Novel multi-transcription factor reporter mice were generated via CRISPR/Cas9 to dissect the developmental progression of ILC and LTi lineages. This study identifies lineage-specified precursors, indicating earlier bifurcation of ILC and LTi lineages than formerly proposed.
Abstract
Transcription factor (TF) reporter mice have proved integral to the characterization of murine innate lymphoid cell (ILC) development and function. Here, we implemented a CRISPR/Cas9-generated combinatorial reporter approach for the simultaneous resolution of several key TFs throughout ILC development in both the fetal liver and adult bone marrow. We demonstrate that the Tcf7-expressing early innate lymphoid precursor (EILP) and the common helper ILC precursor (CHILP) both contain a heterogeneous mixture of specified ILC and lymphoid tissue inducer (LTi) precursors with restricted lineage potential rather than a shared precursor. Moreover, the earliest specified precursor to the LTi lineage was identified upstream of these populations, before Tcf7 expression. These findings match dynamic changes in chromatin accessibility associated with the expression of key TFs (i.e., GATA3 and RORγ(t)), highlighting the distinct origins of ILC and LTi lineages at the epigenetic and functional levels, and provide a revised map for ILC development.
Introduction
Innate lymphoid cells (ILCs) are a diverse lineage classically divided into ILC1, ILC2, ILC3, lymphoid tissue inducer (LTi) cells, and natural killer (NK) cells based on developmental origin, phenotype, and function (Zook and Kee, 2016). Studies identifying restricted precursors provide a basis for the delineation of ILCs into distinct developmental origins, and transcription factor (TF) reporter mice have served as critical tools for these studies (Ishizuka et al., 2016a). LTi precursors (LTiPs) were identified in the fetal liver (FL) of RORγtEGFP mice (Eberl et al., 2004; Possot et al., 2011; Sawa et al., 2010). The ILC precursor (ILCP), which generated ILC1/2/3 but not LTi or NK cells, was characterized in Zbtb16EGFPCre mice (Constantinides et al., 2014). Concurrently, the common helper ILC precursor (CHILP) was described in ID2EGFP mice and generated ILC1/2/3 and LTi but not NK cells (Klose et al., 2014). Early work found that a shared precursor to all ILCs resides downstream of the common lymphoid precursor (CLP) within the α4β7+ lymphoid precursor (αLP; Possot et al., 2011; Seillet et al., 2016; Yoshida et al., 2001; Yu et al., 2014). More recently, however, studies using Tcf7EGFP mice defined an early innate lymphoid precursor (EILP) that resides downstream of the αLP as the common precursor to all ILCs (Fig. S1 A; Harly et al., 2018; Yang et al., 2015).
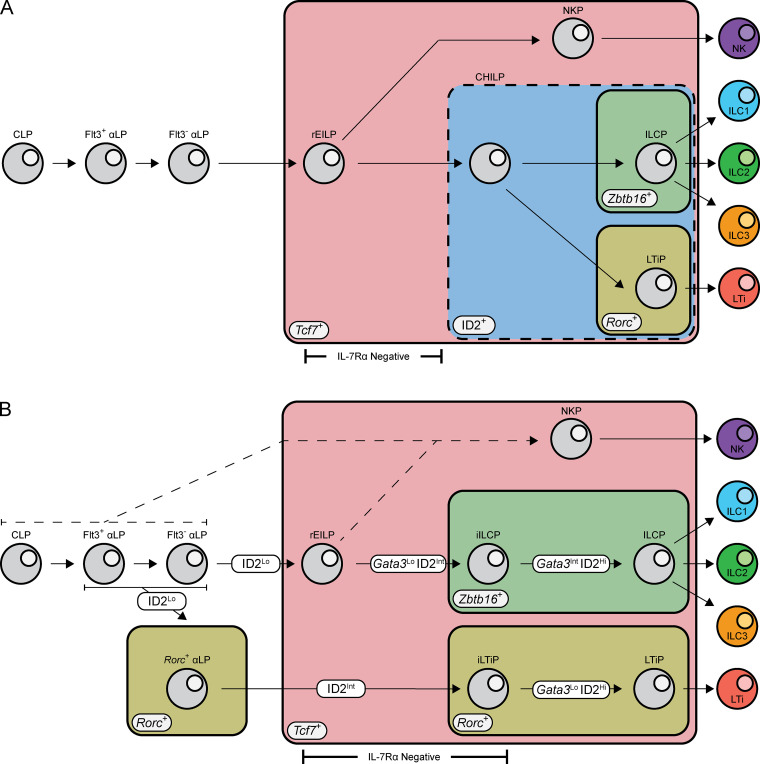
Figure S1.
Current and revised models for ILC development. (A) In the current model, CLP give rise to αLP, gaining α4β7 expression and progressively losing Flt3. The EILP, identified by Tcf7 expression and the lack of IL-7Rα, arises downstream of the αLP generating all known ILC subsets. The CHILP, identified by high ID2 expression, is downstream of the EILP generating ILC and LTi lineage cells. The ILCP, identified by Zbtb16 expression, and LTiP, identified by Rorc, both arise from the CHILP and generate ILC1/2/3 or LTi, respectively. (B) In the revised model, CLP give rise to αLP, gaining α4β7 expression and progressively losing Flt3. Bifurcation into the ILC and LTi lineage occurs within the αLP stage and is marked by low expression of ID2 and the early expression of Rorc within the αLP. Expression of ID2 increases to intermediate levels as cells mature, coinciding with the transient down-regulation of IL-7Rα expression. Expression of Zbtb16 in the iILCP and further up-regulation of Rorc in the iLTiP along with differential Gata3 up-regulation mark the restriction of cellular potential. High expression of ID2 and augmented Gata3 expression coincides with IL-7Rα reexpression as cells further differentiate toward the ILCP and LTiP.
Several models have been proposed for ILC development based on precursor fates; nevertheless, outstanding questions remain regarding precursor ontogeny, heterogeneity, and hierarchy. First, neither the CHILP nor the EILP were characterized in the FL, the dominant source of LTi during ontogeny, and furthermore, LTi were not distinguished from ILC3 in clonal cultures (Harly et al., 2018; Klose et al., 2014; Yang et al., 2015). Second, both the CHILP and EILP are heterogeneous and contain a Zbtb16+ subpopulation (Harly et al., 2018; Harly et al., 2019; Klose et al., 2014). Thus, whether these populations contain a shared precursor to the ILC and LTi lineages (ILC–LTi) or are an amalgamation of distinct precursors remains unresolved. Finally, though studies on ILC progenitor fates have prompted their hierarchical placement, a contemporaneous comparison of precursor potential has not been conducted.
Recent reports have applied polychromatic reporter mice to examine bone marrow (BM) ILC progenitors (Harly et al., 2019; Walker et al., 2019; Xu et al., 2019). To better address the aforementioned questions, we implemented a multi–TF reporter strategy to evaluate ILC progenitors in the FL and BM. We established a method for site-specific integration using CRISPR/Cas9-mediated transgenesis that enables the rapid generation of compatible TF reporter mouse lines expressing distinct reporters (Cong et al., 2013). These novel reporters were combined with preexisting reporters to interrogate ILC developmental stages. Our studies revealed that both the EILP and CHILP were heterogeneous mixtures of ILC and LTi lineage progenitors with restricted lineage potential. Furthermore, we identified a new early LTi-specified precursor upstream of the EILP, before Tcf7 expression. Chromatin accessibility in FL precursors supports the proposed cellular identity for each new ILC and LTi lineage precursor, and TF motif enrichment across developmental trajectories implicates GATA3 and RORγ(t) as major TFs in establishing ILC and LTi lineage identity, respectively. Together, these findings warrant a revised map of ILC and LTi lineage developmental progression and highlight the value of using a combinatorial TF reporter system for resolving complex developmental processes.
Results
Generation of novel TF reporter lines
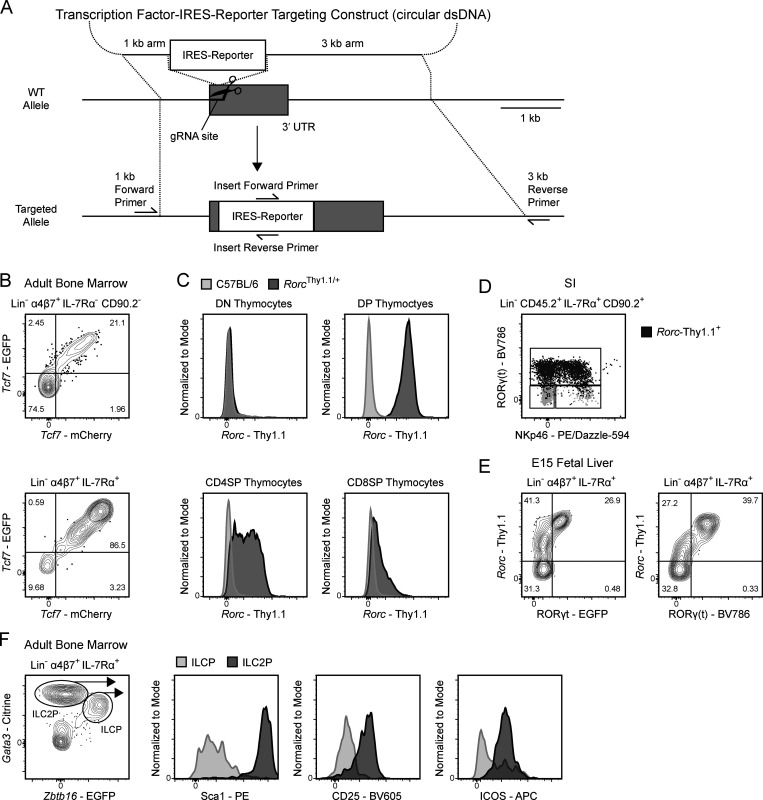
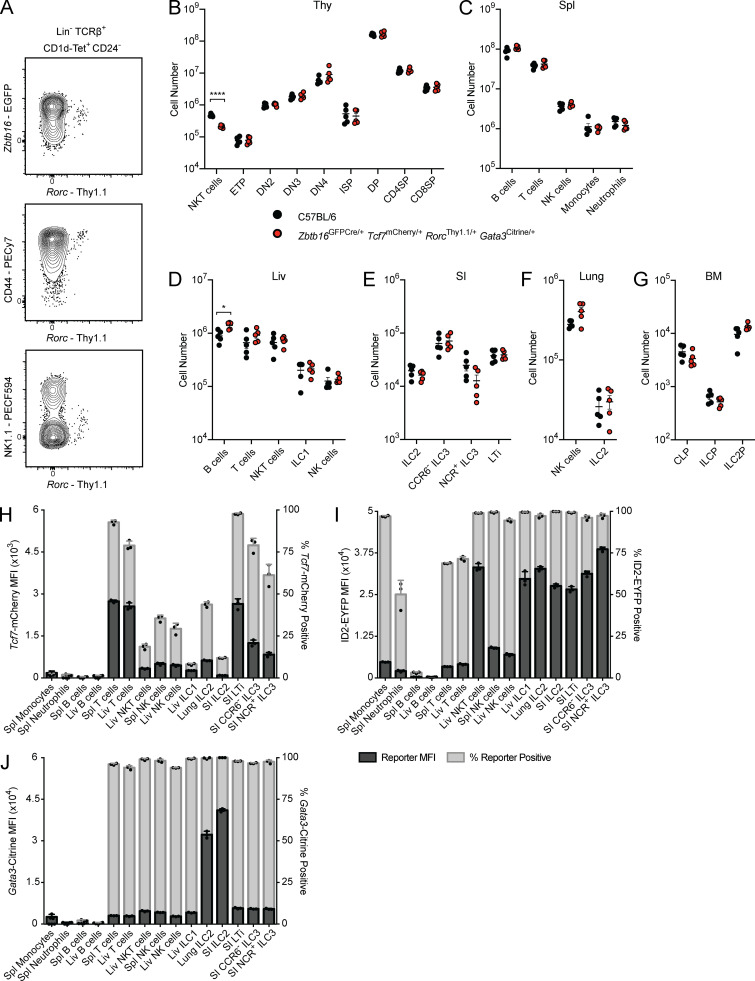
Enhanced characterization of ILC development necessitates the combined use of reporter lines for the simultaneous resolution of key TFs. However, because many previously generated strains used enhanced GFP (EGFP) to report on TF expression, these lines could not be combined for multi-TF analysis (Constantinides et al., 2014; Eberl et al., 2004; Klose et al., 2014; Yang et al., 2015). To overcome this limitation, we employed CRISPR/Cas9-mediated transgenesis for the rapid generation of new designer transgenic lines with distinct reporters (Fig. 1; Cong et al., 2013). TCF1 (Tcf7), RORγ(t) (RORγ and RORγt isoforms, Rorc), and GATA3 (Gata3) were of particular interest for addressing ILC development; therefore, we generated three novel reporter lines: Tcf7mCherry, RorcThy1.1, and Gata3Citrine. An internal ribosome entry site (IRES)–reporter sequence was inserted into the 3′ UTR of each gene; as such, reporter expression closely reflects mRNA expression (Fig. 1 A; Mohrs et al., 2005). To confirm the fidelity of each novel TF reporter, we compared the pattern of reporter expression to previously characterized EGFP reporter lines, intracellular TF staining, and/or known population expression. Tcf7-mCherry expression in lineage-negative (Lin−) α4β7+ populations in the BM was identical to Tcf7-EGFP expression in Tcf7EGFP/mCherry mice (Fig. 1 B; Yang et al., 2015). To circumvent known issues related to hypomorphic expression of RORγt in knock-in knock-out reporters, we elected to report on both RORγ(t) isoforms using a novel RorcThy1.1 reporter (Eberl et al., 2004; Walker et al., 2019; Zhong et al., 2016). Thymocytes from RorcThy1.1/+ mice displayed dynamic reporter expression consistent with RORγtEGFP/+ mice (Fig. 1 C; Eberl et al., 2004). Intracellular staining for RORγ(t) in the small intestine (SI) lamina propria demonstrated that Rorc-Thy1.1 expression coincides with protein expression in mature group 3 ILCs (Fig. 1 D), while FL Rorc-Thy1.1 expression precedes protein expression, likely reflecting the difference in mRNA and protein expression kinetics (Fig. 1 E; Mohrs et al., 2005). Notably, the Rorc-Thy1.1 reporter captures rare thymic NKT17 cells (Fig. S2 A). Lastly, Citrine expression in Gata3Citrine/+ reporter mice brightly marked BM immature ILC2 (Fig. 1 F; Hoyler et al., 2012). To establish parity with wild-type mice, we evaluated major hematopoietic cell populations in C57BL/6 and Zbtb16EGFPCre/+Tcf7mCherry/+RorcThy1.1/+Gata3Citrine/+ mice. No significant differences were observed in cell number apart from the expected reduction in thymic NKT cells, resulting from a hypomorphic Zbtb16 allele, and marginally elevated numbers of liver B cells in compound heterozygotes (Fig. S2, B–G; Park et al., 2019). Overall, the novel Tcf7mCherry, RorcThy1.1, and Gata3Citrine reporters reflect TF expression patterns without altering lymphocyte development.
