Abstract
Background
Glutathione peroxidase 1 (GPX1) is an essential component of the intracellular antioxidant enzyme system, but little is known about the role of GPX1 in the progression of malignancy in gliomas. Using public datasets, this study investigated the prognostic role of GPX1 and immune infiltrates in glioma.
Material/Methods
We investigated GPX1 expression levels in different cancers using the ONCOMINE and Tumor Immune Estimation Resource (TIMER) datasets. We also explored the prognostic landscape of GPX1 in gliomas based on The Cancer Genome Atlas (TCGA) and Chinese Glioma Genome Atlas (CGGA) datasets. Some significant pathways were identified by function enrichment analysis. We then explored the association between GPX1 expression and levels of tumor-infiltrating immune cells based on TIMER and Gene Expression Profiling Interactive Analysis (GEPIA) datasets.
Results
Expression of GPX1 in brain and central nervous system cancers is at a much high level than in normal tissues, and it is higher in glioblastoma (GBM) than in lower-grade glioma (LGG). We found GPX1 expression to be positively correlated with the malignant clinicopathologic characteristics of gliomas. Univariate analysis and multivariate analysis revealed that overexpression of GPX1 was correlated with a worse prognosis in patients, and a nomogram indicated that GPX1 expression can predict clinical prognosis of glioma. Function enrichment analysis showed that some important pathways are related to glioma malignancy. Expression of GPX1 was positively associated with infiltrating levels of 6 types of immune cells and most of their gene markers in GBM and LGG.
Conclusions
These results indicate that GPX1 is an independent prognostic factor and a novel biomarker for predicting the progression of malignancy in gliomas, which is associated with immune infiltration.
MeSH Keywords: Glioma; Glutathione Peroxidase; Immunity, Active; Nutrition Assessment
Background
Glioma is a deadly tumor in the central nervous system [1], while glioblastoma (GBM) is the most malignant type of gliomas [2]. Surgical resection, adjuvant chemotherapy, postoperative radiotherapy, and immunotherapy form the standard treatment for gliomas [3–5]. Despite advances in techniques and equipment in surgery for gliomas, the overall survival (OS) rate of patients with glioma is still poor [6]. Challenges include therapeutic resistance and tumor recurrence, leading to treatment failure and poor prognosis [7]. Therefore, glioma remains one of the most challenging malignant tumors in the world, and there is an urgent need to gain insight into a novel valid biomarker for diagnosis and treatment of patients with glioma.
Glutathione peroxidase 1 (GPX1) is an important antioxidant enzyme, diffusely distributed in the cytoplasm and mitochondria; its main function is the catalytic reduction of hydrogen peroxide to produce water [8,9]. Some studies have indicated that GPX1 plays a significant role in anticancer effects in some tumors. For example, GPX1 overexpression was found to be critically related to malignant clinicopathological features (high grade, nodal metastasis, perineural invasion, depth of tumor invasion, and advanced overall stage) and predicted adverse clinical outcomes of patients in oral squamous cell carcinoma [10]. Similarly, GPX1 overexpression was also found to promote tumor growth and metastasis in a skin cancer mouse model [11]. However, there is evidence indicating that high expression of GPX1 could inhibit pancreatic cancer cell proliferation [12]. Furthermore, in prostate cancer models, tumor cells could enhance radiation-induced micronucleus formation by knocking down GPX1 expression [13]. GPX1 clearly has different functions in different tumors and is significant in the progression of tumor malignance. Some recent studies have shown that glutathione expression levels are correlated with glioma redox regulation [14,15], and some glutathione metabolic pathway can influence the malignancy of gliomas [16–18]. However, there is little research on the biological function of GPX1 and its role in the progression of malignancy in gliomas.
Recent immune infiltration in different tumors can be inferred by using statistical data from the Tumor Immune Estimation Resource (TIMER) dataset (https://cistrome.shinyapps.io/timer/) [19,20]. TIMER is a comprehensive dataset, which includes 10 897 samples from 32 types of tumor collected from The Cancer Genome Atlas (TCGA) dataset (https://portal.gdc.cancer.gov/). It provides a user-friendly web interface for dynamic analysis and visualization of the correlation of immune infiltrates and a wide spectrum of factors. This dataset can be used to analyze the immune infiltration in glioma [21].
Therefore, we aimed to investigate the prognostic role of GPX1 and immune infiltrates in glioma, using public datasets, including ONCOMINE [22] (https://www.oncomine.org/resource/login.html), TIMER, TCGA, Chinese Glioma Genome Atlas (CGGA; http://www.cgga.org.cn/), and Gene Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn/index.html). Furthermore, our results suggested that GPX1 could be an independent prognostic factor in gliomas and have a significant role in glioma malignance, providing a potential mechanism between GPX1 and tumor-immune system interactions. As such, it could be a novel valid biomarker for the treatment of gliomas.
Material and Methods
GPX1 expression based on ONCOMINE and TIMER datasets
GPX1 expression levels in different cancers were analyzed using the ONCOMINE and TIMER datasets. Statistical significance was assumed for P<0.001.
Correlations between GPX1 expression and clinical outcomes and clinicopathologic characteristics in the TCGA and CGGA datasets
The TCGA dataset contains RNA-sequencing and clinicopathological data of 628 patients with glioma, while the CGGA dataset includes data of 298 patients with glioma. The data from the TCGA and CGGA datasets were used to research the prognostic role of GPX1 in gliomas. We attempted to identify correlation between GPX1 expression and clinicopathologic characteristics by t test. Relationships between GPX1 and clinical outcomes were determined via univariate and multivariate Cox regression analyses. The nomograms were formulated with R package “rms” and “foreign.” We evaluated the performance of nomograms via concordance index (C-index) and compared nomogram-predicted estimates with Kaplan-Meier estimates of survival probability. The receiver operating characteristic (ROC) curve was implemented to assess the prediction efficiency of nomograms.
Identification of significant pathways by function enrichment analysis
Based on the TCGA dataset, differentially expressed genes (DEGs) were screened by the R package “limma.” The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were performed based on DEGs by the R packages “clusterProfiler,” “enrichplot,” and “ggplot2.” We used R package “GOplot” and “digest” to perform cluster analysis. Gene set enrichment analysis (GSEA) was further conducted to validate the functions of these DEGs [23]. The GPX1 expression level was identified as a phenotype label, and the nominal value of P and the normalized enrichment score were used to sort the pathways.
