Abstract
Background
Blood clotting in humans is initiated by the binding of tissue factor to activated coagulation factor VII (FVIIa) in the plasma. Previous studies have reported that hepsin and factor VII (FVII)‐activating protease are responsible for generating FVIIa.
Objectives
We aimed to identify other proteases that may activate FVII using zebrafish as a model.
Methods
We screened 179 genes encoding serine protease domains using the piggyback knockdown method to identify genes involved in the activation of zebrafish Fvii. A prolonged kinetic prothrombin time (kPT) assay was used to detect gene knockdown effects.
Results
In the primary screen, 21 genes showed prolonged kPT. In the secondary screen, 14 of 21 genes showed positive results. In the tertiary screen, all 14 genes showed prolonged kPT. These 14 genes were knocked down again to estimate relative levels of zebrafish Fviia. Six genes, including known genes, such as f10 and novel prostasin and hepatocyte growth factor B (hgfb), showed lower Fviia levels. Fvii levels were affected only by the knockdown of f7 and not by the knockdown of the other five genes.
Conclusions
Prostasin and hgfb are involved in generating Fviia. We hypothesize that prostasin exerts serine protease activity directly or indirectly to activate Fvii. As Hgfb has a mutated serine protease domain, it may not cleave Fvii but may bind to Fvii to induce autoactivation. The approach developed here may be extended to design other large‐scale knockdown screens.
Keywords: clotting, Fvii, Fviia, knockdown, serine protease, zebrafish
Essentials.
Blood clotting requires a protein called activated coagulation factor VII (FVIIa), and its generation needs further study.
Piggyback knockdown was used to reduce levels of 179 proteins.
Fourteen of these 179 proteins affected coagulation times.
Two proteins (prostasin, hepatocyte growth factor B) not known to be linked with FVIIa were found.
1. INTRODUCTION
Coagulation factor VII (FVII) is a vitamin K–dependent coagulation factor that triggers the extrinsic pathway of coagulation. 1 Under basal conditions, a small fraction of FVII (<1%) circulates in the form of a two‐chain active protease termed activated coagulation factor VII (FVIIa). 2 FVIIa binds to tissue factor (TF) that is exposed during injury and converts more zymogen FVII to FVIIa, which subsequently cleaves coagulation factor X (FX) and initiates the coagulation cascade. 3 However, the initial generation of FVIIa in vivo is not well understood. Although auto‐activation of zymogen FVII bound to TF has been proposed, 4 , 5 these findings were based on in vitro experiments. It is difficult to imagine that such a reaction could be reproduced with the minute amount of FVIIa (<1%) found in vivo. A variety of coagulation proteases, such as activated forms of prothrombin or coagulation factor II (FII), factor XII (FXII), factor IX (FIX), and factor X (FX), have been shown to activate FVII in vitro. 6 , 7 , 8 , 9 However, activation of FVII by these proteases does not explain the initial generation of FVIIa. Further investigation indicated that 2 serine proteases, factor VII‐activating protease (FSAP) and hepsin, a transmembrane serine protease (TMSP), are essential for FVII activation. 10 , 11 However, other studies reported that FSAP may not be involved in FVII activation. 12 Interestingly, although hepsin produced in cell culture models activated FVII, 11 hepsin knockout in mice showed that it may not play a role in blood clotting. 13 The previous study indicated that hepsin may activate zebrafish Fvii. However, Fsap did not play a role in zebrafish clotting, substantiating the findings in mice. 14 Since both in vitro cell culture model and zebrafish model studies have shown that hepsin activates FVII, the inability of hepsin to activate FVII in mice could possibly be attributed to species‐specific variation. Therefore, it is not possible to exclude a role for hepsin in human FVII activation. As hepsin is generated via activation by TMSPs, we hypothesized that additional proteases may be involved in the generation of FVIIa. We surmised that a whole‐genome knockdown screen of serine proteases and subsequent estimation of Fviia levels in zebrafish plasma may enable the identification of other proteases involved in the Fvii activation pathway. Thus, the current study performed an unbiased large‐scale knockdown of serine proteases and found that Fii, Fvii, Fx, hepsin, prostasin, and hepatocyte growth factor b (Hgfb) showed Fvii activation. Fii, Fvii, Fx, and hepsin are known to be involved in human FVII activation. Thus, the prostasin and Hgfb are novel candidates that showed potential for Fvii activation.
