Abstract
Introduction
Damage to a cell and the loss of integrity of its cell membrane leads to the release of endogenous immunogenic molecules, which together are classified as “damage-associated molecular patterns” (DAMPs). Cell-free DNA (cf-DNA) released from nucleosomes may serve as a procoagulant cofactor and may be an important mediator of immunomodulatory and proinflammatory effects associated with blood transfusion.
Objectives
To assess the levels of cf-DNA in supernatants of stored red cell components and the effect of leukoreduction and gamma irradiation on the release of cf-DNA during storage.
Methods
This is a prospective cohort study on 99 stored red cell components, randomly divided into three groups − buffy coat (BC)-depleted, leuko-filtered (LP), and irradiated (IR) packed red blood cells. Red cell supernatants were drawn over a period of 21 days at three different time points (day 0, 7, and 21) from the study units. cf-DNA extraction was done and quantified by a bench top fluorometer. Change in cf-DNA content, rate of change (μg/day), and percent change were estimated and compared across different groups.
Results
cf-DNA content increased (p = 0.000) with storage duration in the BC (median = 238.66 μg, interquartile range [IQR] = 168.42 on day 21 vs. median = 9.44 μg, IQR = 5.23 on day 0) and IR groups (p = 0.000) (median = 245.55 μg, IQR = 253.88 on day 21 vs. median = 7.07 μg, IQR = 13.58 on day 0), while there was a decreasing trend (p = 0.032) in the LP group (median = 4.55 μg, IQR = 10.73 on day 21 vs. median = 8.66 μg, IQR = 6.56 on day 0). The median rate of change in cf-DNA content (11.13 μg/day) and percent change in cf-DNA content (median = 4,106.16%) was highest in the IR group.
Conclusions
Stored red cell components contain significant amount of cf-DNA. Release of cf-DNA is further aggravated by irradiation while leukoreduction leads to a decrease in cf-DNA content.
Keywords: DAMPs, Cell-free DNA, Red cell components, Gamma irradiation, Immune response, Storage, Leukodepletion
Introduction
Transfusion of blood components has the potential to alter the clinical outcome or confound an already complex clinical situation by way of immunomodulation. There are multiple factors that lead to the immunomodulatory effects associated with blood transfusion and with the extended storage this ability increases [1].
Damage-associated molecular patterns (DAMPs) have been identified [2, 3] as endogenous immunogenic molecules which are released due to damage to a cell and the loss of integrity of its cell membrane. DAMPs act through the same pattern recognition receptors [4, 5] which are involved in immune response mediated by pathogen-associated molecular patterns (PAMPs). The response generated by both DAMPs and PAMPs is therefore very similar at the molecular level. Involvement of DAMPs has been shown in a large number of life-threatening sterile inflammatory conditions, including acute kidney injury, lupus nephritis, and progression of chronic kidney disease in diabetic and nondiabetic glomerulonephritis [6, 7], and release of DAMPs also contributes to tumor growth by mediating skewing of antitumor immunity during so-called immunogenic tumor cell death [8].
Cell-free DNA (cf-DNA), one of the DAMPs, has been identified as an important constituent of neutrophil extracellular traps (NETs), involved in host defense against infection [9]. Patients with various pathologic states including thrombosis, cancer, sepsis, and trauma have been shown to contain increased levels of cf-DNA [10, 11]. In vivo sources of cf-DNA include damaged neutrophils [12, 13, 14], macrophages [15], eosinophils [16], tumor cells [17], and several bacterial agents [18, 19]. In vivo, cf-DNA is involved in a newly described form of cell death known as NETosis [20].
cf-DNA has also been shown to induce in vivo coagulation by serving as a natural foreign surface [21]. Polyanionic molecules (DNA, RNA) activate proteases of the contact pathway of blood coagulation such as factor XII and XI [21, 22]. Similarly, cf-DNA also triggers blood coagulation via the contact pathway [23, 24]. Thus, cf-DNA is capable of triggering inflammatory responses and activating coagulation, and has a potential role in the pathogenesis of sepsis [25, 26].
We hypothesize that stress and sheer force applied on cellular blood components during processing and storage may induce cellular damage and release DAMPs which gradually accumulate in the supernatant of these components. These DAMPs may lead to immunomodulatory and inflammatory response in transfusion recipients. The presence of DAMPs has recently been related to various adverse effects of blood transfusion including TRALI [27, 28, 29, 30] and nonhemolytic transfusion reactions following platelet transfusion. HMGB1 and sRAGE, two of the important DAMPs [28], have not been found to have a role in the development of TRALI in cardiac surgery patients. cf-DNA has the potential to lead to such an adverse outcome.
In this study we have quantified cf-DNA in supernatants of stored red cell components and assessed the effect of leukoreduction and gamma irradiation on the release of cf-DNA during storage.
Materials and Methods
This was a prospective randomized study on stored red cell components at a tertiary care center in North India performed over a period between January 2017 and October 2018 after due approval from the institute's ethics committee.
A total of 450 mL of whole blood collected from nonremunerated voluntary blood donors was processed for the preparation of red cell component by buffy coat (BC) method in the top and bottom quadruple blood bag system with saline-adenine-glucose-mannitol (SAGM) as red cell additive solution (Cat no PB-4BO456J0B; Terumo Penpol, Kerala, India). Female donors and donors with a history of any immune disorders or intake of immunomodulatory drugs during the last year, abnormal total leukocyte count or differential leukocyte count, thrombocytopenia (platelet count <1.5 × 109/L), reactive during screening for transfusion-transmitted infections, and whole blood collection time >12 min were excluded from the study.
Preparation of components was done within a time period of 2 h. Briefly, the blood bags were first given a hard spin (3,800 rpm × 9 min, 22°C) using a refrigerated centrifuge (Cryofuge 6000i, Heraeus; Thermo Scientific, USA), after which plasma and packed red blood cells (PRBC) were separated using TACE-II (Terumo Automatic Component Extractor − II; Terumo Penpol). At this point, 100 mL of SAGM solution was added to prepare the BC-depleted PRBC (BC-PRBC). The residual BC was used for the preparation of platelet concentrate after hanging for a period of at least 1.5 h.
Study Groups
A total of 120 units were randomly assigned into one of the three study groups using computer-generated random sequence (www.random.org). However, 21 units were later excluded due either to low platelet count (n = 19) or a reactive transfusion-transmitted infection screen (n = 2). Thus, a total of 99 units were included in analysis.
Study group 1 (control group) comprised 33 BC-PRBC prepared as described above. Study group 2 (leuko-filtered group, LP-PRBC) comprised 32 BC-PRBC leuko-filtered within 1 h of component preparation using Imugard III-RC 4P (Terumo Corporation, Tokyo, Japan). Under all aseptic conditions, the BC-PRBC bag was attached to the leukofilter using a sterile connecting device (TSCD-II, Terumo Sterile Tubing Welder; Terumo Penpol) which was then attached to a transfer bag. The combination was hung for about 30 min at room temperature to allow the contents of the PRBC unit to pass through the leukofilter and accumulate in the transfer bag. LP-PRBC were separated from the combination using a di-electric tube sealer.