Figure 1.
Generation of the novel TF reporter mice. (A) Design of CRISPR/Cas9 knock-in strategy. (B–F) Representative flow plots of ILC progenitors from a Tcf7EGFP × Tcf7mCherry adult BM (B), thymocytes from a RorcThy1.1 adult thymus (C), mature ILCs from a RorcThy1.1 SI lamina propria with Rorc-Thy1.1+ cells overlaid in dark gray and total ILCs in light gray (D), ILC progenitors from a RorcThy1.1 x RORγtEGFP E15 FL with RORγ(t) intracellular staining (E), and ILC progenitors from Gata3Citrine adult BM (F). Data are representative of at least two independent experiments (B–F). DN, double negative; DP, double positive; dsDNA, double-strand DNA.
Figure S2.
Immune phenotyping of Tcf7mCherry, RorcThy1.1, Gata3Citrine, and ID2EYFP reporters. (A–G) Expression of Rorc-Thy1.1 in thymic NKT cells from Zbtb16EGFPCreRorcThy1.1 adult mice. Cell numbers from adult C57BL/6 and Zbtb16EGFPCre/+Tcf7mCherry/+RorcThy1.1/+Gata3Citrine/+ mice taken from the thymus (Thy; B), spleen (Spl; C), liver (Liv; D), SI (E), lung (F), and BM (G; n = 5). Each symbol represents an individual mouse; data are presented as mean ± SEM. Q-values were calculated with multiple t tests controlling for false discovery rate. (H–J) Reporter MFI and frequency of reporter positive cells in the indicated populations from select tissues of Zbtb16EGFPCreTcf7mCherry (H), Zbtb16EGFPCreTcf7mCherryID2EYFP (I), and Zbtb16EGFPCreTcf7mCherryGata3Citrine (J) adult mice (n = 3). Each symbol represents an individual mouse. Data are representative of (A) or pooled from (B–J) at least two independent experiments. *, Q < 0.05; ****, Q < 0.0001. ETP, early thymic precursor; DN, double negative; ISP, immature single positive; DP, double positive; SP, single positive.
To catalog reporter expression across tissues, we isolated mature ILCs and reference populations from the novel Tcf7mCherry and Gata3Citrine reporters and the previously generated ID2EYFP reporter. The ID2EYFP reporter is a knock-in knock-out expressing enhanced YFP (EYFP), akin to the ID2EGFP reporter used to characterize the CHILP, αLP, and ILC2 precursor (ILC2P; Hoyler et al., 2012; Klose et al., 2014; Rawlins et al., 2009; Seillet et al., 2016; Xu et al., 2019; Yang et al., 2011). Unsurprisingly, peripheral T cells expressed substantial levels of Tcf7-mCherry, though some population-level heterogeneity was observed (Fig. S2 H). Conversely, most mature ILCs, apart from SI LTi, expressed low or negligible levels of Tcf7-mCherry. ID2-EYFP was uniformly expressed across mature ILC populations (Fig. S2 I). However, NK cells displayed markedly lower levels of ID2-EYFP compared with the ILC1/2/3 and LTi. This observation is consistent with the ID2EGFP reporter and the novel ID2TagBFP reporter but contrasts with the recently described Id2RFP reporter, where Id2-RFP expression is equivalent in all ILCs (Hoyler et al., 2012; Walker et al., 2019; Xu et al., 2019). Lastly, Gata3-Citrine expression was predictably high in lung and SI ILC2, while T cells and all other ILCs expressed low to intermediate levels (Fig. S2 J; Hoyler et al., 2012; Zhong et al., 2016). Expectedly, B cells were devoid of reporter expression in Tcf7mCherry, ID2EYFP, and Gata3Citrine mice.
FL EILPs are transcriptionally heterogeneous
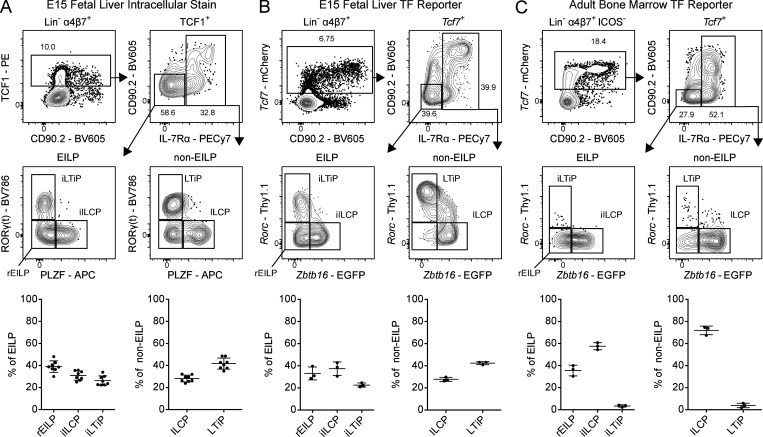
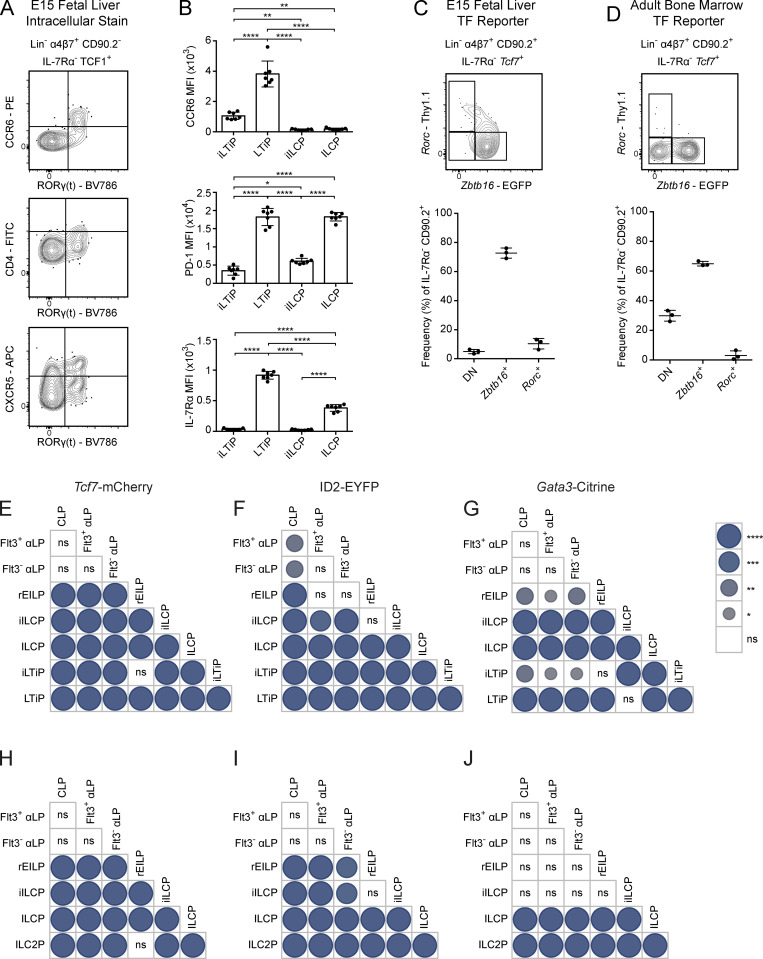
The EILP is the critical stage in which TCF1 expression promotes ILC lineage fate decisions, and was initially overlooked given its unexpected placement at a stage of transient IL-7Rα down-regulation, a marker commonly used to identify mature and developing ILCs (Harly et al., 2018; Harly et al., 2019; Yang et al., 2015). To date, the EILP has only been characterized in the BM, and the existence of an equivalent precursor in the FL has not been reported. Although the BM EILP is considered multipotent for all ILC lineages (Fig. S1 A), its ability to generate LTi at a clonal level remains unresolved, as LTi arise almost exclusively during the fetal period (Constantinides et al., 2014; Eberl et al., 2004; Possot et al., 2011; Sawa et al., 2010; Yang et al., 2015). To this end, we performed intracellular staining for TCF1, PLZF (Zbtb16), and RORγ(t) within embryonic day 15 (E15) FL ILC progenitors and divided Lin−α4β7+TCF1+ cells into non-EILP (IL-7Rα+) and EILP (IL-7Rα−CD90.2−) populations, conforming to the gating strategy used to identify the BM EILP (Fig. 2 A; Yang et al., 2015). This result supports the existence of an EILP within the FL. The non-EILP comprises the previously defined ILCP and LTiP (Fig. 2 A, lower right panel). A fraction of PLZF−RORγ(t)− non-EILP expresses low levels of IL-7Rα and is negative for CD90.2, which may reflect TCF1 expression before IL-7Rα down-regulation. Surprisingly, we observed substantial heterogeneity among the EILP. The EILP displayed a remarkably similar pattern of PLZF and RORγ(t) expression compared with non-EILP cells, mirroring the ILCP and LTiP, respectively (Fig. 2 A, lower left panel). A subset of BM EILP expresses PLZF; however, RORγ(t) expression has not been reported in this population, ostensibly due to the paucity of RORγ(t)+ BM cells (Constantinides et al., 2014; Klose et al., 2014; Walker et al., 2019; Yang et al., 2015). Given the striking similarities of the PLZF+ EILP and RORγ(t)+ EILP with the ILCP and LTiP, respectively, hereafter we refer to the PLZF+ EILP as the incipient ILCP (iILCP), the RORγ(t)+ EILP as the incipient LTiP (iLTiP), and the remaining PLZF−RORγ(t)− EILP as the refined EILP (rEILP).
