Abstract
Metabolic memory, the persistent benefits of early glycemic control on preventing/delaying diabetic complications development, is observed in the Diabetes Control and Complications Trial (DCCT) and in the Epidemiology of Diabetes Interventions and Complications (EDIC) follow-up study, but mechanisms remain unclear. Here, we show the involvement of epigenetic DNA methylation (DNAme) in metabolic memory by examining its associations with preceding glycemic history, and with subsequent development of complications over an 18-year period in blood DNAs of 499 randomly-selected DCCT/EDIC participants with type 1 diabetes. We demonstrate the associations between DNAme near DCCT-closeout and mean HbA1c during DCCT (mean-DCCT-HbA1c) at 186 CpGs (FDR<15%, including 43 at FDR<5%), many of which are located in complications-related genes. Biological function exploration studies reveal these CpGs are enriched in C/EBP transcription factor binding sites, as well as enhancer/transcription regions in blood cells and hematopoietic stem cells, and open chromatin states in myeloid cells. Mediation analyses show that, remarkably, several CpGs in combination explain 68–97% of the association of mean-DCCT-HbA1c with the risk of complications development during EDIC. In summary, DNAme at key CpGs appears to mediate the association between hyperglycemia and complications in metabolic memory, through modifying enhancer activity at myeloid and other cells.
Keywords: epigenetics, DNA methylation, type 1 diabetes, HbA1c, metabolic memory, diabetic complications
INTRODUCTION
Diabetes is associated with significantly increased risk of micro- and macrovascular complications, including diabetic kidney disease (DKD) leading to end-stage renal disease (ESRD), retinopathy leading to blindness, and atherosclerosis leading to cardiovascular diseases. The landmark Diabetes Control and Complications Trial (DCCT, 1983–1993) and the observational follow-up of those subjects in the Epidemiology of Diabetes Interventions and Complications (EDIC, 1994-present) study evaluated the effects of glycemic control on complications development and progression. During DCCT, relative to conventional (CONV) diabetes therapy for glucose control, intensive (INT) therapy significantly reduced the risk of microvascular complications development and progression in type 1 diabetes (T1D)1. Following the DCCT, all participants were encouraged to practice INT therapy, and after the first 4 years of follow-up, the mean HbA1c levels in the two groups were equivalent. After 18 years of follow-up during EDIC, participants randomly assigned to INT therapy during DCCT continued to experience significantly lower rates of retinopathy, neuropathy, and DKD despite no significant differences in HbA1c2–5. Notably, more than 95% of the risk of these complications related to former treatment was mediated by mean HbA1c during DCCT1,3. This suggests that hyperglycemic memory in target cells/tissues has persistent deleterious effects long after glucose normalization, a phenomenon termed “metabolic memory”, for which epigenetic mechanisms have been implicated6–8.
Epigenetics refers to the heritability of gene expression and phenotypes via mitosis or meiosis, which occur without alterations in the underlying DNA sequence9,10. Epigenetics can mediate crosstalk between genes and the environment11,12 and influence the pathogenesis of diabetes and its complications, which are also strongly affected by environmental factors, such as nutrition, infections and lifestyle6,13,14. Several experimental models support the role of epigenetic mechanisms in diabetes complications6,7. Moreover, large scale genome-wide association studies (GWAS) have found few genetic variants associated with diabetes-related complications15–17, suggesting the need to also evaluate epi-mutations including DNA methylation (DNAme), histone post-translational modifications, and non-coding RNAs. DNAme, usually at cytosine-guanine dinucleotides (CpG sites), is a key regulator of gene expression, and several recent epigenome-wide association studies (EWAS)6,8,18–25 have revealed its association with diabetic complications.
Our recent EWAS on a subset of the DCCT/EDIC cohort suggested a role for DNAme in metabolic memory26. The cohort included 32 cases (from the DCCT CONV group, who had high mean-DCCT HbA1c [10.2±0.8%, SD] and developed DKD or retinopathy during EDIC up to EDIC year 10) and 31 controls (from the DCCT INT group who had low mean-DCCT HbA1c, 6.5±0.4% and no complications up to EDIC Year 10). We found that DNAme differences between the cases and controls at various loci persisted in the same participant samples collected 16–17 years apart. However, due to the small sample size and the significant differences in HbA1c and complications between the case and control groups, it was not possible to assess the association of DNAme with both DCCT HbA1c (i.e. preceding glycemic history) and future risk of complications. Therefore, it is unknown what role DNAme plays in the association between HbA1c and complications. These important aspects and mechanistic insights are addressed in the current study which includes a larger, randomly selected, and well-phenotyped cohort from the DCCT/EDIC study. Our new results suggest that DNAme at key CpGs may mediate the association between hyperglycemia and complications in metabolic memory, through modifying enhancer activity at myeloid and other cells.
RESULTS
Cohort Selection
The study design is depicted in Fig 1a. Participants in the DCCT/EDIC study2 have T1D and were randomly assigned to receive either CONV maintaining clinical well-being with no specific glucose targets, or INT maintaining near-normal blood glucose levels. At DCCT entry, participants were enrolled into two cohorts based on microvascular disease severity, primary prevention (PRIM) and secondary intervention (SCND) (see Methods for details). This resulted in 4 design groups: PRIM cohort on CONV therapy (PRIM CONV), or PRIM INT, SCND CONV, and SCND INT. In this study, we used 125 DNA samples randomly-selected from participants in each of these 4 groups (Supplementary Tables 1, 2 and Methods). DNAme was profiled on each sample by Illumina Infinium MethylationEPIC BeadChip (EPIC) arrays. One outlier in the PRIM CONV group identified by cluster analysis of the methylation data (Supplementary Figure 1) was excluded in subsequent analyses. All remaining samples passed quality controls (see Methods). Major demographic and clinical characteristics of the 250 INT (PRIM INT + SCND INT) vs. 249 CONV (PRIM CONV + SCND CONV) participants at blood/DNA sample collection are shown in Supplementary Table 3. For each participant, HbA1c levels during DCCT up to blood/sample collection is presented by mean-DCCT HbA1c, i.e. the mean of HbA1c measured quarterly during the average 5.4-years (range 2.7~9.1, SD=1.6) DCCT period. Although HbA1c levels at DCCT entry were not significantly different between INV and CONV, mean-DCCT HbA1c was, as expected, significantly lower in INT versus CONV (7.04±0.75% versus 8.84±1.20%, p=2.51e-54, two-tailed Wilcoxon rank sum test), with similar differences (7.07±0.95% vs. 8.96±1.41%) in HbA1c at the time of DNA sample collection. Apart from HbA1c, we did not find significant differences in all the other variables between INT vs. CONV [except Body Mass Index (BMI) and severe non-proliferative diabetic retinopathy (SNPDR)] (Supplementary Table 3). The differences in HbA1c history are comparable to the full DCCT/EDIC cohort3 indicating that the sample of 499 is representative of the study. These differences in HbA1c were also seen for INT vs. CONV within each cohort (PRIM and SCND) (Supplementary Table 4).
Figure 1. Identification of HbA1c-assoc CpGs.
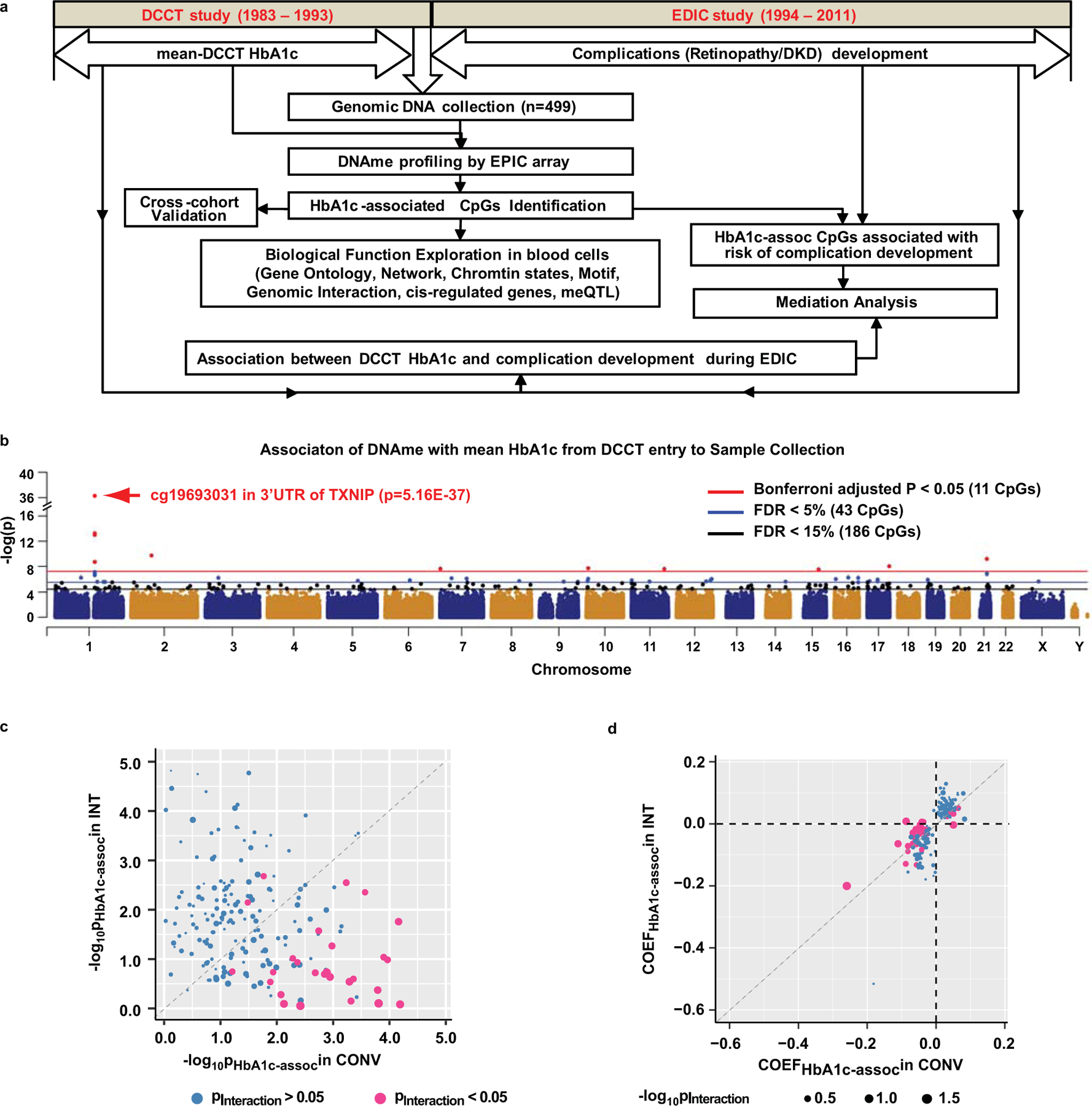
a, Study workflow for identification of CpGs whose DNAme is associated with mean-DCCT HbA1c, subsequent exploration of biological functions of the identified CpGs, and mediation effect of DNAme on HbA1c-associated future complication development. b, Manhattan plot showing genome-wide association of DNAme with mean-DCCT HbA1c across 499 samples. Linear regression adjusted for covariates with two-sided test based on t-statistic was used to study the association of DNAme (M-values) with mean-DCCT HbA1c at each of the 815432 CpGs reliably covered by the Infinium MethylationEPIC array on all samples (n=499, see Methods). Each CpG is presented as a dot across genomic location (x-axis) of 24 chromosomes (alternating dark blue and orange). The significance level (in –log10) of each CpG is represented in the y-axis. 11 CpGs with Bonferroni-adjusted p < 0.05 (p=6.13e-08, red line) are shown as red dots, CpGs identified with FDR < 5% (blue line) as blue dots and those with FDR<15% (black line) as black dots. The top association CpG (cg19693031 at 3’UTR of TXNIP) is highlighted with red arrow. c&d. Comparison of the associations between DNAme and HbA1c in INT vs. CONV groups at 186 HbA1c-assoc CpGs by bubble plots. For each HbA1c-assoc CpG, linear regression model with addition of interaction term between DNAme and treatment (INT or CONV) was applied to the complete dataset (n=499) to identify CpGs having different associations of DNAme with HbA1c in INT vs. CONV. Each CpG is represented by one bubble dot. The size of the dot represents the significance level of interaction between DNAme and treatment (-log10Pinteraction) estimated by linear regression model. Pink represents the CpGs with Pinteraction<0.05 while blue represents CpGs with Pinteraction>0.05. In panel c, the x-axis represents significance level of association estimated by the same linear regression model described in panel b in the CONV group (n=249), and the y-axis represents the INT group (n=250). In panel d, x-axis represents the association coefficients (COEF) in CONV, and y-axis in the INT group. Significance levels obtained in all the multiple linear regression models in panels b-d were determined by two-sided tests based on t-statistic.
Identification of CpGs where DNAme is associated with mean-DCCT HbA1c
To examine hyperglycemia-associated changes in DNAme, we used linear regression models, adjusted for multiple covariates (Methods), including estimated composition of major blood cells (Supplementary Table 5) to determine the association of DNAme with mean-DCCT HbA1c at each of the 815,432 CpGs (in the EPIC array) reliably detected in all the samples.
We thus identified 43 CpGs associated with mean-DCCT HbA1c (HbA1c-assoc CpGs) (23 positively-associated [pos-assoc] and 20 negatively-associated [neg-assoc], Table 1) at a false discovery rate (FDR)<5%, including 11 CpGs (5 pos-assoc and 6 neg-assoc) that remained significant after a Bonferroni adjustment p<0.05 (Fig. 1b, Supplementary Table 6). Interestingly, cg19693031 located in the 3’UTR of Thioredoxin Interacting Protein (TXNIP) was the most significant HbA1c-assoc CpG (p=5.16e-37, FDR=4.21e-31). This was also the most significant differentially-methylated locus (DML) between cases vs. controls in our previous study26. In the current expanded cohort (n=499), we identified 6 additional HbA1c-assoc CpGs near TXNIP, from 4.8kb upstream of the transcription start site (TSS) to 13.5kb downstream of the transcription end (FDR < 5%, Fig. 1b). TXNIP, a ubiquitously expressed protein, can induce oxidative stress and apoptosis by binding to and inhibiting thioredoxin. TXNIP is highly induced by hyperglycemia and associated with islet dysfunction, diabetes and its numerous complications26–30. We also identified other HbA1c-assoc CpGs (FDR<5%) located in or near genes with relevant functions and reported to be genetically or epigenetically associated with blood glucose, diabetes (DQX1, ZCCHC14, GLT1D1, ANKS3, MDN1, FLAD1, NCOR2, TTC7B), chromatin functions (SIRT7, SETD2, GLI3, NCOR2, CHD3), transcriptional activation/repression (MAFG, GLI3, NCOR2), metabolism (PFKFB3, NMNAT2), inflammation (NCOR2, TSLP), and ribosome biogenesis (MDN1, LAS1L) (Supplementary Table 7).
Table 1.
CpGs where DNAme was associated with mean-DCCT HbA1c1 at FDR < 5% (ordered by p-value).
| # | ID | CH2 | Location2 | Gene Symbol3 | EST4 | SE | p | FDR |
|---|---|---|---|---|---|---|---|---|
| 1 | cg19693031 | 1 | 145441552 | TXNIP | −0.24 | 0.02 | 5.2e-37 | 4.2e-31 |
| 2 | cg19266329 | 1 | 145456128 | −0.07 | 0.01 | 5.8e-14 | 2.4e-08 | |
| 3 | cg26974062 | 1 | 145440734 | TXNIP | −0.11 | 0.01 | 1.0e-13 | 2.8e-08 |
| 4 | cg06721411 | 2 | 74753759 | DQX1 | 0.05 | 0.01 | 1.9e-10 | 3.9e-05 |
| 5 | cg08309687 | 21 | 35320596 | LINC00649 | −0.07 | 0.01 | 6.7e-10 | 1.1e-04 |
| 6 | cg02988288 | 1 | 145440445 | TXNIP | −0.11 | 0.02 | 2.0e-09 | 2.7e-04 |
| 7 | cg04568295 | 17 | 79877134 | SIRT7; MAFG | 0.04 | 0.01 | 9.6e-09 | 1.1e-03 |
| 8 | cg26262157 | 10 | 6214079 | PFKFB3 | −0.06 | 0.01 | 2.0e-08 | 2.1e-03 |
| 9 | cg04816311 | 7 | 1066650 | C7orf50 | 0.04 | 0.01 | 2.4e-08 | 2.1e-03 |
| 10 | cg20983494 | 11 | 120627781 | GRIK4 | 0.05 | 0.01 | 2.6e-08 | 2.1e-03 |
| 11 | cg19285358 | 15 | 74928240 | EDC3 | 0.02 | 0.00 | 3.0e-08 | 2.2e-03 |
| 12 | cg26823705 | 1 | 145435523 | −0.06 | 0.01 | 7.9e-08 | 5.4e-03 | |
| 13 | cg27037013 | 21 | 35320667 | LINC00649 | −0.08 | 0.02 | 1.5e-07 | 9.2e-03 |
| 14 | cg20646141 | 1 | 145433673 | −0.03 | 0.01 | 1.7e-07 | 9.9e-03 | |
| 15 | cg05028010 | 1 | 145437567 | TXNIP | −0.07 | 0.01 | 2.1e-07 | 1.2e-02 |
| 16 | cg01902066 | 16 | 50727242 | −0.04 | 0.01 | 5.2e-07 | 2.4e-02 | |
| 17 | cg22761903 | 1 | 94811259 | SETD2 | −0.06 | 0.01 | 5.3e-07 | 2.4e-02 |
| 18 | cg09981464 | 16 | 87441659 | 0.09 | 0.02 | 5.6e-07 | 2.4e-02 | |
| 19 | cg26676775 | 3 | 47159275 | ZCCHC14 | 0.06 | 0.01 | 5.6e-07 | 2.4e-02 |
| 20 | cg06200577 | 7 | 42237350 | GLI3 | 0.04 | 0.01 | 6.5e-07 | 2.7e-02 |
| 21 | cg17540192 | 7 | 97875259 | TECPR1 | 0.04 | 0.01 | 7.5e-07 | 2.9e-02 |
| 22 | cg05014727 | 10 | 6214016 | PFKFB3 | −0.05 | 0.01 | 8.0e-07 | 3.0e-02 |
| 23 | cg16717225 | 12 | 129337768 | LOC100128276; GLT1D1 | −0.03 | 0.01 | 8.5e-07 | 3.0e-02 |
| 24 | cg03497652 | 16 | 4751569 | ANKS3 | 0.03 | 0.01 | 8.8e-07 | 3.0e-02 |
| 25 | cg25425189 | 17 | 37349801 | CACNB1 | 0.04 | 0.01 | 1.1e-06 | 3.5e-02 |
| 26 | cg00994936 | 19 | 1423902 | DAZAP1 | 0.02 | 0.00 | 1.1e-06 | 3.5e-02 |
| 27 | cg16809457 | 6 | 90399677 | MDN1 | 0.05 | 0.01 | 1.4e-06 | 4.0e-02 |
| 28 | cg02304370 | 11 | 587926 | PHRF1 | 0.05 | 0.01 | 1.4e-06 | 4.0e-02 |
| 29 | cg26385126 | 12 | 124912021 | NCOR2 | 0.03 | 0.01 | 1.4e-06 | 4.0e-02 |
| 30 | cg20710777 | 5 | 110411740 | TSLP | −0.06 | 0.01 | 1.7e-06 | 4.5e-02 |
| 31 | cg08025214 | 8 | 101704984 | 0.05 | 0.01 | 1.8e-06 | 4.8e-02 | |
| 32 | cg24129923 | 17 | 7814251 | CHD3 | 0.05 | 0.01 | 1.9e-06 | 4.8e-02 |
| 33 | cg08506528 | 10 | 4093714 | LOC101927964 | −0.07 | 0.01 | 1.9e-06 | 4.8e-02 |
| 34 | cg03743771 | 12 | 108961135 | ISCU | 0.03 | 0.01 | 2.0e-06 | 4.8e-02 |
| 35 | cg00061632 | 17 | 36979013 | CWC25 | 0.03 | 0.01 | 2.1e-06 | 4.8e-02 |
| 36 | cg05525364 | X | 64741336 | LAS1L | 0.04 | 0.01 | 2.1e-06 | 4.8e-02 |
| 37 | cg09593400 | 11 | 66024489 | KLC2 | −0.05 | 0.01 | 2.2e-06 | 4.8e-02 |
| 38 | cg14063347 | 9 | 88207974 | AGTPBP1 | −0.06 | 0.01 | 2.3e-06 | 4.8e-02 |
| 39 | cg04742977 | 14 | 91127460 | TTC7B | 0.05 | 0.01 | 2.3e-06 | 4.8e-02 |
| 40 | cg14334460 | 9 | 140346899 | NSMF | 0.05 | 0.01 | 2.4e-06 | 4.8e-02 |
| 41 | cg16097041 | 1 | 154965544 | LENEP; FLAD1 | 0.03 | 0.01 | 2.4e-06 | 4.8e-02 |
| 42 | cg08844913 | 1 | 177724656 | −0.05 | 2.5e-06 | 4.8e-02 | ||
| 43 | cg16263152 | 1 | 183287819 | NMNAT2 | −0.05 | 2.5e-06 | 4.8e-02 |
Multiple linear regression model was applied to all the samples (two-sided tests based on t-statistic, n=499) using DNAme as dependent variable and mean-DCCT HbA1c as independent variable adjusted for covariates including age at time of DCCT entry, sex, estimated cell composition, diabetes duration at DCCT entry, time from DCCT entry to blood collection, HbA1c at DCCT baseline, array processing time and cohort(PRIM or SCND)
Human Genome Assembly Hg19.