Study group 3 (irradiated group, IR-PRBC) comprised 34 BC-PRBC subjected to 25 Gy of irradiation within 1 h of preparation using a self-contained gamma irradiator (Gammacell 1000-Elite; Nordion International Inc.).
All the study units were stored for a period of 21 days under standard storage conditions considering the total allowable storage of IR-PRBC units of 28 days after irradiation. After completion of sampling from the study units all the units were included in the inventory for transfusion.
Sampling Protocol
Samples from red cell supernatant were withdrawn on the day of preparation (day 0), day 7, and day 21 from individual units with the help of a Luer adapter and sample diversion pouch (Terumo Penpol) to maintain strict aseptic conditions. The PRBC bag was given a soft spin (1,050 rpm × 9 min, 4°C) (Cryofuge 6000i Heraeus; ThermoFisher Scientific, USA) after attaching the sampling assembly using TSCD − II (Terumo Penpol). One mL of supernatant was withdrawn from each bag on each of the designated sampling days under closed sterile conditions in a plain vacutainer through the Luer adapter. The collected red cell supernatant was given hard spin (3,000 rpm × 5 min, 4°C) using a benchtop centrifuge (Heraeus Megafuge 8; ThermoFisher Scientific) to remove any remaining cells from the collected red cell supernatant. The clear cell-free supernatant was collected and stored at or below −40°C until further analysis.
Baseline Investigations on Study Units
A 1-mL representative sample from individual red cell units was withdrawn under strict aseptic conditions after thorough stripping immediately after preparation in the BC-PRBC group and immediately after leuko-filtration or irradiation in the other two groups. White blood cell (WBC) count, platelet count, red cell count, hemoglobin content, and hematocrit content of each sample was performed using an automated cell counter (M16/M20, Medonic; Boule Diagnostics AB, Sweden). WBC count in the LP-PRBC group was manually done using a Nageotte hemocytometer with a large-size cover slip (24 × 24 × 0.4 mm). Briefly, 100 µL of the test sample was added to 40 µL of the lysing agent (Zapoglobin; Coulter Electronics, Hialeah, FL) in a labeled test tube. The mixture was rinsed thoroughly to lyse all red cells, and 360 µL of the Turk's solution (prepared by adding 1 mL of 1% gentian violet and 3 mL of glacial acetic acid diluted to 100 mL with distilled water) was then added to the test tube. The counting chamber was carefully loaded with the diluted sample and the loaded chamber was placed in a moist petri dish for 15 min to allow the leukocytes to settle. The WBCs were counted using a 20× objective lens of a compound microscope within 30 min across 40 rectangular area grids of the counting chamber grid. The leukocyte count was then estimated using the formula:
Leukocyte count (per µL) = number of cells counted/10
Estimation of cf-DNA
cf-DNA extraction from the stored red cell supernatants was carried out using the QIAamp DNA Blood Mini Kit (Qiagen, Germany) as per the manufacturer's instructions [31]. Concentration of cf-DNA was measured by fluorometry (Qubit-2.0; Life Technologies, ThermoFisher Scientific) for measurement of cf-DNA, using the Qubit dsDNA HS Assay Kit.
Statistical Analysis
Data of individual variables/parameters was checked for normality of distribution using the Shapiro-Wilk test. Of all the variables, only donor hematocrit and RBC content of the study units were found to have normal distribution. Hence, the median was used as a measure of central tendency, and percentile values (25th and 75th; 1Q–3Q), range (R), and interquartile range (IQR) were used as a measure of variability. Nonparametric tests (Kruskal-Wallis H test, Mann-Whitney U test, Spearman's rank correlation) were used for data analysis using SPSS statistics software (version 20.0; IBM Corp., New York, NY, USA). A p value of <0.05 was considered significant. Linear regression analysis was performed to assess significant contributing factors determining cf-DNA levels.
For the purpose of analysis, various calculations were done as per the following equations:
WBC content (total number of WBCs in 1 study unit) = WBC count (per μL) × volume of PRBC (mL) × 103
Platelet content (total number of platelets in 1 study unit) = platelet count (per μL) × volume of PRBC (mL) × 103
Red cell content (total number of red cells in 1 study unit) = RBC count (per μL) × volume of PRBC (mL) × 103
Hemoglobin content (total hemoglobin in 1 study unit, in grams) = hemoglobin concentration (g/dL) × volume of PRBC (mL)/100
Volume of red cell supernatant (mL) of individual study unit = ([100 − hematocrit]/100) × volume of PRBC (mL)
cf-DNA content (µg) (total cf-DNA in 1 study unit) = volume of red cell supernatant (mL) × cf-DNA concentration (ng/μL)
Trends in cf-DNA content over the storage period: change in cf-DNA on day 7 and day 21 from baseline levels (day 0) was estimated for individual units as follows:
Change in cf-DNA content (Δ cf-DNA) (µg) = content on day X − content on day Y
Rate of change in cf-DNA content (Δ/t cf-DNA) (µg/day) = Δ cf-DNA/X − Y
Percent change in cf-DNA content (pΔ cf-DNA): (Δ cf-DNA × 100)/cf-DNA content on day Y
(where X is the later day of sampling and Y the earlier day of sampling).
Results
The characteristics of the units included in the study are shown in Table 1. Volume of PRBC, volume of supernatant, RBC content, WBC content, and hemoglobin content were found to be significantly lower in the LP-PRBC group compared to either the BC-PRBC or IR-PRBC groups. Platelet content, however, was slightly higher in the LP-PRBC group compared to the IR-PRBC group (2.24 × 109vs. 2.14 × 109, p = 0.024). Median platelet content in the BC-PRBC group was lower than that in the LP-PRBC group; however, the difference was not statistically significant.
Table 1.
Median values of various hematological parameters of study units
| Parameter | BC-PRBC (n = 33) | LP-PRBC (n = 32) | IR-PRBC (n = 34) | p value | |
|---|---|---|---|---|---|
| Volume, mL | M | 272.00 | 235.00 | 267.00 | 0.000* |
| R | 46 | 31 | 16.70 | 0.000* | |
| IQR | 17 | 13 | 2.95 | 0.055* | |
| 1Q–3Q | 265.00–281.50 | 230.00–242.75 | 265.00–273.50 | ||
| Supernatant volume, mL M | 107.68 | 94.46 | 105.95 | 0.000* | |
| R | 40.50 | 30.85 | 16.28 | 0.000* | |
| IQR | 12.45 | 9.05 | 12.30 | 0.236† | |
| 1Q–3Q | 102.95–115.40 | 99.88–108.93 | 129.20–141.49 | ||
| RBC content, ×1012 | M | 1.80 | 1.54 | 1.77 | 0.000* |
| R | 0.81 | 0.77 | 1.09 | 0.000† | |
| IQR | 0.34 | 0.26 | 0.27 | 0.448‡ | |
| 1Q–3Q | 1.63–1.97 | 1.38–1.64 | 1.61–1.88 | ||
| Platelet content, ×109 | M | 2.11 | 2.24 | 1.87 | 0.78* |
| R | 4.67 | 5.12 | 4.45 | 0.024† | |
| IQR | 1.27 | 0.90 | 0.79 | 0.053‡ | |
| 1Q–3Q | 1.75–3.01 | 1.85–2.75 | 1.35–2.14 | ||
| WBC content, ×106 | M | 812.20 | 0.07 | 712.90 | 0.000* |
| R | 1,989.60 | 0.08 | 2,507.70 | 0.000† | |
| IQR | 588.00 | 0.04 | 609.93 | 0.527‡ | |
| 1Q–3Q | 506.70–1,094.70 | 0.05–0.09 | 339.95–949.88 | ||
| Hemoglobin content, g | M | 51.87 | 44.06 | 51.38 | 0.000* |
| R | 22.22 | 10.26 | 14.23 | 0.000† | |
| IQR | 5.70 | 4.43 | 3.88 | 0.603‡ | |
| 1Q–3Q | 49.47–55.17 | 42.46–46.89 | 49.57–53.45 | ||
| Hematocrit, % | M | 60.00 | 60.80 | 60.50 | 0.783* |
| R | 22.20 | 14.70 | 16.70 | 0.216† | |
| IQR | 4.50 | 3.55 | 2.95 | 0.295‡ | |
| 1Q–3Q | 57.95–62.45 | 58.00–61.55 | 59.33–62.28 | ||
M, median; R, range; IQR, interquartile range; 1Q–3Q, 25th–75th percentile values.
