Abstract
Since intraductal papillary mucinous neoplasms (IPMNs) occasionally contain pancreatic malignancies, it is vital to develop a screening program that can detect IPMNs in the general population and that can identify IPMNs with high malignant potential. The present study investigated whether microRNAs (miRNAs/miRs) in the blood may be diagnostic markers for IPMN screening. Initially, extracellular vesicle-encapsulated miRNAs (EV-miRNAs) in the serum with altered expression between IPMN, IPMN-derived carcinoma (IPMC) and control samples, were identified using microarray analysis. To validate the microarray results, the expression levels of selected EV-miRNAs were detected. Briefly, serum EV-miRNAs were extracted from 38 patients with IPMN (11 patients with IPMC and 27 patients with benign IPMN) and 21 non-tumor controls. The results of the microarray analysis revealed that the expression levels of EV-miR-22-3p, EV-miR-4539 and EV-miR-6132 were higher in the IPMN and IPMC serum samples compared with those in the control samples. With regards to discriminating IPMNs from controls, only miR-4539 exhibited a significant difference (P=0.004). In the comparison between IPMN and IPMC, carcinogenic antigen 19-9 (CA19-9) and EV-miR-6132 exhibited significant differences (P=0.01 and P=0.007, respectively). Receiver operating characteristic (ROC) curve analysis demonstrated that EV-miR-4539 could discriminate patients with IPMNs from control patients, with an area under the curve (AUC) of 0.72. Additionally, ROC analysis indicated that the markers could discriminate patients with IPMC from benign IPMN, with AUC values of 0.77 for EV-miR-6132 and 0.74 for CA19-9. In conclusion, the present study suggested that EV-miRNAs may be used as diagnostic markers for the detection of IPMNs in the general population as well as for identifying IPMNs with high malignant potential.
Keywords: intraductal papillary mucinous neoplasm, intraductal papillary mucinous neoplasm-derived carcinoma, extracellular vesicle, microRNA, screening
Introduction
Intraductal papillary mucinous neoplasms (IPMNs) are epithelial neoplasms composed of mucin-producing columnar cells, and IPMNs are classified as 2 types: Main-duct type (MD-IPMN) and branch-duct type IPMN (BD-IPMN). Since IPMN occasionally develops into malignancy (IPMN-derived carcinoma: IPMC) (MD-IPMN: Range, 36–100%, BD-IPMN: Range, 6.3–46.5%), we need to develop a screening strategy that can detect IPMN in the general population and identify IPMC (1,2). For a reliable screening program, establishment of diagnostic tests with low invasiveness, high diagnostic yield and high reproducibility is essential. In this context, biomarkers from body fluids can play an important role in IPMN screening.
A considerable number of biomarker studies have evaluated markers for use in detecting IPMCs among IPMN patients (3–6). Carcinogenic antigen 19-9 (CA19-9) is the only biomarker described in the guidelines; however, a recent meta-analysis reported its diagnostic performance as highly specific but not sensitive in identifying IPMC in IPMNs (7). Moreover, no biomarker has demonstrated reliable diagnostic ability in detecting IPMNs in the general population. To overcome this challenge, we need to establish new biomarkers for IPMN screening.
Circulating microRNAs (miRNAs) have been shown to have diagnostic potential as blood biomarkers for various tumors (8–11). miRNAs are small, noncoding RNAs (18–25 nucleotides in length) that regulate gene expression at the posttranscriptional level by promoting the degradation of messenger RNAs or by blocking messenger RNA translation. They can be loaded into exosomes or other extracellular vesicles (extracellular vesicle-encapsulated miRNA: EV-miRNA) or exist freely in the blood circulation (circulating miRNA). Among these two forms, EV-miRNA has several advantages over circulating miRNA: i) It can be specifically released for cell-to-cell communication, and ii) lipid membrane coverage protects miRNA from RNase degradation (12).
In this study, we aimed to determine whether serum EV-miRNAs could be diagnostic markers of IPMNs in the general population. In addition, the diagnostic yield of serum EV-miRNAs in distinguishing IPMNs and IPMC was evaluated.
Materials and methods
Patients and treatments
First, we selected 4 patients with IPMNs (2 benign IPMNs and 2 IPMCs) and 4 controls without any type of tumor for microarray analysis to identify differences in the gene expression of serum EV-miRNAs between these groups. To validate the microarray results, we enrolled 38 consecutive patients who were diagnosed with IPMNs by imaging modalities at Fukushima Medical University Hospital and 21 patients without any neoplastic lesions as controls between June 2015 and November 2019. The diagnostic criteria were as follows: i) Dilation of the MD and/or a cystic dilation of the BD, and ii) secretion of mucin from the major or minor papilla identified by endoscopic retrograde cholangiopancreatography or duodenoscopy.
Clinical data including age, sex, background diseases in controls, presence of symptoms (jaundice, body weight loss, etc.), history of diabetes mellitus, pancreatitis, smoking, family history of pancreatic tumor, and serum tumor markers, including carcinoembryonic antigen (CEA) and CA 19-9, were retrieved from electronic medical records. Patients with co-existing tumors or active infection (i.e., cholangitis or cholecystitis) were excluded from the analysis. All recruited patients were evaluated with CT or MRI, and the location of the lesion, maximum diameter of cyst and main pancreatic duct (MPD), and the presence of mural nodules were determined. Medical management of the disease was also evaluated. Classification of BD-IPMN and others was performed according to the 2017 International Consensus Guideline (1).