Genes containing CpGs located in promoters (up to 1500bp relative to transcription start site) and gene bodies (see Methods for details).
Estimated coefficient for association between DNAme and mean-DCCT HbA1c. Pos-assoc CpGs have positive values: neg-assoc CpGs have negative values.
CH: chromosome; SE: standard error; FDR: false discovery rate.
A more relaxed threshold (FDR<15%) was used for subsequent analyses to facilitate exploration of functional and biological significance to T1D and complications. This yielded 186 HbA1c-assoc CpGs, including 107 pos-assoc and 79 neg-assoc (Fig. 1b, Supplementary Table 8).
Sensitivity analyses were also applied to 14 other clinical variables obtained at the time of sample collection (not included as covariates because of very little association significance, see Methods). For each variable, the change in HbA1c-assoc coefficient obtained in the model with and without its addition as covariate was <10% [p<2.69e-05(=0.05/186)] at all 186 CpGs (Extended Data 1 & 2, Supplementary Table 9).
We further examined the impact of DCCT treatment on the association between HbA1c and DNAme at the 186 HbA1c-assoc CpGs (see Methods) and found 28 HbA1c-assoc CpGs showed different associations between CONV and INT group at nominal p<0.05 (Fig. 1c, pink dots). DNAme at 26 out of these 28 CpGs showed more significant association with HbA1c in CONV versus INT. Additionally, 24/28 CpGs had the same direction of association in both treatment groups, although the absolute magnitude in CONV was greater than INT (Fig. 1d). Similar approaches to determine the influence of cohort or sex revealed lesser effects on the HbA1c-DNAme association (Supplementary Figure 2).
From these 186 HbA1c-assoc CpGs, we next identified 11 HbA1c-assoc regions, each containing at least two nearby HbA1c-assoc CpGs within the 500bp window (Supplementary Table 10). We found significant correlation between DNAme at HbA1c-assoc CpGs located within the same region (last column, Supplementary Table 10), suggesting similar regulation of DNAme in each region. Notably, two of these regions (#1 & 2, Supplementary Table 10), were located at/near TXNIP (cg19693031): one ~4.8kb upstream of the TXNIP TSS and the other spanning 5th and 6th exons of the longest TXNIP isoform, NM_006472, further underscoring the significance of the DNAme-HbA1c association at TXNIP and nearby regions.
Validation of HbA1c-assoc CpGs
To validate HbA1c-assoc CpGs, we first used amplicon-seq26, for 96 out of the 499 samples (24 samples/group, clinical characteristics of each group summarized in Supplementary Tables 11–14). We validated DNAme at 4 selected HbA1c-assoc CpGs at nominal p<0.05, cg19693031, cg09981464, cg09641127 and cg26676775 located at TXNIP-3’UTR and 3 other genes related to diabetes or DNAme regulation (Table 2, Supplementary Table 7). Specifically, cg09981464 is one of two HbA1c-assoc CpGs located in 3’UTR of Zinc Finger CCHC-Type Containing14 (ZCCHC14), previously associated with acute insulin response to glucose by GWAS31. Being within a CpG island, amplicon-seq not only validated the positive association at cg09981464 (estimate=0.05, SE=0.04, p=0.023) but also at multiple nearby CpGs in the same island covered in the amplified targeted region (Table 2). cg09641127 and nearby CpG are in the 3’UTR of Kruppel-like factor 11(KLF11), associated with diabetes32,33. Lastly, cg26676775 is intronic in SET Domain Containing 2 (SETD2). SETD2 codes for a histone lysine-methyltransferase known to interact with DNAme and affect transcriptional regulation34.
Table 2.
Validation of candidate HbA1c-assoc CpGs by amplicon-seq1.
| Target regions | CpG to validate | Gene Symbol | Location relative to Gene | Sample size2 | Chrom3 | Genomic Location3 | Estimate4 | SE4 | Pvalue4 |
|---|---|---|---|---|---|---|---|---|---|
| chr1: 145,441,492 – 145,441,619 | cg19693031 | TXNIP | 3’UTR | 96 | chr1 | 145441517 | −0.277 | 0.050 | 3.10e-075 |
| chr1 | 145441526 | −0.278 | 0.049 | 2.26e-075 | |||||
| chr1 | 145441552 | −0.229 | 0.038 | 3.74e-085* | |||||
| chr16: 87,441,505 – 87,441,756 | cg09981464 | ZCCHC14 | 3’UTR | 92 | chr16 | 87441528 | 0.083 | 0.040 | 4.13e-025 |
| chr16 | 87441555 | 0.060 | 0.036 | 9.65e-02 | |||||
| chr16 | 87441562 | 0.079 | 0.037 | 3.57e-025 | |||||
| chr16 | 87441568 | 0.087 | 0.038 | 2.40e-025 | |||||
| chr16 | 87441572 | 0.073 | 0.038 | 5.92e-02 | |||||
| chr16 | 87441594 | 0.088 | 0.039 | 2.77e-025 | |||||
| chr16 | 87441597 | 0.090 | 0.040 | 2.82e-025 | |||||
| chr16 | 87441607 | 0.082 | 0.036 | 2.72e-025 | |||||
| chr16 | 87441616 | 0.080 | 0.038 | 3.90e-025 | |||||
| chr16 | 87441629 | 0.093 | 0.039 | 1.86e-025 | |||||
| chr16 | 87441641 | 0.096 | 0.040 | 1.98e-025 | |||||
| chr16 | 87441659 | 0.090 | 0.039 | 2.35e-025* | |||||
| chr16 | 87441670 | 0.097 | 0.040 | 1.62e-025 | |||||
| chr16 | 87441689 | 0.121 | 0.054 | 2.71e-025 | |||||
| chr2: 10,193,761 – 10,194,017 | cg09641127 | KLF11 | 3’UTR | 95 | chr2 | 10193885 | −0.088 | 0.019 | 2.25e-055 |
| chr2 | 10193895 | −0.081 | 0.019 | 4.48e-055* | |||||
| chr3: 47,159,141 – 47,159,440 | cg26676775 | SETD2 | Intron | 92 | chr3 | 47159275 | 0.046 | 0.021 | 3.18e-025* |
Amplicon-seq was performed in 3 randomly-selected batches containing a total of 96 DNA samples (24 samples from each group: PRIM INT, PRIM CONV, SCND INT and SCND CONV (See Methods and Supplementary Tables 11–14).
Sample size used in validation analyses for each HbA1c-assoc CpG listed. Only samples with aligned reads >2500 are included in the multiple linear regression model for association analyses.
Human Genome Assembly Hg19.
For each CpG site covered by amplicon-seq targeted regions, multiple linear regression model was applied to study the association between DNAme and mean-DCCT HbA1c using DNAme as dependent variable, HbA1c as independent variable adjusting for covariates used in HbA1c-ass CpGs identification (see methods for details).
CpGs whose DNAme is associated with mean-DCCT HbA1c with nominal p< 0.05.
Same CpG as the HbA1c-assoc CpG to be validated.
We next used three separate epigenomic datasets for internal and external cross-cohort validation. The internal validation cohort comprised 41 DCCT subjects (DCCT41) selected from our previous 63-subject study26 after excluding 22 subjects overlapping with the current study (DCCT499). DNAme profiles on whole blood (WB) samples from DCCT41 were generated by 450K arrays. We also utilized two external cohorts for validation. The first comprised published data from 850 participants from the San Antonio Family Heart Study (2002~2006) (SAFHS850), of which ~20% had type 2 diabetes, and an additional ~14% had impaired fasting glucose (pre-diabetes)35. 450K arrays were also used to generate WB DNAme data for this cohort. The second validation cohort comprised 195 T1D subjects selected from the Joslin Kidney Study36, all of whom had proteinuria and impaired kidney function with average estimated glomeruli filtration rate (eGFR) 45.58±12.73ml min–1 per 1.73 m2 at the time of blood collection (Joslin195). The clinical characteristics of Joslin195 are noted in the Methods and Supplementary Table 15.
For validation, we first compared the mean-DCCT HbA1c-assoc coefficients in DCCT499 with related information in each of the 3 validation cohorts using Spearman correlation analyses. We found these coefficients in DCCT499 have highly-significant correlations with: 1) log2 fold-change in DNAme between cases vs. controls (likely due to differences in their HbA1c history) in DCCT41 across 84 HbA1c-assoc CpGs that are covered in both 450K and 850K arrays with rho=0.59 (p=5.67e-9, Fig. 2A)26; 2) coefficients of DNAme with fasting blood glucose (FBG) in SAFHS850 across 77 HbA1c-assoc commonly-covered CpGs [7 out 84 CpGs were excluded for various reasons (see Methods)] with rho=0.58 (p=5.25e-8, Fig. 2b); and 3) coefficients of DNAme with HbA1c at sample collection in Joslin195 across 185 HbA1c-assoc CpGs (DNAme data on one CpG missing due to detection p>0.05) with rho=0.63 (p=2.2e-16, Fig. 2c).
Figure 2. Cross-cohort validation of HbA1c-assoc CpGs in DCCT41, SAFHS850 and Joslin195 and their persistence of DNAme in EDIC61.
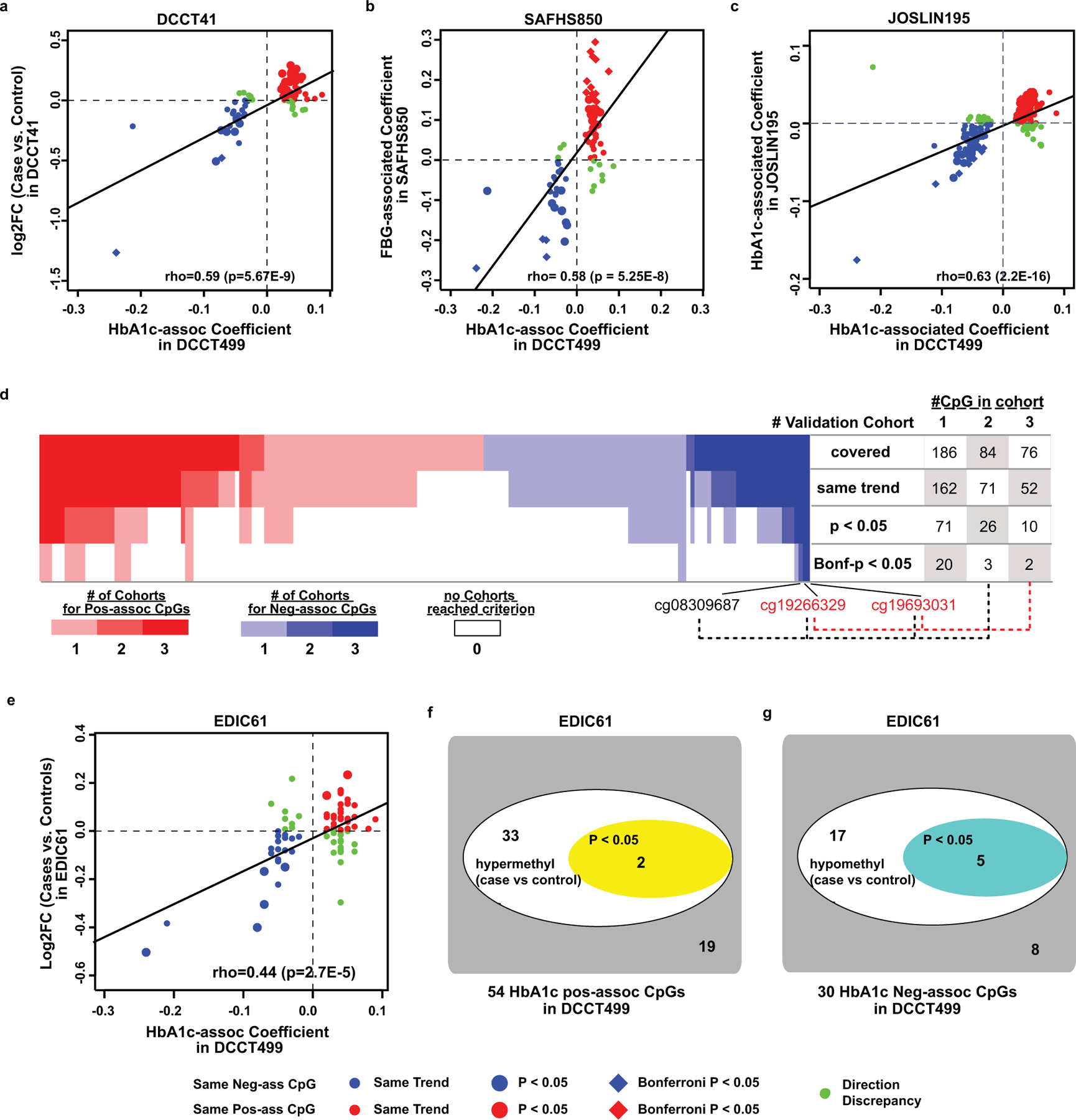
a-c, Spearman correlation of HbA1c-assoc coefficients in DCCT499 (X-axis) versus log2 fold change (log2FC, Y-axis) between cases (history of hyperglycemia) and controls (history of normoglycemia) in the DCCT41 (A), FBG-associated coefficients in SAFHS850 (b), and associations of coefficients of HbA1c at sample collection in Joslin195 (c) across all the HbA1c-assoc CpGs identified in DCCT499 (FDR < 15%) and covered in each validation cohort. Each CpG is represented by one dot. Red/blue represents pos-assoc/neg-assoc CpGs with same trends of associations in both datasets respectively. Those with p < 0.05 in validation cohorts are shown as bigger dots. The remaining CpGs with discrepant association direction are shown in small green dots. The black diagonal line represents linear regression line. d, Heatmap on number (#) of validation cohorts in which each HbA1c-assoc CpG is validated. For each pos-assoc/neg-assoc CpG, total number of validation cohorts (one internal and two external cohorts) which covered the specific CpG and validated at different significance levels (same trend, nominal p < 0.05 or Bonferroni-adjusted p < 0.05) are summarized and depicted as heatmap with each column representing one CpG and each row representing one validation criterion as shown on the right side of the heatmap. Pos-assoc CpGs are shown in red and neg-assoc CpGs in blue as indicated in the color bar. The CpGs not depicting the same trends are shown as white box. The number of CpGs validated in at least 1, 2 or 3 cohorts at each criterion is summarized on the right side of the panel. e, Similar Spearman correlation between DCCT499 (X-axis) and EDIC61(Y-axis) as shown in panel a. f&g, DNAme differences between cases vs. controls in EDIC61 at 54 pos-assoc CpGs (f), and at 30 neg-assoc CpGs (g). Spearman correlation tests were used in panels a-c and e to determine the correlation coefficient and its significance using all the covered CpGs (n=84 in EDIC41, 77 in SAFHS850, 185 in Joslin195, and 84 in EDIC61). In each cohort, coefficients and significance levels of the association between DNAme and glucose-related variables at each covered CpGs were obtained by multiple linear regression model using all the samples in the corresponding cohort (n=41 in EDIC41, n=850 in SAFHS850, n=185 in Joslin195, and n=61 in EDIC61).
We then analyzed the association under various validation levels (same trend, nominal p value and Bonferroni-adjusted p) at the HbA1c-assoc CpGs covered in each of the 3 validation cohorts. For each CpG, the total number of cohorts in which it can be validated is calculated for each validation level and presented as heatmap in Fig. 2d. For the CpGs with same trends of association as in DCCT499, we found 87% of CpGs (162/186) satisfied the criteria in at least one cohort dataset, and 84% (71/84) in at least 2 datasets, and 68% (52/76) in all 3 datasets. At nominal p<0.05, 30.9% (71/186), 31% (26/84) and 13% (10/79) can be validated in at least 1, 2 and 3 cohorts, respectively. At Bonferroni-adjusted p < 0.05, 20 can be validated in at least one validation cohort. Notably, 2 CpGs (cg19693031 and cg19266329) were validated in all 3 cohorts with Bonferroni-adjusted p < 0.05, and cg08309687 in two validation cohorts.
Furthermore, meta-analysis was performed on DCCT499 and Joslin195. Both are T1D cohorts, but differences between them included measurement time for HbA1c (mean-DCCT in DCCT499 vs. at sample collection in Joslin195), HbA1c levels (average of 7.93±1.34% during DCCT period vs 8.81±1.61% at blood collection in Joslin 195) and cohort selection (T1D with limited incidence of DKD or chronic kidney disease in DCCT499 at sample collection vs. T1D with impaired renal function and proteinuria in Joslin195). We compared the association of DNAme with HbA1c obtained by this meta-analysis to that in DCCT499 alone at 185 HbA1c-assoc CpGs. As shown in Extended Data 3, all the 185 CpGs have the same direction of associations as those in the original analysis with DCCT499 alone, with all except 2 CpGs depicting significant association at p<0.05 (orange dots) and 118 reaching Bonferroni-adjusted p<0.05 across 185 CpGs (red dots).
Together, these validation results support the association between DNAme and HbA1c at the HbA1c-assoc CpGs, despite differences between studies, measures of glycemia, and analytical approaches. Details of the HbA1c-assoc CpGs in the 4 studies are in Supplementary Table 16.
Persistence of DNAme at HbA1c-assoc CpGs in monocytes collected at EDIC years 16/17
To examine whether the association of DNAme with mean-DCCT HbA1c at HbA1c-assoc CpGs persists years later during EDIC (consistent with metabolic memory), we evaluated differences in DNAme between cases and controls among 61 monocyte samples (EDIC61) from the same 63-subject cohort as used for DCCT41 (2 had insufficient DNA). These monocytes were collected ~2010 (EDIC years 16/17) when the cases continued to have more complications than controls during EDIC despite reduction in HbA1c differences (depicting metabolic memory)26. We found statistically significant correlation (rho=0.44, p=2.68e-5) between the HbA1c-assoc coefficients in DCCT499 and the difference (log2FC) in cases vs. controls in EDIC61 across 84 commonly-covered HbA1c-assoc CpGs (Fig. 2e). For 54 pos-assoc CpGs, 35 were hypermethylated in cases vs. controls (same trend) in EDIC61, including 2 with p<0.05 (Fig. 2f). For 30 neg-assoc CpGs, 22 were hypomethylated, including 5 with p< 0.05 (Fig. 2g). Notably, these results support a persistence (metabolic memory) of the association of HbA1c with DNAme in samples collected 16/17 year apart, despite cell-type difference (monocytes vs. WB). Details of HbA1c-assoc CpGs in EDIC61 are in Supplementary Table 16.
Genomic locations and Gene ontology analyses of HbA1c-assoc CpGs
To gain functional insights into HbA1c-assoc CpGs, we first examined the genomic locations of the 186 HbA1c-assoc CpGs relative to RefSeq genes, compared to all non-HbA1c-assoc CpGs covered by the EPIC array. We found these 186 CpGs were enriched in introns (odds ratio [OR]=1.64, 95% Confidence Interval [CI]=1.22–2.22, p=7.95e-04, two-tailed Fisher’s exact test) and depleted in proximal promoter-related regions (TSS200 and 5’UTR, OR=0.20, 95% CI=0.05–0.52, p=6.36e-05, Extended Data 4a), similar to our previous findings26 and other diabetes-related studies18,37. Ingenuity Pathway Analysis (IPA) of genes containing HbA1c-assoc CpGs in their promoters or gene bodies revealed the enrichment of canonical pathways (Benjamini-Hochberg [BH]-adjusted p< 0.05) associated with diabetes/complications like NFAT, PPARα/RXRα activation and nutrient-sensing, and a network involving several proteins related to diabetes, its complications and insulin sensitivity, such as nuclear factor-kappa B (NF-kB) and protein kinase B (Extended Data 4b, 4c). Together, these findings suggest functional links between DNAme at these HbA1c-assoc CpGs and disease pathogenesis.