Figure 2.
Characterization of TF expression in the FL EILP. (A) Intracellular staining for TCF1, PLZF, and RORγ(t) TFs in C57BL/6 E15 FL (n = 8). (B) Representative flow cytometry plots of ILC progenitors from a Zbtb16EGFPCreTcf7mCherryRorcThy1.1 E15 FL (n = 3). (C) Representative flow cytometry plots of ILC progenitors from a Zbtb16EGFPCreTcf7mCherryRorcThy1.1 adult BM (n = 3). Associated plots reflect population frequencies; each symbol represents an individual FL or BM; data are presented as mean ± SEM. Data are representative of or pooled from at least two independent experiments.
Phenotypic analysis supported the notion that the iLTiP was more immature than the LTiP. A small fraction of iLTiP express CCR6 and CXCR5 but were devoid of CD4 expression, all of which are markers up-regulated during LTi maturation (Fig. S3 A; Eberl et al., 2004; Ishizuka et al., 2016b; Possot et al., 2011; Sawa et al., 2010). Furthermore, CCR6, PD-1, a marker correlated with ILCP maturation, and IL-7Rα were expressed at lower levels on the iLTiP compared with the LTiP (Fig. S3 B; Mao et al., 2017; Seillet et al., 2016; Yu et al., 2016). These results suggested that the iLTiP could represent an early precursor upstream of the LTiP.
Figure S3.
Analysis of ILC and LTi maturation markers on iILCP and iLTiP. (A) Representative flow cytometry plots of the EILP from a C57BL/6 E15 FL stained for the indicated surface markers and intracellular RORγ(t) (n = 4). (B) MFI of CCR6, PD-1, and IL-7Rα on the iLTiP, LTiP, iILCP, and ILCP from a Zbtb16EGFPCreTcf7mCherryRorcThy1.1 E15 FL (n = 7). Each symbol represents an individual FL; data are presented as mean ± SEM. (C) Representative flow cytometry plots of ILC progenitors from a Zbtb16EGFPCreTcf7mCherryRorcThy1.1 E15 FL (n = 3). (D) Representative flow cytometry plots of ILC progenitors from a Zbtb16EGFPCreTcf7mCherryRorcThy1.1 adult BM (n = 3). Associated plots reflect population frequencies; each symbol represents an individual FL or BM; data are presented as mean ± SEM. (E–J) Pairwise comparisons of Tcf7-mCherry, ID2-EYFP, and Gata3-Citrine reporter expression MFI from Fig. 3 plots for FL (E–G) and BM (H–J) precursors. Circle size and color denote level of significance. Data are representative of (A) or pooled from (B–J) at least two independent experiments. Pairwise comparison P values were calculated by one-way ANOVA with Tukey’s multiple comparisons test *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
Triple transgenic reporter mice reflect EILP heterogeneity
While TF staining facilitates the simultaneous detection of PLZF, RORγ(t), and TCF1, this method precludes live cell sorting for functional analysis. By breeding the novel Tcf7mCherry and RorcThy1.1 mice with existing Zbtb16EGFPCre mice to generate Zbtb16EGFPCreTcf7mCherryRorcThy1.1 mice, we successfully recapitulated FL EILP heterogeneity observed via intracellular staining (Fig. 2 B). We principally observed Zbtb16+ and Zbtb16−Rorc− cells among adult BM EILP and non-EILP, consistent with the diminished capacity of BM precursors to generate ILC3 and LTi (Fig. 2 C; Constantinides et al., 2014; Klose et al., 2014; Possot et al., 2011). Nevertheless, we discerned a rare Rorc-Thy1.1Hi fraction among the non-EILP (Fig. 2 C, lower right plot). Compared with TCF1 in the FL, Tcf7-mCherry expression increased with CD90.2 expression in both FL and BM reporter mice, a facet shared with the Tcf7-EGFP reporter (Fig. 2, A–C, upper left panels; Harly et al., 2018; Harly et al., 2019; Yang et al., 2015). This observation may result from a difference in the half-life of the reporter versus protein (Mohrs et al., 2005). While not noted in previous reports, we consistently observed a population of TCF1+/Tcf7+IL-7Rα−CD90.2+ cells in both the FL and BM (Fig. 2, A–C, upper right panels; Harly et al., 2018; Harly et al., 2019; Yang et al., 2015). These cells were predominantly Zbtb16-EGFP+, though Rorc-Thy1.1+ and double-negative populations were discernable in the FL and BM, respectively (Fig. S3, C and D). Taken together, through the combinatorial use of novel TF reporters, we can reconstruct EILP heterogeneity observed via TF staining.
Given the combinatorial nature of the novel and existing TF reporters, we incorporated Gata3Citrine or ID2EYFP reporters with Zbtb16GFPCreTcf7mCherryRorcThy1.1 mice to gauge the hierarchical placement of the rEILP, iILCP, and iLTiP through TF expression patterns. E15 FL rEILP, iILCP, and iLTiP expressed intermediate levels of Tcf7-mCherry and ID2-EYFP relative to the earliest precursors (CLP, Flt3+ αLP, and Flt3− αLP) and the later ILCP and LTiP (Fig. 3, A and B; and Fig. S3, E and F). A fraction of FL αLP expressed low/intermediate levels of ID2-EYFP compared with the ILCP and LTiP, in accordance with a previous report (Chea et al., 2016). Gata3-Citrine expression was higher in both the iILCP and ILCP compared with the iLTiP and LTiP, respectively, suggesting that early up-regulation of Gata3 coincides with Zbtb16 expression (Fig. 3 C and Fig. S3 G). Tcf7, ID2, and Gata3 expression increases from early to late ILC progenitors, though expression levels vary in mature ILCs (Fig. S2, H–J; Constantinides et al., 2014; Harly et al., 2018; Ishizuka et al., 2016a; Ishizuka et al., 2016b; Seillet et al., 2016; Yu et al., 2016). In support of a linear maturation program, expression of Tcf7-mCherry, ID2-EYFP, and Gata3-Citrine all progressively increased from the rEILP to iILCP to ILCP. Likewise, these reporters increased from the iLTiP to the LTiP. The adult BM displayed comparable Tcf7-mCherry and Gata3-Citrine expression patterns, though low ID2 expressers were absent from the αLP (Fig. 3, D–F; and Fig. S3, H–J). As expected, Tcf7-mCherry expression decreased while Gata3-Citrine expression increased as cells matured into ILC2P (Constantinides et al., 2014; Xu et al., 2019). In summary, the transcriptional states of the rEILP, iILCP, and iLTiP indicate that these precursors may arise before the ILCP and LTiP in developmental time (Fig. S1 B).
Figure 3.
TF reporter expression in FL and adult BM ILC progenitors. (A–F) Representative histogram plots of Tcf7-mCherry, ID2-EYFP, and Gata3-Citrine reporter expression in the indicated ILC progenitors from (A) Zbtb16EGFPCreTcf7mCherryRorcThy1.1 E15 FLs (n = 3), (B) Zbtb16EGFPCreTcf7mCherryRorcThy1.1ID2EYFP E15 FLs (n = 6), (C) Zbtb16EGFPCreTcf7mCherryRorcThy1.1Gata3Citrine E15 FLs (n = 3), (D) Zbtb16EGFPCreTcf7mCherry adult BM, (E) Zbtb16EGFPCreTcf7mCherryRorcThy1.1ID2EYFP adult BM (n = 3), and (F) Zbtb16EGFPCreTcf7mCherryRorcThy1.1Gata3Citrine adult BM (n = 3). Associated plots reflect population median fluorescent intensity (MFI); each symbol represents an individual FL or BM; data are presented as mean ± SEM; pairwise comparison P values are presented in Fig. S3, E–J, and were calculated by one-way ANOVA with Tukey’s multiple comparisons test. Data are representative of or pooled from at least two independent experiments.
The iILCP and iLTiP are restricted precursors to the ILCP and LTiP
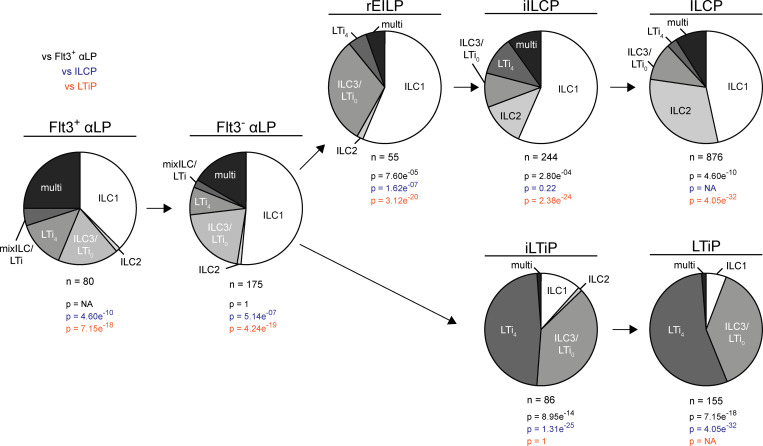
To formally determine the cellular potential of the FL rEILP, iILCP, and iLTiP in comparison with the earlier αLP and the later ILCP and LTiP, we performed clonal analysis using Zbtb16EGFPCreTcf7mCherryRorcThy1.1 mice. Precursors were cultured on OP9 stromal cells and assessed for the ability to differentiate into ILC1/2/3 and/or LTi (Fig. 4 and Table S1, A–G). CCR6 identifies LTi in vivo; however, CCR6 proved unreliable in vitro (unpublished observations). Therefore, we used CD4 as a marker of LTi potential. While CD4 expression is characteristic of a majority of LTi (LTi4), ∼40% of LTi lack CD4 (LTi0) and are indistinguishable from CD4− ILC3 in vitro and in vivo; this uncertainty is acknowledged as ILC3/LTi0 (Ishizuka et al., 2016b; Sawa et al., 2010). Single clones were index sorted from the EILP to facilitate the unbiased assessment of cell potential. In short, EILP were sorted based on the minimal set of identifying markers used in Fig. 2 B (bottom left panel). Following in vitro differentiation, wells were computationally assigned to a subset (rEILP, iILCP, or iLTiP) based on Zbtb16-EGFP and Rorc-Thy1.1 expression at the time of sorting. As expected, the ILCP and LTiP gave rise almost exclusively to ILC1/2 and ILC3/LTi0 or to LTi4 and ILC3/LTi0, respectively. The iLTiP were already largely LTi lineage restricted, displaying a striking bias toward LTi4 and ILC3/LTi0 akin to the LTiP (P = 1). The iILCP exhibited a similar differentiation potential as the ILCP (P = 0.22), generating primarily ILC1/2 and ILC3/LTi0, with a reduced frequency of ILC2, presumably due to the premature interruption of Notch signals required for efficient ILC2 differentiation (Ishizuka et al., 2016b). The iLTiP generated a fraction of ILC1, and the iILCP generated a fraction of LTi4 (∼11% each, Table S1, D and F), distinguishing them from their downstream counterparts, indicating incomplete lineage commitment under our culture conditions. Taken as is, evidence from cell phenotype, transcriptional state, and differentiation potential are consistent with a model wherein the iILCP and iLTiP arise upstream of the ILCP and LTiP and have diminished alternate lineage potential downstream of the common ILC–LTi precursor (Fig. S1 B).
Figure 4.
Clonal potential of FL ILC progenitors. Single-cell potential of the indicated ILC progenitors sorted from Zbtb16EGFPCreTcf7mCherryRorcThy1.1 E14-E16 FLs onto OP9 stromal cells. Pie charts represent the frequency of single ILC1 (NK1.1+ICOS−α4β7−), ILC2 (NK1.1−ICOS+α4β7−), and ILC3/LTi0 (NK1.1−ICOS+/−α4β7+CD4−); two or more ILC subsets (multi); LTi4 (NK1.1−ICOS+/−α4β7+CD4+); and mixed ILC1/2/3/LTi0 and LTi4 (mixILC/LTi). Data are pooled from >20 independent experiments. P values for pie charts were calculated using Chi-Square Test for Independence followed by post-hoc analysis with Bonferroni correction; comparisons were made against the Flt3+ αLP (black), ILCP (blue), and LTiP (orange).