p value between BC-PRBC and LP-PRBC groups;
p value between LP-PRBC and IR-PRBC groups;
p value between BC-PRBC and IR-PRBC groups. Significant p values are highlighted in bold (Mann-Whitney U test).
cf-DNA Content over the Study Period
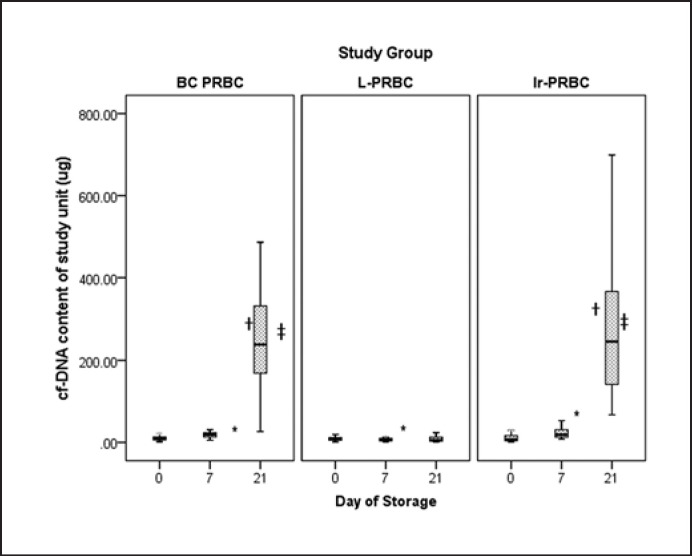
cf-DNA content of individual units on different days of sampling across different study groups was used for analysis instead of cf-DNA concentration (Fig. 1). In the BC-PRBC group, there was a gradual and statistically significant (p = 0.000, Kruskal-Wallis H test) increase in cf-DNA content (µg) ([median 9.44, R = 28.74, IQR = 8.19, 1Q–3Q = 5.23–13.42 on day 0], [median = 19.57, R = 37.35, IQR = 10.16, 1Q–3Q = 13.22–23.38 on day 7], and [median = 238.66, R = 1,428.41, IQR = 168.42, 1Q–3Q = 168.66–337.07 on day 21]) over the study period. In the LP-PRBC group, however, there was a gradual decrease (p = 0.032, Kruskal-Wallis H test) in cf-DNA content (µg) ([median = 8.66, R = 18.06, IQR = 6.56, 1Q–3Q = 5.26–11.82 on day 0], [median = 6.62, R = 24.04, IQR = 5.87, 1Q–3Q = 4.04–9.91 on day 7], and [median = 4.55, R = 1,109.31, IQR = 10.73, 1Q–3Q = 3.52–14.24 on day 21]) over the study period. The decrease from day 7 to day 21, however, was not statistically significant (p = 0.957, Wilcoxon signed-rank test). In the IR-PRBC group, as in the BC-PRBC group, there was a gradual and statistically significant (p = 0.000) increase in cf-DNA content (µg) ([median = 7.07, R = 40.37, IQR = 13.58, 1Q–3Q = 4.08–17.66 on day 0], [median = 19.05, R = 44.89, IQR = 18.16, 1Q–3Q = 12.84 −30.99 on day 7], and [median = 245.55, R = 764.56, IQR = 253.88, 1Q–3Q = 140.7 0–394.58 on day 21]) over the study period. The baseline (day 0) cf-DNA content was comparable across all three groups (p = 0.638, Kruskal-Wallis H test).
Fig. 1.
Median values of cf-DNA content on different days of storage. * Shows p value between day 0 and day 7, †showsp value between day 7 and day 21, ‡shows p value between day 0 and day 21. p values (Wilcoxon signed-rank test): BC-PRBC group (day 0 vs. day 7, p = 0.00; day 7 vs. day 21, p = 0.00; day 0 vs. day 21, p = 0.00), LP-PRBC group (day 0 vs. day 7, p = 0.032; day 7 vs. day 21, p = 0.957; day 0 vs. day 21, p = 0.820), IR-PRBC group (day 0 vs. day 7, p = 0.00; day 7 vs. day 21, p = 0.00, day 0 vs. day 21, p = 0.00).