GPX1 expression associated with immune cells using TIMER and GEPIA datasets
We used the TIMER dataset to infer the immune infiltration in GBM and LGG. Then, we used GEPIA [24] to further confirm critically correlated marks in the TIMER dataset. GEPIA is a new online dataset, which contains 9736 tumor samples and 8587 normal samples from the Genotype-Tissue Expression and TCGA datasets. We investigated the association between GPX1 expression levels and the infiltration levels of 6 types of immune cell (B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells) in GBM and LGG samples using the TIMER dataset. Similarly, we also looked for the correlations between GPX1 expression and tumor purity [25]. Tumor purity can affect the results of tumor immune infiltration level by genomic analysis [26]. Furthermore, the associations between GPX1 and some immune gene markers were also explored to find the latent related subtype of immune cells. We selected gene markers of several immune cells from R&D Systems. The levels of GPX1 expression were plotted on the x-axis, and expression levels of gene markers were plotted on the y-axis. Scatterplots were drawn to determine the relationships between GPX1 expression and marker levels of immune cells. The associations between GPX1 and immune cell markers were also analyzed using the GEPIA dataset, and we used the Spearman method to determine the correlation coefficient.
Statistical analysis
Correlations between expression of GPX1 and clinicopathologic characteristics were analyzed using logistic regression and the Wilcoxon signed-rank test. The Kaplan-Meier method was used to find the correlation between clinicopathologic characteristics and patients’ OS based on the TCGA and CGGA datasets. Furthermore, we compared the influence of GPX1 expression in patients’ OS with other clinical characteristics by univariate and multivariate Cox regression analysis. We used the Spearman method to determine the correlation between GPX1 and immune gene mark. Statistical analysis was performed with SPSS software 26.0 (SPSS Inc.), R software v3.6.3 (http://www.r-project.org/) [27], and Prism 8 (GraphPad Software, Inc). Data were considered significant at P<0.05.
Results
GPX1 Expression Levels in Human Pan-Cancers
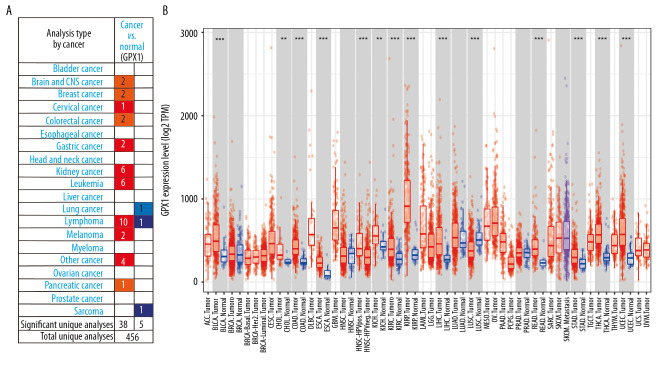
We used the ONCOMINE dataset to determine the variation in GPX1 expression between tumors and normal tissues in multiple human tumor types. This investigation indicated that GPX1 expression in brain and central nervous system cancers was increased compared with GPX1 expression in normal tissues in 2 datasets and did not decrease in any dataset (Figure 1A). Similarly, the expression level of GPX1 was higher in bladder, breast, cervical, colorectal, gastric, kidney, and pancreatic cancers, as well as leukemia, lymphoma tumors, and melanoma. However, the expression level of GPX1 was lower in lung cancer and sarcoma than in normal tissues in some datasets.
Figure 1.
Expression of glutathione peroxidase 1 (GPX1) in multiple human tumor types. Different expression of GPX1 in the dataset of multiple tumors in ONCOMINE (A). Expression of GPX1 in 32 tumor types in Tumor Immune Estimation Resource (TIMER) dataset (B). * P<0.05, ** P<0.01, *** P<0.001.
Furthermore, we checked the expression level of GPX1 using data from the TIMER dataset to estimate differential GPX1 expression levels in multiple tumor types. According to the results of expression levels of GPX1 in 32 types of tumors (Figure 1B), GPX1 was more highly expressed in bladder urothelial carcinoma, cholangiocarcinoma, colon adenocarcinoma, esophageal cancer, kidney chromophobe, liver hepatocellular carcinoma, rectum adenocarcinoma, stomach adenocarcinoma, thyroid carcinoma, and uterine corpus endometrial carcinoma than in normal tissues in the TCGA dataset. Furthermore, based on the TCGA and CGGA datasets, GPX1 was more highly expressed in GBM than in LGG (Figure 2A, 2B).
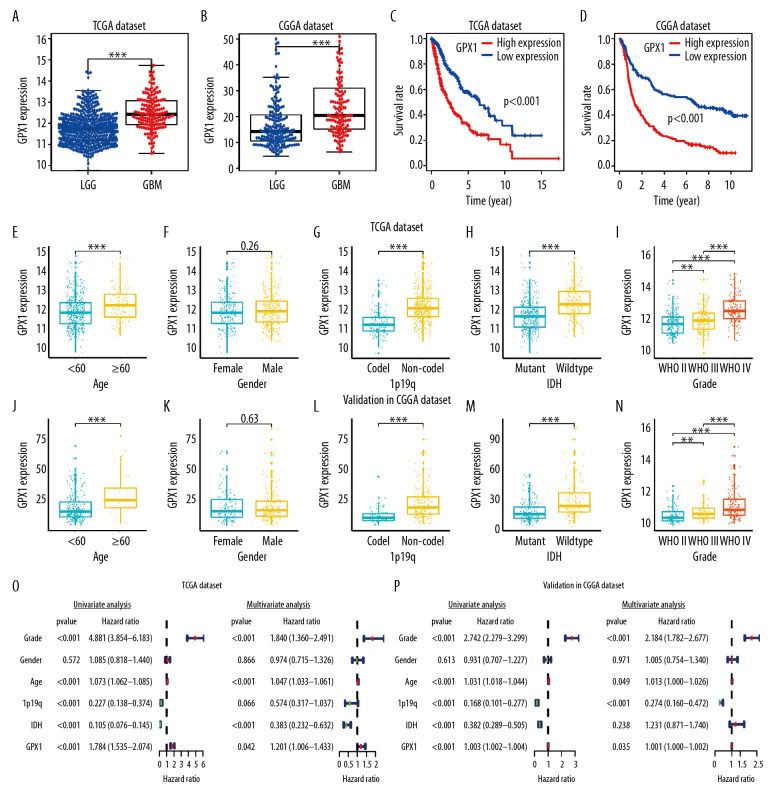
Figure 2.
The correlations between glutathione peroxidase 1 (GPX1) expression, clinical characteristics, and overall survival (OS) based on The Cancer Genome Atlas (TCGA) and Chinese Glioma Genome Atlas (CGGA) datasets. GPX1 expression was higher in glioblastoma than in lower-grade glioma based on the TCGA (A) and CGGA (B) dataset. High expression of GPX1 correlated with poor OS based on the TCGA (C) and CGGA (D) datasets. The associations between GPX1 expression and clinical characteristics: age (E), sex (F), 1p/19q codel status (G), isocitrate dehydrogenase (IDH) status (H), and grade (I) in the TCGA dataset; age (J), sex (K), 1p/19q codel status (L), IDH status (M), and grade (N) in the CGGA dataset. Univariate and multiple Cox regression analysis of clinicopathological features (including GPX1 expression) and OS in the TCGA (O) and CGGA (P) datasets.
