Abstract
Background
The human neutrophil antigen 2 (HNA-2), which is expressed on CD177, is undetectable in 3–5% of the normal population. Exposure of these HNA-2<sub>null</sub> individuals to HNA-2-positive cells can cause immunization and production of HNA-2 antibodies, which can induce immune neutropenia and transfusion-related acute lung injury. In HNA-2-positive individuals, neutrophils are divided into a CD177<sup>pos.</sup> and a CD177<sup>neg.</sup> subpopulation. The molecular background of HNA-2 deficiency and the bimodal expression pattern, however, are not completely decoded.
Study Design
An international collaboration was conducted on the genetic analysis of HNA-2-phenotyped blood samples, including HNA-2-deficient individuals, mothers, and the respective children with neonatal immune neutropenia and regular blood donors.
Results
From a total of 54 HNA-2<sub>null</sub> individuals, 43 were homozygous for the CD177 *787A>T substitution. Six carried the CD177 *c.1291G>A single nucleotide polymorphism. All HNA-2-positive samples with >40% CD177<sup>pos.</sup> neutrophils carried the *787A wild-type allele, whereas a lower rate of CD177<sup>pos.</sup> neutrophils was preferentially associated with *c.787AT heterozygosity. Interestingly, only the *c.787A allele sequence was detected in complementary DNA (cDNA) sequence analysis carried out on all *c.787AT heterozygous individuals. However, cDNA analysis after sorting of CD177<sup>pos.</sup> and CD177<sup>neg.</sup> neutrophil subsets from HNA-2-positive individuals showed identical sequences, which makes regulatory elements within the promoter unlikely to affect CD177 gene transcription in different CD177 neutrophil subsets.
Conclusion
This comprehensive study clearly demonstrates the impact of single nucleotide polymorphisms on the expression of HNA-2 on the neutrophil surface but challenges the hypothesis of regulatory epigenetic effects being implicated in the bimodal CD177 expression pattern.
Keywords: Human neutrophil antigens, Neutropenia, Molecular analysis, Monoallelic expression
Introduction
The human neutrophil antigen 2 (HNA-2) plays an important role in granulocyte immunobiology. Antibodies to HNA-2 can cause severe neonatal immune neutropenia and transfusion-related acute lung injury (TRALI) [1, 2, 3]. HNA-2 is located on CD177, a 56- to 64-kDA glycoprotein that is restricted to neutrophil granulocytes and is anchored to the neutrophil membrane by glycosylphosphatidylinositol (GPI) [4, 5]. The expression can be upregulated by different conditions like bacterial infection, pregnancy, and G-CSF stimulation [6, 7]. At least 3% of Caucasians are completely devoid of HNA-2 (HNA-2null) and only these individuals are prone to develop HNA-2 isoantibodies upon exposure to HNA-2-positive cells [8, 9, 10, 11]. Most HNA-2-positive individuals present a peculiar bimodal expression pattern with 2 neutrophil subsets. One subset is negative whereas the second subset expresses HNA-2 at a high or intermediate level [5, 12]. The latter fraction may vary from person to person between <5% and nearly 100%, but the percentage is a lifelong trait [13].
The molecular background regulating the HNA-2nullphenotype and the bimodal expression pattern in HNA-2-positive individuals is not completely understood. In 2 individuals previously studied, an incorrect splicing was considered as the molecular reason for the HNA-2nullphenotype [14]. Our own unpublished data and observations of others [15], however, demonstrated correctly spliced cDNA in all HNA-2-deficient individuals investigated so far. Numerous CD177 gene single nucleotide polymorphisms (SNPs) are listed in the NCBI dbSNP database. A low or high HNA-2 expression, as well as the presence of 1 or 2 antigen-positive subsets, had initially been attributed to various sequence variations [16, 17, 18]. A homozygous CD177 *c.787A>T substitution within exon 7 introducing a stop codon was identified as the main cause for the HNA-2nullphenotype [13, 15, 19]. The SNP was primarily assigned to either CD177 *c.829A>T [15] or CD177 *c.843A>T [19], but in accordance with the Human Genome Variation Society (HGV) the SNP is now assigned to CD177 *c.787A>T (taking position 1 as the A of the start ATG codon of the CDS) [20]. The mutation was regarded to result from an allelic gene conversion event involving exon 7 of the CD177P1 pseudogene, which would replace the equivalent exon 7 sequence within the CD177gene [13]. CD177P1 is located downstream of the CD177 gene in opposite orientation and comprises exons 4–9 that are highly homologous to the CD177 gene. In rare cases, additional mutations have also been described in HNA-2nullindividuals [15, 19]. Copy number variation has been considered as the reason for both HNA-1 deficiency and expression variation. However, the involvement of copy number variation in the regulation of CD177 gene expression has been excluded [15].
The CD177 *c.787A>T exchange has also been attributed to the percentage of CD177pos.neutrophils in CD177-positive individuals. Heterozygosity for ectopic CD177P1 gene conversion correlated with an increased CD177neg.neutrophil fraction where both alleles were transcribed [13]. Recently, a CD177 *c.1291G>A substitution in combination with the *c.787A>T SNP has been associated with both the HNA-2nullphenotype and the rather rare trimodal expression pattern, depending on the respective haplotype [21]. A monoallelic regulation for CD177 expression on neutrophil subpopulation has been suggested where in HNA-2-positive and HNA-2-negative subpopulations only one allele of CD177 gene is expressed [22].
In order to verify these controversial findings, we initiated an international multicenter study on the molecular basis of CD177 gene expression in HNA-2nulland HNA-2-positive phenotyped donors as well as in immunized women with HNA-2 isoantibodies, including some of their neonates and the respective fathers. This included analysis of the CD177 mRNA content in sorted neutrophil subpopulations of HNA-2-positive donors.
Subjects
In this study, a total of 107 samples were included. Samples were provided by each of the contributing centers: German Red Cross Blood Service West, Hagen and Bad Kreuznach (Germany), University Hospital Heidelberg (Germany), Institute for Clinical Immunology and Transfusion Medicine, Justus-Liebig-University Giessen (Germany), Blood and Tissue Bank, Barcelona (Spain), Sanquin, Amsterdam (The Netherlands), University Hospital, Nantes (France), Karolinska University Hospital, Stockholm (Sweden), and Medical University, Vienna (Austria). The study provided advanced diagnostics in some cases of clinically suspected neonatal alloimmune neutropenia (NIN) when samples were submitted to the respective laboratory. This included samples of 9 mothers, 10 neonates, and 5 respective fathers as well as 3 further patients. Additionally, the cohort comprised 80 regular blood donors and volunteers from the contributing laboratories. All donors gave informed consent.