Trends in cf-DNA Content over the Study Period
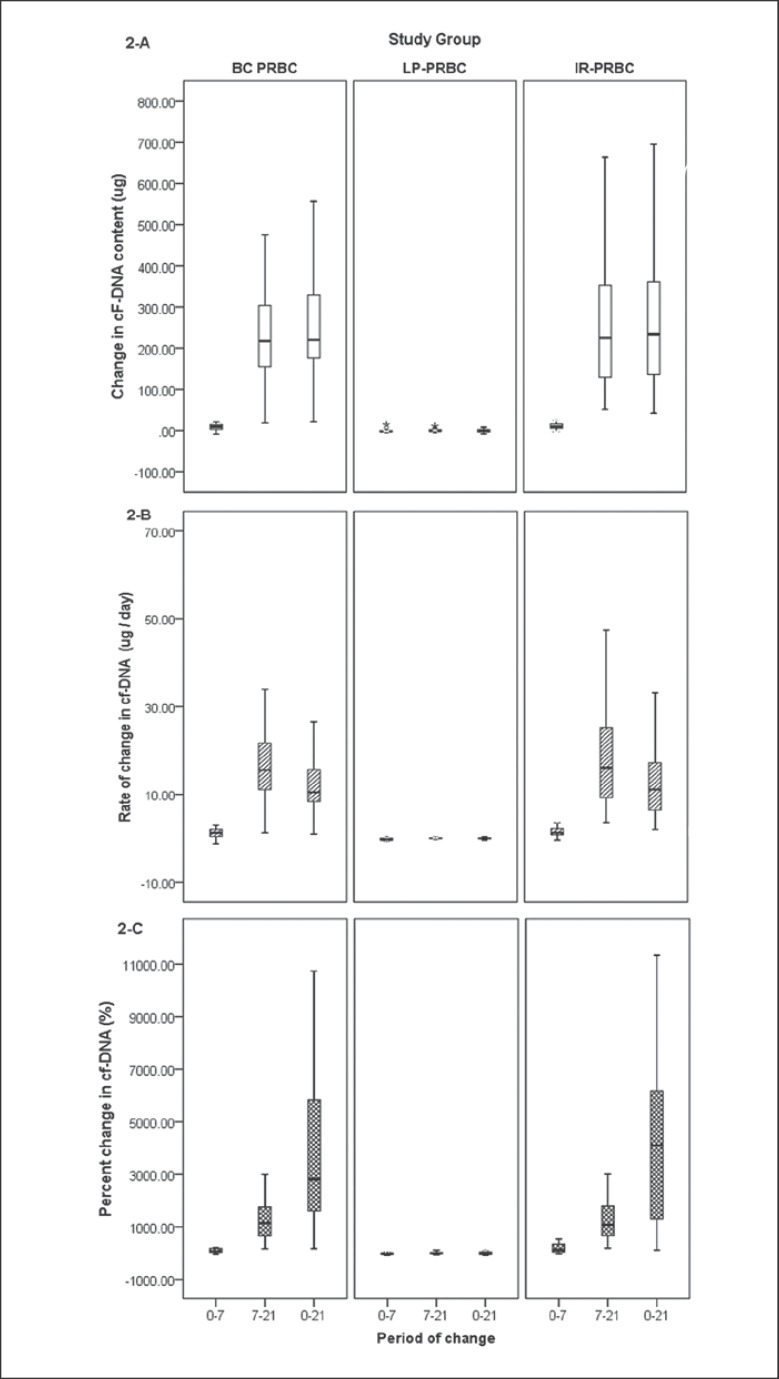
Median Δ cf-DNA over the entire storage period (day 0 vs. day 21) in the IR-PRBC group (233.73 µg) was comparable to that in the BC-PRBC group (220.5 µg) but significantly higher (p = 0.000, Mann-Whitney U test) than that in the LP-PRBC group (–0.50 µg) (Table 2; Fig. 2). Δ cf-DNA was significantly lower during the initial storage period (day 0–day 7) compared to the later phase (day 7–day 21) in both the BC-PRBC (p = 0.000) and IR-PRBC (p = 0.000) groups, while there was a gradual decrease in cf-DNA content reflected by negative Δ cf-DNA in the LP-PRBC group. This decrease was more during the initial phase of storage (day 0–day 7) compared to the later phase, the difference in Δ cf-DNA during both periods in the LP-PRBC group, however, was not statistically significant. Δ/t cf-DNA and pΔ cf-DNA followed the trends as shown by Δ cf-DNA. It was interesting to note that even small differences in Δ cf-DNA seemed to be much more significant when assessed by pΔ cf-DNA estimation.
Table 2.
Change in cf-DNA content of study units with storage
| Storage period (days) | Change in cf-DNA |
|||
|---|---|---|---|---|
| change in content µg, | rate of change, μg/day | percent change, % | ||
| BC-PRBC | ||||
| Initial period (0–7) | M | 9.08 | 1.30 | 76.19 |
| (n = 31) | R | 45.44 | 6.49 | 1,333.26 |
| IQR | 11.56 | 1.65 | 176.43 | |
| 1Q–3Q | 3.04 to 14.60 | 0.43 to 2.09 | 30.39 to 206.82 | |
| Later period (7–21) | M | 217.67* (0.000) | 15.55* (0.000) | 1,175.59* (0.000) |
| (n = 33) | R | 1,418.73 | 101.34 | 7,841.49 |
| IQR | 159.17 | 11.37 | 1,103.32 | |
| 1Q–3Q | 166.06 to 335.46 | 11.10 to 22.47 | 669.23 to 1,772.55 | |
| Total period (0–21) | M | 220.52f(0.000) | 10.50f(0.000) | 2,815.66* (0.000) |
| (n = 31) | R | 1,420.22 | 67.63 | 20,638.26 |
| IQR | 167.41 | 8.07 | 4,706.94 | |
| 1Q–3Q | 155.44 to 314.60 | 7.91 to 15.97 | 1,408.33 to 6,115.28 | |
| LP-PRBC | ||||
| Initial period (0–7) | M | −1.92 | −0.27 | −16.89 |
| (n = 28) | R | 22.72 | 3.25 | 377.83 |
| IQR | 2.88 | 0.41 | 36.03 | |
| 1Q–3Q | −2.92 to −0.04 | −0.42 to −0.005 | −37.04 to −1.07 | |
| Later period (7–21) | M | −0.25 | −0.02 | −3.25 |
| (n = 29) | R | 1,117.13 | 79.80 | 43,310 |
| IQR | 3.20 | 0.23 | 68.11 | |
| 1Q–3Q | −1.35 to 1.85 | −0.10 to 0.13 | −24.59 to 43.53 | |
| Total period (0–21) | M | −0.50 | −0.02 | −4.31 |
| (n = 28) | R | 1,113.89 | 53.04 | 2,056.28 |
| IQR | 6.31 | 0.36 | 94.42 | |
| 1Q–3Q | −3.27 to 3.05 | −0.16 to 0.15 | −48.52 to 45.90 | |
| IR-PRBC | ||||
| Initial period (0–7) | M | 9.20 | 1.31 | 131.21 |
| (n = 34) | R | 52.15 | 7.45 | 1,747.75 |
| IQR | 11.30 | 1.61 | 322.69 | |
| 1Q–3Q | 5.17 to 16.46 | 0.74 to 2.35 | 42.68 to 365.38 | |
| Later period (7–21) | M | 225.49* (0.000) | 16.11* (0.000) | 1,086.34* (0.000) |
| (n = 34) | R | 768.43 | 54.89 | 6,322.03 |
| IQR | 242.55 | 17.33 | 1,173.06 | |
| 1Q–3Q | 129.73 to 372.28 | 9.27 to 26.59 | 650.29 to 1,823.34 | |
| Total period (0–21) | M | 233.72* (0.000) | 11.13* (0.000) | 4,106.16* (0.000) |
| (n = 34) | R | 776.74 | 36.99 | 19,512.64 |
| IQR | 250.06 | 11.91 | 5,199.13 | |
| 1Q–3Q | 134.29 to 384.35 | 6.39 to 18.30 | 1,235.19 to 6,434.32 | |
M, median; R, range; IQR, interquartile range; 1Q–3Q, 25th–75th percentile values. Intragroup comparison: * shows significant difference between initial and later storage periods (day 0–7 vs. day 7–21) within one study group. Change in cf-DNA content, rate of change, and percent change were significantly higher (Wilcoxon signed-rank test) in the BC-PRBC and IR-PRBC groups during the later period (day 7–21) of storage compared to the initial period (day 0–7). There was no statistically significant difference in the LP-PRBC group for all three parameters during the later period compared to the initial period of storage. Intergroup comparison: p values were assessed across various groups for changes in cf-DNA over the total storage duration (day 0–21) only (Mann-Whitney U Test): †shows significant difference between BC-PRBC and LP-PRBC groups; ‡shows significant difference between LP-PRBC and IR-PRBC groups. There was no statistically significant difference between the BC-PRBC and IR-PRBC groups for change in content (p = 1.000), rate of change (p = 0.670), or percent change (p = 0.743). Values in parentheses beside the symbols indicating significant differences indicate the respective p values.