The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the institutional review board of Fukushima Medical University (IRB #2387). All participants provided written informed consent.
Sample collection
To obtain the serum samples, 8 ml of blood was collected and incubated at room temperature for at least 60 min to allow clotting. Samples were then centrifuged at 1,000 × g for 10 min. The serum was collected and stored in aliquots at −80°C.
miRNA preparation and microarrays
Serum miRNA was extracted from serum using the exoRNeasy Serum/Plasma Midi kit (Qiagen) according to the manufacturer's protocol. RNA quantity and quality were determined using an Agilent Bioanalyzer (Agilent Technologies), as recommended. miRNA microarrays were manufactured by Agilent Technologies. Briefly, RNA was dephosphorylated using calf intestinal alkaline phosphatase (CIP) master mix incubated at 37°C for 30 min. Dephosphorylated RNA was denatured with DMSO incubated at 100°C for 5 min and then immediately transferred to ice for 2 min. These products were mixed with a ligation master mix for T4 RNA ligase and Cy3-pCp (Cyanine 3-Cytidine biphosphate) and incubated at 16°C for 2 h. Labeled RNA was dried using a vacuum concentrator at 55°C for 1.5 h. Cy3-pCp-labeled RNA was hybridized on an Agilent SurePrint G3 Human miRNA 8×60K Rel.21 (design ID: 070156) array at 55°C for 20 h. After washing, microarrays were scanned using an Agilent SureScan Microarray Scanner System (G4900DA). The intensity values of each scanned feature were quantified using Agilent Feature Extraction software version 12.1.1.1, which performs background subtractions. We only used features that were flagged as no errors (detected flags) and excluded features that were not positive, not significant, not uniform, not above background, saturated, and population outliers (not detected flags). These expression analyses were performed with Agilent GeneSpring GX version 14.9.1.
Digital PCR
Digital PCR and quantification of the absolute levels of serum miRNAs were performed using the Quant-Studio 3D Digital PCR system (Thermo Fisher Scientific, Inc.). Data were analyzed using QuantStudio 3D Analysis Suite Cloud Software (Thermo Fisher Scientificc, Inc.). The digital PCR mixture contained 5.0 µl of the RT product, 1.0 µl of nuclease-free H2O, 7.50 µl of the QuantStudio™ 3D Digital PCR Master Mix, and 0.75 µl of the TaqMan MiRNA Assay-1 (20X) for let-7d. Samples were individually loaded onto the QuantStudio 3D digital PCR 20K chip kit v2 using the QuantStudio 3D digital PCR Chip Loader. Digital PCR was performed in a Proflex 2X flat block thermal cycler (Applied Biosystems) using standard conditions: 96°C for 10 min followed by 39 cycles of 60°C for 2 min, 98°C for 30 sec, and 60°C for 2 min. Chips were read on the QuantStudio 3D digital PCR instrument, and the number of FAM-positive and FAM-negative (empty) wells was quantified (13,14).
Target gene prediction and pathway enrichment analyses
Target gene prediction was performed using DIANA-miRPath software (15). Specifically, we investigated whether tumor suppressor genes were targeted by these 3 miRNAs because aberrant tumor suppressor gene methylation was observed in malignant IPMN (16).
To predict cell signaling pathways that were potentially influenced by the EV-miRNAs, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed via the Database for Annotation, Visualization and Integrated Discovery (DAVID 6.8; http://david.abcc.ncifcrf.gov/) (17,18). KEGG pathways with a P-value <0.05 were considered significantly enriched.
Statistical analysis
Continuous variables (i.e., age, cyst size, serum CEA and CA 19-9 levels and GS) are reported as the median and range and were compared using Mann-Whitney analysis. Categorical variables (i.e., sex and location of disease) were analyzed using Fisher's exact test. The diagnostic yield of CEA and CA 19-9 levels and EV-miRNAs in distinguishing whole IPMN (benign IPMN and IPMC) from the control and IPMC from benign IPMN was assessed using the area under the receiver operating characteristic (ROC) curve. Statistical analyses were performed using SPSS version 26.0 for Windows (IBM Corp.), and figures were generated with Prism 7 (GraphPad Software, Inc.). P<0.05 was considered to indicate statistical significance.