Chromatin structure at genomic regions containing HbA1c-assoc CpGs
The regulatory effects of DNAme at gene promoters/exons on gene expression are well documented38, but less clear for those located at intronic and intergenic regions. As majority of the HbA1c-assoc CpGs are located in these latter regions (Extended Data 4a), we examined 15 chromatin states in each major blood cell type at HbA1c-assoc CpGs. These states are defined by the NIH Roadmap Epigenetics Program using reference epigenomes of 5 “core” histone modifications (see Methods)39. A heatmap of chromatin states at our 186 HbA1c-assoc CpGs in 5 blood cell types (monocyte, neutrophil, B-cell, T-cell, NK-cell) and hematopoietic stem cells (HSCs) (Fig. 3A) showed a large fraction of HbA1c-assoc CpGs were in states related to TSS (red), transcription (green) and enhancer regions (yellow). Moreover, these states were consistent across cell types, with the same chromatin state observed in at least 4 out of 6 cell types. Relative to non-HbA1c-assoc CpGs covered by the EPIC array, HbA1c-assoc CpGs showed statistically significant enrichments (Extended Data 5) for several chromatin states related to transcription and enhancers. After combining states related to enhancers (Enh, EnhG or EnhBiv) and transcription (TxFlnk, Tx or TxWk), we found HbA1c-assoc CpGs were significantly enriched in both enhancer and transcription regions in 5 major blood cells, and in transcription in HSCs (Fig. 3B). Notably, this enrichment was highly significant at enhancer regions in monocytes (p=9.65e-12) and transcription in HSCs (p=7.00e-15).
Figure 3. Functional explorations of HbA1c-assoc CpGs.
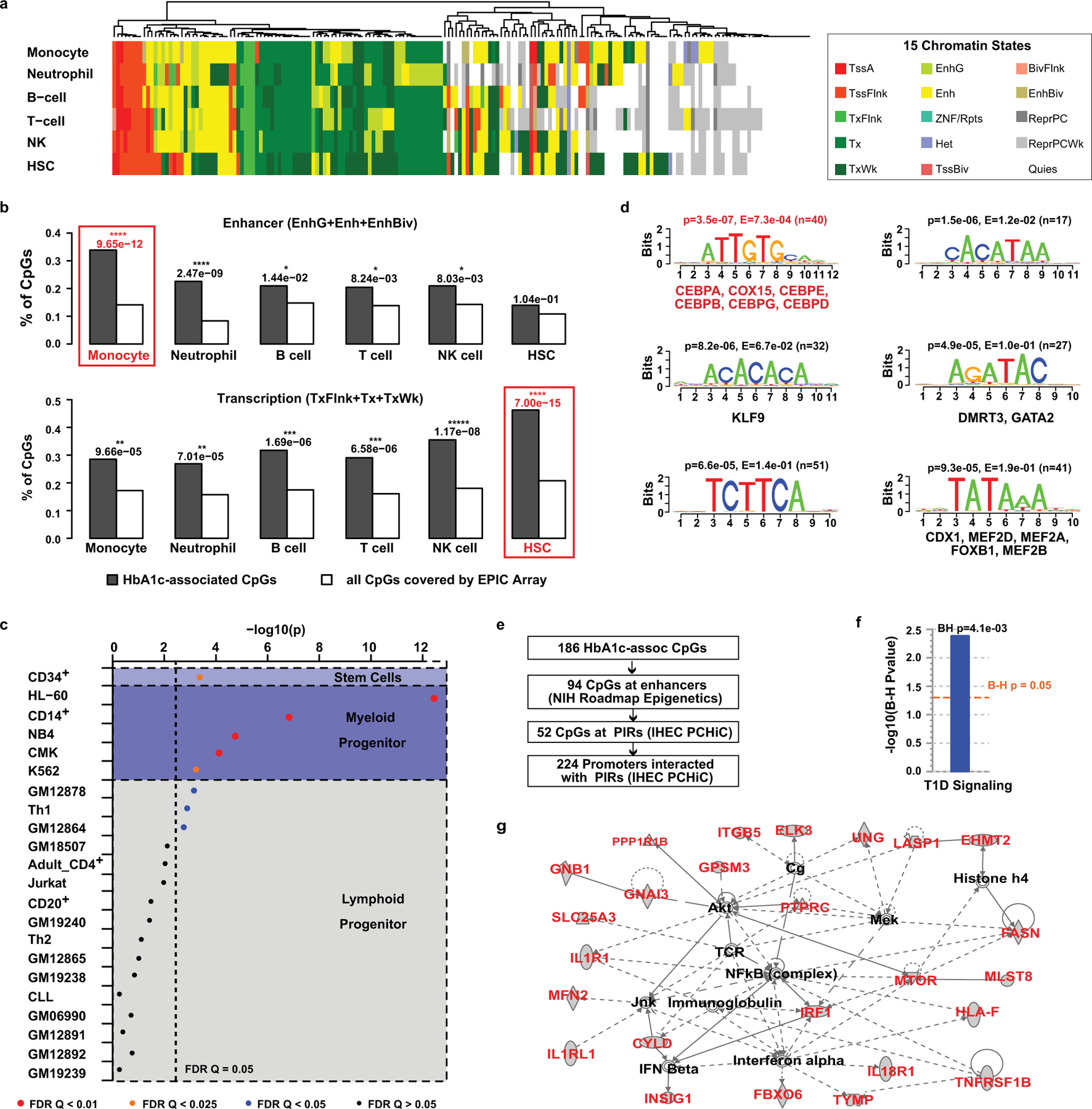
a, Heatmap of chromatin states within 5 major peripheral blood cells and HSCs at 186 HbA1c-assoc CpGs. Fifteen genome-wide chromatin states defined by NIH roadmap Epigenetics Program (https://egg2.wustl.edu/roadmap/web_portal/chr_state_learning.html#core_15state for details) were presented using different colors as depicted in the legend (adjacent box). Each row represents one cell-type and each column represents one CpG. b, Enrichment of states related to enhancer regions (EnhG or Enh or EnhBiv) or transcription (TxFlnk, Tx and TxWk) across different cell types at HbA1c-assoc CpGs versus all CpGs. P values (right-tailed Fisher’s exact test comparing 186 HbA1c-assoc CpGs with all the 815,246 non HbA1c-assoc CpGs reliably covered by the EPIC array in each specific chromatin state) are shown on top of each cell type with the most significant ones emphasized by red boxes. TxFlnk: Flanking Active TSS; Tx: Strong transcription; TxWk: Weak transcription; EnhG: Genic enhancers; Enh: Enhancers; EnhBiv: Bivalent Enhancer. c, Enrichment of DNAse I hypersensitive regions containing HbA1c-assoc CpGs relative to all CpGs in blood cells based on the ENCODE Study (https://www.encodeproject.org). Cell types are classified into 3 categories (stem cells, myeloid progenitors and lymphoid progenitors) shown in different background colors. Each cell type is presented by a dot with –log10p as Y-axis. The color and size of the dots represent the FDR levels (Benjamini-Yekutieli-corrected Q value) as shown in the legend below. The binomial test was used in eFORGE to test 186 HbA1c-assoc CpGs compared to 1,000 matching background CpG sets, each containing the same number of CpGs with matching gene annotation and CpG island. d, The top 6 enriched motifs in the ±250bp regions of neg-assoc CpGs identified by RSAT analysis. For each motif, p value from binomial tests by comparing input sequences vs. background sequences, the corresponding Bonferroni-adjusted p-value (E-value), and number of input sequences containing the specific motif (n in parenthesis) are listed on top of the motif logo. Known JASPAR transcription factor (s) for the motif, if any, is/are listed below the corresponding logo. CEBPs are highlighted in red font. e, Scheme for the identification of promoters interacting with PIRs containing HbA1c-assoc CpGs in enhancers. f&g, IPA pathway (f) and the network (g) characterized using 224 genes identified by steps shown in panel e as input. Right-tailed Fisher’s Exact test on 179 genes (after excluding 45 without match in the IPA datatabase) was used to identify the enriched pathways at B-H adjusted p < 0.05 in panel f.
Similar results were obtained using 18 chromatin states defined by the same consortium after adding one mark for active enhancers (see Methods) in the 4 available blood cells (monocyte, B-cell, T-cell, NK cell), with higher enrichment significance at enhancers compared to 15 states (Extended Data 6a, 6b). Heatmaps at HbA1c-assoc CpGs based on 15 and 18 chromatin states (side-by-side, Extended Data 6c) depicted strong similarity, with some (including cg19693031 at TXNIP 3’UTR) shifting from transcription (green) to enhancer (yellow-green) in 18 states, supporting the increased enhancer enrichment.
We also analyzed open chromatin status/accessibility at HbA1c-assoc CpGs using DNase-1 Hypersensitive Sites defined by ENCODE40 in 22 blood cell types. Interestingly, all 5 blood cell types derived from myeloid progenitors were significantly enriched, FDR Q<=0.0245, including CD14+ monocytes (Q=5.02e-05, eFORGE) (Fig. 3c). The most significant was HL-60 (Q=2.44e-10), a promyelocytic cell line which can differentiate to a neutrophil-like or monocyte-like state. G-CSF-mobilized CD34+ cell was also enriched (Q=0.0196). In contrast, the majority of blood cell types derived from lymphoid progenitors did not depict open chromatin enrichment. These results suggest important roles for DNAme at HbA1c-assoc CpGs in myeloid cells and their precursors, including monocytes, neutrophils and HSCs that have been associated with diabetic complications.
Motif analysis at HbA1c-assoc CpGs
De-novo motif analyses on ±250bp genomic regions relative to HbA1c-assoc CpGs revealed 6 potential transcription factor (TF) binding sites (Fig. 3d) enriched in neg-assoc CpG regions. The most significant one was the Jasper TF binding matrix for the leucine zipper CCAAT-enhancer binding proteins CEBPA, CEBPE, CEBPG, CEBPB, CEBPD. These TFs are widely expressed, including in HSC, where they play important roles in proliferation, differentiation, myelopoiesis, metabolism, and immunity41. This suggests important roles for HbA1c-assoc CpGs in the regulation of white blood cell differentiation and growth.
Genomic Interactions at HbA1c-assoc CpGs in major blood cells
Since our data suggest that HbA1c-assoc CpGs are enriched in regulatory regions, we next asked which genes/regions they might interact with in 3-dimensional space. We utilized published promoter-capture Hi-C (PCHiC) data generated by International Human Epigenome Consortium (IHEC)42. In at least 1 of 5 major blood cells (neutrophil, monocyte, B cell, CD4+, CD8+ T cell), 104 out of 186 HbA1c-assoc CpGs were in regions with significant interactions (ChICAGO score >5) categorized as promoter regions, promoter-interacting regions (PIRs), or both (Supplementary Table 17). Except for cg22223419 (942bp upstream of RORC) on chromosome 1, all had cis-interactions (Supplementary Figure 3). IPA on 224 identified promoters likely regulated by enhancers containing HbA1c-assoc CpGs (pipeline in Fig. 3e) identified enriched signaling pathways related to T1D containing TNFRSF1B, IRF1, IL1RL1, IL1R1, IL18R1 (Fig. 3f) and a network containing proteins related to T1D complications including NF-KB, AKT, MEK, IFN-beta, IFN-alpha, JNK (Fig. 3g). Thus, genomic regions containing HbA1c-assoc CpGs depict interactions with nearby genes related to T1D.
Relationship of DNAme at HbA1c-assoc CpGs with gene expression
Unlike the well-known inverse relationship between promoter DNAme and gene expression, the effects of DNAme at regulatory regions on gene expression are less clear43. As no RNA samples were collected in DCCT when genomic DNA was isolated, we used published datasets containing both DNAme and gene expression profiles from the same blood cells including 1,202 monocyte and 214 CD4+ samples44. Out of 84 commonly-covered HbA1c-assoc CpGs, we identified 24 CpGs having statistically significant association (FDR<5%) with the expression of at least one nearby gene (±500kb relative to CpGs) in monocytes, and 16 CpGs in CD4+ cells (Fig. 4a). Among these, 9 CpGs were in common (Extended Data 7), while 15 and 7 were specific to only monocytes and CD4+, respectively (Extended Data 8–9). The most significantly associated genes for most CpGs (17 and 12 in monocyte and CD4+, respectively) were different from genes harboring the corresponding CpGs (Fig. 4b). Interestingly, significance levels for CpGs stratified based on location showed association with mostly intronic regions in both monocyte and CD4+ cells (Fig. 4c & 4d), suggesting the importance of intronic DNAme on gene expression in both cell-types. Most of these intronic CpGs (both neg-assoc or pos-assoc) were located in enhancer regions (Fig. 4e), highlighting the importance of chromatin-states in DNAme-expression regulation.
Figure 4. Association of DNAme with gene expression and genotypes.
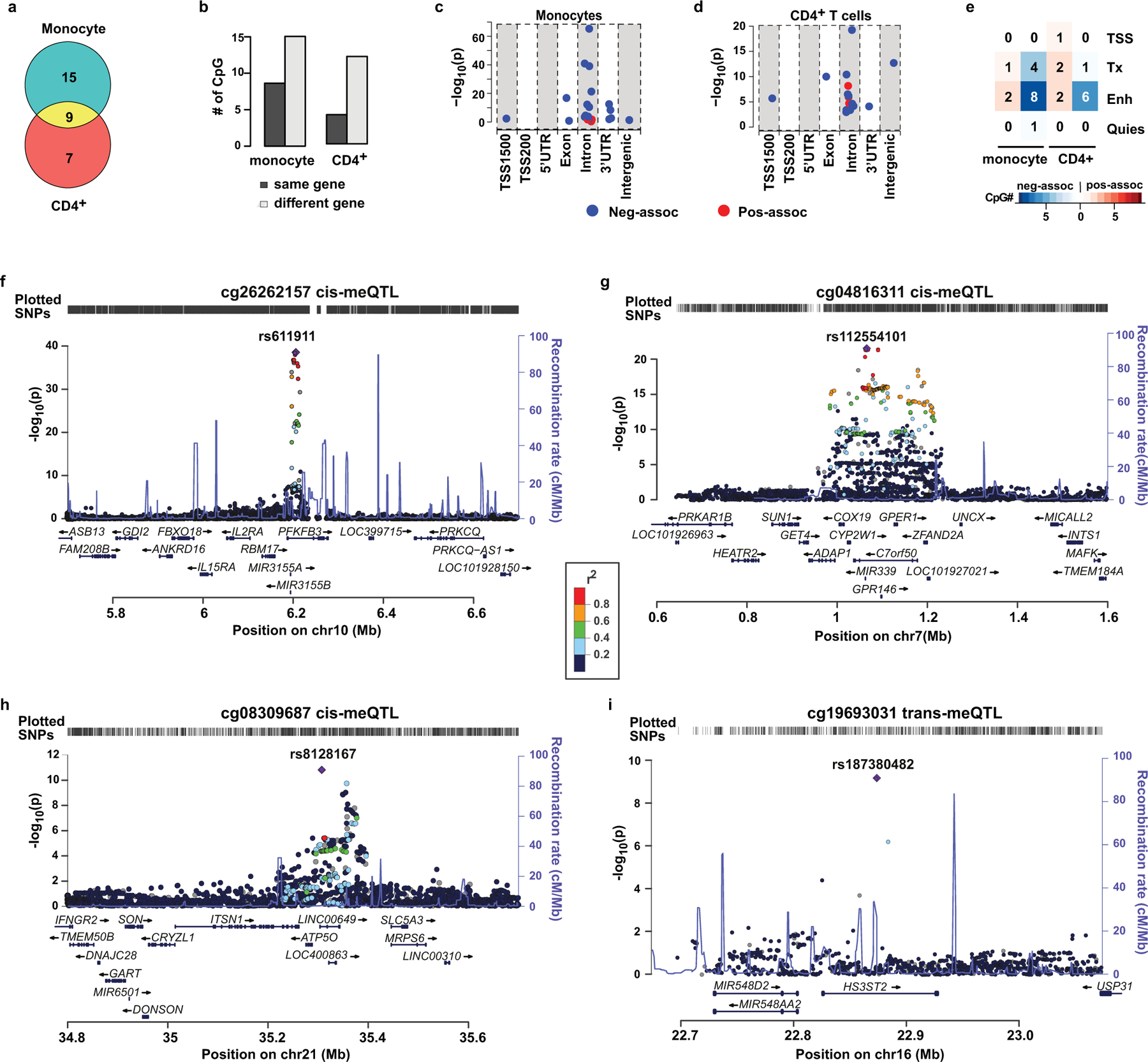
a-e, The associations of DNAme at HbA1c-assoc CpGs with the expression of gene(s) in monocytes or CD4+ T cells were analyzed on each pair of HbA1c-assoc CpGs and its nearby genes within 500kb. Multiple linear regression models (two-sided tests based on t-stastistic, n=1202 for monocytes and n=214 for CD4+ T cells) were applied to published datasets containing both DNAme and gene expression profiles from same sample to identify HbA1c-assoc CpGs whose DNAme is associated with the expression of nearby gene(s) (expression-associated genes) with FDR < 0.05 in monocytes and CD4+ cells. a, Venn diagram of HbA1c-assoc CpGs with expression-associated genes in monocytes and CD4+ cells. b, Bar plots comparing the number (#) of CpGs whose most significant expression-associated gene is the same gene in which the corresponding CpG is located or not. c&d, Dot plots showing the association significance level between DNAme and gene expression at CpGs classified based on their genomic location relative to the expressed gene in monocytes (panel c) and CD4+ T cells (panel d). e, Heatmap of the number of pos-assoc or neg-assoc intronic CpGs with different chromatin states in monocytes and CD4+ cell types. Blue dots indicate neg-assoc CpGs and red dots indicate pos-assoc CpGs. Chromatin states related to transcription start site are represented as TSS, related to transcription as Tx, related to Enhancer as Enh, and quiescent state as Quies. f-i, Regional association plots of CpGs with methylation quantitative trait loci (meQTLs) identified genome-wide. The eight most significant independent HbA1c-assoc CpGs (p < 5e-08) were selected for meQTL analyses. meQTLs were identified from a GWAS by two-sided linear regression under an additive genetic model using DNAme and genotyping data of 474 European DCCT participants included in the current study. The 4 CpGs with statistically significant cis- (f-h) or trans-meQTLs (i) at Bonferroni-adjusted p < 0.05 (p < 5e-08) are shown, with corresponding CpGs listed on top of the plot. In each plot, each dot represents one SNP with left y-axis representing nominal association p value between SNPs and DNAme in –log10 format, right y-axis representing recombination rate, and x-axis representing chromosome coordinates (HG19). The most significant associated SNP is shown as a purple diamond while linkage disequilibrium (LD) between this and the other SNPs is shown in colors defined in the color scheme. The LD measures are based on 1000 Genomes Nov 2014 EUR population. The plot was created using LocusZoom http://locuszoom.sph.umich.edu/locuszoom.
We found the strongest DNAme-nearby gene-expression association in both monocytes and CD4+cells (Extended Data 7a) was the negative association between cg01745539 (located in HLA−DQB1) and HLA-DQB1 expression. Using genotype data for our participants (n=485) from the DCCT genetics study45, we examined the association of DNAme at cg01745539 with genotypes of 13 common SNPs located at probes on both gene expression and DNAme arrays (Supplementary Table 18) and found this highly-significant association might be due to the strong association of DNAme at cg01745539 with SNPs in probes used for the gene expression array (most significant is rs1140342, p=3.62e-38) (Supplementary Table 19).
Relationship between genetics and DNAme at HbA1c-assoc CpGs and identification of methylation quantitative trait loci (meQTLs)
Because our observations on HLA-DQB1 revealed interesting associations of DNAme with SNPs located >3kb away, we next examined this further using data from the previous DCCT genetic study45,46. We first examined if any HbA1c-assoc CpGs are located at/close to the major loci associated with glycemic control previously identified in the DCCT cohort46 and found that none of them was located ±500kb relative to the HbA1c-assoc CpGs identified in CONV or INT group, or both combined.
