Abstract
Objectives
To evaluate and compare the performances of five commercial ELISA assays (EDI, AnshLabs, Dia.Pro, NovaTec, and Lionex) for detecting anti-SARS-CoV-2 IgG.
Methods
Seventy negative control samples (collected before the COVID-19 pandemic) and samples from 101 RT-PCR-confirmed SARS-CoV-2 patients (collected at different time points from symptom onset: ≤7, 8–14 and >14 days) were used to compare the sensitivity, specificity, agreement, and positive and negative predictive values of each assay with RT-PCR. A concordance assessment between the five assays was also conducted. Cross-reactivity with other HCoV, non-HCoV respiratory viruses, non-respiratory viruses, and nuclear antigens was investigated.
Results
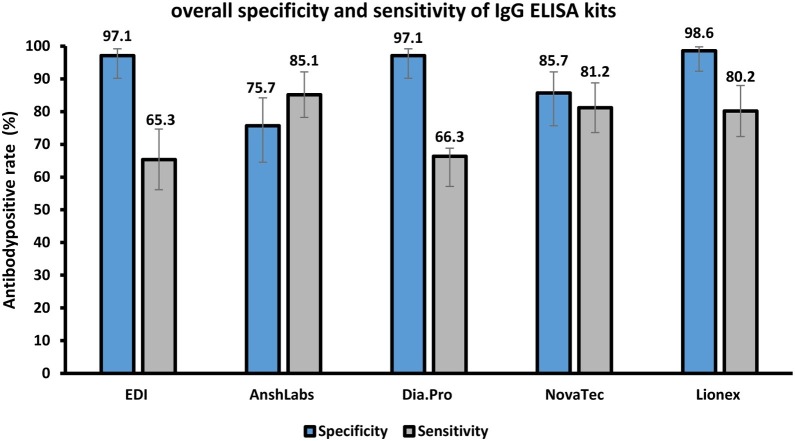
Lionex showed the highest specificity (98.6%; 95% CI 92.3–99.8), followed by EDI and Dia.Pro (97.1%; 95% CI 90.2–99.2), NovaTec (85.7%; 95% CI 75.7–92.1), then AnshLabs (75.7%; 95% CI 64.5–84.2). All ELISA kits cross-reacted with one anti-MERS IgG-positive sample, except Lionex. The sensitivity was low during the early stages of the disease but improved over time. After 14 days from symptom onset, Lionex and NovaTec showed the highest sensitivity at 87.9% (95% CI 72.7–95.2) and 86.4% (95% CI 78.5–91.7), respectively. The agreement with RT-PCR results based on Cohen’s kappa was as follows: Lionex (0.89) > NovaTec (0.70) > Dia.Pro (0.69) > AnshLabs (0.63) > EDI (0.55).
Conclusion
The Lionex and NovaLisa IgG ELISA kits, demonstrated the best overall performance.
Keywords: COVID-19, SARS-CoV-2, Serology, IgG, ELISA, Sensitivity, Specificity
Introduction
Since the start of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) outbreak in December 2019 in Wuhan, China, the virus has rapidly spread and become a major global public health concern (Ong, 2020, Van Elslande, 2020). As of 1 July 2020, the virus had caused more than 10 million confirmed infections and over 500,000 reported deaths (World Health Organization, 2020). Currently, real-time reverse-transcription polymerase chain reaction (RT-PCR) testing is the main technique used for the diagnosis of SARS-CoV-2 infection. However, false-negative RT-PCR results have been found to occur in up to 30% of COVID-19 patients (Wikramaratna et al., 2020, Li et al., 2020, Qin et al., 2020). The reasons for this include poor sample collection techniques, sample collection too late after infection, or disease progression into the lower respiratory tract. Once an individual has been infected for at least seven days, the detection of antibodies is possibly more sensitive than RT-PCR for diagnosing COVID-19 (Zhao et al., 2020). Specific antibodies against SARS-CoV-2 can be detected as early as 4–7 days in approximately 40% of COVID-19 patients, with seroconversion rates reaching >90% by day 14 (Zhao et al., 2020). Therefore, serology could be used as a complementary test to RT-PCR to improve diagnostic sensitivity, particularly in suspected COVID-19 individuals with negative RT-PCR results or those with no respiratory sample collected during the acute phase of illness (Ong, 2020, Van Elslande, 2020).
Serology testing also has other advantages: it is easy to perform and interpret results; it is cheaper and quicker than RT-PCR; it indicates the patient’s immune status and infection stage (sero-survey studies); it facilitates the selection of the best candidate donors (with the highest antibody titers) for plasma exchange; and it aids in assessing the efficacy of vaccines that are in development. Due to urgency and demand in the current crisis, a large number of commercial serological tests have been developed and introduced into the global market, but often with insufficient validation on clinical samples. Hence, there is a pressing need for identifying reliable immunoassays with high sensitivity and specificity for serology testing and surveillance of SARS-CoV-2 infection, as persistent concerns remain regarding the accuracy and reliability of the available SARS-CoV-2 immunoassays.
In order to address this challenge, the present study evaluated the performance of five commercial CE-marked ELISA kits for detecting anti-SARS-CoV-2 IgG antibodies in samples from RT-PCR-confirmed COVID-19 patients. The sensitivity, positive predictive value, negative predictive value, positive percent agreement, and Cohen’s kappa were measured for each assay using samples collected from SARS-CoV-2 RT-PCR-positive patients at different times from symptom onset (≤7, 8–14 and >14 days). The specificity and cross-reactivity were evaluated using pre-pandemic serum samples collected from healthy blood donors. A concordance assessment was conducted to compare the agreement between the kits.