2. MATERIALS AND METHODS
2.1. Zebrafish aquaculture
Maintenance and breeding of zebrafish (purchased from Ekwill farms through Fish n Chirps Pet Centre, Denton, TX, USA) were conducted according to previously published methods. 15 Adult zebrafish were maintained in deionized water supplemented with Instant Ocean at 28.5°C in a fish facility under 14‐hour‐light and 10‐hour‐dark cycle conditions. The fish were fed live brine shrimp and dry flake food. All procedures were approved by the University of North Texas Institutional Animal Care and Use Committee, and animal experiments were performed with humane care in compliance with institutional guidelines.
2.2. Blood collection and plasma preparation
Blood collection and plasma preparation were performed according to a previously published method. 14 Each adult fish was laid flat on a paper towel, and its body wiped with a Kim Wipe. Using dissecting scissors, a lateral incision was made posterior to the dorsal fin, in the region of the dorsal aorta. Two microliters of blood were immediately retrieved with a micropipette and placed in an Eppendorf tube containing 0.5 μL of 3.8% sodium citrate and gently mixed via finger tapping. Next, the preparation was centrifuged at 1000 g for 3 minutes, following which the top layer of citrated plasma was pipetted out without disturbing the blood cell layer. For ELISA assays, zebrafish plasma was prepared using fish injected with 5 μL of 3.8% sodium citrate.
2.3. Piggyback knockdown of serine proteases
Knockdowns of serine proteases were performed via the piggyback method established in our laboratory. 16 First, the National Center for Biotechnology Information (NCBI) and ENSEMBL databases were scanned for genes encoding all serine proteases in the zebrafish genome using the search words “serine protease.” The predicted protein‐coding sequence of each serine protease was used to select the corresponding coding cDNA sequence. Next, a 25‐nucleotide‐long antisense sequence, localized approximately in the middle of the coding sequence, was selected. Then, a 15‐nucleotide‐long sequence, 5′‐TATAAATTGTAACTG‐3′, was added to the 3′‐end of this antisense sequence, resulting in a 40‐nucleotide‐long oligonucleotide. The above 15 nucleotide sequence, base pairs with a vivo morpholino (VMO). The oligonucleotides were purchased from Invitrogen. The VMO (5′‐CCTCTTACCTCAGTTACAATTTATA‐3′) was purchased from Gene‐Tools LLC. The hybrid for intravenous injection into adult zebrafish was prepared by mixing 2.25 μL of 0.5 mmol/L VMO with 2.25 μL of 0.5 mmol/L of the 40‐nucleotide‐long oligonucleotide and 0.5 μL of 10× oligo‐hybridization buffer containing 500 mmol/L NaCl, 10 mmol/L Tris‐HCl (pH 8.0), and 1 mmol/L EDTA (pH 8.0). The above mixture was heated to 90°C and slowly cooled down to room temperature using a Takara PCR Thermal Cycler. Adult zebrafish were anesthetized and injected intravenously with 5 µL of either the above hybrid, or 1× phosphate buffered saline (PBS), using a 27G11/4 needle. Adult zebrafish injected with 1× PBS served as the wild‐type (control) for all experiments.
2.4. Kinetic prothrombin time assay
A kinetic prothrombin time (kPT) assay was performed according to a method established in our laboratory. The thromboplastin reagent was made from zebrafish muscle, as described previously. 17 Briefly, zebrafish muscle tissue (2 g) stored in 0.1 mol/L NaCl and 50 mmol/L Tris at pH 7.5, on ice, was homogenized for 30 seconds. The homogenate was stirred at 37°C for 30 minutes and centrifuged at 11 000 g for 15 minutes. Next, the supernatant, which was treated with EDTA to achieve a concentration of 20 mmol/L, was centrifuged at 4800 g for 60 minutes. The resulting pellet was washed twice and suspended in 1.5 mL of buffer containing 0.1 mol/L NaCl and 50 mmol/L Tris at a pH of 7.5. For the kPT assay, 0.5 μL of citrated zebrafish plasma, 20 μL of 10 mg/mL human fibrinogen (Millipore Sigma, Billerica, MA, USA), 20 μL of the thromboplastin reagent and 53.5 μL of 1× PBS were added to a well in a 96‐well microtiter plate. The plasma was then recalcified using 6 μL of 100 mmol/L CaCl2 and fibrin generation over time was monitored at 405 nm absorbance, using a Synergy H1 Hybrid Multi‐Mode Microplate Reader (BioTek Instruments, Winooski, VT, USA). A sample with all of the above‐mentioned reagents, except plasma, and a sample without CaCl2 were used as negative controls. If the time taken to reach half maximal fibrin formation at 405 nm was longer than 21 minutes, that was considered as indicative of (positive) gene knockdown in the kPT reactions.