BM rEILP generate Zbtb16-EGFP+ cells in short-term culture (Harly et al., 2019). As the FL rEILP appeared to represent a common ILC–LTi precursor downstream of the αLP, we evaluated its clonal potential with respect to the αLP. Previous work from our laboratory suggested that the αLP represents the stage of bifurcation into the ILC and LTi lineages (Ishizuka et al., 2016b). The rEILP generated ILC1/2 and ILC3/LTi0 progeny similar to the iILCP, with a more pronounced decrease in ILC2 (Fig. 4). Few LTi4 arose from the rEILP, and no mixed ILC and LTi lineage wells were observed, which stands in contrast to the Flt3+ αLP and Flt3− αLP (Ishizuka et al., 2016b). Thus, the rEILP likely represents a precursor upstream of the iILCP, before expression of the ILC lineage marker Zbtb16 (Harly et al., 2019). Taken together, these findings indicate that the ILC–LTi precursor resides upstream of the EILP (Fig. S1 B).
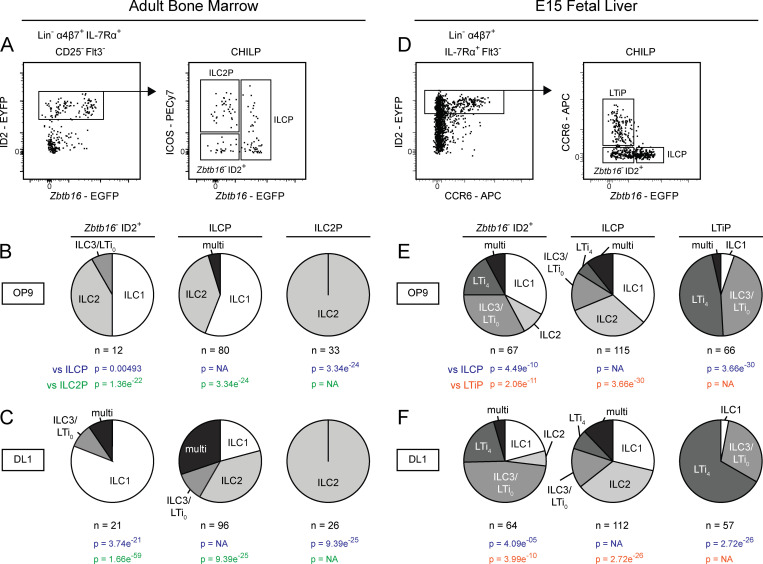
The CHILP is a heterogeneous mixture of ILC and LTi progenitors
The CHILP, characterized in the BM by high ID2 expression upstream of the ILCP, was proposed to be the common ILC–LTi precursor (Fig. S1 A; Ishizuka et al., 2016b; Klose et al., 2014; Zook and Kee 2016). However, whether the CHILP is a shared ILC–LTi precursor or a heterogeneous mixture of ILCP and LTiP has not been tested. As such, we performed index-based clonal analysis on the CHILP population isolated from the BM of Zbtb16EGFPCreID2EYFP mice. Cells were seeded onto OP9 or OP9-DL1 stroma, as Notch ligand is required for efficient generation of group 3 ILCs from BM precursors (Possot et al., 2011). The BM CHILP comprised a mixture of Zbtb16+ ILCP, ICOS+Zbtb16− ILC2P, and Zbtb16−ID2+ cells (Fig. 5 A). The latter subset primarily generated single wells of ILC1, ILC2, or ILC3/LTi0, with a fraction of multi ILC1/2 and ILC3/LTi0 wells arising in cultures with Notch ligand. (Fig. 5, B and C; and Table S1, H–M). Given the limited ability of BM ILC progenitors to generate group 3 ILCs in vitro, we extended our analysis by index sorting the CHILP from the FL of Zbtb16EGFPCreID2EYFP mice. The FL CHILP comprised Zbtb16+ ILCP and Zbtb16−ID2+ cells, similar to the BM, in addition to CCR6-expressing LTiP (Fig. 5 D). FL CHILP subsets did not generate mixed wells of ILC and LTi on either OP9 or OP9-DL1 stroma (Fig. 5, E and F; and Table S1, N–S). Thus, in both the BM and FL the CHILP, like the EILP, represents a mixture of ILC and LTi lineage progenitors downstream of a shared ILC–LTi precursor (Fig. S1, A and B).
Figure 5.
Clonal analysis of the CHILP from adult BM and FL. (A–C) Index gating strategy for the CHILP from Zbtb16EGFPCreID2EYFP adult BM. Pie charts represent the frequency of single ILC1, ILC2, ILC3/LTi0; two or more ILC subsets (multi); LTi4; and mixed ILC1/2/3/LTi0 and LTi4 (mixILC/LTi) on OP9 (B) and OP9-DL1 (C) stromal cells. (D) Index gating strategy for the CHILP from Zbtb16EGFPCreID2EYFP E14-E16 FL. (E and F) Pie charts represent the frequency of single ILC1, ILC2, ILC3/LTi0; two or more ILC subsets (multi); LTi4; and mixed ILC1/2/3/LTi0 and LTi4 (mixILC/LTi) on (E) OP9 and (F) OP9-DL1 stromal cells. Early T cells were identified in OP9-DL1 cultures by high expression of CD25+; T cell–only wells were excluded from pie charts, while ILC+T cell wells were included with the appropriate ILC classification. Data are representative of (A and D) or pooled from (B, C, E, and F) four independent experiments. P values for pie charts were calculated using Chi-Square Test for Independence followed by post-hoc analysis with Bonferroni correction; comparisons were made against the ILCP (blue), ILC2P (green), and LTiP (orange).
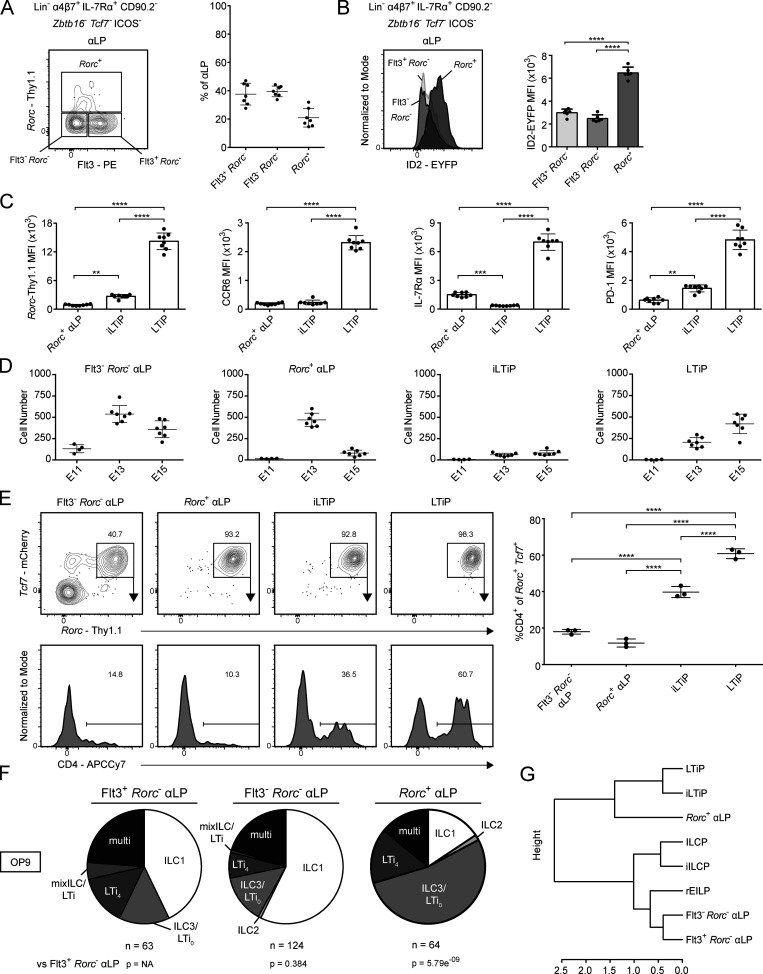
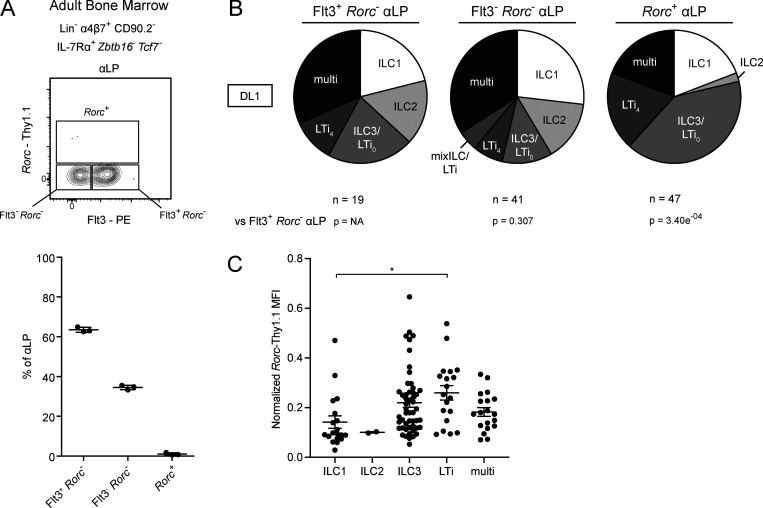
An early Rorc-expressing αLP marks the bifurcation between ILC and LTi lineages
As both the EILP and the CHILP contain separate precursors to the ILC and LTi lineages, we examined the upstream αLP for the presence of an earlier precursor in Zbtb16EGFPCreTcf7mCherryRorcThy1.1 mice. A subset of αLP expressed Rorc-Thy1.1 (Rorc+ αLP), despite lacking Tcf7-mCherry expression (Fig. 6 A). This population was absent from the adult BM (Fig. S4 A). The Rorc+ αLP represented the majority of ID2-EYFPLo/Int cells among the FL αLP (Fig. 6 B and Fig. 3 B). Compared with the iLTiP and LTiP, the Rorc+ αLP expressed less Rorc-Thy1.1 and PD-1, while being low for CCR6 and intermediate for IL-7Rα (Fig. 6 C). We then examined the ontogeny and immediate potential of the Rorc+ αLP. The iLTiP was detectable as early as E13 alongside the LTiP, and both increased until E15 (Fig. 6 D). The Rorc+ αLP was maximally present at E13, in greater numbers than either the iLTiP or LTiP, but decreased by E15, while the Flt3−Rorc− αLP was detectable as early as E11. The phenotype and rapid expansion and contraction indicated that the Rorc+ αLP might precede the iLTiP and LTiP in a developmental wave during ontogeny. In a short-term in vitro culture assay, the Rorc+ αLP, iLTiP, and LTiP rapidly and almost exclusively generated Tcf7+Rorc+ cells; however, only the iLTiP and LTiP generated a substantial frequency of LTi4 cells by 2 d (Fig. 6 E). Notably, while both the Flt3−Rorc− αLP and Rorc+ αLP promptly up-regulated Tcf7-mCherry expression, only a fraction of Flt3−Rorc− αLP expressed Rorc-Thy1.1. These results suggested that the Rorc+ αLP could represent a very early precursor biased toward the LTi lineage.
Figure 6.
Identification of Rorc expression in the FL αLP. (A) Representative flow cytometry plot of αLP from Zbtb16EGFPCreTcf7mCherryRorcThy1.1 E15 FL (n = 7). Adjacent plot reflects population frequencies. (B) Representative histogram plot of ID2-EYFP expression in the indicated ILC progenitors from Zbtb16EGFPCreTcf7mCherryRorcThy1.1ID2EYFP E15 FL (n = 6). Associated plots reflect population MFI. (C) MFI of Rorc-Thy1.1, CCR6, IL-7Rα, and PD-1 on Rorc+ αLP, iLTiP, and LTiP from Zbtb16EGFPCreTcf7mCherryRorcThy1.1 E15 FL (n = 8). (D) Quantification of Flt3− Rorc− αLP, Rorc+ αLP, iLTiP, and LTiP from Zbtb16EGFPCreTcf7mCherryRorcThy1.1 FL at E11, E13, and E15. (E) Representative flow plots and histograms of bulk cultured Flt3−Rorc− αLP, Rorc+ αLP, iLTiP, and LTiP from Zbtb16EGFPCreTcf7mCherryRorcThy1.1 E15 FL cultured for 2 d on OP9 stromal cells (n = 3). Adjacent plot reflects CD4+ frequency among Rorc+Tcf7+ cells. (F) Single-cell potential of the indicated αLP precursors sorted from Zbtb16EGFPCreTcf7mCherryRorcThy1.1 E15 FL onto OP9 stromal cells. (G) Hierarchical clustering dendrogram of ILC progenitor clonal potential on OP9 stromal cells calculated from Pearson correlation. For plots, each symbol represents an individual FL; data are presented as mean ± SEM; and P values were determined by one-way ANOVA with Tukey’s multiple comparisons test. P values for pie charts were calculated using Chi-Square Test for Independence followed by post-hoc analysis with Bonferroni correction; comparisons were made against the Flt3+ Rorc− αLP (black). Data are pooled from at least two independent experiments. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Figure S4.