Fig. 2.
Mathematical derivatives to depict change in cf-DNA content across different groups on different days of storage. AChange in cF-DNA content (Δ cf-DNA). BRate of change in cf-DNA content (Δ/t cf-DNA). CPercent change in cf-DNA content (pΔ cf-DNA). BC-PRBC, buffy coat-depleted packed red blood cells; LP-PRBC, leuko-filtered packed red blood cells; IR-PRBC, irradiated packed red blood cells.
Regarding variables affecting cf-DNA content, no correlation was observed between cf-DNA content on day 0 across all study groups with any of the hematological variables included in the study. However, all the hematological variables except the hematocrit unit were significantly correlated with pΔ cf-DNA (Table 3).
Table 3.
Predictors of percent change in cf-DNA content over the study period (n = 93)
| Parameter | Correlation |
Linear regression analysis |
|||
|---|---|---|---|---|---|
| coefficient (r) | p value | r 2 | β | p value | |
| PRBC volume | 0.446 | 0.000* | NS | ||
| Supernatant volume | 0.223 | 0.002* | NS | ||
| RBC content | 0.321 | 0.000* | 0.092 | 0.221 | 0.000* |
| Platelet content | −0.374 | 0.001* | 0.160 | −0.248 | 0.000* |
| WBC content | 0.498 | 0.000* | 0.182 | 0.165 | 0.000 |
| Hemoglobin content | 0.403 | 0.000* | NS | ||
| Hematocrit unit | 0.104 | 0.159 | NS | ||
NS, not significant. Correlation coefficient and p value according to Spearman's rank correlation.
Statistically significant.
Regression analysis of significantly correlated hematological parameters revealed that pΔ cf-DNA was dependent on three hematological parameters: RBC content (r 2= 0.092, β = 0.22, p = 0.000), WBC content (r 2= 0.182, β = 0.165, p = 0.000), and platelet content (r 2= 0.160, β = −0.248, p = 0.000).
Quantification of cf-DNA could not be done on 9 samples from a total of 7 study units; 5 of these units belonged to the LP-PRBC group and 2 belonged to the BC-PRBC group. On 5 occasions the sample belonged to day 0, on 3 occasions the sample belonged to day 7, and on 1 occasion it belonged to day 21.
Discussion
In this study we were able to demonstrate the presence of cf-DNA in the supernatant of stored red cell components which gradually increased over the storage period (day 0–day 21) except for the LP-PRBC group. For the purpose of statistical analysis of this study we concentrated on unit characteristics and the total content of relevant variables per unit. The reason behind such an approach was to avoid bias in cf-DNA concentration due to the hematocrit variable of the individual units. Correlation of study variables with cf-DNA concentration could have given false results as 2 units with the same cf-DNA concentration may have different cf-DNA content depending upon hematocrit or red cell supernatant volume. It is also noteworthy that only by calculating the total content per unit one can establish the total dose that is being administered to the transfused patient.
In our study we have excluded units collected from female donors to avoid confounding variables as far as possible. Moreover, at our center, we collect 350 mL of blood from female donors irrespective of the weight of the donor, unlike male donors where 450 mL of blood is collected if the donor weighs more than 55 kg. Inclusion of units from female donors would therefore have created a heterogeneous group leading to erroneous results. Gender, however, is a biological variable that affects the functions of the immune system with differences in both innate and adaptive immune responses. Sex chromosome genes and sex hormones as well as environmental factors (like nutrition status and the composition of the microbiome) alter the development and functioning of the immune system differently in males and females [32]. The gender differences in transfusion recipients is highlighted in a study by Caram-Deelder et al. [33], who reported that among patients who received RBC transfusions, receipt of a transfusion from an ever-pregnant female donor, compared with a male donor, was associated with increased all-cause mortality among male recipients but not among female recipients. The authors have attributed this difference to possible sex differences in immunologic mechanisms. Kanias et al. [34] in their study correlating gender with the propensity of RBCs to hemolyze, have reported that RBCs collected from women were more resilient to mechanical and osmotic stresses. In view of the above, exclusion of female donors may have implications on the generalizability of our study results and further research including female donors needs to be done.
All the unit hematological parameters were significantly lower in the LP-PRBC group compared to the other groups due to loss incurred during leuko-filtration. In an earlier study from our center [35], we have reported a PRBC volume loss of approximately 50 mL after leuko-reduction. Lesser volume reduction (37 mL in LP-PRBC vs. BC-PRBC group) observed in the present study may be attributed to a smaller length of tubing due to use of a sterile connecting device and improvement in the quality of the leukofilter used.
The decreased amount of cf-DNA in the LP-PRBC group may be related to the reduction of WBC by filtration. Fuchs et al. [36] have reported a similar finding in their study on leuko-reduced RBC units.
Out of the total 99 study units, cf-DNA could not be quantified in 7 units. The low level of cf-DNA in these samples may partially be attributed to low WBC content as 5 such samples belonged to the LP-PRBC group. Apart from the difference in WBC content of the study units, we did not find any difference in donor characteristics (age, BMI, frequency of donation) or other unit characteristics (hematocrit, hemoglobin content, platelet content, volume of PRBC unit, volume of red cell supernatant) in these units compared to other units where cf-DNA could be quantified (data not shown). Factors affecting the quality during the component preparation process or during storage of red cell components need to be studied in order to delineate the possible causes leading to too low levels in these assay tubes.
Jaax et al. [37] have reported that normal healthy individuals have extracellular nucleic acid concentrations in plasma ranging from 0 to >1 ng/μL. Accordingly, 450 mL of whole blood unit (assuming a hematocrit of 45%) will have about 247.5 mL of plasma and a cf-DNA content between 0 and >247.5 μg. During the process of component preparation, plasma is separated from the PRBC and only a limited amount of cf-DNA will remain in the PRBC unit. During component preparation itself, we believe that a certain amount of cf-DNA will be released from the cellular constituents due to the sheer and stress of centrifugation. The cf-DNA content on day 0 will therefore include the residual cf-DNA of donor origin and cf-DNA released during component preparation. cf-DNA released during storage will be over and above this level. This also explains the lack of correlation of various hematological parameter units with cf-DNA content on day 0 of storage as observed in our study. Estimation of cf-DNA level in plasma-containing components is therefore important to ascertain the amount of cf-DNA actually released during storage. Waldvogel Abramowski et al. [38] in their study on PRBC units at different times of storage showed an average double-stranded DNA concentration of 290 ± 120 ng/mL (0.29 ± 0.12 ng/μL). They found no correlation between the time of storage and the normalized amount of cf-DNA in PRBC units. The increase in cf-DNA content in the BC-PRBC and IR-PRBC groups in our study may be attributed to the release of cf-DNA from WBC during storage. Fuchs et al. [36] analyzed supernatants and blood smears of human RBC units that were either leuko-reduced or not leuko-reduced before storage for markers of NETs. They identified extracellular DNA, which was associated with histones and myeloperoxidase, a marker of neutrophil granules, in supernatants and blood smears of non-leuko-reduced RBC units. These markers of NETs were absent in leuko-reduced RBC units. They also concluded that NETs are liberated during storage of non-leuko-reduced RBC units.
