Abstract
Background
Stripe rust (yellow rust) is a significant disease for bread wheat (Triticum aestivum L.) worldwide. A genome-wide association study was conducted on 240 Chinese wheat cultivars and elite lines genotyped with the wheat 90 K single nucleotide polymorphism (SNP) arrays to decipher the genetic architecture of stripe rust resistance in Chinese germplasm.
Results
Stripe rust resistance was evaluated at the adult plant stage in Pixian and Xindu in Sichuan province in the 2015–2016 cropping season, and in Wuhan in Hubei province in the 2013–2014, 2016–2017 and 2018–2019 cropping seasons. Twelve stable loci for stripe rust resistance were identified by GWAS using TASSEL and GAPIT software. These loci were distributed on chromosomes 1B, 1D, 2A, 2B, 3A, 3B, 4B (3), 4D, 6D, and 7B and explained 3.6 to 10.3% of the phenotypic variation. Six of the loci corresponded with previously reported genes/QTLs, including Sr2/Yr30/Lr27, while the other six (QYr.hbaas-1BS, QYr.hbaas-2BL, QYr.hbaas-3AL, QYr.hbaas-4BL.3, QYr.hbaas-4DL, and QYr.hbaas-6DS) are probably novel. The results suggest high genetic diversity for stripe rust resistance in this population. The resistance alleles of QYr.hbaas-2AS, QYr.hbaas-3BS, QYr.hbaas-4DL, and QYr.hbaas-7BL were rare in the present panel, indicating their potential use in breeding for stripe rust resistance in China. Eleven penta-primer amplification refractory mutation system (PARMS) markers were developed from SNPs significantly associated with seven mapped QTLs. Twenty-seven genes were predicted for mapped QTLs. Six of them were considered as candidates for their high relative expression levels post-inoculation.
Conclusion
The resistant germplasm, mapped QTLs, and PARMS markers developed in this study are resources for enhancing stripe rust resistance in wheat breeding.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12870-020-02693-w.
Keywords: Marker-trait association, Single nucleotide polymorphism (SNP), Triticum aestivum, Yellow rust
Background
Stripe rust (yellow rust), incited by obligate biotroph fungus Puccinia striiformis Westend. f. sp. tritici (Pst), is a significant disease of bread wheat (Triticum aestivum L.) worldwide. An outbreak will fast destroy green leaves and, in turn, dramatically reduce photosynthesis, resulting in stunted and weakened plants, reduced grain numbers per spike, shriveled grains, and lower grain weights. Grain yield losses of 100% can occur in fields sown to susceptible cultivars [1]. Annual grain yield losses caused by this disease are currently estimated to be 5.47 million tons [2].
The use of resistant cultivars is an environmentally friendly, economical, and effective way to manage this disease [3]. Thus, genetic studies on stripe rust resistance are of great significance. Host resistance is categorized into seedling or all-stage resistance and adult-plant resistance (APR) [4]. To date, 82 stripe rust resistance genes have been permanently designated [5], and many more have been given temporary names or quantitative trait locus (QTL) designations [6]. These genes were summarized in 2013 to more than 140 QTLs identified in various studies [7]. Out of the identified resistance genes, eight genes were isolated (Yr36 [8], Yr18 [9], Yr10 [3], Yr46 [10], Yr5/YrSP, Yr7 [11], Yr15 [12], and YrAS2388R [13]).
With the co-evolution of host and pathogen, epidemics of stripe rust follow a boom-and-bust cycle. Extensive use of single resistance genes usually leads to the emergence of new virulent Pst races, followed by subsequent disease epidemics. The breakdown of resistance genes Yr1 in Bima 1 and Yr9 in 1BL.1RS wheat cultivars caused huge economic losses in China’s wheat production [6]. Continuous gene discovery is urgently needed to enrich the resistance gene diversity to slow the boom-and-bust cycle. For the longer-term, quantitative resistances controlled by minor QTLs are encouraged to be pyramided to achieve broad-spectrum and durable resistance to stripe rust.
As a powerful tool for QTL mining, genome-wide association studies (GWAS) have been widely used for gene discovery of multiple traits of wheat, including stripe rust resistance in a worldwide collection of spring wheat landraces [14], Ethiopian wheat [15], International Maize and Wheat Improvement Center (CIMMYT) elite wheat [16], European winter wheat [17], northern Chinese wheat landraces [18], and Sichuan wheat [19].
In this study, we undertook a GWAS on stripe rust resistance in 240 wheat accessions to: 1) study the phenotypic variance of stripe rust response and 2) detect the genetic loci underlying the stripe rust resistance. The results should help to understand the genetic basis of stripe rust resistance in Chinese modern wheat cultivars and facilitate the improvement of stripe rust resistance through marker-assisted selection (MAS).
Results
Phenotypic variation
The best linear unbiased predictor (BLUP) values of stripe rust maximum disease severity (MDS) across five environments ranged from 8.8 to 89.1%, with an average of 49.8% (Additional file 1; Additional file 2), presenting a wide range of disease responses across the population. The levels of stripe rust symptoms varied across environments. The mean stripe rust MDS of the GWAS population were 43.6, 34.9, 71.2, 58.1 and 41.0% in Wuhan 2014, Wuhan 2017, Wuhan 2019, Xindu 2016, and Pixian 2016, respectively. The highest MDS was observed in Wuhan 2019 and disease severity in Wuhan 2017 was lighter than other environments. This might be attributed to not only the different weather conditions during inoculation and disease development but also the amount of pathogen in the natural environments. The variation of symptom levels might cause differences in the frequency distributions of the stripe rust MDS. Analysis of variance (ANOVA) revealed highly significant differences among genotypes, environments, and genotype × environment interactions (Table 1). Broad-sense heritability (H2) for MDS was estimated to be 0.91, indicating less environmental variance than genotypic variance; thus, this panel was suitable for further genetic analysis. High Pearson’s correlation coefficients (0.62–0.79) of stripe rust MDS were observed across five environments (Fig. 1). The highest correlation coefficient was observed between Pixian 2016 and Xindu 2016, and the lowest was between Wuhan 2017 and Xindu 2016.
Table 1.
Analysis of variance of stripe rust severity of 240 wheat accessions
| Source | Df | Sum squares | Mean squares | P value |
|---|---|---|---|---|
| Genotype | 239 | 1,420,723.0 | 5944.4 | < 0.0001 |
| Environment | 4 | 412,868.0 | 103,217.0 | < 0.0001 |
| Genotype×Environment | 912 | 494,173.6 | 541.9 | < 0.0001 |
| Replication/Year | 5 | 26,315.7 | 5263.1 | < 0.0001 |
H2 = 0.91
Fig. 1.
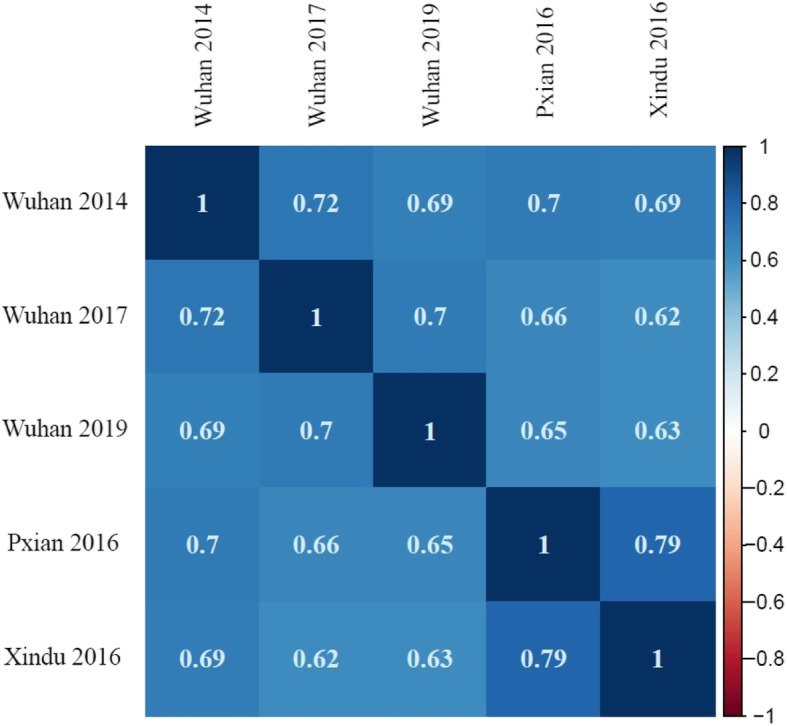
The pearson correlation coefficients of stripe rust severity in the natural populaiton among five environments
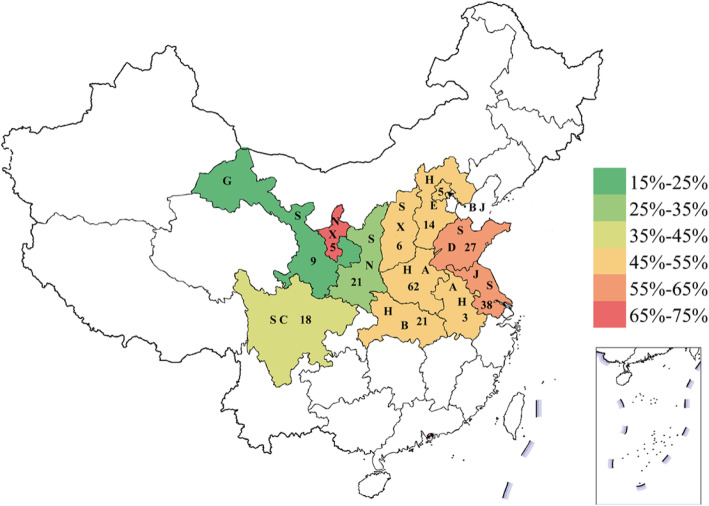
Cultivars from Gansu had the highest stripe rust resistance with an average MDS of 20.0%, followed by Shaanxi and Sichuan at 35.4 and 37.0%, respectively. The cultivars from Henan (55.1%), Shandong (57.5%), and Jiangsu (59.5%) tended to be more susceptible (Fig. 2). CIMMYT lines presented good resistance to stripe rust with an average MDS of 28.6%.