Potential prognostic value of GPX1 in gliomas
Kaplan-Meier analysis was used to explore the association between GPX1 expression and OS of patients with gliomas, using the TCGA and CGGA dataset. These results indicated that high expression levels of GPX1 correlated with a poor OS of patients with glioma (Figure 2C, 2D). Furthermore, we investigated the associations between GPX1 expression and some clinical characteristics, such as grade, age, sex, isocitrate dehydrogenase (IDH) status, and 1p/19q codel status. GPX1 expression was sharply higher in high-grade, 1p/19q non-codel status, and IDH-wild-type status gliomas and in older patients; there was no statistically significant sex-based differences based on the TCGA (Figure 2E–2I) and CGGA (Figure 2J–2N) datasets. Our findings suggested that high GPX1 expression is correlated with the malignant clinical character of gliomas.
To establish whether GPX1 expression represented an independent prognostic index, we performed univariate and multivariate Cox regression analyses in both the TCGA and CGGA datasets. Univariate Cox regression analysis based on the TCGA dataset showed that the WHO grade, age, IDH status, 1p/19q status, and GPX1 expression were strongly associated with OS of patients. Multivariate Cox regression analysis revealed that WHO grade (P<0.001), age (P<0.001), IDH (P<0.001), and GPX1 expression (P<0.05) remained significantly correlated with OS (Figure 2O). Similar conclusions were reached from multivariate Cox regression analysis based on the CGGA dataset (Figure 2P), which showed significant correlations of WHO grade (P<0.001), 1p19q status (P<0.001), and GPX1 expression (P<0.05) with OS. These results suggest that GPX1 expression has a strong prognostic value in gliomas.
Establishment of a nomogram for predicting clinical prognosis
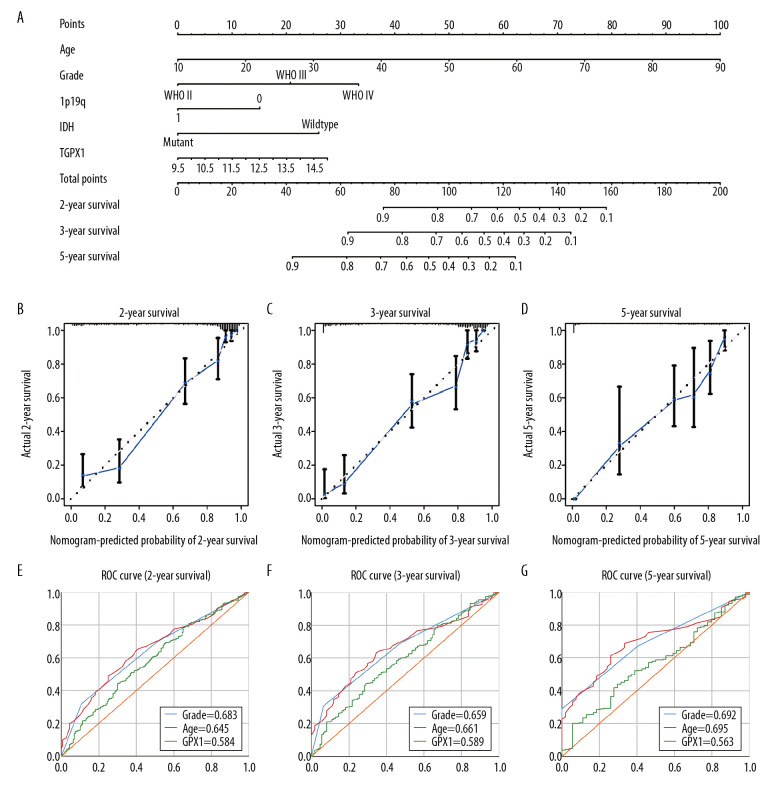
We then developed a quantitative model for predicting clinical prognosis. The nomogram integrated age, grade, 1p19q status, IDH status, and GPX1 expression for gliomas in the TCGA dataset (Figure 3A), and the C-index was 0.861. The calibration curve for survival probability at 2, 3, and 5 years revealed an optimal agreement between the nomogram prediction and the actual observed outcomes (Figure 3B–3D). The area under the ROC curve for OS was 0.584 at 2 years, 0.589 at 3 years, and 0.563 at 5 years, showing a reliable predictive ability in the TCGA dataset (Figure 3E–3G).
Figure 3.
Construction and assessment of nomogram for predicting patients’ overall survival. Nomogram based on the clinical characteristics and glutathione peroxidase 1 expression for predicting patient survival (A). The calibration curve for predicting patient survival at 2 years (B), 3 years (C), and 5 years (D). The predictive efficiency of risk score, World Health Organization grade, and age showed by the receiver operating characteristic curves based on 2-, 3-, and 5-year survival rates (E–G).
Molecular mechanisms of GPX1 contributing to malignant progression in glioma
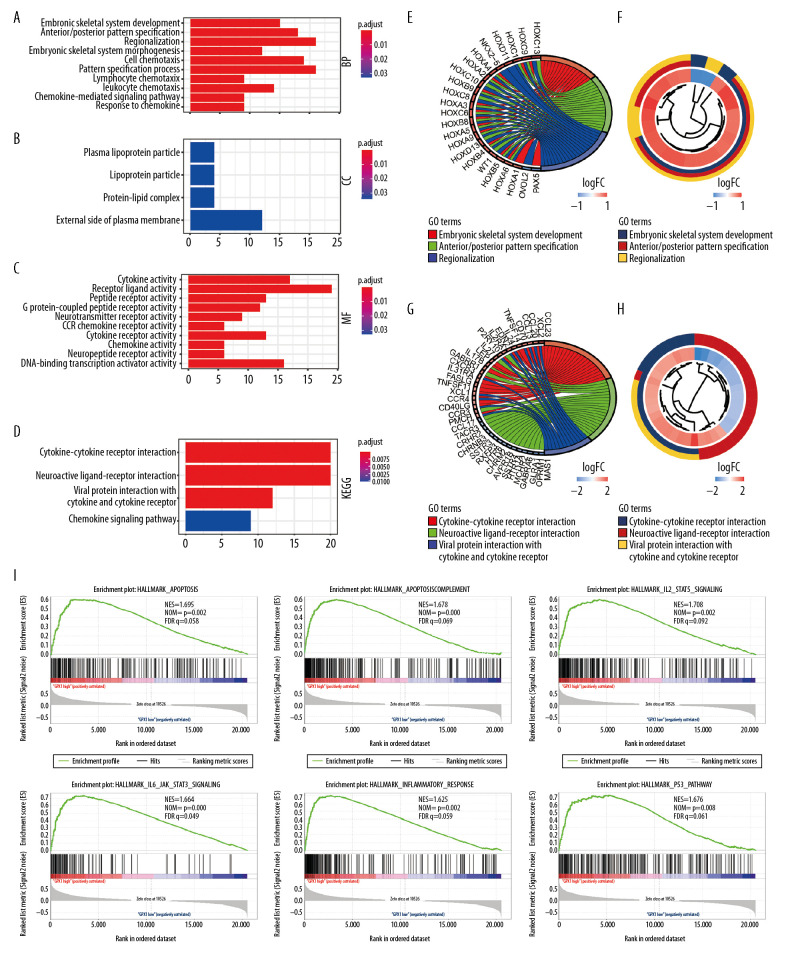
A total of 243 DEGs were identified in glioma data through DEG analysis. GO analysis demonstrated that the biological processes of these DEGs were significantly enriched in embryonic skeletal system development, cell chemotaxis, lymphocyte chemotaxis, and chemokine-mediate signaling pathways (Figure 4A). Cell components were mainly concentrated in the plasma lipoprotein portion and protein-lipid complexes (Figure 4B). Molecular functions were obviously enriched in cytokine activity, receptor ligand activity, chemokine activity, and CCR chemokine receptor binding (Figure 4C). KEGG pathway analysis revealed that DEGs were obviously enriched in cytokine-cytokine receptor interaction, neuroactive ligand-receptor interaction, and chemokine signaling pathways (Figure 4D). Meanwhile, we explored significant enriched DEGs and conducted cluster analyses in the top GO and KEGG pathways (Figure 4E–4H). In addition, we identified hallmarks of malignant tumors with the aid of GSEA. Our results suggest that apoptosis (NES=1.695, normalized P=0.002), complement (NES=1.678, normalized P<0.001), interleukin-2 (IL2) STAT signaling (NES=1.708, normalized P=0.002), and IL JAK/STAT signaling pathways (NES=1.664, normalized P<0.001) are differentially enriched in high-expression GPX1 phenotypes (Figure 4I).