Methods
Study design, ethical compliance and sample collection
The performance of five CE-marked ELISA assays (EDI, Dia.Pro, AnshLabs, NovaTec, and Lionex) for detecting anti-SARS-CoV-2 IgG antibodies was evaluated. The performance was assessed using anonymous samples collected from RT-PCR-confirmed SARS-CoV-2 patients admitted to Hamad Medical Corporation (HMC), the main public healthcare provider and the nationally designated provider for COVID-19 healthcare needs, with different COVID-19 clinical outcomes. Pre-pandemic serum samples collected from blood donors before 2019 were selected for the negative control group. IRB approval for this study was obtained from HMC (HMC-IRB# MRC-01-20-145, HMC-IRB# MRC-05-003 and HMC-IRB# MRC-05-007) and Qatar University (QU-IRB # QU-IRB 804-E/17).
Serum samples
Serum samples were selected from 101 RT-PCR-confirmed COVID-19 patients, including: ICU-admitted patients (n = 35), hospitalized non-ICU patients (n = 45) and convalescent samples collected from COVID-19-recovered patients by the Qatar Communicable Disease Center (CDC) at HMC (n = 21). Clinical records of the patients were reviewed to determine the time from symptom onset to collection, and categorized into three groups: Group 1, ≤7 days; Group 2, 8–14 days; Group 3, >14 days. RNA was extracted from nasopharyngeal swab specimens using the Qiagen extraction kit. The extracted RNA was tested for SARS-CoV-2 using the SuperscriptIII OneStep RT-PCR kit, as recommended by the manufacturer’s instruction (Cat No. 12594100, ThermoFisher, USA). Each sample was tested using three sets of primers targeting the E gene for screening, and confirmed with two different sets of primers targeting the RdRp gene, as described in (Corman et al., 2020). CT values <32 were considered positive. Characteristics of the 101 COVID-19 patients, including the demographic data and classification, are summarized in Table 1 . The patients had a median age (IQR) of 48.0 years (40.0–57.0), of which 89.1% were male and 4.9% were female. Patients in the three time points (≤7, 8–14 and >14 days) had a median age of 50.0 (39.3–56.8), 49.0 (41.3–58.5) and 46.0 (34.3–55.5) years, respectively.
Table 1.
Demographic data and clinical characteristics of COVID-19 patients and control group.
| Characteristic | All COVID-19 patients | Days between symptom onset and sample collection |
Control group |
||
|---|---|---|---|---|---|
| ≤7 days | 8–14 days | >14 days | |||
| N | 101 | 28 (27.7%) | 40 (39.6%) | 33 (32.7%) | 70 |
| Age median (IQR) | 48.0 (40.0–57.0) |
50.0 (39.3–56.8) | 49.0 (41.3–58.5) | 46.0 (34.3–55.5) | 36.0 (30.3–45.0) |
| Gender | |||||
| Male | 76 (89.1%) | 25 (89.3%) | 40 (100%) | 11 (33.3%) | 58 (82.9%) |
| Female | 4 (4.9%) | 3 (10.7%) | – | 1 (3.0%) | 6 (8.6%) |
| N/A | 21 (20.8%) | – | – | 21 (63.6%) | 6 (8.6%) |
| Sample source | |||||
| Hospitalized, non-ICU patients | 45 (44.6%) | 17 (60.7%) | 23 (57.5%) | 5 (15.2%) | – |
| ICU-admitted patients | 35 (34.7%) | 11 (39.3%) | 17 (42.5%) | 7 (21.2%) | – |
| Recovered convalescent plasma donors | 21 (20.8%) | – | – | 21 (63.6%) | – |
Samples from healthy blood donors collected before 2019 and used in previous studies were utilized for the control group (Nasrallah et al., 2017, Smatti et al., 2017, Humphrey et al., 2019, Al-Qahtani et al., 2016, Dargham et al., 2018, Nasrallah et al., 2018, Smatti et al., 2020, Al Kahlout et al., 2019). The healthy blood donors had a median age of 36.0 (30.3–45.0) years, with 82.9% male and 8.6% female. Details about the collection, transport and storage methods of the control samples were described elsewhere (Nasrallah et al., 2017, Smatti et al., 2017, Humphrey et al., 2019, Al-Qahtani et al., 2016, Dargham et al., 2018, Nasrallah et al., 2018, Smatti et al., 2020, Al Kahlout et al., 2019). The control group included samples that were seropositive for various viruses, including all other human coronaviruses (HCoV). Further details about the control samples can be found in Table 1 and Table S1.
IgG ELISA kits
Commercial ELISA kits from five different companies were used for the qualitative detection of anti-SARS-CoV-2 IgG antibodies against the Spike (S) or the Nucleocapsid (N) proteins in the sera of the COVID-19 patient and control groups. These kit were: (i) Epitope Diagnostic (EDI™) novel coronavirus COVID-19 IgG (Ref. no. KT-1032, USA); (ii) AnshLabs SARS-CoV-2 IgG (Ref. no. AL-1001-I, USA); (iii) Diagnostic Bioprobes (Dia.Pro) COVID-19 IgG (Ref. no. COV19G.CE, Italy); (iv) NovaTec (NovaLisa®) SARS-COV-2 IgG (Ref. no. COVG0940, Germany); and (v) Lionex COVID-19 ELISA-human IgG (Ref. no. LIO-COV19-IgG, Germany). More details about the ELISA kits–including specifications, reported sensitivity, and specificity–are shown in Table S2. All tests were carried out manually, according to the manufacturers’ instructions. A microplate reader, Epoch 2 microplate spectrophotometer (Bio-Tek, Italy) was used to read the optical density (OD) in all ELISA reactions. Borderline results were considered positive (Meyer, 2020, Van Elslande, 2020).
