Abstract
Background
PGF and TNFAIP2 are important angiogenic factors, which were abnormal expression in cervical cancer (CC). However, there is currently no report investigating the relationship of PGF and TNFAIP2 gene polymorphisms to CC risk.
Methods
We conducted a case-control study of 342 CC patients and 498 cancer-free controls in a Chinese Uygur female population. Three SNPs (PGF rs8019391, PGF rs2268615, and TNFAIP2 rs710100) were selected and genotyped to assess the possible association of PGF and TNFAIP2 polymorphisms with CC susceptibility. Logistic regression analysis adjusted by age was used.
Results
PGF rs2268615 (OR = 1.39, 95% CI = 1.04–1.86, p = 0.024) and TNFAIP2 rs710100 (OR = 1.44, 95% CI =1.07–1.95, p = 0.018) polymorphisms were associated with the increased risk of CC. Moreover, T allele of PGF rs8019391 was highly represented in patients with stage III–IV compared with stage I-II (OR = 2.17, p = 4.58 × 10− 4). MDR analysis revealed a positive interaction between the SNPs.
Conclusion
Our data indicated that PGF rs2268615, and TNFAIP2 rs710100 polymorphisms might be risk factors for CC susceptibility, which contributed to the increased risk of CC.
Trial registration
Not applicable.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12881-020-01144-5.
Keywords: PGF, TNFAIP2, Polymorphism, Cervical cancer
Background
Worldwide, cervical cancer (CC) is the fourth most common cancer, with an estimated 570,000 cases and 311,000 deaths in 2018 [1]. Cervical cancer is the most common cancer of female genital system in China and the incidence of CC tends to be younger [2]. A wide range of inter-individual genetic variability and other pathogeneses might contribute to cervical carcinogenesis. Accumulating evidence suggested that single nucleotide polymorphisms (SNPs) in tumor-associated genes played important role in the genetic susceptibility to CC, such as FAS, IL-17A, IL-17F and HOTAIR polymorphisms [3–5].
PGF (placental growth factor) gene, also named PLGF, encodes a homologous of vascular endothelial growth factor. PGF has been reported as a potent stimulator in cancer invasion by activating angiogenesis [6]. In addition, the overexpression of PGF is correlated with tumor stage, cancer progression and metastasis [7]. TNFAIP2 (TNF alpha induced protein 2) is a primary response gene of TNFα, and the expression of TNFAIP2 is regulated by various transcription factors and signaling pathways, including NF-κB, KLF5 and retinoic acid pathways [8]. TNFAIP2 is an important angiogenic factor, and is significantly associated with intratumoral microvessel density [9]. In addition, previous studies reported the abnormal expression of PGF and TNFAIP2 in human cancer, including CC [6, 10]. However, there is no report about the associations between PGF and TNFAIP2 polymorphisms and CC risk. We hypothesized that genetic variants in PGF and TNFAIP2 could contribute to CC susceptibility.
Here, PGF rs8019391 and rs2268615, and TNFAIP2 rs710100 were selected as candidate SNPs from the NCBI dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP) and the 1000 Genomes Project data (http://www.internationalgenome.org/), based on minor allele frequency (MAF) of at least 5% in Chinese populations, with a pairwise r2 > 0.80 and the call rate of genotyping > 95%. The aim of our study was to investigate the possible association between three SNPs (PGF rs8019391, PGF rs2268615, and TNFAIP2 rs710100) and the risk of CC in Chinese Uygur female population.
Subjects and methods
Study participants
This study protocol was approved by the Ethics Committee of the People’s Hospital of Xinjiang Uygur Autonomous Region and was in accordance with the guidelines of the Declaration of Helsinki. Written informed consents were obtained from all participants.
Three hundred fourty-two cervical cancer patients (cases, 43.27 ± 11.78 years) and 498 age-matched cancer-free individuals (controls, 43.46 ± 13.03 years) were enrolled from the People’s Hospital of Xinjiang Uygur Autonomous Region, as shown in Table 1. All recruited subjects were unrelated ethnic Han Chinese females. Patients were newly diagnosed and histopathologically confirmed primary cervical cancer according to the clinical staging standards of the International Federation of Gynecology and Obstetrics (FIGO). Among patients, 132 cases were stage I–II, 80 cases were stage III–IV and 130 cases were missing. Patients with any history of other cancers, radiotherapy, chemotherapy, or surgery and inflammatory diseases were excluded. The controls were recruited from the health checkup in the same hospital during the same period. Controls had no any history of cancers and diseases of the liver, kidneys, heart, brain, and vascular system, as well as gynecological inflammatory diseases. Demographic characteristics and clinical information were collected from the standardized questionnaires.
