Single-cell analysis reveals an accumulation of short-lived disease-specific as well as non–disease-specific intestinal plasma cells in untreated and short-term–treated celiac disease. Plasma cells differ transcriptionally depending on specificity, longevity, and disease status and may contribute to formation of the celiac lesion.
Abstract
Disease-specific plasma cells (PCs) reactive with transglutaminase 2 (TG2) or deamidated gluten peptides (DGPs) are abundant in celiac disease (CeD) gut lesions. Their contribution toward CeD pathogenesis is unclear. We assessed expression of markers associated with PC longevity in 15 untreated and 26 treated CeD patients in addition to 13 non-CeD controls and performed RNA sequencing with clonal inference and transcriptomic analysis of 3,251 single PCs. We observed antigen-dependent V-gene selection and stereotypic antibodies. Generation of recombinant DGP-specific antibodies revealed a key role of a heavy chain residue that displays polymorphism, suggesting that immunoglobulin gene polymorphisms may influence CeD-specific antibody responses. We identified transcriptional differences between CeD-specific and non–disease-specific PCs and between short-lived and long-lived PCs. The short-lived CD19+CD45+ phenotype dominated in untreated and short-term–treated CeD, in particular among disease-specific PCs but also in the general PC population. Thus, the disease lesion of untreated CeD is characterized by massive accumulation of short-lived PCs that are not only directed against disease-specific antigens.
Graphical Abstract
Introduction
A feature of autoimmune diseases is the secretion of specific autoantibodies by terminally differentiated B cells, plasma cells (PCs). IgA- and IgM-secreting PCs are abundant in the lamina propria in the gut, where antibodies are secreted and transported into the intestinal lumen. Traditionally, gut PCs were thought to be short-lived antibody factories, and other potential roles of PCs in health and autoimmune diseases have not been under much investigation. However, expression of HLA class II molecules and costimulatory molecules such as CD40, CD80, and CD86, as well as cytokine secretion, suggests that PCs can have additional functions (Ellyard et al., 2004; Fillatreau, 2015; Fritz et al., 2012; Høydahl et al., 2019; Pelletier et al., 2010). Recently, based on cell-turnover analysis in gut transplants and by retrospective cell-birth dating measuring carbon-14 in genomic DNA, three subsets of short-, intermediate-, and long-lived lamina propria PCs with distinct expression of cell surface markers CD19 and CD45 were described (Landsverk et al., 2017). The longevity profile of human gut PCs in a disease setting is yet uncharacterized.
Celiac disease (CeD) is associated with a highly specific autoantibody response. The patients have autoantibodies to the enzyme transglutaminase 2 (TG2) as well as antibodies to deamidated gluten peptides (DGPs; Iversen and Sollid, 2020). The disease is driven by an immune response to cereal gluten proteins, which explains the presence of the DGP antibodies. TG2 is involved in the pathogenesis by catalyzing deamidation of gluten peptides. The patients have CD4+ T cells that recognize DGP bound to disease-associated HLA-DQ molecules (Molberg et al., 1998). These T cells likely provide T cell help to DGP-specific B cells as well as TG2-specific B cells; the latter by involvement of TG2-gluten peptide hapten-carrier–like molecules (Iversen and Sollid, 2020). The disease lesion in the small intestine is characterized by blunting of villi and infiltration of inflammatory cells, including PCs. The active lesion typically has a two- to threefold increased density of IgA and IgM PCs in lamina propria (Baklien et al., 1977), with ∼10% of the PCs being specific for TG2 and 1% being specific for DGP (Di Niro et al., 2010; Steinsbø et al., 2014). The only available treatment for CeD is adherence to a strict gluten-free diet (GFD). After commencing a GFD, the concentration of disease-specific serum antibodies (Sugai et al., 2010) and number of disease-specific PCs in lamina propria drop within months (Di Niro et al., 2016).
The role of PCs in CeD is currently unknown, and it is uncertain to what extent the secreted autoantibodies play a pathogenic role (Iversen et al., 2014; Lindfors et al., 2009). Recent reports have shown that intestinal PCs may have additional functions to antibody secretion, such as antigen presentation and cytokine secretion (Høydahl et al., 2019; Snir et al., 2019), leading to the hypothesis that disease-specific PCs are playing an active role in the pathogenesis of CeD, including having a role as APCs for gluten-specific T cells. Identifying functions of gut PCs could provide more knowledge about disease mechanisms and point out disease-specific PCs as therapeutic targets.
High-throughput sequencing technologies have revolutionized the profiling of immune receptor repertoires in health and disease (AIRR-seq; Benichou et al., 2012; Yaari and Kleinstein, 2015), and we have previously applied these technologies to dissect the repertoire of disease-specific PCs in CeD (Di Niro et al., 2012; Iversen et al., 2017; Roy et al., 2017; Snir et al., 2017; Snir et al., 2015; Steinsbø et al., 2014). However, little is known about the transcriptional state of these cells. The development of single-cell RNA-sequencing (scRNA-seq) protocols provides a unique tool to identify and characterize cell subsets according to their full transcriptional profile (Buettner et al., 2015; Kolodziejczyk and Lönnberg, 2018; Stubbington et al., 2017). Additionally, recent computational tools for reconstruction of antigen receptor sequences from scRNA-seq data (Afik et al., 2019; Afik et al., 2017; Canzar et al., 2017; Eltahla et al., 2016; Lindeman et al., 2018; Rizzetto et al., 2018; Stubbington et al., 2016; Upadhyay et al., 2018) now allow us to link the transcriptional profile of individual PCs with information about their B cell receptor (BCR) and thus clonal relatedness.
Here, we used scRNA-seq and BCR reconstruction and clonotype inference by the computational tool BraCeR (Lindeman et al., 2018) to characterize PCs in the healthy small intestine and in an autoimmune setting, specifically CeD. We observed antigen-dependent variable (V)-gene selection, and we identified a novel stereotypic DGP-specific antibody. We further observed that disease-specific gut PCs in celiac lesions of untreated CeD (UCeD) or short-term–treated CeD (TCeD) patients rarely carried a phenotype previously shown to be associated with long life span (Landsverk et al., 2017), whereas PCs of this phenotype were more abundant in TCeD patients treated for long term. PCs of different longevity phenotype differed in their transcriptomic profiles. Differentially expressed genes included genes involved in metabolism and regulation of apoptosis. The dominance of the short-lived CD19+CD45+ phenotype in UCeD and short-term TCeD, both among disease-specific PCs but also in the general PC population, suggests that new and short-lived PCs massively accumulate in the UCeD lesion.
Results
Study participants
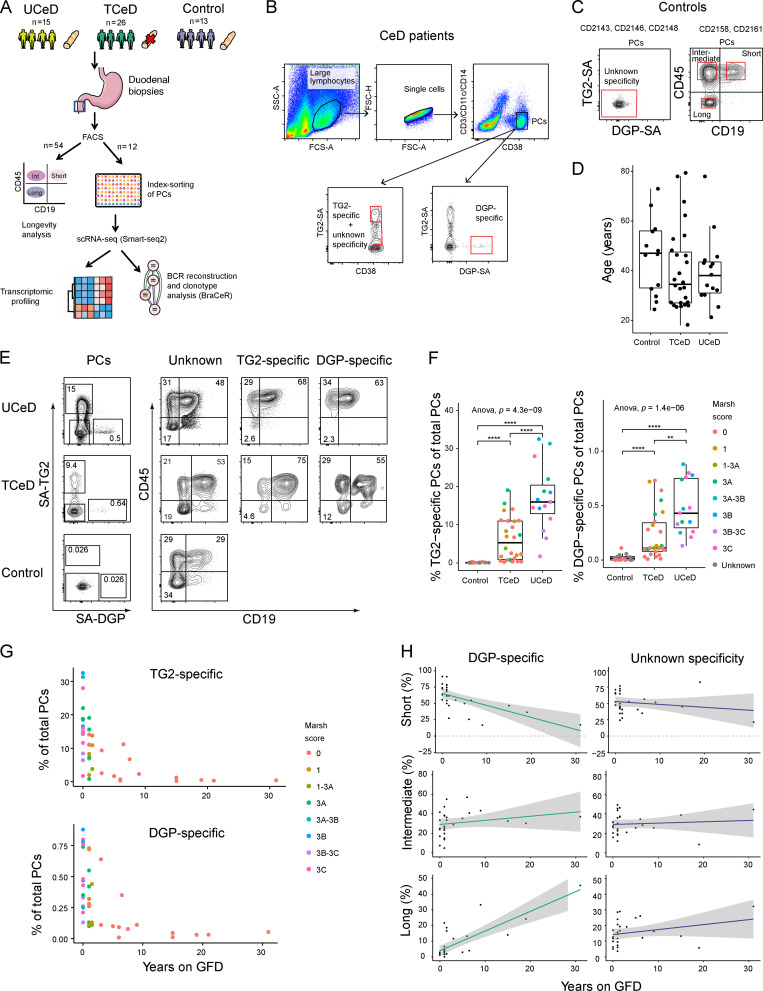
We collected duodenal biopsies from 15 UCeD patients, 26 TCeD patients, and 13 non-CeD controls and performed flow cytometry to analyze the longevity of PCs (large CD3−CD11c−CD14−CD38high) in each individual (Fig. S1 A). For four UCeD patients, three TCeD patients, and five controls, we additionally sorted PCs of interest by FACS based on antigen specificity or the longevity markers CD19 and CD45 (Fig. S1, B and C) and performed scRNA-seq. Information about the study participants is presented in Table S1. Subjects in the TCeD group had been on a GFD for 1–31 yr (median 2.25 yr). Non-CeD controls had no known autoimmune diseases or intestinal inflammation. The median ages of the participants were 38 (UCeD), 34.5 (TCeD), and 47 yr (controls; Fig. S1 D).
Figure S1.
Analysis of PC longevity and specificity by flow cytometry. (A) Experimental approach. The red cross indicates a GFD. The blue square indicates location of biopsy sampling (duodenum). (B) Single-cell FACS index sorting based on antigen specificity. TG2- and DGP-specific PCs were identified by fluorescent tetramers (SA) loaded with TG2 or DGP. Representative flow cytometry plots and sorting strategy is shown for one CeD patient. Gates used for sorting are outlined in red. (C) Single-cell FACS index sorting for control subjects. PCs of unknown specificity were sorted with a natural composition of PC longevity populations for three controls (left). Equal numbers of short-, intermediate-, and long-lived PCs were sorted from two controls (right) based on CD45/CD19 surface marker expression. Flow cytometry plots from one control are shown. Gates used for sorting are outlined in red. (D) Age distribution in patient groups. (E) Representative flow cytometry plots showing percent TG2-specific and DGP-specific PCs and the longevity markers CD19/CD45 for each specificity for one UCeD, one TCeD, and one control subject. PCs were gated as large, single CD3−CD11c−CD14−CD38high cells. (F) Percentage of PCs identified as being specific for TG2 (left) or DGP (right) in UCeD (n = 15), TCeD (n = 26), and controls (n = 13) by flow cytometry, colored according to the Marsh score of intestinal inflammation (Oberhuber et al., 1999). P values were calculated with an unpaired Wilcoxon rank-sum test. Adjusted P values are shown. **, P < 0.01; ****, P < 0.0001. (G) Percentage of PCs identified as being specific for TG2 (top) or DGP (bottom) in UCeD (n = 15) and TCeD patients (n = 26) as a function of time on a GFD. (H) Percentage of PCs belonging to the short-, intermediate- (CD19−CD45+), and long-lived compartments for DGP-specific PCs and other PCs as a function of treatment duration (years on GFD). UCeD patients are plotted as 0 yr on GFD. The colored lines represent fitted trend using linear regression; shades indicate the 95% confidence interval of the fitted line. Only patients with at least 30 recorded DGP-specific PCs are included (n = 27).
TG2-specific and DGP-specific gut PCs have a short- or intermediate-lived phenotype in UCeD and short-term TCeD
Using flow cytometry, we found that 17.0 ± 8.7% (mean ± SD) of the intestinal PCs from UCeD patients were TG2-specific, and 0.48 ± 0.25% were specific for DGP (Fig. S1, E and F; and Table S2). TCeD patients had 6.2 ± 5.7% TG2-specific and 0.23 ± 0.22% DGP-specific PCs persisting after minimum a year on GFD, with decreasing populations of TG2- and DGP-specific PCs in more long-term TCeD patients (Fig. S1 G). The controls had negligible frequencies of TG2-specific (0.08 ± 0.09%) and DGP-specific (0.02 ± 0.03%) PCs.