Statistical analysis
The sensitivity, specificity, positive predictive value, negative predictive value, positive percent agreement, and Cohen’s Kappa were calculated to assess the performance of each assay with the positive SARS-CoV-2 RT-PCR patients (Whitman et al., 2020, Beavis et al., 2020). Specificity and cross-reactivity of each assay were assessed using the pre-pandemic control samples. Data were summarized by number and percentage of positive results for each assay. Borderline results were considered positive. Samples were categorized into three groups according to the time between collection and the onset of symptoms (≤7, 8–14 and >14 days), and all parameters were calculated for each group. Concordance assessment between the ELISA kits was conducted to assess the agreement between the kits. These concordance measures included overall, positive and negative percent agreement, as well as Cohen’s kappa statistic. The latter measure is a standard and robust metric that estimates the level of agreement (beyond chance) between two diagnostic tests. Ranging between 0 and 1, a kappa value ≤0.40 denotes poor agreement, a value between 0.40 and 0.75 denotes fair/good agreement, and a value ≥0.75 denotes excellent agreement (Anon, 2020). The significance level was indicated at 5%, and a 95% confidence interval (CI) was reported for each metric. All calculations were performed using Microsoft Excel 2016. The Chi–squared test was used to calculate the significance between the performances of ELISA kits. Significance was (*) = p < 0.05; (**) = p < 0.01; (***) = p < 0.001. Further details about the statistical analysis and calculations can be found in Table S4.
Results
Diagnostic assessment of the IgG ELISA kits according to the time of sample collection after symptom onset (n = 101)
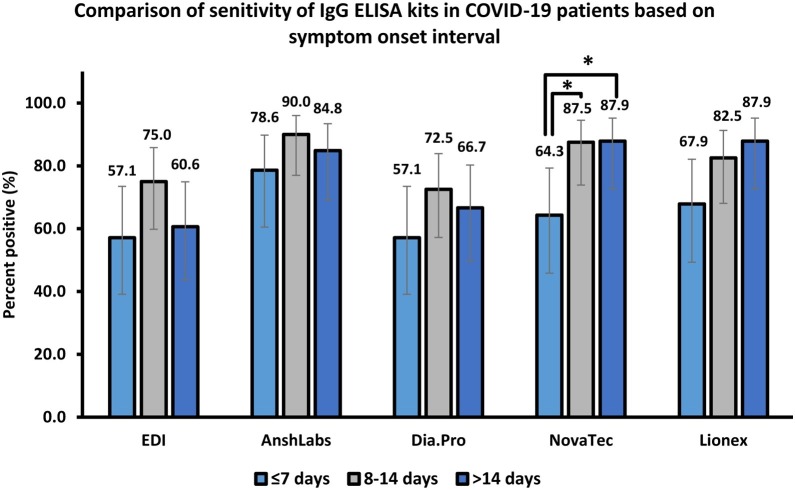
The diagnostic assessment of all ELISA kits according to each time-point after symptom onset (≤7 days, 8–14 days and >14 days) is summarized in Table 2 , Figure 1 and Figure 2 .
Table 2.
The diagnostic assessment of the different commercial IgG ELISA kits according to time of sample collection after symptoms onset.
| Days after symptoms onset | ELISA kit | Sensitivity % (95% CI) |
Positive predictive value % (95% CI) |
Negative predictive value % (95% CI) |
Positive percent agreement % (95% CI) |
Cohen's kappa k (95% CI) |
|---|---|---|---|---|---|---|
| ≤7 days (n = 28) | EDI | 57.1 (39.1–73.5) | 88.9 (67.2–96.9) | 85.0 (75.6–91.2) | 85.7 (77.4–91.3) | 0.61 (0.49–0.72) |
| AnshLabs | 78.6 (60.5–89.8) | 56.4 (41.0–70.7) | 89.8 (79.5–95.3) | 76.5 (67.2–83.8) | 0.49 (0.36–0.61) | |
| Dia.Pro | 57.1 (39.1–73.5) | 88.9 (67.2–96.9) | 85.0 (75.6–91.2) | 85.7 (77.4–91.3) | 0.61 (0.49–0.72) | |
| NovaTec | 64.3 (45.8–79.3) | 64.3 (45.8–79.3) | 85.7 (75.7–92.1) | 79.6 (70.6–86.4) | 0.50 (0.38–0.62) | |
| Lionex | 67.9 (49.3–82.1) | 95.0 (76.4–99.1) | 88.5 (79.5–93.8) | 89.8 (82.2–94.4) | 0.73 (0.63–0.82) | |
| 8–14 days (n = 40) | EDI | 75.0 (59.8–85.8) | 93.8 (79.9–98.3) | 87.2 (78.0–92.9) | 89.1 (81.9–93.7) | 0.75 (0.67–0.84) |
| AnshLabs | 90.0 (77.0–96.0) | 67.9 (54.5–78.9) | 93.0 (83.3–97.2) | 80.9 (72.6–87.2) | 0.61 (0.51–0.72) | |
| Dia.Pro | 72.5 (57.2–83.9) | 93.5 (79.3–98.2) | 86.1 (76.8–92.1) | 88.2 (80.8–93.0) | 0.73 (0.64–0.82) | |
| NovaTec | 87.5 (73.9–94.5) | 77.8 (63.7–87.5) | 92.3 (83.2–96.7) | 86.4 (78.7–91.6) | 0.71 (0.62–0.81) | |
| Lionex | 82.5 (68.1–91.3) | 97.1 (85.1–99.5) | 90.8 (82.2–95.5) | 92.7 (86.3–96.3) | 0.84 (0.77–0.91) | |
| >14 days (n = 33) | EDI | 60.6 (43.7–75.3) | 90.9 (72.2–97.5) | 84.0 (74.5–90.4) | 85.4 (77.4–91.0) | 0.63 (0.53–0.74) |
| AnshLabs | 84.8 (69.1–93.4) | 62.2 (47.6–74.9) | 91.4 (81.4–96.3) | 78.6 (69.8–85.5) | 0.55 (0.44–0.67) | |
| Dia.Pro | 66.7 (49.6–80.3) | 91.7 (74.2–97.7) | 86.1 (76.8–92.1) | 87.4 (79.6–92.5) | 0.69 (0.59–0.79) | |
| NovaTec | 87.9 (72.7–95.2) | 74.4 (58.9–85.4) | 93.8 (85.0–97.5) | 86.4 (78.5–91.7) | 0.70 (0.60–0.80) | |
| Lionex | 87.9 (72.7–95.2) | 96.7 (83.3–99.4) | 94.5 (86.7–97.9) | 95.1 (89.1–97.9) | 0.89 (0.82–0.95) | |
| Overall (n = 101) | EDI | 65.3 (56.1–74.6) | 97.1 (94.5–99.6) | 66.0 (58.9–73.1) | 78.4 (72.2–84.5) | 0.58 (0.46–0.70) |