The finding of our study in terms of the increasing trends in these two groups is in contrast to those observed by Shih et al. [39]. They observed that fresh PRBC units overall had a significantly higher concentration of cf-DNA compared to older PRBC units using spectrophotometry (3.77 ± 1.66 μg/mL vs. 3.02 ± 1.44 μg/mL, p = 0.0031) and (3.77 ± 1.66 ng/μL vs. 3.02 ± 1.44 ng/μL), respectively. They observed no significant difference when comparing cf-DNA quantified by PicoGreen in fresh compared to older units. The difference in the results of the current study may be attributed to a difference in the methods used for quantification of cf-DNA as the usable concentration range for spectrophotometer is 0.4–15,000 ng/μL [40] and inaccurate estimation may be encountered in samples having a low concentration (<10 ng/µL) of nucleic acid [41].
Many of the recent studies have focused on mitochondria DNA rather than total cf-DNA as mitochondrial DNA (mtDNA) is thought to be more immunogenic with higher procoagulant potential. Mitochondria contain approximately 1% of total cellular DNA content while the remaining 99% is packed in the form of nuclear DNA. Bakkour et al. [42] have assessed the levels of mtDNA and extracellular vesicles in the supernatant of stored red cell components prepared by nine different manufacturing methods (including two semi-automated buffy coat methods, two semi-automated whole blood filtration methods, two manual whole blood processing methods, and three automated apheresis methods). Bakkour et al. [42] have quantified mtDNA in copies/µL by the rt-PCR method (one copy of mtDNA corresponds to 1.8 × 10−8ng). Our results for the BC-PRBC group (median cf-DNA concentration 0.191 ng/µL on day 7) are comparable to their results for the non-leuko-reduced group (estimated mean mtDNA concentration 0.0095 ng/µL on day 5). Similar to our findings, the authors reported a gradual increase in mtDNA concentration during the initial phase of red cell storage.
In another study conducted by Cognasse et al. [43], in a series of reported adverse effects after platelet concentrate transfusion, the authors found that mtDNA was significantly (p < 0.05) elevated in single donor apheresis platelet concentrates, which produced adverse effects in the recipients, than in the matched control platelet counts (platelet concentrates). The levels of mtDNA were elevated on day 4 (122.55 ± 52.64 ng/L) compared with days 1–3 in the control, nonpathogenic platelet concentrates. A similar profile was observed in platelet counts that were associated with adverse effects, but the peak mtDNA was on day 3 (484 ± 313.45 ng/L) and not day 4 (233.95 ± 109.75 ng/L).
On comparing the three groups in terms of the total amount of cf-DNA present on day 21, we can see that the LP-PRBC group had the least amount of cf-DNA at the end of the study duration with a median of 5.55 μg, whereas the IR-PRBC group had the highest amount of cf-DNA at the end of the study duration with a median of 245.55 μg. The BC-PRBC group had a median of 238.66 μg. These findings suggest that WBCs are the main source of cf-DNA in red cell supernatant during storage. The higher amount of cf-DNA observed in the IR-PRBC group may be due to the cell damage caused by radiation and consequently greater release from the cells. It is therefore reasonable to conclude that pre-storage leuko-filtration of the unit provides the patient with the added benefit of lower cf-DNA levels in the transfused component.
We observed that with duration, not only was the amount of cf-DNA in the BC-PRBC and IR-PRBC groups rising, but also, as the storage period was prolonged, the rate at which cf-DNA was released also increased in an exponential manner. The rate of change between day 0 and day 7 was 1.30 μg/day for the BC-PRBC group and 1.31 μg/day for the IR-PRBC group, whereas between day 7 and day 21 the rate of change was 15.55 μg/day for the BC-PRBC group and 16.11 μg/day for the IR-PRBC group. This may be due to more and more cells undergoing storage-related degenerative changes and necrosis and hence both an increased amount and rate of release with duration of storage.
Both the RBC and WBC content of the unit were found to have a positive correlation with percent change in cf-DNA content. Positive correlation with red cell content is difficult to explain as red cells themselves do not contain DNA. However, we hypothesize that red cells might have an inhibitory effect on enzymes responsible for the breakdown of DNA, thereby increasing the levels of cf-DNA. Another possible mechanism may be the release of more bioactive inflammatory mediators from the increased number of red cells in individual units, leading to accelerated damage of WBC during storage.
The negative correlation of platelet content with percent change in cf-DNA content may be related to the expression of platelet hyaluronidase-2 (HYAL2) enzyme on the surface of the activated platelets. Storage of PRBC at 4°C may increase the activation of platelets as demonstrated by Kattlove et al. [44]. Recently, Albeiroti et al. [45] have demonstrated that platelet HYAL2, stored in α-granules, becomes surface expressed upon activation and functions to degrade extracellular matrix and may also be involved in the sequestration of bacterial DNA. This same effect of sequestration of cf-DNA on the platelet surface may decrease the amount of cf-DNA in the red cell supernatant.
Leukocyte reduction by means of filtration can be performed at three different time points − before storage, after storage but before issue from the blood bank, and at the bedside. Pre-storage leuko-filtration provides an advantage over bedside leuko-filtration as it is more effective in preventing febrile nonhemolytic transfusion reactions and HLA allo-immunization due to the fact that it does not allow accumulation of cytokines released from leukocytes during storage [46, 47]. It also prevents solubilization of HLA antigens from leukocyte membranes during storage which may then pass through the leukofilter and immunize the recipient [48, 49]. Furthermore, the release of enzymes by leukocytes during storage may be detrimental to red cell metabolism and viability [50, 51]. In addition, pre-storage leuko-filtration also permits quality control checks and leads to more uniform results [52]. Our findings also support the need for leukocyte reduction/depletion as part of the production of PRBC instead of filtration at bedside.
The clinical correlation of potential adverse events due to the immunomodulatory and procoagulant activity of cf-DNA in transfused components requires follow-up of blood components (with known cf-DNA content) of transfusion recipients for the occurrence of any adverse outcome in terms of transfusion reactions, in-hospital mortality rates, and length of hospital stay. Another way may be analysis of cf-DNA in blood products associated with adverse effects such as TRALI or nonhemolytic transfusion reactions.
Conclusions
cf-DNA is released in the supernatant of stored red cell components which gradually increases over the storage period (day 0–day 21), except in the LP-PRBC group. WBCs seem to be the main source of cf-DNA during storage. The rate of release of cf-DNA was exponentially related to the storage duration of the red cell unit.
Statement of Ethics
All the authors undertake that the study complies with the guidelines for human studies, and the research was conducted ethically in accordance with the World Medical Association Declaration of Helsinki after due approval of the study protocol from the institute's research committee and ethics committee on human research. Written informed consent was obtained from all donors for participation in the study.
Disclosure Statement
All the authors hereby declare that they do not have any actual or potential financial or personal relationships with other people or organizations that could inappropriately influence (bias) this work. None of the authors has received any form of support or has been financially involved (e.g., employment, consultancies, honoraria, stock ownership and options, expert testimony, grants or patents received or pending, royalties) in the past 3 years. The authors have no conflicts of interest to declare.
Funding Sources
The study was conducted as part of an intramural research project funded by the Sanjay Gandhi Postgraduate Institute of Medical Sciences (SGPGI).