Fig. 2.
Sample numbers and averaged stripe rust severity of wheat accessions from different provinces of China. BJ, Beijing, 52.43%; HE, Hebei, 51.96%; SD, Shandong, 57.54%; SX, Shanxi, 51.04%; SN, Shaanxi, 35.38%; NX, Ningxia, 68.39%; GS, Gansu, 19.95%; JS, Jiangsu, 59.47%; AH, Anhui, 49.45%; HA, Henan, 55.06%; HB, Hubei, 49.86%; SC, Sichuan, 37.00%. Numeral in each province represents the number of cultivars (lines) sampled. Different colors represent corresponding stripe rust severity according to the legend. Map of China was obtained from http://bzdt.ch.mnr.gov.cn
Marker coverage, genetic diversity, and population structure
After removing SNPs (single nucleotide polymorphism) with minor allele frequency (MAF) < 5% and missing rate > 20%, 14,578 SNPs were used for subsequent analyses. Of these, 5778 (39.6%), 6588 (45.2%), and 2212 (15.2%) were from the A, B and D genomes. The A (1.2 SNPs/Mb) and B (1.3 SNPs/Mb) genomes had higher marker density than the D (0.5 SNP/Mb) genome. Similarly, the A and B genomes had higher genetic diversity and polymorphism information content (PIC) values than the D genome (Additional file 3). A weak kinship was observed among cultivars in the association population [20]. The population was grouped into three sub-populations. The result was consistent with the geographic origin and pedigree. Most of the samples from Zone I (Northern Winter Wheat Zone), II (Yellow and Huai River Valleys Facultative Wheat Zone), VIII (Northwestern Spring Wheat Zone), and CIMMYT were grouped in sub-population I (Additional file 4), most of the Zone IV (Southwestern Autumn-Sown Spring Wheat Zone) cultivars in sub-population II, and the majority from Zone III in sub-population III. Some of the accessions from Zone II were also allocated to subgroups II and III indicating the germplasm exchange among different agro-ecological zones.
Maker-trait associations (MTAs) and geographical distribution of favorable alleles
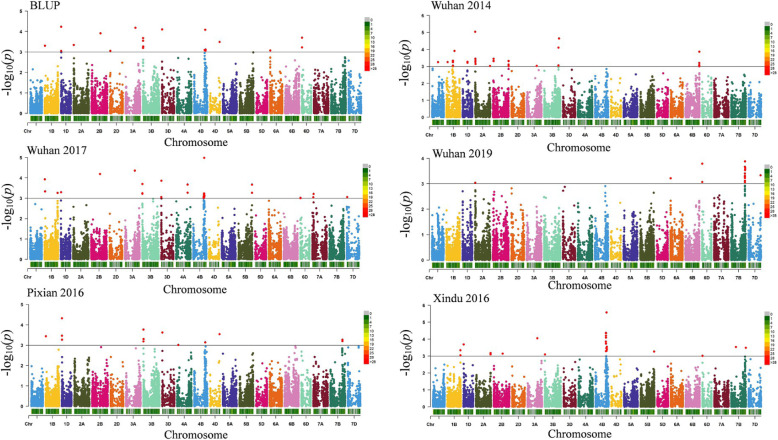
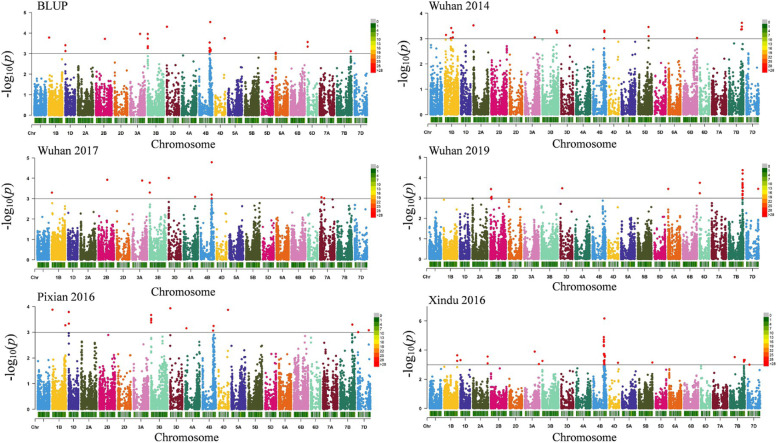
Twelve stable loci for stripe rust resistance were identified (Table 2). These loci were distributed on chromosomes 1B, 1D, 2A, 2B, 3A, 3B, 4B (3), 4D, 6D, and 7B, explaining 3.6 to 10.3% of the phenotypic variation. Each locus was detected in two or more environments. Except for the stable loci, twenty-one QTLs were detected only in one environment on chromosomes 1B (3), 2A (2), 3B, 4A (2), 4B (4), 5B, 6B, 7B (6), and 7D (Additional file 5). TASSEL and Genomic Association and Prediction Integrated Tool (GAPIT) produced similar results (Fig. 3; Fig. 4). In contrast, more MTAs were detected by TASSEL than by GAPIT. The Q-Q plots obtained from the mixed linear model (MLM) in TASSEL v5.2.53 and GAPIT showed a well-controlled false-positive in this study, indicating the reliability of the MTAs (Additional file 6; Additional file 7).
Table 2.