| AnshLabs | 85.1 (78.2–92.1) | 83.5 (77.9–89.1) | 77.9 (71.7–84.2) | 81.3 (75.4–87.1) | 0.61 (0.49–0.73) | |
| Dia.Pro | 66.3 (57.1–75.6) | 97.1 (94.6–99.6) | 66.7 (59.6–73.7) | 78.9 (72.8–85.1) | 0.59 (0.48–0.71) | |
| NovaTec | 81.2 (73.6–88.8) | 89.1 (84.5–93.8) | 75.9 (69.5–82.4) | 83.0 (77.4–88.7) | 0.66 (0.54–0.77) | |
| Lionex | 80.2 (72.4–88.0) | 98.8 (97.1–100) | 77.5 (71.2–83.8) | 87.7 (82.8–92.6) | 0.76 (0.66–0.85) |
Figure 1.
Comparison of overall sensitivity (n = 101) and specificity (n = 70) of each IgG ELISA kit.
Figure 2.
Proportion of samples testing positive for anti-SARS-CoV-2 IgG antibodies.
Samples were stratified based on the time of collection after symptoms onset (≤7 days, n = 28; 8–14 days, n = 40; >14 days, n = 33).
*p < 0.05.
In the first week of symptom onset (≤7 days), the sensitivity (95% CI) ranged from 57.1% (39.1–73.5) to 78.6% (60.5–89.8) for EDI and AnshLabs, respectively. The highest positive and negative predictive values were estimated at 95.0% (76.4–99.1) for Lionex and 89.8% (79.5–95.3) for AnshLabs, respectively. The best agreement with RT-PCR was observed in Lionex, with 89.8% (82.2–94.4) positive percent agreement and Cohen’s kappa index of 0.73 (0.63–0.82). The lowest agreement was observed in AnshLabs, with 76.5% (67.2–83.8) positive percent agreement and a kappa index of 0.49 (0.36–0.61).
In the second week of symptom onset (8–14 days), all parameters increased compared with the first week, where the highest sensitivity was scored by AnshLabs at 90.0% (77.0–96.0). The highest positive and negative predictive values were estimated at 97.1% (85.1–99.5) for Lionex and 93.0% (83.3–97.2) for AnshLabs, respectively. The lowest agreement with RT-PCR was observed in AnshLabs, with 80.9% (72.6–87.2) positive percent agreement and a kappa index of 0.61 (0.51–0.72), while the highest agreement was scored by Lionex, with 92.7% (86.3–96.3) positive percent agreement and a kappa index of 0.84 (0.77–0.91).
The performance of the evaluated IgG ELISA kits varied after 14 days of symptom onset. Compared with the second week, the sensitivity decreased in EDI, AnshLabs and Dia.Pro down to 60.6% (43.7–75.3), 84.8 (69.1–93.4) and 66.7% (49.6–80.3), respectively (Figure 2). However, the sensitivity slightly increased for NovaTec and Lionex, where both assays showed the highest sensitivity at 87.9% (72.7–95.2). Also, Lionex showed the highest positive and negative predictive values at 96.7% (83.3–99.4) and 94.5% (86.7–97.9), respectively. The positive percent agreement of EDI, AnshLabs and Dia.Pro also slightly dropped to 85.4% (77.4–91.0), 78.6% (69.8–85.5) and 87.4% (79.6–92.5), respectively. Whilst no change was observed in the positive percent agreement of NovaTec, it slightly increased in Lionex to 95.1% (89.1–97.9) with a Kappa index of 0.89 (0.82–0.95).
Assay specificity according to the negative control subgroups (n = 70)
All assays showed acceptable overall specificity, ranging from 85.7 to 98.6%, except Anshlabs, which had a 75.7% (53/70; 64.5–84.2) specificity. Lionex showed the highest specificity at 98.6% (69/70; 92.3–99.8), followed by EDI and Dia.Pro at 97.1% (68/70; 90.2–99.2), and then NovaTec with 85.7% (60/70; 75.7–92.1) specificity (Table 3 and Figure 1). The specificity of each kit in relation to sample cross-reactivity with other viruses (Table 3 and Table S1) was also calculated. All assays cross-reacted with other human coronaviruses (HCoVs), except Lionex, which had a 100% specificity (20/20; 83.9–100) in this sub-group. AnshLabs, NovaTec and Dia.Pro showed false-positive results with non-HCoVs respiratory viruses (RSV and influenza), with a specificity of 60.0% (9/15; 35.8–80.2), 73.3% (11/15; 48.1–89.1) and 93.3% (14/15; 70.2–98.8), respectively. Only EDI and Lionex did not show cross-reactivity with non-HCoV respiratory viruses, with 100% (15/15; 79.6–100) specificity for this subgroup. All assays showed some cross-reactivity with non-respiratory viruses, except Dia.Pro, which had 100% (33/33; 93.9–100) specificity. Finally, all assays showed no cross-reactivity with antinuclear antibody samples, except AnshLabs, which cross-reacted with one control sample [50.0% (1/2; 9.5–90.6)]. However, the sample size was very small, as two specimens positive for antinuclear antibodies were used.