Results
Identification of miRNAs for IPMN screening
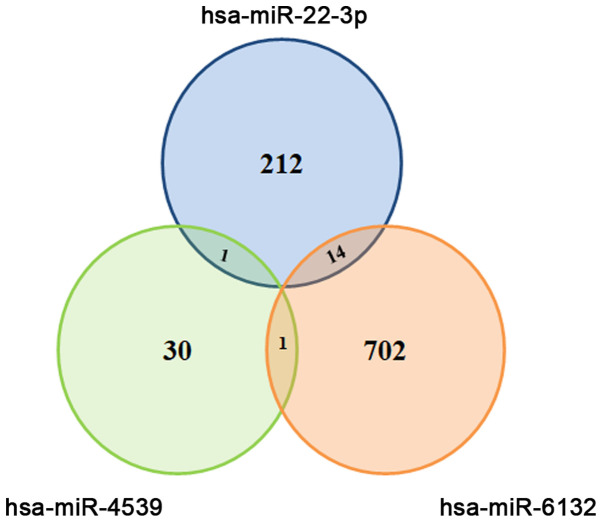
We first identified miRNAs showing altered expression in 2 IPMNs,2 IPMCs and 4 controls (Table I). Among the 2588 mature miRNAs evaluated in the microarray, we found 3 EV-miRNAs [hsa-miR-6132 (EV-miR-6132), hsa-miR-22-3p (EV-miR-22) and hsa-miR-4539 (EV-miR-4539)] that could discriminate IPMN, IPMC and control (Table II). As shown in Fig. 1, target prediction using DIANA-miRPath software revealed that these 3 miRNAs could target various genes. Regarding tumor suppressor genes, we found that miR-22-3p could target TP53INP1 and mir-6132 could target 3 genes (tuberous sclerosis complex 2: TSC2, tumor protein p53-inducible protein 11: TP53I11 and protein phosphatase 2 regulatory subunit B'Delta: PPP2R5D).
Table I.
Background disease of patients in microRNA array analysis.
| Sample nos. | Disease | Group name |
|---|---|---|
| 1-4 | Control | A1 |
| 5 and 6 | IPMC | A2 |
| 7 and 8 | IPMN | A3 |
IPMN, intraductal papillary mucinous neoplasm; IPMC, IPMN-derived carcinoma.
Table II.
Different expressions of miRNAs between each group.
| Systematic name | FC [(A2) vs. (A3)] | Log FC [(A2) vs. (A3)] | FC [(A2) vs. (A1)] | Log FC [(A2) vs. (A1)] | FC [(A3) vs. (A1)] | Log FC [(A3) vs. (A1)] |
|---|---|---|---|---|---|---|
| hsa-miR-6132 | 4.55 | 2.19 | 21.55 | 4.43 | 4.74 | 2.24 |
| hsa-miR-22-3p | 2.92 | 1.55 | 7.54 | 2.91 | 2.58 | 1.37 |
| hsa-miR-4539 | 2.94 | 1.56 | 7.54 | 2.91 | 2.56 | 1.36 |
| hsa-miR-6732-3p | 9.31 | 3.22 | 7.54 | 2.91 | −1.23 | −0.30 |
| hsa-miR-4534 | 2.16 | 1.11 | 2.30 | 1.20 | 1.06 | 0.09 |
| hsa-miR-3679-5p | 1.83 | 0.87 | 1.60 | 0.67 | −1.15 | −0.20 |
miR/miRNA, microRNA; FC, fold change.
Figure 1.
Selection of candidate EV-miRNAs for identifying benign IPMNs and IPMC. Target gene prediction using DIANA-miRPath software revealed that EV-miR-22-3p could target 227 genes, EV-miR-4539 could target 32 genes and EV-miR-6132 could target 717 genes. Moreover, several genes were targeted by 2 EV-miRNAs. EV, extracellular vesicle; miRNA/miR, microRNA; IPMN, intraductal papillary mucinous neoplasm; IPMC, IPMN-derived carcinoma.
In addition, pathway enrichment analyses found that these 3 miRNAs could influence the NOD-like receptor signaling pathway (6 target genes involved), glycosphingolipid biosynthesis-lacto and neolacto series-(1 target gene involved) and fat digestion and absorption (5 target genes involved) (Table III).
Table III.
Results of KEGG pathway analysis.
| KEGG pathways | P-value | Involved genes |
|---|---|---|
| KEGG pathway: miR-22-3p and miR-6132 | ||
| NOD-like receptor signaling pathway | 0.04 | NF-kappa-B inhibitor beta (NFKBIB), Caspase 1 (CASP1), NACHT, LRR and PYD domains-containing protein 3 (NLRP3), mitogen-activated protein kinase 11 (MAPK11), Suppressor of G2 allele of SKP1 homolog (SUGT1), mitogen-activated protein kinase 1 (MAPK1) |
| KEGG pathway: miR-22-3p and miR-4539 | ||
| Glycosphingolipid biosynthesis-lacto and neolacto series | 0.002 | Fucosyltransferase 9 (FUT9) |
| KEGG pathway: miR-4539 and miR-6132 | ||
| Fat digestion and absorption | 0.04 | Fatty acid binding protein 1 (FABP1), Apolipoprotein A4 (APOA4), CD36, microsomal triglyceride transfer protein (MTTP), Phospholipase A2 Group IIC (PLA2G2C) |
KEGG, Kyoto Encyclopedia of Genes and Genomes; miR, microRNA.