Table 1.
Characteristics of patients with cervical cancer and controls
| Characteristics | Cases (n = 342) |
Controls (n = 498) |
p |
|---|---|---|---|
| Age | |||
| Mean ± SD (years) | 43.27 ± 11.78 | 43.46 ± 13.03 | 0.832 |
| > 43 | 176 (51.5%) | 263 (52.8%) | |
| ≤ 43 | 166 (48.5%) | 235 (47.2%) | |
| HPV status | |||
| Negative | 51 (14.9%) | ||
| Positive | 195 (57.0%) | ||
| Missing | 96 (28.1%) | ||
| Stage | |||
| I-II | 132 (38.6%) | ||
| III-IV | 80 (23.4%) | ||
| Missing | 130 (38.0%) | ||
SNP selection and genotyping
Peripheral venous blood samples (5 mL) were obtained from all subjects and stored in EDTA-coated tubes. Genomic DNA was extracted using the GoldMag DNA Purification Kit (GoldMag Co. Ltd., Xi′an City, China) according to the manufacturer’s protocol, then quantified by NanoDrop 2000 (Thermo Scientifc, Waltham, MA, USA), and stored at − 20°Cfor further experiments. PGF rs8019391 and rs2268615, and TNFAIP2 rs710100 were selected as candidate from the NCBI dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP) and the 1000 Genomes Project data (http://www.internationalgenome.org/), based on minor allele frequency (MAF) of at least 5% in Chinese populations, with a pairwise r2 > 0.80, and the call rate of genotyping > 95%. SNPs. miRNASNP_v2 database (http://bioinfo.life.hust.edu.cn/ miRNASNP2/index.php) and HaploReg v4.1 (https://pubs.broadinstitute.org/mammals/haploreg/ haploreg.php) were used to predict the potential function of these polymorphisms (Supplementary Table 1). These candidate SNPs were genotyped with Agena MassARRAY system (Agena, San Diego, CA, U.S.A.) as described previously [11, 12], and performed by two laboratory technicians in a double-blinded fashion. The primers for PCR amplification and single base extension were designed using the Assay Design 3.0 software (Supplementary Table 2). For quality control, approximately 10% of the samples were randomly selected and repeated genotyping, and 100% concordance rate was observed.
Data analysis
All statistical analysis was performed using SPSS version 18.0 (SPSS Inc., Chicago, IL, USA) and PLINK software. A student’s t-test was performed to analyze the differences in the age distribution between patients and controls. The Hardy–Weinberg equilibrium (HWE) was tested by a goodness-of-fit χ2 test for each SNP among the control subjects. The genotype and allele frequencies between two groups were compared usingχ2 test. The association of candidate SNPs with CC risk was assessed by odds ratios (ORs) and 95% confidence intervals (CIs) using logistic regression analysis for both combined and respective genotype [13]. Stratification analysis by demographic and clinic variables was also performed to assess the genetic association. Multifactor dimensionality reduction (MDR) (version 3.0.2) was used to evaluate the SNP–SNP interactions in CC risk [14]. A two-tailed p-value < 0.05 was considered to be statistically significant for all the analyses.
Results
The results of genotyping was shown in Additional file 2. In Supplementary Table 3, the MAF of PGF rs8019391 and rs2268615, and TNFAIP2 rs710100 between the case and control groups were listed. The genotype distribution of these SNPs in controls were in accordance with the Hardy-Weinberg equilibrium (p > 0.05). The call rate for rs8019391, rs2268615 and rs710100 were 100, 99.7 and 99.1%, respectively. The MAFs distribution of PGF rs2268615-A allele and TNFAIP2 rs710100-A allele were higher in the case group, which increased the risk of CC (rs2268615, A vs C, OR = 1.27, 95% CI = 1.03–1.58, p = 0.029; and rs710100, A vs G, OR = 1.23, 95% CI = 1.01–1.50, p = 0.043).