QTLs for stripe rust resistance detected by association study using TASSEL and GAPIT
| QTL | Representative SNP | Position (Mb) | Allele a | Environment | Method | P-value | R2 b(%) |
|---|---|---|---|---|---|---|---|
| QYr.hbaas-1BS | IWB45438 | 9.7 | G/A | BLUP | GAPIT | 1.65E-04 | 4.7 |
| Pixian 2016 | GAPIT | 1.33E-04 | 5.5 | ||||
| Wuhan 2017 | GAPIT | 5.09E-04 | 4.3 | ||||
| BLUP | TASSEL | 4.94E-04 | 5.7 | ||||
| Pixian 2016 | TASSEL | 3.65E-04 | 6.2 | ||||
| Wuhan 2017 | TASSEL | 1.17E-04 | 6.7 | ||||
| QYr.hbaas-1DS | IWA1787 | 8.6 | C/T | BLUP | TASSEL | 9.00E-04 | 4.9 |
| Pixian 2016 | TASSEL | 3.43E-04 | 6 | ||||
| QYr.hbaas-2AS | IWB25308 | 13.8 | C/T | BLUP | TASSEL | 4.58E-04 | 5.5 |
| Wuhan 2014 | TASSEL | 5.54E-04 | 6 | ||||
| Wuhan 2019 | TASSEL | 9.11E-04 | 4.8 | ||||
| QYr.hbaas-2BL | IWA586 | 453.3 | T/C | BLUP | GAPIT | 1.91E-04 | 4.6 |
| Wuhan 2017 | GAPIT | 1.19E-04 | 5.3 | ||||
| BLUP | TASSEL | 1.23E-04 | 6.5 | ||||
| Wuhan 2017 | TASSEL | 6.44E-05 | 6.9 | ||||
| Xindu 2016 | TASSEL | 7.13E-04 | 5 | ||||
| QYr.hbaas-3AL | IWB70396 | 502.8 | A/G | BLUP | GAPIT | 1.10E-04 | 5 |
| Wuhan 2014 | GAPIT | 8.97E-04 | 4.4 | ||||
| Wuhan 2017 | GAPIT | 1.29E-04 | 5.2 | ||||
| Xindu 2016 | GAPIT | 1.27E-04 | 5.4 | ||||
| BLUP | TASSEL | 6.60E-05 | 7.4 | ||||
| Wuhan 2014 | TASSEL | 9.30E-04 | 5.3 | ||||
| Wuhan 2017 | TASSEL | 4.39E-05 | 7.4 | ||||
| Xindu 2016 | TASSEL | 8.84E-05 | 7.1 | ||||
| QYr.hbaas-3BS | IWB12253 | 9.1 | C/T | BLUP | GAPIT | 4.40E-04 | 4.1 |
| Pixian 2016 | GAPIT | 2.98E-04 | 4.9 | ||||
| Xindu 2016 | GAPIT | 5.50E-04 | 4.4 | ||||
| BLUP | TASSEL | 5.39E-04 | 5.2 | ||||
| Pixian 2016 | TASSEL | 4.98E-04 | 5.5 | ||||
| Xindu 2016 | TASSEL | 8.05E-04 | 4.9 | ||||
| QYr.hbaas-4BL.1 | IWB73717 | 531.3 | C/T | BLUP | GAPIT | 8.36E-04 | 3.7 |
| Xindu 2016 | GAPIT | 2.79E-05 | 6.6 | ||||
| Xindu 2016 | TASSEL | 8.16E-05 | 6.9 | ||||
| QYr.hbaas-4BL.2 | IWB63337 | 558.1 | T/C | Wuhan 2017 | GAPIT | 6.44E-04 | 4.1 |
| Xindu 2016 | GAPIT | 9.20E-04 | 4 | ||||
| Wuhan 2017 | TASSEL | 5.80E-04 | 5.1 | ||||
| QYr.hbaas-4BL.3 | IWB32927 | 579.4 | T/C | BLUP | GAPIT | 6.35E-04 | 3.9 |
| Wuhan 2014 | GAPIT | 4.80E-04 | 4.9 | ||||
| Xindu 2016 | GAPIT | 2.91E-04 | 4.9 | ||||
| BLUP | TASSEL | 7.67E-04 | 5.2 | ||||
| Xindu 2016 | TASSEL | 3.88E-04 | 5.8 | ||||
| QYr.hbaas-4DL | IWB44356 | 477.9 | A/C | BLUP | GAPIT | 1.77E-04 | 4.7 |
| Pixian 2016 | GAPIT | 1.35E-04 | 5.5 | ||||
| BLUP | TASSEL | 3.24E-04 | 5.9 | ||||
| Pixian 2016 | TASSEL | 2.87E-04 | 6.2 | ||||
| QYr.hbaas-6DS | IWB60233 | 6.0 | A/G | BLUP | GAPIT | 4.57E-04 | 4.1 |
| Wuhan 2019 | GAPIT | 5.82E-04 | 4.2 | ||||
| BLUP | TASSEL | 5.97E-04 | 5.2 | ||||
| Wuhan 2017 | TASSEL | 9.68E-04 | 4.7 | ||||
| Wuhan 2019 | TASSEL | 8.57E-04 | 4.9 | ||||
| QY.rhbaas-7BL | IWB64121 | 704.3 | T/C | BLUP | GAPIT | 7.65E-04 | 3.8 |
| Wuhan 2019 | GAPIT | 2.24E-04 | 4.8 | ||||
| Xindu 2016 | GAPIT | 6.17E-04 | 4.3 | ||||
| Wuhan 2019 | TASSEL | 4.90E-04 | 5.5 |
a Resistance alleles are underlined; b Phenotypic variation explained by the QTL
Fig. 3.
Manhattan plots for stripe rust severity of 240 wheat accessions performed by the mixed linear model with Tassel. The horizontal line indicates the threshold for significance
Fig. 4.
Manhattan plots for stripe rust severity of 240 wheat accessions performed by the mixed linear model with GAPIT. The horizontal line indicates the threshold for significance
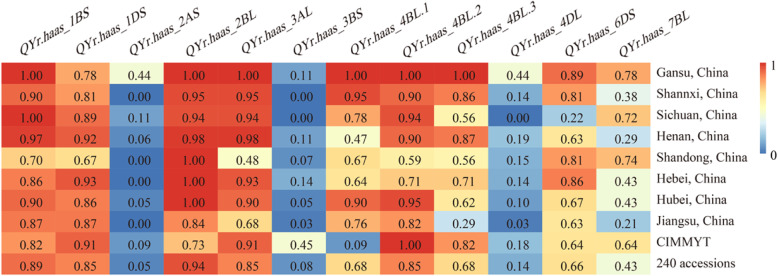
The favorable alleles of QYr.hbaas-2AS, QYr.hbaas-3BS, QYr.hbaas-4DL, and QYr.hbaas-7BL were rare in the present panel, with frequencies ranging from 0.05 to 0.43. The frequencies of the other seven QTLs were above 0.60. Relatively higher favorable allele frequencies (0.44) of QYr.hbaas-2AS and QYr.hbaas-4DL were observed in Gansu cultivars, compared with 0–0.19 in cultivars from other provinces. The favorable allele of QYr.hbaas-3BS was rare in Chinese wheat (0–0.14) compared with lines from CIMMYT (0.45). In contrast, QYr.hbaas-4BL.1 was widely adapted in Chinese wheat but rare in CIMMYT lines (Fig. 5).
Fig. 5.
Allelic frequencies of the 12 stable QTLs in wheat cultivars from different provinces in China and CIMMYT lines. Provinces with number of wheat cultivars less than 9 were not shown
Fifty-one and 192 wheat accessions from the GWAS panel were speculated to contain Yr62 and Yr64, respectively based on linked SSR markers analysis (Additional file 1). Six and 52 wheat accessions were tested to contain YrSP and Yr7, respectively using gene-specific markers. None of the wheat samples contains Yr5 and Yr15 indicating a lot of room for stripe rust resistance improvement in China.
Relationship between stripe rust MDS and the number of favorable alleles
To illustrate the pyramiding effects of favorable alleles in different QTLs, we examined the number of favorable alleles of 12 mapped loci in each accession. The number of favorable alleles ranged from 1 to 12 (Additional file 1). Linear regression (r2 = 0.87) showed the dependence of disease severity on the number of favorable alleles (Fig. 6). Accessions with more favorable alleles, such as Lantian 15 (11 favorable alleles), Lantian 26 (11), Lantian 21 (10), Lantian 12 (10), and Zhongmai 12 (10) exhibited strong stripe rust resistance.
Fig. 6.
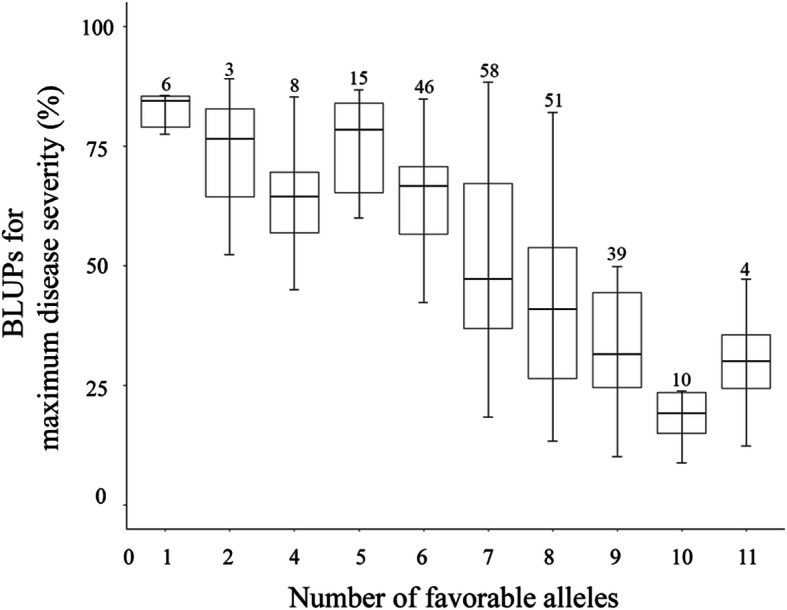
Averaged stripe rust maximum disease severity of lines with different number of favorable alleles of mapped QTLs. Arabic numerals on the bars indicates the number of accessions corresponding to each class
PCR-based markers for mapped loci
A set of 11 penta-primer amplification refractory mutation system (PARMS) markers were successfully developed to detect the presence of stripe rust resistance QTLs (Additional file 8). Primers for these 11 markers are given in Additional file 9, and the protocols are described in Additional file 10. Developed markers were validated using the 240 GWAS accessions; the results produced very low frequencies of inconsistency (3.7–5.9%) with chip data.
Candidate genes predicted
With TPMs (transcripts per kilobase million) above 0.5, three genes encoding NBS-LRR (nucleotide-binding site-leucine-rich repeat) proteins and two genes encoding receptor-like kinase (RLK) were identified in the region of QYr.hbaas-1BS (Additional file 11). Genes encoding disease resistance protein RPM1 and NBS-LRR might be candidates for QYr.hbaas-1DS. In the QYr.hbaas-2AS region, we identified four NBS-LRR genes. A TIR-NBS-LRR gene was found near QYr.hbaas-2BL. Kinase genes might be candidates for QYr.hbaas-3AL, QYr.hbaas-4BL.3, and QYr.hbaas-4DL. Two RLK genes were also identified at QYr.hbaas-4DL. Four genes encoding glycosyltransferase and an ATP-binding cassette gene were identified at QYr.hbaas-3BS. For QYr.hbaas-6BS, we identified three RLK genes and one gene encoding stress response NST1-like protein. The highest relative expression levels of these candidate genes ranged from 0.7 to 6.0. Compared to the expression at 0 hpi, the highest relative expression levels of TraesCS1B01G020600, TraesCS1B01G020900 (QYr.hbaas-1BS), TraesCS3B01G021100, TraesCS3B01G022000 (QYr.hbaas-3BS), TraesCS6D01G013600, TraesCS6D01G014300 (QYr.hbaas-6DS) exceeded 1.5 which indicates their strong candidacy.