2.5. Fviia and Fvii assays by ELISA
Fviia assays were performed according to published protocols 14 using an IMUBIND Factor VIIa ELISA kit (American Diagnostica Inc, Stamford, CT, USA). Briefly, microtiter plate wells coated with a capture monoclonal antibody specific to human FVII/FVIIa were used. Plasma from adult zebrafish was incubated with the biotinylated enzyme inhibitor for FVIIa (a probable TF pathway inhibitor‐related product supplied in the kit), added to these wells, and incubated for 30 minutes at 20°C. After washing, streptavidin‐conjugated horseradish peroxidase (HRP) was added. The mixture was then incubated for 30 minutes at 20°C and washed thrice briefly. The reaction was detected using 3, 3′, 5, 5′‐tetramethylbenzidine (TMB). The enzymatic reaction was quenched by the addition of 50 μL of 0.5 N H2SO4, following which color development was quantified using a Synergy H1 Hybrid Multi‐Mode Microplate Reader at 450 nm. Total Fviia concentrations were interpolated from standard curves generated for each assay, as described below. The standard curve for Fviia estimation was generated using human FVIIa standards. For these curves, the standards were serially diluted to generate concentrations of 200, 100, 50, 25, 12.5, 6.25, and 3.12 ng/mL. The FVIIa standards were captured using antibodies against human FVIIa, and their concentrations detected as described above.
To measure Fvii levels, microtiter plates (96 wells/plate) were coated with 100 μL of 10 μg/mL rabbit anti‐zebrafish Fvii peptide antibodies, used in previous studies. 14 Zebrafish plasma was added to the above‐coated plate and incubated at 4°C for 1 hour. After washing briefly, anti‐Gla antibodies, which bind the mouse Gla domain, were used to detect total Fvii. After washing, anti‐mouse antibodies conjugated with HRP were added to the preparation, which was then incubated for 30 minutes at room temperature, followed by three brief washes. Reaction was detected using 3, 3′, 5, 5′‐TMB. The enzymatic reaction was quenched by adding 50 μL of 0.5 N H2SO4, following which color development was quantified, using a Synergy H1 Hybrid Multi‐Mode Microplate Reader at 450 nm, as mentioned above. Total Fvii concentration was extrapolated from standard curves generated each time the assay was performed. The standard curve for Fvii estimation was generated using the human FVII standard. The FVII standard was serially diluted to generate concentrations of 200, 100, 50, 25, 12.5, 6.25, and 3.12 ng/mL. The generated FVII standards were captured by antibodies against human FVII, where Gla antibodies were used to detect FVII concentration.
The amount of Fviia, or Fvii, in wild‐type (control) samples was considered to be 100%. The concentrations of Fviia, or Fvii that were obtained following the knockdown of genes were plotted as relative values compared to those of wild‐type (control) samples.
2.6. Statistical analysis
Statistical analysis was performed using Sigma Plot 10 with Sigma Stat integration software (Systat Software, Chicago, IL, USA). Statistical significance was assessed by one‐way analysis of variance and set at P < .05. Differences in significance between means were tested via Tukey’s post hoc analysis. The data were expressed as mean ± SD.
