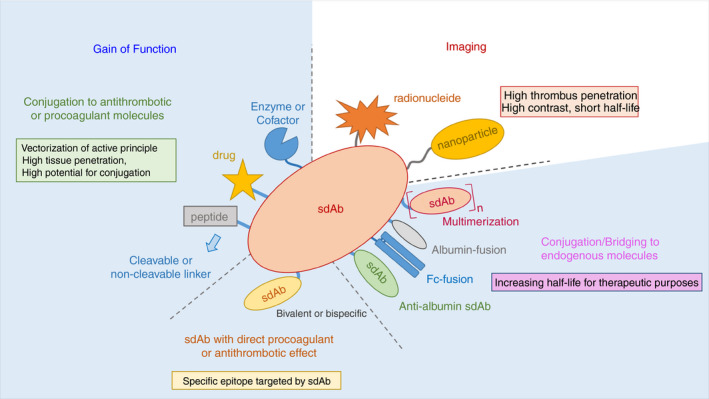
Examples of potential applications of sdAbs in relation to thrombosis and hemostasis. The ease of manipulation of sdAbs makes them an ideal choice for the design of therapeutic and diagnostic tools. sdAbs can be incorporated in larger structure to participate in the gain‐of‐function of therapeutic compound, conjugated by cleavable or noncleavable linker. Due to their small size (associated with a high tissue penetration) and fast clearance from the bloodstream, sdAbs are suitable for the design of imaging probes. This can be achieved by the direct conjugation of target‐specific sdAbs with radionucleides, or by incorporating target‐specific sdAbs in nanoparticles. In the case of sdAbs with direct pro‐ or anticoagulant properties, increasing their half‐lives requires their fusion to other proteins such as albumin or albumin adaptors, or to the Fc domain of conventional IgGs. For example, bivalent constructs bridging a target to an albumin molecule would potentially increase the half‐life of the target protein to that of albumin
6.1. sdAbs and therapeutic design
An important challenge in hemostasis is to maintain the delicate balance between bleeding and thrombosis. For patients suffering from an excess or lack of coagulation, replacement therapy is often the main option, but the ideal would be to direct the missing factor where it is needed—to the bleeding site or to the thrombus site. So far, such treatments are administered systemically, thereby increasing the risk of unwanted events. Due to their small size and low complexity, sdAbs are easy to combine, incorporate into fusion molecules, and can be functionalized. Therefore, the use of sdAbs for targeting active pro‐ or anticoagulant molecules at the site where they are needed is an attractive option. Moreover, sdAbs present a high tissue penetration capacity, which is of tremendous interest for thrombotic events. 12 , 89 The field of fusion protein design with sdAbs can build on what has already been done with scFvs. Such examples include the fusion protein between a scFv against the activated form of α2bβ3 and a FXa inhibitor peptide, 90 a scFv targeting α2bβ3 to a modified plasminogen, 91 or an anti–platelet endothelial cell adhesion molecule (PECAM‐1) scFv linked to a single‐chain pro‐urokinase plasminogen activator. 92 In all these examples, the fusion between the scFv and the active peptide or molecule has resulted in successful cellular targeting of the fusion protein. Compared to scFv, sdAbs will be easier to use for protein engineering due to their high solubility, stability, and high yield of production. 93
Besides protein fusion, another interesting possibility relies on the potential incorporation of sdAbs in the capsid of gene‐delivery vectors such as AAVs. This was recently reported by Eichhoff et al, 94 who elegantly generated AAV variants whose capsid VP1 protein was fused with receptor‐specific sdAbs. These AAV variants demonstrated high specificity for the cells expressing the respective sdAbs targeted receptor. Future development of such strategies will help generate highly tissue‐specific AAV vectors that would increase gene‐therapy efficiency. This would be particularly interesting in the hemophilia field where improvement in gene‐therapy efficiency could lead to lower doses of virus particles injected and therefore to fewer adverse effects such as liver toxicity.