Clinical characteristics of the patients
To validate whether the 3 miRNAs could be diagnostic markers for IPMN, we conducted a validation study using digital PCR. The clinical characteristics of patients in the IPMN (n=38) and control (n=21) groups are presented in Table IV. Briefly, there were no significant differences in age, sex, absence of symptoms, history of diabetes, pancreatitis, smoking, or family history of pancreatic cancer between the IPMN group and the control group. With regard to management, a total of 9 patients underwent surgery among 38 IPMN patients. We confirmed the pathological diagnosis of the resected cases according to the latest classification (1): Low-grade dysplasia (n=4), high-grade dysplasia (n=1), and invasive carcinoma (n=4). Twenty-eight patients did not require surgery and were continuously observed. One patient was treated with chemotherapy.
Table IV.
Clinical characteristics of patients with IPMN and controls.
| Characteristics | Control (n=21) | IPMN (n=38) | P-value |
|---|---|---|---|
| Median age, years (range) | 71.0 (46.0–89.0) | 74.0 (47.0–91.0) | 0.51 |
| Sex, male (%) | 13 (61.9) | 19 (50.0) | 0.42 |
| Background disease | |||
| Bile duct stone, n (%) | 14 (66.7) | ||
| Chronic pancreatitis, n (%) | 7 (33.3) | ||
| Presence of symptoms, n (%) | 3 (14.2) | 7 (18.4) | 0.99 |
| Hx of diabetes mellitus, n (%) | 4 (19.0) | 11 (28.9) | 0.53 |
| Hx of pancreatitis, n (%) | 2 (9.5) | 2 (5.3) | 0.61 |
| Hx of smoking, never/ever/current | 9/6/6 | 19/9/10 | 0.29 |
| Family Hx of pancreatic tumor (%) | 0 (0.0) | 2 (5.3) | 0.53 |
| IPMN subtype, branch-duct, n (%) | 25 (65.8) | ||
| IPMC, n (%) | 11 (28.9) | ||
| Management, n (%) | |||
| Follow-up | 21 (100.0) | 28 (73.6) | |
| Surgical resection | 9 (23.6) | ||
| Chemotherapy | 1 (2.6) |
Hx, history; IPMN, intraductal papillary mucinous neoplasm; IPMC, IPMN-derived carcinoma.
We also divided IPMNs into 2 groups, namely, IPMC (n=11) and benign IPMN without any suspicious findings of malignancy (n=27), and compared clinical characteristics as shown in Table V. Clinical symptoms were observed more frequently in IPMC than IPMN (36.3 vs. 0.0%, P=0.0001). Additionally, the median diameter of the MPD was significantly larger in IPMC than in IPMN (11.3 vs. 4.4 mm, P=0.0003).
Table V.
Clinical characteristics of patients with IPMC and IPMN.
| Characteristics | Benign IPMN (n=27) | IPMC (n=11) | P-value |
|---|---|---|---|
| Median age, years (range) | 69.5 (48.0–89.0) | 74.5 (47.0–91.0) | 0.71 |
| Sex, male (%) | 11 (40.7) | 8 (63.6) | 0.15 |
| Presence of symptoms, n (%) | 0 (0.0) | 7 (36.3) | 0.0001 |
| Hx of diabetes mellitus, n (%) | 7 (33.3) | 4 (33.3) | 0.99 |
| Hx of pancreatitis, n (%) | 2 (7.4) | 0 (0.0) | 0.99 |
| Hx of smoking, never/ever/current | 15/5/7 | 5/5/1 | 0.29 |
| Family Hx of pancreatic tumor (%) | 1 (3.7) | 1 (9.1) | 0.50 |
| Location of the lesion, Ph/Pb/Pt/diffuse | 12/7/4/4 | 6/3/0/2 | 0.60 |
| Median cyst diameter, mm (range) | 24.5 (12.0–63.0) | 31.0 (10.0–170.0) | 0.09 |
| Median MPD diameter, mm (range) | 4.4 (1.5–13.0) | 11.2 (2.2–33.0) | 0.0003 |
| Median size of mural nodule, mm (range) | 8.1 (3.0–15.0) | 18.0 (4.0–50) | 0.02 |
Hx, history; Ph/Pb/Pt, pancreatic head, body and tail; MPD, main pancreatic duct; IPMN, intraductal papillary mucinous neoplasm; IPMC, IPMN-derived carcinoma.
miRNAs as diagnostic markers for IPMN
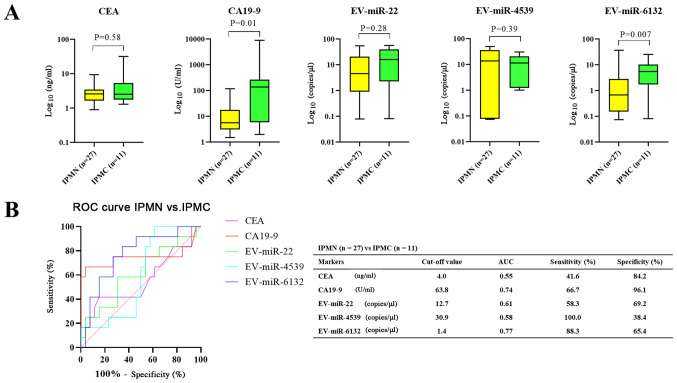
Fig. 2 shows the differences in each biomarker between the control and IPMN (both benign IPMN or IPMC) groups (Fig. 2) and between benign IPMN and IPMC groups (Fig. 3). With regard to discriminating IPMNs from control, only EV-miR-4539 showed a significant difference (P=0.004) (Fig. 2A). ROC analysis showed that 5 markers could discriminate patients with IPMN and from control patients, with areas under the curve (AUCs) of 0.72 for EV-miR-4539 (95% confidence interval [CI]: 0.59–0.85), 0.55 for CEA (95% CI, 0.39–0.71), 0.55 for CA19-9 (95% CI, 0.41–0.71), 0.59 for EV-miR-22 (95% CI, 0.42–0.76) and 0.64 for EV-miR-6132 (95% CI, 0.47–0.81). As shown in Fig. 2B, EV-miR-4539 had the highest diagnostic yield compared with other markers at the cutoff value of 3.2 copies/µl, and the sensitivity and specificity were 60.5 and 95.2%, respectively (Fig. 2B).