3. RESULTS
To perform an unbiased large‐scale knockdown of serine proteases, 179 genes encoding proteins containing serine protease domains were extracted from the NCBI and ENSEMBL databases. Proteins encoded by these genes were subjected to Basic Local Alignment Search Tool (BLAST) analysis to identify the catalytic triad. The list of genes, the ENSEMBL ID/Gene ID, the gene name, the presence or absence of human homologs, and the presence or absence of catalytic triads are shown (Table S1). For each of these 179 genes, we custom‐synthesized 40‐nucleotide‐long oligonucleotides in such a manner that 15 nucleotides at the 3′‐end of the oligonucleotide hybridized to the VMO, while the remaining 25 nucleotides, which were antisense to corresponding mRNA, were located near the middle of the coding sequence (Figure 1A). 16 The list of antisense primer sequences is shown (Table S1). Each of these 40‐nucleotide‐long oligonucleotides was mixed with VMO, and the hybrid was prepared by heating and cooling this mixture. Each hybrid was injected intravenously into a single zebrafish, which was injected once more after 24 hours. Twenty‐four hours after the second injection, plasma was prepared and used in a kPT assay. 18 Since we had previously shown that Fviia levels may affect kPT, 14 we chose kPT as a rapid screening assay. The mean time taken to half maximal fibrin formation in 12 wild‐types (controls) was found to be 18 minutes ± SD of 1 minute. Therefore, if time taken to reach half maximal fibrin formation was greater than 21 minutes, that was considered as positive for prolonged kPT. Compared with wild‐type (control) (Figure 1B), the primary knockdown screen of all 179 genes yielded 21 hybrids showing prolonged kPT, taking >21 minutes to reach half maximal fibrin formation at 405 nm (Figure S1; certain representative curves are shown).
FIGURE 1.
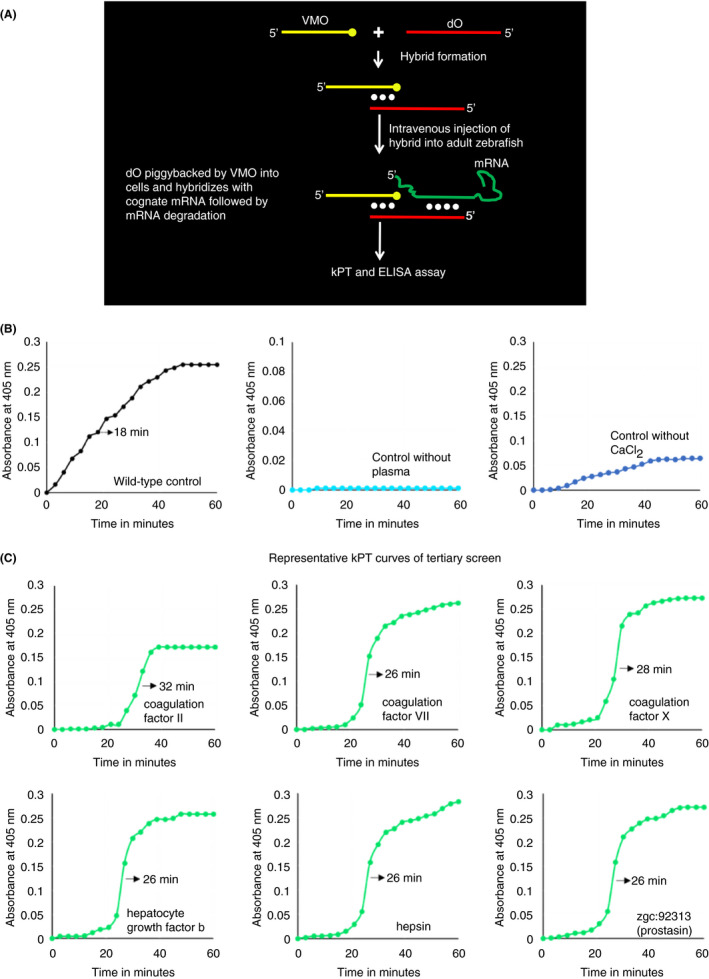
Piggyback knockdown screening and kinetic prothrombin time assay. (A) Schematic diagram of the piggyback knockdown screening method. A 25 nucleotide VMO (shown as a yellow line with a solid yellow circle representing the octa‐guanidine dendrimer at the 3′ end) is mixed with 40‐nucleotide‐long antisense deoxyoligonucleotide (dO), shown as red line, and consists of 25‐nucleotide‐long antisense sequence and 15 nucleotide long sequence, that hybridizes with the VMO. Hybrid formation is shown by base pairing (represented by solid white circles). Following intravenous injections, the hybrid enters the cells and the antisense portion of the oligonucleotide pairs with the cognate mRNA. This RNA:DNA hybrid is recognized by RNA degradation machinery, which degrades mRNA, resulting in knockdown of the gene. At 24 hour after the second injection, blood was collected, plasma prepared, and kPT/ELISA assay was performed. (B) Controls for the knockdown kPT screens. The first panel from left shows a representative kPT curve obtained using wild‐type control zebrafish plasma after recalcification. The second panel shows a representative kPT curve obtained without zebrafish plasma. The third panel shows a representative kPT curve obtained using zebrafish plasma without the addition of CaCl2. (C) Representative tertiary knockdown screen. The 14 genes that showed prolonged kPT after secondary knockdown screen were subjected to the tertiary knockdown screen by the piggyback knockdown method. Plasma from zebrafish was obtained after knockdown hybrid injection and used in kPT assay. Representative kPT assays are shown. The gene and the corresponding curve are shown in each panel. The arrow shows the time taken to reach half maximal fibrin formation at 405 nm. kPT, kinetic prothrombin time; VMO, vivo morpholino.