The results of multiple genetic model adjusted by age revealed PGF rs2268615 and TNFAIP2 rs710100 conferred to the increased CC risk (Table 2). PGF rs2268615 was associated with an increased risk of CC under heterozygote (OR = 1.39, 95% CI = 1.04–1.86, p = 0.024), dominant (OR = 1.40, 95% CI = 1.06–1.84, p = 0.018) and log-additive (OR = 1.29, 95% CI =1.03–1.61, p = 0.027) models. For rs710100 in TNFAIP2, compared with GG genotype, GA genotype (OR = 1.44, 95% CI =1.07–1.95, p = 0.018) and GA + AA genotype (OR = 1.42, 95% CI = 1.07–1.89, p = 0.016) increased 1.44-fold and 1.42-fold CC risk, respectively. Moreover, the result of the additive model also showed an increased risk of CC (rs710100, OR = 1.23, 95% CI = 1.00–1.50, p = 0.046). However, there was no significant association between PGF rs8019391 and CC susceptibility.
Table 2.
Relationships between the candidate SNPs and cervical cancer risk
| Gene SNP ID |
Model | Genotype | Case | Control | Adjusted by age and gender | |
|---|---|---|---|---|---|---|
| OR (95%CI) | p | |||||
|
PGF rs8019391 |
Genotype | CC | 208 | 327 | 1.00 | |
| CT | 119 | 145 | 1.29 (0.96–1.74) | 0.093 | ||
| TT | 15 | 26 | 0.91 (0.47–1.76) | 0.777 | ||
| Dominant | CC | 208 | 327 | 1.00 | 0.150 | |
| CT-TT | 134 | 171 | 1.23 (0.93–1.64) | |||
| Recessive | CC-CT | 327 | 472 | 1.00 | 0.585 | |
| TT | 15 | 26 | 0.83 (0.43–1.6) | |||
| Log-additive | – | – | – | 1.13 (0.89–1.42) | 0.324 | |
|
PGF rs2268615 |
Genotype | CC | 160 | 273 | 1.00 | |
| CA | 156 | 191 | 1.39 (1.04–1.86) | 0.024 | ||
| AA | 26 | 31 | 1.43 (0.82–2.49) | 0.209 | ||
| Dominant | CC | 160 | 273 | 1.00 | 0.018 | |
| CA-AA | 182 | 222 | 1.40 (1.06–1.84) | |||
| Recessive | CC-CA | 316 | 464 | 1.00 | 0.453 | |
| AA | 26 | 31 | 1.23 (0.72–2.11) | |||
| Log-additive | – | – | – | 1.29 (1.03–1.61) | 0.027 | |
|
TNFAIP2 rs710100 |
Genotype | GG | 118 | 210 | 1.00 | |
| GA | 171 | 211 | 1.44 (1.07–1.95) | 0.018 | ||
| AA | 53 | 69 | 1.37 (0.89–2.08) | 0.150 | ||
| Dominant | GG | 118 | 210 | 1.00 | 0.016 | |
| GA-AA | 224 | 280 | 1.42 (1.07–1.89) | |||
| Recessive | GG-GA | 289 | 421 | 1.00 | 0.576 | |
| AA | 53 | 69 | 1.12 (0.76–1.65) | |||
| Log-additive | – | – | – | 1.23 (1.00–1.50) | 0.046 | |
SNP single nucleotide polymorphism, OR odds ratio; 95% CI 95% confidence interval
p values were calculated by logistic regression analysis with adjustments for age and gender
p < 0.05 means the data is statistically significant
Age stratification displayed that PGF rs2268615 and TNFAIP2 rs710100 increased the risk of CC among women at age ≤ 43 years (Table 3). After calculating the ORs for the allele (OR = 1.38, p = 0.041 and OR = 1.42, p = 0.018, respectively), genotype (CA vs CC, OR = 1.55, p = 0.039; and AA vs GG, OR = 1.97, p = 0.031, respectively), dominant (OR = 1.56, p = 0.030; and OR = 1.57, 95%, p = 0.034, respectively), and log-additive (OR = 1.40, p = 0.042; and OR = 1.42, p = 0.020) genetic models, they all displayed the genetic association of PGF rs2268615 and TNFAIP2 rs710100 with CC susceptibility.
Table 3.