Table 3.
The specificity of the five evaluated IgG ELISA kits according to the negative control subgroups (n = 70).
| Subgroup with IgG/IgM antibodies against | No. of samples | Specificity (%, 95% confidence interval) |
||||
|---|---|---|---|---|---|---|
| EDI | AnshLabs | Dia.Pro | NovaTec | Lionex | ||
| Other coronaviruses (SARS-CoV, MERS-CoV, HCoV-229E, NL63, OC43, and HKU1)* | 20 | 19/20 (95.0%; 76.4–99.1%) | 17/20 (85.0%; 64.0–94.8%) | 19/20 (95.0%; 76.4–99.1%) | 17/20 (85.0%; 64.0–94.8%) | 20/20 (100%; 83.9–100%) |
| Non-CoV respiratory viruses (Influenza and RSV)* | 15 | 15/15 (100%; 79.6–100%) | 9/15 (60.0%; 35.8–80.2%) | 14/15 (93.3%; 70.2–98.8%) | 11/15 (73.3%; 48.1–89.1%) | 15/15 (100%; 79.6–100%) |
| Non-respiratory viruses (HEV, HGV, HCV, HBV, DENV, WNV, CHIKV, B19, HSV-1, HSV-2, EBV, HHV-6, and HHV-8)* | 33 | 32/33 (97.0%; 91.1–100%) | 25/33 (75.8%; 61.1–90.4%) | 33/33 (100%; 93.9–100%) | 30/33 (90.9%; 81.1–100%) | 32/33 (97.0%; 91.1–100%) |
| Nuclear antigens (ANAs) | 2 | 2/2 (100%; 34.2–100%) | 1/2 (50.0%; 9.5–90.6%) | 2/2 (100%; 34.2–100%) | 2/2 (100%; 34.2–100%) | 2/2 (100%; 34.2–100%) |
| Overall specificity | 70 | 68/70 (97.1%; 90.2–99.2) | 53/70 (75.7%; 64.5–84.2) | 68/70 (97.1%; 90.2–99.2) | 60/70 (85.7%; 75.7–92.1) | 69/70 98.6%; 92.3–99.8) |
Abbreviations: *MERS, middle east respiratory syndrome coronavirus; SARS-CoV, severe acute respiratory syndrome coronavirus; RSV, respiratory syncytial virus; HSV-1, herpes simplex virus 1; HSV-2, herpes simplex virus 2; HHV-6, human herpes virus-6; HHV-8, human herpes virus-8; EBV, Epstein-Barr virus; HBV, hepatitis B virus; HCV, hepatitis C virus; HEV, hepatitis E virus; HGV, hepatitis G virus; B19, Parvovirus B19; WNV, West Nile virus.
Concordance assessment between IgG ELISA kits
Table 4 shows the concordance assessment between the different IgG ELISA kits. The overall percent agreement ranged from 79.5% (72.9–84.9) for AnshLabs/EDI test combination to 97.1% (93.3–98.8) for Dia.Pro/EDI test combination. The positive percent agreement ranged from 66.0% (56.4–74.4) for EDI vs. AnshLabs and Dia.Pro vs. AnshLabs to 100% (94.7–100) for AnshLabs vs. EDI, NovaTec vs. EDI and NovaTec vs. Dia.Pro. The negative percent agreement ranged from 65.7% (56.1–74.2) for AnshLabs vs. Dia.Pro to 100% (95.4–100) for EDI vs. NovaTec and Dia.Pro vs. NovaTec, and also 100% (94.7–100) for EDI vs. AnshLabs. Importantly, Cohen’s Kappa statistic denoted fair/good to excellent agreement and ranged between 0.59 (0.51–0.68) for AnshLabs/Dia.Pro test combination and 0.94 (0.90–0.98) for Dia.Pro/EDI test combination.
Table 4.
Concordance assessment between the commercial IgG ELISA kits.