Figure 4.
Functional annotations and predicting signaling pathways. Biological process (A), cell component (B), and molecular function (C) enrichment. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway (D). Significant different expression genes enriched in The Gene Ontology (GO) (E) and KEGG (G) pathway; GO (F) and KEGG (H) pathway cluster analysis. Malignant hallmarks enriched in high-expression glutathione peroxidase 1 phenotypes determined using gene set enrichment analysis (I).
GPX1 expression is associated with 6 types of infiltrating immune cell in gliomas
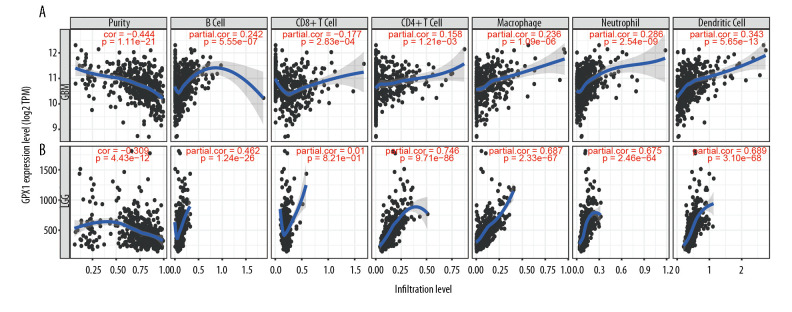
Tumor-infiltrating lymphocytes can affect a patient’s OS [28], and our results indicate that GPX1 expression plays a significant role in the malignancy of gliomas. Hence, we explored the association between GPX1 expression and immune infiltration levels of 6 types of immune cells, namely, B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells in GBM and LGG using the TIMER dataset. These analyses revealed that high expression levels of GPX1 could critically increase immune infiltrating levels of B cells (R=0.242, P=5.55e-07), CD4+ cells (R=0.158, P=1.21e-03), macrophages (R=0.236, P=1.09e-06), neutrophils (R=0.286, P=2.54e-09), and dendritic cells (R=0.343, P=5.65–13), while negative associations were found for levels of CD8+ T cells (R=−0.1777, P=2.83e-04) in GBM (Figure 5A). In LGG, the relationships between GPX1 expression and infiltration levels of the 6 types of immune cells were little different (Figure 5B), and infiltration levels of CD8+ T cells were not significantly correlated with GPX1 expression levels (P=0.821). Moreover, there were negative relationships between GPX1 expression levels and tumor purity in GBM (R=−0.444, P=1.11e-21) and LGG (R=−0.309, P=4.43e-12) (Figure 5A, 5B). The results indicate that GPX1 has a significant role in immune infiltration in GBM and LGG; in particular, there were significantly positive correlations with dendritic cells, neutrophils, and macrophages.
Figure 5.
Associations between the expression of glutathione peroxidase 1 (GPX1) and immune infiltration in glioblastoma (GBM) and lower-grade glioma (LGG). Expression of GPX1 is significantly negatively associated with tumor purity and is significantly positively associated with infiltrations with the level of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells in GBM (A). Expression of GPX1 is significantly negatively associated with tumor purity, and it is significantly positively associated with infiltrations with the level of B cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells. It has no significant associations with infiltrations with level of CD8+ T cells in LGG (B).
Associations between expression of GPX1 and immune gene markers
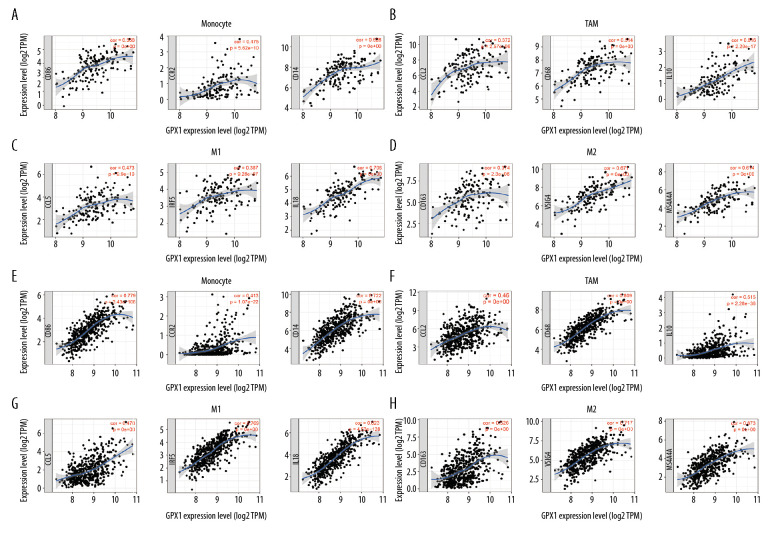
To comprehensively investigate the correlations of GPX1 and infiltrating immune cells in GBM and LGG, we explored the relationships between expression of GPX1 and levels of immune gene markers, using the TIMER and GEPIA datasets. These markers were used to characterize specific immune cells, including B cells, T cells (general), CD8+ T cells, monocytes, tumor-associated macrophages (TAMs), M1 and M2 macrophages, neutrophils, natural killer cells, dendritic cells, helper T cells, and regulatory T cells in GBM and LGG. The findings indicated positive associations between GPX1 expression and most immune markers of various immune cells in GBM and LGG after adjustment by purity (Table 1). Furthermore, we found that high GPX1 expression levels were related to high infiltration levels of most markers of monocytes, TAMs, M1 macrophages, and M2 macrophages in GBM and LGG, such as CD86, CCR2, and CD14 of monocytes; CCL2, CD68, and IL10 of TAMs; CCL5, IRF5, and IL18 of M1 macrophages; and CD163, VSIG4, and MS4A4A of M2 macrophages (Figure 6A–6H, Table 1). Interestingly, our results further indicated that GPX1 expression level was critically correlated with these markers, as identified using the GEPIA dataset, including GBM and LGG (Table 2). These findings further reveal that GPX1 can regulate macrophage polarization in gliomas.