Discussion
Phenotypic variation and genetic diversity of stripe rust resistance
The levels of resistance to stripe rust in different provinces could be assessed with the mean stripe rust MDS. Samples from Gansu, Shaanxi, and Sichuan had the highest levels of resistance to stripe rust in China. This result is in accordance with the severe occurrence of stripe rust and substantial resistance-breeding efforts in these three provinces.
Additionally, CIMMYT germplasm showed high level of resistance to stripe rust. This is partly due to the long history of breeding for durable resistance to stripe rust at CIMMYT. The pathologists and wheat breeders from CIMMYT have focused on partial resistance for more than 30 years. This resistance source has been successfully used in the spring wheat region in China, such as Sichuan province [19]. The different resistance backgrounds of Chinese wheat in the CIMMYT germplasm suggest its usefulness in wheat breeding in China.
According to the favorable alleles frequency analysis, resistant stocks from Gansu, Shaanxi, Sichuan and CIMMYT carry different resistance loci. The high resistance level and diversity of resistance genes make them valuable sources of stripe rust resistance in breeding. This diversity also allows wheat breeders to pyramid various QTLs to achieve high level, broad-spectrum, and even durable resistance to stripe rust.
Novelty of the mapped QTLs
QYr.hbaas-1BS was mapped at the distal region of 1BS (9.7 Mb). This chromosome arm is rich with stripe rust resistance genes/QTLs, including Yr64 [21] and Yr15 [12]. Most of the previously mapped loci are far from QYr.hbaas-1BS based on physical position (Additional file 12). The genotype of linked marker for Yr64 is inconsistent with QYr.hbaas-1BS suggesting they are not the same loci. QYr.hbaas-1BS is different from Yr15 either as no sample in the GWAS panel was detected to contain this gene based on gene-specific markers. Zegeye et al. [22] detected a QTL on 1BS (9.1 Mb) for stripe rust resistance by GWAS in synthetic hexaploid wheat, which is close to QYr.hbaas-1BS. However, this resistance source is unlikely to be widely adopted in Chinese wheat, and might differ from QYr.hbaas-1BS as the frequency of QYr.hbaas-1BS is very high in this wheat panel. Thus, QYr.hbaas-1BS is considered a new locus for stripe rust resistance.
QYr.hbaas-1DS overlapped QYr.nwafu-1DS.1, QYrst.orr-1DS, and QYr.sun-1D [23–25] with distances of 6.1, 8.2, and 8.3 Mb, respectively. Further studies are needed to clarify the relationships among the these QTLs on 1DS (Additional file 12).
Numerous genes/QTLs for stripe rust resistance on chromosome 2AS (Additional file 12) have been reported. Most of them are located in a 3.8–19.1 Mb interval, including Traes_2AS_6BC67DD45, Traes_2AS_A477CDA77, Traes_2AS_6CE6AB560 [16], Yr2A.1PBL, Yr2A.2PBL, Yr2A.3PBL, Yr2A.4PBL [26], QYr.tam-2A [27], QYr.tsw-2A.3 [28], YrCH86 [29], YrZM175 [30], QYr.sun-2A [31], QYr.ufs-2A [32], YrR61 [33] and a QTL linked to IWB57199 [15]. QYr.hbaas-2AS mapped in the present study is also located in this region.
On 2BL, Yr5/YrSP, Yr7 [34, 35], and many other loci were mapped or isolated (Additional file 12). Among them, a QTL associated with wsnp_JD_c744_1111659 is the closest to QYr.hbaas-2BL, with a distance of 126.7 Mb. The distinct locations between QYr.hbaas-2BL and other reported loci on 2BL indicated that QYr.hbaas-2BL is a new QTL for stripe rust resistance. The genotypes of gene-specific markers for Yr5, YrSP, and Yr7 were distinct from QYr.hbaas-2BL further indicating their difference.
QYr.dms-3A [36], QYrPI182103.wgp-3AL [37], QYrdr.wgp-3AL [38], QYr.nwafu-3AL [23], QTLs linked to wsnp_JD_c14691_14352459 [39], IWB11855 [40], and wsnp_RFL_Contig4814_5829093 [22] were mapped on 3AL (Additional file 12). Of these, the QTL linked to wsnp_JD_c14691_14352459 is the closest to QYr.hbaas-3AL, with a distance of 30.2 Mb. This distance indicates that QYr.hbaas-3AL might be a new QTL for stripe rust resistance.
QYr.hbaas-3BS is close to a well-characterized pleiotropic locus Sr2/Yr30/Lr27 [41] with a distance of 3 Mb. Sr2 has been extensively used at CIMMYT to achieve durable resistance to multiple diseases. The highest frequency of QYr.hbaas-3BS in the CIMMYT accessions also indicates that this QTL might be Sr2/Yr30/Lr27.
On wheat chromosome 4BL, QYrhm.nwafu-4B [42] and Yr62 [43] overlapped QYr.hbaas-4BL.1 and QYr.hbaas-4BL.2, whereas no reported QTL was associated with QYr.hbaas-4BL.3 based on physical position. Thus, QYr.hbaas-4BL.3 is considered a new locus for stripe rust resistance. However, the genotypes of the representative SNPs for QYr.hbaas-4BL.1 and QYr.hbaas-4BL.2 were not consistent with the linked marker Xgwm251 for Yr62 with very low R2.
On 4DL, a QTL linked to Xwmc399 (484.7 Mb) in oligoculm wheat [44], was close to QYr.hbaas-4DL. Most of the varieties used in China come from CIMMYT, while the oligoculm variety is from Israel; it is unlikely that oligoculm is used in China, so this QTL may not be the QTL associated in this study. Although the two QTLs were mapped to similar regions, they appear to have different origins. Thus, QYr.hbaas-4DL is probably new.
The QTL linked with IWA167 [45], QYr.sicau-6D [46], and QYr.ufs-6D [32] were mapped on chromosome 6DS. None was close to QYr.hbaas-6DS, indicating that QYr.hbaas-6DS is probably a new locus.
Several stripe rust QTLs were mapped near QYr.hbaas-7BL, including QYr.nwafu-7BL [47], Qyr.saas-7B [48], Qyrsicau-7BL [19], QYr.nwafu-7BL [23], QYr.caas-7BL.2 [49], and a QTL flanked by wPt-4342 and wPt-8921 [50].
Application of resistant germplasm and MTAs in breeding for stripe rust resistance
The GWAS population represents a large proportion of the diversity of recently cultivated wheat in China. Fifty-four cultivars from the population have achieved a peak annual acreage of 1 × 105 ha in the period 2000–2016 (http://202.127.42.47:6006/Home/BigDataIndex). Nine of them show good resistance to stripe rust with BLUE values of MDS under 30%. Interestingly, five of the nine resistant cultivars, Zhoumai 18, Zhoumai 22, Aikang 58, Jimai 22, and Liangxing 99, have also been used as founder parents in corresponding regions in recent years. This indicates that resistance is vital for parent selection in stripe rust-prone areas.
Average stripe rust MDS decreases dramatically when QTL number increases according to the regression of QTL number and the phenotype with a slop of − 5.4. This result indicates a promising effect of detected QTLs for stripe rust resistance. However, validations are needed in future studies. To further validate the effect of the mapped QTLs, the linked markers can be tested in different natural and bi-parental populations. Resistant accessions with potentially new QTLs could be used to produce bi-parental populations for further studies.
Grain yield and end-use quality are the main objectives for most breeding programs, however, resistance or tolerance to abiotic and biotic stresses is also a major concern for grain yield stabilization. As a minor QTL alone does not provide adequate resistance, pyramiding minor loci is necessary to achieve an acceptable level of resistance. For the long term, resistance QTLs are worth being gradually pyramided into founder parents for further improvement in stripe rust prevalent regions. PARMS markers developed in the present study will expedite this procedure with advantages of high throughput, low cost, and stability. Furthermore, the DNA template for PARMS can be prepared with alkaline lysis, which is convenient and time-saving [51]. In the stripe rust prevalent area, minor QTLs can also be pyramided through phenotypic selection which was verified to be feasible at CIMMYT [52]. Further, well-characterized resistance genes Yr5 and Yr15 effective against prevalent Chinese Pst races have not been incorporated in Chinese wheat cultivars. Combining these genes with minor QTLs will be another solution for stripe rust resistance breeding.