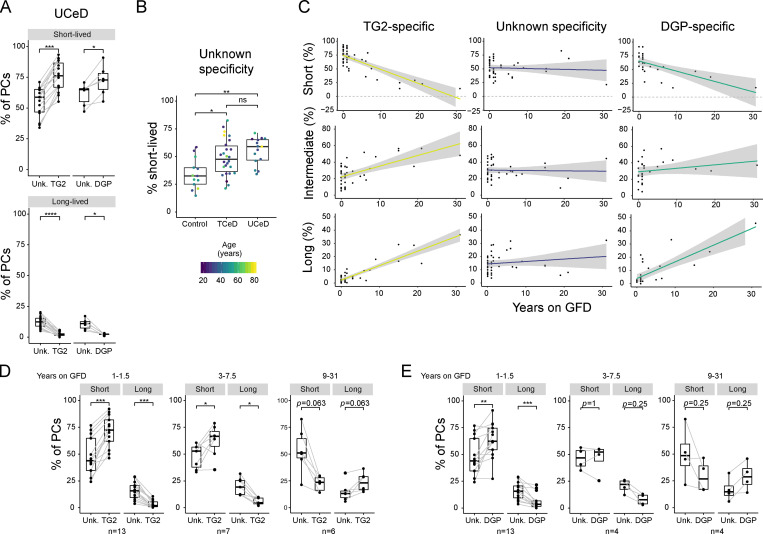
To assess the longevity of PCs in the gut of CeD patients and non-CeD controls, we analyzed the surface expression of CD45 and CD19 (Fig. 1, Fig. S1 E, and Table S2). TG2-specific and DGP-specific PCs from CeD patients largely belonged to the short-lived CD45+CD19+ or intermediate-lived CD45+CD19− compartments (Fig. 1). Significantly more CeD-specific PCs in UCeD patients belonged to the short-lived compartment compared with PCs of unknown specificity from the same patients (Fig. 1 A). Interestingly, even when looking only at PCs of unknown specificity, UCeD patients had a significantly higher percentage of short-lived PCs compared with the controls (Fig. 1 B).
Figure 1.
Analysis of PC longevity by flow cytometry. Small intestinal lamina propria CD38high PCs from UCeD, TCeD, and control patients were analyzed for cell surface expression of CD19 and CD45 by flow cytometry as a proxy for PC longevity. Each data point represents an individual; gray lines connect data from the same patients for different PC specificities (A, D, and E). (A) Summarized data for short-lived (CD19+CD45+) and long-lived (CD19−CD45−) PCs specific for TG2 or DGP compared with PCs of unknown specificity in UCeD patients. All 15 UCeD patients are shown for the analysis of TG2-specific PCs; patients with at least 30 recorded DGP-specific PCs are included in the comparisons between DGP-specific PCs and PCs of unknown specificity (n = 6). All samples were analyzed in independent experiments (n = 15). P values were calculated with a paired Wilcoxon signed-rank test. (B) Percentage of PCs of unknown specificity belonging to the short-lived population in UCeD (n = 15), TCeD (n = 26), and controls (n = 13). Data were collected from, in total, 35 independent experiments. P values were calculated with an unpaired Student’s t test. (C) Percentage of PCs belonging to the short-, intermediate- (CD19−CD45+), and long-lived compartments for TG2-specific or DGP-specific PCs and PCs of unknown specificity as a function of treatment duration (years on GFD). UCeD patients are plotted as 0 yr on GFD. The colored lines represent fitted trend using linear regression; shades indicate the 95% confidence interval of the fitted line. All CeD patients are included for TG2-specific PCs and PCs of unknown specificity (left and central panel; n = 42), while only patients with at least 30 recorded DGP-specific PCs are included for DGP-specific PCs (right panel; n = 27). Data were collected from, in total, 27 independent experiments. (D and E) Percentage of PCs belonging to the short- and long-lived compartments for TG2-specific (D) or DGP-specific (E) PCs compared with PCs of unknown specificity. All TCeD patients are included in D (n = 26 samples, 12 independent experiments). Only patients with at least 30 recorded DGP-specific PCs are included in E (n = 21). TCeD patients are grouped according to treatment length. P values were calculated with a paired Wilcoxon signed-rank test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****; P < 0.0001; ns, not significant; Unk., unknown.
Next, we investigated whether the distribution between short-, intermediate-, and long-lived PCs in CeD patients changes with the duration of treatment (years on a GFD). The percentage of short-lived PCs decreased with time on GFD for both TG2- and DGP-specific PCs, while no such trend was observed for PCs of unknown specificity (Fig. 1 C and Fig. S1 H). Conversely, the percentage of long-lived PCs among the TG2- and DGP-specific cells increased with time on a GFD. By grouping the TCeD patients according to treatment duration, we found that short-term TCeD patients (treated up to 7.5 yr) had a composition of PC longevities similar to UCeD patients, in that the TG2-specific and to some extent the DGP-specific PCs were enriched for short-lived and depleted of long-lived PCs (Fig. 1, C–E). We observed the opposite trend for long-term TCeD patients (treated for 9–31 yr), which showed a marked decrease in short-lived and increase in long- and intermediate-lived PCs among CeD-specific PCs compared with PCs of unknown specificity (Fig. 1, D and E).
Taken together, our data demonstrate that CeD-specific PCs are skewed toward a short-lived phenotype in UCeD and short-term TCeD patients and suggest that intermediate- and long-lived PCs are relatively more prevalent within the CeD-specific PC populations with time on a GFD.
Intestinal PCs express mRNA transcripts for certain HLA class II molecules, costimulatory molecules, and cytokines
A recent bulk RNA-seq study from our group revealed antibody-independent functions of intestinal PCs in health and in CeD (Snir et al., 2019). However, in this study, contamination from other cell types could not be completely ruled out, as only mean gene expression within populations was assessed. To confirm previous findings and further investigate the transcriptional profiles of intestinal PCs at a single-cell resolution, we performed scRNA-seq of intestinal PCs from CeD patients and controls (Fig. S2). A total of 3,251 PCs (Table S3) were analyzed for clonal inference and transcriptional profile. In agreement with previous reports that small intestinal lamina propria antibody-secreting cells do not proliferate and have a PC-like phenotype (Di Niro et al., 2010; Di Niro et al., 2012; Mesin et al., 2011), we did not detect gene expression of the proliferation marker Ki67, indicating that no plasmablasts were present in our data.
Figure S2.
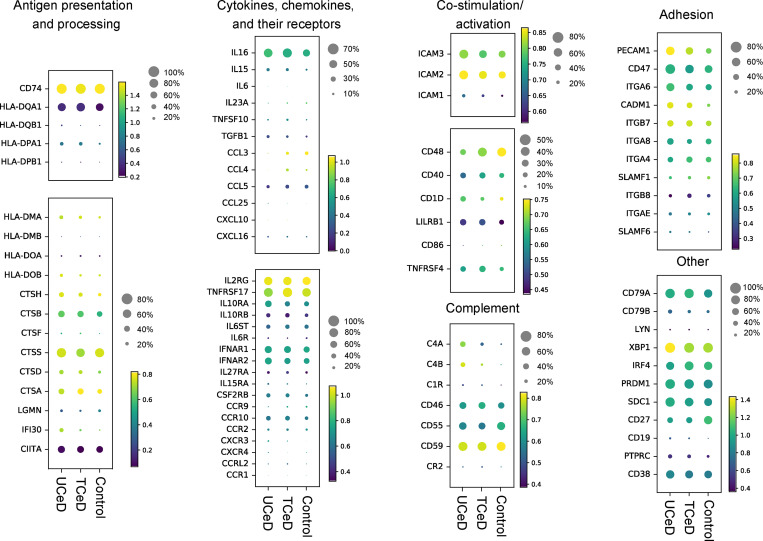
Transcriptional profiling of intestinal PCs using scRNA-seq. scRNA-seq of PCs from UCeD (n = 4), TCeD (n = 3), and control (n = 5) patients was performed following the Smart-seq2 protocol. After quality control, cells from all individuals, specificities, and longevities were pooled together (Table S3). Scaled expression data of genes of interest are shown in heatmaps according to disease status. Colors represent mean expression of each gene among cells in which the gene was expressed. Dot size represents proportion of cells with nonzero expression of each gene.
We first looked for expression of genes associated with antigen presentation and detected expression of CD74 and HLA class II genes, in particular HLA-DQA1, in a large part of the cells regardless of PC specificity, longevity, or disease status. Notably, we also detected expression of the costimulatory molecules CD40, CD48, and to a small extent CD86, but not CD80, in a population of the PCs. Additionally, ICAM1, ICAM2, and ICAM3 transcripts were widely expressed.
Next, we looked for evidence of cytokine and/or chemokine secretion. Indeed, we detected expression of mRNA transcripts for IL-15, IL-16, and/or TGF-β1 in a substantial population of cells in all patients and controls. Expression of the chemokine genes CCL3, CCL4, and CXCL16 was also detected in some of the PCs. Finally, we found wide expression of genes encoding various cytokine/chemokine receptors, such as TNFRSF17 (B cell maturation antigen) and IL2RG. Our transcriptional analysis of intestinal PCs thus supports the notion that PCs have additional functions to antibody secretion.
Short- and long-lived intestinal PCs differ transcriptionally
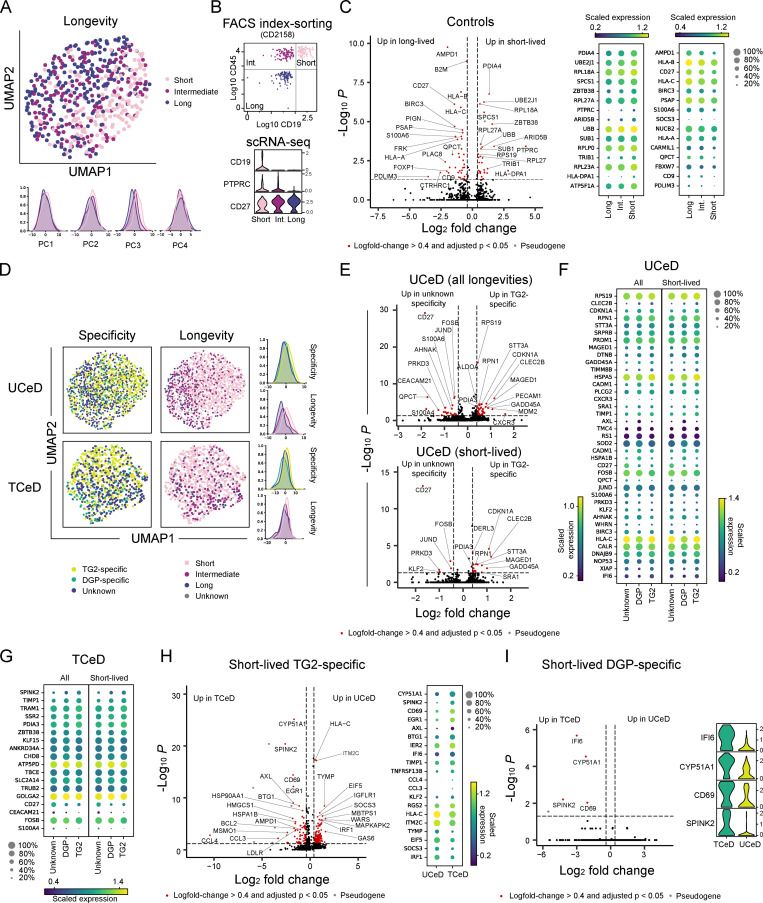
Having looked at the general transcriptional phenotype of intestinal PCs, we next investigated transcriptional differences between short-, intermediate-, and long-lived PCs. As few long-lived PCs were index sorted by FACS for the first three controls, we strictly gated on each longevity population and sorted equal numbers of these from two additional controls in order to compare these clearly defined subsets (Fig. S1 C).
scRNA-seq followed by principal component analysis (PCA) and uniform manifold approximation and projection for dimension reduction (UMAP) revealed that short-lived PCs to some extent differ transcriptionally from long- and intermediate-lived PCs in controls (Fig. 2 A). As expected, CD19 was exclusively detected in short-lived (CD19+) PCs, while PTPRC (CD45) was enriched in the short- and intermediate-lived subsets (CD45+; Fig. 2 B). To identify which genes explain the transcriptional differences between the cell populations, we performed differential expression analysis. Our analysis revealed that CD27 was differentially expressed between all three subpopulations, with highest expression in long-lived PCs and lowest expression in short-lived PCs (Fig. 2, B and C). Among the genes that were more highly expressed in long-lived compared with short-lived PCs, we found genes involved in antigen presentation on MHC class I (HLA-A, HLA-B, and HLA-C), metabolism (AMPD1 and QPCT), inhibition of apoptosis (BIRC3), and CD9. Genes that were overexpressed in the short-lived PCs were heavily enriched for genes encoding ribosomal proteins and genes involved in metabolism (PDIA4, UBE2J1, SPCS1, UBB, TRIB1, ATP5F1A), possibly reflecting higher antibody secretion in the short-lived PCs (Landsverk et al., 2017). The highest ranking differentially expressed genes (DEGs) were also identified when looking at each patient individually. Thus, we demonstrate that PCs of different longevities exhibit distinct transcriptional profiles.