Table 1.
Correlation analysis between GPX1 and gene markers of immune cells in TIMER.
| Description | Gene markers | GBM | LGG | ||||||
|---|---|---|---|---|---|---|---|---|---|
| None | Purity | None | Purity | ||||||
| Cor | P | Cor | P | Cor | P | Cor | P | ||
| CD8+ T cell | CD8A | 0.225 | * | 0.151 | 0.079 | 0.126 | * | 0.03 | 0.514 |
| CD8B | 0.388 | *** | 0.307 | ** | 0.158 | ** | 0.068 | 0.137 | |
| T cell (general) | CD3D | 0.563 | *** | 0.461 | *** | 0.453 | *** | 0.406 | *** |
| CD3E | 0.408 | *** | 0.287 | ** | 0.459 | *** | 0.436 | *** | |
| CD2 | 0.501 | *** | 0.38 | *** | 0.434 | *** | 0.41 | *** | |
| B cell | CD19 | 0.187 | * | 0.17 | 0.047 | 0.416 | *** | 0.376 | *** |
| CD79A | 0.164 | * | 0.16 | 0.062 | 0.374 | *** | 0.397 | *** | |
| Monocyte | CD86 | 0.475 | *** | 0.392 | *** | 0.779 | *** | 0.754 | *** |
| CCR2 | 0.377 | *** | 0.313 | ** | 0.413 | *** | 0.394 | *** | |
| CD14 | 0.528 | *** | 0.357 | *** | 0.722 | *** | 0.708 | *** | |
| TAM | CCL2 | 0.372 | *** | 0.183 | 0.032 | 0.46 | *** | 0.423 | *** |
| CD68 | 0.514 | *** | 0.339 | *** | 0.809 | *** | 0.794 | *** | |
| IL10 | 0.616 | *** | 0.438 | *** | 0.515 | *** | 0.469 | *** | |
| M1 Macrophage | CCL5 | 0.473 | *** | 0.355 | *** | 0.476 | *** | 0.453 | *** |
| IRF5 | 0.387 | *** | 0.175 | 0.040 | 0.769 | *** | 0.745 | *** | |
| IL18 | 0.706 | *** | 0.573 | *** | 0.823 | *** | 0.806 | *** | |
| M2 Macrophage | CD163 | 0.374 | *** | 0.204 | 0.017 | 0.526 | *** | 0.527 | *** |
| VSIG4 | 0.671 | *** | 0.544 | *** | 0.717 | *** | 0.694 | *** | |
| MS4A4A | 0.614 | *** | 0.472 | *** | 0.673 | *** | 0.67 | *** | |
| Neutrophils | CD66b (CEACM8) | −0.23 | * | −0.296 | ** | −0.004 | 0.92 | −0.012 | 0.791 |
| CD11b (ITGAM) | 0.304 | ** | 0.063 | 0.464 | 0.687 | *** | 0.645 | *** | |
| CCR7 | 0.305 | ** | 0.214 | 0.011 | 0.274 | *** | 0.26 | *** | |
| Natural killer cell | NCR3 | 0.308 | * | 0.238 | 0.005 | 0.198 | *** | 0.184 | ** |
| CD127 | 0.291 | * | 0.132 | 0.124 | 0.234 | *** | 0.192 | *** | |
| KIR2DL1 | 0.089 | 0.273 | 0.043 | 0.618 | −0.021 | 0.633 | 0.004 | 0.923 | |
| Dendritic cell | HLA-DPB1 | 0.521 | *** | 0.386 | *** | 0.677 | *** | 0.653 | *** |
| HLA-DRA | 0.64 | *** | 0.527 | *** | 0.666 | *** | 0.642 | *** | |
| HLA-DPA1 | 0.478 | *** | 0.362 | *** | 0.617 | *** | 0.591 | *** | |
| BDCA-1(CD1C) | 0.361 | *** | 0.191 | 0.025 | 0.339 | *** | 0.328 | *** | |
| Th1 | CD4 | 0.436 | *** | 0.242 | * | 0.754 | *** | 0.727 | *** |
| CD103 | 0.34 | *** | 0.413 | *** | 0.147 | ** | 0.168 | ** | |
| TNF-α(TNF) | 0.266 | ** | 0.118 | 0.169 | 0.231 | *** | 0.181 | *** | |
| Th2 | CD14 | 0.528 | *** | 0.357 | *** | 0.722 | *** | 0.708 | *** |
| IL13 | −0.243 | 0.025 | −0.209 | 0.014 | 0.045 | 0.303 | 0.048 | 0.291 | |
| Tfh | BCL6 | −0.355 | *** | −0.405 | *** | 0.145 | ** | 0.176 | ** |
| IL21 | −0.014 | 0.868 | 0.021 | 0.807 | 0.095 | 0.031 | 0.073 | 0.113 | |
| Th17 | STAT3 | −0.361 | *** | −0.371 | *** | 0.328 | *** | 0.332 | *** |
| IL17A | −0.058 | 0.476 | −0.146 | 0.089 | −0.025 | 0.578 | −0.042 | 0.365 | |
| Treg | CD103 | 0.34 | *** | 0.413 | *** | 0.147 | ** | 0.168 | ** |
| CCR8 | 0.254 | * | 0.177 | 0.039 | 0.162 | ** | 0.166 | ** | |
| STAT5B | −0.606 | *** | −0.525 | *** | −0.185 | *** | −0.127 | * | |
| T cell exhaustion | PD-1(PDCD1) | 0.284 | ** | 0.216 | 0.011 | 0.538 | *** | 0.522 | *** |
| TIM-3 (HAVCR2) | 0.62 | *** | 0.479 | *** | 0.805 | *** | 0.788 | *** | |
| GZMB | 0.369 | *** | 0.237 | * | 0.182 | *** | 0.202 | *** | |
TIMER – Tumor Immune Estimation Resource; TAM – tumor-associated macrophage; Th – T helper cell; Tfh – follicular helper T cell; Treg – regulatory T cell; Cor – R value of Spearman’s correlation; None – correlation without adjustment; Purity – correlation adjusted by purity.
P<0.01;
P<0.001;
P<0.0001.
Figure 6.