Analysis of Rorc+ αLP. (A) Representative flow cytometry plot of αLP from Zbtb16EGFPCreTcf7mCherryRorcThy1.1 adult BM (n = 3). Associated plot reflects population frequencies; each symbol represents an individual BM; data are presented as mean ± SEM. (B) Single-cell potential of the indicated αLP precursors sorted from Zbtb16EGFPCreTcf7mCherryRorcThy1.1 E15 FL onto OP9-DL1 stromal cells. Early T cells were identified in OP9-DL1 cultures by high expression of CD25+; T cell–only wells were excluded from charts, while ILC+T cell wells were included with the appropriate ILC. (C) Normalized Rorc-Thy1.1 MFI of index-sorted single Rorc+ αLP categorized by in vitro potential on OP9 and OP9-DL1. Data were normalized by experiment to average LTiP Rorc-Thy1.1 MFI; data are presented as mean ± SEM; and P values were determined by one-way ANOVA with Tukey’s multiple comparisons test. Data are pooled from two (A and B) or four (C) independent experiments. P values for pie charts were calculated using Chi-Square Test for Independence followed by post-hoc analysis with Bonferroni correction; comparisons were made against the Flt3+ Rorc− αLP (black). *, P < 0.05.
We further addressed the clonal potential of the Rorc+ αLP through index-based sorting of the αLP onto OP9 and OP9-DL1 stromal cells, subdividing into Flt3+Rorc− αLP, Flt3−Rorc− αLP, and Rorc+ αLP. Regardless of the culture condition, the Rorc+ αLP predominantly generated ILC3/LTi0 and LTi4, though the large fraction of ILC3/LTi0 may indicate that additional signals are required for efficient CD4 expression (Fig. 6 F, Fig. S4 B, and Table S1, T–Y). Nevertheless, the Rorc+ αLP retained multipotentiality similar to the Flt3+Rorc− and Flt3−Rorc− αLP. Multipotentiality was independent of initial Rorc-Thy1.1 expression level in index-sorted Rorc+ αLP (Fig. S4 C). Hierarchical clustering by clonal potential on OP9 stroma revealed that the Rorc+ αLP was more similar to the iLTiP and LTiP than it was to the Flt3+Rorc− and Flt3−Rorc− αLP (Fig. 6 G). Based on these results, the Rorc+ αLP appears to be an early LTi-specified precursor before LTi lineage restriction.
ATAC-seq reinforces ILC and LTi lineage progression and reveals differential use of TF modules
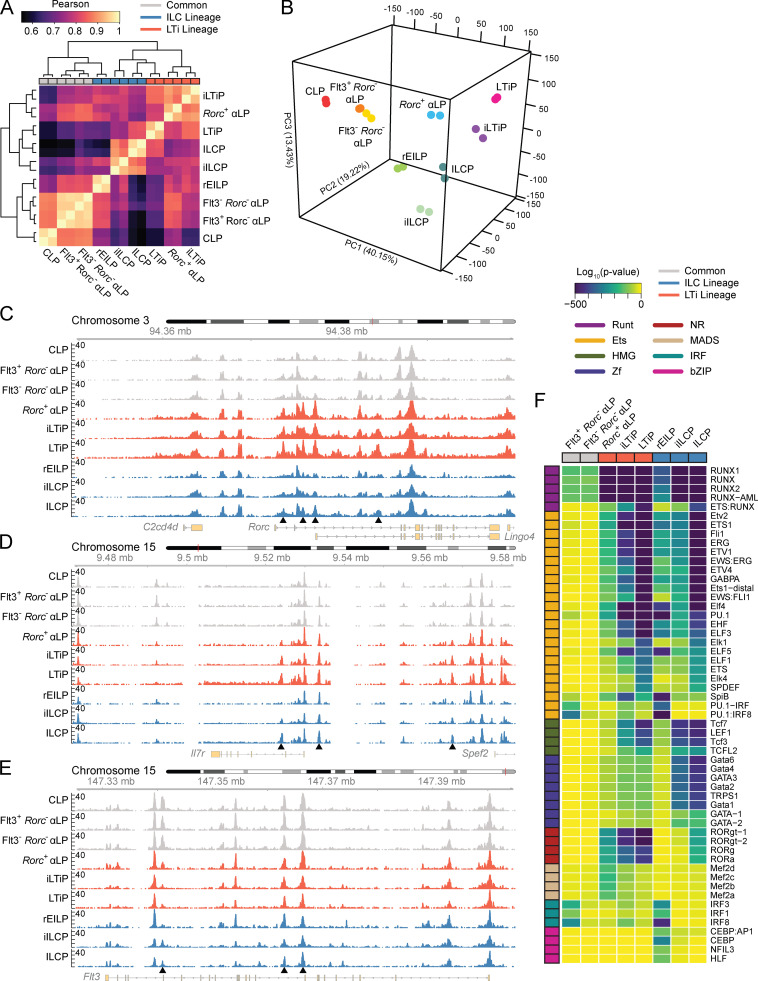
To gain further insight into the lineage relationship between the newly identified FL ILC and LTi lineage intermediates, we profiled their chromatin accessibility landscape using ATAC-seq. Unsupervised hierarchical clustering separated the progenitors into roughly three groups. Common precursors (CLP, Flt3+Rorc− αLP, and Flt3−Rorc− αLP) clustered together away from ILC and LTi lineage progenitors (Fig. 7 A). Consistent with its transitional placement, the Rorc+ αLP clustered with the iLTiP while exhibiting high correlation with the common αLPs. Notably, ILC and LTi lineage progenitors did not cleanly separate into two groups. Though the iILCP and ILCP formed a distinct cluster separate from the Rorc+ αLP and iLTiP, the LTiP clustered with the ILCP, despite strong correlation with LTi lineage progenitors. Clustering of the LTiP with the ILCP may result from an oversized influence of principal component 1 on the correlation analysis that reflects lineage maturation and/or the appearance of common type 3 signatures in a heterogeneous ILCP (Fig. 7 B; Ishizuka et al., 2016b). Additionally, the rEILP clustered with the common precursors away from the iILCP and ILCP, mirroring the hierarchical clustering based on differentiation potential (Fig. 6 G).
Figure 7.
Chromatin accessibility of FL ILC progenitors. (A) Correlation heatmap for FL ILC progenitor ATAC-seq samples (n = 2). Precursors are categorized by in vitro clonal potential. (B–E) 3D principal component (PC) analysis of ATAC-seq data. Accessibility coverage tracks at the indicated loci in FL ILC progenitors: Rorc (C), Il7r (D), and Flt3 (E). Black arrow heads indicate regions of dynamic chromatin accessibility. (F) Developmental trajectory heatmap of TF motif enrichment derived from comparing the indicated FL ILC progenitor to the CLP. Motifs were plotted if they exhibited a log10(P value) ≤ −100 and a signal-to-noise ratio ≥1.5 in at least one comparison. Data represents two samples per cell type and were collected across seven independent experiments.
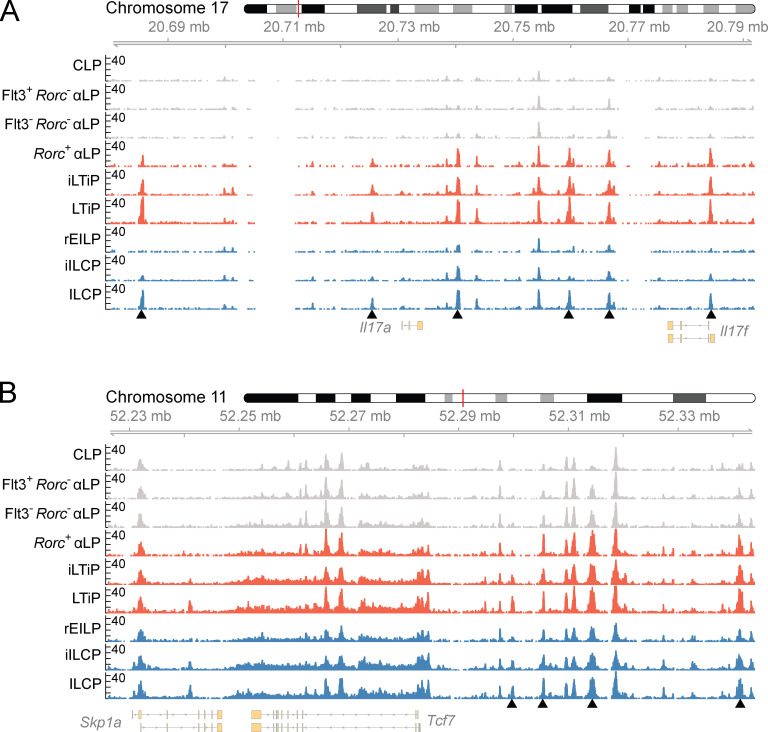
We then examined chromatin accessibility data tracks near genes associated with ILC and LTi lineage differentiation. Though the Rorc+ αLP is only specified toward the LTi lineage, accessibility at the Rorc locus was comparable to that observed in the iLTiP and LTiP (Fig. 7 C). ILC lineage progenitors, in contrast, only showed increased accessibility at the Rorc locus within the ILCP, the stage in which multi-transcriptional priming and ILC1/2/3 subset differentiation occurs (Ishizuka et al., 2016b). This pattern of accessibility was mirrored at the Il17a/Il17f locus (Fig. S5 A). Increasing accessibility was observed in the Il7r locus at two sites bound by TCF1 (−3.6 kb) and GATA3 (+5.3 kb), tracking IL-7Rα reexpression (Fig. 7 D; Yang et al., 2013; Zhong et al., 2016). Similarly, accessibility at a Tcf7 enhancer (−31 kb) progressively increased before the expression of Tcf7 (Fig. S5 B and Fig. 3 A). Examination of the Flt3 locus showed that regions accessible in common precursors were either unchanged or moderately reduced in accessibility within the Rorc+ αLP and rEILP but were diminished in later progenitors (Fig. 7 E). Taken together, comparison of the chromatin accessibility landscape between FL precursors reinforces the hierarchical placement of each precursor during ILC development (Fig. S1 B).
Figure S5.
Chromatin accessibility profiles of FL ILC progenitors. (A and B) Accessibility coverage tracks at the (A) Il17a/Il17f and (B) Tcf7 loci in FL ILC progenitors. Black arrow heads indicate regions of dynamic chromatin accessibility. Data represents two samples per cell type and were collected across seven independent experiments.
The molecular determinants that govern the distinct differentiation of the ILC and LTi lineages are poorly understood. To determine the TFs important for ILC and LTi lineage differentiation across the FL developmental trajectory, we identified statistically enriched TF motifs in ATAC-seq peaks with increased accessibility in each common αLP, ILC, or LTi lineage progenitor in comparison to the CLP. Peaks were robustly and predictably enriched for Runt and ETS motifs in both the ILC and LTi lineages, starting in the common αLPs and progressively increasing in enrichment along each developmental trajectory (Fig. 7 F; Ebihara et al., 2015; Mao et al., 2017; Zook et al., 2016). While the rEILP showed similar enrichment for Runt and ETS motifs, there was distinguishable motif enrichment for the bZIP member CEBP, several IRF factors, and myeloid-expressed ETS factors (i.e., PU.1 and SpiB), consistent with a recent report demonstrating that this population expresses several of these factors and retains dendritic cell (DC) potential (Harly et al., 2019). Binding motifs for TCF1 and other high-mobility group family members were comparably enriched in both the ILC and LTi lineages; motif enrichment followed the known pattern of Tcf7 expression in these cells (Fig. 3 A). More notable was the differential enrichment for zinc finger and nuclear receptor TF motifs between the ILC and LTi lineage, respectively. Whereas motifs for GATA3 and similar zinc finger family members were greatly enriched in the ILC lineage, maximally within the ILCP, the LTi lineage displayed a markedly lower enrichment, mirroring Gata3 expression (Fig. 3 C). Conversely, the LTi lineage was significantly enriched in nuclear receptor motifs comprising the TFs RORγ, RORγt, and RORα, while these motifs only showed enrichment late in ILC lineage development in the ILCP. In summary, the patterns uncovered through TF motif enrichment analysis provide insight into the shared and distinct usage of TFs throughout ILC and LTi lineage development.