The 21 above‐stated genes (zgc:123295, zgc:136461, f2, coagulation factor IX like [f9‐like], f7, f10, elastase3 like, chymotrypsin like protease CTRL‐1, acrosin like, si:dkeyp‐93a5.3, hgfb, magi3a [membrane associated guanylate kinase, WW and PDZ domain containing 3a], prss56 [serine protease 56], si:dkey‐32n7.7, si:dkey‐21e2.12, si:dkey‐33m11.7 [trypsin 3 like], complement factor b like, hepsin, prss23 [serine protease 23], tmprss12 [transmembrane serine protease 12], and zgc:92313 [prostasin]) were subjected to secondary knockdown. A new batch of fish was injected with each hybrid for the secondary knockdown screen in the same manner that was described in the primary screen. In this secondary screen, 14 of 21 gene knockdowns (Table S2) yielded prolonged kPT (Figure S2; only certain representative curves are shown). Subsequently, a tertiary knockdown of these 14 genes (zgc:123295, zgc:136461, f2, f7, f10, acrosin like, hgfb, magi3a, prss56, si:dkey‐32n7.7, si:dkey‐21e2.12, hepsin, tmprss12, and zgc:92313) was performed using another new batch of fish as described above and screened with the kPT assay again. All 14 gene knockdowns continued to yield prolonged kPTs (Figure 1C and Figure S3). Thus, of 179 genes coding for serine proteases, 14 knocked‐down genes were found to affect the extrinsic coagulation cascade. Furthermore, we analyzed the oligonucleotide sequences and found that the sequences were correct and were antisense to serine protease gene sequences.
To elucidate their role in Fvii activation, knockdown hybrids of each of these 14 genes were again injected into 6 separate zebrafish, and plasma from these fish were analyzed via the Fviia ELISA assay. Six (f2, f7, f10, hepsin, prostasin, and hgfb) of the 14 knocked‐down genes resulted in lower levels of Fviia antigen compared to wild‐type (controls) (Figure 2A). Next, we tested whether the knockdown of these six genes resulted in the alteration of Fvii levels. The knockdown hybrids for each of the above six genes were again injected into six separate zebrafish, and plasmas from these fish were analyzed with the Fvii ELISA assay. Knockdown of f7 resulted in a significant reduction of Fvii levels. However, knockdown of hgfb and prostasin resulted in a modest reduction in Fvii levels (Figure 2B), which were significant compared to wild‐type (controls).
FIGURE 2.