Glutathione peroxidase 1 (GPX1) expression is associated with macrophage polarization in glioblastoma (GBM) and lower-grade glioma (LGG). Association between GPX1 and biomarkers of monocytes (A), tumor-associated macrophages (TAMs) (B), M1 macrophages (C), and M2 macrophages (D) in GBM. Association between GPX1 and biomarkers of monocytes (E), TAM (F), M1 macrophages (G), and M2 macrophages (H) in LGG.
Table 2.
Correlation analysis between GPX1 and gene markers of macrophages in GEPIA.
| Description | Gene markers | GBM | LGG | ||
|---|---|---|---|---|---|
| R | P | R | P | ||
| Monocyte | CD86 | 0.66 | *** | 0.65 | *** |
| CCR2 | 0.28 | ** | 0.38 | *** | |
| CD14 | 0.55 | *** | 0.57 | *** | |
| TAM | CCL2 | 0.35 | *** | 0.25 | *** |
| CD68 | 0.5 | *** | 0.14 | *** | |
| IL10 | 0.56 | *** | 0.37 | *** | |
| M1 Macrophage | CCL5 | 0.33 | *** | 0.47 | *** |
| IRF5 | 0.44 | *** | 0.72 | *** | |
| IL18 | 0.76 | *** | 0.75 | *** | |
| M2 Macrophage | CD163 | 0.45 | *** | 0.41 | *** |
| VSIG4 | 0.66 | *** | 0.58 | *** | |
| MS4A4A | 0.57 | *** | 0.5 | *** | |
GEPIA – Gene Expression Profiling Interactive Analysis; TAM – tumor-associated macrophage.
P<0.01;
P<0.001;
P<0.0001.
Our analyses indicated that significantly positive correlations existed between GPX1 expression and infiltration levels of dendritic cells in GBM and LGG; we also found that GPX1 expression was strongly associated with dendritic cells markers, such as HLA-DRA and HLA-DPA1 (Table 1). These results suggest that GPX1 expression may strongly correlate with infiltration by dendritic cells. Expression of GPX1 was also positively associated with Treg cell markers (CD103 and STAT5B) in GBM and LGG. Moreover, the significant correlation between GPX1 and several markers (CD4, CD103, CD14, BCL6, STAT3, TIM-3, and GZMB) of helper T cells and T-cell exhaustion were found in GBM and LGG (Table 1). The TIM-3 marker, which plays a significant role in regulating T-cell exhaustion [29], was positively associated with the expression level of GPX1, indicating that GPX1 might be an essential factor in mediating TIM-3 and T-cell exhaustion. In summary, these findings comprehensively indicate strong positive associations between GPX1 expression and the infiltration of immune cells in GBM and LGG.
Discussion
Reactive oxygen species (ROS), which include hydroxyl radicals, superoxide, and hydrogen peroxide, can be produced by enzymatic and mitochondrial sources in normal and tumor cells [30,31]. If they continue to accumulate in cytoplasm and mitochondria, they may cause oxidative damage to proteins, DNA, RNA, and organelles [32,33]. Normal cells can clear ROS through the antioxidant enzyme system, but ROS accumulates in tumor cells because of their more active metabolism and relative hypoxia-induced mitochondrial dysfunction [34]. Some studies have identified relationships between the accumulation of ROS and tumor transformation, tumor initiation, and chemotherapy tolerance in tumor cells. Nevertheless, the accumulation of ROS can be counteracted by intracellular antioxidant enzyme systems, which can reduce the damage of oxidative stress to intracellular DNA and proteins [35,36]. GPX1, an essential antioxidant enzyme, is an important component of the intracellular antioxidant enzyme system. Some studies have shown that GPX1 is related to tumor initiation and transformation in tumor cells. However, few studies have reported the expression of GPX1 and its role in the progression of malignancy in gliomas.
In this investigation, we used the ONCOMINE and TIMER datasets to determine the differences in GPX1 expression between tumors and normal tissues in multiple human tumor types. We found that expression of GPX1 was higher in bladder, breast, cervical, colorectal, gastric, kidney, and pancreatic cancer, as well as leukemia, lymphoma tumor, and melanoma, relative to normal tissues, according to the ONCOMINE dataset. Furthermore, GPX1 was highly expressed in bladder urothelial carcinoma, cholangiocarcinoma, colon adenocarcinoma, esophageal cancer, chromophobe renal cell carcinoma, hepatocellular carcinoma, rectal adenocarcinoma, stomach adenocarcinoma, and thyroid carcinoma, as well as in uterine corpus endometrial carcinoma. Moreover, GPX1 was more highly expressed in GBM than in LGG, as determined from the TCGA and CGGA datasets. The analysis indicated positive associations existed between the expression level of GPX1 and worse OS as a result of gliomas. We also explored the associations of GPX1 expression levels with different clinicopathological features in patients, using the TCGA and CGGA datasets. The GPX1 expression level was positively correlated with older patients, and high-grade, 1p19q non-codel, and IDH-wild-type status glioma; this correlation may imply a previously unidentified molecular mechanism of GPX1 in glioma malignancies. Furthermore, univariate analysis indicated a positive association between GPX1 expression and poor prognosis of patients with glioma, as found using the TCGA and CGGA datasets. In addition, some clinical characteristics also had a positive correlation with worse OS; these included patient age, glioma grade, and 1p/19q codel and IDH status. Furthermore, multivariate analysis revealed that high GPX1 expression levels remained independently correlated with poor OS. In addition, we constructed a nomogram for predicting prognosis of glioma. The C-index for survival prediction and the AUC for OS showed our nomogram was reliable in the TCGA dataset, which further indicated that GXP1 expression correlated with worse OS. These results indicate that GPX1 is an independent prognostic factor for the diagnosis of glioma and plays a significant role in the malignancy of gliomas.
In addition, we also found positive relationships between expression levels of GPX1 and infiltration levels of immune cells in GBM and LGG. These findings demonstrated that a high GPX1 expression phenotype is positively correlated with cell chemotaxis, lymphocyte chemotaxis, chemokine-mediate signaling pathway, apoptosis, complement, IL2 STAT signaling and IL JAK/STAT signaling pathways, using function enrichment analysis. We hypothesized that these signaling pathways have a critical role in malignancy progression of gliomas, and high GPX1 expression is correlated with these critical inflammatory signal pathways. Furthermore, these findings revealed that the expression level of GPX1 was moderately positively correlated with infiltration level of macrophages, neutrophils, and dendritic cells, while strongly positively associated with the infiltration level of B cells, CD8+ T cells, and CD4+ T cells in GBM and LGG.
Finding associations between GPX1 expression and immune gene markers can further reveal the regulation of GPX1 in tumor-related immune mechanisms in gliomas. Interestingly, we found that expression levels of GPX1 were strongly associated with markers of monocytes, TAMs, M1 macrophages, and M2 macrophages. Our findings indicate that GPX1 is involved in regulating the polarization of TAMs. Similarly, our results also reveal that GPX1 participates in activating dendritic cells and Treg cells. Increasing numbers of Treg cells and dendritic cells can promote tumor metastasis [37], which means that GPX1 may play an important role in the mediation of tumor metastasis by dendritic cells in gliomas. In addition, the high expression levels of GPX1 were also positively associated with T helper cells and their gene markers. These associations can reveal a potential molecular mechanism for the regulation of T cells in GBM and LGG by GPX1. All of these results show that GPX1 has a vital role in the recruitment and regulation of immune cells in the microenvironment of gliomas.