Methods
Serological Assays
Maternal sera of NIN newborns were tested for the presence of granulocyte-specific antibodies in the laboratories of the contributing centers, applying validated and standardized granulocyte immunobiology methods. These included the granulocyte agglutination test (GAT) [23], granulocyte immunofluorescence test (GIFT) [24, 25] with either microscopic or flow cytometric evaluation (Flow GIFT) [26], and the monoclonal antibody immobilization of granulocyte antigen (MAIGA) test [25, 27] as well as simultaneous analysis of specific granulocyte antigens (SASGA) assay [28] based on HNA-typed neutrophil cell panels and CD177-specific monoclonal antibodies for the MAIGA test.
HNA-2 phenotypes were only available from blood donors and volunteers because shipping of patient samples after 24 h of blood withdrawal impedes the isolation of intact neutrophils. Individuals with proven HNA-2 isoantibody were assessed as HNA-2 negative without serological phenotyping. Phenotyping of blood donors was performed in the GIFT on either freshly isolated neutrophils or EDTA anticoagulated blood using CD177-specific FITC-labeled mouse monoclonal antibody (moab), clone MEM-166 (Serotec distributed by BioRad, Munich, Germany), and CD16-specific PE/CY7-labeled moab, clone 3G8 (BioLegend, San Diego, CA, USA) to gate the neutrophils followed by flow cytometric evaluation (Beckman Coulter Epics XL; Beckman Coulter, Krefeld, Germany). In Vienna, phenotyping was performed by Flow-GIFT [26] using human allo-anti-HNA-2 and Alexa Fluor 488® goat anti-human IgG Fab Fragment (Jackson ImmunoResearch, New Market, UK). In few further cases fluorescence was assessed microscopically.
Genomic DNA Sequencing
DNA was isolated by the contributing laboratories. The CD177 gene was separated from the CD177P1 pseudogene by long-range PCR amplification (Long-Range PCR kit; Qiagen, Hilden, Germany) as described previously [19]. Genomic typing was conducted based on a template-specific long-range amplification of the whole CD177 coding region encompassing exons 1–9 (8729 bp or alternatively 9323 bp). All amplification and sequencing primers are listed in Table 1. Sequencing reaction in most genomic DNA (gDNA) samples covered only exon 7, which comprises the CD177 *c.787A>T substitution. Some samples were subjected to sequencing of all CD177 exons, including exon 9, which contains the CD177 *c.1291G>A substitution. Sequencing was done using a cycle sequencing kit followed by electrophoretical separation in the ABI Prism 310 DNA Analyzer (Applied Biosystems, ABI, Weiterstadt, Germany).
Table 1.
Amplification and sequencing primers
| Name | Position | Usage | Sequence (5′–3′) | Direction | Ref. |
|---|---|---|---|---|---|
| CD177_LR_s | 5′ UTR, −39 to −8 | gDNA long-range PCR a | ctgaaaaagcagaaagagattaccagccacag | Sense | 19 |
| CD177_LR_as | 3′ UTR, +115 to +144 | gDNA long-range PCR a | gtccaaggccattaggttatgaggtcaga | Anti-sense | 19 |
| CD177_LR2_s | 5′ UTR, −71 to −43 | gDNA long-range PCR b | cttaagggctggtataaaggactt | Sense | |
| CD177_LR2_as | 3′ UTR, +534 to 562 | gDNA long-range PCR b | cgctacaatgttcctatggtcataaaatc | Anti-sense | |
| CD177R_1F | 5′ UTR, −24 to −7 | cDNA nested PCR 1; Seq. exon 1+2, cDNA+gDNA |
gagattaccagccacaga | Sense | 8 |
| CD177R_1R | 3′UTR, +53 to +29 | cDNA nested PCR1 | ggaggttgagtgtgggtggtcagca | Anti-sense | 8 |
| CD177R_9F | 5′UTR, −6 to 15 | cDNA nested PCR2 | cgggtcATGAGCCCGGTATTA | Sense | 8 |
| CD177R_11R | 1,289 to 1,311 | cDNA nested PCR2; Seq. exon 9, cDNA | GCAGGAAGGGCAAACCACTCCCC | Anti-sense | |
| CD177R_3F | 11 to 30 | Seq. exon 2+3, cDNA + gDNA | TATTACTGCTGGCCCTCCTG | Sense | 16 |
| CD177R_4F | 163 to 183 | Seq. exon 3+4, cDNA + gDNA | TGCCAGGACACGTTGATGCTC | Sense | 16 |
| CD177R_4R | 404 to 385 | Seq. exon 3, cDNA | ACTGGGCACCTCAAGGATCC | Anti-sense | 16 |
| CD177R_5F | 680 to 699 | Seq. exon 6+7, cDNA | CCACTGATTGGACCACATCG | Sense | 16 |
| CD177R_6F | Intron 5, −86 to −67 | Seq. exon 7–9, cDNA | gacctgtgcaatagtgccagc | Sense | 16 |
| CD177R_7F | 380 to 399 | Seq. exon 4–6, cDNA | CAGAAGAGATCTGCCCCAAG | Sense | |
| CD177_E7F | Intron 6, −134 to −115 | Seq. exon 7+8, gDNA | tgacccagcagttgtgatca | Sense | |
| CD177g_E3F | Intron 2, −18 to exon 3 194 | Seq. exon 3, gDNA | ctccctctttcggtccagG | Sense | |
| CD177g_E4-5R | Intron 5, 82 to 63 | Seq. exon 4+5, gDNA | ttggtgtgatggctctggat | Anti-sense | |
| CD177g_E6F | Intron 5, −86 to −67 | Seq. exon 6, gDNA | tgtgatcaccttccctagcc | Sense | |
| CD177g_E9F | Intron 8, −61 to −42 | Seq. exon 9, gDNA | gggtttacaacttggctggg | Sense |
Position numbers within exons refer to CDS (NC_000019.10), whereas UTR and intron positions refer to the nearest exon. Intronic and UTR sequences are printed in lower-case letters.