Relationships between the candidate SNPs and cervical cancer risk according to the stratification by age
| SNP ID | Model | Genotype | > 43 years | ≤ 43 years | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | OR (95%CI) | p | Case | Control | OR (95%CI) | p | |||
|
PGF rs2268615 |
Allele | C | 253 | 393 | 1.00 | 0.303 | 223 | 344 | 1.00 | 0.041 |
| A | 99 | 131 | 1.17 (0.87–1.59) | 109 | 122 | 1.38 (1.01–1.88) | ||||
| Genotype | CC | 89 | 147 | 1.00 | 71 | 126 | 1.00 | |||
| CA | 75 | 99 | 1.25 (0.84–1.86) | 0.282 | 81 | 92 | 1.55 (1.02–2.36) | 0.039 | ||
| AA | 12 | 16 | 1.22 (0.55–2.70) | 0.626 | 14 | 15 | 1.62 (0.74–3.56) | 0.228 | ||
| Dominant | CC | 89 | 147 | 1.00 | 0.269 | 71 | 126 | 1.00 | 0.030 | |
| CA-AA | 87 | 115 | 1.24 (0.85–1.82) | 95 | 107 | 1.56 (1.05–2.33) | ||||
| Recessive | CC-CA | 164 | 246 | 1.00 | 0.795 | 152 | 218 | 1.00 | 0.481 | |
| AA | 12 | 16 | 1.11 (0.51–2.41) | 14 | 15 | 1.31 (0.61–2.81) | ||||
| Log-additive | – | – | – | 1.17 (0.86–1.60) | 0.317 | – | – | 1.40 (1.01–1.92) | 0.042 | |
|
TNFAIP2 rs710100 |
Allele | G | 217 | 331 | 1.00 | 0.597 | 190 | 300 | 1.00 | 0.018 |
| A | 135 | 191 | 1.08 (0.82–1.43) | 142 | 158 | 1.42 (1.06–1.90) | ||||
| Genotype | GG | 64 | 111 | 1.00 | 54 | 99 | 1.00 | |||
| GA | 89 | 109 | 1.41 (0.93–2.14) | 0.105 | 82 | 102 | 1.46 (0.94–2.27) | 0.092 | ||
| AA | 23 | 41 | 0.96 (0.53–1.74) | 0.890 | 30 | 28 | 1.97 (1.07–3.63) | 0.031 | ||
| Dominant | GG | 64 | 111 | 1.00 | 0.208 | 54 | 99 | 1.00 | 0.034 | |
| GA-AA | 112 | 150 | 1.29 (0.87–1.91) | 112 | 130 | 1.57 (1.03–2.38) | ||||
| Recessive | GG-GA | 153 | 220 | 1.00 | 0.418 | 136 | 201 | 1.00 | 0.103 | |
| AA | 23 | 41 | 0.80 (0.46–1.38) | 30 | 28 | 1.59 (0.91–2.79) | ||||
| Log-additive | – | – | – | 1.07 (0.81–1.41) | 0.635 | – | – | 1.42 (1.06–1.90) | 0.020 | |
SNP single nucleotide polymorphism, OR odds ratio, 95% CI 95% confidence interval
p values were calculated by logistic regression analysis with adjustments for age
p < 0.05 indicates statistical significance
Subsequently, stratification analysis by tumor stage showed that the risk effect for PGF rs8019391 appeared to be more prominent in the subset of patients with stage III + IV (Table 4). Compared with the C allele, rs8019391 T allele was highly represented in patients with stage III–IV as compared to patients with stage I–II under the allele (OR = 2.17, p = 4.58 × 10− 4), heterozygote (OR = 2.34, p = 0.005), homozygote (OR = 5.76, p = 0.015), dominant (OR = 2.59, p = 0.001), recessive (OR = 4.13, p = 0.045), and log-additive models (OR = 2.36, p < 0.001).
Table 4.