Figure 2.
Comparison of serum biomarkers between the control and IPMN groups. (A) Only EV-miR-4539 showed a statistically significant difference. (B) EV-miR-4539 showed the highest diagnostic yield among the markers. Data were presented in box-and-whisker plot and error bars in figures were presented with range (minimum to maximum). EV, extracellular vesicle; miRNA/miR, microRNA; IPMN, intraductal papillary mucinous neoplasm; CEA, carcinoembryonic antigen; CA19-9, carcinogenic antigen 19-9; ROC, receiver operating characteristic; AUC, area under the curve.
Figure 3.
Comparison of serum biomarkers between benign IPMN and IPMC groups. (A) CA19-9 EV-miR-6132 showed a significant difference. (B) miR-6132 had the highest diagnostic yield among the markers. Data were presented in box-and-whisker plot and error bars in figures were presented with range (minimum to maximum). EV, extracellular vesicle; miRNA/miR, microRNA; IPMN, intraductal papillary mucinous neoplasm; IPMC, IPMN-derived carcinoma; CEA, carcinoembryonic antigen; CA19-9, carcinogenic antigen 19-9; ROC, receiver operating characteristic; AUC, area under the curve.
In the comparison between benign IPMN and IPMC, CA19-9 and EV-miR-6132 showed significant differences (P=0.01 and 0.007, respectively) (Fig. 3A). ROC analysis showed that the markers could discriminate patients with IPMC from patients with benign IPMN, with an AUC of 0.77 for EV-miR-6132 (95% CI, 0.61–0.93), 0.74 for CA19-9 (95% CI, 0.52–0.97), 0.61 for EV-miR-22 (95% CI, 0.41–0.81), 0.58 for EV-miR-4539 (95% CI, 0.40–0.77) and 0.55 for CEA (95% CI, 0.34–0.77). EV-miR-6132 showed the highest diagnostic yield compared with other markers at the cutoff value of 1.4 copies/µl and the sensitivity and specificity were 88.3 and 65.4%, respectively (Fig. 3B).
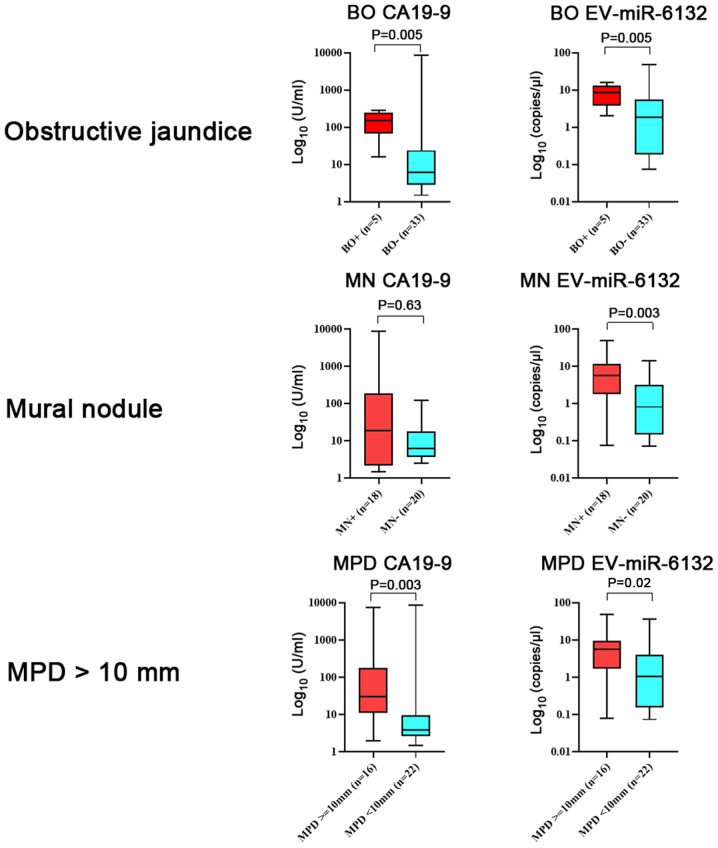
With regard to the serum levels of biomarkers and the existence of high-risk indicators, the serum level of CA19-9 was higher in patients with obstructive jaundice (Fig. 4). The serum level of EV-miR-6132 was higher in patients with high-risk indicators (mural nodules and MPD diameter more than 10 mm) than in patients without them.