Sorting of HNA-2-Positive and HNA-2-Negative Subpopulations
Neutrophils were isolated from 2 CD177pos.individuals by dextran sedimentation. Isolated neutrophils were stained with FITC-labeled MEM-166 or mouse IgG. The stained CD177pos.and CD177neg.neutrophil subpopulations were then sorted using FACSAria (Becton Dickinson, Heidelberg, Germany). The neutrophil subpopulation with mean fluorescence intensity values identical to those of mIgG was considered as the CD177neg.subpopulation.
Isolation and Reverse Transcription of RNA from Peripheral Blood Cells
RNA was isolated either from human whole blood or isolated neutrophils. Whole blood was taken in PAXgene storage tubes (Becton Dickinson Diagnostics, Heidelberg, Germany) followed by mRNA isolation using the PAXgene mRNA kit (Qiagen) according to the manufacturer's instructions. Neutrophil granulocytes were isolated from EDTA anticoagulated blood as described elsewhere [29]. The purity of the preparations, determined microscopically from stained smears, was about 95%. RNA was isolated from up to 107isolated neutrophils using the RNeasy Mini Kit (Qiagen). Reverse transcription into cDNA was performed with the Omniscript RT kit (Qiagen) according to the instructions of the manufacturer by use of an oligo-dT-primer (TibMolbiol, Berlin, Germany). A CD177 -specific fragment was amplified from the first-strand cDNA template in a nested PCR reaction covering the 5′ UTR to the 3′ end of exon 9 as described elsewhere, with slight modifications [8]. In the first PCR reaction, denaturation for 3 min at 95°C was followed by 25 cycles with 30 s at 95°C, 30 s at 58°C, 180 s at 72°C, and a final elongation of 7 min at 72°C. The products were checked electrophoretically for the presence of clearly visible bands and afterwards 2 µL were subjected to the 2nd PCR reaction, which was performed in analogy to the first one with the exception of 40 cycles und otherwise unchanged conditions. The products were subjected to cycle sequencing applying primers covering the complete coding region (CDS; Table 1).
Results
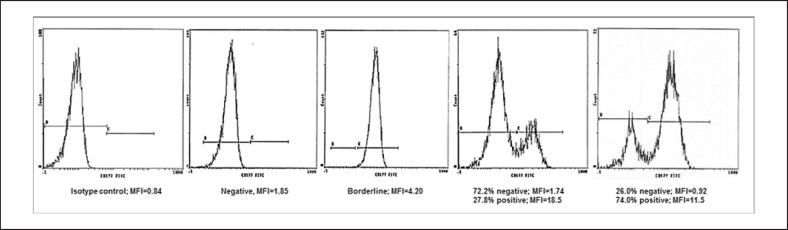
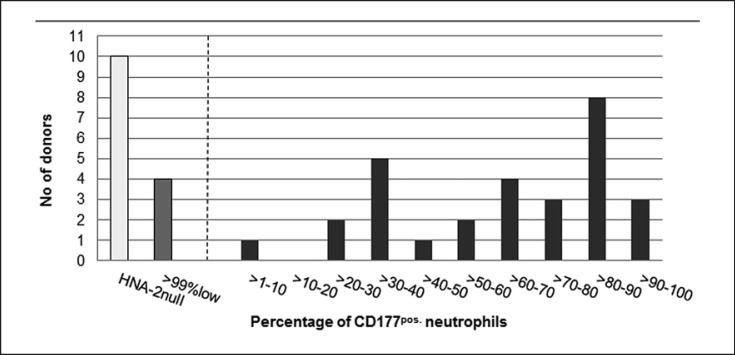
The study comprised 107 individuals, including 80 regular blood donors from different centers, 9 maternal samples corresponding to NIN cases, 10 of the respective children, and 5 fathers as well as 3 individuals with HNA-2 isoantibodies. Indeed, the plasma of one of the donors included in this study was involved in a case of TRALI. The selection of blood donors in the whole panel was not random because only the transfusion centers Giessen and Bad Kreuznach added samples of blood donors with an HNA-2-positive phenotype whereas the focus of the other centers was put on HNA-2nullindividuals. Donor samples were assigned to 4 groups, with (1) an HNA-2nullphenotype (≤1% CD177pos.neutrophils), (2) HNA-2 low expression (>99% neutrophils with a borderline or questionable expression), (3) ≤40% clearly CD177pos.neutrophils, and (4) >40% clearly CD177pos.neutrophils (Fig. 1, 2). Overall, 29 of the 43 phenotyped blood donors from Bad Kreuznach were clearly CD177pos.with a bimodal expression and a mean positive subset of 63.5%. Four had low expression of HNA-2 (Table 2) and 10 were negative. No trimodal expression pattern with 2 CD177pos.subsets was detected. Samples from blood donors of the other centers were assigned as either negative (n = 32) or questionable (in cases of a borderline fluorescence intensity; n = 5) according to the protocols and defined classifications of the respective laboratories.
Fig. 1.
Neutrophil CD177 expression patterns in HNA-2nulland HNA-2-positive individuals demonstrated by flow cytometry.
Fig. 2.
Range of CD177pos.neutrophil subsets in HNA-2-positive donors as well as the number of negative and borderline phenotypes from 43 donors of the Transfusion Center Bad Kreuznach. The number of individuals with the respective percentage of CD177pos.neutrophils is shown.
Table 2.
Distribution of the genomic CD177*787A.T variation within the phenotyped blood donors, patients, NIN mothers, and their neonates
| HNA-2 phenotype/HNA-2 immunization |
CD177*
c.787A |
CD177*
c.787A+T |
CD177*
c.787T |
Total |
|---|---|---|---|---|
| Donors | 80 | |||
| HNA-2 negative | 3 | 7 | 32a | 42 |
| Borderline/questionable | 3 | 4 | 2 | 9 |
| ≤40% CD177pos.neutrophils | 3 | 5 | 0 | 8 |
| <40% CD177pos.neutrophils | 21 | 0 | 0 | 21 |
| Patients with HNA-2 alloantibody | 0 | 0 | 3 | 3 |
| NIN mothers with HNA-2 alloantibody | 0 | 1 | 8 | 9 |
| NIN neonates | 4 | 6 | 0 | 10 |
| Fathers of the neonates | 4 | 1 | 0 | 5 |
Values indicate the number of subjects.
Three of these had HNA-2 antibody, and the blood product of one was involved in a case of TRALI.