Still with the aim of achieving a better targeted therapy, sdAbs can be incorporated into drug delivery systems such as liposomes, micelles, or nanoparticles. 95 , 96 , 97
6.2. sdAbs and bioimaging
To date, thrombus characterization suffers from the lack of precise tools for structure and composition analysis, which is crucial to make a medical decision. This is particularly true in stroke where “time is brain” but where recanalization treatment will be more or less successful according to the etiology of the thrombus. 98 Thus, markers of thrombus composition and also of thrombus maturity are critically needed to decide whether anticoagulation, pharmacological thrombolysis, mechanical thrombolysis, or a combination of these is required. 99 The recent availability of thrombi obtained by thrombectomy has only further highlighted the differences in thrombus composition and therefore the need for more adapted and targeted therapies. 100 , 101 However, the imaging technologies that are currently available such as single‐photon emission computed tomography, positron emission tomography, magnetic resonance imaging and optical imaging techniques, despite their advantages, do not yet allow the precise characterization of a thrombus composition that would be needed upon patient arrival. This is why mixed imaging techniques that can combine molecular/functional/anatomic imaging are gaining increasing attention. They rely on the accuracy and precision of radiolabeled markers and sdAbs have many of the desired qualities required to constitute excellent markers for imaging. Their small molecular weight (about 15 kDa) is well below the renal elimination cutoff (~50 kDa). As a result, sdAbs size favors their tissue penetration and allows for their rapid renal clearance. This leads to a high signal‐to‐noise ratio and rapid imaging after administration, as demonstrated in the case of tumor imaging. 102 , 103 It is therefore possible to imagine using sdAbs fused to imaging probes for detection of fibrin/red blood cells/VWF or activated platelets in order to gain knowledge in thrombus structure. Again, the feasibility of such an approach was demonstrated using scFv. A synthetic protein‐based nanoparticle containing a near infrared fluorescent molecule and functionalized with a scFv specific for activated platelets was engineered by Bonnard et al. 104 This construct exhibited a strong potential as biocompatible molecular imaging probe in an in vivo model of carotid arterial thrombosis.
7. CONCLUSION
sdAbs have only begun to demonstrate their utility in biomedical research. The thrombosis and hemostasis community has rapidly seized their potential by developing very interesting biological tools against complex proteins and by using them to tackle unanswered biochemical issues. Notably, so far, no sdAbs against platelet receptors have been developed. Importantly, the first clinically approved sdAb is in the thrombosis and hemostasis field, demonstrating the swift embracement of this technology by our community. However, as a laboratory that has invested in sdAb technology for the past 6 years, we can testify that, although they constitute great tools, not all sdAbs are equal and working with some of them can prove challenging. Affinity maturation steps to optimize sdAbs are often necessary and can be time consuming. Only future will tell whether sdAbs will be the magic bullets of next‐generation immunotherapy strategies.
AUTHOR CONTRIBUTIONS
All authors have contributed to the writing of the review and approved the final manuscript.
RELATIONSHIP DISCLOSURE
PJL, ODC, and CVD are co‐inventors on four patents related to the use of single‐domain antibodies for therapeutic purposes.
Peyron I, Kizlik‐Masson C, Dubois M‐D, et al. Camelid‐derived single‐chain antibodies in hemostasis: Mechanistic, diagnostic, and therapeutic applications. Res Pract Thromb Haemost. 2020;4:1087–1110. 10.1002/rth2.12420
Handling Editor: Alisa Wolberg
Contributor Information
Ivan Peyron, @IvanPeyron.
Cécile V. Denis, Email: cecile.denis@inserm.fr, @cecile_denis.
Peter J. Lenting, @LentingPeter.
Caterina Casari, @caterinacasari.
Olivier D. Christophe, @ODChristophe1.
REFERENCES