| Reference test | Compared to | Overall percent agreement | Positive percent agreement | Negative percent agreement | Cohen's kappa |
|---|---|---|---|---|---|
| % (95% CI) | % (95% CI) | % (95% CI) | k (95% CI) | ||
| EDI | AnshLabs | 79.5 (72.9–84.9) | 100 (94.7–100) | 66.0 (56.4–74.4) | 0.60 (0.52–0.69) |
| Dia.Pro | 97.1 (93.3–98.8) | 97.1 (89.9–99.2) | 97.1 (91.8–99.0) | 0.94 (0.90–0.98) | |
| NovaTec | 86.0 (80.0–90.4) | 100 (94.7–100) | 76.7 (67.7–83.8) | 0.72 (0.65–0.80) | |
| Lionex | 86.0 (80.0–90.4) | 92.6 (83.9–96.8) | 81.6 (73.0–87.9) | 0.72 (0.64–0.79) | |
| AnshLabs | EDI | 79.5 (72.9–84.9) | 66.0 (56.4–74.4) | 100 (94.7–100) | 0.60 (0.52–0.69) |
| Dia.Pro | 78.9 (72.2–84.4) | 66.0 (56.4–74.4) | 98.5 (92.1–99.7) | 0.59 (0.51–0.68) | |
| NovaTec | 91.2 (86.0–94.6) | 87.4 (79.6–92.5) | 97.1 (89.9–99.2) | 0.82 (0.76–0.88) | |
| Lionex | 80.7 (74.1–85.9) | 73.8 (64.6–81.3) | 91.2 (82.1–95.9) | 0.62 (0.53–0.70) | |
| Dia.Pro | EDI | 97.1 (93.3–98.8) | 95.7 (88.0–98.5) | 98.0 (93.1–99.5) | 0.94 (0.90–0.98) |
| AnshLabs | 78.9 (72.2–84.4) | 98.6 (92.2–99.7) | 65.7 (56.1–74.2) | 0.59 (0.51–0.68) | |
| NovaTec | 86.5 (80.6–90.9) | 100 (94.7–100) | 100 (95.4–100) | 0.73 (0.66–0.81) | |
| Lionex | 85.4 (79.3–89.9) | 91.2 (82.3–96.0) | 77.5 (68.4–84.5) | 0.71 (0.63–0.78) | |
| NovaTec | EDI | 86.0 (80.0–90.4) | 73.9 (64.1–81.8) | 100 (95.4–100) | 0.72 (0.65–0.80) |
| AnshLabs | 91.2 (86.0–94.6) | 97.8 (92.4–99.4) | 83.5 (73.9–90.1) | 0.82 (0.76–0.88) | |
| Dia.Pro | 86.5 (80.6–90.9) | 75.0 (65.3–82.7) | 93.3 (86.1–96.9) | 0.73 (0.66–0.81) | |
| Lionex | 83.6 (77.4–88.4) | 79.3 (70.0–86.4) | 81.4 (72.7–87.7) | 0.67 (0.60–0.75) | |
| Lionex | EDI | 86.0 (80.0–90.4) | 76.8 (66.6–84.6) | 94.4 (87.5–97.6) | 0.72 (0.64–0.79) |
| AnshLabs | 80.7 (74.1–85.9) | 92.7 (84.9–96.6) | 69.7 (59.5–78.2 | 0.62 (0.53–0.70) | |
| Dia.Pro | 85.4 (79.3–89.9) | 76.8 (66.6–84.6) | 78.7 (69.1–85.9) | 0.71 (0.63–0.78) | |
| NovaTec | 83.6 (77.4–88.4) | 89.0 (80.4–94.1) | 88.6 (79.8–93.9) | 0.67 (0.60–0.75) |
Discussion
This study evaluated the performances of five CE-marked ELISA kits using 101 samples collected from SARS-CoV-2 RT-PCR-confirmed patients and 70 pre-pandemic control samples collected from healthy blood donors. The sensitivity, specificity, agreement, and positive and negative predictive values were calculated at different time points from symptom onset (≤7, 8–14 and >14 days) for each kit (Table 2). The overall agreement and Cohen’s kappa were also calculated to compare the assays (Table 4).
The results showed that most of the evaluated assays demonstrated a very good performance during the first week after symptom onset compared with other studies (Adams et al., 2020, Bundschuh et al., 2020, Colavita et al., 2020, Lassaunière et al., 2020). Expectedly, the agreement between the outcome of each ELISA kit and RT-PCR increased with time after symptom onset, which was consistent with a time lag between the onset of infection and the development of detectable antibodies. High rates of positive results were reached after the first week of clinical illness. This increase was observed with the sensitivity, positive and negative predictive values, positive percent agreement and Cohen’s Kappa (Table 2). Even though the sensitivity was lower during the early stages of the disease, it was greatly improved 8–14 days after symptom onset. AnshLabs showed the highest sensitivity in patients tested within the first two weeks of symptom onset (Figure 2). However, AnshLabs had the lowest specificity compared with the other kits. After 14 days of symptom onset, the sensitivity slightly decreased in all assays, except NovaTec and Lionex. This could be because 63.6% (21/33) of the samples in this time point were collected by the CDC from recovered patients, for whom there were clinical data, including the severity of the disease, whether they developed symptoms or not, and the exact day of sample collection. Hence, these patients might not have elicited enough antibody response to be detected by most of the assays (Table S4, group 3 sample Nos. 4, 11, 15, and 26). Surprisingly, one of the ICU-admitted patients did not show a detectable antibody response by all ELISA assays (Table S3, group 3 sample No. 33), which needs further investigation by other highly sensitive assays. Typically, if borderline results were obtained in ELISA testing, another sample was taken from the patient 1–2 weeks later for re-testing. However, this was not possible, as sensitivity and specificity were being tested in specific time frames. Considering that these borderline samples were collected from RT-PCR-positive patients, borderline results were considered positive, which was consistent with similar studies (Meyer, 2020, Van Elslande, 2020). Interestingly, one pre-pandemic sample with positive anti-MERS-CoV IgG antibodies was found to be positive by four ELISA kits (Table 3 and Table S3), except Lionex, which demonstrated the highest specificity at 98.6% (Figure 1).
Another interesting finding was the heterogeneity of IgG antibody response in COVID-19 patients. Most developed serology assay targets for either the spike (S) or the nucleocapsid (N) protein of SARS-CoV-2. Previous studies performed on other HCoV suggested that the anti-nucleocapsid (anti-N) antibody response may appear earlier than the anti-spike (anti-S) response and may wane more rapidly (Coste et al., 2020, Chia et al., 2020). In the current study, it was expected that the differences in sensitivity between ELISA kits would depend on the targeted protein used in each assay. It was noticed that there was a decline in the sensitivity of the ELISA kits targeting the N protein. However, the sensitivity increased in the kit that solely targets the S1 protein (Lionex). Therefore, a possible explanation for this is that the level of anti-N and anti-S antibodies may be similar during the acute phase of COVID-19 illness, but anti-N antibodies could be waning after the second week (Coste et al., 2020, Chia et al., 2020). Moreover, this could also explain the high specificity of Lionex compared with the other assays (Figure 1). That is, Lionex targets the S1 protein, which is smaller and less conserved across different families of viruses than the N protein. Therefore, detection of anti-N antibodies may be useful in distinguishing more recent antibody responses, while anti-S antibody may be used during the early and convalescent phases. However, this does not explain why the sensitivity of NovaTec, which targets the N protein, remained steady after the second week compared with EDI, which also targets the same protein (N).