Some studies have found potential molecule mechanisms explaining the association between expression levels of GPX1 and immune infiltration. It has been reported that the NF-κB signaling pathway, as a nuclear factor that can activate B-cell signaling, can be regulated by GPX1 [38,39]. The NF-κB signaling pathway has a significant role in the modulation of a variety of genes that participate in activation of the immune and inflammatory responses, including pro-inflammatory cytokines, such as IL-1β, TNF-α, and IL-6, and enzymes that can produce secondary inflammatory mediators, such as cyclooxygenase-2 and inducible NO synthase [40,41]. Furthermore, some studies have demonstrated that most inducers of NF-κB rely on the production of intracellular ROS, suggesting that low intracellular ROS levels could inhibit the activation of the NF-κB-IκB-α complex and the degradation of IκB-α protein [42,43]. Moreover, IκB-α degradation can precede NF-κB activation, but can also be transactivated by NF-κB, promoting IκB-α to restore the inhibited state. The intracellular levels of ROS activate or inactivate some specific phosphatases to IκB-α to control the level of the b phosphor-isoform of this protein [44]. Hence, GPX1 can mediate low levels of ROS, promoting NF-κB activation by inhibiting IκB-α phosphorylation. Therefore, interactions between the GPX1 and NF-κB pathway might provide a novel molecular mechanism for GPX1 involvement in the activation of immune infiltration in gliomas.
Although we integrate information across multiple datasets, there are still some limitations in this work. First, a large proportion of the microarray and sequencing data are collected by analyzing tumor tissue information; therefore, there is some introduced systematic bias in the cell-level analysis of immune cell markers. Second, these datasets do not include data reflecting the posttranslational modification of GPX1, which can interfere with the molecular function. Third, our findings were inferred from transcriptome data alone, and the findings should be supported with immunohistochemistry, immunofluorescence, and flow cytometry studies. Fourth, although we reveal that expression of GPX1 is associated with the prognosis of patient and immune infiltration in gliomas, we cannot prove that GPX1 affects prognosis through immune infiltration.
Conclusions
These findings demonstrate that overexpression of GPX1 affects malignancy of gliomas and has a strong positive association with the poor OS of patients with glioma. Moreover, we also found that high GPX1 expression increases immune infiltration levels in GBM and LGG, which means that GPX1 is involved in the activation of immune infiltration in gliomas. These results suggest that GPX1 could be an independent prognostic factor and a novel potential biomarker for predicting the progression of malignancy in gliomas, associated with immune infiltration in gliomas, and could inform the design of new drugs and strategies for gliomas.
Footnotes
Source of support: This study was supported by the National Natural Science Foundation of China, Grant/Award number: 81660420 and 81760446, and the Jiangxi Province Department of Education Science and technology research project, Grant/Award number: GJJ190018
References
- 1.Lin L, Cai J, Jiang C. Recent advances in targeted therapy for glioma. Curr Med Chem. 2017;24(13):1365–81. doi: 10.2174/0929867323666161223150242. [DOI] [PubMed] [Google Scholar]
- 2.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang T, Mao Y, Ma W, et al. CGCG clinical practice guidelines for the management of adult diffuse gliomas. Cancer Lett. 2016;375(2):263–73. doi: 10.1016/j.canlet.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 4.Jhaveri J, Liu Y, Chowdhary M. Is less more? Comparing chemotherapy alone with chemotherapy and radiation for high-risk grade 2 glioma: An analysis of the National Cancer Data Base. Cancer. 2018;124(6):1169–78. doi: 10.1002/cncr.31158. [DOI] [PubMed] [Google Scholar]
- 5.Xu S, Tang L, Li X, et al. Immunotherapy for glioma: Current management and future application. Cancer Lett. 2020;476:1–12. doi: 10.1016/j.canlet.2020.02.002. [DOI] [PubMed] [Google Scholar]
- 6.Gehring K, Stuiver MM, Visser E, et al. A pilot randomized controlled trial of exercise to improve cognitive performance in patients with stable glioma: A proof of concept. Neuro Oncol. 2020;22(1):103–15. doi: 10.1093/neuonc/noz178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersen BM, Miranda C, Hatzoglou V, et al. Leptomeningeal metastases in glioma: The Memorial Sloan Kettering Cancer Center experience. Neurology. 2019;92(21):e2483–91. doi: 10.1212/WNL.0000000000007529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lubos E, Loscalzo J, Handy DE. Glutathione peroxidase-1 in health and disease: From molecular mechanisms to therapeutic opportunities. Antioxid Redox Signal. 2011;15(7):1957–97. doi: 10.1089/ars.2010.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ekoue DN, He C, Diamond AM, et al. Manganese superoxide dismutase and glutathione peroxidase-1 contribute to the rise and fall of mitochondrial reactive oxygen species which drive oncogenesis. Biochim Biophys Acta Bioenerg. 2017;1858(8):628–32. doi: 10.1016/j.bbabio.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee JR, Roh JL, Lee SM, et al. Overexpression of glutathione peroxidase 1 predicts poor prognosis in oral squamous cell carcinoma. J Cancer Res Clin Oncol. 2017;143(11):2257–65. doi: 10.1007/s00432-017-2466-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu YP, Lou YR, Yen P, et al. Enhanced skin carcinogenesis in transgenic mice with high expression of glutathione peroxidase or both glutathione peroxidase and superoxide dismutase. Cancer Res. 1997;57(8):1468–74. [PubMed] [Google Scholar]
- 12.Liu J, Hinkhouse MM, Sun W, et al. Redox regulation of pancreatic cancer cell growth: Role of glutathione peroxidase in the suppression of the malignant phenotype. Hum Gene Ther. 2004;15(3):239–50. doi: 10.1089/104303404322886093. [DOI] [PubMed] [Google Scholar]
- 13.Baliga MS, Diwadkar-Navsariwala V, Koh T, et al. Selenoprotein deficiency enhances radiation-induced micronuclei formation. Mol Nutr Food Res. 2008;52(11):1300–4. doi: 10.1002/mnfr.200800020. [DOI] [PubMed] [Google Scholar]