We next performed more extensive analyses to identify meQTLs related to the 11 most significant HbA1c-assoc CpGs (Bonferroni-adjusted p< 0.05). Out of these 11 HbA1c-assoc CpGs, we tested 8 CpGs depicting independent signals (see Methods). Using both DNAme and genotyping data from same European subjects (N = 474), we performed GWAS for each of the 8 CpGs to identify meQTLs. We found multiple significant (p<5e-08) cis-meQTLs for three CpGs (cg08309687, cg04816311, cg26262157) (Fig. 4f–4h), and a single trans-meQTL for cg19693031 (Fig. 4i, Supplementary Table 20). No significant meQTLS were identified for the other 4 CpGs.
DNAme mediation in HbA1c-associated complication development
The DCCT/EDIC study clearly demonstrated that hyperglycemia (higher HbA1c) increases the risk of complications, including retinopathy and DKD. We therefore performed in-depth analyses to determine whether DNAme could play a role in mediating the association between HbA1c during DCCT and subsequent complications during EDIC in the 499 participants, following the steps shown in Extended Data 10. Complications included proliferative diabetic retinopathy (PDR), SNPDR, clinically significant macular edema (CSME); albumin excretion rate >300mg/24h (AER300), eGFR < 60 ml/min/1.73 m2 (GFR60), and eGFR slope (representing kidney function decline) for DKD.
After excluding participants who developed the specific complication during DCCT, there were 96 out of 482 (19.9%), 92/473 (19.4%), 108/464 (23.3%), 30/485 (6.2%) and 23/498 (4.6%) who developed PDR, SNPDR, CSME, AER300, and GFR60, respectively during EDIC (Supplementary Table 21). The survival plots for retinopathy/DKD during EDIC and the cumulative distribution of normalized eGFR slope (see Methods) by group all demonstrated the expected trend of increased incidence of complications in our 499 subcohort across the 4 groups in order of PRIM INT, SCND INT, PRIM CONV and SCND CONV, and more negative eGFR slope in CONV vs. INT (Fig. 5a). We also confirmed the significant association between mean-DCCT HbA1c and risk of complications development or eGFR slope in our subcohort (Fig. 5b, Supplementary Table 22).
Figure 5. The association of mean-DCCT HbA1c and DNAme at DCCT closeout with retinopathy/DKD development during EDIC.
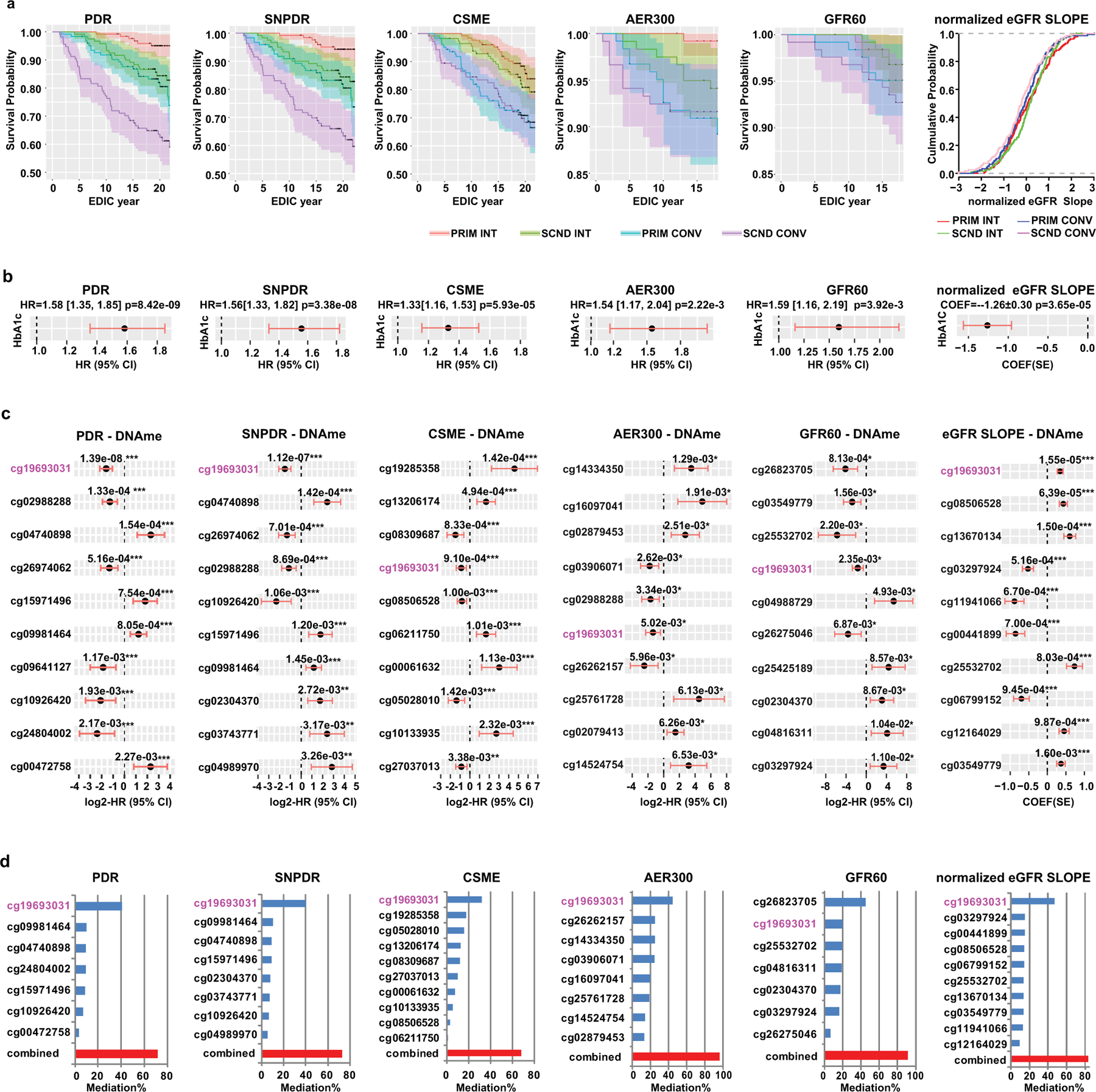
a, Survival plots of retinopathy (PDR, SNPDR and CSME) or DKD (AER300, GFR60) and cumulative distribution of normalized eGFR slopes during EDIC are stratified based on group. The follow-up period for retinopathy is through EDIC year 18 (interval censored data due to staggered clinic visit intervals, up to EDIC year 23) and, for DKD up to EDIC year 18. Each group (PRIM INT, SCND INT, PRIM CONV and SCND CONV) was plotted in different colors as indicated in the legend with 95% confidence interval (CI) depicted by the shaded area. b, Forest plots for association of mean-DCCT HbA1c (10% increase from previous time point) with risk of complications development (as indicated on the top of each panel), or with normalized eGFR slopes. Black dots with red lines represent either hazard ratios (HR) with 95% CI in forest plots, or coefficient (COEF) with standard error (SE) in dot plot. c, Forest plots for the association of DNAme with risk of complication development or with eGFR slope at HbA1c-assoc CpGs. The top 10 most significant HbA1c-assoc CpGs for each complication/manifestation are shown in order of significance level from high to low, each labeled with CpG ID on the left side and nominal p values listed on top of the plot. FDR is estimated by BH-adjustment over 186 HbA1c-assoc CpGs. *** FDR < 5%; ** FDR < 10%; * nominal P < 0.05. d, Percentage of association (mediation %) between history of HbA1c and complications that is explained by DNAme at the indicated HbA1c-assoc CpGs alone (each labeled with CpG ID), or in combination, using mediation analyses. The best combination of CpGs (indicated by red bar) which explains the association between mean DCCT HbA1c and risk of complications development for each indicated complication was identified from the top 10 CpGs among the HbA1c-assoc CpGs associated with complications. The explanation percentages of each CpG (up to 10 CpGs) in the combination are also presented as blue bars. cg19693031 located in TXNIP-3’UTR is shown in pink font. In a-d, participants who did not develop the corresponding retinopathies or DKDs during DCCT were included in the analyses for each complication (PDR: n=482; SNPDR: n=473; CSME: n=464; AER300: n=485; and GFR60: n=498). For eGFR slope, all the participants (n=499) were included in the analyses. In panel b-d, two-sided CoxPH for analyses related to each complication and linear regression models for eGFR slope adjusting for covariates were applied. See Methods for details.
Next, the 186 (FDR<0.15) HbA1c-assoc CpGs were tested for association with risk of each complication development during EDIC (EDIC baseline to year 18, i.e. after sample collection for DNAme profiling). Supplementary Table 23 shows 10, 7, and 9 HbA1c-assoc CpGs that were associated with risk of PDR, SNPDR, and CSME development at FDR< 5%; 24, 16, and 14 at FDR<10%; and 48, 46 and 41 at nominal p<0.05, respectively. For AER300 and GFR60, no HbA1c-assoc CpGs indicated association at FDR<10%, while 36 and 32 were identified with a nominal p<0.05. However, a strong association between DNAme at HbA1c-assoc CpGs and eGFR slope during EDIC was observed, with 23, 43 and 60 CpGs identified at FDR<5%, <10% and p<0.05, respectively. For each phenotype, the top 10 HbA1c-assoc CpGs depicting the most significant association with the risk of complication development or normalized eGFR slope are shown in Fig. 5c.
For mediation analyses, performed as shown in Supplementary Table 24 using cg19693031 as example, each of these top 10 HbA1c-assoc CpGs was added as covariate in the CoxPH or linear models examining the association between mean-DCCT HbA1c and complications. Interestingly, DNAme at cg19693031 (3’UTR of TXNIP) alone could explain 32–41% of the association between mean-DCCT HbA1c and retinopathy (41%, 40% and 32% for PDR, SNPDR and CSME respectively) and explain ~45% of the association between HbA1c and DKD/slope (45%, 45% and 47% for AER300, GFR60 and eGFR slopes respectively). Moreover, the best combinations of multiple CpGs (selected from the top 10 HbA1c-assoc CpGs based on association significances with risk of disease development for each complication using Brute-force approach) (Fig. 5c) could explain up to 71%, 73% and 68% of the association between mean-DCCT HbA1c and PDR, SNPDR and CSME, respectively, and 97%, 92% and 84% for AER300, GFR60, and eGFR slope, respectively (Fig. 5d). The detailed annotations of the specific combinations of mediatory CpGs for retinopathies and DKDs are shown in Supplementary Tables 25 and 26. These results suggest for the first time that DNAme at several CpGs across the genome can play a mediation role between HbA1c and future disease development, and combinations of multiple CpGs capture 68–97% (Fig. 5d, red bars) of the DCCT mean-HbA1c effect on complications development, further strengthening the connection between DNAme and metabolic memory.
We also examined the potential causal effect of DNAme on future complications development using Mendelian Randomization (MR) for the 4 HbA1c-assoc CpGs that we identified to have meQTLs (Fig. 4f–i and Supplementary Table 20). Using two-sample MR analyses and data from the largest available meta-GWAS study for DKD (GFR60 and AER300)25 and for retinopathies (PDR vs. non-PDR or no-retinopathy [NR], and PDR vs. NR)16, we detected a causal effect of cg08309687 on GFR60 development (Supplementary Tables 27& 28).
Functional exploration of the TXNIP region
As DNAme at TXNIP and nearby regions likely plays an important role in metabolic memory, we examined this interesting genomic region (Fig. 6a) highlighted by red dashed-line box in Fig. 6b. Besides the top HbA1c-assoc CpG cg19693031 and 2 HbA1c-assoc regions (500bp window, pink background), 4 additional CpGs at ~2.9kb, 0.9kb upstream of TSS, and 13.5kb, 15.0kb downstream of TXNIP were identified (FDR<15%). Among these 9 CpGs, except for 2 located at two ends of the region, all showed significant association with mean-DCCT HbA1c, FDR<5%. Notably, the majority of this region had enhancer features (yellow), apart from promoter (red) and transcription (green) defined in 15 chromatin states (upper panel, Fig. 6c). The same region containing cg19693031 was re-classified as enhancer (yellow green) in 18 chromatin states (lower panel, Fig.6c). Furthermore, this region spans both TXNIP promoter and adjacent PIR (containing 3’UTR) defined by PCHiC (Fig. 6d). The fact that both regions interact with similar genomic regions (mainly promoters of nearby genes including POLR3GL, LIX1L, Fig. 6e) suggests a high probability of interactions between TXNIP promoter and its 3’UTR.
Figure 6. Functional exploration of DNAme at the TXNIP genomic region.
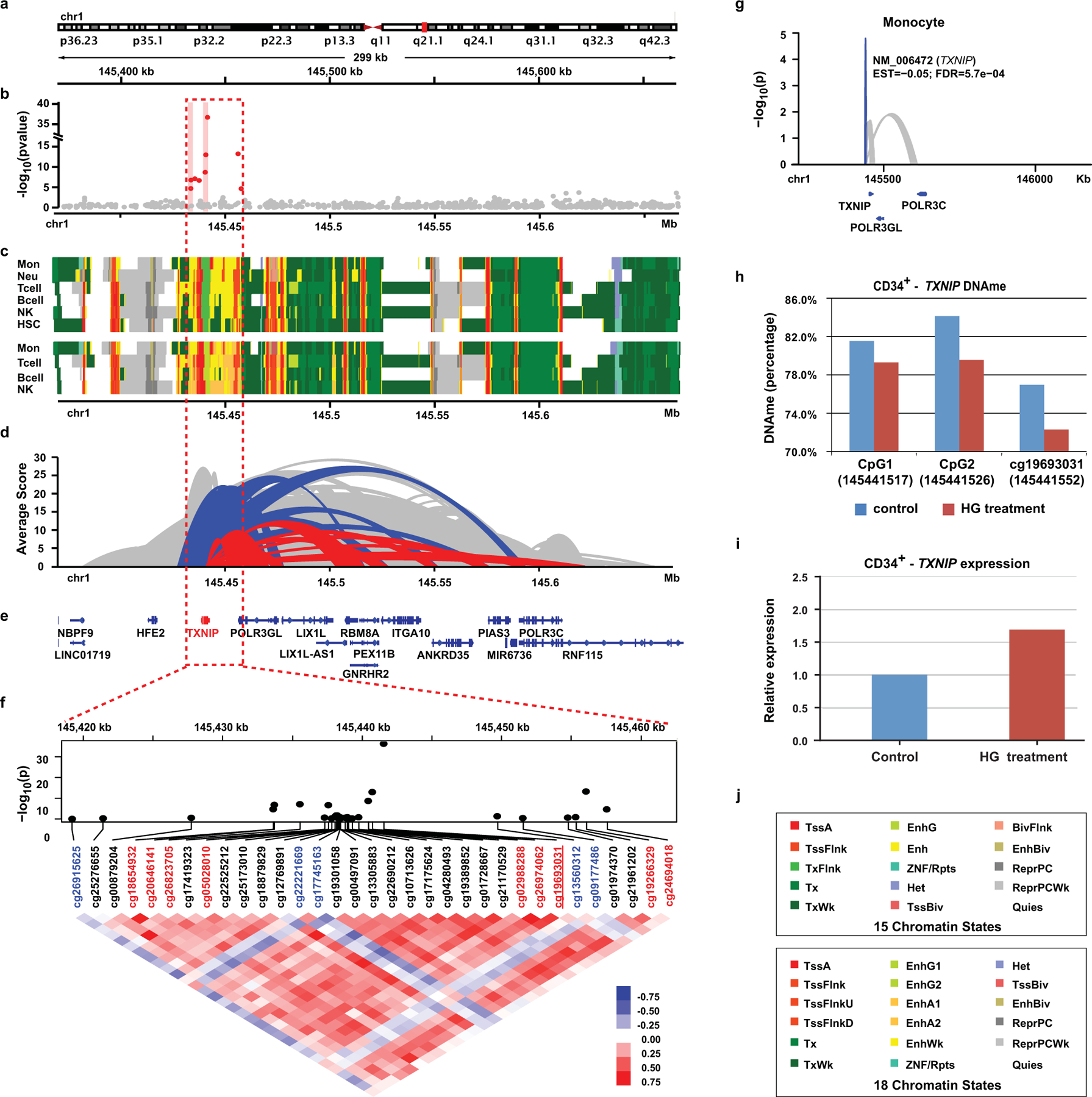
a, Genomic location of the depicted region at/near TXNIP. b, Manhattan plots of association between mean-DCCT HbA1c and DNAme at the covered CpGs. Multiple linear regression models adjusting for covariates were applied to each CpG across all the samples (two-sided tests based on t-statistic, n=499). X-axis represents CpG location and Y-axis represents –log(p). Two HbA1c-assoc regions are highlighted with pink background. c, Heatmap showing the 15 chromatin states (top) and 18 states (bottom). Colors are shown in the legends (panel j). d, Ribbon plots representing the genomic interactions identified by IHEC PCHi-C data. Red represents the interactions involved in PIRs containing TXNIP 3’UTR. Blue represents interactions containing TXNIP promoter. Height of each ribbon represents the average CHiCAGO score (Y-axis) of the corresponding interaction across 5 major blood cells including monocytes, neutrophils, CD4+ T-cells, CD8+ T-cells and B-cells. e, Annotations of the RefSeq genes in the depicted region. f, Pearson correlation of DNAme at 33 CpGs in TXNIP and its 25kb flanking region. The associations of DNAme with mean-DCCT HbA1c at each CpG are obtained by the same linear regression model (n=499) indicated in panel b and shown as a Manhattan plot in the upper panel. HbA1c-assoc CpGs (FDR < 15%) are labeled in red font with the most significant cg19693031 underlined. Pair-wise correlations of DNAme of these CpGs were analyzed by Pearson correlation with coefficients shown as heatmap. Blue/red represents negative/positive correlation, respectively. The majority of the CpGs depict positive correlations including all 9 HbA1c-assoc CpGs (in red), while 5 CpGs (in blue) showed little to negative correlation with other CpGs. g, Ribbon plot representing the association of DNAme at cg19693031with expression of its nearby genes (including TXNIP) in monocytes. Multiple linear regression models adjusting for covariates based were applied to DNAme at cg19693031and expression of each nearby gene within 500 bp distance (n=1202 monocytes) to identify DNAme-assoc genes with nominal p < 0.05 (two-sided tests based on t-statistic). Blue indicates negative association at FDR < 0.05, while grey indicates associations with p < 0.05. h, In-vitro DNA hypomethylation at 3 CpGs in the 3’UTR of TXNIP (including cg19693031) induced by high glucose (HG) treatment of human primary bone marrow CD34+ cells. DNAme was measured by amplicon-seq. i, Upregulation of TXNIP expression induced by HG treatment of human primary bone marrow CD34+ cells. Cells were cultured in medium (see Methods) containing 25mmol/L glucose (control), or same medium with the addition of 20mmol/L glucose (45mmol/L total) for 72 hours (HG treatment). RT-PCRs were performed in triplicate, and the data shown represent means of triplicates from one experiment. j, Color codes of chromatin states in the heatmaps for 15 and 18 chromatin states shown in panel c.
Pearson correlation of DNAme at all CpGs in the region showed mostly positive correlations (Fig. 6f), especially with cg19693031. In addition, conditional analyses (Supplementary Table 29 legend for details) showed that HbA1c-DNAme association at all the CpGs was dependent on cg19693031 except cg19266329 (nominal p=1.95e-02). The negative association between TXNIP expression and DNAme at cg19693031 in monocytes (FDR=5.8e-04, Fig. 6g & Extended Data 8g) provided additional evidence of this CpGs’ regulatory role. Together, these results further underscore the important role of DNAme at the TXNIP locus and nearby regions in T1D and complication development through enhancer regulation.
Our in silico data implied that DNAme in HSCs at HbA1c-assoc CpG regions such as TXNIP (enriched in enhancer/transcription states) might be transmitted during differentiation to myeloid cells to confer memory. To support this, we tested human primary bone marrow CD34+ cells cultured in vitro with or without high glucose (HG) treatment for 72 hours. This extra glucose induced hypomethylation at 3 CpGs including cg19693031 in the TXNIP-3’UTR (Fig. 6h), and parallel upregulation of TXNIP expression (Fig. 6i) in these progenitor cells.