Concordance assessment between the different assays showed good to excellent agreement between the kits. EDI and Dia.Pro had the best overall agreement (97.1%) and kappa index (0.94). However, both assays demonstrated the lowest sensitivity in all time points compared with the assays, despite having a very high specificity (97.1%). Therefore, these two assays are the least recommended for diagnosis and clinical relevance. NovaTec and AnshLabs also showed an excellent positive percent agreement (91.2%) and a kappa index (0.82), where both assays had comparable overall sensitivity and specificity. Lionex, however, showed a variation in the agreement with the other ELISA kits, which could be due to the fact that Lionex is the only kit that targets the S1 protein.
From an epidemiological perspective, high sensitivity of the assay in combination with robust specificity is desirable. Here, Lionex and NovaTec ELISA kits showed the best overall performance in terms of specificity, sensitivity, agreement with RT-PCR, and positive and negative predictive values compared with the other assays. The overall performance of both NovaTec and Lionex IgG manual ELISA was comparable with other detection methods, including automated tests, reported elsewhere (Chew, 2020; Poljak, 2020). Both assays showed a diagnostic sensitivity of 87.9% after 14 days of symptom onset compared with Abbott Architect (84.2%) and Cobas 6800 systems (95.2%). The specificity of Lionex (98.6%) was also comparable with the aforementioned automated assays (100% and 99.3%, respectively) (Chew, 2020; Poljak, 2020).
A strength of this study was the use of a diverse control group to evaluate cross reactivity with antibodies against various viruses, including MERS, SARS-CoV, endemic coronaviruses, respiratory viruses, and other viruses. One of the limitations of this study was that the clinical details of the patients were unavailable, which are important to understand why some of them did not develop an antibody response detectable by the evaluated kits. It would be very beneficial to perform a new study using a large sample size collected from patients with known disease severity outcomes (e.g. critical, severe, moderate, mild, and asymptomatic), to have a better implication about each assay performance and clinical practice relevance.
In conclusion, two ELISA kits (NovaLisa and Lionex) showed promising overall results, which could be used in the future for clinical testing. Further, all assays showed acceptable specificity, ranging from 85.7 to 98.6%, except for the AnshLabs ELISA. Finally, although serological assays do not replace molecular tests in diagnosing active infection, they serve as an essential tool with which to accurately estimate the seroprevalence of SARS-CoV-2 in the general population and to quantify the level of herd immunity (Winter and Hegde, 2020). This could help ease the restrictions on human mobility and interactions without provoking a significant resurgence of transmission and mortality. However, it is still unclear whether positive results by serology reflect a protective immune response against infection (Melgaço et al., 2020). Further studies are essential to distinguish functional antibodies from total binding antibodies using virus neutralization assays.
Conflict of interest
All authors have no conflict of interest to declare.
Funding
This work was made possible by grant No. RRC-2-032 from the Q atar National Research Fund(a member of Qatar Foundation). The statements made herein are solely the responsibility of the authors. GKN would like to acknowledge funds from Qatar University's internal grant QUERG-CMED-2020-2.
CRediT authorship contribution statement
Hadi M. Yassine: Conceptualization, Writing - review & editing, Supervision, Funding acquisition. Hadeel Al-Jighefee: Methodology, Validation, Investigation, Data curation, Writing - original draft, Writing - review & editing, Visualization. Duaa W. Al-Sadeq: Methodology, Validation, Data curation, Project administration. Soha R. Dargham: Formal analysis, Investigation, Writing - original draft, Writing - review & editing. Salma N. Younes: Methodology, Validation, Data curation. Farah Shurrab: Methodology, Validation, Data curation. Reham M. Marei: Methodology. Ali Ait. Hssain: Methodology. Sara Taleb: Methodology. Hashim Alhussain: Methodology. Maryam A. Al-Nesf: Methodology. Abdullatif Al-Khal: Methodology. Hamda Qotba: Investigation, Resources. Asmaa A. Althani: Investigation, Resources. Patrick Tang: Investigation, Writing - original draft, Writing - review & editing, Funding acquisition. Laith J. Abu-Raddad: Conceptualization, Formal analysis, Investigation, Writing - original draft, Writing - review & editing, Funding acquisition. Gheyath K. Nasrallah: Conceptualization, Formal analysis, Investigation, Resources, Data curation, Writing - original draft, Writing - review & editing, Visualization, Supervision, Project administration, Funding acquisition.
Acknowledgments
We would like to acknowledge Ms. Enas S. Al-Absi for her help in sorting the negative control samples.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.ijid.2020.10.042.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Adams E. Evaluation of antibody testing for SARS-Cov-2 using ELISA and lateral flow immunoassays. medRxiv. 2020 [Google Scholar]
- Al Kahlout R.A. Comparative serological study for the prevalence of anti-MERS coronavirus antibodies in high- and low-risk groups in Qatar. J Immunol Res. 2019:1386740. doi: 10.1155/2019/1386740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Qahtani A.A. Prevalence of anelloviruses (TTV, TTMDV, and TTMV) in healthy blood donors and in patients infected with HBV or HCV in Qatar. Virology. 2016;13:208. doi: 10.1186/s12985-016-0664-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Measurement of Interrater Agreement, in Statistical Methods for Rates and Proportions. PP. 598–626.