Figure 4.
Expression levels of biomarkers per high-risk indicator. CA19-9 was highly expressed in patients with BO (P=0.005). Data were presented in box-and-whisker plot and error bars in figures were presented with range (minimum to maximum). CA19-9, carcinogenic antigen 19-9l EV, extracellular vesicle; miRNA/miR, microRNA; BO, biliary obstruction; MN, mural nodule; MPD, main pancreatic duct.
Discussion
In the present study, we aimed to confirm the potentially useful EV-miRNAs (EV-miR-22, EV-miR-4539 and EV-miR-6132) for IPMN and IPMC detection in microarray analysis. To the best of our knowledge, this is the first study that suggested the usefulness of EV-miRNAs in IPMN screening. It was found that EV-miR-4539 was better at distinguishing IPMNs from non-tumor controls than other biomarkers. While the diagnostic yield of EV-miR-6132 to discriminate IPMNs and IPMC was comparable to that of CA19-9, it had the advantage that the expression level was not influenced by biliary obstruction.
miRNAs can play multifunctional roles in cancer progression. With regard to IPMN, Habbe et al first reported abnormal miRNA expression in IPMN (19). In this study, the expression of miR-155 was found to be elevated in both IPMN tissue and pancreatic juice. Authors suggested that miRNA could be a diagnostic marker for pancreatic neoplasms. Subsequently, multiple studies focused on the identification of high-risk IPMN, and the results suggested that many miRNAs in the blood, pancreatic juice, and cystic fluid could be useful diagnostic markers. More recently, some researchers have attracted the attention of miRNAs encapsulated in EV-like exosomes since they are supposed to be stabilized to avoid degradation in the blood and highly enriched compared to circulating miRNAs. Goto et al (20) compared the diagnostic yield of serum circulating miRNAs and exosomal miRNAs in distinguishing control tissues from IPMNs. Their study was quite informative as the diagnostic yield of exosomal miRNAs was found to be 5–20% superior to that of serum circulating miRNAs (e.g., exosomal miR-21: AUC 0.826, accuracy 80%). Circulating miRNA-21: AUC 0.653, accuracy 62.3%). Although they did not focus on discriminating IPMCs from benign IPMNs, in contrast to our study, the results indicated the superiority of encapsulated miRNAs over circulating miRNAs as diagnostic markers.
While we cannot find any relevant data regarding miR-6132 and tumors, the expression of miR-22 and miR-4539 in cancer tissue has been studied in gastric cancer, rhabdomyosarcoma, breast cancer, prostate cancer, osteosarcoma and papillary thyroid cancer (21–30). Most studies have reported that, in contrast to their expression levels in the blood, the expression levels of these miRNAs are decreased in cancer tissue. Moreover, in vivo and in vitro studies regarding the biophysical properties of those miRNAs encapsulated in EVs have not been conducted. Hence, the mechanism by which the 3 EV-miRNAs in the blood contribute to IPMN progression has not been elucidated. As in silico analysis suggests, inhibition of the NOD-like receptor signaling pathway may be a cause of IPMN progression. NOD-like receptors are genetically conserved proteins that belong to the cellular pattern recognition receptor protein family. They are important components in the innate immune system of mammals, regulating the immune response and inflammatory response. Recently, accumulative evidence has extended the concept that the NOD-like receptor signaling pathway contributes to the activation of the antitumor immune response by priming antitumor CD4+ and CD8+ T cells (31). Considering this result, increased expression of EV-miRNAs might be attributable to suppression of the antitumor immune response and progression of IPMN. Regarding the other 2 pathways, glycosphingolipid biosynthesis-lacto and neolacto series- and fat digestion and absorption, we could not find any relevant studies.
Regarding progression of IPMN to IPMC, EV-miR-6132 may play an important role because its serum levels are higher in IPMC patients than in benign IPMN patients. Further evaluation revealed that 3 tumor suppressor genes (TSC2, TP53I11 and PPP2R5D) were found among 717 target genes. Aberrant tumor suppressor gene methylation which lead to gene suppression just like miRNA is often found in IPMC, and we speculate that the high level of EV-miR-6132 in the serum of IPMC patients may influence tumor suppressor gene activity as well (16).
Several limitations were found in this study. First, this study was conducted in a single referral center, and the results may not be generalizable to all patients with IPMNs and IPMCs. The relatively small sample size also limited the reliability of our statistical analysis. Second, only 9 of 38 IPMN patients underwent surgical resection. The remaining 29 patients were diagnosed with a benign IPMN or an IPMC based on the imaging findings and clinical outcomes. Third, we did not investigate the expression levels of the 3 miRNAs in the tumor tissue, and we do not know whether the miRNAs directly contribute to carcinogenesis or tumor progression. Therefore, we must conduct a further study that includes a large number of patients with histopathological evaluation.
In conclusion, we found that EV-miRNAs can be diagnostic markers for use in detecting IPMNs in the general population as well as in identifying IPMNs with high malignant potential.