When selecting the resistance alleles of stripe rust QTLs through MAS, a large part of the respective chromosome will be fixed as identical by descent (IBD) which means the surrounding genes from the donor will be introduced along with the target QTLs. From this point of view, the resistant donors should be carefully selected not only for the target trait but also the agronomic performance to minimize linkage drag. Compared with introduced germplasm, using widely planted cultivars in the corresponding region may lower the risks of linkage drag. To avoid narrowing down the genetic diversity of stripe rust resistance, introducing resistant cultivars from other agro-ecological regions can enrich the resistance gene pool. When using these unadapted donors, 1–2 backcross with large populations will help getting rid of undesired traits.
Breeding for quantitative resistance is a challenge due to its complex inheritance. Genomic selection can facilitate parental selection, potentially accurately predict the phenotypes, and increase the genetic gain. Mean prediction accuracies reached 0.34 to 0.71 for stripe rust APR using different genomic prediction models [53] indicating its effectiveness in wheat breeding for stripe rust resistance. Markers for validated QTLs and well-characterized genes can used as fixed effects in genomic selection models to increases the prediction accuracy.
Conclusions
In the present study, we identified 12 stable stripe rust resistance loci using the wheat 90 K SNP assay. Six stripe rust resistance loci QYr.hbaas-1BS, QYr.hbaas-2BL, QYr.hbaas-3AL, QYr.hbaas-4BL.3, QYr.hbaas-4DL, and QYr.hbaas-6DS are probably novel. Eleven PARMS markers associated with seven mapped QTLs were developed. The resistant germplasm, mapped QTLs, and developed PARMS markers could be employed in stripe rust resistance breeding.
Methods
Plant materials
The wheat association panel comprises 240 geographically diverse cultivars and elite lines (Additional file 1; Fig. 2), of which, 229 were from 12 provinces from China: Henan (62), Jiangsu (38), Shandong (27), Hubei (21), Shannxi (21), Sichuan (18), Hebei (14), Gansu (9), Shanxi (6), Beijing (5), Ningxia (5), and Anhui (3). This collection represents the currently cultivated wheat cultivars in China with more than 50 cultivars planted on at least 100,000 ha per year, including predominant cultivars Aikang 58, Jimai 22, Xinong 979, and Zhoumai 22. These Chinese wheat accessions were collected by Lijun Yang and Zhanwang Zhu from corresponding institutions with their permissions. The other 11 are ten elite lines from CIMMYT and an Australian cultivar, which were provided by David Bonnett under permission from CIMMYT.
Disease assessment
All 240 accessions were evaluated for their response to stripe rust at the adult plant stage in Pixian (30°05′N, 102°54′E) and Xindu (30°83′N, 104°15′E) in Sichuan province, China, during the 2015–2016 cropping season (hereafter referred to as Pixian 2016 and Xindu 2016, respectively) and the Nanhu Experimental Station (30°48′N, 114°32′E) in Wuhan in Hubei province, China, during the 2013–2014, 2016–2017, 2018–2019 cropping seasons (noted as Wuhan 2014, Wuhan 2017, and Wuhan 2019, respectively). The field trials were conducted with randomized complete blocks with two replications. Each plot was sown in 1 m long paired rows. Stripe rust susceptible cultivar Mingxian 169 was planted around the nurseries as a disease spreader to facilitate uniform disease development. The disease spreader was inoculated with a mixture of prevalent Pst races CYR 32 and CYR 33 in the 2013–2014 cropping season in Wuhan and CYR 32 and CYR 34 in the other environments at the wheat shooting stage. CYR34 is the most virulent strain documented in China with an avirulence/virulence formula of Yr5, Yr15 / Yr1, Yr2, Yr3, Yr4, Yr6, Yr7, Yr8, Yr9, Yr10, Yr17, Yr18, Yr24/Yr26, Yr25, Yr27, Yr29, Yr30, Yr32, YrSP, YrA, YrSk. CYR32 and CYR33 are avirulent to Yr10 and Yr24/Yr26 besides Yr5 and Yr15 compared with CYR34 [54]. Field studies were conducted in accordance with local practices.
The stripe rust response was scored as MDS when the severity of the susceptible cultivar Mingxian 169 reached the maximum level. The MDS under five environments and the BLUP across environments (hereafter referred to as BLUP) were used for subsequent analysis.
Statistical analysis
The correlation coefficients were calculated using corrgram (https://github.com/kwstat/corrgram) package in R language (www.r-project.org). ANOVA was conducted with the ANOVA function in IciMapping v4.1 [55]. Broad-sense heritability (H2) for different traits was calculated using the following formula: H2 = /( + /e + /(re)), where is genotypic effect, is genotype by environment effect, is residual error, e is number of environments, and r is number of replications. BLUP values were calculated using ‘lme4’ package with R.
Genotyping
The 240 wheat accessions were genotyped with an Illumina 90 K SNP array [56]. Calling and filtering of SNPs, kinship, and population structure analysis are described in our previous study [20]. Physical positions of SNPs were referred to the Chinese Spring reference genome sequences RefSeq v1.0 (http://www.wheatgenome.org). Genetic diversity [57], PIC, and MAF were computed by PowerMarker v3.25 [58].
Linked SSR markers Xgwm251 [43] and Xgdm33 [59] were used to test Yr62 and Yr64, respectively in the GWAS panel. The PCR products were detected by 12% polyacrylamide gel electrophoresis. Gene-specific markers Y15K1 [12] for Yr15 and Yr5-Insertion for Yr5 [11] were used to test corresponding genes. The PCR product was detected by 1.0% agarose gel electrophoresis. Linked KASP marker Sr2_ger9 3p was used to test Yr30 [60]. Gene-specific KASP markers Yr7-A, Yr7-D for Yr7 and YrSP for YrSP [11] were used to test corresponding genes.
Genome-wide association analysis
GWAS was conducted using the MLM (PCA + K) in TASSEL v5.2.53 (http://www.maizegenetics.net/tassel) and GAPIT (http://zzlab.net/GAPIT). Due to the higher extent of linkage disequilibrium (LD) in wheat and the complex genetic architecture of stripe rust resistance, markers with an adjusted –log10 (P) ≥ 3.0 were regarded as significant for the trait. This threshold was also used in previous GWAS on stripe rust in bread wheat [17–19, 61]. Loci significant in at least two environments were reported in the results and considered stable QTLs. ‘CMplot’ package (https://github.com/YinLiLin/R-CMplot) was used to draw the Manhattan plots and quantile-quantile (Q-Q) plots with R. Alleles leading to lower MDS were referred to as favorable, whereas those leading to higher MDS were unfavorable. The frequencies of favorable alleles and their allelic effects were calculated based on the representative SNPs associated with the resistance loci.
Development of PARMS markers for significant SNPs
To facilitate the use of mapped QTLs, significantly associated SNPs were developed as PARMS markers [51]. Primers were designed using PolyMarker (http://polymarker.tgac.ac.uk). Fluorescence signals were screened with PHERAstarPlus (BMG LABTECH) and analyzed with KlusterCaller (LGC Genomics).
Prediction of candidate genes
Annotated high confidence genes with a distance no more than 1 Mb from the representative SNPs of mapped QTLs were examined to consider the candidacy. The annotations of high confidence genes were referred to IWGSC RefSeq Annotation v1.0 (www.wheatgenome.org). Expressions of disease-related genes were analyzed in the public wheat expression database Triticeae Multi-omics Center (http://202.194.139.32) using the study performed by Zhang et al. (2014) [62]. Genes with a TPM > 0.5 were reported as the candidate genes for mapped QTLs. The highest expressions of these genes post-inoculation were also compared to those before inoculation to further confirm their candidacy.
Supplementary information
Additional file 1. Names and origins of 240 wheat accessions.
Additional file 2. Frequency distributions for stripe rust maximum disease severity (MDS) of 240 wheat accessions in five environments.
Additional file 3. Marker density of the 240 wheat accessions genotyped with the 90 K SNP arrary.
Additional file 4. Principal components analysis (PCA) plots of 240 wheat accessions.
Additional file 5. Quantitative trait loci (QTLs) for stripe rust resistance significant only in one environment detected by genome-wide association study by TASSEL and GAPIT.
Additional file 6. Quantile-quantile plot for stripe rust maximum disease severity in 240 wheat accessions analyzed using the mixed linear model with Tassel.
Additional file 7. Quantile-quantile plot for stripe rust maximum disease severity in 240 wheat accessions analyzed using the mixed linear model with GAPIT.
Additional file 8. Profiles of 11 penta-primer amplification refractory mutation system markers for mapped stripe rust resistance loci.
Additional file 9. Primers for 11 penta-primer amplification refractory mutation system markers for seven stripe rust resistance loci.
Additional file 10. Protocols for 11 developed markers for seven stripe rust resistance loci.
Additional file 11. Candidate genes for stripe rust resistance QTLs.
Additional file 12. Reported QTLs/genes, linked markers and their physical positions used to align with QTLs mapped in the present study.
Acknowledgments
We are grateful to Dr. Chengdao Li from Murdoch University and Dr. Xiwen Cai from North Dakota State University for their critical reviews on this manuscript. We thank Drs. Xianchun Xia, Chengdao Li, Yucai Liao, and Jianbing Yan for their suggestions on designing the experiment.