Discussion
Using a novel combination of CRISPR/Cas9-generated TF reporter lines, we have identified and resolved population-level heterogeneity within FL ILC progenitors. The EILP is present within the FL and comprises a previously unappreciated mixture of PLZF- and RORγ(t)-expressing cells, herein termed the iILCP and iLTiP, respectively. Although PLZF expression in the BM EILP has been described before, RORγ(t) expression in the FL EILP was overlooked due to the exclusive focus on the adult BM (Harly et al., 2018; Harly et al., 2019; Yang et al., 2015). Functional assessment of precursor potential using the combination reporter mice revealed that the EILP and CHILP contained discrete precursors to the ILC and LTi lineages. The sensitivity and deliberate selection of our reporter combination also enabled the identification of a Rorc+ population before Tcf7 expression, marking the earliest known precursor to the LTi lineage. Analysis of the chromatin accessibility landscape in precursors reenforces the lineages distinctions reached through phenotypic, transcriptional, and in vitro clonal analysis and implicates the differential usage of GATA3 and RORγ(t) in the development of the ILC and LTi lineages, respectively. These results warranted a revised model for ILC development that accounts for the newly discovered heterogeneity and places the common ILC–LTi precursor upstream of its previously assumed position within the Tcf7+ compartment (Fig. S1 B).
Conventional TF staining captures the cellular heterogeneity present within the FL EILP, yet functional analysis necessitated the use of TF reporters. Compared with previous reports that have relied on single TF reporters and surface markers to make claims about ILC development, the multi-TF reporter system enables the simultaneous resolution of several TFs throughout development while permitting the functional assessment of lineage potential. Several recent reports have implemented combinatorial reporters, generated through standard transgenic methods, to interrogate ILC development in the adult BM (Harly et al., 2019; Walker et al., 2019; Xu et al., 2019). The novel application of CRISPR/Cas9-mediated transgenesis in this study stands out as a rapid and flexible method for the generation of custom reporter lines that can be used to evaluate or discover cellular complexity.
The EILP and the CHILP were identified in previous reports using Tcf7EGFP and ID2EGFP mice respectively, and both precursors were reported to generate restricted ILC subsets (Fig. S1 A; Harly et al., 2018; Klose et al., 2014; Yang et al., 2015). The observations made herein suggest a different model for ILC development (Fig. S1 B). Clonal analysis performed with Zbtb16EGFPCreTcf7mCherryRorcThy1.1 mice demonstrates that the FL EILP is not a precursor to all ILCs but instead contains distinct intermediates to the ILC and LTi lineages upstream of the ILCP and LTiP. Our discovery of an iILCP and iLTiP within the EILP suggested several surprising implications. First, precursors to the ILC and LTi lineage both transiently down-regulate IL-7Rα expression (Harly et al., 2018). The consequence of reduced IL-7Rα expression during ILC and LTi development remains to be determined. Continuous IL-7Rα expression via transgene only marginally impairs thymocyte development, potentially by inhibiting TCF1 up-regulation, though group 3 ILCs appear unperturbed by sustained IL-7Rα expression (Allman et al., 2003; Yu et al., 2004). Second, the existence of intermediate precursors to the ILCP and LTiP intermediate for ID2 expression called into question the identity of the CHILP. Using Zbtb16EGFPCreID2EYFP reporter mice to isolate the Zbtb16−ID2+ fraction of the CHILP, we provide evidence through index-based clonal analysis that the CHILP is not a shared ILC–LTi precursor as formerly suggested, but actually comprises an admixture of restricted precursors in both the BM and FL, similar to the EILP, and is unlikely to be a discrete precursor.
Another inference that can be made from EILP heterogeneity is that the bifurcation of the ILC and LTi must occur earlier in development. Despite looking like a shared ILC–LTi precursor, the rEILP appears to represent a stage before the iILCP, preceding Zbtb16 expression with negligible LTi potential. This observation is in line with the recent study that found that a BM-specified EILP, equivalent to the rEILP here, generated Zbtb16+ cells in short-term culture (Harly et al., 2019). Instead, we discerned an earlier multipotent Rorc+ αLP in the FL of Zbtb16EGFPCreTcf7mCherryRorcThy1.1 mice that was devoid of several maturation markers, including Tcf7, but was already biased toward LTi differentiation. Sawa et al. (2010) proposed the existence of two FL α4β7+RORγt-EGFP+ populations that generated LTi almost exclusively. RORγt-EGFPhigh cells noted in that study likely represent the LTiP, while the RORγt-EGFPmid cells likely represent a mixture of both iLTiP and Rorc+ αLP, as these two populations were indistinguishable in the absence of Tcf7 and IL-7Rα assessment. The absence of Rorc+ αLP and diminished output of LTi from adult BM may result from a failure to up-regulate ID2 in αLP (Fig. 3 E), as sequential expression of ID2 and RORγ(t) was proposed to facilitate LTi development (Cherrier et al., 2012). Taken together, our results suggest a model in which a shared ILC–LTi precursor within the αLP expresses Rorc, diminishing ILC lineage potential, followed closely by Tcf7 expression and down-regulation of IL-7Rα giving rise to the iLTiP. In parallel, Tcf7 and subsequent Zbtb16 and Gata3 expression in the iILCP defines the reduction in LTi lineage potential.
ATAC-seq assessment of FL ILC progenitors reinforced the cellular identity and lineage classification of the newly characterized rEILP, iILCP, Rorc+ αLP, and iLTiP. The Rorc+ αLP categorically presented a transitional chromatin accessibility landscape between common precursors and the more mature iLTiP and LTiP. ILC lineage and LTi lineage progenitors largely clustered together apart from two exceptions. First, the rEILP clustered with common precursors and displayed aspects of alternate lineage potential, including CEBP, IRF8, PU.1, and SpiB motif enrichment. Harly et al. (2019) found that rEILP from BM generated DCs in vitro under permissive conditions and established TCF1 as the key factor in driving ILC lineage commitment. Our results indicate that, like BM rEILP, FL rEILP may retain DC potential, emulating in the ILC lineage the progressive loss of B cell and then myeloid cell potential observed during thymocyte development (Allman et al., 2003; Yoshida et al., 2001). Second, notwithstanding the disparate phenotype, transcriptional state, and cellular potential of the ILCP and LTiP, clustering of these precursors revealed that a similar accessibility landscape was acquired during cellular maturation, and that only a fraction of accessible regions contributed to their distinct cellular identity. The similarity may result from the appearance of shared type 3 signatures, as the ILCP contains ILC3-biased cells (Ishizuka et al., 2016b). Future characterization of the chromatin accessibility landscape will benefit from the isolation of homogenous populations of ILC1, ILC2, and ILC3 precursors. Nevertheless, TF motif enrichment across the developmental trajectories suggests a markedly differential usage of GATA3 and RORγ(t) in establishing ILC and LTi lineage cellular identity, respectively. Moreover, our Gata3Citrine reporter indicated that expression of Gata3 in the LTi lineage is delayed when comparing the iLTiP to the iILCP and does not reach the expression level observed in the ILCP. Zhong et al. (2020) demonstrated a differential reliance on the TF GATA3 for the development of ILC1/2/3 and LTi cells; whereas ILC1/2/3 require GATA3 for development, LTi develop in the absence of GATA3, albeit while exhibiting functional defects. The results presented herein highlight the role GATA3 plays as a defining factor governing the distinct development of these two lineages.
The model proposed herein conforms with our earlier observations using single-cell analysis that implicated the αLP as the point of lineage bifurcation between the ILC and LTi lineage, based on the generation of mixed colonies (Ishizuka et al., 2016b). The frequency of mixed colonies was notably low, possibly resulting from a lack of critical factors necessary for the overall survival of early multipotent αLPs given that our culture system supports ample LTi generation. Furthermore, the window to capture large numbers of ILC–LTi precursors among the αLP may be earlier than E14–16, as suggested by the early wave of Rorc+ αLP at E13 (Fig. 6 D). In stark contrast to previous findings and the work presented here, Walker et al. (2019) generated a novel PLZFtdTomato reporter and observed PLZF-tdTomato expression in peripheral LTi-like cells and in CD4+ LTiP from the FL (Constantinides et al., 2014; Ishizuka et al., 2016b). These incongruous findings may be explained by the design of each Zbtb16 reporter. The Zbtb16EGFPCre reporter was constructed by inserting an IRES-EGFPCre sequence downstream of the Zbtb16 stop codon (Constantinides et al., 2014). The PLZFtdTomato reporter, on the other hand, was constructed by inserting a T2A-tdTomato polyA sequence before the Zbtb16 stop codon (Walker et al., 2019). While the T2A sequence enables precise expression of tdTomato during protein translation, the incorporation of an exogenous polyA bypasses the endogenous polyA signal sequence and may disrupt any 3′ UTR regulatory mechanisms that could govern the stability or turnover of Zbtb16 mRNA (Mayr, 2017; Park et al., 2019). Altered post-transcriptional regulation of Zbtb16 mRNA, potentially resulting in exaggerated expression, may explain the observation that the PLZFtdTomato reporter does not reflect Zbtb16EGFPCre fate mapping and known intracellular staining patterns for PLZF in the LTiP, ILC2P, or mature ILC populations (Constantinides et al., 2014; Ishizuka et al., 2016b; Klose et al., 2014; Seillet et al., 2016; Zhong et al., 2020).
In summary, our study has highlighted the value of applying a multi-TF reporter approach, specifically to the dissection of ILC and LTi lineage progression. Moreover, we have identified several unappreciated precursors along both the ILC and LTi development pathways. Delineation of these early intermediates will facilitate the molecular interrogation of factors and mechanisms that govern ILC development and lineage progression.
Materials and methods
Mice
Zbtb16EGFPCre mice were generated as previously described (Constantinides et al., 2014). ID2EYFP mice were a gift from Ananda Goldrath (University of California, San Diego, La Jolla, CA) and were described previously (Yang et al., 2011). RORγtEGFP mice were described previously (Eberl et al., 2004). Tcf7EGFP mice were a gift from Avinash Bhandoola (National Institutes of Health, Bethesda, MD) and were described previously (Yang et al., 2015). C57BL/6J mice were maintained in-house. For FL experiments, the morning after timed pregnancy setup was considered as E0. All mice were maintained on the C57BL/6J background and were bred and housed in the specific pathogen-free facility at the University of Chicago. Experiments were conducted under the guidelines of the University of Chicago Institutional Animal Care and Use Committee.
Generation of reporter mice
Tcf7mCherry, RorcThy1.1, and Gata3Citrine mice were generated via CRISPR/Cas9-mediated insertion. Guide sites targeting the 3′ UTR of each gene were designed using the Broad Institute or IDT design tools. Regions flanking the cut site were amplified to generate 1-kb and 3-kb asymmetric homology arms. Homology arms were cloned into a pUC19 vector along with a minimal IRES sequence and reporter sequence using NEB Hi-Fi DNA Assembly Master Mix (NEB). The vector was amplified, purified with the Thermo Purelink HiPure Plasmid Filter Kit (ThermoFisher), and the sequence was verified by Sanger sequencing. Alt-R CRISPR-Cas9 crRNA, tracrRNA, and Cas9 nuclease were purchased from IDT. The following sequences were used for crRNA: 5′-CGACCTGAGAATGTTGGTGC-3′ (Tcf7), 5′-GTCCTACAAGGCAAGCCTAG-3′ (Rorc), and 5′-GGAGGAACTCTTCGCACACT-3′ (Gata3). For microinjections, gRNA was assembled with crRNA and tracrRNA according to the manufacturer’s instructions and added to the final injection mix at 20 ng/µl along with 50 ng/µl Cas9 nuclease and 12.5 ng/µl plasmid in embryo-grade water. Mixes were injected into the nuclei of C57BL/6J embryos. Successful integrations were determined by targeting PCR using two primer sets that generate a product spanning the insert out beyond the homology arms, which was sequence verified.