Sequencing of gDNA
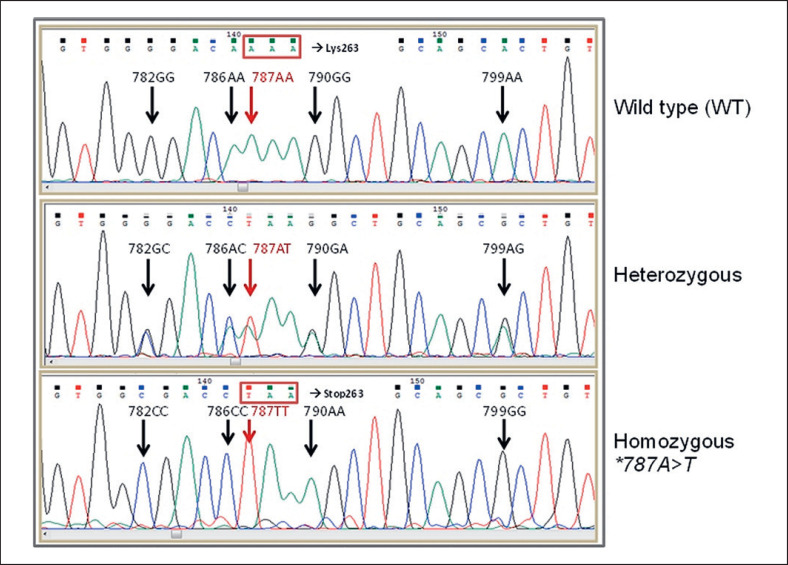
Genomic sequencing of CD177 exon 7 and flanking intron sequences in all individuals studied so far has detected 2 haplotypes; wild-type (WT) *c.782G, 786A, 787A, 790G, and 799A or the mutated *c.782A, 786C, 787T, 790A, and 799G, or heterozygosity for both (Fig. 3). In 90 out of 105 samples, an intron 7+19c>t substitution, located 178 nucleotides downstream of CD177 *c.787, was detected. This substitution was associated with the mutant haplotype in exon 7. Genomic sequencing of all 9 CD177 exons in 16 individuals (11 CD177neg.or questionable and 5 CD177pos.) demonstrated a number of different polymorphisms (at positions CD177 *c.92, 114, 551, 610, 614, 751, 1038, 1042, 1097, 1178, and 1291) that were not necessarily in the same haplotype as the exon 7 mutations and did not exhibit a uniform pattern (data not shown).
Fig. 3.
DNA sequencing of CD177 exon 7 covering position *c.787 in donors with different genotypes.
In total, 79.6% of all individuals with an HNA-2nullphenotype, including 8 of the 9 NIN mothers but only 2 of 9 donors with borderline or questionable CD177 expression, were homozygous for the CD177 *c.787A>T substitution (Table 2) whereas all donors with a >40% CD177pos.neutrophil subset were homozygous for the WT allele. A low percentage of ≤40% CD177pos.neutrophils preferentially went along with heterozygosity for the mutation (Table 2).
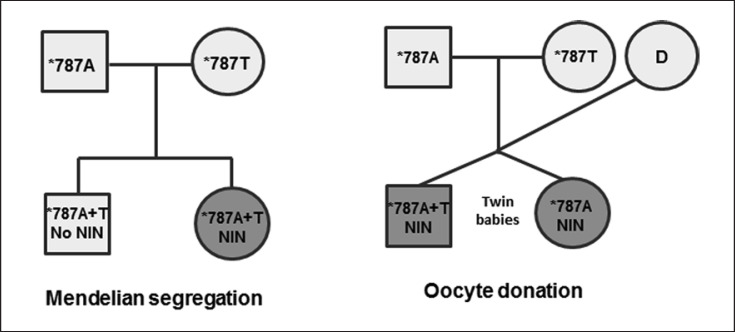
Fourteen of the total 15 individuals with HNA-2 isoantibodies, including the 9 NIN mothers, 3 immunized donors, and 3 patients with HNA-2 isoantibodies, were homozygous for the CD177 *c.787A>T (*787TT) mutation (Table 2). One NIN mother, however, was heterozygous for the *c.787A>T mutation within exon 7 and negative for the *c.955delG and c.*1291G>A mutations within exons 8 and 9, respectively. For each of the 8 homozygous *c.787TT mothers, pedigrees were available, demonstrating that only 6 of 10 neonates were heterozygous (Table 2; Fig. 4). The 5 available paternal samples that were included to complete the pedigrees carried at least one WT allele. In 1 family with affected twin babies, the pregnancy resulted from an oocyte donation. The immunized gestational mother exclusively carried the *c.787T allele whereas both neonates were homozygous for the WT alleles that had been inherited by the genetic parents (Fig. 4). Maternal inheritance was unresolved in 2 additional cases.
Fig. 4.
Pedigrees with differential segregation of the CD177 *c.787 SNP in cases of NIN. In the right pedigree only CD177 typing of the gestational mother but not of the oocyte donor (D) was available.
Further sequencing analysis, spanning all CD177 gene exons was carried out with DNA samples from 12 donors with CD177nullor questionable phenotype and 1 NIN mother without the homozygous CD177 *c.787A>T substitution. In 3 CD177 *c.787A homozygous and 1 *c.787AT heterozygous donor samples, a homozygous CD177 *c.1291G>A substitution was observed. In 2 other CD177nulldonors, heterozygosity for both *c.787AT and *c.1291GA was detected. The remaining 7 individuals carried the *c.1291G WT allele in combination with further heterozygous mutations within exon 5 (*c.551G>T →p.Gly184Val, *c.610A>G →p.Asn204Asp, and *c.614 G>T →p.Arg205Met), exon 6 (*c.751C>A →p.Leu251Ile), exon 8 (*c.1042G>A →p.Ala348Thr), and exon 9 (*c.1178G>C →p.Arg393Pro).