- 1. Kohler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–7. [DOI] [PubMed] [Google Scholar]
- 2. Gruel Y, Kizlik‐Masson C, Lenting P. Therapeutic antibodies in hemostasis. From the past to the future. Med Sci (Paris). 2019;35:1022–5. [DOI] [PubMed] [Google Scholar]
- 3. EPIC Investigators . Use of a monoclonal antibody directed against the platelet glycoprotein IIb/IIIa receptor in high‐risk coronary angioplasty. N Engl J Med. 1994;330:956–61. [DOI] [PubMed] [Google Scholar]
- 4. Coller BS. A new murine monoclonal antibody reports an activation‐dependent change in the conformation and/or microenvironment of the platelet glycoprotein IIb/IIIa complex. J Clin Invest. 1985;76:101–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Muczynski V, Christophe OD, Denis CV, Lenting PJ. Emerging therapeutic strategies in the treatment of hemophilia A. Semin Thromb Hemost. 2017;43:581–90. [DOI] [PubMed] [Google Scholar]
- 6. Weyand AC, Pipe SW. New therapies for hemophilia. Blood. 2019;133:389–98. [DOI] [PubMed] [Google Scholar]
- 7. Kitazawa T, Shima M. Emicizumab, a humanized bispecific antibody to coagulation factors IXa and X with a factor VIIIa‐cofactor activity. Int J Hematol. 2020;111:20–30. [DOI] [PubMed] [Google Scholar]
- 8. Lebozec K, Jandrot‐Perrus M, Avenard G, Favre‐Bulle O, Billiald P. Design, development and characterization of ACT017, a humanized Fab that blocks platelet's glycoprotein VI function without causing bleeding risks. mAbs. 2017;9:945–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Voors‐Pette C, Lebozec K, Dogterom P, Jullien L, Billiald P, Ferlan P, et al. Safety and tolerability, pharmacokinetics, and pharmacodynamics of ACT017, an antiplatelet GPVI (glycoprotein VI) fab. Arterioscler Thromb Vasc Biol. 2019;39:956–64. [DOI] [PubMed] [Google Scholar]
- 10. DeLoughery EP, Olson SR, Puy C, McCarty OJT, Shatzel JJ. The safety and efficacy of novel agents targeting factors XI and XII in early phase human trials. Semin Thromb Hemost. 2019;45:502–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Larsson M, Rayzman V, Nolte MW, Nickel KF, Bjorkqvist J, Jamsa A, et al. A factor XIIa inhibitory antibody provides thromboprotection in extracorporeal circulation without increasing bleeding risk. Sci Transl Med. 2014;6:222ra17. [DOI] [PubMed] [Google Scholar]
- 12. Debie P, Lafont C, Defrise M, Hansen I, van Willigen DM, van Leeuwen FWB, et al. Size and affinity kinetics of nanobodies influence targeting and penetration of solid tumours. J Control Release. 2020;317:34–42. [DOI] [PubMed] [Google Scholar]
- 13. Jovcevska I, Muyldermans S. The therapeutic potential of nanobodies. BioDrugs. 2020;34:11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Khodabakhsh F, Behdani M, Rami A, Kazemi‐Lomedasht F. Single‐domain antibodies or nanobodies: a class of next‐generation antibodies. Int Rev Immunol. 2018;37:316–22. [DOI] [PubMed] [Google Scholar]
- 15. Konning D, Zielonka S, Grzeschik J, Empting M, Valldorf B, Krah S, et al. Camelid and shark single domain antibodies: structural features and therapeutic potential. Curr Opin Struct Biol. 2017;45:10–6. [DOI] [PubMed] [Google Scholar]
- 16. Hamers‐Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Songa EB, et al. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–8. [DOI] [PubMed] [Google Scholar]
- 17. Nguyen VK, Hamers R, Wyns L, Muyldermans S. Loss of splice consensus signal is responsible for the removal of the entire C(H)1 domain of the functional camel IGG2A heavy‐chain antibodies. Mol Immunol. 1999;36:515–24. [DOI] [PubMed] [Google Scholar]
- 18. Arbabi Ghahroudi M, Desmyter A, Wyns L, Hamers R, Muyldermans S. Selection and identification of single domain antibody fragments from camel heavy‐chain antibodies. FEBS Lett. 1997;414:521–6. [DOI] [PubMed] [Google Scholar]
- 19. van der Linden RH, Frenken LG, de Geus B, Harmsen MM, Ruuls RC, Stok W, et al. Comparison of physical chemical properties of llama VHH antibody fragments and mouse monoclonal antibodies. Biochim Biophys Acta. 1999;1431:37–46. [DOI] [PubMed] [Google Scholar]
- 20. Decanniere K, Desmyter A, Lauwereys M, Ghahroudi MA, Muyldermans S, Wyns L. A single‐domain antibody fragment in complex with RNase A: non‐canonical loop structures and nanomolar affinity using two CDR loops. Structure. 1999;7:361–70. [DOI] [PubMed] [Google Scholar]