DISCUSSION
Metabolic memory in T1D was first described by DCCT/EDIC2 in which a history of high HbA1c was an independent risk factor for complication development, in addition to current glycemia47,48. Although epigenetic mechanisms have been implicated6,26, it is unclear whether epigenetic DNAme plays a mediatory role in glucose-induced complication development even after restoration of normoglycemia. This temporal connection is addressed in the current study using blood DNA samples from a large DCCT cohort (n=499), a rich database with ~6 years of quarterly HbA1c measurements on each participant (prior to blood collection for DNA isolation), and detailed clinical/complications information collected during the subsequent 18+ years of follow-up in the EDIC study.
We observed associations between DNAme near DCCT-closeout and long-term preceding glycemic history using mean-DCCT HbA1c. To our knowledge, this is the first report on DNAme changes/accumulation after a long-term history of hyperglycemia, as previous studies only reported the association of DNAme with HbA1c or blood glucose (including FBS) at blood draw in non-diabetic cohorts49–51, or T2D35,52. Even in our previous smaller DCCT study, although differentially methylated loci reported were likely associated with previous history of HbA1c, a firm conclusion could not be made due to limitations of the study design26. However, we cannot fully differentiate our observed DNAme changes from those induced by recent hyperglycemia (close to blood collection) because of the high correlation between mean-DCCT HbA1c and blood-draw HbA1c (rho=0.88, p< 2.2e-16). Among the 186 HbA1c-assoc CpGs we identified, the top significant CpG showing association with previous history of HbA1c was cg19693031 in TXNIP-3’UTR. Of note, similar negative associations at this same CpG with HbA1c in the general population, with T2D and T2D-related metabolic traits have been reported earlier35,52–56. All these findings highlight the importance of DNAme in TXNIP-3’UTR in diabetes. Furthermore, by integrating with GWAS data, we found significant meQTLs associated with 4 out of the top 8 most significant HbA1c-assoc CpGs, suggesting potential associations between DNAme and genetic variants at some of these HbA1c-assoc CpGs. We also subsequently detected a causal effect of cg08309687 on GFR60 development by MR. These data suggest that, apart from genetics, epiphenomena dictated by as-yet unknown biological mechanisms also play a key role in the DNAme association with HbA1c.
We also found that, at specific CpGs, the effect size of the HbA1c association was larger in the CONV than INT. As CONV has significantly higher mean and variance of DCCT HbA1c relative to INT, this result suggests changes in DNAme at some HbA1c-assoc CpGs are triggered only when HbA1c is above some threshold and maintained for a certain time period, if not due to the greater DCCT HbA1c range/variance in CONV (6.28%~13.85%/1.44%) than in INT (5.32%~9.99%/0.56%).
The HbA1c-assoc CpGs were validated using one internal and two external cohorts. These validations also suggest potential common mechanisms for DNAme changes induced by hyperglycemia in T1D (our study and Joslin) and in the SAFHS850 cohort where 34% of individuals had T2D/impaired glucose tolerance. Although all these cohorts do not represent ideal replications of our study, a more appropriate T1D cohort with comparable sample size and clinical manifestations (including complications) was unavailable. We realize that, in the absence of DNA samples or DNAme profiles from earlier time(s) during DCCT, we cannot determine causality, but we speculate HbA1c exhibits causal effects on DNAme because HbA1c measures preceded time of sample collection for DNAme. Our in-vitro experiments also implicated a causal effect of HG on DNAme at TXNIP-3’UTR, also in line with other studies showing HG-induced DNAme changes6,8,26.
The persistence of DNAme differences at 186 HbA1c-assoc CpGs was also observed in EDIC61, consistent with our previous findings from profiling DNAme at two time points 16/17 years apart, in both monocytes and lymphocytes26. But the question remains: how do these DNAme changes accumulate and persist in peripheral blood over several years, especially in neutrophils and monocytes that have limited life spans in the circulation? We speculate that the DNAme changes might occur in HSC progenitors of these white blood cells, which then persist via maintenance methylation during expansion and differentiation26. This is supported not only by our own in-vitro data (with CD34+ cells in this study and monocytes in our previous study26), but also by reports showing hyperglycemia in vitro causes “memorized” DNAme changes in CD34+ HSCs that contribute to endothelial dysfunction57.
Notably, the majority of the HbA1c-assoc CpGs were significantly enriched in enhancer and transcription-related regions in blood monocytes and HSCs, with C/EBP binding sites being the most enriched motifs in these regions. C/EBP TFs regulate energy metabolism, immunity, inflammation, and adipogenesis, all related to diabetes/complications58. Moreover, C/EBPs play critical roles in hematopoiesis and myeloid cell (monocyte/neutrophil) differentiation41,58, suggesting C/EBP-dependent links between DNAme and HSC differentiation, especially myelopoiesis. This is further supported by our observation that HbA1c-assoc CpGs were enriched in open chromatin regions in HSCs or cells derived from myeloid progenitors, as well as reports demonstrating hyperglycemia-induced myelopoiesis in animal models59 and modulation of myeloid progenitors during trained immunity60. These data, along with our results from HG-treated CD34+ cells, substantiate the importance of hyperglycemia-associated enhancer DNAme in myeloid differentiation, T1D complications and metabolic memory. Further studies are needed to confirm this hypothesis.
Importantly, we provide the first evidence in humans supporting a mediation role for DNAme, especially at TXNIP-3’UTR, in the association between prior history of blood glucose and future complication development, a critical matter in the epigenetic explanation of metabolic memory. Strikingly, we found combinations of DNAme at several HbA1c-assoc CpGs could explain at least 68% of risk attributed to mean-DCCT-HbA1c for retinopathies and 84% for DKDs. These data suggest DNAme at multiple sites can “co-operate” to augment mechanisms involved in metabolic memory possibly by affecting different biological pathways related to complications. These pathways are related to oxidative stress, innate response and inflammation in blood cells as well as target cells/tissues affected by complications (Supplementary Table 30). However, our mediation analyses could not differentiate whether DNAme at specific CpGs play causal effects on complication development. Involvement of TXNIP in complications development is supported by data showing its up-regulation by HG in-vivo and in-vitro in eye and kidney cells associated with retinopathy and DKD respectively29,30, and in kidney tubuli from subjects with DKD61. This suggests potential pleiotropy of TXNIP DNAme for the development of multiple subsequent complications. Our detection of a causal effect of cg08309687, the CpG validated in all 3 validation cohorts, besides cg19693031 (Fig. 2C), on GFR60 development by MR also suggests causal mediation roles of DNAme at some CpGs. Nevertheless, we also cannot rule out causal effects of DNAme at other CpGs undetected by MR, which could be due to limited power (for binary outcomes, largely based on the proportion of cases, odds ratio of complications per standard deviation of DNAme adjusted for confounders, and the proportion of DNAme variance explained by the genetic instrument). Moreover, biological mechanisms due to epiphenomena are also likely, as recently noted55. Given our exciting findings, future studies with larger EWAS cohorts and greater disease incidence (longer follow-up) can help further validate our findings and increase power for MR to evaluate causality.
Our data suggests that DNAme changes at HbA1c-assoc CpGs impact blood cells such as myeloid cells (monocytes and macrophages). These cells, well-known to promote inflammation, infiltrate and accumulate in target tissues during diabetes and are involved in most diabetic complications62. Notably, reports showing that non-diabetic rodents developed DKD after bone marrow transplant from diabetic donors62 not only suggest the potential role of HSCs in metabolic memory, but also emphasize the importance of HSC-derived differentiated blood cells in complication development. Hence, epigenetic changes in blood and myeloid cells could serve as good proxies for processes in target organs affected by diabetes/complications. Interestingly, none of our HbA1c-assoc CpGs are similar to CpGs previously associated with kidney function decline or interstitial fibrosis24, suggesting possible blood-cell specific functions also for these CpGs. This is supported by reports in which many genes containing the mediatory HbA1c-assoc CpGs were connected to innate immune response and inflammation (Supplementary Table 30), along with those having potential functions in affected target tissues (eg kidney, retina, nerves, heart). DNAme at some of the HbA1c-assoc CpGs in the target cells/tissues may be similar to those in blood cells, or even more pronounced because of the cell-type specific nature of epigenetic changes, as well as their longer lifespans relative to blood cells. In fact, in-vitro and in-vivo studies support the involvement of DNAme in target cells/tissues in complication development6. When EWAS data from large numbers of target tissues from people with diabetes become available in the future, we can examine whether they depict similar results as ours.
We are also aware of other limitations. First, because DCCT is a non-random sample of people with T1D with extensive inclusion/exclusion criteria63, our results might not be fully representative of other cohorts. Second, the published datasets we examined in blood cells or HSCs are mainly from healthy donors, and not T1D subjects.
Taken together, our EWAS of DCCT samples has uncovered key functions for DNAme in diabetic complications and metabolic memory during EDIC. Notably, for the first time, our results reveal a mediation role for DNAme in the association between HbA1c and future development of complications. Thus, prior history of hyperglycemia may induce persistent DNAme changes at key loci, including TXNIP, in various target cells, and in HSCs, which are epigenetically retained in differentiated myeloid (and other) cells to facilitate metabolic memory, likely through modifying enhancer activity at nearby genes.
Methods
Human subjects
The study protocol was approved by the Institutional Review Board (IRB) at the City of Hope (COH) Medical Center and at each of the DCCT/EDIC clinics. Our study was performed in compliance with ethical regulations. WB genomic samples (1419) were collected at DCCT closeout and archived in the EDIC Central Biochemistry lab from the 1991–1993 DCCT Family study. After filtering the samples based on criteria, including adult vs. adolescent, informed consent, and sufficient DNA (listed in Supplementary Table 2), 1042 DNA samples were used to randomly select 500 WB samples from four DCCT/EDIC groups/strata (125 per group): PRIM INT, SCND INT, PRIM CONV, SCND CONV. The PRIM cohort included participants with T1D duration of 1–5 years and no retinopathy at study entry (DCCT baseline). The SCND cohort included subjects who had T1D for 1 to 15 years and mild to moderate retinopathy at DCCT baseline. Patients in INT group received three or more daily insulin injections to maintain daily glucose level at 3.9–6.7 mmol/L (70–120mg/dL) before meals and peak levels <10.0 mmol/L (180 mg/dl) after meals, and monthly HbA1c within normal range (< 6.05%). Patients in CONV group received one or two daily injections of insulin with no glucose goals except the prevention of symptoms of hyperglycemia and hypoglycemia1. Power analysis was performed at the time this study was designed to determine the sample size. Specifically, we estimated the power for detecting an association of DNAme with the risk of complications based on clinical data up to EDIC year 10. The projected number of ~100 incident cases for retinopathy provides very good power (85%) to detect a hazard ratio (HR) of 1.4 with a two-sided test at the 0.05 level, while a much lower incidence of kidney disease (≤ 30) provides modest to low power (43% ~ 68%) to detect a HR of 1.6 at the same significance level.
DNAme profiling and data processing
To minimize batch variations or systemic bias among the 4 groups introduced during processing of DNAme arrays, all 500 samples were classified into fifteen 32-sample batches (batch 1–15) plus one 20-sample batch (batch 16). Each 32-sample batch was comprised of 8 samples from each group, and the 20-sample batch had 5 samples each. The samples were randomly assigned to different batches by the EDIC Data Coordinating Center, and the batch number of each sample was released without group identity (blinded information). 1ug of each WB sample (estimated by picogreen) was sorted by batch number and processed at the University of Southern California (USC) Molecular Genetics Center Core where samples were bisulfite-treated using the EZ DNA methylation kit and subsequently profiled for DNAme using Infinium MethylationEPIC Beadchips (WG-317–1003, Illumina, CA). Samples were processed in batches (1–6 batches) in each run. Specifically, probes for internal controls designed in the EPIC array were used to monitor each step during array processing, detection p-values were also examined to ensure reliable DNAme values for each sample. Moreover, 5 samples processed in different runs were repeated for batch variation control resulting in 505 datasets in total. Normal-exponential out-of-band (noob) normalization (within-array normalization) was applied to each sample for background correction and dye-bias normalization64, followed by stratified quantile normalization65 across all samples for both within- and between-array normalization to correct both type 1 and type 2 probes. Detection p-values on all 866,554 CpGs covered by the EPIC array (manifest v1.0-B4) were determined, and CpGs with detection p > 0.05 in any one sample were filtered out. Cluster analyses on the resulting datasets, after excluding CpGs located on the X or Y chromosomes were subsequently performed. The pre-processing and QC steps verified that batch variation was minimal after normalization, with all 5 replicated samples processed in different runs clustering with each other (Supplementary Figure 1). Moreover, one outlier was also detected based on cluster analyses (likely due to large differences in estimated white cell composition from other samples) and was excluded from the complete dataset (Supplementary Figure 1). All subsequent analyses were performed on the remaining 499 samples. The percentages of CpGs with detection p < 0.05 are all < 5%. PCA analyses of these samples using data on both sex chromosomes and autosomes (Supplementary Figure 4) revealed clear expected separation on sex using sex chromosomes, and no clusters or outliers based on cohort-treatment groups. After excluding one outlier and 5 repeated samples, the raw data was re-processed (normalization and filtration based on detection P) using the same procedures as mentioned previously. The final normalized dataset included 815,432 CpGs across each of 499 samples. All the analyses mentioned above were performed using functions provided in the R bioconductor package “Minfi”65.
Identification of HbA1c-assoc CpGs
Multiple linear regression models were used to evaluate the association of DNAme, in the form of normalized M values (logarithm transformed ratio of methylated signal versus unmethylated signal), as the dependent variable and the mean-DCCT HbA1c as the independent variable, across all samples. Variables known to impact DNAme levels or measurement, namely sex, age, cell composition (Supplementary Table 5) and array processing time (batch effects) were included as covariates. Blood-glucose related variables, such as DCCT baseline HbA1c, diabetes duration prior to DCCT baseline, and duration from DCCT baseline to sample collection were included in the model to account for their potential effects on DNAme. Cohort was also included as a covariate in the analyses of all 499 samples in either PRIM or SCND cohorts. As cell compositions were not measured on these WB samples when they were collected by DCCT, the compositions of major blood cells (granulocytes, monocyte, B-cell, CD4+, CD8+ and NK cell) were estimated for each sample using the Houseman deconvolution method66 based on normalized beta values [methylated /(methylated + unmethylated)] after excluding CpGs on chromosomes X and Y (Supplementary Table 5). The DNAme dataset in the Bioconductor R package “FlowSorted.blood.450K” was used as the reference dataset. Specifically, the raw reference dataset was processed using the same pipeline (noob+quantile normalization, detection P value filtration and sex chromosome exclusion) as done in the current study. Beta values on CpGs covered in both datasets were retrieved and subjected to deconvolution using functions provided in Minfi. Significance levels were estimated by two-sided tests based on t-stastitic in linear regression model. BH-adjusted p values (FDR) for each CpG were estimated using the p.adjust function in the R base package “stats” under R v3.4.3. HbA1c-assoc CpGs at different confidence level (FDR < 5% and FDR < 15%) were then identified.
Fourteen clinical variables (at the time of sample collection) not included as covariates for the identification of HbA1c-assoc CpGs were also tested to determine if they have any significant associations with DNAme at each HbA1c-assoc CpG with FDR < 15%. These include race, smoking, systolic blood pressure (SBP), diastolic blood pressure (DBP), BMI, cholesterol (CHL), triglyceride (TRG), low-density lipoprotein (LDL), high-density lipoprotein (HDL), and any complications that developed during DCCT including PDR, SNPDR, CSME, AER300, and cardiovascular disease. We found that very few of these additional variables have significant association with DNAme, as shown in Supplementary Figure 5. Hence these additional clinical variables were not included in the model as covariates.
HbA1c-assoc CpGs depicting different associations between DNAme and mean-DCCT HbA1c in the INT treatment group compared to CONV group were identified by adding an interaction term between treatment group (CONV or INT) and DNAme in the multiple linear regression models. The HbA1c-assoc CpGs with interaction p < 0.05 were considered to have different association between the two groups. Similar analyses were also applied to identify HbA1c-assoc CpGs with different associations between the two cohorts (PRIM and SCND) and between the two gender groups (male and female). HbA1c-assoc regions were classified as those having at least 2 HbA1c-assoc CpGs within a 500bp window.
Sensitivity analyses of HbA1c-assoc CpGs
Sensitivity analyses were performed on 14 additional clinical variables (described in the previous paragraph) at time of blood collection, which were not included in the original model for HbA1c-assoc CpGs identification. The original multiple linear regression model with addition of each of these clinical variables one at a time was applied to the 186 CpGs. The association of DNAme with mean-DCCT HbA1c (including both significance and coefficients) obtained with this “new” model (with an additional variable) was then compared to the original model (without the additional variable).
Amplicon-seq and data analysis
Amplicon-seq pools were prepared on 96-well plate. Briefly, for each genomic DNA sample, 500 ng DNA was treated with bisulfite and dissolved in 40 ul water. For each targeted region, primers were designed using MethPrimer v1.0 (Applied Biosystems, MA) and each pair was tested to function optimally using both bisulfite treated and untreated genomic DNA. PCRs (25ul volume) were then performed with 1 ul of bisulfite-treated sample. The candidate target genes and their corresponding primers used are listed in Supplementary Table 31. An unique 6-mer barcode was encoded in primers for each PCR-amplified sample to distinguish patient samples. PCR products were verified on 1.5% agarose gels. The amplified DNA fragments were pooled and cleaned with DNA clean & concentrator kits (#D4003, Zymo Research, CA) and sent to the City of Hope Integrative Genomics Core where libraries were constructed using PCR free library preparation method. Libraries were then sequenced (paired-end, 100bp) on the Illumina Hiseq 2500. After trimming of barcodes, each sample was aligned to the targeted genomic sequences by Bismark67 v0.16.3 (Babraham Bioinformatics, Cambridge, UK). At each CpG site covered in the targeted region, methylated and unmethylated reads were summarized, and the DNAme levels (in M values) were calculated. For validation purpose, amplicon-seqs were applied on 96 samples (3 randomly selected batches, each containing 8 samples from each group). Samples with aligned reads > 2500 were used to study the association with mean-DCCT HbA1c at each CpG covered in the targeted regions using the same model as that used for HbA1c-assoc CpGs identification.
Cross-cohort (internal and external) validation
Three cohorts, namely DCCT41, SAFHS850 and Joslin195 were used to validate HbA1c-assoc CpGs, with the former one as an internal, and the latter two as external validation cohorts. For DCCT41, DNAme profile data (GEO#GSE76169) on 41 WB DNA samples collected at DCCT closeout (after excluding 22 samples that overlapped with current study, as both studies used the samples from the same sample collection) were previously performed in our lab using Infinium human methylation 450k beadchips26. The raw data were normalized using the same normalization method as in our current study (noob+quantile). At each of the HbA1c-assoc CpGs also covered by the 450K array, the same linear regression model as published (DNAme as dependent variable and group as independent variable, adjusted for covariates26) was applied to compare DNAme between cases and controls. The difference between cases and controls at each candidate CpG was used to validate the association of DNAme with mean-DCCT HbA1c at the same CpG in our current study.
The first external validation cohort (SAFHS850) was taken from the San Antonio Family Heart Study which was conducted to prospectively evaluate genetic risk of heart disease in Mexican-American families from San Antonio. Datasets from this cohort included those collected from 850 participants, of which ~20% had type-2 Diabetes (ADA criteria of fasting glucose ≥126 mg/dL or those receiving anti-diabetic medication) and an additional ~14% had impaired fasting glucose (prediabetes). DNAme profiling data was obtained with 450K arrays on WB samples collected during 2002 ~2006 (SAFHS850) and processed and analyzed using inverse-normalization followed by polygenic regression35. Using published data, the association between DNAme and FBG (the only available variable related to blood glucose in SAFHS850) was assessed at 77 HbA1c-assoc CpGs covered in the original study (7 CpGs were excluded due to detection P values > 0.05, location on sex chromosomes, or/and inability to calculate heritability) and used to validate the association of DNAme with mean-DCCT HbA1c in our study.