Preparation of cell suspensions
FLs were passed through a 70-µm cell strainer and resuspended in FACS buffer (1× HBSS [Gibco], 0.25% BSA [Millipore-Sigma], 50 ng/ml DNase, and 5 mM sodium azide [Millipore-Sigma]). BM was collected from the femur, tibia, and ilium (six bones in total), crushed with the syringe plunger flange, passed through a 70-µm cell strainer, and resuspended in FACS buffer. Spleens and thymi were prepared in the same manner as FLs. Adult livers were passed through a 70-µm cell strainer and resuspended in FACS buffer. Fat was removed by centrifugation in 0.45% Percoll (Millipore-Sigma). RBCs were removed with 1× RBC lysis buffer (ThermoFisher). The lung was perfused with 1× PBS (Corning), chopped into small pieces, and digested by shaking (250 rpm) at 37°C for 15 min in RPMI (Hyclone) with 650 U/ml collagenase A (Roche) and 0.01% DNase (Millipore-Sigma). The SI lamina propria was prepared by removing Peyer’s patches, washing out intestinal contents with ice-cold 1× PBS, and cutting length- and widthwise into small pieces. Intraepithelial lymphocytes were removed by two 37°C 15-min shakes (250 rpm) in RPMI with 5 mM EDTA (ThermoFisher) and 1% FBS (Atlanta Biologicals), and lamina propria was digested by shaking (250 rpm) at 37°C for 30 min in RPMI with 20% FBS, 0.5 mg/ml collagenase A, and 0.17 mg/ml DNase. Digests for the lung and lamina propria were passed through 70-µm or 100-µm cell strainers, respectively, followed by fat removal and RBC lysis as above.
Flow cytometry
Single-cell suspensions were incubated with BD FcBlock for 15 min on ice. All FL cells were lineage depleted with CD3ε, CD11c, CD19, GR-1, NK1.1, TCRβ, and Ter119, while BM depletions additionally contained B220, CD4, CD8α, and CD11b. Lineage antibodies conjugated to APC-Cy7 or Biotin were incubated with cells for 30 min on ice, then subsequently incubated with anti-Cy7 or anti-streptavidin microbeads (Miltenyi Biotec) for an additional 30 min. Bead-bound cells were depleted with an autoMACS (Miltenyi Biotec) using the depletion-sensitive program. The following fluorochrome- or biotin-conjugated antibodies were used: α4β7 (DATK32), CCR6 (29-2L17), CD11c (N418), CD19 (6D5), CD90.2 (Thy-1.2; 53–2.1), CD127 (IL-7Rα; A7R34), CXCR5 (L138D7), GR-1 (RB6-8C5), ICOS (C398.4A), PD-1 (29F.1A12), Ter119 (TER-119), mouse IgG1 κ-chain (MG1-45), mouse IgG2a κ-chain (MG2a-53), and rat IgG2b κ-chain (RTK45-30; all from BioLegend); B220 (RA3-6B2), CD3ε (145-2C11), CD4 (GK1.5), CD8α (53–6.7), CD11b (M1/70), Flt3 (A2F10.1), NK1.1 (PK136), TCRβ (H57-597), Thy1.1 (OX-7), RORγt (Q31-378), and PLZF (R17-809; all from BD Biosciences); TCF1 (Cell Signaling Technologies); and polyclonal goat α-Rb IgG (Abcam). To exclude dead cells, Zombie NIR Fixable Viability Dye (BioLegend) was added to live cells. Intracellular staining was performed using the Foxp3 Transcription Factor Staining Buffer Kit (ThermoFisher) according to the manufacturer’s instructions. Cells were blocked with unlabeled isotype-matched antibody (above) and negative controls were prepared using 20-fold excess unlabeled antibody to the TF (cold competitor). Samples were acquired on a four-laser LSRII (BD Biosciences) or sorted on a FACSAriaII or FACSAria Fusion (BD Biosciences) and analyzed using FlowJo software (TreeStar). Unless otherwise indicated, cell populations were lineage negative and identified as follows: ILC2P (α4β7+IL-7Rα+CD90.2+ICOS+Zbtb16−), ILCP (α4β7+IL-7Rα+CCR6−Zbtb16+ or α4β7+IL-7Rα+CCR6−Tcf7+Zbtb16+), LTiP (α4β7+IL-7Rα+CCR6+Zbtb16− or α4β7+IL-7Rα+CCR6+Tcf7+Zbtb16−Rorc+), iILCP (α4β7+IL-7Rα−CD90.2−Tcf7+Zbtb16+Rorc−), iLTiP (α4β7+IL-7Rα−CD90.2−Tcf7+Zbtb16−Rorc+), rEILP (α4β7+IL-7Rα−CD90.2−Tcf7+Zbtb16−Rorc−), Rorc+ αLP (α4β7+IL-7Rα+ CD90.2−PD1−Tcf7−Zbtb16−Rorc+), Flt3− αLP (α4β7+IL-7Rα+CD90.2−PD1−Flt3−Tcf7−Zbtb16−), Flt3+ αLP (α4β7+IL-7Rα+CD90.2−PD1−Flt3+Tcf7−Zbtb16−), CLP (α4β7−IL-7Rα+CD90.2−PD1−Flt3+), and CHILP (α4β7+IL-7Rα+ID2+CD25−Flt3−).
OP9 and OP9-DL1 stromal cell culture
Stocks of OP9 and OP9-DL1 stromal cells were a gift from J.C. Zúñiga-Pflücker (University of Toronto, Toronto, Canada). All experiments were performed in Opti-MEM with GlutaMAX (Gibco) containing heat-inactivated 10% FBS, 1% penicillin-streptomycin (Gibco), and 60 µM 2-mercaptoethanol (Millipore-Sigma) and were maintained in a 37°C incubator (ThermoFisher) with 5% CO2. Stromal cells were plated at a density of 10,000 cells per well of a 96-well plate, allowed to grow overnight, and then irradiated (1500 rad). Before sorting lymphocytes, the media was replaced and supplemented with murine stem cell factor (25 ng/ml; BioLegend), IL-7 (25 ng/ml; BioLegend), and IL-2 (25 ng/ml; BioLegend). FL cells were lineage depleted as described, stained, and single cells were sorted. Cultures were analyzed after 6 d, and colonies with five or more CD45+ cells were considered. Cell populations were defined as follows for single-cell cultures: ILC1 (NK1.1+ICOS−α4β7−), ILC2 (NK1.1−ICOS+α4β7−), ILC3/LTi0 (NK1.1−ICOS+/−α4β7+CD4−), and/or LTi4 (NK1.1−ICOS+/−α4β7+CD4+). Wells were categorized as LTi4 if >8% of NK1.1−ICOS+/−α4β7+ cells were CD4+; no frequency was used to categorize ILC1, ILC2, or ILC3/LTi0. Early T cells were identified in OP9-DL1 cultures by high expression of CD25. For bulk cultures, stromal cells were plate at a density of 24,000 cells per well of a 48-well plate, grown overnight, and irradiated as above. Media was replaced and supplemented with stem cell factor (25 ng/ml), IL-7 (25 ng/ml), IL-2 (25 ng/ml), and Flt3-L (25 ng/ml; BioLegend). Cells were sorted, and 500 cells of each precursor population were plated and allowed to grow for 48 h before analysis. CD45+ cell populations were defined as follows for bulk cell cultures: ILC1 (NK1.1+ICOS−), ILC2 (NK1.1−ICOS+), ILC3/LTi0 (NK1.1−ICOS−Rorc-Thy1.1+CD4−), and/or LTi4 (NK1.1−ICOS−Rorc-Thy1.1+CD4+).
ATAC-seq sample preparation
ATAC-seq was performed as described in the published Omni-ATAC protocol (Corces et al., 2017). Briefly, 5,000–10,000 cells were sorted into FACS buffer and washed twice with 1 ml ice-cold ATAC-seq resuspension buffer (RSB; 10 mM Tris, pH 7.4 [Invitrogen], 10 mM NaCl [Millipore-Sigma], 3 mM MgCl2 [Millipore-Sigma]) by centrifugation at 0.5 ×g for 5 min at 4°C. After centrifugation, the supernatant was carefully removed to avoid the cell pellet, which was then gently resuspended in 50 µl ATAC-seq RSB containing 0.1% NP-40 (Roche), 0.1% Tween-20 (Millipore-Sigma), and 0.1% digitonin (Millipore-Sigma). The lysis reaction was incubated on ice for 3 min, 1 ml ATAC-seq RSB containing 0.1% Tween-20 was added, and the nuclei were centrifuged at 0.5 ×g for 10 min at 4°C. The supernatant was subsequently removed, and the pelleted nuclei were gently resuspended in 50 µl transposition mix (25 µl 2× Tagment DNA buffer [Illumina], 2.5 µl Tn5 transposase [Illumina], 16.5 µl 1× PBS, 0.5 µl 1% digitonin, 0.5 µl 1% Tween-20, and 5 µl water). Transposition reactions were incubated for 30 min in a 37°C water bath. Zymo DNA Clean and Concentrator kits were used to clean up the transposition reactions, and libraries were prepared as previously described (Buenrostro et al., 2015a). Libraries were sequenced by 50 bp single-end, dual-index sequencing on an Illumina HiSeq 4000.
ATAC-seq analysis
ATAC-seq reads were aligned to the mouse genome (mm10) with Bowtie, and duplicate reads were removed using Picard Tools. A pooled bam file was generated from all samples, including replicates (n = 2), and peaks were called on the pooled bam file with MACS2 to control for type I error (Lun and Smyth, 2014). Called peaks were filtered through a modified blacklist, and mitochondrial and Y chromosome peaks were removed (n = 214,321; Buenrostro et al., 2015b). ATAC-seq read counts for all samples, including replicates, were determined via bedtools multiBamCov, and peaks with low read counts were removed (n = 122,250). Quantile normalization, GC content normalization, and peak length normalization were performed using the CQN R package (Corces et al., 2016). Peaks were filtered to select for the top peaks (n = 100,000) with the highest variance. Heatmap generation and principal component analysis were performed on normalized data. Generation of normalized bigWig files for data visualization was performed by first generating normalized bedGraph files with bedtools genomecov, where the sum of reads within peaks associated with transcription start sites was used as a scaling factor, followed by file conversion using the University of California, Santa Cruz, bedGraphToBigWig program. Motif enrichment analysis was performed using HOMER (Heinz et al., 2015). The R package Gviz was used for coverage track visualization.
Statistical analysis
Single-cell indexing, Chi-Square Test for Independence followed by post-hoc analysis with Bonferroni correction, hierarchical clustering by Pearson correlation as the distance metric, heatmap generation, and principal component analysis were all performed in R. One-way ANOVA with Tukey’s multiple comparisons test and multiple t tests controlling for false discovery rate were performed in GraphPad Prism 8.
Accession code
For ATAC-seq data, see GEO accession no. GSE150763.
Online supplemental material
Fig. S1 provides a current and revised model of ILC development. Fig. S2 shows phenotyping of the novel TF reporters. Fig. S3 shows the evaluation of ILC maturation markers and statistical analysis for Fig. 3. Fig. S4 provides additional analysis of the Rorc+ αLP. Fig. S5 displays chromatin accessibility coverage tracks at Il17a/Il17f and Tcf7 loci. Table S1 provides granular information for pie charts of cell potential in Figs. 4, 5, and 6.
Supplementary Material
gives granular information for pie charts of cell potential in Figs. 4, 5, and 6.
Acknowledgments
We thank A. Goldrath for the ID2EYFP mice and A. Bhandoola for the Tcf7EGFP mice; J.C. Zúñiga-Pflücker for stocks of OP9 and OP9-DL1 stromal cells; S. Erickson (University of Chicago) for productive discussion of CRISPR/Cas9 methodology; and B. Kee (University of Chicago) and F. Gounari (University of Chicago) for critical reading of the manuscript. We are grateful to L. Degenstein (University of Chicago) and the University of Chicago Transgenics Core for support with microinjections; the University of Chicago DNA Sequencing and Genotyping Core for sequencing of reporter plasmid constructs; the University of Chicago Flow Cytometry Core for technical assistance with cell sorting; and the University of Chicago Genomics Core for sequencing of ATAC-seq samples.