- 21. Conrath K, Vincke C, Stijlemans B, Schymkowitz J, Decanniere K, Wyns L, et al. Antigen binding and solubility effects upon the veneering of a camel VHH in framework‐2 to mimic a VH. J Mol Biol. 2005;350:112–25. [DOI] [PubMed] [Google Scholar]
- 22. Vincke C, Loris R, Saerens D, Martinez‐Rodriguez S, Muyldermans S, Conrath K. General strategy to humanize a camelid single‐domain antibody and identification of a universal humanized nanobody scaffold. J Biol Chem. 2009;284:3273–84. [DOI] [PubMed] [Google Scholar]
- 23. De Genst E, Saerens D, Muyldermans S, Conrath K. Antibody repertoire development in camelids. Dev Comp Immunol. 2006;30:187–98. [DOI] [PubMed] [Google Scholar]
- 24. Muyldermans S. Nanobodies: natural single‐domain antibodies. Annu Rev Biochem. 2013;82:775–97. [DOI] [PubMed] [Google Scholar]
- 25. Li X, Duan X, Yang K, Zhang W, Zhang C, Fu L, et al. Comparative analysis of immune repertoires between bactrian Camel's conventional and heavy‐chain antibodies. PLoS One. 2016;11:e0161801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mitchell LS, Colwell LJ. Comparative analysis of nanobody sequence and structure data. Proteins. 2018;86:697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nguyen VK, Hamers R, Wyns L, Muyldermans S. Camel heavy‐chain antibodies: diverse germline V(H)H and specific mechanisms enlarge the antigen‐binding repertoire. EMBO J. 2000;19:921–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sircar A, Sanni KA, Shi J, Gray JJ. Analysis and modeling of the variable region of camelid single‐domain antibodies. J Immunol. 2011;186:6357–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vu KB, Ghahroudi MA, Wyns L, Muyldermans S. Comparison of llama VH sequences from conventional and heavy chain antibodies. Mol Immunol. 1997;34:1121–31. [DOI] [PubMed] [Google Scholar]
- 30. Baghban R, Gargari SL, Rajabibazl M, Nazarian S, Bakherad H. Camelid‐derived heavy‐chain nanobody against Clostridium botulinum neurotoxin E in Pichia pastoris. Biotechnol Appl Biochem. 2016;63:200–5. [DOI] [PubMed] [Google Scholar]
- 31. Frenken LG, van der Linden RH, Hermans PW, Bos JW, Ruuls RC, de Geus B, et al. Isolation of antigen specific llama VHH antibody fragments and their high level secretion by Saccharomyces cerevisiae . J Biotechnol. 2000;78:11–21. [DOI] [PubMed] [Google Scholar]
- 32. Ta DT, Redeker ES, Billen B, Reekmans G, Sikulu J, Noben JP, et al. An efficient protocol towards site‐specifically clickable nanobodies in high yield: cytoplasmic expression in Escherichia coli combined with intein‐mediated protein ligation. Protein Eng Des Sel. 2015;28:351–63. [DOI] [PubMed] [Google Scholar]
- 33. Bannas P, Hambach J, Koch‐Nolte F. Nanobodies and nanobody‐based human heavy chain antibodies as antitumor therapeutics. Front Immunol. 2017;8:1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. De Vlieger D, Ballegeer M, Rossey I, Schepens B, Saelens X. Single‐domain antibodies and their formatting to combat viral infections. Antibodies (Basel). 2018;8:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jank L, Pinto‐Espinoza C, Duan Y, Koch‐Nolte F, Magnus T, Rissiek B. Current approaches and future perspectives for nanobodies in stroke diagnostic and therapy. Antibodies (Basel). 2019;8:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Arbabi‐Ghahroudi M. Camelid single‐domain antibodies: historical perspective and future outlook. Front Immunol. 2017;8:1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hoogenboom HR. Selecting and screening recombinant antibody libraries. Nat Biotechnol. 2005;23:1105–16. [DOI] [PubMed] [Google Scholar]
- 38. Moutel S, Bery N, Bernard V, Keller L, Lemesre E, de Marco A, et al. NaLi‐H1: a universal synthetic library of humanized nanobodies providing highly functional antibodies and intrabodies. eLife. 2016;5:e16228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pardon E, Laeremans T, Triest S, Rasmussen SG, Wohlkonig A, Ruf A, et al. A general protocol for the generation of nanobodies for structural biology. Nat Protoc. 2014;9:674–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Macfarlane RG. An enzyme cascade in the blood clotting mechanism, and its function as a biochemical amplifier. Nature. 1964;202:498–9. [DOI] [PubMed] [Google Scholar]