- Beavis K.G. Evaluation of the EUROIMMUN anti-SARS-CoV-2 ELISA assay for detection of IgA and IgG antibodies. J Clin Virol. 2020;129:104468. doi: 10.1016/j.jcv.2020.104468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundschuh C. Evaluation of the EDI enzyme linked immunosorbent assays for the detection of SARS-CoV-2 IgM and IgG antibodies in human plasma. Clin Chim Acta. 2020;509:79–82. doi: 10.1016/j.cca.2020.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew K.L. Clinical evaluation of serological IgG antibody response on the Abbott Architect for established SARS-CoV-2 infection. Clin Microbiol Infect. 2020;26(9):1256.e9–1256.e11. doi: 10.1016/j.cmi.2020.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia W.N. Serological differentiation between COVID-19 and SARS infections. Emerg Microbes Infect. 2020;9(1):1497–1505. doi: 10.1080/22221751.2020.1780951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colavita F. Evaluation of ELISA tests for the qualitative determination of IgG, IgM and IgA to SARS-CoV-2. MedRxiv. 2020;2020(May):20111682. [Google Scholar]
- Corman V.M. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveillance. 2020;25(3):2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste A.T. Comparison of SARS-CoV-2 serological tests with different antigen targets. MedRxiv. 2020;2020(July):20149864. doi: 10.1016/j.jcv.2020.104690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dargham S.R. Herpes simplex virus type 2 seroprevalence among different national populations of Middle East and North African Men. Sex Transm Dis. 2018;45(7):482–487. doi: 10.1097/OLQ.0000000000000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey J.M. Dengue and chikungunya seroprevalence among Qatari nationals and immigrants residing in Qatar. PLoS One. 2019;14 doi: 10.1371/journal.pone.0211574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassaunière R. Evaluation of nine commercial SARS-CoV-2 immunoassays. MedRxiv. 2020;2020(April):20056325. [Google Scholar]
- Li D. False-negative results of real-time reverse-transcriptase polymerase chain reaction for severe acute respiratory syndrome coronavirus 2: role of deep-learning-based ct diagnosis and insights from two cases. Korean J Radiol. 2020;21(4):505–508. doi: 10.3348/kjr.2020.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melgaço J.G., Azamor T., Ano Bom A.P.D. Protective immunity after COVID-19 has been questioned: what can we do without SARS-CoV-2-IgG detection? Cell Immunol. 2020;353:104114. doi: 10.1016/j.cellimm.2020.104114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B. Validation of a commercially available SARS-CoV-2 serological Immunoassay. medRxiv. 2020;2020(May):20080879. doi: 10.1016/j.cmi.2020.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrallah G.K. Seroprevalence of hepatitis E virus among blood donors in Qatar (2013–2016) Transfusion. 2017;57(7):1801–1807. doi: 10.1111/trf.14116. [DOI] [PubMed] [Google Scholar]
- Nasrallah G.K. Estimating seroprevalence of herpes simplex virus type 1 among different Middle East and North African male populations residing in Qatar. J Med Virol. 2018;90(1):184–190. doi: 10.1002/jmv.24916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong D.S.Y. Comparison of diagnostic accuracies of rapid serological tests and ELISA to molecular diagnostics in patients with suspected coronavirus disease 2019 presenting to the hospital. Clin Microbiol Infect. 2020;26(8):1094.e7–1094.e10. doi: 10.1016/j.cmi.2020.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poljak M. Clinical evaluation of the cobas SARS-CoV-2 test and a diagnostic platform switch during 48 hours in the Midst of the COVID-19 pandemic. J Clin Microbiol. 2020;58(6) doi: 10.1128/JCM.00599-20. e00599–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C. 18 F-FDG PET/CT findings of COVID-19: a series of four highly suspected cases. Eur J Nuclear Med Mol Imag. 2020:1–6. doi: 10.1007/s00259-020-04734-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smatti M.K. Prevalence and molecular profiling of Epstein Barr virus (EBV) among healthy blood donors from different nationalities in Qatar. PLoS One. 2017;12(12) doi: 10.1371/journal.pone.0189033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smatti M.K. Measuring influenza hemagglutinin (HA) stem-specific antibody-dependent cellular cytotoxicity (ADCC) in human sera using novel stabilized stem nanoparticle probes. Vaccine. 2020;38(4):815–821. doi: 10.1016/j.vaccine.2019.10.093. [DOI] [PubMed] [Google Scholar]
- Van Elslande J. Diagnostic performance of seven rapid IgG/IgM antibody tests and the Euroimmun IgA/IgG ELISA in COVID-19 patients. Clin Microbiol Infect. 2020;26(8):1082–1087. doi: 10.1016/j.cmi.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman J.D. Evaluation of SARS-CoV-2 serology assays reveals a range of test performance. Nat Biotechnol. 2020;38(10):1174–1183. doi: 10.1038/s41587-020-0659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikramaratna P. Estimating false-negative detection rate of SARS-CoV-2 by RT-PCR. medRxiv. 2020 doi: 10.2807/1560-7917.ES.2020.25.50.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter A.K., Hegde S.T. The important role of serology for COVID-19 control. Lancet Infect Dis. 2020;20(7):758–759. doi: 10.1016/S1473-3099(20)30322-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . 2020. WHO Coronavirus Disease (COVID-19) Dashboard. Available from: https://covid19.who.int/ [cited 1 July 2020] [Google Scholar]
- Zhao J. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.