Acknowledgements
The authors would like to thank Ms. Chikako Sato and Ms. Rie Hikichi (Department of Gastroenterology, Fukushima Medical University School of Medicine) for their assistance in the experiments.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during the present study are included in the published article.
Authors' contributions
YS, RS and HO conceived and designed the experiments. YS and RS performed the experiments. YS, RS, TT, MS and HO collected and analyzed the data. YS, RS, TT, MS and HO interpreted the results and wrote the manuscript. All authors read and approved the manuscript, and agree to be accountable for all aspects of the research and to guarantee for the accuracy and integrity of any part of the work.
Ethics approval and consent to participate
The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the institutional review board of Fukushima Medical University (IRB #2387). All participants provided written informed consent for participation.
Patient consent for publication
All participants provided written informed consent for publication.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Tanaka M, Fernandez-Del Castillo C, Kamisawa T, Jang JY, Levy P, Ohtsuka T, Salvia R, Shimizu Y, Tada M, Wolfgang CL. Revisions of international consensus Fukuoka guidelines for the management of IPMN of the pancreas. Pancreatology. 2017;17:738–753. doi: 10.1016/j.pan.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 2.Oyama H, Tada M, Takagi K, Tateishi K, Hamada T, Nakai Y, Hakuta R, Ijichi H, Ishigaki K, Kanai S, et al. Long-term risk of malignancy in branch-duct intraductal papillary mucinous neoplasms. Gastroenterology. 2020;158:226–237 e5. doi: 10.1053/j.gastro.2019.08.032. [DOI] [PubMed] [Google Scholar]
- 3.Al-Haddad M, DeWitt J, Sherman S, Schmidt CM, LeBlanc JK, McHenry L, Coté G, El Chafic AH, Luz L, Stuart JS, et al. Performance characteristics of molecular (DNA) analysis for the diagnosis of mucinous pancreatic cysts. Gastrointest Endosc. 2014;79:79–87. doi: 10.1016/j.gie.2013.05.026. [DOI] [PubMed] [Google Scholar]
- 4.Matthaei H, Wylie D, Lloyd MB, Dal Molin M, Kemppainen J, Mayo SC, Wolfgang CL, Schulick RD, Langfield L, Andruss BF, et al. mRNA biomarkers in cyst fluid augment the diagnosis and management of pancreatic cysts. Clin Cancer Res. 2012;18:4713–4724. doi: 10.1158/1078-0432.CCR-12-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takano S, Fukasawa M, Maekawa S, Kadokura M, Miura M, Shindo H, Takahashi E, Sato T, Enomoto N. Deep sequencing of cancer-related genes revealed GNAS mutations to be associated with intraductal papillary mucinous neoplasms and its main pancreatic duct dilation. PLoS One. 2014;9:e98718. doi: 10.1371/journal.pone.0098718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moris D, Damaskos C, Spartalis E, Papalampros A, Vernadakis S, Dimitroulis D, Griniatsos J, Felekouras E, Nikiteas N. Updates and critical evaluation on novel biomarkers for the malignant progression of intraductal papillary mucinous neoplasms of the pancreas. Anticancer Res. 2017;37:2185–2194. doi: 10.21873/anticanres.11553. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka M, Heckler M, Liu B, Heger U, Hackert T, Michalski CW. Cytologic analysis of pancreatic juice increases specificity of detection of malignant IPMN-A systematic review. Clin Gastroenterol Hepatol. 2019;17:2199–2211 e21. doi: 10.1016/j.cgh.2018.12.034. [DOI] [PubMed] [Google Scholar]
- 8.Reddy KB. MicroRNA (miRNA) in cancer. Cancer Cell Int. 2015;15:38. doi: 10.1186/s12935-015-0185-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng Y, Croce CM. The role of microRNAs in human cancer. Signal Transduct Target Ther. 2016;1:15004. doi: 10.1038/sigtrans.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. 2012;6:590–610. doi: 10.1016/j.molonc.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol Med. 2014;20:460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 12.de Miguel Perez D, Rodriguez Martinez A, Ortigosa Palomo A, Delgado Ureña M, Garcia Puche JL, Robles Remacho A, Exposito Hernandez J, Lorente Acosta JA, Ortega Sánchez FG, Serrano MJ. Extracellular vesicle-miRNAs as liquid biopsy biomarkers for disease identification and prognosis in metastatic colorectal cancer patients. Sci Rep. 2020;10:3974. doi: 10.1038/s41598-020-60212-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conte D, Verri C, Borzi C, Suatoni P, Pastorino U, Sozzi G, Fortunato O. Novel method to detect microRNAs using chip-based QuantStudio 3D digital PCR. BMC Genomics. 2015;16:849. doi: 10.1186/s12864-015-2097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki R, Asama H, Waragai Y, Takagi T, Hikichi T, Sugimoto M, Konno N, Watanabe K, Nakamura J, Kikuchi H, et al. Fibrosis-related miRNAs as serum biomarkers for pancreatic ductal adenocarcinoma. Oncotarget. 2017;9:4451–4460. doi: 10.18632/oncotarget.23377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T, Dalamagas T, Hatzigeorgiou AG. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43((W1)):W460–W466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.House MG, Guo M, Iacobuzio-Donahue C, Herman JG. Molecular progression of promoter methylation in intraductal papillary mucinous neoplasms (IPMN) of the pancreas. Carcinogenesis. 2003;24:193–198. doi: 10.1093/carcin/24.2.193. [DOI] [PubMed] [Google Scholar]