- 41. Deupi X, Standfuss J. Structural insights into agonist‐induced activation of G‐protein‐coupled receptors. Curr Opin Struct Biol. 2011;21:541–51. [DOI] [PubMed] [Google Scholar]
- 42. Ries J, Kaplan C, Platonova E, Eghlidi H, Ewers H. A simple, versatile method for GFP‐based super‐resolution microscopy via nanobodies. Nat Methods. 2012;9:582–4. [DOI] [PubMed] [Google Scholar]
- 43. Gueorguieva D, Li S, Walsh N, Mukerji A, Tanha J, Pandey S. Identification of single‐domain, Bax‐specific intrabodies that confer resistance to mammalian cells against oxidative‐stress‐induced apoptosis. FASEB J. 2006;20:2636–8. [DOI] [PubMed] [Google Scholar]
- 44. Yamaguchi N, Colak‐Champollion T, Knaut H. zGrad is a nanobody‐based degron system that inactivates proteins in zebrafish. eLife. 2019;8:e43125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kooijmans SAA, Gitz‐Francois J, Schiffelers RM, Vader P. Recombinant phosphatidylserine‐binding nanobodies for targeting of extracellular vesicles to tumor cells: a plug‐and‐play approach. Nanoscale. 2018;10:2413–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yamagata M, Sanes JR. Reporter‐nanobody fusions (RANbodies) as versatile, small, sensitive immunohistochemical reagents. Proc Natl Acad Sci U S A. 2018;115:2126–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Persson E, Olsen OH. Allosteric activation of coagulation factor VIIa. Front Biosci (Landmark Ed). 2011;16:3156–63. [DOI] [PubMed] [Google Scholar]
- 48. Giansily‐Blaizot M, Schved JF. Recombinant human factor VIIa (rFVIIa) in hemophilia: mode of action and evidence to date. Ther Adv Hematol. 2017;8:345–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ferriere S, Kawecki C, Ottavi JF, Denis CV, Kauskot A, Christophe OD, et al. A single‐domain antibody that blocks factor VIIa activity in the absence but not presence of tissue factor. J Thromb Haemost. 2019;17:2035–46. [DOI] [PubMed] [Google Scholar]
- 50. Renne T, Pozgajova M, Gruner S, Schuh K, Pauer HU, Burfeind P, et al. Defective thrombus formation in mice lacking coagulation factor XII. J Exp Med. 2005;202:271–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Revenko AS, Gao D, Crosby JR, Bhattacharjee G, Zhao C, May C, et al. Selective depletion of plasma prekallikrein or coagulation factor XII inhibits thrombosis in mice without increased risk of bleeding. Blood. 2011;118:5302–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. de Maat S, van Dooremalen S, de Groot PG, Maas C. A nanobody‐based method for tracking factor XII activation in plasma. Thromb Haemost. 2013;110:458–68. [DOI] [PubMed] [Google Scholar]
- 53. Plug T, Meijers JC. Structure‐function relationships in thrombin‐activatable fibrinolysis inhibitor. J Thromb Haemost. 2016;14:633–44. [DOI] [PubMed] [Google Scholar]
- 54. Buelens K, Hassanzadeh‐Ghassabeh G, Muyldermans S, Gils A, Declerck PJ. Generation and characterization of inhibitory nanobodies towards thrombin activatable fibrinolysis inhibitor. J Thromb Haemost. 2010;8:1302–12. [DOI] [PubMed] [Google Scholar]
- 55. Hendrickx ML, De winter A, Buelens K, Compernolle G, Hassanzadeh‐ghassabeh G, Muyldermans S, et al. TAFIa inhibiting nanobodies as profibrinolytic tools and discovery of a new TAFIa conformation. J Thromb Haemost. 2011;9:2268–77. [DOI] [PubMed] [Google Scholar]
- 56. Mishra N, Buelens K, Theyskens S, Compernolle G, Gils A, Declerck PJ. Increased zymogen activity of thrombin‐activatable fibrinolysis inhibitor prolongs clot lysis. J Thromb Haemost. 2012;10:1091–9. [DOI] [PubMed] [Google Scholar]
- 57. Zhou X, Weeks SD, Ameloot P, Callewaert N, Strelkov SV, Declerck PJ. Elucidation of the molecular mechanisms of two nanobodies that inhibit thrombin‐activatable fibrinolysis inhibitor activation and activated thrombin‐activatable fibrinolysis inhibitor activity. J Thromb Haemost. 2016;14:1629–38. [DOI] [PubMed] [Google Scholar]