Sequencing of cDNA
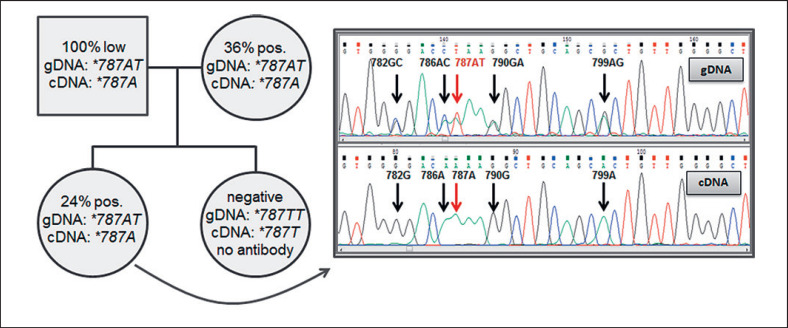
Complementary DNA was available for sequencing from 28 blood donors. Full-length CD177 cDNA was detected in all samples but consistent genotypes with gDNA sequencing were only obtained in 19 cases (Table 3). Nine samples with heterozygosity for *c.787A and T on gDNA only showed the WT *c.787A allele on cDNA level. This finding included 1 donor family with an HNA-2nulldaughter where both parents and her sibling with low CD177pos.expression exhibited heterozygosity for *c.787A and T on gDNA but exclusively *c.787A on cDNA (Fig. 5).
Table 3.
Comparison of gDNA and cDNA sequencing results in blood donors in relation to the HNA-2 phenotype
| gDNA/cDNA HNA-2 expression | *787TT/*787T | *787AT/*787AT | *787AT/*787A | *787AA/*787A | Total |
|---|---|---|---|---|---|
| HNA-2null | 9 | 0 | 1 | 0 | 10 |
| Questionable/borderline | 1 | 0 | 4 | 3 | 8 |
| >40% CD177pos. | 0 | 0 | 1 | 5 | 6 |
| ≤40% CD177pos. | 0 | 0 | 3 | 1 | 4 |
| Total | 10 | 0 | 9 | 9 | 28 |
Values indicate the number of subjects. In order to conform to a possible MAE only one allele is given for cDNA expression.
Fig. 5.
Pedigree of a donor family with differential cDNA and gDNA allele expression. The antigen-negative daughter was not immunized against HNA-2.
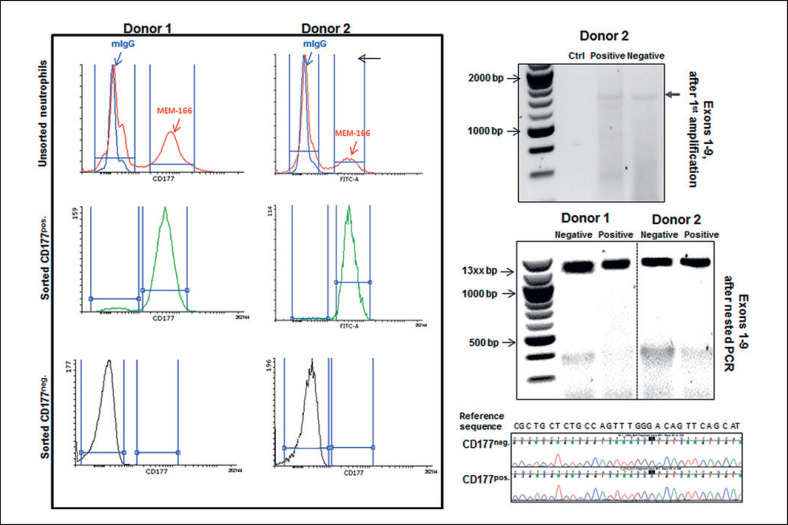
Analysis of cDNA in Sorted CD177pos.and CD177neg.Neutrophil Subpopulations
Recently, monoallelic expression (MAE) regulation for CD177 on neutrophil subsets has been suggested [22]. To clarify whether CD177 MAE in neutrophil subpopulations is responsible for the discrepancy between gDNA and cDNA analysis, the isolated neutrophils from 2 CD177pos.phenotyped individuals were stained with FITC-labeled MEM166 antibody. The CD177pos.and CD177neg.subpopulations were gated. The neutrophil subpopulation with a peak congruent with that of the negative control was considered as the HNA-2neg.subpopulation (Fig. 6, left panel). CD177 cDNA content of both neutrophil subpopulations was amplified for CD177 exons 1–9 and the products of the first PCR reaction, which exhibited a comparably weak band intensity after electrophoretic separation was used as a template for the nested PCR reaction (amplification of exons 1–9) in order to obtain enough template for subsequent cDNA sequencing.
Fig. 6.
Analysis of sorted HNA-2-positive and HNA-2-negative neutrophil subpopulations. Neutrophils were labeled with FITC-MEM166 or FITC-mIgG and the subsets were sorted to positive and negative subpopulations according to mean fluorescent intensity (left panel). mRNA content of sorted subpopulations was isolated and converted to cDNA. CD177 cDNA was amplified by primers flanking exons 1–9. The very weak bands representing the products of the first PCR reaction covering exons 1–9 of the CD177pos.and the CD177neg.fraction of one donor as well as an aqua control are shown in the upper right panel. After the 2nd round of amplification of the nested PCR, CD177 cDNA is presented in the CD177neg.and CD177pos.fractions of 2 donors. The sequencing chromatograms exemplarily show sequence homology in the exon 2 amplicons of both CD177neg.and CD177pos.subpopulations of donor 2 with the CD177 reference sequence (NM_020406.4) for comparison.
According to MAE, it was expected that methylation mechanisms would suppress CD177 expression in the CD177neg.subpopulation, which would consequently restrict the presence of CD177 cDNA to CD177pos.positive neutrophil subpopulations. In contrast, our results showed the presence of full-length CD177 cDNA in both CD177pos.and CD177neg.subpopulations at comparable amounts (according to band intensities), indicating active CD177 gene transcription in both neutrophil subpopulations (Fig. 6, right panel).
Sequencing analysis of amplified bands in neutrophil subpopulations demonstrated identical sequences in both negative and positive subpopulations (Fig. 6, right panel).