Estimation of Fviia and Fvii in zebrafish plasma via ELISA. (A) The 14 genes that were found to delay fibrin formation following the tertiary knockdown screen, were analyzed for their effect on Fviia levels, after knocking these genes down individually. Six fishes were injected with the piggyback knockdown hybrids for each gene and plasma was obtained from individual fishes for the estimation of Fviia levels. The average of 6 individual plasma samples plotted as relative concentration of Fviia in percentage (SD) is shown. The statistical significance between wild‐type control and knockdown sample is shown by * in the graph (P = <.001). (B) The six genes, the individual knockdown of which lowered Fviia levels, were analyzed for Fvii levels. Six fishes were injected with the piggyback knockdown hybrids for each gene and plasma was obtained from individual fishes for the estimation of Fvii levels. The average of six individual plasma samples plotted as relative concentration of Fvii in percentage (SD) is shown. The statistical significance between wild‐type control and knockdown sample is shown by * in the graph (P = <.001)
4. DISCUSSION
The current study established a large‐scale knockdown screening method for 179 proteins containing serine protease domains. To select this list, we used annotated information from the NCBI/ENSEMBL database. The possibility exists that the names cited in these annotations may not be accurate, but they do carry serine protease domains. Knockdown of f2, f7, f10, hepsin, prostasin, and hgfb led to a reduction in Fviia levels. Of the above genes, f2, f7, and f10 gene products are known to be involved in human FVII activation. Our laboratory identified hepsin product as showing potential for zebrafish Fvii activation. 14 This indicates that the remaining two genes, prostasin and hgfb, may play a role in the Fvii activation pathway of zebrafish. Prostasin encodes a GPI‐anchored protein containing a catalytic triad for trypsin‐like serine protease activity, while hgfb encodes the hepatocyte growth factor b that lacks this catalytic triad.
The knockdown of zgc:123295, zgc:136461, acrosin like, magi3a, serine protease 56, si:dkey‐32n7.7, si:dkey‐21e2.12, serine protease 23, and tmprss12 did not affect Fviia levels. The mechanism underlying the effect of the knockdown of these genes on kPT remains unknown. The gene MAGI3 is involved in increasing the levels of TF. Thus, it may positively contribute to Fviia production. The gene, TMPRSS12, encodes a hepsin‐like protease. Since tmprss12 in zebrafish is not completely characterized, it is likely that this may correspond to original hepsin in humans, even though synteny argues against it. Furthermore, zgc:123295 is an uncharacterized serine protease; zgc:136461 is a chymotrypsinogen 2–like precursor; serine protease 56 is again a novel serine protease; si:dkey‐32n7.7 appears to be an orthologue of serine protease 56; and finally serine protease 23 is a novel serine protease. Although these proteases may not be involved in the direct activation of coagulation factors vii, x, ii and fibrinogen, they may activate receptors on cells that produce the above factors and thereby govern their activity indirectly. For example, si:dkey‐21e2.12 encodes a granzyme‐like protein, and granzyme has been shown to cleave protease‐activated receptor 1. 19 Future studies are needed to establish these roles.
Fviia and Fvii levels were evaluated using ELISAs. In these assays, human FVIIa and FVII were used to prepare standard curves. This was due to the lack of zebrafish Fviia and Fvii standards. Therefore, the estimated levels of proteins were relative and not expressed as absolute units, and are thus presented as relative percent concentrations of Fviia or Fvii. Despite these estimates, our screening approach is substantiated by the fact that genes known to regulate Fviia levels, such as f2, f7, f10, and hepsin, were identified by the present unbiased screen. Spontaneous bleeding in embryos/larvae was observed in a previous study of ours that investigated f2 knockdown. 20 However, such spontaneous bleeding in adult zebrafish was not observed following the knockdown of either f2 or any of the other serine proteases mentioned above. This difference may be due to the transparency of the embryos, which made them easily visible in our previous study, whereas in adults, minor bleeding may not be immediately obvious.
The gene hgfb may exert effects that are similar to those exerted by the hepatocyte growth factor, which correlates with f7 transcription and, thereby affects Fviia levels. 21 However, knockdown of hgfb did not reduce Fvii levels drastically, compared to Fviia in zebrafish plasma, suggesting that Fviia levels may be regulated via a different mechanism. Since the serine protease domain is mutated and inactive in Hgfb, it is unable to cleave Fvii. However, it is possible that Hgfb binds to Fvii, thereby inducing the autoactivation of Fvii. This mechanism remains to be proven. Furthermore, regulation of Fviia levels by prostasin may be considered a novel finding. In this case, Fvii levels were not drastically reduced compared to Fviia levels. Thus, in addition to other proteases, prostasin also appears to be involved in Fvii activation. However, the fact that Fvii levels were only slightly reduced due to the knockdown of both hgfb and prostasin is difficult to explain. It is probable that, in addition to their role in Fviia generation, they may also be partially affecting Fvii levels via an unknown mechanism. Reportedly, prostasin, which contains a catalytically active site, has been shown to regulate epithelial cell sodium channels and modulate epidermal growth factor receptor signaling. 22 Its soluble form is present in seminal fluid. It is also expressed in the liver, where FVII is synthesized. Thus, it is possible that the soluble form of prostasin present in plasma may indirectly exert protease activity on FVII when secreted to generate FVIIa. However, the mechanism of FVII activation by prostasin is poorly understood and needs further exploration.