Figure 2.
Transcriptional differences between PC longevities in control subjects and PC specificities in CeD. For A–C, scRNA-seq analysis was performed by pooling all PCs from the two controls (CD2158 and CD2161) for which equal numbers of cells were sorted for each longevity phenotype (gated as in Fig. S1 C, right panel; in total, 162 short-, 160 intermediate-, and 194 long-lived PCs). Variation explained by sequencing library or patient was regressed out, while retaining variation explained by PC longevity. (A) UMAP projection based on the first five principal components (top) and individual principal components, showing variation explained by longevity. (B) FACS index-sorting values for cell surface markers CD19 and CD45 for sorted PCs from one of the controls (CD2158; top). Negative values were converted to 1 before plotting on a logarithmic scale. scRNA-seq expression of CD19, PTPRC (CD45), and CD27 are shown below. (C) Volcano plot (left) showing DEGs between the short- and long-lived PC populations in A and B. Mean expression level of DEGs of interest are shown (right), with dot sizes indicating fraction of cells with nonzero expression. (D) PCs from CeD patients were pooled together according to disease status (four UCeD patients, three TCeD patients) for scRNA-seq analysis. 606 TG2-specific, 132 DGP-specific, and 657 PCs of unknown specificity were included in the analysis for UCeD; 351 TG2-specific, 99 DGP-specific, and 384 PCs of unknown specificity were included for TCeD. Variation explained by patient (and sequencing library for TCeD) was regressed out, retaining variation explained by PC specificity. UMAP projections (left) of PCs from UCeD patients (top) and TCeD patients (bottom) are based on the first 10 (UCeD) or 5 (TCeD) principal components. The individual principal components (PC1 for UCeD, PC2 for TCeD) with the most variation explained by specificity or longevity are shown (right). (E) DEGs between all (top) or only short-lived (bottom) TG2-specific PCs and PCs of unknown specificity in the UCeD patients included in D. 437 TG2-specific and 383 PCs of unknown specificity were included in the analysis of short-lived PCs. (F) Mean expression levels of DEGs of interest between TG2- or DGP-specific PCs and PCs of unknown specificity in UCeD as identified in E. (G) Mean expression levels of DEGs of interest between TG2- or DGP-specific PCs and PCs of unknown specificity in the TCeD patients shown in D. 270 TG2-specific, 62 DGP-specific, and 229 PCs of unknown specificity were included in the analysis of short-lived PCs. (H) DEGs in short-lived TG2-specific PCs between UCeD (437 cells, four patients) and TCeD (270 cells, three patients). Mean expression levels of the most interesting genes are shown (right). (I) DEGs in short-lived DGP-specific PCs between UCeD (78 cells, two patients) and TCeD (62 cells, two patients).
TG2-specific gut PCs have characteristic transcriptional features
Next, we set out to investigate whether TG2-specific or DGP-specific PCs are transcriptionally distinct from PCs of unknown specificity in CeD. When looking at the overall transcriptional landscape of the PCs by dimension reduction with PCA and UMAP, TG2-specific and DGP-specific PCs occupied a slightly different space compared with PCs of unknown specificity, with the TG2-specific cells having the most distinct profile (Fig. 2 D). As for the controls, we also found PC longevity to be a driver of transcriptional heterogeneity in CeD patients.
To identify particular genes characteristic of the different specificities, we performed differential expression analysis for UCeD and TCeD patients separately (Fig. 2, E–G). Strikingly, CD27 was up-regulated in PCs of unknown specificity compared with TG2- or DGP-specific PCs both in UCeD and TCeD. Genes involved in metabolism (STT3A, MDM2, PDIA3, ALDOA), regulation of cell cycle (CDKN1A), and the adhesion molecule CD31 (PECAM1) were up-regulated among TG2-specific PCs compared with other PCs in UCeD patients.
To investigate whether the transcriptional differences between PCs with different specificities could be explained by the skewed composition of longevity phenotypes, we repeated the differential expression analysis for cells of short longevity only. Notably, we found the majority of the most highly up-regulated genes in TG2-specific cells to be differentially expressed also when looking at short-lived PCs only, indicating that the differential gene expression between TG2-specific and other PCs cannot be explained by differences in longevity phenotype only.
Lastly, we explored potential differences in gene expression between UCeD and TCeD patients and identified DEGs for short-lived PCs specific for TG2 (Fig. 2 H) or DGP (Fig. 2 I). Many of the genes that were differentially expressed between antigen-specific PCs in UCeD and TCeD were also differentially expressed between PCs of unknown specificity (see below).
PCs of CeD patients show altered expression of several genes
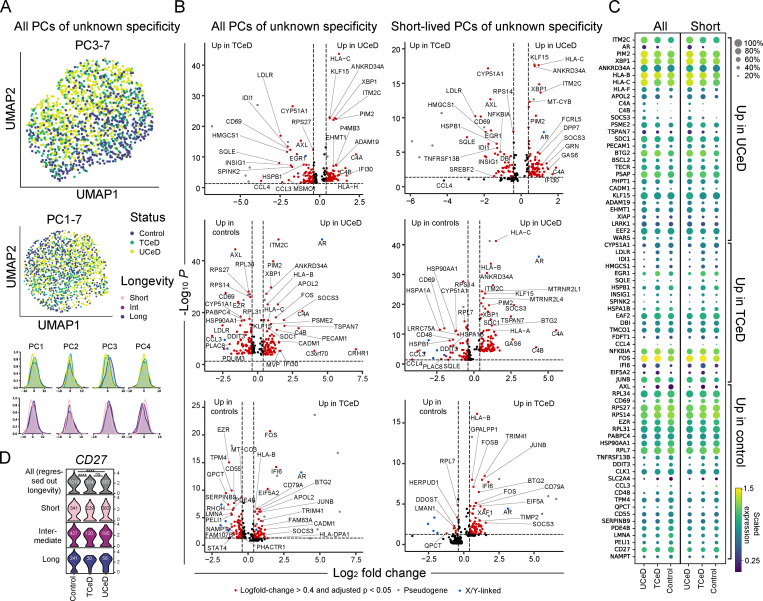
Having compared TG2- and DGP-specific PCs to PCs of unknown specificity in CeD, we next explored the possibility that intestinal PCs of CeD patients in general differ from PCs from healthy individuals. To test this hypothesis, we compared PCs of unknown specificity from CeD patients to PCs of controls. PCA revealed that principal component 3 (PC3) was the main driver of variation between controls, TCeD and UCeD patients, as visualized by UMAP (Fig. 3 A). Longevity also explained some of the variation, but seemingly to a lesser degree than disease status.
Figure 3.
Transcriptional differences between PCs of unknown specificity in CeD patients and controls. (A) UMAP and individual principal component distributions of all PCs of unknown specificities. PCs of unknown specificity from all UCeD (four patients, 657 cells), TCeD (three patients, 384 cells), and control patients for which we sorted a natural composition of longevities (three patients, 503 cells, sorted as outlined in Fig. S1 C) were sequenced in nine independent libraries in order to perform scRNA-seq analysis. (B) Volcano plots showing DEGs between all (left) and only short-lived (right) PCs of unknown specificity. The analysis of short-lived PCs was done by pooling short-lived PCs from all UCeD (four patients, 383 cells), TCeD (three patients, 229 cells), and all controls (five patients, 341 cells). (C) Mean expression levels of the most highly DEGs of interest between all (left) and short-lived PCs (right) of unknown specificity shown in B. Dot sizes indicate fraction of cells with nonzero expression. (D) Transcription of CD27 in PCs of unknown specificity in UCeD (four subjects), TCeD (three subjects), and controls (five subjects). Effects of different compositions of PC longevities were regressed out (top), or cells of each longevity were plotted separately (bottom). Cell numbers are indicated for each violin plot. P values were calculated with an unpaired Wilcoxon rank-sum test, adjusted for multiple hypothesis testing; ****, P < 0.0001; ns, not significant. No statistically significant differences were found when analyzing each PC longevity separately.
We then looked into individual genes that were differentially expressed between PCs of unknown specificity in UCeD patients, TCeD patients, and controls (Fig. 3, B and C). Genes encoding complement proteins C4A and C4B, HLA class I genes, as well as PECAM1, ITM2C, XBP1, APOL2, SOCS3, and SDC1 were more highly expressed in CeD patients (in particular in UCeD) compared with the controls. Interestingly, we also observed some DEGs between UCeD and TCeD patients, including XBP1, ITM2C, CD69, C4A, and C4B, with TCeD patients showing expression levels more similar to those of control subjects (Fig. 3 C). Moreover, CD69, QPCT, TNFRSF13B (encoding TACI), CCL3, and CD27 were up-regulated in controls compared with CeD patients (Fig. 3, B–D). The majority of the genes that were differentially expressed between the disease statuses were also differentially expressed when looking only at short-lived cells, demonstrating that the transcriptional differences between disease statuses are not mainly due to a skewed composition of cell longevities. Our single-cell analysis of intestinal PCs in CeD and controls thus indicates that intestinal PCs, in general, differ between CeD patients and healthy individuals.
DGP-specific PCs derive from a few heavily expanded stereotypical clonotypes
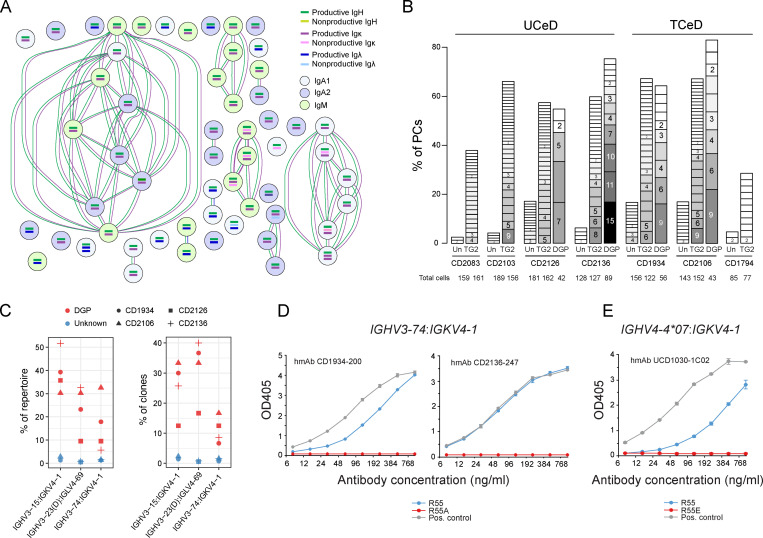
To identify clonally related PCs of each specificity, we computationally reconstructed the paired heavy and light chain sequences of each BCR from our scRNA-seq data using BraCeR (Lindeman et al., 2018). We observed very little clonal expansion in the PC populations of unknown specificity, while there were many clone groups of moderate sizes for TG2-specific PCs and very large clonal expansions for the DGP-specific PCs (Fig. 4, A and B). Although the BCR repertoires of TG2-specific PCs in CeD have been extensively studied (Iversen et al., 2017; Roy et al., 2017; Snir et al., 2015), fewer and more small-scale studies have been done for DGP-specific PCs (Snir et al., 2017; Steinsbø et al., 2014).
Figure 4.