Relationship of clinical stage with PGF rs8019391 polymorphism in cervical cancer patients adjusted by age
| SNP ID | Model | Genotype | I-II | III-IV | OR (95%CI) | p |
|---|---|---|---|---|---|---|
| rs8019391 | Allele | C | 110 | 220 | 1.00 | 4.58 × 10−4 |
| T | 50 | 44 | 2.27 (1.43–3.62) | |||
| Codominant | CC | 37 | 91 | 1.00 | ||
| CT | 36 | 38 | 2.34 (1.29–4.25) | 0.005 | ||
| TT | 7 | 3 | 5.76 (1.41–23.52) | 0.015 | ||
| Dominant | CC | 37 | 91 | 1.00 | 0.001 | |
| CT-TT | 43 | 41 | 2.59 (1.46–4.60) | |||
| Recessive | CC-CT | 73 | 129 | 1.00 | 0.045 | |
| TT | 7 | 3 | 4.13 (1.04–16.45) | |||
| Log-additive | – | – | – | 2.36 (1.45–3.86) | < 0.001 |
SNP single nucleotide polymorphism, OR odds ratio, 95% CI 95% confidence interval
p values were calculated by logistic regression analysis with adjustments for age
p < 0.05 indicates statistical significance
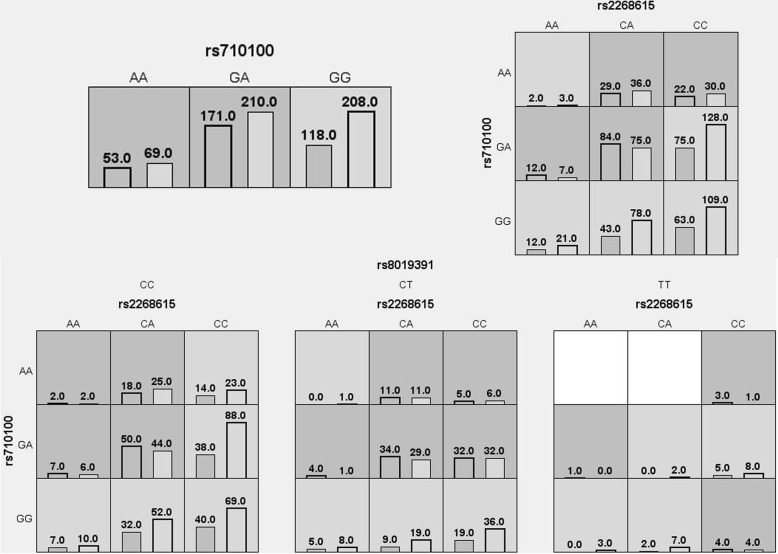
Subsequently, MDR analysis was implemented to assess the impact of SNP-SNP interaction. Association of higher order interaction with CC risk was summarized in Fig. 1. The result revealed the additive effect between TNFAIP2 rs710100-GA, PGF rs2268615-CA, and PGF rs8019391-CT on conferring risk towards the susceptibility to CC. The result of dendrogram and the Fruchterman-Reingold (Fig. 2) showed that PGF rs2268615, TNFAIP2 rs710100, PGF rs8019391 exhibited a strong synergy effect on CC risk. Table 5 showed that TNFAIP2 rs710100 was the best single-locus model to predict the risk of CC (testing accuracy = 0.508, CVC = 6/10, p = 0.014). The best two-locus model was the combination of PGF rs2268615 and TNFAIP2 rs710100 (testing accuracy = 0.536, CVC = 9/10, p < 0.0001. The three-locus model included TNFAIP2 rs710100, PGF rs2268615, and PGF rs8019391 (testing accuracy = 0.550, CVC = 10/10, p < 0.0001).
Fig. 1.
Summary of MDR gene-gene interaction. Each cell shows counts of “case” on left and “control” on right
Fig. 2.
SNP-SNP interaction dendrogram and Fruchterman-Reingold
Table 5.
SNP–SNP interaction models of the PGF and TNFAIP2 genes analyzed by the MDR method
| Model | Training Bal. Acc. | Testing Bal. Acc. | CVC | OR (95% CI) | p |
|---|---|---|---|---|---|
| TNFAIP2 rs710100 | 0.544 | 0.508 | 6/10 | 1.46 (1.08–1.97) | 0.014 |
| PGF rs2268615, TNFAIP2 rs710100 | 0.564 | 0.536 | 9/10 | 1.91 (1.41–2.59) | < 0.0001 |
| PGF rs2268615, TNFAIP2 rs710100, PGF rs8019391 | 0.587 | 0.550 | 10/10 | 2.11 (1.56–2.84) | < 0.0001 |
MDR multifactor dimensionality reduction, Bal. Acc. balanced accuracy, CVC cross–validation consistency, OR odds ratio, CI confidence interval
p values were calculated using χ2 tests
p < 0.05 indicates statistical significance
Discussion
In this case–control study, we assessed the association between three SNPs (PGF rs8019391, PGF rs2268615, and TNFAIP2 rs710100) and CC susceptibility in the Chinese Uygur female population. We found that PGF rs2268615, and TNFAIP2 rs710100 were associated with the increased risk of CC. Our findings also suggested some possible interaction between genetic variations (PGF rs2268615 and TNFAIP2 rs710100) and age in CC risk. Moreover, the risk effect for PGF rs8019391 appeared to be more prominent in patients with stage III + IV. To the best of our knowledge, this is the first report describing that PGF and TNFAIP2 polymorphisms might be risk factor for CC.