This study generated the first large‐scale knockdown screen using zebrafish. Each of the primary, secondary, and tertiary knockdown screens involved a single fish receiving a hybrid injection, wherein a different batch of fishes was used for each screen. The very nature of such a design reduces the probability of finding background mutations in the wild‐type population using three sequential screens to virtually nil, thereby ruling out the chances of finding a false positive. Thus, this type of screening may be considered as the ultimate method for finding positives because a gene is scored as positive, only when it turns out to be positive in three rounds of screening. In this rapid screening approach using single fish injections, there is a chance of missing genes. For example, although we found that the f9‐like gene knockdown resulted in prolonged kPT in the primary screen, this result was not repeated in the secondary screen, resulting in f9‐like being eliminated without being considered for the tertiary screen (Table S1). This is because intravenous injections are not successful 100% of the time. Moreover, variations that occur during blood collection may cause differences in the kPT assay. This discrepancy may be alleviated by using 6‐12 fish injections for each hybrid in future screens to obtain statistically significant results.
In summary, knockdown of large‐scale serine proteases in zebrafish genome using piggyback technology, along with screening via kPT, Fviia and Fvii ELISA assays suggested a novel role for prostasin and Hgfb in the Fvii activation pathway. This may mark the beginning of large‐scale piggyback knockdown screening in zebrafish hemostasis studies. The current study indicated that, in addition to previously known factors, prostasin and Hgfb also play a role in activating Fvii. Thus, the initial generation of Fviia appears to be a multifactorial event.
RELATIONSHIP DISCLOSURE
The authors declare no conflicts of interest.
AUTHOR CONTRIBUTIONS
GK maintained zebrafish, designed primers, performed piggyback knockdowns, coagulation assays, analyzed the data, and participated in discussions. NI performed factor VII assays, prepared the tables and figures, and participated in discussions. PJ designed the research, analyzed the data, and wrote the paper.
Supporting information
Fig S1
Fig S2
Fig S3
Table S1
Table S2
Fig S1‐S3‐Cap
ACKNOWLEDGMENTS
The work was supported by the funds from NIH grants HL077910 and DK117384.
Khandekar G, Iyer N, Jagadeeswaran P. Prostasin and hepatocyte growth factor B in factor VIIa generation: Serine protease knockdowns in zebrafish. Res Pract Thromb Haemost. 2020;4:1150–1157. 10.1002/rth2.12428
Handling Editor: Alisa Wolberg
Contributor Information
Gauri Khandekar, @gauri_khandekar.
Neha Iyer, @NehaIyer3.
Pudur Jagadeeswaran, Email: jag@unt.edu, @PuurJ.
REFERENCES
- 1. Mackman N, Tilley RE, Key NS. Role of the extrinsic pathway of blood coagulation in hemostasis and thrombosis. Arterioscler Thromb Vasc Biol. 2007;27(8):1687–93. [DOI] [PubMed] [Google Scholar]
- 2. Hedner U. Recombinant activated factor VII as a universal haemostatic agent. Blood Coagul Fibrinolysis. 1998;9(Suppl 1):S147–S152. [PubMed] [Google Scholar]
- 3. Lu G, Broze GJ Jr, Krishnaswamy S. Formation of factors IXa and Xa by the extrinsic pathway: differential regulation by tissue factor pathway inhibitor and antithrombin III. J Biol Chem. 2004;279(17):17241–9. [DOI] [PubMed] [Google Scholar]
- 4. Pedersen AH, Lund‐Hansen T, Bisgaard‐Frantzen H, Olsen F, Petersen LC. Autoactivation of human recombinant coagulation factor VII. Biochemistry. 1989;28(24):9331–6. [DOI] [PubMed] [Google Scholar]