Discussion
In this study on a relevant number of blood donor and clinical samples, we tried to corroborate known information on the complex genetic basis of HNA-2 phenotypes and to add further data. The CD177 *c.787A>T substitution that creates a premature stop codon within the CD177 coding region was identified as the main cause for complete HNA-2 deficiency only within 79.6% of HNA-2nullindividuals who were homozygous for this mutation [15]. The *c.787 SNP was in complete linkage with the nucleotides at positions 782, 786, 790, and 799 [13, 15] but not constantly with additional SNPs within other exons or flanking intron sequences, for example, at positions CD177 *c.551, 610, 614, 751, 1,042, and 1,178. The CD177 *c.787>T substitution has also recently been found in a TRALI case caused by a male donor that carried HNA-2 isoantibodies [30]. However, this mutation cannot explain the missing HNA-2 expression in all HNA-2nullphenotyped blood donors tested so far, as discussed before [15, 19]. However, a definite discrimination between the true HNA-2nullphenotype on the one hand and a very small CD177pos.neutrophil subset or a borderline mean fluorescence intensity on the other hand is sometimes challenging such that samples might be falsely phenotyped as HNA-2null[11, 16]. An unequivocal HNA-2nullphenotype is only given in immunized individuals with an HNA-2 isoantibody, either neutropenic patients, mothers of NIN babies, or donors without clinical symptoms, which was the reason to include 15 immunized individuals. But even in this group, 1 of 15 samples carried only one allele with the CD177 *c.787A>T mutation and did not exhibit an additional c.1291G>A or c.955delG mutation as observed before in other studies [13, 15]. Compound CD177 *c.787A>T and CD177 *c.955delG (originally assigned to position *c.997) heterozygosity was not detected in any of our samples, which might be due to differential ethnicities of the probands [15, 19].
In 6 of 12 donor samples with negative or borderline CD177 expression, an additional CD177 *c.1291G>A substitution leading to a p.Gly431Arg exchange might explain the CD177neg.phenotype when the premature stop caused by CD177 *c.787A>T is missing or only found in a heterozygous manner. The CD177 *c.1291G>A substitution abrogates the GPI binding site of the CD177 GP so that the molecule is synthesized by the neutrophil but cannot be fixed to the membrane [21]. Three of our HNA-2nullsamples and one with borderline expression were homozygous for CD177 *c.1291A, which can explain the HNA-2-deficient phenotype. Provided that within the 2 samples that were heterozygous for CD177 *c.787A and *c.1291A both forms are located in trans, this could also abolish CD177 expression. However, 2 further HNA-2nulldonors and 4 with borderline expression did not carry the CD177 *c.1291G>A mutation. Unfortunately, no mRNA was available from these individuals to test for a potential MAE. In one of them an additional heterozygous CD177 *c.1178G>C (p.Arg393Pro) substitution might account for the deficiency or a substantially reduced expression in the case of compound heterozygosity for both mutations. The effect of further single nucleotide polymorphisms on the CD177 expression remains unclear because there was no definite pattern of the SNPs at positions CD177 *c.551, 610, 614, 751, and 1042.
Unexpectedly, gDNA of only 6 of 10 NIN neonates was heterozygous for the CD177 *c.787A>T substitution, while 4 babies only carried the WT allele although their mothers were homozygous for the CD177 *c.787A>T substitution. In a twin baby pair this could be explained by the fact they resulted from an oocyte donation where the *c.787A WT twins immunized the gestational mother who was homozygous for the *c.787A>T mutation. In 2 further cases no information on potential oocyte donations was available, leaving the cases unresolved.
The CD177 *c.787A>T substitution was not only associated with neutrophil HNA-2 deficiency but also affected the percentage of CD177neg.neutrophils in individuals with a bimodal expression pattern. Consistent with previous findings, heterozygosity for the mutation was preferentially associated with a borderline HNA-2 expression or a small CD177pos.neutrophil subset, whereas all 21 donors with >40% CD177pos.neutrophils carried the WT form, which is consistent with previous findings [15].
Most interesting were the differential typing results of gDNA and cDNA sequencing obtained in 9/28 blood donors whose gDNA was heterozygous for the CD177 *c.787T haplotype within exon 7 but only exhibited the WT haplotype within cDNA. The data sets of the remaining 19 donors concordantly either showed the CD177 *c.787 WT or the mutated allele. Different approaches try to explain this effect. Monoallelic gene expression is the phenomenon of gene expression when only 1 of the 2 gene copies is actively transcribed. An unstable nature of mRNA from the CD177 *c.787T allele that is rapidly degraded by the mechanism of nonsense-mediated mRNA decay and thus undetectable by cDNA sequencing has been discussed as the reason of MAE [15], but this is rebutted by the status of the 10 CD177 *c.787T homozygous donors (gDNA) where the respective cDNA was detected. Second, gene silencing by DNA methylation in the CD177 promoter region may induce the MAE of the CD177 gene, which consequently leads to the presence of only one allele in mRNA analysis [22]. In a HeLa cell model this epigenetic mechanism was associated with the CD177neg.fraction, whereas CpG demethylation converted monoallelic into biallelic expression within the respective fraction. During differentiation of CD34+ hematopoietic stem cells into neutrophils 1 of 2 different parental alleles was silenced, thus inducing MAE, which also might explain our own findings [22]. CD177 mRNA expression has been reported to increase during differentiation of neutrophils, whereas there is no de novo synthesis of protein in mature neutrophils [31]. A possible mechanism for MAE could be a preferential usage of the CD177 *c.787A allele for transcription during maturation so that only one mRNA version is found in mature neutrophils. Another mechanism that might explain the discrepancy between gDNA and cDNA is the nonspecific amplification of the CD177 pseudogene along with the CD177 gene. However, this fault was eliminated by the amplification of a CD177 gene-specific template for the downstream application [19].
To clarify whether the MAE mechanism is regulating CD177 expression on neutrophil subpopulations, the mRNA extracted from sorted CD177pos.and CD177neg.neutrophils was analyzed. Analysis of the CD177 mRNA content of both sorted CD177pos.and CD177neg.granulocytes showed the presence of identical CD177 cDNA sequences in both subpopulations. As demonstrated semiquantitatively by agarose gel electrophoresis after both the 1st and the 2nd round of amplification of the nested PCR reaction, product quantities were comparable in the CD177pos.and CD177neg.neutrophil subpopulations. Although it is always possible to argue that the CD177 mRNA from the CD177neg.fraction was derived from a minute contamination by CD177pos.neutrophils, we interpret our data as an indication of the presence of an active CD177 gene in CD177neg.neutrophils, which at least indirectly would exclude gene silencing by methylation of CpG regions as the regulatory mechanism implicated in the bi- or even tri-modal CD177 expression. Thus, other not yet identified upstream regulatory mechanisms are likely responsible for the formation of the CD177neg.neutrophil subset. On the contrary, in another report not only CD177 protein but also CD177 mRNA expression was restricted to the CD177pos.subset, whereas no CD177 mRNA was detected within the negative subset [22]. Further experimental data are needed to resolve this seeming discrepancy.