PGF, located at 14q24.3, belongs to the vascular endothelial growth factor family and presents on various cell types. The expression of PGF in tissue or plasma of cancer patients was upregulated in most human tumor types, including gallbladder, gastric and prostate cancers. PGF regulates certain cellular processes such as survival, vascular endothelial cell growth, invasiveness, and PGF is also involved in pathological angiogenesis and metastasis [15–17]. Previous researches indicated that PGF was overexpressed in CC tissues, serum and vaginal lavage compared with adjacent normal tissues or normal women group [6, 18]. PGF promotes migration by regulating the expression of epithelial-mesenchymal transition-related protein in CC [6]. These evidence led us to propose the hypothesis that PGF could be of importance in CC occurrence. In our study, we firstly assess the association of PGF polymorphisms (rs8019391 and rs2268615) with CC susceptibility, and found that rs2268615 conferred the increased risk of CC, and rs8019391 was a risk factor for patients with stage III-IV. A retrospective population-based study showed that 5-year relative survival rates of CC were 90.9, 71.0, 41.7, and 7.8% for the stage I, II, III, and IV, respectively [19]. Therefore, it is highly speculated that PGF rs8019391 polymorphism may affect CC progression. Further large-scale studies are needed to verify our findings.
TNFAIP2 gene (also named B94) is located on chromosome 14q32, and encodes TNFα-inducible protein 2. TNFAIP2 participates in the NFκB and KLF5 signaling pathway to regulate cell inflammatory, angiogenesis, cell proliferation, migration and invasion [20, 21]. The expression of TNFAIP2 was found to be abnormal in various cancers, including breast cancer, esophageal squamous cell carcinoma, and glioma [20, 22, 23]. The expression of TNFAIP2 was significantly increased in CC tissues compared with normal tissues based on TCGA (The Cancer Genome Atlas) database [8]. In addition, the disruption of TNFAIP2 cytokine/retinoic acid-inducible gene through viral integration contributed to the rapid progression of CC [10]. These evidence suggested that TNFAIP2 might play an important role in the progression of CC. In this study, we found that TNFAIP2 rs710100 was associated with an increased risk of CC. Rs710100 in the 3′UTR of TNFAIP2, located at the predicted miRNAs-binding sites, was related to an increased risk of CC. Specifically, rs1064607 putatively affects the binding of miR-155 [24], whose abnormal expression in CC was correlated with FIGO stage, lymph nodes metastasis, and vascular invasion [25]. Therefore, we proposed that rs710100 in 3′UTR of TNFAIP2 might affect TNFAIP2 expression in CC by disturbing mRNA stability or miRNA binding activity, thus causing a higher risk of CC. However, it should be confirmed in further functional studies.
In spite of interesting findings on the association of PGF and TNFAIP2 polymorphisms with CC risk, several limitations need to be addressed. First, there may be selection and information bias since the retrospective study was designed as a hospital-based case–control study. Second, due to insufficient data of HPV screening results and lifestyle data (e.g., smoking), we could not evaluate HPV infection and lifestyle as the potential confounder in the risk estimation of CC.
Conclusion
In conclusion, our findings indicated a relationship between PGF rs2268615, and TNFAIP2 rs710100 and the increased CC susceptibility in the Chinese Uygur females. Considering that this is the first report on the association of PGF and TNFAIP2 polymorphisms with CC risk, well-designed large and prospective studies are required to validate our findings.