Clonal expansion, repertoire analysis, and site-directed mutagenesis of antibodies of PCs in CeD. (A) Example of clonal expansion of DGP-specific PCs from a TCeD patient (CD1934) as inferred by BraCeR. Circles represent individual cells, and connections indicate clonal relationship based on both heavy and light chains. (B) Clone group sizes for each specificity in each CeD patient shown as percentage of PCs. Darker shades of gray and larger boxes indicate bigger clones, with number of cells in each clone group shown. Clones with two or more cells are shown. Un, unknown specificity. (C) Percentage of total DGP-specific PCs (left) or DGP-specific clone groups (right) in each patient using the three dominating V-gene chain pairings. IGHV3-23 and IGHV3-23D are collectively referred to as IGHV3-23(D). (D and E) Importance of heavy chain R55 for DGP binding assessed by ELISA. Two patient-derived DGP-specific hmAbs using IGHV3-74:IGKV4-1 reported in this study (D) and one previously reported DGP-specific hmAb (UCD1030-1C02; Steinsbø et al., 2014) using IGHV4-4*07:IGKV4-1 (E) were analyzed with or without R in position 55. A previously characterized DGP-specific hmAb (UCD1002-1E03; Steinsbø et al., 2014) using IGHV3-15:IGKV4-1 was used as positive control for comparison. Error bars indicate SD based on sample duplicates.
Here, by looking at the reconstructed BCR sequences of TG2-specific and DGP-specific PCs, we observed a similar skewing of V-gene usage as previously reported (Iversen et al., 2017; Roy et al., 2017; Snir et al., 2017; Snir et al., 2015; Steinsbø et al., 2014). TG2-specific PCs preferentially used IGHV5-51, IGHV1-69, and IGHV3-43, with 66 ± 3.9% of unique clones pairing with Igκ (in particular, V-genes IGKV1-5 and IGKV3-11). For DGP-specific PCs, we identified stereotypical V-gene pairing between IGHV3-74 and IGKV4-1, which has not been previously reported.
Importance of heavy chain residue R55, occasionally polymorphic, for binding to DGP
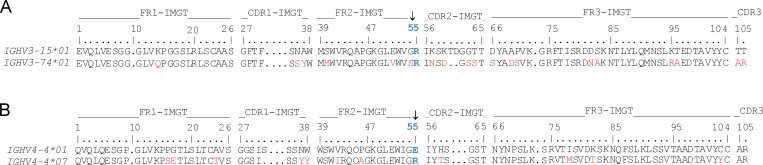
The novel stereotypical pairing of IGHV3-74 and IGKV4-1, in addition to the previously reported IGHV3-15:IGKV4-1 and IGHV3-23(D):IGLV4-69 (Snir et al., 2017), made up almost the entire repertoire of the DGP-specific PCs in all four CeD patients for which DGP-specific BCR sequences were available (Fig. 4 C). Interestingly, IGHV3-15 and IGHV3-74 share a conserved arginine residue at IMGT (the international ImMunoGeneTics information system) position 55 (R55; Fig. S3 A), a residue which plays a key role in binding DGP as shown by x-ray crystallography and by binding studies with site-directed antibody mutants (Snir et al., 2017). R55 is only present as a nonpolymorphic residue in five IGHV genes but occurs as an allelic variant of seven additional IGHV genes (Lefranc et al., 2015). R55 has previously been demonstrated to be essential for binding of DGP-specific antibodies using IGHV3-15:IGKV4-1 (Snir et al., 2017). We therefore hypothesized that IGHV3-74:IGKV4-1 antibodies could bind DGP in a similar manner to that of IGHV3-15:IGKV4-1 antibodies.
Figure S3.
IGHV sequence alignments related to Fig. 4, D and E. (A) Alignment of the amino acid sequences of IGHV3-15 and IGHV3-74 according to the IMGT numbering. The conserved R residue in position 55 is highlighted in blue and with an arrow. Positions that differ between the two gene segments are shown in red. (B) Alignment of the amino acid sequences of IGHV4-4*01 and IGHV4-4*07 according to the IMGT numbering. The polymorphic R/E residue in position 55 is highlighted in blue and with an arrow. Other residues that differ between the two alleles are shown in red.
To test the importance of R55 in the heavy chain of IGHV3-74:IGKV4-1 antibodies for binding to DGP, we expressed two different human mAbs (hmAbs) based on the BCR sequences of patient-derived DGP-specific PCs using the IGHV3-74:IGKV4-1 chain pairing. We expressed each antibody in two variants with either R or A in position 55 of the heavy chain (R55 or R55A) and tested for reactivity to DGP in ELISA. While both hmAbs harboring R55 demonstrated reactivity to DGP, the R55A mutation led to a complete loss of DGP reactivity (Fig. 4 D).
Interestingly, IMGT position 55 is polymorphic in IGHV4-4, with R55 present in the *07 allele and E55 in alleles *01 through *06 (Fig. S3 B). Since Ig polymorphisms can influence antibody repertoires (Avnir et al., 2016), we performed site-directed mutagenesis of R55 in a previously reported (Steinsbø et al., 2014) patient-derived DGP-specific hmAb using IGHV4-4*07:IGKV4-1, and showed that the R55E mutation disrupted binding to DGP (Fig. 4 E).
CeD-specific clonal lineage trees contain PCs spanning different isotypes and longevities
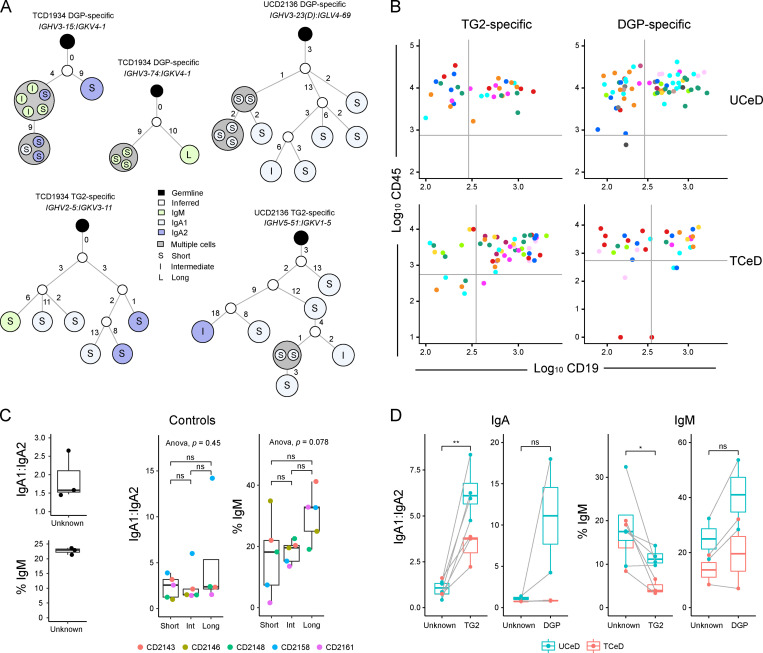
In addition to facilitating clonal inference and repertoire analysis, reconstruction of BCR sequences also allowed us to accurately determine the isotype of each PC. Inspection of individual clone groups and lineage trees revealed that several clone groups contained PCs of different isotypes and longevities (Fig. 5, A and B).
Figure 5.
Clonality and isotype usage spanning different PC longevity populations. (A) Examples of lineage trees created based on reconstructed BCR sequences for DGP- or TG2-specific clones containing different isotypes and longevities from an untreated (UCD2136) and treated (TCD1934) patient. IGHV3-23 and IGHV3-23D are grouped together as IGHV3-23(D). (B) Log10 index-sorting values for CD19 and CD45 for highlighted TG2-specific (left) and DGP-specific (right) clone groups for two representative patients (CD2136 and CD2106). Negative values were converted to 1 before they were logarithmized. Only the largest clones are shown, with members of each clone in the same color. (C) IgA1:IgA2 ratio and IgM usage in controls for which a natural composition of PC longevities was sorted (left, n = 3). IgA1:IgA2 ratio (center) and percent IgM PCs (right) between PCs of different longevities in all controls (n = 5) are color coded for each individual. No IgA2 PCs were detected among the only nine long-lived PCs from CD2146, and no IgA1:IgA2 ratio was plotted for long-lived PCs for this patient. P values were calculated with a paired Student’s t test. (D) IgA subclass usage (left) and IgM usage (right) between disease-specific and other PCs in CeD. Each point represents IgA1:IgA2 ratio (left) or percent IgM PCs (right) in an individual based on reconstructed BCR sequences; data from the same individual are connected with a gray line. All four UCeD and three TCeD patients were included in the comparisons between TG2-specific PCs and PCs of unknown specificity; only patients from which DGP-specific PCs were sorted (two UCeD patients, two TCeD patients) were included in the comparisons including DGP-specific PCs. P values were calculated with a paired Student’s t test. *, P < 0.05; **, P < 0.01; ns, not significant.
It has long been known that IgA1 is preferentially used over IgA2 in the upper small intestine (Chiba et al., 1987). When looking at isotype usage in the controls, we observed this preferred usage of IgA1 (Fig. 5 C). The proportion of PCs expressing IgM was also in line with previous observations (Brandtzaeg and Johansen, 2005). While short-lived intestinal PCs have been shown to be skewed toward IgA usage (Landsverk et al., 2017), no reports of IgA subclass usage have been made. We therefore tested for differences in IgA1/IgA2 ratio between the three PC longevity populations in the controls but found no statistically significant differences.
For both UCeD and TCeD, TG2-specific PCs had significantly lower usage of the IgM isotype (and higher usage of IgA) compared with PCs of unknown specificity from the same individuals (Fig. 5 D). In line with previous observations, DGP-specific PCs seemed less restricted to the IgA-compartment than TG2-specific PCs (Steinsbø et al., 2014; Fig. 5 D). Notably, we also observed a significantly higher ratio of IgA1/IgA2 usage in TG2-specific PCs compared with PCs of unknown specificity, in line with a previous study (Iversen et al., 2017).
Somatic hypermutation (SHM) rates vary with isotype subclass, longevity, specificity and V-gene usage
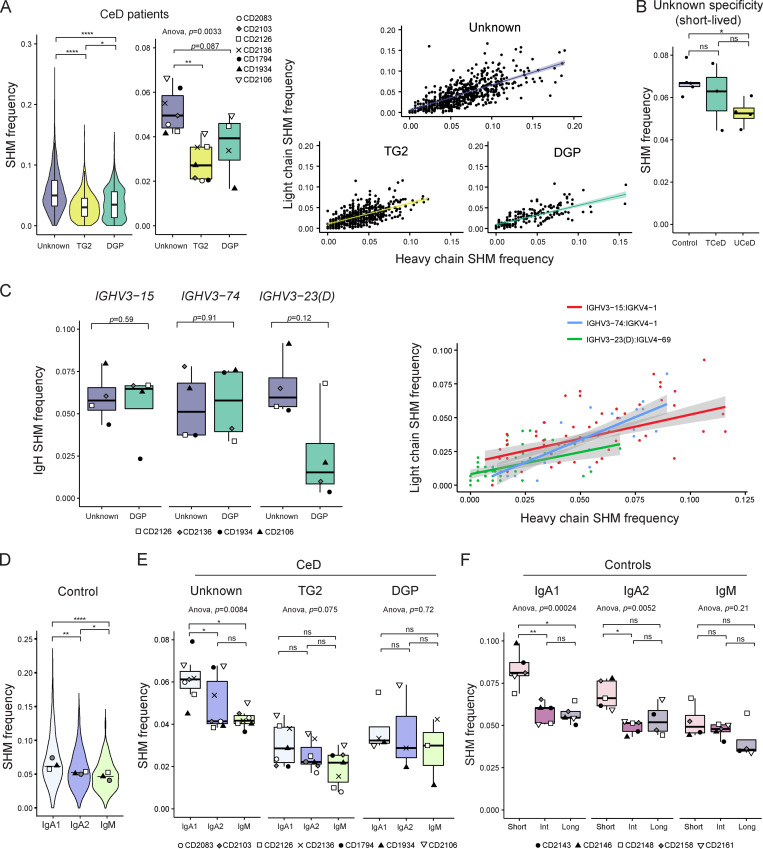
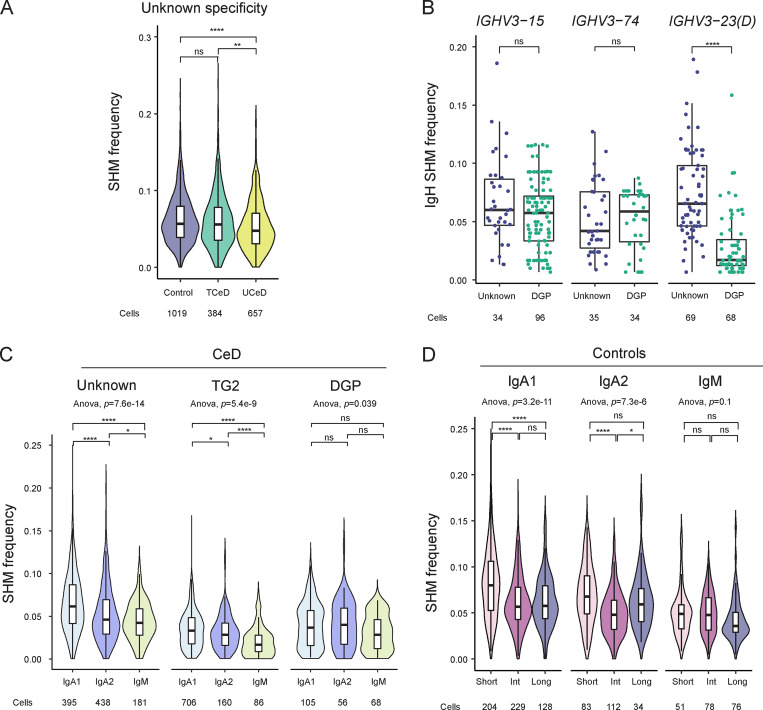
Lastly, we looked into the SHM frequency in each reconstructed BCR sequence (Fig. 6 and Fig. S4). In line with previous reports (Di Niro et al., 2012; Roy et al., 2017; Snir et al., 2017; Snir et al., 2015; Steinsbø et al., 2014), TG2- and DGP-specific PCs had fewer somatic mutations compared with PCs of unknown specificity, and the number of mutations in heavy and light chains correlate (Fig. 6 A). Unexpectedly, we also detected fewer somatic mutations in PCs of unknown specificity in UCeD patients compared with TCeD patients or controls (Fig. 6 B and Fig. S4 A).