- 58. Kromann‐Hansen T, Louise Lange E, Peter Sorensen H, Hassanzadeh‐Ghassabeh G, Huang M, Jensen JK, et al. Discovery of a novel conformational equilibrium in urokinase‐type plasminogen activator. Sci Rep. 2017;7:3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kromann‐Hansen T, Oldenburg E, Yung KW, Ghassabeh GH, Muyldermans S, Declerck PJ, et al. A camelid‐derived antibody fragment targeting the active site of a serine protease balances between inhibitor and substrate behavior. J Biol Chem. 2016;291:15156–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sillen M, Weeks SD, Zhou X, Komissarov AA, Florova G, Idell S, et al. Molecular mechanism of two nanobodies that inhibit PAI‐1 activity reveals a modulation at distinct stages of the PAI‐1/plasminogen activator interaction. J Thromb Haemost. 2020;18:681–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Medcalf RL. Fibrinolysis: from blood to the brain. J Thromb Haemost. 2017;15:2089–98. [DOI] [PubMed] [Google Scholar]
- 62. Lenting PJ, Casari C, Christophe OD, Denis CV. von Willebrand factor: the old, the new and the unknown. J Thromb Haemost. 2012;10:2428–37. [DOI] [PubMed] [Google Scholar]
- 63. Ayme G, Adam F, Legendre P, Bazaa A, Proulle V, Denis CV, et al. A novel single‐domain antibody against von Willebrand factor A1 domain resolves leukocyte recruitment and vascular leakage during inflammation‐brief report. Arterioscler Thromb Vasc Biol. 2017;37:1736–40. [DOI] [PubMed] [Google Scholar]
- 64. Denorme F, Martinod K, Vandenbulcke A, Denis CV, Lenting PJ, Deckmyn H, et al. The von Willebrand Factor A1 domain mediates thromboinflammation, aggravating ischemic stroke outcome in mice. Haematologica. 2020; haematol.2019.241042. 10.3324/haematol.2019.241042. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Springer TA. von Willebrand factor, Jedi knight of the bloodstream. Blood. 2014;124:1412–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hulstein JJ, de Groot PG, Silence K, Veyradier A, Fijnheer R, Lenting PJ. A novel nanobody that detects the gain‐of‐function phenotype of von Willebrand factor in ADAMTS13 deficiency and von Willebrand disease type 2B. Blood. 2005;106:3035–42. [DOI] [PubMed] [Google Scholar]
- 67. Groot E, de Groot PG, Fijnheer R, Lenting PJ. The presence of active von Willebrand factor under various pathological conditions. Curr Opin Hematol. 2007;14:284–9. [DOI] [PubMed] [Google Scholar]
- 68. Tischer A, Brehm MA, Machha VR, Moon‐Tasson L, Benson LM, Nelton KJ, et al. Evidence for the misfolding of the A1 domain within multimeric von Willebrand factor in type 2 von Willebrand disease. J Mol Biol. 2020;432:305–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Federici AB, Mannucci PM, Castaman G, Baronciani L, Bucciarelli P, Canciani MT, et al. Clinical and molecular predictors of thrombocytopenia and risk of bleeding in patients with von Willebrand disease type 2B: a cohort study of 67 patients. Blood. 2009;113:526–34. [DOI] [PubMed] [Google Scholar]
- 70. Levy GG, Nichols WC, Lian EC, Foroud T, McClintick JN, McGee BM, et al. Mutations in a member of the ADAMTS gene family cause thrombotic thrombocytopenic purpura. Nature. 2001;413:488–94. [DOI] [PubMed] [Google Scholar]
- 71. Hulstein JJ, van Runnard Heimel PJ, Franx A, Lenting PJ, Bruinse HW, Silence K, et al. Acute activation of the endothelium results in increased levels of active von Willebrand factor in hemolysis, elevated liver enzymes and low platelets (HELLP) syndrome. J Thromb Haemost. 2006;4:2569–75. [DOI] [PubMed] [Google Scholar]
- 72. de Mast Q, Groot E, Lenting PJ, de Groot PG, McCall M, Sauerwein RW, et al. Thrombocytopenia and release of activated von Willebrand factor during early Plasmodium falciparum malaria. J Infect Dis. 2007;196:622–8. [DOI] [PubMed] [Google Scholar]
- 73. Hulstein JJ, Lenting PJ, de Laat B, Derksen RH, Fijnheer R, de Groot PG. beta2‐Glycoprotein I inhibits von Willebrand factor dependent platelet adhesion and aggregation. Blood. 2007;110:1483–91. [DOI] [PubMed] [Google Scholar]
- 74. Chen J, Hobbs WE, Le J, Lenting PJ, de Groot PG, Lopez JA. The rate of hemolysis in sickle cell disease correlates with the quantity of active von Willebrand factor in the plasma. Blood. 2011;117:3680–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hyseni A, Kemperman H, de Lange DW, de Groot PG, Linssen M, Kesecioglu J, et al. Increased mortality in systemic inflammatory response syndrome patients with high levels of coagulation factor VIIa. J Thromb Haemost. 2013;11:2111–7. [DOI] [PubMed] [Google Scholar]