Supplementary information
Additional file 1: Supplementary Table 1. Predicted stability, duplex formation, and functional effect of 3′-UTR polymorphisms in miRNA-binding sites of selected genes. Supplementary Table 2. Primers sequence of PCR and UEP used in this study. Supplementary Table 3. The information and HWE about the candidate SNPs.
Acknowledgements
The authors thank all the participants in this study.
Abbreviations
- CC
Cervical cancer
- SNPs
Single nucleotide polymorphisms
- PGF
Placental growth factor
- TNFAIP2
TNF alpha induced protein 2
Authors’ contributions
ZA, RM and LLH designed this study protocol and drafted the manuscript. JL1, SH and JL2 performed the data analysis. RM, MN and LLH conceived and supervised the study. All of these authors contributed to this article. The author(s) read and approved the final manuscript.
Funding
This article was financially supported by the National Natural Science Foundation of China (No. 81760467).
Availability of data and materials
The datasets generated and/or analysed during the current study are available in the Additional file 2.
Ethics approval and consent to participate
This study protocol was approved by the Ethics Committee of the People’s Hospital of Xinjiang Uygur Autonomous Region and under the guidelines of the Declaration of Helsinki. Written informed consent was obtained from all participants.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zumurelaiti Ainiwaer and Reyilanmu Maisaidi are co-first authors.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Jiang X, Tang H. Epidemiology of gynecologic cancers in China. J Gynecol Oncol. 2018;29(1):e7. [DOI] [PMC free article] [PubMed]
- 3.Tan SC, Ismail MP, Duski DR, Othman NH, Ankathil R. FAS c.-671A>G polymorphism and cervical cancer risk: a case-control study and meta-analysis. Cancer Genet. 2017;211:18–25. doi: 10.1016/j.cancergen.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 4.Lv Q, Zhu D, Zhang J, Yi Y, Yang S, Zhang W. Association between six genetic variants of IL-17A and IL-17F and cervical cancer risk: a case-control study. Tumour Biol. 2015;36(5):3979–3984. doi: 10.1007/s13277-015-3041-y. [DOI] [PubMed] [Google Scholar]
- 5.Guo L, Lu X, Zheng L, Liu X, Hu M. Association of Long non-Coding RNA HOTAIR polymorphisms with cervical Cancer risk in a Chinese population. PLoS One. 2016;11(7):e0160039. doi: 10.1371/journal.pone.0160039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang W, Zhu S, Liu Q, Li C, Li L. Placenta growth factor promotes migration through regulating epithelial-mesenchymal transition-related protein expression in cervical cancer. Int J Clin Exp Pathol. 2014;7(12):8506–8519. [PMC free article] [PubMed] [Google Scholar]
- 7.Dewerchin M, Carmeliet P. Placental growth factor in cancer. Expert Opin Ther Targets. 2014;18(11):1339–1354. doi: 10.1517/14728222.2014.948420. [DOI] [PubMed] [Google Scholar]
- 8.Jia L, Shi Y, Wen Y, Li W, Feng J, Chen C. The roles of TNFAIP2 in cancers and infectious diseases. 2018;22(11):5188–95. [DOI] [PMC free article] [PubMed]
- 9.Chen LC, Chen CC, Liang Y, Tsang NM, Chang YS, Hsueh C. A novel role for TNFAIP2: its correlation with invasion and metastasis in nasopharyngeal carcinoma. Mod Pathol. 2011;24(2):175–184. doi: 10.1038/modpathol.2010.193. [DOI] [PubMed] [Google Scholar]
- 10.Einstein MH, Cruz Y, El-Awady MK, Popescu NC, DiPaolo JA, van Ranst M, et al. Utilization of the human genome sequence localizes human papillomavirus type 16 DNA integrated into the TNFAIP2 gene in a fatal cervical cancer from a 39-year-old woman. Clin Cancer Res. 2002;8(2):549–554. [PubMed] [Google Scholar]
- 11.Xia P, Li B, Geng T, Deng Z, Dang C, Chang D, et al. FGFR2 gene polymorphisms are associated with breast cancer risk in the Han Chinese population. Am J Cancer Res. 2015;5(5):1854–1861. [PMC free article] [PubMed] [Google Scholar]