Figure 6.
SHM frequency for PCs according to specificity, disease status, V-gene usage, isotype, and longevity. SHM load in the V-region of BCR sequences reconstructed by BraCeR was calculated with IMGT/HighV-QUEST and normalized by sequence length. Average SHM frequencies for heavy and light chain are shown, unless only one of the chains was reconstructed or otherwise stated. (A) SHM frequency in all BCR sequences from all CeD patients (n = 7) pooled together, stratified by specificity (left). Median SHM frequencies for each patient according to antigen specificity are shown in the central boxplot. P values were calculated with an unpaired Wilcoxon rank-sum test (left) or paired Student’s t test (center). Correlation between heavy and light chain SHM frequencies stratified by PC specificity are shown in the right panel for patients with sorted DGP-specific PCs (n = 4). The colored lines represent fitted trends using linear regression; shades indicate the 95% confidence interval of the fitted line. (B) SHM frequency of short-lived PCs of unknown specificity for all UCeD patients (n = 4), TCeD patients (n = 3), and controls (n = 5). Median values for each individual are shown. P values were calculated with a paired Student’s t test. (C) Heavy chain (IgH) SHM frequency of DGP-specific PCs and PCs of unknown specificity from patients for which DGP-specific PCs were sorted (n = 4), stratified by usage of the three stereotypical IGHV genes used by DGP-specific PCs (left). P values were calculated with a paired Student’s t test. Correlation between heavy and light chain SHM frequencies in the three most common chain pairings among DGP-specific PCs are shown to the right. IGHV3-23 and IGHV3-23D are collectively referred to as IGHV3-23(D). (D) SHM frequency of isotype (sub)classes from controls. Only controls with a natural composition of PC longevities are shown and were pooled together (n = 3). Median values for each individual have been added to the violin plots. P values were calculated with an unpaired Wilcoxon rank-sum test based on the pooled sequences; horizontal lines represent median values based on the pooled sequences. (E) SHM frequency of isotype (sub)classes for each PC specificity in all CeD patients (n = 7). Each data point represents the median SHM frequency for one patient. Only patients with at least three unique BCR sequences in a given group are included. P values were calculated with a paired Student’s t test. (F) SHM frequency of isotype (sub)classes and PC longevities from all controls (n = 5). Only patients with at least three unique BCR sequences in a given group are included. P values were calculated with a paired Student’s t test. Adjusted P values are shown in all figures. *, P < 0.05; **, P < 0.01; ****, P < 0.0001; ns, not significant.
Figure S4.
SHM frequency for PCs according to specificity, disease status, V-gene usage, isotype, and longevity. SHM load in the V-region of BCR sequences reconstructed by BraCeR was calculated with IMGT/HighV-QUEST and normalized by sequence length. Average SHM frequencies for heavy and light chain are shown, unless only one of the chains was reconstructed or otherwise stated. P values were calculated with an unpaired Wilcoxon rank-sum test in all subfigures. Adjusted P values are shown. *, P < 0.05; **, P < 0.01; ****, P < 0.0001; ns, nonsignificant. (A) SHM frequency in all BCR sequences from PCs of unknown specificity pooled together according to disease status (UCeD patients n = 4, TCeD patients n = 3, and controls n = 5). (B) Heavy chain (IgH) SHM frequency of DGP-specific PCs and PCs of unknown specificity from patients for which DGP-specific PCs were sorted (n = 4), stratified by usage of the three stereotypical IGHV genes used by DGP-specific PCs. IGHV3-23 and IGHV3-23D are collectively referred to as IGHV3-23(D). (C) SHM frequency of pooled BCR sequences from all CeD patients (n = 7), stratified by specificity and isotype sub(class). (D) SHM frequency of pooled BCR sequences from all controls (n = 5), stratified by isotype (sub)class and PC longevity: short- (CD19+CD45+), intermediate (Int)- (CD19−CD45+), or long lived (CD19−CD45−).
DGP-specific PCs using IGHV3-23(D) have previously been shown to accumulate significantly fewer mutations than IGHV3-23(D) PCs of unknown specificity (Snir et al., 2017). We therefore investigated if this was the case also for DGP-specific PCs using IGHV3-15:IGKV4-1 or IGHV3-74:IGKV4-1. In contrast to IGHV3-23(D):IGLV4-69, we found no significant differences in mutational load between DGP-specific PCs and PCs of unknown specificity using IGHV3-15 or IGHV3-74 (Fig. 6 C, left panel; and Fig. S4 B). This finding indicates that the lower SHM frequency of DGP-specific PCs compared with PCs of unknown specificity is coupled to the usage of V-genes. Again, the frequencies of mutations in heavy and light chains were correlated (Fig. 6 C, right panel).
We observed a higher rate of SHM in PCs of the IgA1 subclass compared with IgA2 and IgM in the controls (Fig. 6 D), in line with what has previously been reported for peripheral blood mononuclear cells (PBMCs; Kitaura et al., 2017). This trend was also evident for PCs of unknown specificity and TG2-specific PCs in the CeD patients (Fig. 6 E and Fig. S4 C). We then investigated whether SHM rates differ between PC longevities. To remove bias in the form of different isotype preferences between the longevities, we looked at each isotype subclass separately. Interestingly, when looking at the most abundant isotype subclass, IgA1, short-lived PCs had significantly more mutations than intermediate- or long-lived PCs (Fig. 6 F and Fig. S4 D).
Discussion
This study provides information on the longevity profile of disease-specific PCs of CeD. We found that a substantial population of disease-specific PCs remained in patients even after at least 1 yr on a GFD, which is in keeping with a previous observation (Di Niro et al., 2016). Conceivably, this feature could imply that the cells that remain are mainly long lived. This appears to be the case only in very long-term TCeD patients, as we demonstrate that in UCeD and in short-term TCeD patients most of the TG2- and DGP-specific PCs have the phenotype of short-lived CD19+CD45+ PCs, with a substantial population having an intermediate-lived CD19−CD45+ phenotype. This finding can explain why the TG2-specific PC population decreases already the first months after a patient starts on a GFD (Di Niro et al., 2016). While we found that the frequency of PCs with intermediate- and long-lived phenotype among the disease-specific PCs increased with time on GFD, the presence of disease-specific short-lived PCs even in long-term TCeD patients indicates that a significant proportion of such PCs must be continuously regenerated even on a GFD. A reason underlying this observation can be that most patients have occasional sporadic intake of small amounts of gluten (Freeman, 2017). The disease-specific PCs with short-lived phenotype could potentially be formed by exposure to trace amounts of gluten. We also observe that PCs of different longevities exist within the same clonal lineages. This suggests that at least some of the short-lived PCs have been generated from memory B cells, as would fit with the observation of clonal relatedness between TG2-specific PCs and CD27+ memory B cells sampled from peripheral blood (Snir et al., 2015). Importantly, we observed a dominance of short-lived CD19+CD45+ phenotype in UCeD and short-term TCeD, both among disease-specific PCs but also in the general PC population. These data indicate that new and short-lived PCs massively accumulate in the lesion of UCeD and that the antigen targeting of many of these PCs is not directed to the disease-specific antigens. The exact molecular mechanisms underlying the accumulation of PCs with antigen specificities not related to CeD remain unclear. Notwithstanding, these cells, as demonstrated here, have immunologically active profiles suggesting that they may well partake in processes relevant to formation of the gut lesion in CeD.
Our transcriptional analysis of PC populations with different longevities revealed interesting differences. Short-lived PCs exhibited higher expression of a range of genes encoding ribosomal proteins and various genes involved in metabolism. Other genes involved in metabolism, CD27, and HLA class I genes were up-regulated in long-lived PCs. These differences in gene expression may thus reflect previously reported differences in metabolic activity or antibody production between short- and long-lived PCs (Lam et al., 2018; Landsverk et al., 2017).
Functional roles of PCs that go beyond antibody secretion have been suggested over the last years, including reports of cytokine production (Fillatreau, 2015). Our results and our previously reported bulk RNA-seq data (Snir et al., 2019) indicate that intestinal PCs produce mRNA transcripts for cytokines such as IL-15 and IL-16. IL-15 has been implicated in many autoimmune diseases and is a known player in CeD (Jabri and Abadie, 2015). It is not known, however, if all the cytokines for which we identified mRNA are actually translated and produced in the PCs. In the case of IL-15, for instance, it is known that its translation is under extensive regulation of a range of control elements, often resulting in repressed translation (Bamford et al., 1998; Mishra et al., 2014; Nishimura et al., 2000). Furthermore, two isoforms of IL-15 exist, one of which is not secreted but rather stored intracellularly (Nishimura et al., 2000; Tagaya et al., 1997).
Conventionally, PCs are not thought to be APCs for T cells. However, the retained cell surface expression of a functional BCR in IgA- and IgM-expressing PCs (Blanc et al., 2016; Di Niro et al., 2010; Pinto et al., 2013) make these cells able to fulfil the key hallmark of B cells as APCs having a cell surface receptor for antigen binding and uptake. In line with previously published bulk RNA-seq data (Snir et al., 2019), we detected intermediate to low expression of HLA class II genes, in particular HLA-DQA1, and costimulatory molecules in a large fraction of the PCs. Indeed, a recent study found PCs to be the main cell type displaying DGP on disease-associated HLA class II molecules in the CeD lesion (Høydahl et al., 2019).
We also showed, for the first time on a single-cell level, that TG2-specific PCs, DGP-specific PCs, and PCs of unknown specificity display some transcriptional differences. A few of these differences can likely be ascribed to PC longevity. However, we also observed transcriptional differences between PC specificities when longevity was removed as a covariate, suggesting that differences not related to potential longevity of the PCs may exist.
By comparing PCs of unknown specificity from CeD patients and controls, our work supports the recent finding that PCs of CeD patients generally may differ from PCs of healthy individuals (Snir et al., 2019). Notably, we also detected some DEGs between TCeD and UCeD patients, with TCeD patients often exhibiting intermediate expression values compared with UCeD or controls. Looking further into such differences may shed light on the potentially pathogenic roles of PCs in CeD, including the possibility that non–disease-specific PCs may also contribute to CeD.