- 76. Campuzano S, Salema V, Moreno‐Guzman M, Gamella M, Yanez‐Sedeno P, Fernandez LA, et al. Disposable amperometric magnetoimmunosensors using nanobodies as biorecognition element. Determination of fibrinogen in plasma. Biosens Bioelectron. 2014;52:255–60. [DOI] [PubMed] [Google Scholar]
- 77. Salema V, Lopez‐Guajardo A, Gutierrez C, Mencia M, Fernandez LA. Characterization of nanobodies binding human fibrinogen selected by E. coli display. J Biotechnol. 2016;234:58–65. [DOI] [PubMed] [Google Scholar]
- 78. Kylilis N, Riangrungroj P, Lai HE, Salema V, Fernandez LA, Stan GV, et al. Whole‐cell biosensor with tunable limit of detection enables low‐cost agglutination assays for medical diagnostic applications. ACS Sens. 2019;4:370–8. [DOI] [PubMed] [Google Scholar]
- 79. Barbon E, Ayme G, Mohamadi A, Ottavi JF, Kawecki C, Casari C, et al. Single‐domain antibodies targeting antithrombin reduce bleeding in hemophilic mice with or without inhibitors. EMBO Mol Med. 2020;12:e11298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ulrichts H, Silence K, Schoolmeester A, de Jaegere P, Rossenu S, Roodt J, et al. Antithrombotic drug candidate ALX‐0081 shows superior preclinical efficacy and safety compared with currently marketed antiplatelet drugs. Blood. 2011;118:757–65. [DOI] [PubMed] [Google Scholar]
- 81. Kooijmans SA, Aleza CG, Roffler SR, van Solinge WW, Vader P, Schiffelers RM. Display of GPI‐anchored anti‐EGFR nanobodies on extracellular vesicles promotes tumour cell targeting. J Extracell Vesicles. 2016;5:31053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Peyvandi F, Scully M, Kremer Hovinga JA, Cataland S, Knobl P, Wu H, et al. Caplacizumab for acquired thrombotic thrombocytopenic purpura. N Engl J Med. 2016;374:511–22. [DOI] [PubMed] [Google Scholar]
- 83. Scully M, Cataland SR, Peyvandi F, Coppo P, Knobl P, Kremer Hovinga JA, et al. Caplacizumab treatment for acquired thrombotic thrombocytopenic purpura. N Engl J Med. 2019;380:335–46. [DOI] [PubMed] [Google Scholar]
- 84. Momi S, Tantucci M, Van Roy M, Ulrichts H, Ricci G, Gresele P. Reperfusion of cerebral artery thrombosis by the GPIb‐VWF blockade with the Nanobody ALX‐0081 reduces brain infarct size in guinea pigs. Blood. 2013;121:5088–97. [DOI] [PubMed] [Google Scholar]
- 85. Whyte CS, Morrow GB, Mitchell JL, Chowdary P, Mutch NJ. Fibrinolytic abnormalities in acute respiratory distress syndrome (ARDS) and versatility of thrombolytic drugs to treat COVID‐19. J Thromb Haemost. 2020;18(7):1548–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Larios Mora A, Detalle L, Gallup JM, Van Geelen A, Stohr T, Duprez L, et al. Delivery of ALX‐0171 by inhalation greatly reduces respiratory syncytial virus disease in newborn lambs. mAbs. 2018;10:778–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Van Heeke G, Allosery K, De Brabandere V, De Smedt T, Detalle L, de Fougerolles A. Nanobodies(R) as inhaled biotherapeutics for lung diseases. Pharmacol Ther. 2017;169:47–56. [DOI] [PubMed] [Google Scholar]
- 88. Muczynski V, Casari C, Moreau F, Ayme G, Kawecki C, Legendre P, et al. A factor VIII‐nanobody fusion protein forming an ultrastable complex with VWF: effect on clearance and antibody formation. Blood. 2018;132:1193–7. [DOI] [PubMed] [Google Scholar]
- 89. De Vos J, Devoogdt N, Lahoutte T, Muyldermans S. Camelid single‐domain antibody‐fragment engineering for (pre)clinical in vivo molecular imaging applications: adjusting the bullet to its target. Expert Opin Biol Ther. 2013;13:1149–60. [DOI] [PubMed] [Google Scholar]