The second external validation cohort was selected from adult T1D participants from the Joslin Kidney Study (JKS), a longitudinal observation study led by Dr. Andrzej Krolewski (Joslin Diabetes Center and Harvard Medical School, Boston, Massachusetts). JKS aims to investigate determinants/biomarkers and to describe the natural history of renal function decline in T1D36. Subjects for the current study were selected from the JKS participants who had proteinuria and impaired renal function at enrollment (Supplementary Table 15). These subjects were recently used for a proteomics study68. Out of 219 T1D subjects with proteinuria and impaired renal function (average eGFR of 45.6±12.7ml/min/1.73 meters2) at enrollment68, 200 subjects were selected which included half (n=100) that developed ESRD during 7–15 years of follow-up, while the other half did not progress to ESRD during the same period of follow-up. The informed consent, recruitment and examination protocols for the JKS on Human Studies are approved by the Joslin Diabetes Center Committee, and protocol for DNAme profiling are approved by the IRB at City of Hope. DNAme profiles on their WB DNA samples were measured and processed using the same approaches as those used for DCCT499. Five samples, including one that failed QC and four that did not have some clinical information (at blood collection) needed for identification of HbA1c-assoc CpGs, were excluded from further analyses, resulting in the DNAme dataset from 195 Joslin study participants (Joslin195). The association of DNAme with HbA1c at the time of blood collection across 195 samples at 186 HbA1c-assoc CpGs were analyzed using multiple linear regression models with DNAme (M value) as dependent variable and HbA1c as independent variable, adjusted for covariates at time of blood collection including age, sex, diabetic duration, estimated cell composition, array processing time, hypertension, log-transformed eGFR and ACR. Fixed meta-analyses on both DCCT499 and Joslin195 was performed by “Bacon” R package version 1.10.0 using association coefficients (effect-size) and standard errors separately obtained in DCCT499 and Joslin195.
Analyses to examine DNAme persistence at HbA1c-assoc CpGs
The DNAme dataset (GEO#GSE75170) previously generated on 61 monocyte samples collected in EDIC years 16/17 (EDIC61)26 was used to study the DNAme persistence at HbA1c-assoc CpGs identified in EDIC499 with EDIC61. The same approaches as those adopted for DCCT41 were applied for data preprocessing and group comparison. The associations of DNAme with HbA1c were compared to DNAme differences between 31 cases and 30 controls in EDIC61.
Annotation of HbA1c-assoc CpGs
Each HbA1c-assoc CpG was annotated based on its genomic location relative to RefSeq Genes downloaded from the UCSC genome browser (Human Genome Assembly Hg19). Six regions were defined for each transcript; 200~1500bp upstream to TSS (TSS1500), 0~200bp upstream to TSS (TSS200), 5’UTR, coding exon, intron, and 3’UTR. For each CpG, transcripts which contain HbA1c-assoc CpGs in any of the 6 regions were identified. CpGs located in multiple transcripts were randomly assigned to one transcript. Those CpGs not annotated to any gene were considered to be in intergenic regions. R package “GenomicRange” v.1.30.1 was used for annotation. IPA of HbA1c-assoc CpGs was performed on all the unique genes annotated to contain these CpGs using IPA © 2000–2019 (QIAGEN Inc., MD, https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis) to identify enriched biological pathways or networks using Ingenuity Knowledge Base.
Functional exploration of HbA1c-assoc CpGs in blood cells
Genome-wide chromatin states representing various functions related to gene transcription were defined by the NIH roadmap Epigenetics Program as reference epigenomes based on Chromatin-immunoprecipitation-sequencing (ChIP-seq) profiles of representative histone modification marks across 111 human tissues and cells types39. Fifteen chromatin states were defined based on 5 “core” chromatin marks: histone H3 lysine4 trimethylation (H3K4me3) enriched in promoters; H3 lysine-4 monomethylation (H3K4me1) in enhancers; H3K36 trimethylation (H3K36me3) in transcribed regions; H3K27me3 in polycomb repressed regions; and H3K9me3 in heterochromatin regions. The states consist of active TSS (TssA), proximal promoter (TssAFlnk), transcription at the 5′ and 3′ end of genes (TxFlnk), actively transcribed (Tx, TxWk), enhancer (Enh, EnhG), associated with zinc finger protein genes (ZNF/Rpts), constitutive heterochromatin (Het), bivalent regulatory (TssBiv, BivFlnk, EnhBiv), repressed Polycomb (ReprPC, ReprPCWk), and quiescent (Quies) features39. Detailed information is provided in the website https://egg2.wustl.edu/roadmap/web_portal/chr_state_learning.html#core_15state. Chromatin states in each of the major peripheral blood cells (monocyte, neutrophil, B-cell, T-cell and NK cells) and HSCs at CpGs of interest were retrieved. For each state, its enrichment at our HbA1c-assoc CpGs among all the 815432 CpGs was analyzed using one-tailed Fisher’s exact test. Eighteen chromatin states (https://egg2.wustl.edu/roadmap/web_portal/chr_state_learning.html#core_18state for details) were defined by the same NIH Consortium in various blood cell types using the original 5 core histone modifications marks plus a 6th one, H3K27 acetylation (H3K27Ac), for active enhancer regions. The 18 states include 4 TSS-related (TssA, TssFlnk, TssFlnkU and TssFlnkD), 2 transcription-related (Tx and TxWk), 5 enhancer-related (EnhG1, EnhG2, EnhA1, EnhA2 and EnhWk) and 7 others (same as in 15 states). The enrichment of each of the 18 states was similarly analyzed39 in monocytes, B-cell, T-cell and NK cells. Neutrophils and HSCs (analyzed for 15 chromatin states) were not analyzed due to unavailability of H3K27Ac ChIP-seq data.
Enrichment of open chromatin (DNA accessibility) at HbA1c-assoc CpGs was analyzed using DNase 1 Hypersensitive Sites defined by ENCODE using the online tool eFORGE v.2.0 (https://eforge.altiusinstitute.org/) with default settings. Detailed cell type description for each of the 22 blood samples was obtained from https://genome.ucsc.edu/encode/cellTypes.html. For each blood cell type, binomial tests were used to test enrichment of HbA1c-assoc CpGs located in open chromatin regions, compared to 1,000 matching background CpG sets each containing same number of CpGs with matching gene annotation and CpG island.
De novo motif analysis was performed using 79 neg-assoc CpGs and their ±250bp flanking regions. Genomic regions for CpGs located within ± 500bp were merged. The sequence of the resulting 74 genomic regions with 501 to 790 bp width were used as input in Regulatory Sequence Analysis Tools (RSAT Metazoa, http://rsat.sb-roscoff.fr/peak-motifs_form.cgi) to detect enriched motifs with hexanucleotides (k = 6) and heptanucleotides (k = 7) using default settings. Background sequences were generated using 50k randomly-selected CpGs and their ±250kb flanking regions followed by merging overlapped sequences, which resulted in a total of 46,187 sequences with 501 bp to 1626 bp width. Top 10 enriched motifs were identified based on binomial tests comparing input vs. background sequences. The detected motifs with E value (Bonferroni-adjusted p across all the identified motifs) < 0.2 were then searched against JASPAR core non-redundant vertebrates 2018 to identify any known transcription binding sites at normalized Pearson correlation coefficients <0.60.
Genomic interactions in primary peripheral blood cells were defined by the IHEC using PCHiC data42. Interactions with CHiCAGO score > 5 in at least one of the major blood cells (neutrophils, monocytes, total B cells, total CD4+ T cells and CD8+ T cells) were retrieved. Interactions with either bait regions (promoter) or PIRs containing CpGs of interests (e.g. HbA1c-assoc CpGs) were identified. The annotation of baits or PIR regions was based on the annotations of genes provided in the original study42.
Analyses of DNAme-associated gene expression at HbA1c-assoc CpGs
Methylomes profiled by Illumina HumanMethylation450 BeadChip and transcriptomes profiled by Illumina HumanHT-12 v4 Expression BeadChip on monocyte samples from the same 1202 human subjects were previously published44. The participants were randomly selected from 6,814 subjects from the Multi-Ethnic Study of Atherosclerosis (MESA) study of subclinical cardiovascular disease. The purified monocyte samples were obtained from randomly selected MESA participants (55–94-year-old, Caucasian (47%), African American (21%) and Hispanic (32%); female (51%)) from four MESA field centers. Normalized DNAme data (GEO accession# GSE56046) and gene expression data (GSE56045) with related phenotype data were downloaded to examine the association of DNAme at each HbA1c-assoc CpG with the expression of genes located nearby in monocytes. In addition, DNAme (GSE56581) and gene expression data (GSE56580) using the same platforms were also obtained in 214 CD4+ T cell samples randomly selected from participants with purified monocytes, to study the association between DNAme and gene expression in CD4+ T cells. To identify genes whose expressions is associated with HbA1c-assoc CpGs, all nearby RefSeq genes (Hg19, downloaded from UCSC genome browser) for each HbA1c-assoc CpG were first identified if the TSS was located within the ±500kb flanking region relative to CpGs. The association of DNAme at HbA1c-assoc CpGs with the expression of the corresponding nearby genes was analyzed one by one using multiple linear regression models with gene expression as the dependent variable and DNAme of CpG as the independent variable. The covariates adjusted in our model were age, race, gender, site, beadchip ID, beadchip position, and other cell content, same as that used in the published study44. FDR of the associations between CpGs and the nearby genes were estimated using all the association p values between DNAme and gene expression (two-sided tests based on t-statistic in multiple linear regression model) by adjust.p function across all pairs of CpGs and nearby genes analyzed. FDR < 5% was used to select DNAme-associated genes for HbA1c-associated CpGs. Among these, the most significant expression-associated gene for each CpG was then identified based on association p-value among all its nearby genes.
Identification of meQTLs of HbA1c-assoc CpGs
Genotyping data from 474 European DCCT subjects45 was used in the analyses. Illumina HumanCoreExome BeadArrays (Illumina, San Diego, CA, USA) were used to generated the data and un-genotyped autosomal SNPs were imputed using the 1000 Genomes project data (phase 3, v5)45. GWAS was performed on each of the 8 CpGs associated with mean-DCCT HbA1c at Bonferroni-adjusted p < 0.05 (p < 5e-08) (namely cg19693031, cg06721411, cg04816311, cg26262157, cg20983494, cg19285358, cg04568295 and cg08309687, Table 1) after removing 3 CpGs which had the same dependent association signals as cg19693031 (Supplementary Table 29). SNPs with minor allele frequency (MAF) >0.01 and high imputation quality (R2 > 0.5) were included in the analysis. SNPs (dosage) were tested for association with methylation levels (beta values) using linear regression under an additive genetic model adjusted for age, sex, batch and 5 predicted blood cell proportions (monocyte, B-cell, CD4+, CD8+, and NK cell) which were included as covariates in the model. For each CpG tested, SNPs whose genotypes were associated with its DNAme at p < 5e-08 were identified as meQTLs.
Analyses of the connections between DNAme at HbA1c-assoc CpGs and the risk of future development of diabetic complications
During the DCCT and EDIC study, the development of multiple complications of diabetes was carefully monitored in each participant3,5. Specifically, retinopathy was assessed by standardized 7-field stereoscopic fundus photographs obtained every 6 months during DCCT and in one quarter of the cohort each year during EDIC. Photographs were centrally graded with standardized methods using the Early Treatment Diabetic Retinopathy Study (ETDRS) scale. SNPDR was defined as any ETDRS score ≥10, PDR was defined as any ETDRS score ≥12 or receipt of scatter laser treatment during DCCT/EDIC, and CSME was defined as any score ≥20 or receipt of focal laser or anti-VEGF treatment. eGFRs were calculated from serum creatinine measured annually during DCCT/EDIC. AERs were measured annually during DCCT and in alternate years during EDIC. DKDs were defined as any impaired eGFR (<60 mL/min/1.73m2) on ≥2 consecutive visits during DCCT/EDIC (GFR60), or macro-albuminuria (any AER ≥300 mg/24 during DCCT/EDIC, AER300). The follow-up period for retinopathy is through EDIC year 18 (interval censored data due to staggered clinic visit intervals, up to EDIC year 23) and, for DKD up to EDIC year 18. In addition, based on eGFR data collected annually from EDIC baseline to EDIC year 18, the decline in kidney function was estimated for each participant by determining the coefficients of eGFR slopes using linear regression models. The estimate slopes coefficients were then rank-based inverse-normal transformed and used in subsequent analyses to avoid bias introduced by abnormality of the raw slope data.
Mediation analyses were performed in four steps to evaluate the role of DNAme in mediating the association between HbA1c and the risk of development of each complication in participants not having the complication during DCCT, as shown schematically in Extended Data 10. First (step 1), the association of mean-DCCT HbA1c and complications was analyzed using CoxPH regression models (function provided by R package “survival” v2.41–3) with fixed and time-dependent covariates. A multiple linear regression model was used for the normalized eGFR slope. In the CoxPH models, fixed covariates included DCCT baseline HbA1c, age, sex, diabetes duration at DCCT entry, time period from DCCT entry to blood collection, 850K array processing time, cohort assignment (PRIM or SCND), and blood sample cell composition. To adjust for baseline renal function, the log-transformed AER at EDIC baseline was added as a fixed covariate to the AER300 model, and eGFR at EDIC baseline to the GFR60 model. Log-transformed annual HbA1c values during EDIC (up to the end of follow-up or complication development) were included in each CoxPH model as a time-dependent covariate. For retinopathy, interval censored data through EDIC year 18 were used. Since AER measurements were taken every 2 years during EDIC, models for AER300 were stratified by collection year (odd vs. even). The covariates included in the linear model for eGFR slope were the same as the fixed covariates used for GFR60 plus mean HbA1c from EDIC entry to EDIC year 18. Second (step 2), for each complication, we used a similar model as the first step, except the mean-DCCT HbA1c was replaced with DNAme in each model to identify HbA1c-assoc CpGs which are also associated with the risk of complication development. Third (step 3), both mean-DCCT HbA1c and DNAme were included in the model for each of the CpGs associated with both HbA1c and the complication. The percentage of statistic score change (z-score in CoxPH models and t-score in linear regression) on mean-DCCT HbA1c from step 3 versus step 1 was then calculated to obtain the percentage of DNAme that explains the association between HbA1c and complications (defined from step 1). The example data (step 1 to step 3) on mediation analysis at CpG levels is presented in Supplementary Table 24. Finally (step 4), similar mediation analyses were applied to test all possible combinations of the top 10 HbA1c-assoc CpGs that were also associated with disease (ordered by association between DNAme and complications). Only top 10 HBA1c-assoc CpGs were considered to capture the major mediation effect while maintaining reasonable computational complexity (which increases dramatically with increase in the number of CpGs included using this Brute-force approach). Specifically, for each CpG combination, all the CpGs were included in the model with mean-DCCT HbA1c and other covariates (as done in step 3) and the explanation percentage was calculated using statistic scores obtained from the model of the step 4 versus step 1 as done in step 3. The best CpG combinations that explained the association between HbA1c and risk of development of a specific complication was identified as the one with highest explanation percentage across all combinations.
MR analysis was performed to investigate the causal effect of methylation levels at CpGs on diabetic complications including DKDs and retinopathies. Analyses were restricted to the 4 top HbA1c-assoc CpGs which had either cis- or trans- meQTLs. Since the SNPs associated with methylation levels at each of these CpGs were in linkage disequilibrium (LD), MR was only performed for the SNP with lowest p-value for each CpG. Summary statistics for the association of the selected SNPs with DKD including GFR60 and AER300 determined by meta-analyses on 19,406 individuals of European descent with type 1 diabetes, and retinopathy (PDR vs. non-PDR/NR and PDR vs. NR) by meta-analyses using eight European cohorts (n = 3,246) were obtained from the largest available meta-GWAS16,25. For the SNPs showing at least nominal association with nephropathy or retinopathy, two-sample MR using Wald ratio test implemented in R package TwoSampleMR version 0.4.2269 was applied for each CpG.
In-vitro experiments with CD34+ cells
Human primary bone marrow CD34+ cells (1M-101C, Lonza, Switzerland,) were cultured in growth medium (#09605, STEMCELL Technologies, MA), STEMSPAN II plus StemSpan™ CD34+ Expansion Supplement (#02691, STEMCELL Technologies, MA) containing 25mmol/L glucose (control), or same medium with the addition of 20mmol/L glucose for 72 hours (HG treatment). Cells were collected for preparation of genomic DNA and total RNA.
Data visualization
All plots, unless otherwise mentioned, were generated in R v3.4.3 (https://cran.r-project.org). Specifically, heatmap.2 function in package “gplots” v3.0.1 was used to generate heatmaps. Manhattan plots were generated using Manhattan function in package “qqman” v0.1.4. Bubble plots were generated using ggplot function in package “ggplot2” v3.2.1. Circular plots were generated using package “circlize” v0.4.3. Ribbon plots were generated using package “Sushi” v.1.16.0. Logos for transcription factor motifs were generated based on position-specific scoring matrices of each motif using package “seqLogo” v1.44.0. Density plots, dot plots and bar plots were generated using functions provided by the package “graphics” v3.4.3.
Statistical Analysis
Detailed information about the Statistical methods/models used for each of the assays reported in the manuscript are provided under the respective sections, including Methods, Results, Legends and Supplement.
Extended Data
Extended Data Fig. 1. Sensitivity analyses of additional clinical variables on the association significance between mean-DCCT HbA1c and DNAme at the HbA1c-assoc CpGs.
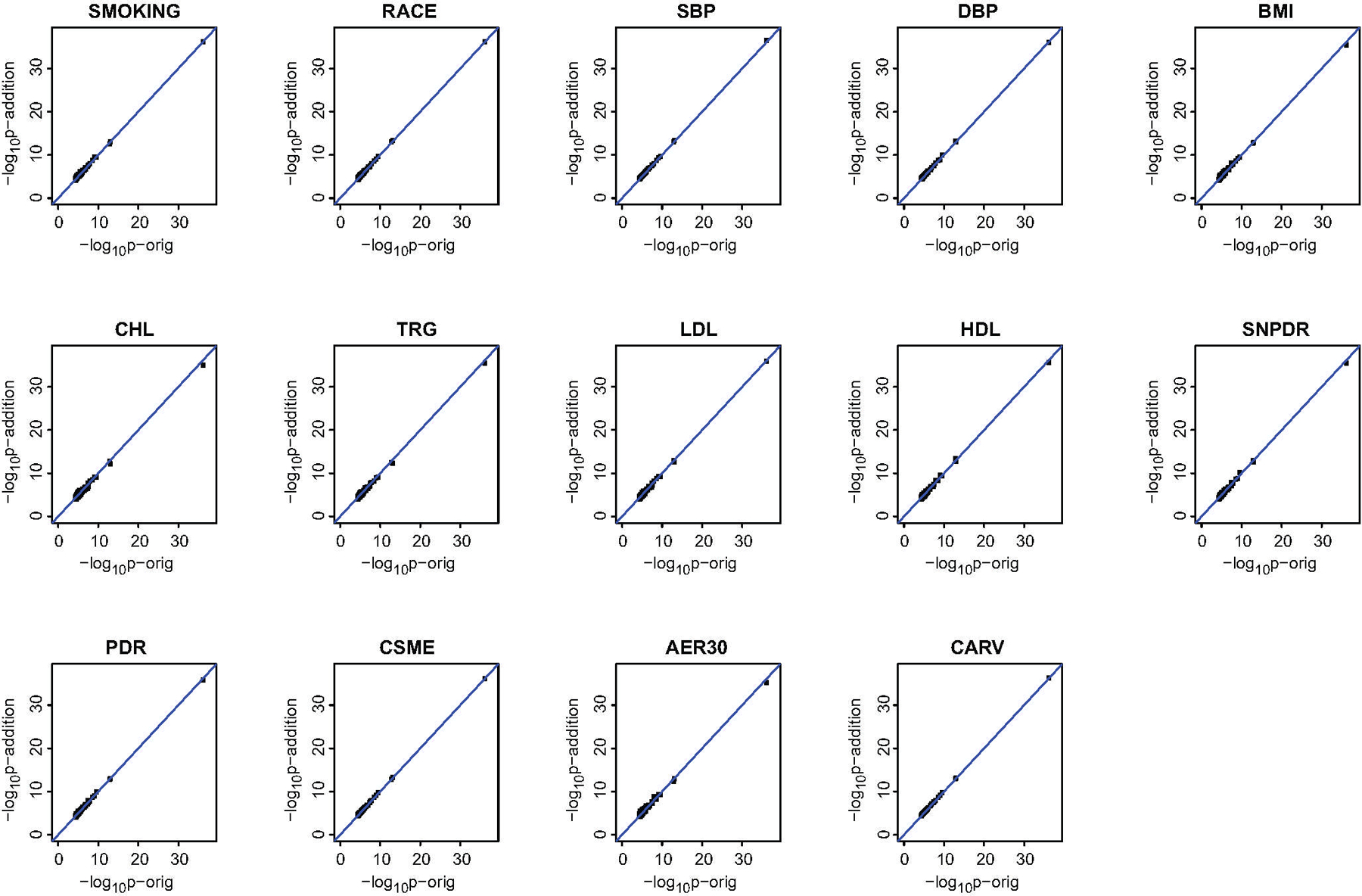
For each HbA1c-assoc CpG, one additional clinical variable (indicated on top of each plot) was added as covariate in the multiple linear regression model used for the identification of HbA1c-assoc CpGs using all the samples (two-sided tests based on t-statistic, n=499). The resulting association significance in –log10p (y-axis) was compared with that obtained from the model without the specific variable (x-axis) and shown as one dot in the scatter plots. The blue line represents all the dots with same significance values. Abbreviations: SBP- systolic blood pressure; DBP-diastolic blood pressure; BMI-body mass index; CHL-cholesterol; TRG-triglyceride; LDL-low density lipoprotein; HDL-high density lipoprotein; PDR- proliferative diabetic retinopathy; SNPDR-severe nonproliferative diabetic retinopathy; CSME- clinically significant macular edema; AER30-AER > 30mg/24h; CARV-cardiovascular disease.