PCs are generally thought to express high levels of CD27 on their surface. Interestingly, we found that CD27 was up-regulated in long-lived versus short-lived PCs, PCs of unknown specificity versus TG2-specific and DGP-specific PCs, and in controls compared with CeD patients. Up-regulation of CD27 in non–TG2-specific compared with TG2-specific PCs has also been observed in bulk RNA-seq (Snir et al., 2019). Heterogeneous expression of CD27 has previously been reported in multiple myeloma (Guikema et al., 2003), and plasmablasts/PCs in patients with hemorrhagic fever with renal syndrome were found to have very low surface expression of CD27 (Kerkman et al., 2019 Preprint).
We observed a somewhat higher frequency of TG2-specific PCs in UCeD and TCeD patients than previously reported (Di Niro et al., 2012; Snir et al., 2015; Steinsbø et al., 2014). This discrepancy could be due to the fact that previous studies have focused on CD27+ IgA PCs, while we here made no such restrictions, and many of the TCeD patients in this study had been on a GFD for little more than one year. While we found only ∼12% of TG2-specific PCs to be of the IgM isotype in UCeD, DGP-specific PCs seemed less restricted in their isotype usage, with only IgM PCs being detected in several clones. Unlike IgA antibodies, these IgM autoantibodies could be pathogenic through activation of complement via the classical pathway, as previously suggested (Halstensen et al., 1992).
The preferential usage of IgA1 over IgA2 that we observed in PCs of the controls was already well established (Chiba et al., 1987). Beyond this known preference, we observed a further skewed IgA1/IgA2 usage in TG2-specific PCs compared with PCs of unknown specificity in line with a previous report from our group (Iversen et al., 2017). While many clone groups contained only IgA1- or IgA2-expressing PCs, we also observed several clones spanning IgA1, IgA2, and even IgM. Although isotype class switching has been studied over the years (Horns et al., 2016), many details are still unclear. It has previously been suggested that switching to IgA2 could be a feature of the inducing antigen, as locations rich in bacteria often have a higher ratio of IgA2 cells (Kett et al., 1995). This seems unlikely to be the case for TG2 or DGP, as members of each clone group have been induced by the same antigen, but we still see isotype switch to both IgA subclasses in several clones.
IgA1 PBMCs are known to generally acquire more mutations than IgA2 or IgM PBMCs (Kitaura et al., 2017). Here, we show that this is the case also for intestinal PCs. Moreover, we found that PCs of different longevities may acquire different numbers of mutations. When looking at IgA1 PCs, short-lived PCs had significantly more mutations compared with intermediate- or long-lived PCs in the controls. This is surprising, as TG2-specific PCs in CeD have strikingly low numbers of mutations (Di Niro et al., 2012; Roy et al., 2017; Snir et al., 2015) despite being prone to more SHM due to their mainly short-lived IgA1 phenotype. This finding further emphasizes the functional importance of limited SHM in CeD-specific antibodies.
The finding of biased usage of IGHV3-74:IGKV4-1 for recognition of DGP also underscores the importance of germline-encoded residues in CeD-specific antibodies. Like in a previously identified stereotypic CeD antibody IGHV3-15:IGKV4-1 (Snir et al., 2017), the IGHV3-74:IGKV4-1 antibody utilizes heavy chain residue R55 for recognition of DGP. This canonical function of R55, as we here demonstrate, was also observed for an IGHV4-4*07:IGKV4-1 CeD antibody, where R55 is a polymorphic residue. While the latter is one antibody from one patient, and the overall impact of Ig polymorphisms for the formation of the CeD antibodies remains unexplored, this finding illustrates the principle that germline polymorphisms can indeed influence disease-specific antibody responses in CeD. More studies on the importance of Ig polymorphism for the humoral response in CeD are warranted.
Materials and methods
Ethics statement
This study was approved by the Regional Ethics Committee of South-Eastern Norway (REK 6544). Informed, written consent was obtained from all patients before sample collection.
Sample collection
We collected intestinal biopsies from 15 patients with UCeD, 26 CeD patients on GFD for a minimum of 1 yr (TCeD), and 13 non-CeD controls (no known autoimmune diseases or intestinal inflammation) coming in for gastroduodenoscopy at the Department of Gastroenterology, Oslo University Hospital (OUH) Rikshospitalet, Norway, or Department of Gastroenterology, Akershus University Hospital (AUH), Norway. CeD diagnosis was given following the British Society of Gastroenterology guidelines (Ludvigsson et al., 2014). Pathological and laboratory data (Table S1) are taken from the hospital records. Duodenal biopsies were collected in ice-cold RPMI 1640 medium (samples from OUH) or PBS (samples from AUH). Samples from all individuals were used for flow cytometry analysis, while four UCeD patients, three TCeD patients, and five controls also were analyzed by scRNA-seq.
Sample processing
Intestinal biopsies were treated twice with 2 mM EDTA in PBS with 2% FCS at 37°C under constant rotation for 10 min in order to remove the epithelium and intraepithelial lymphocytes. The biopsies were subsequently digested with 1 mg/ml collagenase type H (Sigma) in 2% FCS in PBS at 37°C for 35 min under constant rotation. The digested tissue was homogenized with a syringe and needle and filtered through a 40-μm cell strainer and washed with PBS. Samples were then immediately used for analysis and/or sorting. 16 of the samples had been cryopreserved before flow cytometry analysis (Table S1).
FACS
We used fluorescently labeled tetramers loaded with TG2 and DGPs in order to identify CeD-specific cells. BirA-tagged TG2 was produced in Sf9 insect cells and N-terminally biotinylated (Stamnaes et al., 2016). The immunodominant deamidated gluten epitope PLQPEQPFP (DGP) was obtained as biotinylated peptide (GL Biochem). TG2 and DGP were multimerized on PE-labeled streptavidin (SA; Invitrogen) and APC-SA (PhycoLink), respectively, on ice for at least 45 min at a 4:1 ratio of TG2/DGP:PE/APC-SA. The single-cell suspension was stained with TG2-SA-PE, DGP-SA-APC, and the following anti-human antibodies: CD3-BV605 (UCHT1, Biolegend) or CD3-Super Bright 600 (UCHT1, eBioscience), CD11c-BV605 (3.9, Biolegend), CD14-BV605 (M5E2, Biolegend), CD45-APC-Cy7 (H130, Biolegend), CD19-Pacific Blue (HIB19, Biolegend), and CD38-FITC (HB7, eBioscience) in PBS with 2% FCS for 30 min on ice, washed with 2% FCS PBS and filtered before cell sorting using a BD FACSAria II (OUH Radiumhospitalet) or BD FACSAria III instrument (OUH Rikshospitalet) with FACS Diva software. Flow cytometry data were analyzed with FlowJo (BD) v10.
Single, large CD38highCD3−CD11c−CD14− PCs were randomly index sorted according to antigen specificity (Fig. S1, B and C, left). We aimed at obtaining approximately equal numbers of TG2-specific PCs and PCs of unknown specificity on each plate for CeD patients. DGP-specific PCs, being rare, were also selectively sorted for some patients. For two of the controls, we sorted PCs based on their CD19/CD45 phenotype as a proxy for cell longevity (Fig. S1 C, right). Cells were sorted into 96-well plates containing 2 μl lysis buffer per well (0.2% vol/vol Triton X-100 [Sigma] in H2O with 2 U/μl RNase inhibitor), immediately spun down after sorting and placed on dry ice before being stored at −70°C until further processing. Cells of different specificities and longevities were sorted on the same plate to minimize batch effects.
scRNA-seq
We performed scRNA-seq of in total 3,743 intestinal PCs. The cells were subjected to reverse transcription and cDNA preamplification according to a modified version of the Smart-seq2 protocol (Picelli et al., 2014b), using 0.5 μl/well SMARTScribe Reverse transcription (Clontech) for reverse transcription. The cDNA was preamplified with 21 PCR cycles and purified with 20 μl Ampure XP beads per well (Agencourt). Amplified cDNA was tagmented using an in-house–produced Tn5 transposase (Picelli et al., 2014a) and dual indexed with Nextera (XT) N7xx and S5xx index primers at a final concentration of 125 nM. Four plates were pooled together and sequenced at the Norwegian Sequencing Centre on a NextSeq500 instrument with 75-bp paired-end reads in high-output mode. Average number of read pairs per cell was around one million.
scRNA-seq quality control
Raw reads were trimmed for adapter sequences and low-quality sequences using Cutadapt v.1.18 (Martin, 2011) through Trim Galore v0.6.1 in paired-end mode. We quantified transcript expression for the scRNA-seq data with Salmon v0.11.3 (Patro et al., 2017) using cDNA sequences from GRCh38.94 and a k-mer length of 25 for creating the salmon index. Transcripts were aggregated to gene level, and a transcript length–corrected gene expression matrix for all the cells was constructed using tximport v1.8.0 (Soneson et al., 2015). The single cells were subsequently filtered based on the following quality measures to discard low-quality cells: number of detected genes, number of reads, percent mitochondrial genes, percent reads mapping to the reference transcriptome, percent Ig genes, and whether a BCR heavy chain could be reconstructed by BraCeR. Library-specific quality threshold values are shown in Table S3. Quality control was performed in R version 3.5.3 using the scater package (McCarthy et al., 2017). In total, 3,251 cells remained after quality control. Demultiplexed reads for each cell have been deposited with regulated access at the European Genome-phenome Archive under accession no. EGAS00001004623.
scRNA-seq batch correction and normalization
Since a large part of the PC transcriptome (67.7 ± 8.4% in our data) consists of Ig genes, and these genes would dominate downstream analyses and mask non–Ig-related transcriptional differences between cell populations, all Ig genes were discarded from the analysis before normalization. After removing Ig genes, genes with >3 reads detected in >20 cells were retained. For each analysis, the gene expression matrix was subset according to the cells of interest and normalized by total counts per cell using scanpy v1.4.4 (Wolf et al., 2018) in python 3.7. The normalized expression matrix was then logarithmized as Highly variable genes were identified with the highly_variable_genes function of scanpy with the following parameters: min_mean = 0.1, max_mean = 10, min_disp = 0.25. Unwanted variation, such as the number of genes and reads detected and percent mitochondrial genes, was then regressed out using NaiveDE v1.2.0 (https://github.com/Teichlab/NaiveDE) and the highly variable genes, while retaining the variation of interest for each analysis (see figure legends for analysis-specific details). The expression matrix was scaled using sklearn (scikit-learn v0.21.3; Pedregosa et al., 2011).
PCA and UMAP
We performed PCA of the scaled expression matrices using scanpy and investigated which variables drove the different principal components. The principal components explaining most of the variation in the variable of interest were then used as input for UMAP (McInnes et al., 2018 Preprint).
Identifying DEGs
DEGs were identified with the scanpy function rank_genes_groups using the corrected gene expression matrix as input and Wilcoxon rank-sum test, correcting for multiple testing with the Benjamini-Hochberg method. Only genes with an adjusted P value < 0.05 were retained. Volcano plots were created based on DEGs using the EnhancedVolcano v.1.0.1 R package, with adjusted P value = 0.05 and logfold-change = 0.4 as thresholds for statistically significant DEGs. DEGs of interest were plotted using the scanpy function dotplot.
BCR reconstruction with BraCeR
Paired BCR heavy and light chains were reconstructed for each cell using BraCeR in assembly mode with all raw reads as input. BraCeR assembly was run with --threshold 5000 in order to filter out lowly expressed reconstructed BCRs that may arise from contamination between wells or errors in indices. Clonally related PCs were determined separately for each patient and each PC specificity for cells passing scRNA-seq quality control using BraCeR in summary mode with the following parameters: --include_multiplets --infer_lineage. Potential cell multiplets were manually inspected and removed as previously outlined (Lindeman and Stubbington, 2019). Somatic mutations in the variable regions were analyzed with IMGT/HighV-QUEST (Brochet et al., 2008), and SHM rates were calculated by dividing the number of point mutations by sequence length. Reconstructed BCR sequences for each cell passing quality control have been deposited at the European Genome-phenome Archive under accession no. EGAS00001004623.