- 90. Hanjaya‐Putra D, Haller C, Wang X, Dai E, Lim B, Liu L, et al. Platelet‐targeted dual pathway antithrombotic inhibits thrombosis with preserved hemostasis. JCI Insight. 2018;3:e99329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Bonnard T, Tennant Z, Niego B, Kanojia R, Alt K, Jagdale S, et al. Novel thrombolytic drug based on thrombin cleavable microplasminogen coupled to a single‐chain antibody specific for activated GPIIb/IIIa. J Am Heart Assoc. 2017;6:e004535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Ding BS, Gottstein C, Grunow A, Kuo A, Ganguly K, Albelda SM, et al. Endothelial targeting of a recombinant construct fusing a PECAM‐1 single‐chain variable antibody fragment (scFv) with prourokinase facilitates prophylactic thrombolysis in the pulmonary vasculature. Blood. 2005;106:4191–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Schumacher D, Helma J, Schneider AFL, Leonhardt H, Hackenberger CPR. Nanobodies: chemical functionalization strategies and intracellular applications. Angew Chem Int Ed Engl. 2018;57:2314–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Eichhoff AM, Borner K, Albrecht B, Schafer W, Baum N, Haag F, et al. Nanobody‐enhanced targeting of AAV gene therapy vectors. Mol Ther Methods Clin Dev. 2019;15:211–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Altintas I, Heukers R, van der Meel R, Lacombe M, Amidi M, van Bergen en Henegouwen PMP, et al. Nanobody‐albumin nanoparticles (NANAPs) for the delivery of a multikinase inhibitor 17864 to EGFR overexpressing tumor cells. J Control Release. 2013;165:110–8. [DOI] [PubMed] [Google Scholar]
- 96. Oliveira S, Schiffelers RM, van der Veeken J, van der Meel R, Vongpromek R, van Bergen en Henegouwen PMP, et al. Downregulation of EGFR by a novel multivalent nanobody‐liposome platform. J Control Release. 2010;145(2):165–75. [DOI] [PubMed] [Google Scholar]
- 97. Talelli M, Rijcken CJ, Oliveira S, van der Meel R, van Bergen en Henegouwen PMP, Lammers T, et al. Reprint of “Nanobody–shell functionalized thermosensitive core‐crosslinked polymeric micelles for active drug targeting”. J Control Release. 2011;153:93–102. [DOI] [PubMed] [Google Scholar]
- 98. Le Behot A, Gauberti M, Martinez De Lizarrondo S, Montagne A, Lemarchand E, Repesse Y, et al. GpIbalpha‐VWF blockade restores vessel patency by dissolving platelet aggregates formed under very high shear rate in mice. Blood. 2014;123:3354–63. [DOI] [PubMed] [Google Scholar]
- 99. Czaplicki C, Albadawi H, Partovi S, Gandhi RT, Quencer K, Deipolyi AR, et al. Can thrombus age guide thrombolytic therapy? Cardiovasc Diagn Ther. 2017;7:S186–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Di Meglio L, Desilles JP, Ollivier V, Nomenjanahary MS, Di Meglio S, Deschildre C, et al. Acute ischemic stroke thrombi have an outer shell that impairs fibrinolysis. Neurology. 2019;93:e1686–e1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Staessens S, Denorme F, Francois O, Desender L, Dewaele T, Vanacker P, et al. Structural analysis of ischemic stroke thrombi: histological indications for therapy resistance. Haematologica. 2020;105:498–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Huang L, Gainkam LO, Caveliers V, Vanhove C, Keyaerts M, De Baetselier P, et al. SPECT imaging with 99mTc‐labeled EGFR‐specific nanobody for in vivo monitoring of EGFR expression. Mol Imaging Biol. 2008;10:167–75. [DOI] [PubMed] [Google Scholar]
- 103. Vaneycken I, Devoogdt N, Van Gassen N, Vincke C, Xavier C, Wernery U, et al. Preclinical screening of anti‐HER2 nanobodies for molecular imaging of breast cancer. FASEB J. 2011;25:2433–46. [DOI] [PubMed] [Google Scholar]
- 104. Bonnard T, Jayapadman A, Putri JA, Cui J, Ju Y, Carmichael C, et al. Low‐fouling and biodegradable protein‐based particles for thrombus imaging. ACS Nano. 2018;12:6988–96. [DOI] [PubMed] [Google Scholar]
- 105. Zhou X, Hendrickx ML, Hassanzadeh‐Ghassabeh G, Muyldermans S, Declerck PJ. Generation and in vitro characterisation of inhibitory nanobodies towards plasminogen activator inhibitor 1. Thromb Haemost. 2016;116:1032–40. [DOI] [PubMed] [Google Scholar]
- 106. Kawecki C, Aymonnier K, Ferriere S, Venisse L, Arocas V, Boulaftali Y, et al. Development and characterization of single‐domain antibodies neutralizing protease nexin‐1 as tools to increase thrombin generation. J Thromb Haemost. 2020. 10.1111/jth.14940. [DOI] [PubMed] [Google Scholar]
- 107. Hendrickx ML, Zatloukalova M, Hassanzadeh‐Ghassabeh G, Muyldermans S, Gils A, Declerck PJ. Identification of a novel, nanobody‐induced, mechanism of TAFI inactivation and its in vivo application. J Thromb Haemost. 2014;12:229–36. [DOI] [PubMed] [Google Scholar]