Author Contributions
N.G., D.K., and R.C. conceptualized and designed the study, and performed the data analysis. N.G. and J.S.S. performed the laboratory tests. The manuscript was prepared and approved by all the authors before sending for publication.
Acknowledgments
The authors thank all the blood donors who kindly consented to participate in this study.
References
- 1.Vamvakas EC, Blajchman MA. Transfusion-related immunomodulation (TRIM): an update. Blood Rev. 2007 Nov;21((6)):327–48. doi: 10.1016/j.blre.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Garg AD, Nowis D, Golab J, Vandenabeele P, Krysko DV, Agostinis P. Immunogenic cell death, DAMPs and anticancer therapeutics: an emerging amalgamation. Biochim Biophys Acta. 2010 Jan;1805((1)):53–71. doi: 10.1016/j.bbcan.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Krysko DV, Agostinis P, Krysko O, Garg AD, Bachert C, Lambrecht BN, et al. Emerging role of damage-associated molecular patterns derived from mitochondria in inflammation. Trends Immunol. 2011 Apr;32((4)):157–64. doi: 10.1016/j.it.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 4.Chen GY, Nuñez G. Sterile inflammation: sensing and reacting to damage. Nat Rev Immunol. 2010 Dec;10((12)):826–37. doi: 10.1038/nri2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010 Mar;140((6)):805–20. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 6.Rosin DL, Okusa MD. Dangers within: DAMP responses to damage and cell death in kidney disease. J Am Soc Nephrol. 2011 Mar;22((3)):416–25. doi: 10.1681/ASN.2010040430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anders HJ, Schaefer L. Beyond tissue injury-damage-associated molecular patterns, toll-like receptors, and inflammasomes also drive regeneration and fibrosis. J Am Soc Nephrol. 2014 Jul;25((7)):1387–400. doi: 10.1681/ASN.2014010117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hou W, Zhang Q, Yan Z, Chen R, Zeh Iii HJ, Kang R, et al. Strange attractors: DAMPs and autophagy link tumor cell death and immunity. Cell Death Dis. 2013 Dec;4((12)):e966. doi: 10.1038/cddis.2013.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gould TJ, Lysov Z, Liaw PC. Extracellular DNA and histones: double-edged swords in immunothrombosis. J Thromb Haemost. 2015 Jun;13(Suppl 1):S82–91. doi: 10.1111/jth.12977. [DOI] [PubMed] [Google Scholar]
- 10.Butt AN, Swaminathan R. Overview of circulating nucleic acids in plasma/serum. Ann N Y Acad Sci. 2008 Aug;1137((1)):236–42. doi: 10.1196/annals.1448.002. [DOI] [PubMed] [Google Scholar]
- 11.Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001 Feb;61((4)):1659–65. [PubMed] [Google Scholar]
- 12.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, et al. Neutrophil extracellular traps kill bacteria. Science. 2004 Mar;303((5663)):1532–5. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 13.Clark SR, Ma AC, Tavener SA, McDonald B, Goodarzi Z, Kelly MM, et al. Platelet TLR4 activates neutrophil extracellular traps to ensnare bacteria in septic blood. Nat Med. 2007 Apr;13((4)):463–9. doi: 10.1038/nm1565. [DOI] [PubMed] [Google Scholar]
- 14.Urban CF, Reichard U, Brinkmann V, Zychlinsky A. Neutrophil extracellular traps capture and kill Candida albicans yeast and hyphal forms. Cell Microbiol. 2006 Apr;8((4)):668–76. doi: 10.1111/j.1462-5822.2005.00659.x. [DOI] [PubMed] [Google Scholar]
- 15.Choi JJ, Reich CF, 3rd, Pisetsky DS. The role of macrophages in the in vitro generation of extracellular DNA from apoptotic and necrotic cells. Immunology. 2005 May;115((1)):55–62. doi: 10.1111/j.1365-2567.2005.02130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yousefi S, Gold JA, Andina N, Lee JJ, Kelly AM, Kozlowski E, et al. Catapult-like release of mitochondrial DNA by eosinophils contributes to antibacterial defense. Nat Med. 2008 Sep;14((9)):949–53. doi: 10.1038/nm.1855. [DOI] [PubMed] [Google Scholar]
- 17.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008 Sep;14((9)):985–90. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002 Feb;295((5559)):1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 19.Moscoso M, García E, López R. Biofilm formation by Streptococcus pneumoniae: role of choline, extracellular DNA, and capsular polysaccharide in microbial accretion. J Bacteriol. 2006 Nov;188((22)):7785–95. doi: 10.1128/JB.00673-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kannemeier C, Shibamiya A, Nakazawa F, Trusheim H, Ruppert C, Markart P, et al. Extracellular RNA constitutes a natural procoagulant cofactor in blood coagulation. Proc Natl Acad Sci USA. 2007 Apr;104((15)):6388–93. doi: 10.1073/pnas.0608647104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morrissey JH, Choi SH, Smith SA. Polyphosphate: an ancient molecule that links platelets, coagulation, and inflammation. Blood. 2012 Jun;119((25)):5972–9. doi: 10.1182/blood-2012-03-306605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swystun LL, Mukherjee S, Liaw PC. Breast cancer chemotherapy induces the release of cell-free DNA, a novel procoagulant stimulus. J Thromb Haemost. 2011 Nov;9((11)):2313–21. doi: 10.1111/j.1538-7836.2011.04465.x. [DOI] [PubMed] [Google Scholar]
- 23.Gould TJ, Vu TT, Swystun LL, Dwivedi DJ, Mai SH, Weitz JI, et al. Neutrophil extracellular traps promote thrombin generation through platelet-dependent and platelet-independent mechanisms. Arterioscler Thromb Vasc Biol. 2014 Sep;34((9)):1977–84. doi: 10.1161/ATVBAHA.114.304114. [DOI] [PubMed] [Google Scholar]
- 24.Levi M, van der Poll T. Inflammation and coagulation. Crit Care Med. 2010 Feb;38((2 Suppl)):S26–34. doi: 10.1097/CCM.0b013e3181c98d21. [DOI] [PubMed] [Google Scholar]
- 25.Robboy SJ, Major MC, Colman RW, Minna JD. Pathology of disseminated intravascular coagulation (DIC). Analysis of 26 cases. Hum Pathol. 1972 Sep;3((3)):327–43. doi: 10.1016/s0046-8177(72)80034-0. [DOI] [PubMed] [Google Scholar]
- 26.Shimamura K, Oka K, Nakazawa M, Kojima M. Distribution patterns of microthrombi in disseminated intravascular coagulation. Arch Pathol Lab Med. 1983 Oct;107((10)):543–7. [PubMed] [Google Scholar]
- 27.Land WG. Transfusion-Related Acute Lung Injury: the Work of DAMPs. Transfus Med Hemother. 2013 Feb;40((1)):3–13. doi: 10.1159/000345688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Müller MC, Tuinman PR, Vlaar AP, Tuip AM, Maijoor K, Achouiti A, et al. Contribution of damage-associated molecular patterns to transfusion-related acute lung injury in cardiac surgery. Blood Transfus. 2014 Jul;12((3)):368–75. doi: 10.2450/2014.0184-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee YL, King MB, Gonzalez RP, Brevard SB, Frotan MA, Gillespie MN, et al. Blood transfusion products contain mitochondrial DNA damage-associated molecular patterns: a potential effector of transfusion-related acute lung injury. J Surg Res. 2014 Oct;191((2)):286–9. doi: 10.1016/j.jss.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garraud O, Tariket S, Sut C, Haddad A, Aloui C, Chakroun T, et al. Transfusion as an Inflammation Hit: knowns and Unknowns. Front Immunol. 2016 Nov;7:534. doi: 10.3389/fimmu.2016.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Available from https://www.qiagen.com/us/resources/resourcedetail?id=62a200d6-faf4-469b-b50f-2b59cf738962&lang=en.