Expression of mAbs, site-directed mutagenesis, and ELISA
BCR sequences obtained from two DGP-reactive PCs using the IGHV3-74:IGKV4-1 gene pair were expressed as recombinant human IgG1 molecules as previously described (Smith et al., 2009). Briefly, synthetic DNA (GenScript) encoding the antibody variable regions with either R or A in IMGT position 55 of the heavy chain were cloned into expression vectors and used for transfection of HEK 293-F cells. The secreted antibodies were subsequently purified from the supernatant on HiTrap Protein G columns (GE Healthcare). Plasmid DNA encoding a gliadin-reactive IGHV4-4:IGKV4-1 antibody with R in position 55 (corresponding to the IGHV4-4*07 allele) was already available in the laboratory (Steinsbø et al., 2014). In this case, an R to E (found in alleles *01 to *06) substitution was introduced by overlap extension PCR. The reactivity of all antibody variants with biotinylated, synthetic DGP was assessed by ELISA. Briefly, the peptide (50 nM) was immobilized in SA-coated microplates (Thermo Scientific) followed by incubation with different concentrations of recombinant antibodies in TBS supplemented with 0.1% (vol/vol) Tween 20. Bound antibody was detected with alkaline phosphatase-conjugated goat anti-human IgG (Southern Biotech).
Statistical analysis
The statistical tests applied for each analysis are detailed in the figure legends. In brief, differences between population distributions were tested using a paired Wilcoxon signed-rank test or paired two-sample t test for data in which, for example, TG2-specific, DGP-specific, and PCs of unknown specificity were compared against each other within each patient. To test differences in distribution between unpaired samples, such as between CeD patients and controls, an unpaired Wilcoxon rank-sum test or unpaired two-sample t test was used. Results of the statistical tests are throughout this paper given as P value significance levels (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001) after adjusting for multiple testing. Global differences between means were calculated with one-way ANOVA and reported in addition to individual adjusted P values when appropriate for tests between independent groups. P values were calculated and added to plots using ggpubr v0.2.4. Linear regression was performed in R with the lm (linear model) method.
Online supplemental material
Table S1 provides clinical data for patients included in the study. Table S2 contains data from longevity analysis by flow cytometry for all patients. Table S3 provides quality-control threshold values and cell numbers for scRNA-seq data. Fig. S1 presents approach, gating strategy, and supplemental results for analysis of PC longevity and specificity by flow cytometry. Fig. S2 provides mean gene expression values for selected markers for all PCs analyzed by scRNA-seq. Fig. S3 shows IGHV sequence alignments related to Fig. 4, D and E. Fig. S4 presents additional SHM frequency data.
Supplementary Material
provides clinical data for patients included in the study.
contains data from longevity analysis by flow cytometry for all patients.
provides quality-control threshold values and cell numbers for scRNA-seq data.
Acknowledgments
We would like to thank all patients donating samples, as well as C. Hinrichs, S. Furholm, M. Valde, S. Isaksen and F. van Megen (all OUH, Oslo, Norway) for assistance with obtaining patient samples and clinical information. Furthermore, we thank M.K. Johannesen, B. Simonsen, and Ł. Wyrożemski for technical assistance with experiments. We also thank the Flow Cytometry Core Facility at OUH, in particular H. Notø, for assistance with flow cytometry, and R. Sandberg and G. Winberg at the Karolinska Institute (Solna, Sweden) for the generous gift of the plasmid encoding Tn5 transposase. Sequencing was performed at the Norwegian Sequencing Centre at Ullevål, Oslo, Norway, and raw sequencing data were stored and analyzed on the Tjeneste for Sensitive Data facilities, owned by the University of Oslo.
This work was funded by the University of Oslo world-leading research program on human immunology and Stiftelsen Krisitan Gerhard Jebsen (project SKGJ-MED-017) to L.M. Sollid, and a Single Cell Gene Expression Atlas grant from the Wellcome Trust (108437/Z/15/Z) to Z. Miao. Elements of Fig. S1 A were modified from Servier Medical Art, licensed under a Creative Commons Attribution 3.0 Unported License.
Author contributions: I. Lindeman, C. Zhou, L.M. Eggesbø, J. Polak, and S.-W. Qiao performed experiments. I. Lindeman, C. Zhou, and Z. Miao analyzed the data. K.E.A. Lundin and J. Jahnsen provided patient samples. I. Lindeman drafted the manuscript. I. Lindeman, L.M. Sollid, R. Iversen, C. Zhou, K.E.A. Lundin, and S.-W. Qiao revised the manuscript. All authors approved the revised manuscript. L.M. Sollid supervised the project.
References
- Afik, S., Yates K.B., Bi K., Darko S., Godec J., Gerdemann U., Swadling L., Douek D.C., Klenerman P., Barnes E.J., et al. . 2017. Targeted reconstruction of T cell receptor sequence from single cell RNA-seq links CDR3 length to T cell differentiation state. Nucleic Acids Res. 45:e148 10.1093/nar/gkx615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afik, S., Raulet G., and Yosef N.. 2019. Reconstructing B-cell receptor sequences from short-read single-cell RNA sequencing with BRAPeS. Life Sci. Alliance. 2:e201900371 10.26508/lsa.201900371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avnir, Y., Watson C.T., Glanville J., Peterson E.C., Tallarico A.S., Bennett A.S., Qin K., Fu Y., Huang C.Y., Beigel J.H., et al. . 2016. IGHV1-69 polymorphism modulates anti-influenza antibody repertoires, correlates with IGHV utilization shifts and varies by ethnicity. Sci. Rep. 6:20842 10.1038/srep20842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baklien, K., Brandtzaeg P., and Fausa O.. 1977. Immunoglobulins in jejunal mucosa and serum from patients with adult coeliac disease. Scand. J. Gastroenterol. 12:149–159. 10.1080/00365521.1977.12031127 [DOI] [PubMed] [Google Scholar]
- Bamford, R.N., DeFilippis A.P., Azimi N., Kurys G., and Waldmann T.A.. 1998. The 5¢ untranslated region, signal peptide, and the coding sequence of the carboxyl terminus of IL-15 participate in its multifaceted translational control. J. Immunol. 160:4418–4426. [PubMed] [Google Scholar]
- Benichou, J., Ben-Hamo R., Louzoun Y., and Efroni S.. 2012. Rep-Seq: uncovering the immunological repertoire through next-generation sequencing. Immunology. 135:183–191. 10.1111/j.1365-2567.2011.03527.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc, P., Moro-Sibilot L., Barthly L., Jagot F., This S., de Bernard S., Buffat L., Dussurgey S., Colisson R., Hobeika E., et al. . 2016. Mature IgM-expressing plasma cells sense antigen and develop competence for cytokine production upon antigenic challenge. Nat. Commun. 7:13600 10.1038/ncomms13600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandtzaeg, P., and Johansen F.-E.. 2005. Mucosal B cells: phenotypic characteristics, transcriptional regulation, and homing properties. Immunol. Rev. 206:32–63. 10.1111/j.0105-2896.2005.00283.x [DOI] [PubMed] [Google Scholar]
- Brochet, X., Lefranc M.P., and Giudicelli V.. 2008. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 36(Web Server):W503-W508 10.1093/nar/gkn316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buettner, F., Natarajan K.N., Casale F.P., Proserpio V., Scialdone A., Theis F.J., Teichmann S.A., Marioni J.C., and Stegle O.. 2015. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat. Biotechnol. 33:155–160. 10.1038/nbt.3102 [DOI] [PubMed] [Google Scholar]
- Canzar, S., Neu K.E., Tang Q., Wilson P.C., and Khan A.A.. 2017. BASIC: BCR assembly from single cells. Bioinformatics. 33:425–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba, M., Ohta H., Yagisawa H., and Masamune O.. 1987. IgA1 & IgA2 distribution in the intestine. Gastroenterol. Jpn. 22:18–23. 10.1007/BF02806330 [DOI] [PubMed] [Google Scholar]
- Di Niro, R., Mesin L., Raki M., Zheng N.Y., Lund-Johansen F., Lundin K.E., Charpilienne A., Poncet D., Wilson P.C., and Sollid L.M.. 2010. Rapid generation of rotavirus-specific human monoclonal antibodies from small-intestinal mucosa. J. Immunol. 185:5377–5383. 10.4049/jimmunol.1001587 [DOI] [PubMed] [Google Scholar]
- Di Niro, R., Mesin L., Zheng N.Y., Stamnaes J., Morrissey M., Lee J.H., Huang M., Iversen R., du Pré M.F., Qiao S.W., et al. . 2012. High abundance of plasma cells secreting transglutaminase 2-specific IgA autoantibodies with limited somatic hypermutation in celiac disease intestinal lesions. Nat. Med. 18:441–445. 10.1038/nm.2656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Niro, R., Snir O., Kaukinen K., Yaari G., Lundin K.E., Gupta N.T., Kleinstein S.H., Cols M., Cerutti A., Mäki M., et al. . 2016. Responsive population dynamics and wide seeding into the duodenal lamina propria of transglutaminase-2-specific plasma cells in celiac disease. Mucosal Immunol. 9:254–264. 10.1038/mi.2015.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellyard, J.I., Avery D.T., Phan T.G., Hare N.J., Hodgkin P.D., and Tangye S.G.. 2004. Antigen-selected, immunoglobulin-secreting cells persist in human spleen and bone marrow. Blood. 103:3805–3812. 10.1182/blood-2003-09-3109 [DOI] [PubMed] [Google Scholar]
- Eltahla, A.A., Rizzetto S., Pirozyan M.R., Betz-Stablein B.D., Venturi V., Kedzierska K., Lloyd A.R., Bull R.A., and Luciani F.. 2016. Linking the T cell receptor to the single cell transcriptome in antigen-specific human T cells. Immunol. Cell Biol. 94:604–611. 10.1038/icb.2016.16 [DOI] [PubMed] [Google Scholar]
- Fillatreau, S. 2015. Regulatory plasma cells. Curr. Opin. Pharmacol. 23:1–5. 10.1016/j.coph.2015.04.006 [DOI] [PubMed] [Google Scholar]
- Freeman, H.J. 2017. Dietary compliance in celiac disease. World J. Gastroenterol. 23:2635–2639. 10.3748/wjg.v23.i15.2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz, J.H., Rojas O.L., Simard N., McCarthy D.D., Hapfelmeier S., Rubino S., Robertson S.J., Larijani M., Gosselin J., Ivanov I.I., et al. . 2012. Acquisition of a multifunctional IgA+ plasma cell phenotype in the gut. Nature. 481:199–203. 10.1038/nature10698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guikema, J.E., Hovenga S., Vellenga E., Conradie J.J., Abdulahad W.H., Bekkema R., Smit J.W., Zhan F., Shaughnessy J. Jr., and Bos N.A.. 2003. CD27 is heterogeneously expressed in multiple myeloma: low CD27 expression in patients with high-risk disease. Br. J. Haematol. 121:36–43. 10.1046/j.1365-2141.2003.04260.x [DOI] [PubMed] [Google Scholar]
- Halstensen, T.S., Hvatum M., Scott H., Fausa O., and Brandtzaeg P.. 1992. Association of subepithelial deposition of activated complement and immunoglobulin G and M response to gluten in celiac disease. Gastroenterology. 102:751–759. 10.1016/0016-5085(92)90155-R [DOI] [PubMed] [Google Scholar]
- Horns, F., Vollmers C., Croote D., Mackey S.F., Swan G.E., Dekker C.L., Davis M.M., and Quake S.R.. 2016. Lineage tracing of human B cells reveals the in vivo landscape of human antibody class switching. eLife. 5:e16578 10.7554/eLife.16578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Høydahl, L.S., Richter L., Frick R., Snir O., Gunnarsen K.S., Landsverk O.J.B., Iversen R., Jeliazkov J.R., Gray J.J., Bergseng E., et al. . 2019. Plasma cells are the most abundant gluten peptide MHC-expressing cells in inflamed intestinal tissues from patients with celiac disease. Gastroenterology. 156:1428–1439.e10. 10.1053/j.gastro.2018.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iversen, R., and Sollid L.M.. 2020. Autoimmunity provoked by foreign antigens. Science. 368:132–133. 10.1126/science.aay3037 [DOI] [PubMed] [Google Scholar]