- 12.Liang J, Kang X, Halifu Y, Zeng X, Jin T, Zhang M, et al. Secreted frizzled-related protein promotors are hypermethylated in cutaneous squamous carcinoma compared with normal epidermis. BMC Cancer. 2015;15:641. doi: 10.1186/s12885-015-1650-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li S, Jin T, Zhang J, Lou H, Yang B, Li Y, et al. Polymorphisms of TREH, IL4R and CCDC26 genes associated with risk of glioma. Cancer Epidemiol. 2012;36(3):283–287. doi: 10.1016/j.canep.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 14.Leem S, Park T. An empirical fuzzy multifactor dimensionality reduction method for detecting gene-gene interactions. BMC Genomics. 2017;18(Suppl 2):115. doi: 10.1186/s12864-017-3496-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Jin Y, Hu Y, Jiang L, Liu F, Zhang Y, et al. The PLGF/c-MYC/miR-19a axis promotes metastasis and stemness in gallbladder cancer. Cancer Sci. 2018;109(5):1532–44. [DOI] [PMC free article] [PubMed]
- 16.Mahmoodi F, Akrami H. PlGF knockdown decreases Tumorigenicity and Stemness properties of spheroid body cells derived from gastric Cancer cells. J Cell Biochem. 2017;118(4):851–9. [DOI] [PubMed]
- 17.Zins K, Thomas A, Lucas T, Sioud M, Aharinejad S, Abraham D. Inhibition of stromal PlGF suppresses the growth of prostate cancer xenografts. Int J Mol Sci. 2013;14(9):17958–17971. doi: 10.3390/ijms140917958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang S, Cheng H, Cai J, Cai L, Zhang J, Wang Z. PlGF expression in pre-invasive and invasive lesions of uterine cervix is associated with angiogenesis and lymphangiogenesis. Apmis. 2009;117(11):831–838. doi: 10.1111/j.1600-0463.2009.02538.x. [DOI] [PubMed] [Google Scholar]
- 19.Cheung FY, Mang OW, Law SC. A population-based analysis of incidence, mortality, and stage-specific survival of cervical cancer patients in Hong Kong: 1997-2006. Hong Kong Med J. 2011;17(2):89–95. [PubMed] [Google Scholar]
- 20.Jia L, Zhou Z, Liang H, Wu J, Shi P, Li F, et al. KLF5 promotes breast cancer proliferation, migration and invasion in part by upregulating the transcription of TNFAIP2. Oncogene. 2016;35(16):2040–2051. doi: 10.1038/onc.2015.263. [DOI] [PubMed] [Google Scholar]
- 21.Thair SA, Topchiy E, Boyd JH, Cirstea M, Wang C, Nakada TA, et al. TNFAIP2 inhibits early TNFalpha-induced NF-x03BA;B signaling and decreases survival in septic shock patients. J Innate Immun. 2016;8(1):57–66. doi: 10.1159/000437330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie Y, Wang B. Downregulation of TNFAIP2 suppresses proliferation and metastasis in esophageal squamous cell carcinoma through activation of the Wnt/beta-catenin signaling pathway. Oncol Rep. 2017;37(5):2920–2928. doi: 10.3892/or.2017.5557. [DOI] [PubMed] [Google Scholar]
- 23.Cheng Z, Wang HZ, Li X, Wu Z, Han Y, Li Y, et al. MicroRNA-184 inhibits cell proliferation and invasion, and specifically targets TNFAIP2 in Glioma. J Exp Clin Cancer Res. 2015;34:27. doi: 10.1186/s13046-015-0142-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Z, Wei S, Ma H, Zhao M, Myers JN, Weber RS, et al. A functional variant at the miR-184 binding site in TNFAIP2 and risk of squamous cell carcinoma of the head and neck. Carcinogenesis. 2011;32(11):1668–1674. doi: 10.1093/carcin/bgr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fang H, Shuang D, Yi Z, Sheng H, Liu Y. Up-regulated microRNA-155 expression is associated with poor prognosis in cervical cancer patients. Biomed Pharmacother. 2016;83:64–69. doi: 10.1016/j.biopha.2016.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Table 1. Predicted stability, duplex formation, and functional effect of 3′-UTR polymorphisms in miRNA-binding sites of selected genes. Supplementary Table 2. Primers sequence of PCR and UEP used in this study. Supplementary Table 3. The information and HWE about the candidate SNPs.
Data Availability Statement
The datasets generated and/or analysed during the current study are available in the Additional file 2.