- 17.Huang da W, Sherman BT, Zheng X, Yang J, Imamichi T, Stephens R, Lempicki RA. Extracting biological meaning from large gene lists with DAVID. Curr Protoc Bioinformatics Chapter. 2009;13 doi: 10.1002/0471250953.bi1311s27. Unit 13 11. doi: 10.1002/0471250953.bi1311s27. [DOI] [PubMed] [Google Scholar]
- 18.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Habbe N, Koorstra JB, Mendell JT, Offerhaus GJ, Ryu JK, Feldmann G, Mullendore ME, Goggins MG, Hong SM, Maitra A. MicroRNA miR-155 is a biomarker of early pancreatic neoplasia. Cancer Biol Ther. 2009;8:340–346. doi: 10.4161/cbt.8.4.7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goto T, Fujiya M, Konishi H, Sasajima J, Fujibayashi S, Hayashi A, Utsumi T, Sato H, Iwama T, Ijiri M, et al. An elevated expression of serum exosomal microRNA-191, −21, −451a of pancreatic neoplasm is considered to be efficient diagnostic marker. BMC Cancer. 2018;18:116. doi: 10.1186/s12885-018-4006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pasqualini L, Bu H, Puhr M, Narisu N, Rainer J, Schlick B, Schäfer G, Angelova M, Trajanoski Z, Börno ST, et al. mR-22 and miR-29a are members of the androgen receptor cistrome modulating LAMC1 and Mcl-1 in prostate cancer. Mol Endocrinol. 2015;29:1037–1054. doi: 10.1210/me.2014-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang G, Shen N, Cheng L, Lin J, Li K. Downregulation of miR-22 acts as an unfavorable prognostic biomarker in osteosarcoma. Tumour Biol. 2015;36:7891–7895. doi: 10.1007/s13277-015-3379-1. [DOI] [PubMed] [Google Scholar]
- 23.Bersani F, Lingua MF, Morena D, Foglizzo V, Miretti S, Lanzetti L, Carrà G, Morotti A, Ala U, Provero P, et al. Deep sequencing reveals a novel miR-Regulatory network with therapeutic potential in rhabdomyosarcoma. Cancer Res. 2016;76:6095–6106. doi: 10.1158/0008-5472.CAN-16-0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koufaris C, Valbuena GN, Pomyen Y, Tredwell GD, Nevedomskaya E, Lau CH, Yang T, Benito A, Ellis JK, Keun HC. Systematic integration of molecular profiles identifies miR-22 as a regulator of lipid and folate metabolism in breast cancer cells. Oncogene. 2016;35:2766–2776. doi: 10.1038/onc.2015.333. [DOI] [PubMed] [Google Scholar]
- 25.Jafarzadeh-Samani Z, Sohrabi S, Shirmohammadi K, Effatpanah H, Yadegarazari R, Saidijam M. Evaluation of miR-22 and miR-20a as diagnostic biomarkers for gastric cancer. Chin Clin Oncol. 2017;6:16. doi: 10.21037/cco.2017.03.01. [DOI] [PubMed] [Google Scholar]
- 26.Fu Q, Liu CJ, Zhang X, Zhai ZS, Wang YZ, Hu MX, Xu XL, Zhang HW, Qin T. Glucocorticoid receptor regulates expression of microRNA-22 and downstream signaling pathway in apoptosis of pancreatic acinar cells. World J Gastroenterol. 2018;24:5120–5130. doi: 10.3748/wjg.v24.i45.5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sui J, Liu Q, Zhang H, Kong Y. Deep integrative analysis of microRNA-mRNA regulatory networks for biomarker and target discovery in chondrosarcoma. J Cell Biochem. 2019;120:9631–9638. doi: 10.1002/jcb.28241. [DOI] [PubMed] [Google Scholar]
- 28.Wang D, Guo C, Kong T, Mi G, Li J, Sun Y. Serum miR-22 may be a biomarker for papillary thyroid cancer. Oncol Lett. 2019;17:3355–3361. doi: 10.3892/ol.2019.10011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pellatt DF, Stevens JR, Wolff RK, Mullany LE, Herrick JS, Samowitz W, Slattery ML. Expression profiles of miRNA subsets distinguish human colorectal carcinoma and normal colonic mucosa. Clin Transl Gastroenterol. 2016;7:e152. doi: 10.1038/ctg.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Medarova Z, Pantazopoulos P, Yoo B. Screening of potential miRNA therapeutics for the prevention of multi-drug resistance in cancer cells. Sci Rep. 2020;10:1970. doi: 10.1038/s41598-020-58919-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garaude J, Kent A, van Rooijen N, Blander JM. Simultaneous targeting of toll- and nod-like receptors induces effective tumor-specific immune responses. Sci Transl Med. 2012;4:120ra116. doi: 10.1126/scitranslmed.3002868. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during the present study are included in the published article.