This work was supported by the National Institutes of Health (R37 AI127518, R01 AI144094, and U01 AI125250) and support funds from the University of Chicago dean’s office.
Author contributions: D.N. Kasal and A. Bendelac conceived the study and wrote the manuscript. D.N. Kasal designed experiments; generated reporter constructs; performed flow cytometry, single-cell sorting, and culture experiments; and conducted ATAC-seq computational analysis. A. Bendelac supervised experiments.
References
- Allman, D., Sambandam A., Kim S., Miller J.P., Pagan A., Well D., Meraz A., and Bhandoola A.. 2003. Thymopoiesis independent of common lymphoid progenitors. Nat. Immunol. 4:168–174. 10.1038/ni878 [DOI] [PubMed] [Google Scholar]
- Buenrostro, J.D., Wu B., Chang H.Y., and Greenleaf W.J.. 2015a. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109:1–9. 10.1002/0471142727.mb2129s109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro, J.D., Wu B., Litzenburger U.M., Ruff D., Gonzales M.L., Snyder M.P., Chang H.Y., and Greenleaf W.J.. 2015b. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 523:486–490. 10.1038/nature14590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chea, S., Schmutz S., Berthault C., Perchet T., Petit M., Burlen-Defranoux O., Goldrath A.W., Rodewald H.-R., Cumano A., and Golub R.. 2016. Single-Cell Gene Expression Analyses Reveal Heterogeneous Responsiveness of Fetal Innate Lymphoid Progenitors to Notch Signaling. Cell Rep. 14:1500–1516. 10.1016/j.celrep.2016.01.015 [DOI] [PubMed] [Google Scholar]
- Cherrier, M., Sawa S., and Eberl G.. 2012. Notch, Id2, and RORγt sequentially orchestrate the fetal development of lymphoid tissue inducer cells. J. Exp. Med. 209:729–740. 10.1084/jem.20111594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong, L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., and Zhang F.. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science. 339:819–823 https://doi.org. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinides, M.G., McDonald B.D., Verhoef P.A., and Bendelac A.. 2014. A committed precursor to innate lymphoid cells. Nature. 508:397–401. 10.1038/nature13047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces, M.R., Buenrostro J.D., Wu B., Greenside P.G., Chan S.M., Koenig J.L., Snyder M.P., Pritchard J.K., Kundaje A., Greenleaf W.J., et al. . 2016. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat. Genet. 48:1193–1203. 10.1038/ng.3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces, M.R., Trevino A.E., Hamilton E.G., Greenside P.G., Sinnott-Armstrong N.A., Vesuna S., Satpathy A.T., Rubin A.J., Montine K.S., Wu B., et al. . 2017. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods. 14:959–962. 10.1038/nmeth.4396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl, G., Marmon S., Sunshine M.-J., Rennert P.D., Choi Y., and Littman D.R.. 2004. An essential function for the nuclear receptor RORgamma(t) in the generation of fetal lymphoid tissue inducer cells. Nat. Immunol. 5:64–73. 10.1038/ni1022 [DOI] [PubMed] [Google Scholar]
- Ebihara, T., Song C., Ryu S.H., Plougastel-Douglas B., Yang L., Levanon D., Groner Y., Bern M.D., Stappenbeck T.S., Colonna M., et al. . 2015. Runx3 specifies lineage commitment of innate lymphoid cells. Nat. Immunol. 16:1124–1133. 10.1038/ni.3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harly, C., Cam M., Kaye J., and Bhandoola A.. 2018. Development and differentiation of early innate lymphoid progenitors. J. Exp. Med. 215:249–262. 10.1084/jem.20170832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harly, C., Kenney D., Ren G., Lai B., Raabe T., Yang Q., Cam M.C., Xue H.-H., Zhao K., and Bhandoola A.. 2019. The transcription factor TCF-1 enforces commitment to the innate lymphoid cell lineage. Nat. Immunol. 20:1150–1160. 10.1038/s41590-019-0445-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz, S., Romanoski C.E., Benner C., and Glass C.K.. 2015. The selection and function of cell type-specific enhancers. Nat. Rev. Mol. Cell Biol. 16:144–154. 10.1038/nrm3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyler, T., Klose C.S.N., Souabni A., Turqueti-Neves A., Pfeifer D., Rawlins E.L., Voehringer D., Busslinger M., and Diefenbach A.. 2012. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity. 37:634–648. 10.1016/j.immuni.2012.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka, I.E., Constantinides M.G., Gudjonson H., and Bendelac A.. 2016a. The innate lymphoid cell precursor. Annu. Rev. Immunol. 34:299–316. 10.1146/annurev-immunol-041015-055549 [DOI] [PubMed] [Google Scholar]
- Ishizuka, I.E., Chea S., Gudjonson H., Constantinides M.G., Dinner A.R., Bendelac A., and Golub R.. 2016b. Single-cell analysis defines the divergence between the innate lymphoid cell lineage and lymphoid tissue-inducer cell lineage. Nat. Immunol. 17:269–276. 10.1038/ni.3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose, C.S.N., Flach M., Möhle L., Rogell L., Hoyler T., Ebert K., Fabiunke C., Pfeifer D., Sexl V., Fonseca-Pereira D., et al. . 2014. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell. 157:340–356. 10.1016/j.cell.2014.03.030 [DOI] [PubMed] [Google Scholar]
- Lun, A.T.L., and Smyth G.K.. 2014. De novo detection of differentially bound regions for ChIP-seq data using peaks and windows: controlling error rates correctly. Nucleic Acids Res. 42:e95 10.1093/nar/gku351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, A.-P., Ishizuka I.E., Kasal D.N., Mandal M., and Bendelac A.. 2017. A shared Runx1-bound Zbtb16 enhancer directs innate and innate-like lymphoid lineage development. Nat. Commun. 8:863 10.1038/s41467-017-00882-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr, C. 2017. Regulation by 3′-Untranslated Regions. Annu. Rev. Genet. 51:171–194. 10.1146/annurev-genet-120116-024704 [DOI] [PubMed] [Google Scholar]
- Mohrs, K., Wakil A.E., Killeen N., Locksley R.M., and Mohrs M.. 2005. A two-step process for cytokine production revealed by IL-4 dual-reporter mice. Immunity. 23:419–429. 10.1016/j.immuni.2005.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J.-Y., DiPalma D.T., Kwon J., Fink J., and Park J.-H.. 2019. Quantitative Difference in PLZF Protein Expression Determines iNKT Lineage Fate and Controls Innate CD8 T Cell Generation. Cell Rep. 27:2548–2557.e4. 10.1016/j.celrep.2019.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possot, C., Schmutz S., Chea S., Boucontet L., Louise A., Cumano A., and Golub R.. 2011. Notch signaling is necessary for adult, but not fetal, development of RORγt(+) innate lymphoid cells. Nat. Immunol. 12:949–958. 10.1038/ni.2105 [DOI] [PubMed] [Google Scholar]
- Rawlins, E.L., Clark C.P., Xue Y., and Hogan B.L.M.. 2009. The Id2+ distal tip lung epithelium contains individual multipotent embryonic progenitor cells. Development. 136:3741–3745. 10.1242/dev.037317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa, S., Cherrier M., Lochner M., Satoh-Takayama N., Fehling H.J., Langa F., Di Santo J.P., and Eberl G.. 2010. Lineage relationship analysis of RORgammat+ innate lymphoid cells. Science. 330:665–669. 10.1126/science.1194597 [DOI] [PubMed] [Google Scholar]
- Seillet, C., Mielke L.A., Amann-Zalcenstein D.B., Su S., Gao J., Almeida F.F., Shi W., Ritchie M.E., Naik S.H., Huntington N.D., et al. . 2016. Deciphering the Innate Lymphoid Cell Transcriptional Program. Cell Rep. 17:436–447. 10.1016/j.celrep.2016.09.025 [DOI] [PubMed] [Google Scholar]
- Walker, J.A., Clark P.A., Crisp A., Barlow J.L., Szeto A., Ferreira A.C.F., Rana B.M.J., Jolin H.E., Rodriguez-Rodriguez N., Sivasubramaniam M., et al. . 2019. Polychromic Reporter Mice Reveal Unappreciated Innate Lymphoid Cell Progenitor Heterogeneity and Elusive ILC3 Progenitors in Bone Marrow. Immunity. 51:104–118.e7. 10.1016/j.immuni.2019.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, W., Cherrier D.E., Chea S., Vosshenrich C., Serafini N., Petit M., Liu P., Golub R., and Di Santo J.P.. 2019. An Id2RFP-Reporter Mouse Redefines Innate Lymphoid Cell Precursor Potentials. Immunity. 50:1054–1068.e3. 10.1016/j.immuni.2019.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C.Y., Best J.A., Knell J., Yang E., Sheridan A.D., Jesionek A.K., Li H.S., Rivera R.R., Lind K.C., D’Cruz L.M., et al. . 2011. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat. Immunol. 12:1221–1229. 10.1038/ni.2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Q., Monticelli L.A., Saenz S.A., Chi A.W.-S., Sonnenberg G.F., Tang J., De Obaldia M.E., Bailis W., Bryson J.L., Toscano K., et al. . 2013. T cell factor 1 is required for group 2 innate lymphoid cell generation. Immunity. 38:694–704. 10.1016/j.immuni.2012.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Q., Li F., Harly C., Xing S., Ye L., Xia X., Wang H., Wang X., Yu S., Zhou X., et al. . 2015. TCF-1 upregulation identifies early innate lymphoid progenitors in the bone marrow. Nat. Immunol. 16:1044–1050. 10.1038/ni.3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, H., Kawamoto H., Santee S.M., Hashi H., Honda K., Nishikawa S., Ware C.F., Katsura Y., and Nishikawa S.-I.. 2001. Expression of α(4)β(7) integrin defines a distinct pathway of lymphoid progenitors committed to T cells, fetal intestinal lymphotoxin producer, NK, and dendritic cells. J. Immunol. 167:2511–2521. 10.4049/jimmunol.167.5.2511 [DOI] [PubMed] [Google Scholar]
- Yu, Q., Erman B., Park J.-H., Feigenbaum L., and Singer A.. 2004. IL-7 receptor signals inhibit expression of transcription factors TCF-1, LEF-1, and RORγt: impact on thymocyte development. J. Exp. Med. 200:797–803. 10.1084/jem.20032183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, X., Wang Y., Deng M., Li Y., Ruhn K.A., Zhang C.C., and Hooper L.V.. 2014. The basic leucine zipper transcription factor NFIL3 directs the development of a common innate lymphoid cell precursor. eLife. 3:e04406 10.7554/eLife.04406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y., Tsang J.C.H., Wang C., Clare S., Wang J., Chen X., Brandt C., Kane L., Campos L.S., Lu L., et al. . 2016. Single-cell RNA-seq identifies a PD-1hi ILC progenitor and defines its development pathway. Nature. 539:102–106. 10.1038/nature20105 [DOI] [PubMed] [Google Scholar]
- Zhong, C., Cui K., Wilhelm C., Hu G., Mao K., Belkaid Y., Zhao K., and Zhu J.. 2016. Group 3 innate lymphoid cells continuously require the transcription factor GATA-3 after commitment. Nat. Immunol. 17:169–178. 10.1038/ni.3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, C., Zheng M., Cui K., Martins A.J., Hu G., Li D., Tessarollo L., Kozlov S., Keller J.R., Tsang J.S., et al. . 2020. Differential Expression of the Transcription Factor GATA3 Specifies Lineage and Functions of Innate Lymphoid Cells. Immunity. 52:83–95.e4. 10.1016/j.immuni.2019.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zook, E.C., and Kee B.L.. 2016. Development of innate lymphoid cells. Nat. Immunol. 17:775–782. 10.1038/ni.3481 [DOI] [PubMed] [Google Scholar]
- Zook, E.C., Ramirez K., Guo X., van der Voort G., Sigvardsson M., Svensson E.C., Fu Y.-X., and Kee B.L.. 2016. The ETS1 transcription factor is required for the development and cytokine-induced expansion of ILC2. J. Exp. Med. 213:687–696. 10.1084/jem.20150851 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gives granular information for pie charts of cell potential in Figs. 4, 5, and 6.