Taken together, these results prove the multiple complex mechanisms regulating CD177 expression on the neutrophil surface as described before. The CD177 *c.787A>T -induced premature stop codon is definitely the main reason for the HNA-2nullphenotype and a low proportion of the CD177neg.neutrophil subset. CD177 *c.1291G>A accounts for the absence of CD177 expression in most of the remaining cases. However, a definite identification of all molecular mechanisms is indispensable for molecular typing of HNA-2, which moreover is aggravated by the fact that an additional long-range PCR reaction is necessary to discriminate a CD177 -specific template from the CD177 pseudogene [19]. Recently, a fast screening method based on TaqManTM-PCR was introduced for the identification of HNA-2nullindividuals homozygous for the *c.787A>T mutation in both CD177 and CD177P1 genes [32]. However, HNA-2nullindividuals who are able to develop the HNA-2 isoantibody cannot be reliably identified by molecular methods until the molecular basis has been entirely decoded.
Statement of Ethics
The study was approved by the ethics boards of the contributing institutions. All donors gave informed consent.
Disclosure Statement
The authors disclose no conflicts of interest.
Funding Sources
The study was supported by in-house grants from the German Red Cross Blood Service West.
Author Contributions
B.K.F. initiated and coordinated the study, characterized and analyzed samples, and wrote the manuscript. B.B. added new features, analyzed samples, and actively contributed to the manuscript. A.R., N.N., C.C., P.B., T.J.S., E.H., L.P., P.H., P.R., M.S., H.K., and J.K. all characterized and provided samples, discussed the topic, and carefully revised the manuscript.
Acknowledgments
The authors are grateful to Anne Janson, Monika Steitz, Heike Berghöfer, and Gabriela Michel for excellent technical expertise.
References
- 1.Lalezari P, Murphy GB, Allen FH., Jr NB1, a new neutrophil-specific antigen involved in the pathogenesis of neonatal neutropenia. J Clin Invest. 1971 May;50((5)):1108–15. doi: 10.1172/JCI106582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sachs UJ, Hattar K, Weissmann N, Bohle RM, Weiss T, Sibelius U, et al. Antibody-induced neutrophil activation as a trigger for transfusion-related acute lung injury in an ex vivo rat lung model. Blood. 2006 Feb;107((3)):1217–9. doi: 10.1182/blood-2005-04-1744. [DOI] [PubMed] [Google Scholar]
- 3.Bux J, Becker F, Seeger W, Kilpatrick D, Chapman J, Waters A. Transfusion-related acute lung injury due to HLA-A2-specific antibodies in recipient and NB1-specific antibodies in donor blood. Br J Haematol. 1996 Jun;93((3)):707–13. doi: 10.1046/j.1365-2141.1996.d01-1703.x. [DOI] [PubMed] [Google Scholar]
- 4.Stroncek DF, Skubitz KM, McCullough JJ. Biochemical characterization of the neutrophil-specific antigen NB1. Blood. 1990 Feb;75((3)):744–55. [PubMed] [Google Scholar]
- 5.Goldschmeding R, van Dalen CM, Faber N, Calafat J, Huizinga TW, van der Schoot CE, et al. Further characterization of the NB 1 antigen as a variably expressed 56-62 kD GPI-linked glycoprotein of plasma membranes and specific granules of neutrophils. Br J Haematol. 1992 Jul;81((3)):336–45. doi: 10.1111/j.1365-2141.1992.tb08237.x. [DOI] [PubMed] [Google Scholar]
- 6.Göhring K, Wolff J, Doppl W, Schmidt KL, Fenchel K, Pralle H, et al. Neutrophil CD177 (NB1 gp, HNA-2a) expression is increased in severe bacterial infections and polycythaemia vera. Br J Haematol. 2004 Jul;126((2)):252–4. doi: 10.1111/j.1365-2141.2004.05027.x. [DOI] [PubMed] [Google Scholar]
- 7.Wolff JC, Goehring K, Heckmann M, Bux J. Sex-dependent up regulation of CD 177-specific mRNA expression in cord blood due to different stimuli. Transfusion. 2006 Jan;46((1)):132–6. doi: 10.1111/j.1537-2995.2005.00676.x. [DOI] [PubMed] [Google Scholar]
- 8.Kissel K, Santoso S, Hofmann C, Stroncek D, Bux J. Molecular basis of the neutrophil glycoprotein NB1 (CD177) involved in the pathogenesis of immune neutropenias and transfusion reactions. Eur J Immunol. 2001 May;31((5)):1301–9. doi: 10.1002/1521-4141(200105)31:5<1301::AID-IMMU1301>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 9.Bierling P, Poulet E, Fromont P, Seror T, Bracq C, Duedari N. Neutrophil-specific antigen and gene frequencies in the French population. Transfusion. 1990 Nov-Dec;30((9)):848–9. doi: 10.1046/j.1537-2995.1990.30991048794.x. [DOI] [PubMed] [Google Scholar]
- 10.Bux J. Molecular genetics of granulocyte polymorphisms. Vox Sang. 2000;78(Suppl 2):125–30. [PubMed] [Google Scholar]
- 11.Matsuo K, Lin A, Procter JL, Clement L, Stroncek D. Variations in the expression of granulocyte antigen NB1. Transfusion. 2000 Jun;40((6)):654–62. doi: 10.1046/j.1537-2995.2000.40060654.x. [DOI] [PubMed] [Google Scholar]
- 12.Stroncek DF, Plachta LB, Herr GP, Dalmasso AP. Analysis of the expression of neutrophil-specific antigen NB1: characterization of neutrophils that react with but are not agglutinated by anti-NB1. Transfusion. 1993 Aug;33((8)):656–60. doi: 10.1046/j.1537-2995.1993.33893342747.x. [DOI] [PubMed] [Google Scholar]