- Iversen, R., Mysling S., Hnida K., Jørgensen T.J., and Sollid L.M.. 2014. Activity-regulating structural changes and autoantibody epitopes in transglutaminase 2 assessed by hydrogen/deuterium exchange. Proc. Natl. Acad. Sci. USA. 111:17146–17151. 10.1073/pnas.1407457111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iversen, R., Snir O., Stensland M., Kroll J.E., Steinsbø Ø., Korponay-Szabó I.R., Lundin K.E.A., de Souza G.A., and Sollid L.M.. 2017. Strong clonal relatedness between serum and gut IgA despite different plasma cell origins. Cell Rep. 20:2357–2367. 10.1016/j.celrep.2017.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabri, B., and Abadie V.. 2015. IL-15 functions as a danger signal to regulate tissue-resident T cells and tissue destruction. Nat. Rev. Immunol. 15:771–783. 10.1038/nri3919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkman, P., Dernstedt A., Tadala L., Mittler E., Dannborg M., Sundling C., Maleki K., Tauriainen J., Tuiskunen-Bäck A., Wigren B.J., et al. . 2019. The generation of plasma cells and CD27-IgD- B cells during Hantavirus infection are associated with distinct pathological findings. bioRxiv. 723585 (Preprint posted August 2, 2019) 10.1101/723585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kett, K., Baklien K., Bakken A., Kral J.G., Fausa O., and Brandtzaeg P.. 1995. Intestinal B-cell isotype response in relation to local bacterial load: evidence for immunoglobulin A subclass adaptation. Gastroenterology. 109:819–825. 10.1016/0016-5085(95)90389-5 [DOI] [PubMed] [Google Scholar]
- Kitaura, K., Yamashita H., Ayabe H., Shini T., Matsutani T., and Suzuki R.. 2017. Different somatic hypermutation levels among antibody subclasses disclosed by a new next-generation sequencing-based antibody repertoire analysis. Front. Immunol. 8:389 10.3389/fimmu.2017.00389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziejczyk, A.A., and Lönnberg T.. 2018. Global and targeted approaches to single-cell transcriptome characterization. Brief. Funct. Genomics. 17:209–219. 10.1093/bfgp/elx025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, W.Y., Jash A., Yao C.H., D’Souza L., Wong R., Nunley R.M., Meares G.P., Patti G.J., and Bhattacharya D.. 2018. Metabolic and transcriptional modules independently diversify plasma cell lifespan and function. Cell Rep. 24:2479–2492.e6. 10.1016/j.celrep.2018.07.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsverk, O.J., Snir O., Casado R.B., Richter L., Mold J.E., Réu P., Horneland R., Paulsen V., Yaqub S., Aandahl E.M., et al. . 2017. Antibody-secreting plasma cells persist for decades in human intestine. J. Exp. Med. 214:309–317. 10.1084/jem.20161590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc, M.P., Giudicelli V., Duroux P., Jabado-Michaloud J., Folch G., Aouinti S., Carillon E., Duvergey H., Houles A., Paysan-Lafosse T., et al. . 2015. IMGT®, the international ImMunoGeneTics information system® 25 years on. Nucleic Acids Res. 43(D1):D413–D422. 10.1093/nar/gku1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeman, I., and Stubbington M.J.T.. 2019. Antigen receptor sequence reconstruction and clonality inference from scRNA-seq data. In Computational methods for single-cell data analysis. Yuan G.C., editor. Humana Press, New York: 223–249. 10.1007/978-1-4939-9057-3_15 [DOI] [PubMed] [Google Scholar]
- Lindeman, I., Emerton G., Mamanova L., Snir O., Polanski K., Qiao S.W., Sollid L.M., Teichmann S.A., and Stubbington M.J.T.. 2018. BraCeR: B-cell-receptor reconstruction and clonality inference from single-cell RNA-seq. Nat. Methods. 15:563–565. 10.1038/s41592-018-0082-3 [DOI] [PubMed] [Google Scholar]
- Lindfors, K., Kaukinen K., and Mäki M.. 2009. A role for anti-transglutaminase 2 autoantibodies in the pathogenesis of coeliac disease? Amino Acids. 36:685–691. 10.1007/s00726-008-0127-5 [DOI] [PubMed] [Google Scholar]
- Ludvigsson, J.F., Bai J.C., Biagi F., Card T.R., Ciacci C., Ciclitira P.J., Green P.H., Hadjivassiliou M., Holdoway A., van Heel D.A., et al. British Society of Gastroenterology . 2014. Diagnosis and management of adult coeliac disease: guidelines from the British Society of Gastroenterology. Gut. 63:1210–1228. 10.1136/gutjnl-2013-306578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17:10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- McCarthy, D.J., Campbell K.R., Lun A.T.L., and Wills Q.F.. 2017. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics. 33:1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes, L., Healy J., and Melville J.. 2018. Umap: Uniform manifold approximation and projection for dimension reduction. arXiv. 1802.03426. (Preprint posted December 6, 2018)
- Mesin, L., Di Niro R., Thompson K.M., Lundin K.E., and Sollid L.M.. 2011. Long-lived plasma cells from human small intestine biopsies secrete immunoglobulins for many weeks in vitro. J. Immunol. 187:2867–2874. 10.4049/jimmunol.1003181 [DOI] [PubMed] [Google Scholar]
- Mishra, A., Sullivan L., and Caligiuri M.A.. 2014. Molecular pathways: interleukin-15 signaling in health and in cancer. Clin. Cancer Res. 20:2044–2050. 10.1158/1078-0432.CCR-12-3603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molberg, O., Mcadam S.N., Körner R., Quarsten H., Kristiansen C., Madsen L., Fugger L., Scott H., Norén O., Roepstorff P., et al. . 1998. Tissue transglutaminase selectively modifies gliadin peptides that are recognized by gut-derived T cells in celiac disease. Nat. Med. 4:713–717. 10.1038/nm0698-713 [DOI] [PubMed] [Google Scholar]
- Nishimura, H., Yajima T., Naiki Y., Tsunobuchi H., Umemura M., Itano K., Matsuguchi T., Suzuki M., Ohashi P.S., and Yoshikai Y.. 2000. Differential roles of interleukin 15 mRNA isoforms generated by alternative splicing in immune responses in vivo. J. Exp. Med. 191:157–170. 10.1084/jem.191.1.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhuber, G., Granditsch G., and Vogelsang H.. 1999. The histopathology of coeliac disease: time for a standardized report scheme for pathologists. Eur. J. Gastroenterol. Hepatol. 11:1185–1194. 10.1097/00042737-199910000-00019 [DOI] [PubMed] [Google Scholar]
- Patro, R., Duggal G., Love M.I., Irizarry R.A., and Kingsford C.. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 14:417–419. 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa, F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., and Dubourg V.. 2011. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 12:2825–2830. [Google Scholar]
- Pelletier, N., McHeyzer-Williams L.J., Wong K.A., Urich E., Fazilleau N., and McHeyzer-Williams M.G.. 2010. Plasma cells negatively regulate the follicular helper T cell program. Nat. Immunol. 11:1110–1118. 10.1038/ni.1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli, S., Björklund Å.K., Reinius B., Sagasser S., Winberg G., and Sandberg R.. 2014a. Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res. 24:2033–2040. 10.1101/gr.177881.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli, S., Faridani O.R., Björklund Å.K., Winberg G., Sagasser S., and Sandberg R.. 2014b. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9:171–181. 10.1038/nprot.2014.006 [DOI] [PubMed] [Google Scholar]
- Pinto, D., Montani E., Bolli M., Garavaglia G., Sallusto F., Lanzavecchia A., and Jarrossay D.. 2013. A functional BCR in human IgA and IgM plasma cells. Blood. 121:4110–4114. 10.1182/blood-2012-09-459289 [DOI] [PubMed] [Google Scholar]
- Rizzetto, S., Koppstein D.N.P., Samir J., Singh M., Reed J.H., Cai C.H., Lloyd A.R., Eltahla A.A., Goodnow C.C., and Luciani F.. 2018. B-cell receptor reconstruction from single-cell RNA-seq with VDJPuzzle. Bioinformatics. 34:2846–2847. 10.1093/bioinformatics/bty203 [DOI] [PubMed] [Google Scholar]
- Roy, B., Neumann R.S., Snir O., Iversen R., Sandve G.K., Lundin K.E.A., and Sollid L.M.. 2017. High-throughput single-cell analysis of B cell receptor usage among autoantigen-specific plasma cells in celiac disease. J. Immunol. 199:782–791. 10.4049/jimmunol.1700169 [DOI] [PubMed] [Google Scholar]
- Smith, K., Garman L., Wrammert J., Zheng N.Y., Capra J.D., Ahmed R., and Wilson P.C.. 2009. Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat. Protoc. 4:372–384. 10.1038/nprot.2009.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snir, O., Mesin L., Gidoni M., Lundin K.E., Yaari G., and Sollid L.M.. 2015. Analysis of celiac disease autoreactive gut plasma cells and their corresponding memory compartment in peripheral blood using high-throughput sequencing. J. Immunol. 194:5703–5712. 10.4049/jimmunol.1402611 [DOI] [PubMed] [Google Scholar]
- Snir, O., Chen X., Gidoni M., du Pré M.F., Zhao Y., Steinsbø Ø., Lundin K.E., Yaari G., and Sollid L.M.. 2017. Stereotyped antibody responses target posttranslationally modified gluten in celiac disease. JCI Insight. 2:e93961 10.1172/jci.insight.93961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snir, O., Kanduri C., Lundin K.E.A., Sandve G.K., and Sollid L.M.. 2019. Transcriptional profiling of human intestinal plasma cells reveals effector functions beyond antibody production. United European Gastroenterol. J. 7:1399–1407. 10.1177/2050640619862461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson, C., Love M.I., and Robinson M.D.. 2015. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000 Res. 4:1521 10.12688/f1000research.7563.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamnaes, J., Cardoso I., Iversen R., and Sollid L.M.. 2016. Transglutaminase 2 strongly binds to an extracellular matrix component other than fibronectin via its second C-terminal beta-barrel domain. FEBS J. 283:3994–4010. 10.1111/febs.13907 [DOI] [PubMed] [Google Scholar]
- Steinsbø, Ø., Henry Dunand C.J., Huang M., Mesin L., Salgado-Ferrer M., Lundin K.E.A., Jahnsen J., Wilson P.C., and Sollid L.M.. 2014. Restricted VH/VL usage and limited mutations in gluten-specific IgA of coeliac disease lesion plasma cells. Nat. Commun. 5:4041 10.1038/ncomms5041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbington, M.J.T., Lönnberg T., Proserpio V., Clare S., Speak A.O., Dougan G., and Teichmann S.A.. 2016. T cell fate and clonality inference from single-cell transcriptomes. Nat. Methods. 13:329–332. 10.1038/nmeth.3800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbington, M.J.T., Rozenblatt-Rosen O., Regev A., and Teichmann S.A.. 2017. Single-cell transcriptomics to explore the immune system in health and disease. Science. 358:58–63. 10.1126/science.aan6828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugai, E., Nachman F., Váquez H., González A., Andrenacci P., Czech A., Niveloni S., Mazure R., Smecuol E., Cabanne A., et al. . 2010. Dynamics of celiac disease-specific serology after initiation of a gluten-free diet and use in the assessment of compliance with treatment. Dig. Liver Dis. 42:352–358. 10.1016/j.dld.2009.07.011 [DOI] [PubMed] [Google Scholar]
- Tagaya, Y., Kurys G., Thies T.A., Losi J.M., Azimi N., Hanover J.A., Bamford R.N., and Waldmann T.A.. 1997. Generation of secretable and nonsecretable interleukin 15 isoforms through alternate usage of signal peptides. Proc. Natl. Acad. Sci. USA. 94:14444–14449. 10.1073/pnas.94.26.14444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay, A.A., Kauffman R.C., Wolabaugh A.N., Cho A., Patel N.B., Reiss S.M., Havenar-Daughton C., Dawoud R.A., Tharp G.K., Sanz I., et al. . 2018. BALDR: a computational pipeline for paired heavy and light chain immunoglobulin reconstruction in single-cell RNA-seq data. Genome Med. 10:20 10.1186/s13073-018-0528-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, F.A., Angerer P., and Theis F.J.. 2018. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19:15 10.1186/s13059-017-1382-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaari, G., and Kleinstein S.H.. 2015. Practical guidelines for B-cell receptor repertoire sequencing analysis. Genome Med. 7:121 10.1186/s13073-015-0243-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
provides clinical data for patients included in the study.
contains data from longevity analysis by flow cytometry for all patients.
provides quality-control threshold values and cell numbers for scRNA-seq data.