Abbreviations
- ANOVA
Analysis of variance
- APR
Adult-plant resistance
- BLUP
Best linear unbiased predictor
- CIMMYT
International Maize and Wheat Improvement Center
- GAPIT
Genomic Association and Prediction Integrated Tool
- GWAS
Genome-wide association study
- H2
Broad-sense heritability
- IBD
Identical by descent
- MAF
Minor allele frequency
- MAS
Marker-assisted selection
- MDS
Maximum disease severity
- MLM
Mixed linear model
- MTA
Maker-trait association
- NBS-LRR
Nucleotide-binding site-leucine-rich repeat
- PARMS
Penta-primer amplification refractory mutation system
- PIC
Polymorphism information content
- Pst
Puccinia striiformis f. sp. Tritici
- QTL
Quantitative trait locus
- RLK
Receptor-like kinase
- SNP
Single nucleotide polymorphism
- TPM
Transcripts per kilobase million
Authors’ contributions
ZZ and CG designed the experiment. MJ, JL, and ZZ performed the experiment. LC and WZ contributed to genotyping the wheat panel. LY, GR, EY, WW, YL, HT, WH, and YZ participated in field trials. MJ and ZZ analyzed the data. MJ and ZZ wrote the paper. All authors read the final version of this manuscript and approved it for publication.
Funding
The development of the PARMS (penta-primer amplification refractory mutation system) markers was financially supported by the National Key Research and Development Program of China (2016YFD0101802). Genotyping of the association population with wheat 90 K SNP array was supported by the National Natural Science Foundation of China (31301306). The Technology Innovation Program of Hubei Province (2016AHB022) and the China Agriculture Research System (CARS-03) supported stripe rust resistance assessment in field trials.
Availability of data and materials
The genotypic data generated from the 90 K wheat SNP array for the 240 wheat accessions is in the European Nucleotide Archive, PRJEB36125, https://www.ebi.ac.uk/ena/data/view/PRJEB36125. Other data supporting the results of this publication are included within the article and its additional files.
Ethics approval and consent to participate
We declare that these experiments comply with the ethical standards and legislation in China, and all wheat cultivars or breeding lines were collected in accordance with national and international guidelines.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhanwang Zhu, Email: zhuzhanwang@163.com.
Chunbao Gao, Email: gcbgybwj@163.com.
References
- 1.Chen X. Pathogens which threaten food security: Puccinia striiformis, the wheat stripe rust pathogen. Food Secur. 2020;12:239–251. doi: 10.1007/s12571-020-01016-z. [DOI] [Google Scholar]
- 2.Beddow JM, Pardey PG, Chai Y, Hurley TM, Kriticos DJ, Braun HJ, Park RF, Cuddy WS, Yonow T. Research investment implications of shifts in the global geography of wheat stripe rust. Nat Plants. 2015;1:15132. doi: 10.1038/nplants.2015.132. [DOI] [PubMed] [Google Scholar]
- 3.Liu W, Frick M, Huel R, Nykiforuk CL, Wang X, Gaudet DA, Eudes F, Conner RL, Kuzyk A, Chen Q, Kang Z, Laroche A. The stripe rust resistance gene Yr10 encodes an evolutionary-conserved and unique CC-NBS-LRR sequence in wheat. Mol Plant. 2014;7(12):1740–1755. doi: 10.1093/mp/ssu112. [DOI] [PubMed] [Google Scholar]
- 4.Chen X. Epidemiology and control of stripe rust on wheat. Can J Plant Pathol. 2005;27:314–337. doi: 10.1080/07060660509507230. [DOI] [Google Scholar]
- 5.Pakeerathan K, Bariana H, Qureshi N, Wong D, Hayden M, Bansal U. Identification of a new source of stripe rust resistance Yr82 in wheat. Theor Appl Genet. 2019;132(11):3169–3176. doi: 10.1007/s00122-019-03416-y. [DOI] [PubMed] [Google Scholar]
- 6.McIntosh R, Mu J, Han D, Kang Z. Wheat stripe rust resistance gene Yr24/Yr26: a retrospective review. Crop J. 2018;6(4):3–11. doi: 10.1016/j.cj.2018.02.001. [DOI] [Google Scholar]
- 7.Rosewarne GM, Herrera-Foessel SA, Singh RP, Huerta-Espino J, Lan C, He Z. Quantitative trait loci of stripe rust resistance in wheat. Theor Appl Genet. 2013;126:2427–2449. doi: 10.1007/s00122-013-2159-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu D, Uauy C, Distelfeld A, Blechl A, Epstein L, Chen X, Sela H, Fahima T, Dubcovsky J. A kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science. 2009;323(5919):1357–1360. doi: 10.1126/science.1166289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krattinger SG, Lagudah ES, Spielmeyer W, Singh RP, Huerta-Espino J, McFadden H, Bossolini E, Selter LL, Keller B. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science. 2009;323(5919):1360–1363. doi: 10.1126/science.1166453. [DOI] [PubMed] [Google Scholar]
- 10.Moore JW, Herrera-Foessel S, Lan C, Schnippenkoetter W, Ayliffe M, Huerta-Espino J, Lillemo M, Viccars L, Milne R, Periyannan S, Kong X, Spielmeyer W, Talbot M, Bariana H, Patrick JW, Dodds P, Singh R, Lagudah E. A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat Genet. 2015;47:1494–1498. doi: 10.1038/ng.3439. [DOI] [PubMed] [Google Scholar]
- 11.Marchal C, Zhang J, Zhang P, Fenwick P, Steuernagel B, Adamski NM, Boyd L, McIntosh R, Wulff BBH, Berry S, Lagudah E, Uauy C. Bed-domain-containing immune receptors confer diverse resistance spectra to yellow rust. Nat Plants. 2018;4(9):662–668. doi: 10.1038/s41477-018-0236-4. [DOI] [PubMed] [Google Scholar]
- 12.Klymiuk V, Yaniv E, Huang L, Raats D, Fatiukha A, Chen S, Feng L, Frenkel Z, Krugman T, Lidzbarsky G, Chang W, Jääskeläinen MJ, Schudoma C, Paulin L, Laine P, Bariana H, Sela H, Saleem K, Sørensen CK, Hovmøller MS, Distelfeld A, Chalhoub B, Dubcovsky J, Korol AB, Schulman AH, Fahima T. Cloning of the wheat Yr15 resistance gene sheds light on the plant tandem kinase-pseudokinase family. Nat Commun. 2018;9(1):3735. doi: 10.1038/s41467-018-06138-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang C, Huang L, Zhang H, Hao Q, Lyu B, Wang M, Epstein L, Liu M, Kou C, Qi J, Chen F, Li M, Gao G, Ni F, Zhang L, Hao M, Wang J, Chen X, Luo M, Zheng Y, Wu J, Liu D, Fu D. An ancestral NB-LRR with duplicated 3′UTRs confers stripe rust resistance in wheat and barley. Nat Commun. 2019;10(1):4023. doi: 10.1038/s41467-019-11872-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kankwatsa P, Singh D, Thomson PC, Ebrahiem M, Babiker EM, Bonman JM, Maria NM, Park RF. Characterization and genome-wide association mapping of resistance to leaf rust, stem rust and stripe rust in a geographically diverse collection of spring wheat landraces. Mol Breeding. 2017;37(9):113. doi: 10.1007/s11032-017-0707-8. [DOI] [Google Scholar]