- 14.Tang X, Fu X, Liu Y, et al. Blockade of glutathione metabolism in IDH1-mutated glioma. Mol Cancer Ther. 2020;19(1):221–30. doi: 10.1158/1535-7163.MCT-19-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu D, Liu Y, Zhou Y, et al. Triptolide suppresses IDH1-mutated malignancy via Nrf2-driven glutathione metabolism. Proc Natl Acad Sci USA. 2020;117(18):9964–72. doi: 10.1073/pnas.1913633117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poore B, Yuan M, Arnold A, et al. Inhibition of mTORC1 in pediatric low-grade glioma depletes glutathione and therapeutically synergizes with carboplatin. Neuro Oncol. 2019;21(2):252–63. doi: 10.1093/neuonc/noy150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tjiong R, Stavrinou P, Rohn G, et al. Heterogeneity of human gliomas: Glutathione-s-transferase expression profile during disease progression and under systemic therapy. Anticancer Res. 2019;39(4):1795–805. doi: 10.21873/anticanres.13286. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y, Lu Y, Celiku O, et al. Targeting IDH1-mutated malignancies with NRF2 blockade. J Natl Cancer Inst. 2019;111(10):1033–41. doi: 10.1093/jnci/djy230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li T, Fan J, Wang B, et al. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–10. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li B, Severson E, Pignon JC, et al. Comprehensive analyses of tumor immunity: Implications for cancer immunotherapy. Genome Biol. 2016;17(1):174. doi: 10.1186/s13059-016-1028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Liu S, Zhao X, et al. Magnetic resonance imaging-based radiomic features for extrapolating infiltration levels of immune cells in lower-grade gliomas. Strahlenther Onkol. 2020 doi: 10.1007/s00066-020-01584-1. [Online ahead of print] [DOI] [PubMed] [Google Scholar]
- 22.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9(2):166–80. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang Z, Li C, Kang B, et al. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aran D, Sirota M, Butte AJ. Systematic pan-cancer analysis of tumour purity. Nat Commun. 2015;6:8971. doi: 10.1038/ncomms9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshihara K, Shahmoradgoli M, Martinez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ozuna A, Liberto D, Joyce RM, et al. baerhunter: An R package for the discovery and analysis of expressed non-coding regions in bacterial RNA-seq data. Bioinformatics. 2020;36(3):966–69. doi: 10.1093/bioinformatics/btz643. [DOI] [PubMed] [Google Scholar]
- 28.Azimi F, Scolyer RA, Rumcheva P, et al. Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. J Clin Oncol. 2012;30(21):2678–83. doi: 10.1200/JCO.2011.37.8539. [DOI] [PubMed] [Google Scholar]
- 29.Huang YH, Zhu C, Kondo Y, et al. CEACAM1 regulates TIM-3-mediated tolerance and exhaustion. Nature. 2015;517(7534):386–90. doi: 10.1038/nature13848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamauchi S, Mano S, Oikawa K, et al. Autophagy controls reactive oxygen species homeostasis in guard cells that is essential for stomatal opening. Proc Natl Acad Sci USA. 2019;116(38):19187–92. doi: 10.1073/pnas.1910886116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu X, Yan B, Li Y, et al. Graphene oxide-grafted magnetic nanorings mediated magnetothermodynamic therapy favoring reactive oxygen species-related immune response for enhanced antitumor efficacy. ACS Nano. 2020;14(2):1936–50. doi: 10.1021/acsnano.9b08320. [DOI] [PubMed] [Google Scholar]
- 32.Tafani M, Sansone L, Limana F, et al. The interplay of reactive oxygen species, hypoxia, inflammation, and sirtuins in cancer initiation and progression. Oxid Med Cell Longev. 2016;2016 doi: 10.1155/2016/3907147. 3907147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu H, Wang C, Sun J, et al. Self-assembled and self-monitored sorafenib/indocyanine green nanodrug with synergistic antitumor activity mediated by hyperthermia and reactive oxygen species-induced apoptosis. ACS Appl Mater Interfaces. 2019;11(47):43996–4006. doi: 10.1021/acsami.9b18086. [DOI] [PubMed] [Google Scholar]
- 34.Godet I, Shin YJ, Ju JA, et al. Fate-mapping post-hypoxic tumor cells reveals a ROS-resistant phenotype that promotes metastasis. Nat Commun. 2019;10(1):4862. doi: 10.1038/s41467-019-12412-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L, Chen X, Du Z, et al. Curcumin suppresses gastric tumor cell growth via ROS-mediated DNA polymerase gamma depletion disrupting cellular bioenergetics. J Exp Clin Cancer Res. 2017;36(1):47. doi: 10.1186/s13046-017-0513-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lisboa da Motta L, Muller CB, De Bastiani MA, et al. Imbalance in redox status is associated with tumor aggressiveness and poor outcome in lung adenocarcinoma patients. J Cancer Res Clin Oncol. 2014;140(3):461–70. doi: 10.1007/s00432-014-1586-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sawant A, Hensel JA, Chanda D, et al. Depletion of plasmacytoid dendritic cells inhibits tumor growth and prevents bone metastasis of breast cancer cells. J Immunol. 2012;189(9):4258–65. doi: 10.4049/jimmunol.1101855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang Z, Liu Y, Huang Z, et al. 1,25-Dihydroxyvitamin D3 alleviates salivary adenoid cystic carcinoma progression by suppressing GPX1 expression through the NF-kappaB pathway. Int J Oncol. 2016;48(3):1271–79. doi: 10.3892/ijo.2016.3341. [DOI] [PubMed] [Google Scholar]
- 39.Yin J, Wu M, Duan J, et al. Pyrrolidine dithiocarbamate inhibits NF-kappaB activation and upregulates the expression of Gpx1, Gpx4, Occludin, and ZO-1 in DSS-induced colitis. Appl Biochem Biotechnol. 2015;177(8):1716–28. doi: 10.1007/s12010-015-1848-z. [DOI] [PubMed] [Google Scholar]
- 40.Kim D, Nam HJ, Lee W, et al. PKCalpha-LSD1-NF-kappaB-signaling cascade is crucial for epigenetic control of the inflammatory response. Mol Cell. 2018;69(3):398–411. doi: 10.1016/j.molcel.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 41.Mann M, Mehta A, Zhao JL, et al. An NF-kappaB-microRNA regulatory network tunes macrophage inflammatory responses. Nat Commun. 2017;8(1):851. doi: 10.1038/s41467-017-00972-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Q, Sun Y, Liu B, et al. ACT001 modulates the NF-kappaB/MnSOD/ROS axis by targeting IKKbeta to inhibit glioblastoma cell growth. J Mol Med (Berl) 2020;98(2):263–77. doi: 10.1007/s00109-019-01839-0. [DOI] [PubMed] [Google Scholar]
- 43.Li D, Wang X, Huang Q, et al. Cardioprotection of CAPE-oNO2 against myocardial ischemia/reperfusion induced ROS generation via regulating the SIRT1/eNOS/NF-kappaB pathway in vivo and in vitro. Redox Biol. 2018;15:62–73. doi: 10.1016/j.redox.2017.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cui Q, Chen FY, Chen HY, et al. Benzo[a]pyrene (BaP) exposure generates persistent reactive oxygen species (ROS) to inhibit the NF-kappaB pathway in medaka (Oryzias melastigma) Environ Pollut. 2019;251:502–9. doi: 10.1016/j.envpol.2019.04.063. [DOI] [PubMed] [Google Scholar]