- 13.Wu Z, Liang R, Ohnesorg T, Cho V, Lam W, Abhayaratna WP, et al. Heterogeneity of Human Neutrophil CD177 Expression Results from CD177P1 Pseudogene Conversion. PLoS Genet. 2016 May;12((5)):e1006067. doi: 10.1371/journal.pgen.1006067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kissel K, Scheffler S, Kerowgan M, Bux J. Molecular basis of NB1 (HNA-2a, CD177) deficiency. Blood. 2002 Jun;99((11)):4231–3. doi: 10.1182/blood.v99.11.4231. [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Mair DC, Schuller RM, Li L, Wu J. Genetic mechanism of human neutrophil antigen 2 deficiency and expression variations. PLoS Genet. 2015 May;11((5)):e1005255. doi: 10.1371/journal.pgen.1005255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moritz E, Chiba AK, Kimura EY, Albuquerque D, Guirão FP, Yamamoto M, et al. Molecular studies reveal that A134T, G156A and G1333A SNPs in the CD177 gene are associated with atypical expression of human neutrophil antigen-2. Vox Sang. 2010 Feb;98((2)):160–6. doi: 10.1111/j.1423-0410.2009.01233.x. [DOI] [PubMed] [Google Scholar]
- 17.Wolff J, Brendel C, Fink L, Bohle RM, Kissel K, Bux J. Lack of NB1 GP (CD177/HNA-2a) gene transcription in NB1 GP- neutrophils from NB1 GP-expressing individuals and association of low expression with NB1 gene polymorphisms. Blood. 2003 Jul;102((2)):731–3. doi: 10.1182/blood-2002-09-2831. [DOI] [PubMed] [Google Scholar]
- 18.Caruccio L, Walkovich K, Bettinotti M, Schuller R, Stroncek D. CD177 polymorphisms: correlation between high-frequency single nucleotide polymorphisms and neutrophil surface protein expression. Transfusion. 2004 Jan;44((1)):77–82. doi: 10.1046/j.0041-1132.2004.00606.x. [DOI] [PubMed] [Google Scholar]
- 19.Bayat B, Bein G, Sachs UJ. A sequence-specific polymerase chain reaction method for HNA-2 genotyping: homozygous c.843A[{GT}]T mutation predicts the absence of CD177. Transfusion. 2016 Aug;56((8)):2127–32. doi: 10.1111/trf.13689. [DOI] [PubMed] [Google Scholar]
- 20.Flesch BK, Reil A. A.: Molecular genetics of the human neutrophil antigens. Transfus Med Hemother. 2018 Oct;45((5)):300–9. doi: 10.1159/000491031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu J, Li Y, Schuller RM, Li L, Litmeyer AS, Bein G, et al. The nonconservative CD177 single-nucleotide polymorphism c.1291G[{GT}]A is a genetic determinant for human neutrophil antigen-2 atypical/low expression and deficiency. Transfusion. 2019 May;59((5)):1836–42. doi: 10.1111/trf.15222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eulenberg-Gustavus C, Bähring S, Maass PG, Luft FC, Kettritz R. Gene silencing and a novel monoallelic expression pattern in distinct CD177 neutrophil subsets. J Exp Med. 2017 Jul;214((7)):2089–101. doi: 10.1084/jem.20161093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lalezari P, Bernard GE. Identification of a specific leukocyte antigen: another presumed example of 5b. Transfusion. 1965 Mar-Apr;5((2)):135–42. doi: 10.1111/j.1537-2995.1965.tb01148.x. [DOI] [PubMed] [Google Scholar]
- 24.Verheugt FW, von dem Borne AE, Décary F, Engelfriet CP. The detection of granulocyte alloantibodies with an indirect immunofluorescence test. Br J Haematol. 1977 Aug;36((4)):533–44. doi: 10.1111/j.1365-2141.1977.tb00994.x. [DOI] [PubMed] [Google Scholar]
- 25.Reil A, Bux J. Geno- and phenotyping of human neutrophil antigens. Methods Mol Biol. 2015;1310:193–203. doi: 10.1007/978-1-4939-2690-9_16. [DOI] [PubMed] [Google Scholar]
- 26.Nguyen XD, Flesch B, Sachs UJ, Kroll H, Klüter H, Müller-Steinhardt M. Rapid screening of granulocyte antibodies with a novel assay: flow cytometric granulocyte immunofluorescence test. Transfusion. 2009 Dec;49((12)):2700–8. doi: 10.1111/j.1537-2995.2009.02330.x. [DOI] [PubMed] [Google Scholar]
- 27.Bux J, Kober B, Kiefel V, Mueller-Eckhardt C. Analysis of granulocyte-reactive antibodies using an immunoassay based upon monoclonal-antibody-specific immobilization of granulocyte antigens. Transfus Med. 1993 Jun;3((2)):157–62. doi: 10.1111/j.1365-3148.1993.tb00055.x. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen XD, Scherpf R, Sassenhof F, Flesch B, Klüter H. Detection of granulocyte antibodies using simultaneous analysis of specific granulocyte antibodies assay (SASGA) Vox Sang. 2011 Aug;101((2)):147–53. doi: 10.1111/j.1423-0410.2011.01470.x. [DOI] [PubMed] [Google Scholar]
- 29.Flesch BK, Achtert G, Bauer F, Neppert J. The NA“null” phenotype of a young man is caused by an Fc gammaRIIIB gene deficiency while the products of the neighboring Fc gammaRIIA and Fc gammaRIIIA genes are present. Ann Hematol. 1998 May;76((5)):215–20. doi: 10.1007/s002770050392. [DOI] [PubMed] [Google Scholar]
- 30.Nguyen XD, Schulze TJ, Bugert P, Lauber-Härtl S, Schulz-Linkholt M, González-Schulze K, et al. Granulocyte antibodies in male blood donors: can they trigger transfusion-related acute lung injury? Transfusion. 2018 Aug;58((8)):1894–901. doi: 10.1111/trf.14630. [DOI] [PubMed] [Google Scholar]
- 31.Hu N, Mora-Jensen H, Theilgaard-Mönch K, Doornbos-van der Meer B, Huitema MG, Stegeman CA, et al. Differential expression of granulopoiesis related genes in neutrophil subsets distinguished by membrane expression of CD177. PLoS One. 2014 Jun;9((6)):e99671. doi: 10.1371/journal.pone.0099671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Portegys J, Rink G, Bloos P, Scharberg EA, Klüter H, Bugert P. Towards a Regional Registry of Extended Typed Blood Donors: Molecular Typing for Blood Group, Platelet and Granulocyte Antigens. Transfus Med Hemother. 2018 Oct;45((5)):331–40. doi: 10.1159/000493555. [DOI] [PMC free article] [PubMed] [Google Scholar]