- 32.Klein SL, Flanagan KL. Sex differences in immune responses. Nat Rev Immunol. 2016 Oct;16((10)):626–38. doi: 10.1038/nri.2016.90. [DOI] [PubMed] [Google Scholar]
- 33.Caram-Deelder C, Kreuger AL, Evers D, de Vooght KM, van de Kerkhof D, Visser O, et al. Association of Blood Transfusion From Female Donors With and Without a History of Pregnancy With Mortality Among Male and Female Transfusion Recipients. JAMA. 2017 Oct;318((15)):1471–8. doi: 10.1001/jama.2017.14825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanias T, Lee J, Yazer MH, Triulzi DJ, Lippert A, Gladwin MT. Correlation Between Female Gender and the Red Blood Cell Propensity to Hemolyze Under Various Stresses. Blood. 2011;118((21)):2325. [Google Scholar]
- 35.Sonker A, Dubey A, Chaudhary R. Evaluation of a red cell leukofilter performance and effect of buffy coat removal on filtration efficiency and post filtration storage. Indian J Hematol Blood Transfus. 2014 Dec;30((4)):321–7. doi: 10.1007/s12288-013-0257-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fuchs TA, Alvarez JJ, Martinod K, Bhandari AA, Kaufman RM, Wagner DD. Neutrophils release extracellular DNA traps during storage of red blood cell units. Transfusion. 2013 Dec;53((12)):3210–6. doi: 10.1111/trf.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaax ME, Krauel K, Marschall T, Brandt S, Gansler J, Fürll B, et al. Complex formation with nucleic acids and aptamers alters the antigenic properties of platelet factor 4. Blood. 2013 Jul;122((2)):272–81. doi: 10.1182/blood-2013-01-478966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waldvogel Abramowski S, Tirefort D, Lau P, Guichebaron A, Taleb S, Modoux C, et al. Cell-free nucleic acids are present in blood products and regulate genes of innate immune response. Transfusion. 2018 Jul;58((7)):1671–81. doi: 10.1111/trf.14613. [DOI] [PubMed] [Google Scholar]
- 39.Shih AW, Bhagirath VC, Heddle NM, Acker JP, Liu Y, Eikelboom JW, et al. Quantification of Cell-Free DNA in Red Blood Cell Units in Different Whole Blood Processing Methods. J Blood Transfus. 2016;2016:9316385. doi: 10.1155/2016/9316385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.NanoDrop nucleic acid quantification. [cited 2018 November 14] Available from: https://www.thermofisher.com/in/en/home/industrial/spectroscopy-elemental-isotope-analysis/molecular-spectroscopy/ultraviolet-visible-visible-spectrophotometry-uv-vis-vis/uv-vis-vis-instruments/nanodrop-microvolume-spectrophotometers/nanodrop-nucleic-acid-quantification.html.
- 41.Interpretation of nucleic acid 260/280 ratios. [cited 2018 November 14]. Available from: http://tools.thermofisher.com/content/sfs/brochures/T123-NanoDrop-Lite-Interpretation-of-Nucleic-Acid-260-280-Ratios.pdf.
- 42.Bakkour S, Acker JP, Chafets DM, Inglis HC, Norris PJ, Lee TH, et al. Manufacturing method affects mitochondrial DNA release and extracellular vesicle composition in stored red blood cells. Vox Sang. 2016 Jul;111((1)):22–32. doi: 10.1111/vox.12390. [DOI] [PubMed] [Google Scholar]
- 43.Cognasse F, Aloui C, Anh Nguyen K, Hamzeh-Cognasse H, Fagan J, Arthaud CA, et al. Platelet components associated with adverse reactions: predictive value of mitochondrial DNA relative to biological response modifiers. Transfusion. 2016 Feb;56((2)):497–504. doi: 10.1111/trf.13373. [DOI] [PubMed] [Google Scholar]
- 44.Kattlove HE, Alexander B, White F. The effect of cold on platelets. II. Platelet function after short-term storage at cold temperatures. Blood. 1972 Nov;40((5)):688–96. [PubMed] [Google Scholar]
- 45.Albeiroti S, Ayasoufi K, Hill DR, Shen B, de la Motte CA. Platelet hyaluronidase-2: an enzyme that translocates to the surface upon activation to function in extracellular matrix degradation. Blood. 2015 Feb;125((9)):1460–9. doi: 10.1182/blood-2014-07-590513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stack G, Snyder EL. Cytokine generation in stored platelet concentrates. Transfusion. 1994 Jan;34((1)):20–5. doi: 10.1046/j.1537-2995.1994.34194098597.x. [DOI] [PubMed] [Google Scholar]
- 47.Heddle NM, Klama L, Singer J, Richards C, Fedak P, Walker I, et al. The role of the plasma from platelet concentrates in transfusion reactions. N Engl J Med. 1994 Sep;331((10)):625–8. doi: 10.1056/NEJM199409083311001. [DOI] [PubMed] [Google Scholar]
- 48.Blajchman MA, Bardossy L, Carmen RA, Goldman M, Heddle NM, Singal DP. An animal model of allogeneic donor platelet refractoriness: the effect of the time of leukodepletion. Blood. 1992 Mar;79((5)):1371–5. [PubMed] [Google Scholar]
- 49.Ramos RR, Curtis BR, Duffy BF, Chaplin H. Low retention of white cell fragments by polyester fiber white cell-reduction platelet filters. Transfusion. 1994 Jan;34((1)):31–4. doi: 10.1046/j.1537-2995.1994.34194098599.x. [DOI] [PubMed] [Google Scholar]
- 50.Brecher ME, Pineda AA, Torloni AS, Harbaugh CA, Emery RL, Moore SB, et al. Prestorage leukocyte depletion: effect on leukocyte and platelet metabolites, erythrocyte lysis, metabolism, and in vivo survival. Semin Hematol. 1991 Jul;28((3 Suppl 5)):3–9. [PubMed] [Google Scholar]
- 51.Davey RJ, Carmen RA, Simon TL, Nelson EJ, Leng BS, Chong C, et al. Preparation of white cell-depleted red cells for 42-day storage using an integral in-line filter. Transfusion. 1989 Jul-Aug;29((6)):496–9. doi: 10.1046/j.1537-2995.1989.29689318446.x. [DOI] [PubMed] [Google Scholar]
- 52.Freedman JJ, Blajchman MA, McCombie N. Canadian Red Cross Society symposium on leukodepletion: report of proceedings. Transfus Med Rev. 1994 Jan;8((1)):1–14. doi: 10.1016/s0887-7963(94)70093-6. [DOI] [PubMed] [Google Scholar]