Extended Data Fig. 2. Sensitivity analyses of additional clinical variables on the percentage of association coefficients changes between mean-DCCT HbA1c and DNAme at the HbA1c-assoc CpGs.
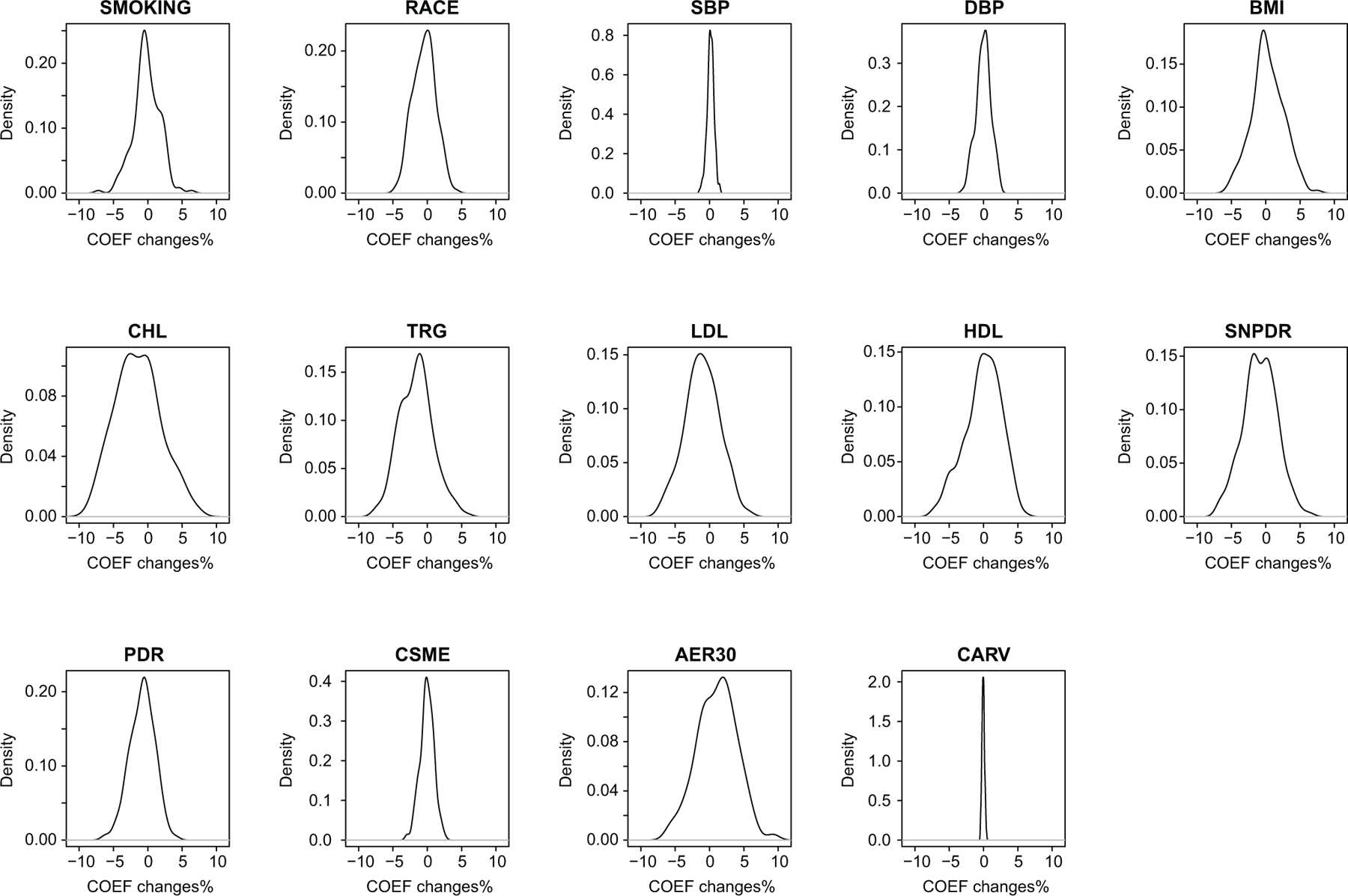
For each HbA1c-assoc CpG, one additional clinical variable (indicated on top of each plot) was added as covariate in the multiple linear regression model with two-sided tests based on t-statistic used for identification of HbA1c-assoc CpGs using all the samples (n=499). For each clinical variable, the percentage change of the resulting association coefficients versus the coefficients estimated using the original model without the specific variable was calculated. For each additional variable, the distribution of the calculated changes was plotted using density plots. Abbreviations: COEF-coefficient; SBP- systolic blood pressure; DBP-diastolic blood pressure; BMI-body mass index; CHL-cholesterol; TRG-triglyceride; LDL-low density lipoprotein; HDL-high density lipoprotein; PDR- proliferative diabetic retinopathy; SNPDR-Severe nonproliferative diabetic retinopathy; CSME- clinically significant macular edema; AER30-AER > 30mg/24h; CARV-cardiovascular disease.
Extended Data Fig. 3. Validation of HbA1c-assoc CpGs identified in DCCT499 in a meta-analysis of DCCT499 and Joslin195.
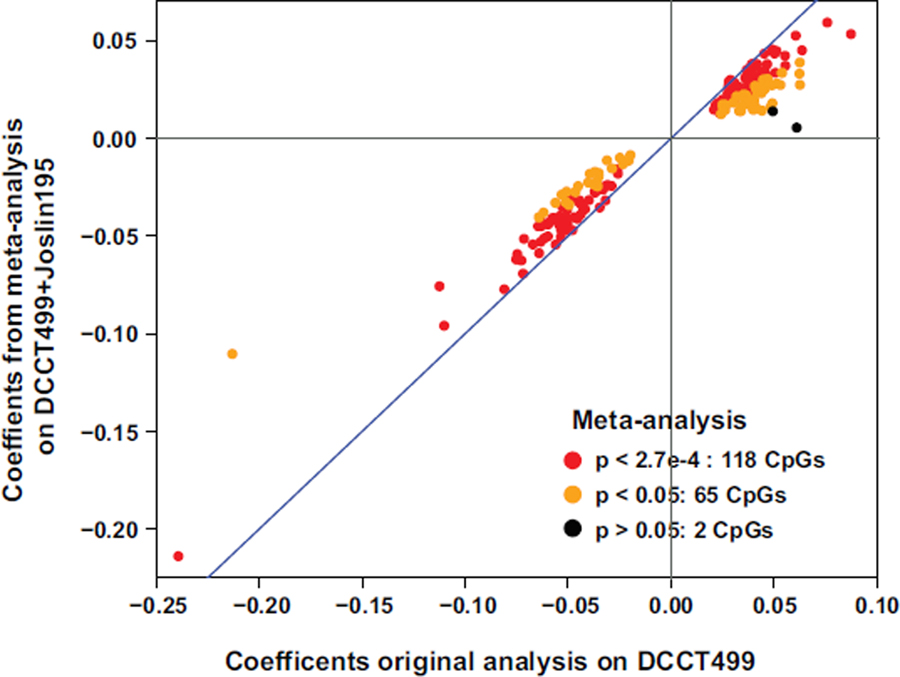
Fixed meta-analysis was performed using Bacon R package based on coefficients and standard error of CpGs obtained in both datasets (n=499 in DCCT499 and n=195 in Joslin195). Scatter plot is shown to compare the association estimates generated from meta-analysis and original models across 185 HbA1c-assoc CpGs, after excluding one HbA1c-assoc CpG not covered reliably in Joslin195 dataset due to detection p > 0.05 in at least one sample. Each dot represents one CpG with color indicating the significance levels obtained from meta-analysis. Red dots represent Bonferroni-adjusted p <0.05 (cutoff p=0.05/185=2.7e-4), orange dots represent nominal p < 0.05, black dots represent remaining CpGs. Blue line represents all the dots with same association estimates obtained from meta-analysis and original model.
Extended Data Fig. 4. Genomic locations and IPA analyses of HbA1c-assoc genes.
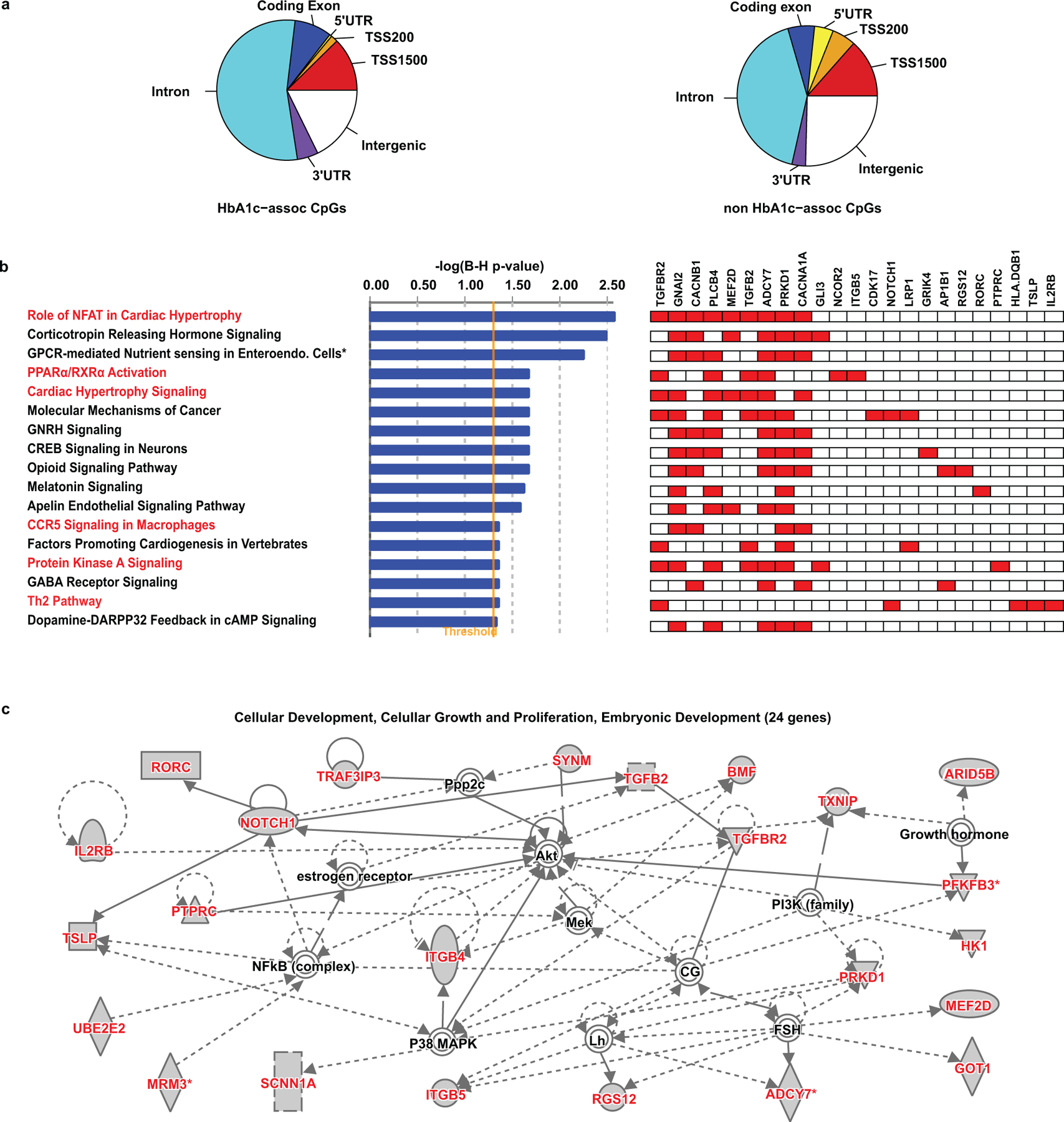
a, Genomic locations of HbA1c-assoc CpGs and non-associated CpGs relative to Refseq genes depicted using pie charts. TSS, transcription start site; TSS1500, distal promoter region from 1500bp upstream of TSS to TSS; TSS200, proximal promoter from 200bp upstream to 1500bp upstream of TSS; 5’UTR, 5’ untranslated region; 3’UTR, 3’ untranslated region; intergenic, genomic regions excluding TSS1500, TSS200 and gene body. b, Enriched canonical pathways (Benjamini-Hochberg adjusted P < 0.05) identified among annotated genes of HbA1c-assoc CpGs. Right-tailed Fisher’s exact test was used in IPA analysis. Total of 143 unique annotated genes were used as input for IPA, and 141 genes after excluding 2 that did not match in the IPA database were finally included in the analysis. Pathway names are listed on the left with corresponding significance level in –log10 form presented in the middle as bar plot. The pathways labeled in red font are known to be associated with diabetes and its complications. HbA1c-assoc genes identified in each pathway are shown on the right. Each row represents one pathway and each column represents one HbA1c-assoc gene found in the enriched pathway. Red indicates the corresponding specific gene (with name shown on the top of the column) that is involved in the pathway specified on the left of the panel. The majority of them contain several NFAT pathway genes, including TGFBR2, GNAI2, CACNB1, PLCB4, MEF2D, TGFB2, ADCY7, PRKD1 and CACNA1A. c, Top network related to cellular growth, proliferation and embryonic development. The 24 HbA1c-assoc genes labeled in red font are those identified to depict interactions with several proteins related to diabetes, its complications and insulin sensitivity including nuclear factor kappaB (NF-kB), protein kinaseB (PKB), phosphoinositide 3-kinase (PI3K), p38 mitogen-activated protein kinase (P38MAPK) and protein phosphatase2 catalytic (PPP2C) subunits.
Extended Data Fig. 5. Enrichment of 15 chromatin states at 186 HbA1c-assoc CpGs compared to all the non-HbA1c-assoc CpGs.
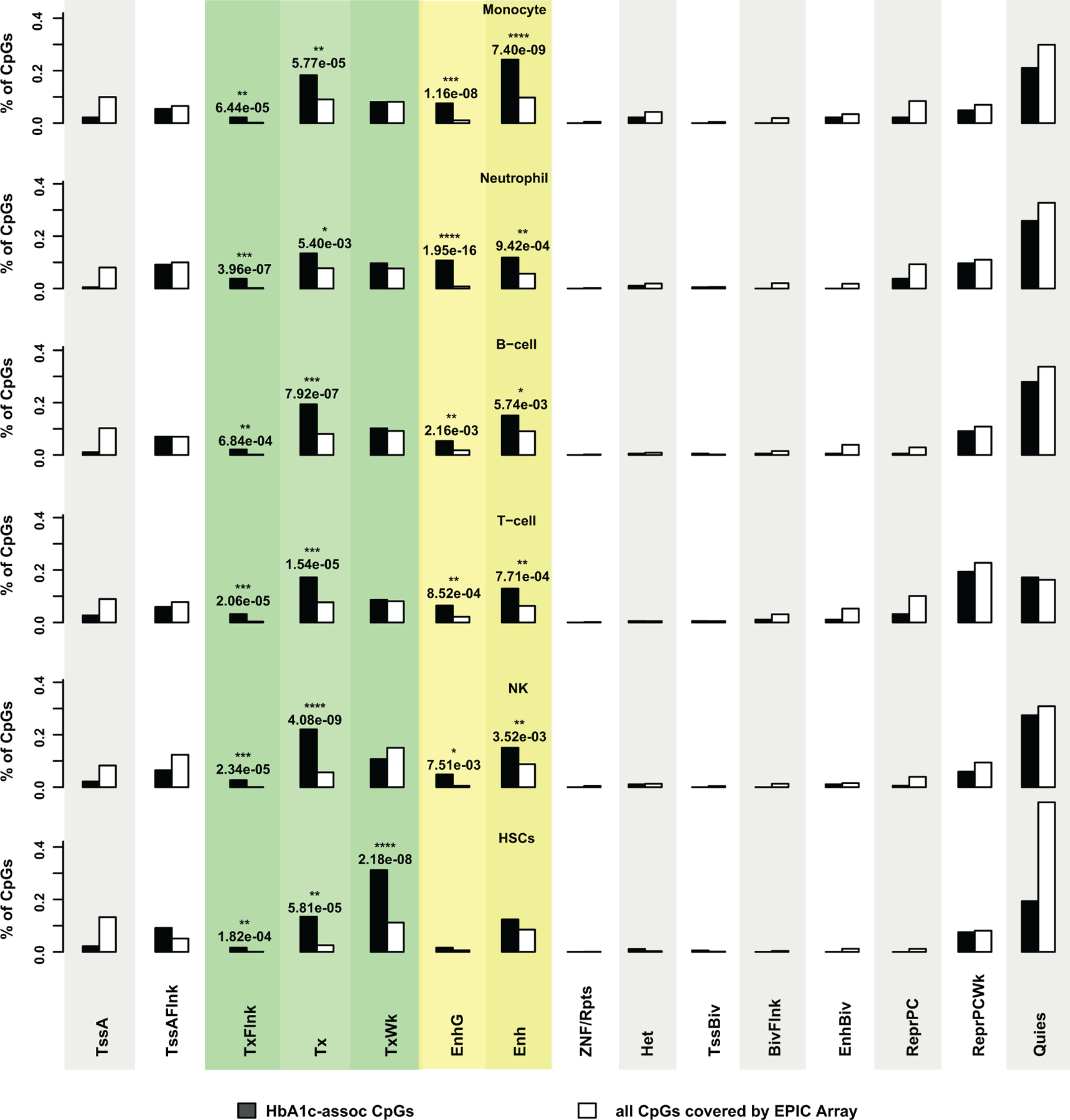
Fifteen genome-wide chromatin states were defined in 5 major blood cell types and hematopoietic stem cell (HSC) by the NIH roadmap Epigenetics Program (https://egg2.wustl.edu/roadmap/web_portal/chr_state_learning.html#core_15state for details). For each cell type, the percentage of CpGs located in the genomic regions depicting each chromatin state among HbA1c-assoc CpGs (black bars) versus all CpGs covered by the EPIC array (named as “all CpGs”, white bars) are shown side by side using bar plots. The enrichment significances of HbA1c-assoc CpGs versus the other CpGs were tested by right-tailed Fisher’s exact tests on 186 HbA1c-assoc CpGs vs. all the 815,246 non HbA1c-assoc CpGs reliably covered by the EPIC array in each specific chromatin state. **** p < 5e-08; *** p < 5e-05; ** p < 5e-03 and * p < 5e-02. States with statistically significant enrichment in at least one cell type are highlighted with colored background/shading: green for transcription-related (TxFlnk, Tx and TxWk) and yellow for enhancer-related (EnhG or Enh). The other states are shown in alternating grey and white shades for better visualization. Abbreviations: HSC-hematopoietic stem cell; TSS-transcription start site; TssA-Active TSS; TssAFlnk-Flanking Active TSS; TxFlnk-Transcribed state at gene 5’ and 3’; Tx-Strong transcription; TxWk-Weak transcription; EnhG-Genic enhancers; Enh-Enhancers; ZNF/Rpts- ZNF genes & repeats;Het- Heterochromatin; TssBiv- Bivalent/Poised TSS; BivFlnk- Flanking Bivalent TSS/Enh; EnhBiv- Bivalent Enhancer; ReprPC- Repressed PolyComb; ReprPCWk- Weak Repressed PolyComb; Quies-Quiescent/Low.
Extended Data Fig. 6. Enrichment of chromatin (epigenetic) states at HbA1c-assoc CpG sites in blood cells.