- 15.Muleta KT, Rouse MN, Rynearson S, Chen X, Buta BG, Pumphrey MO. Characterization of molecular diversity and genome-wide mapping of loci associated with resistance to stripe rust and stem rust in Ethiopian bread wheat accessions. BMC Plant Biol. 2017;17(1):134. doi: 10.1186/s12870-017-1082-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Juliana P, Singh RP, Singh PK, Poland JA, Bergstrom GC, Huerta-Espino J, Bhavani S, Crossa J, Sorrells ME. Genome-wide association mapping for resistance to leaf rust, stripe rust and tan spot in wheat reveals potential candidate genes. Theor Appl Genet. 2018;131:1405–1422. doi: 10.1007/s00122-018-3086-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miedaner T, Akel W, Flath K, Jacobi A, Taylor M, Longin F, Würschum T. Molecular tracking of multiple disease resistance in a winter wheat diversity panel. Theor Appl Genet. 2019;133(2):419–431. doi: 10.1007/s00122-019-03472-4. [DOI] [PubMed] [Google Scholar]
- 18.Yao F, Zhang X, Ye X, Li J, Long L, Yu C, Li J, Wang Y, Wu Y, Wang J, Jiang Q, Li W, Ma J, Wei Y, Zheng Y, Chen G. Characterization of molecular diversity and genome-wide association study of stripe rust resistance at the adult plant stage in northern Chinese wheat landraces. BMC Genet. 2019;20(1):38. doi: 10.1186/s12863-019-0736-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye X, Li J, Cheng Y, Yao F, Long L, Yu C, Wang Y, Wu Y, Li J, Wang J, Jiang Q, Li W, Ma J, Wei Y, Zheng Y, Chen G. Genome-wide association study of resistance to stripe rust (Puccinia striiformis f. sp. tritici) in Sichuan wheat. BMC Plant Biol. 2019;19(1):147. doi: 10.1186/s12870-019-1764-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu Z, Chen L, Zhang W, Yang L, Zhu W, Li J, Liu Y, Tong H, Fu L, Liu J, Rasheed A, Xia X, He Z, Hao Y, Gao C. Genome-wide association analysis of Fusarium head blight resistance in Chinese elite wheat lines. Front Plant Sci. 2020;11:206. doi: 10.3389/fpls.2020.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qie Y, Liu Y, Li X, Wang M, See DR, An D, Chen X. Development, validation, and re-selection of wheat lines with pyramided genes Yr64 and Yr15 linked on the short arm of chromosome 1B for resistance to stripe rust. Plant Dis. 2018;103(1):51–58. doi: 10.1094/PDIS-03-18-0470-RE. [DOI] [PubMed] [Google Scholar]
- 22.Zegeye H, Rasheed A, Makdis F. Badebo, Ogbonnaya FC. Genome-wide association mapping for seedling and adult plant resistance to stripe rust in synthetic hexaploid wheat. PLoS One. 2014;9(8):e105593. doi: 10.1371/journal.pone.0105593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu J, Wang X, Chen N, Wang H, Yu R, Yu S, Wang Q, Huang S, Singh R, Bhavani S, Kang Z, Han D, Zeng Q. Association analysis identifies new loci for resistance to Chinese Yr26-virulent races of the stripe rust pathogen in a diverse panel of wheat germplasm. Plant Dis. 2020. 10.1094/PDIS-12-19-2663-RE. [DOI] [PubMed]
- 24.Vazquez DM, Peterson JC, Lizarazu RO, Chen XM, Heesacker A, Ammar K, Crossa J, Mundt CC. Genetic analysis of adult plant, quantitative resistance to stripe rust in wheat cultivar ‘Stephens’ in multi-environment trials. Theor Appl Genet. 2012;124(1):1–11. doi: 10.1007/s00122-011-1681-x. [DOI] [PubMed] [Google Scholar]
- 25.Zwart RS, Thompson JP, Milgate AW, Bansal U, Williamson PM, Raman H, Bariana HS. QTL mapping of multiple foliar disease and root-lesion nematode resistances in wheat. Mol Breeding. 2010;26(1):107–124. doi: 10.1007/s11032-009-9381-9. [DOI] [Google Scholar]
- 26.Ramírez LL, Moya ES, Iturriaga G, Sehgal D, Reyes-Valdes MH, Tavera VM, Sansaloni C, Burgueño J, Ortiz C, Aguirre-Mancilla CL, Ramírez-Pimentel JG, Vikram P, Singh S. GWAS to identify genetic loci for resistance to yellow rust in wheat pre-breeding lines derived from diverse exotic crosses. Front Plant Sci. 2019;10:1390. doi: 10.3389/fpls.2019.01390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Basnet BR, Ibrahim AMH, Chen XM, Singh RP, Mason ER, Bowden RL, Liu S, Hays DB, Devkota RN, Subramanian NK, Rudd JC. Molecular mapping of stripe rust resistance in hard red winter wheat TAM 111 adapted to the U.S. high plains. Crop Sci. 2015;54(4):1361. doi: 10.2135/cropsci2013.09.0625. [DOI] [Google Scholar]
- 28.Godoy JG, Rynearson S, Chen X, Pumphrey M. Genome-wide association mapping of loci for resistance to stripe rust in north American elite spring wheat germplasm. Phytopathology. 2017;108(2):234–245. doi: 10.1094/PHYTO-06-17-0195-R. [DOI] [PubMed] [Google Scholar]
- 29.Hou L, Jia J, Zhang X, Li X, Yang Z, Ma J, Guo H, Zhan H, Qiao L, Chang Z. Molecular mapping of the stripe rust resistance gene Yr69 on wheat chromosome 2AS. Plant Dis. 2016;100(8):1717–1724. doi: 10.1094/PDIS-05-15-0555-RE. [DOI] [PubMed] [Google Scholar]
- 30.Lu JL, Chen C, Liu P, He ZH, Xia XC. Identification of a new stripe rust resistance gene in Chinese winter wheat Zhongmai 175. J Integr Agric. 2016;15(11):2461–2468. doi: 10.1016/S2095-3119(16)61379-5. [DOI] [Google Scholar]
- 31.Bansal UK, Kazi AG, Singh B, Hare RA, Bariana HS. Mapping of durable stripe rust resistance in a durum wheat cultivar Wollaroi. Mol Breeding. 2013;33:51–59. doi: 10.1007/s11032-013-9933-x. [DOI] [Google Scholar]
- 32.Agenbag GM, Pretorius ZA, Boyd LA, Bender CM, Prins R. Identification of adult plant resistance to stripe rust in the wheat cultivar Cappelle-Desprez. Theor Appl Genet. 2012;125(1):109–120. doi: 10.1007/s00122-012-1819-5. [DOI] [PubMed] [Google Scholar]
- 33.Hao Y, Chen Z, Wang Y, Bland D, Buck JW, Brown-Guedira G, Johnson J. Characterization of a major QTL for adult plant resistance to stripe rust in US soft red winter wheat. Theor Appl Genet. 2011;123(8):1401–1411. doi: 10.1007/s00122-011-1675-8. [DOI] [PubMed] [Google Scholar]
- 34.Feng J, Wang M, Chen X, See D, Zheng Y, Chao S, Wan A. Molecular mapping of YrSP and its relationship with other genes for stripe rust resistance in wheat chromosome 2BL. Phytopathology. 2015;105(9):1206–1212. doi: 10.1094/PHYTO-03-15-0060-R. [DOI] [PubMed] [Google Scholar]
- 35.Bariana HS, Hayden MJ, Ahmed NU, Bell JA, Sharp PJ, McIntosh RA. Mapping of durable adult plant and seedling resistances to stripe rust and stem rust diseases in wheat. Aust J Agric Res. 2001;52(12):1247–1255. doi: 10.1071/AR01040. [DOI] [Google Scholar]
- 36.Bemister DH, Semagn K, Iqbal M, Randhawa H, Strelkov SE, Spaner DM. Mapping QTL associated with stripe rust, leaf rust, and leaf spotting in a Canadian spring wheat population. Crop Sci. 2019;59(2). 10.2135/cropsci2018.05.0348.