- 5. Nakagaki T, Foster DC, Berkner KL, Kisiel W. Initiation of the extrinsic pathway of blood coagulation: evidence for the tissue factor dependent autoactivation of human coagulation factor VII. Biochemistry. 1991;30(45):10819–24. [DOI] [PubMed] [Google Scholar]
- 6. Kisiel W, Fujikawa K, Davie EW. Activation of bovine factor VII (proconvertin) by factor XIIa (activated Hageman factor). Biochemistry. 1977;16(19):4189–94. [DOI] [PubMed] [Google Scholar]
- 7. Butenas S, Mann KG. Kinetics of human factor VII activation. Biochemistry. 1996;35(6):1904–10. [DOI] [PubMed] [Google Scholar]
- 8. Wildgoose P, Kisiel W. Activation of human factor VII by factors IXa and Xa on human bladder carcinoma cells. Blood. 1989;73(7):1888–95. [PubMed] [Google Scholar]
- 9. Radcliffe R, Nemerson Y. Activation and control of factor VII by activated factor X and thrombin. Isolation and characterization of a single chain form of factor VII. J Biol Chem. 1975;250(2):388–95. [PubMed] [Google Scholar]
- 10. Romisch J. Factor VII activating protease (FSAP): a novel protease in hemostasis. Biol Chem. 2002;383(7–8):1119–24. [DOI] [PubMed] [Google Scholar]
- 11. Kazama Y, Hamamoto T, Foster DC, Kisiel W. Hepsin, a putative membrane‐associated serine protease, activates human factor VII and initiates a pathway of blood coagulation on the cell surface leading to thrombin formation. J Biol Chem. 1995;270(1):66–72. [DOI] [PubMed] [Google Scholar]
- 12. Stavenuiter F, Dienava‐Verdoold I, Boon‐Spijker MG, Brinkman HJ, Meijer AB, Mertens K. Factor seven activating protease (FSAP): does it activate factor VII? J Thromb Haemost. 2012;10(5):859–66. [DOI] [PubMed] [Google Scholar]
- 13. Wu Q, Yu D, Post J, Halks‐Miller M, Sadler JE, Morser J. Generation and characterization of mice deficient in hepsin, a hepatic transmembrane serine protease. J Clin Invest. 1998;101(2):321–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Khandekar G, Jagadeeswaran P. Role of hepsin in factor VII activation in zebrafish. Blood Cells Mol Dis. 2014;52(1):76–81. [DOI] [PubMed] [Google Scholar]
- 15. Westerfield M. The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio). 5th ed. Eugene, OR: University of Oregon Press; 2007. [Google Scholar]
- 16. Sundaramoorthi H, Khandekar G, Kim S, Jagadeeswaran P. Knockdown of αIIb by RNA degradation by delivering deoxyoligonucleotides piggybacked with control vivo‐morpholinos into zebrafish thrombocytes. Blood Cells Mol Dis. 2015;54(1):78–83. [DOI] [PubMed] [Google Scholar]
- 17. Jagadeeswaran P, Sheehan JP. Analysis of blood coagulation in the zebrafish. Blood Cells Mol Dis. 1999;25(3–4):239–49. [DOI] [PubMed] [Google Scholar]
- 18. Jagadeeswaran P, Gregory M, Johnson S, Thankavel B. Haemostatic screening and identification of zebrafish mutants with coagulation pathway defects: an approach to identifying novel haemostatic genes in man. Br J Haematol. 2000;110(4):946–56. [DOI] [PubMed] [Google Scholar]
- 19. Lee PR, Johnson TP, Gnanapavan S, Giovannoni G, Wang T, Steiner JP, et al. Protease activated receptor‐1 activation by granzyme B causes neurotoxicity that is augmented by interleukin‐1beta. J Neuroinflammation. 2017;14(1):131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Day K, Krishnegowda N, Jagadeeswaran P. Knockdown of prothrombin in zebrafish. Blood Cells Mol Dis. 2004;32(1):191–8. [DOI] [PubMed] [Google Scholar]
- 21. Chiu B, Melin‐Aldana H, Pillai S, Chu F, Superina RA. Factor VII transcription correlates with hepatocyte proliferation and hepatocyte growth factor expression in a rodent extrahepatic portal vein obstruction model. J Am Coll Surg. 2007;205(2):277–83. [DOI] [PubMed] [Google Scholar]
- 22. Aggarwal S, Dabla PK, Arora S. Prostasin: an epithelial sodium channel regulator. J Biomark. 2013;2013:179864. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Fig S3
Table S1
Table S2
Fig S1‐S3‐Cap