- 37.Feng J, Wang M, See D, Chao S, Zheng Y, Chen X. Characterization of novel gene Yr79 and four additional QTL for all-stage and high-temperature adult-plant resistance to stripe rust in spring wheat PI 182103. Phytopathology. 2018;108(6):737–747. doi: 10.1094/PHYTO-11-17-0375-R. [DOI] [PubMed] [Google Scholar]
- 38.Hou L, Chen X, Wang M, See DR, Chao S, Bulli P, Jing J. Mapping a large number of QTL for durable resistance to stripe rust in winter wheat druchamp using simple sequence repeat (SSR) and SNP markers. PLoS One. 2015;10(5):e0126794. doi: 10.1371/journal.pone.0126794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jighly A, Oyiga BC, Makdis F, Nazari K, Youssef O, Tadesse W, Abdalla O, Ogbonnaya FC. Genome-wide DArT and SNP scan for QTL associated with resistance to stripe rust (Puccinia striiformis f. sp. tritici) in elite ICARDA wheat (Triticum aestivum L.) germplasm. Theor Appl Genet. 2015;128(7):1277–1295. doi: 10.1007/s00122-015-2504-2. [DOI] [PubMed] [Google Scholar]
- 40.Liu W, Naruoka Y, Miller K, Garland-Campbell KA, Carter A. Characterizing and validating stripe rust resistance loci in US Pacific Northwest Winter Wheat Accessions (Triticum aestivum L.) by genome-wide association and linkage mapping. Plant Genome. 2018;11(1). 10.3835/plantgenome2017.10.0087. [DOI] [PMC free article] [PubMed]
- 41.Mago R, Tabe L, McIntosh RA, Pretorius Z, Kota R, Paux E, Wicker T, Breen J, Lagudah ES, Ellis JG, Spielmeyer W. A multiple resistance locus on chromosome arm 3BS in wheat confers resistance to stem rust (Sr2), leaf rust (Lr27) and powdery mildew. Theor Appl Genet. 2011;123(4):615–623. doi: 10.1007/s00122-011-1611-y. [DOI] [PubMed] [Google Scholar]
- 42.Yuan F, Zeng Q, Wu J, Wang Q, Yang Z, Liang B, Kang Z, Chen X, Han D. QTL mapping and validation of adult plant resistance to stripe rust in Chinese wheat landrace Humai 15. Front Plant Sci. 2018;9:968. doi: 10.3389/fpls.2018.00968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu Y, Wang M, Chen X, See D, Chao S, Jing J. Mapping of Yr62 and a small-effect QTL for high-temperature adult-plant resistance to stripe rust in spring wheat PI 192252. Theor Appl Genet. 2014;127(6):1449–1459. doi: 10.1007/s00122-014-2312-0. [DOI] [PubMed] [Google Scholar]
- 44.Suenaga K, Singh RP, Huerta-Espino J, William HM. Microsatellite markers for genes Lr34/Yr18 and other quantitative trait loci for leaf rust and stripe rust resistance in bread wheat. Phytopathology. 2003;93(7):881–890. doi: 10.1094/PHYTO.2003.93.7.881. [DOI] [PubMed] [Google Scholar]
- 45.Maccaferri M, Zhang J, Bulli P, Abate Z, Chao S, Cantu D, Bossolini E, Chen X, Pumphrey M, Dubcovsky J. A genome-wide association study of resistance to stripe rust (Puccinia striiformis f. sp. tritici) in a worldwide collection of hexaploid spring wheat (Triticum aestivum L.) G3 (Bethesda) 2015;5(3):449–465. doi: 10.1534/g3.114.014563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng Y, Li J, Yao F, Long L, Wang Y, Wu Y, Li J, Ye X, Wang J, Jiang Q, Kang H, Li W, Qi P, Liu Y, Deng M, Ma J, Jiang Y, Chen X, Zheng Y, Wei Y, Chen G. Dissection of loci conferring resistance to stripe rust in Chinese wheat landraces from the middle and lower reaches of the Yangtze River via genome-wide association study. Plant Sci. 2019;287:110204. doi: 10.1016/j.plantsci.2019.110204. [DOI] [PubMed] [Google Scholar]
- 47.Huang S, Wu J, Wang X, Mu J, Xu Z, Zeng Q, Liu S, Wang Q, Kang Z, Han D. Utilization of the genome-wide wheat 55K SNP array for genetic analysis of stripe rust resistance in common wheat line P9936. Phytopathology. 2019;109(5):819–827. doi: 10.1094/PHYTO-10-18-0388-R. [DOI] [PubMed] [Google Scholar]
- 48.Yang M, Li G, Wan H, Li L, Li J, Yang W, Pu Z, Yang Z, Yang E. Identification of QTLs for stripe rust resistance in a recombinant inbred line population. Int J Mol Sci. 2019;20(14):3410. doi: 10.3390/ijms20143410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ren Y, He Z, Li J, Lillemo M, Wu L, Bai B, Lu Q, Zhu H, Zhou G, Du J, Lu Q, Xia X. QTL mapping of adult-plant resistance to stripe rust in a population derived from common wheat cultivars Naxos and Shanghai 3/catbird. Theor Appl Genet. 2012;125(6):1211–1221. doi: 10.1007/s00122-012-1907-6. [DOI] [PubMed] [Google Scholar]
- 50.Rosewarne GM, Singh RP, Espino JH, Herrera-Foessel SA, Forrest KL, Hayden MJ, Rebetzke GJ. Analysis of leaf and stripe rust severities reveals pathotype changes and multiple minor QTLs associated with resistance in an avocet × Pastor wheat population. Theor Appl Genet. 2012;124(7):1283–1294. doi: 10.1007/s00122-012-1786-x. [DOI] [PubMed] [Google Scholar]
- 51.Lu J, Hou J, Ouyang Y, Luo H, Zhao J, Mao C, Han M, Wang L, Xiao J, Yang Y, Li X. A direct PCR–based SNP marker–assisted selection system (D-MAS) for different crops. Mol Breeding. 2020;40:9. doi: 10.1007/s11032-019-1091-3. [DOI] [Google Scholar]
- 52.Singh RP, Huerta-Espino J, Rajaram S. Achieving near-immunity to leaf and stripe rusts in wheat by combining slow rusting resistance genes. Acta Phytopathol et Entomol Hungarica. 2000;35:133–139. [Google Scholar]
- 53.Juliana P, Singh RP, Singh PK, Crossa J, Huerta-Espino J, Lan C, Bhavani S, Rutkoski JE, Poland JA, Bergstrom GC, Sorrells ME. Genomic and pedigree-based prediction for leaf, stem, and stripe rust resistance in wheat. Theor Appl Genet. 2017;130:1415–1430. doi: 10.1007/s00122-017-2897-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu B, Liu T, Zhang Z, Jia Q, Wang B, Gao L, Peng Y, Jin S, Chen W. Discovery and pathogenicity of CYR34, a new race of Puccinia striiformis f. sp. Tritici in China. Acta Phytopathol Sin. 2017;5:681–687. [Google Scholar]
- 55.Wang J, Li H, Zhang L, Meng L (2016) Users’ manual of QTL IciMapping. The quantitative genetics group, Institute of Crop Science, Chinese Academy of Agricultural Sciences (CAAS), Beijing 100081, China, and genetic resources program, international maize and wheat improvement center (CIMMYT), Apdo. Postal 6-641, 06600 Mexico, D.F., Mexico.
- 56.Wang S, Wong D, Forrest K, Allen A, Chao S, Huang BE, Maccaferri M, Salvi S, Milner SG, Cattivelli L, Mastrangelo AM, Whan A, Stephen S, Barker G, Wieseke R, Plieske J, International Wheat Genome Sequencing Consortium. Lillemo M, Mather D, Appels R, Dolferus R, Brown-Guedira G, Korol A, Akhunova AR, Feuillet C, Salse J, Morgante M, Pozniak C, Luo MC, Dvorak J, Morell M, Dubcovsky J, Ganal M, Tuberosa R, Lawley C, Mikoulitch I, Cavanagh C, Edwards KJ, Hayden M, Akhunov E. Characterization of polyploid wheat genomic diversity using a high-density 90000 single nucleotide polymorphism array. Plant Biotechnol J. 2014;12(6):787–796. doi: 10.1111/pbi.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Botstein D, White RL, Skolnick M, Davis RW. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet. 1980;32(3):314–331. [PMC free article] [PubMed] [Google Scholar]
- 58.Liu K, Muse SV. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21(9):2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- 59.Cheng P, Xu L, Wang M, See D. R., Chen X. molecular mapping of genes Yr64 and Yr65 for stripe rust resistance in hexaploid derivatives of durum wheat accessions PI 331260 and PI 480016. Theor Appl Genet. 2014;127:2267–2277. doi: 10.1007/s00122-014-2378-8. [DOI] [PubMed] [Google Scholar]
- 60.Mago R, Brown-Guedira G, Dreisigacker S, Breen J, Jin Y, Singh R, Appels R, Lagudah ES, Ellis J, Spielmeyer W. An accurate DNA marker assay for stem rust resistance gene Sr2 in wheat. Theor Appl Genet. 2011;122:735–744. doi: 10.1007/s00122-010-1482-7. [DOI] [PubMed] [Google Scholar]
- 61.Kumar D, Kumar A, Chhokar V, Gangwar O, Bhardwaj S, Sivasamy M, Prasad S, Prakasha T, Khan H, Singh R, Sharma P, Sheoran S, Iquebal M, Jaiswal S, Angadi U, Singh G, Rai A, Singh G, Kumar D, Tiwari R. Genome-wide association studies in diverse spring wheat panel for stripe, stem, and leaf rust resistance. Front Plant Sci. 2020;11:748. doi: 10.3389/fpls.2020.00748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang H, Yang Y, Wang C, Liu M, Li H, Fu Y, Wang Y, Nie Y, Liu X, Ji W. Large-scale transcriptome comparison reveals distinct gene activations in wheat responding to stripe rust and powdery mildew. BMC Genomics. 2014;15(1):898. doi: 10.1186/1471-2164-15-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Names and origins of 240 wheat accessions.
Additional file 2. Frequency distributions for stripe rust maximum disease severity (MDS) of 240 wheat accessions in five environments.
Additional file 3. Marker density of the 240 wheat accessions genotyped with the 90 K SNP arrary.
Additional file 4. Principal components analysis (PCA) plots of 240 wheat accessions.
Additional file 5. Quantitative trait loci (QTLs) for stripe rust resistance significant only in one environment detected by genome-wide association study by TASSEL and GAPIT.
Additional file 6. Quantile-quantile plot for stripe rust maximum disease severity in 240 wheat accessions analyzed using the mixed linear model with Tassel.
Additional file 7. Quantile-quantile plot for stripe rust maximum disease severity in 240 wheat accessions analyzed using the mixed linear model with GAPIT.
Additional file 8. Profiles of 11 penta-primer amplification refractory mutation system markers for mapped stripe rust resistance loci.
Additional file 9. Primers for 11 penta-primer amplification refractory mutation system markers for seven stripe rust resistance loci.
Additional file 10. Protocols for 11 developed markers for seven stripe rust resistance loci.
Additional file 11. Candidate genes for stripe rust resistance QTLs.
Additional file 12. Reported QTLs/genes, linked markers and their physical positions used to align with QTLs mapped in the present study.
Data Availability Statement
The genotypic data generated from the 90 K wheat SNP array for the 240 wheat accessions is in the European Nucleotide Archive, PRJEB36125, https://www.ebi.ac.uk/ena/data/view/PRJEB36125. Other data supporting the results of this publication are included within the article and its additional files.