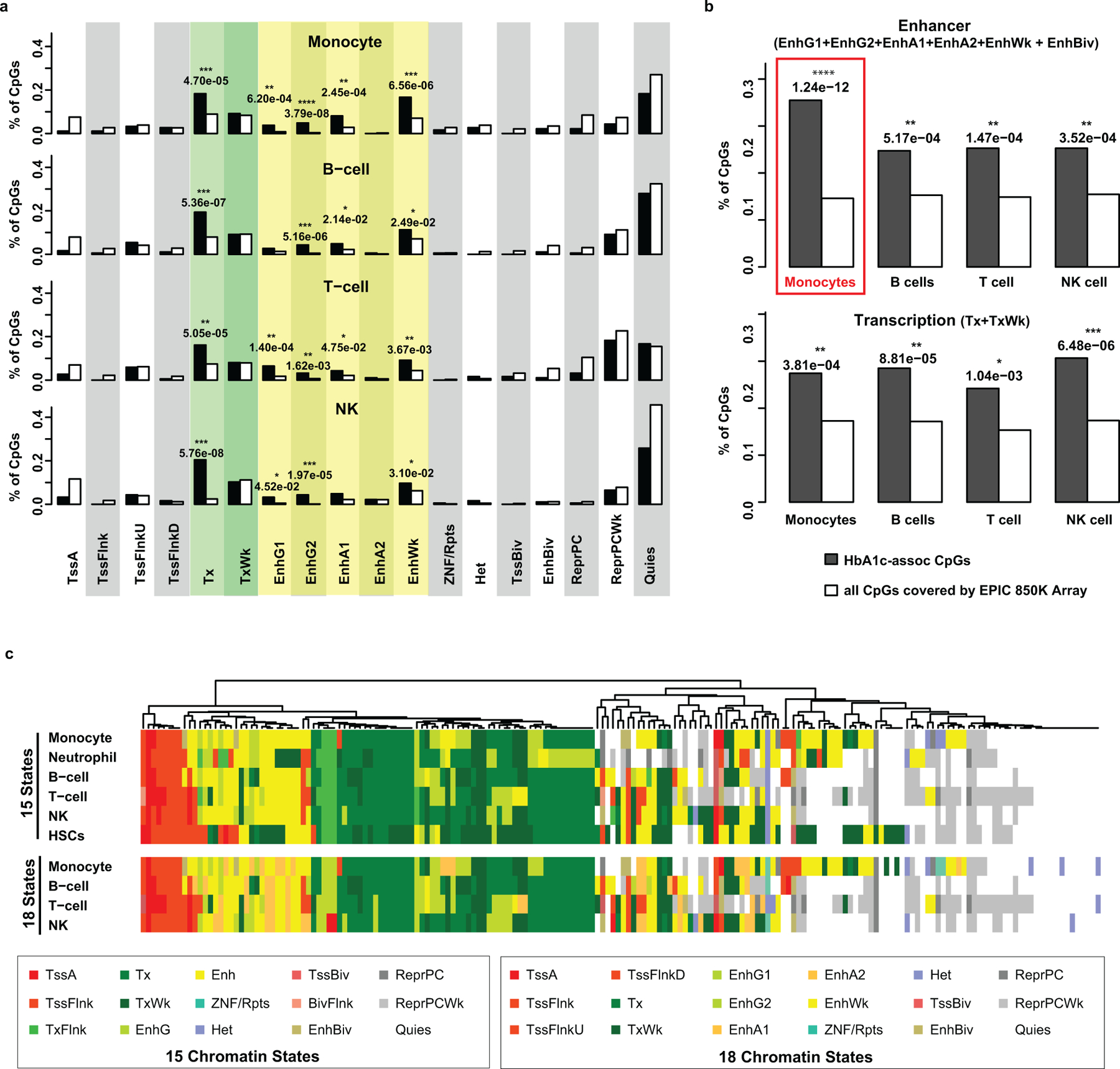
a, Enrichment of chromatin states in each cell-type at the 186 HbA1c-assoc CpGs. For each cell-type, the percentage of CpGs located in genomic regions enriched with 18 chromatin states (labeled under the plots) at HbA1c-assoc CpGs (dark bars) versus all CpGs covered by the EPIC array (white bars) in 4 major blood cell-types are shown side-by-side in the bar plots. Right-tailed Fisher’s exact tests were conducted to identify the significance of enrichment of each state in 186 HbA1c-assoc CpGs relative to all the 815,246 non HbA1c-assoc CpGs reliably covered by EPIC array. States with significant enrichment in at least one cell-type are highlighted with colored background/shading: green for transcription-related (Tx and TxWk) and yellow for enhancer-related states (EnhG1, EnhG2, EnhA1, EnhA2 or EnhWk). P values listed on top of each significantly enriched state.**** p< 5e-08; *** p< 5e-05; ** p< 5e-03 and * p< 5e-02. The other states are shown in alternating grey and white shades for better visualization. b, Enrichment of enhancer- or transcription-related states across 4 different cell-types in HbA1c-assoc CpGs versus all CpGs. The states for each plot are shown in the heading. P-values were determined using same tests and sample sizes as in panel a. c, Comparison of heatmaps of 15 chromatin states in 6 blood cell-types (top) with heatmaps of 18 states at 4 blood cell-types (bottom) at HbA1c-assoc CpGs. These states are defined by the NIH Roadmap Epigenetics Program. Unsupervised hierarchical analysis was performed on data from 15 states and visualized in the top panel using colors depicted in the boxed legend for 15 states (bottom left). 18-chromatin states at HbA1c-assoc CpGs, presented in the same order as 15 states, are shown in the lower panel with corresponding color legends in the bottom right box. Each row represents one cell-type and each column, one CpG.
Extended Data Fig. 7. Ribbon plots for the association between DNAme at candidate loci (HbA1c-assoc CpGs) and the expression of genes located within 500kb (FDR < 0.05) that were observed in both monocytes and CD4+ cells.
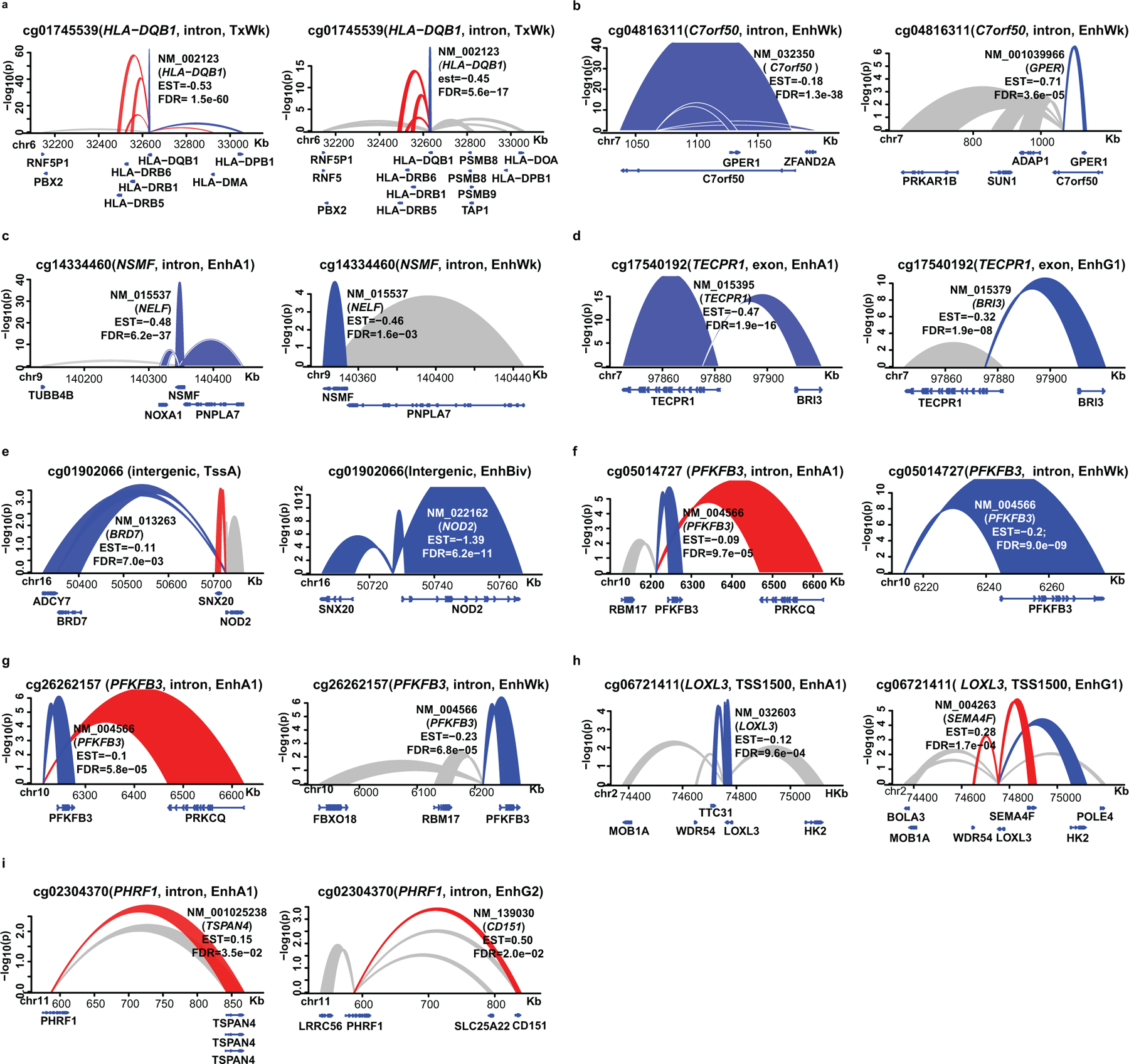
The associations of DNAme at HbA1c-assoc CpGs with the expression of gene (s) in monocytes or CD4+ T cells were analyzed on each pair of HbA1c-assoc CpGs and corresponding nearby genes within 500kb. Multiple linear regression models using two-sided tests based on t-statistic (n=1202 for monocytes and n=214 for CD4+ T cells) were applied to published datasets containing both DNAme and gene expression profiles from the same samples to identify HbA1c-assoc CpGs whose DNAme is associated with the expression of nearby gene (s) (expression-associated genes) with FDR < 0.05 in monocytes and CD4+ cells. The associations for each CpG in both monocytes (left) and CD4+ cells (right) are shown side-by-side within each panel. The order of CpGs is based on the significance level of the CpG with its most significantly expression-associated gene in both cell types. The height of the ribbon represents the significance level in –log10(p). Blue indicates negative association while red indicates positive association. Grey represents associations with nominal p < 0.05 but FDR >0.05. Most of the 9 common CpGs are in enhancers, suggesting a more ubiquitous regulatory role for DNAme at enhancers across different blood cell types.
Extended Data Fig. 8. Ribbon plots for the association between DNAme at candidate loci (HbA1c-assoc CpGs) and expression of genes located within 500bp (FDR < 0.05) that were observed only in monocytes (not in CD4+ cells).
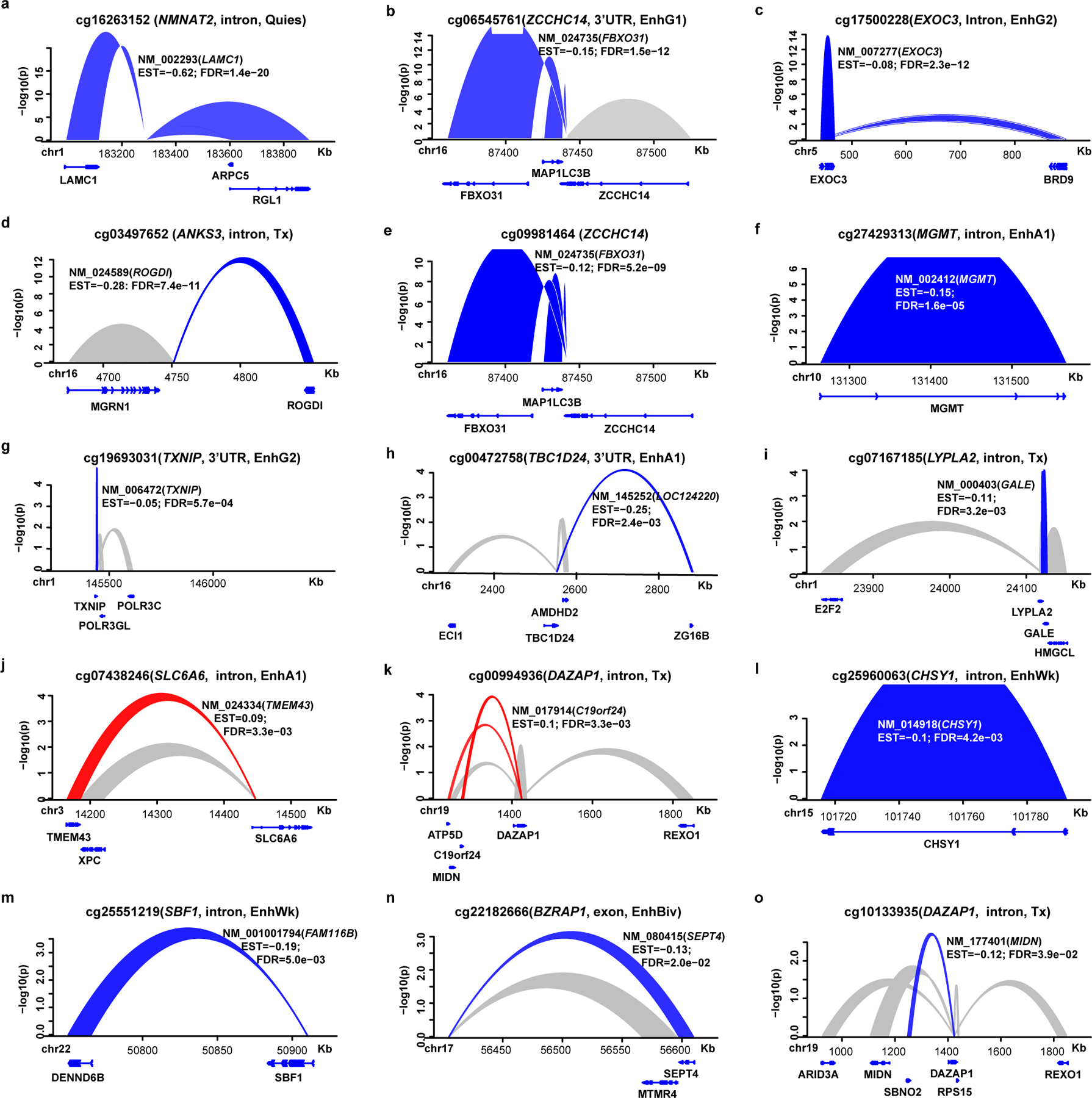
The associations of DNAme at HbA1c-assoc CpGs with the expression of gene (s) in monocytes were analyzed on each pair of HbA1c-assoc CpGs and corresponding nearby genes within 500kb. Multiple linear regression models using two-sided tests based on t-statistic were applied to published datasets containing both DNAme and gene expression profiles from same monocyte samples (n=1202) to identify HbA1c-assoc CpGs whose DNAme is associated with the expression of nearby gene (s) (expression-associated genes) with FDR < 0.05. The order of CpGs is based on the significance level of the CpG with its most significantly expression-associated gene. The height of the ribbon represents the significance level in –log10(p). Blue indicates negative association while red indicates positive association. Grey represents the associations with nominal p < 0.05 but FDR >0.05.
Extended Data Fig. 9. Ribbon plots for the association between DNAme at candidate loci (HbA1c-assoc CpGs) and expression of genes located within 500bp (FDR < 0.05) that were observed only in CD4+ cells (not in monocytes).
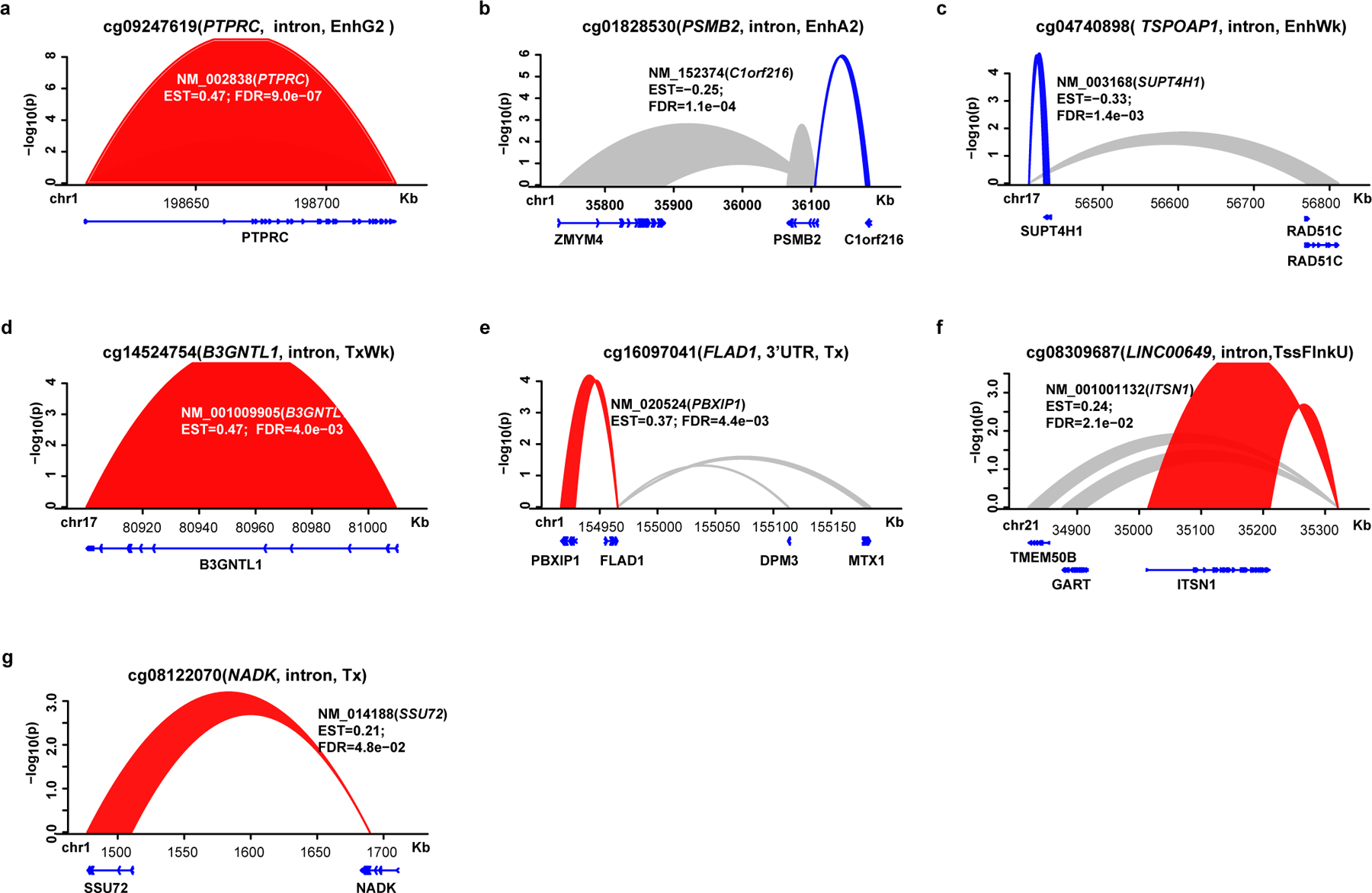
The associations of DNAme at HbA1c-assoc CpGs with the expression of gene(s) in CD4+ T cells were analyzed on each pair of HbA1c-assoc CpGs and corresponding nearby genes within 500kb. Multiple linear regression models using two-sided tests based on t-statistic were applied to published datasets containing both DNAme and gene expression profiles from same CD4+ T cell samples (n=214) to identify HbA1c-assoc CpGs whose DNAme is associated with the expression of nearby gene(s) (expression-associated genes) with FDR < 0.05. The order of the CpGs is based on significance level of the CpG with its most significantly associated gene (expression data). The height of the ribbon represents the significance level in –log10(p). Blue indicates negative association while red indicates positive association. Grey represents the associations with nominal p < 0.05 but FDR >0.05.
Extended Data Fig. 10. Pipeline depicting the step-wise sequence used for selecting CpGs for mediation analyses of DNAme in between mean-DCCT HbA1c and complication development during EDIC.
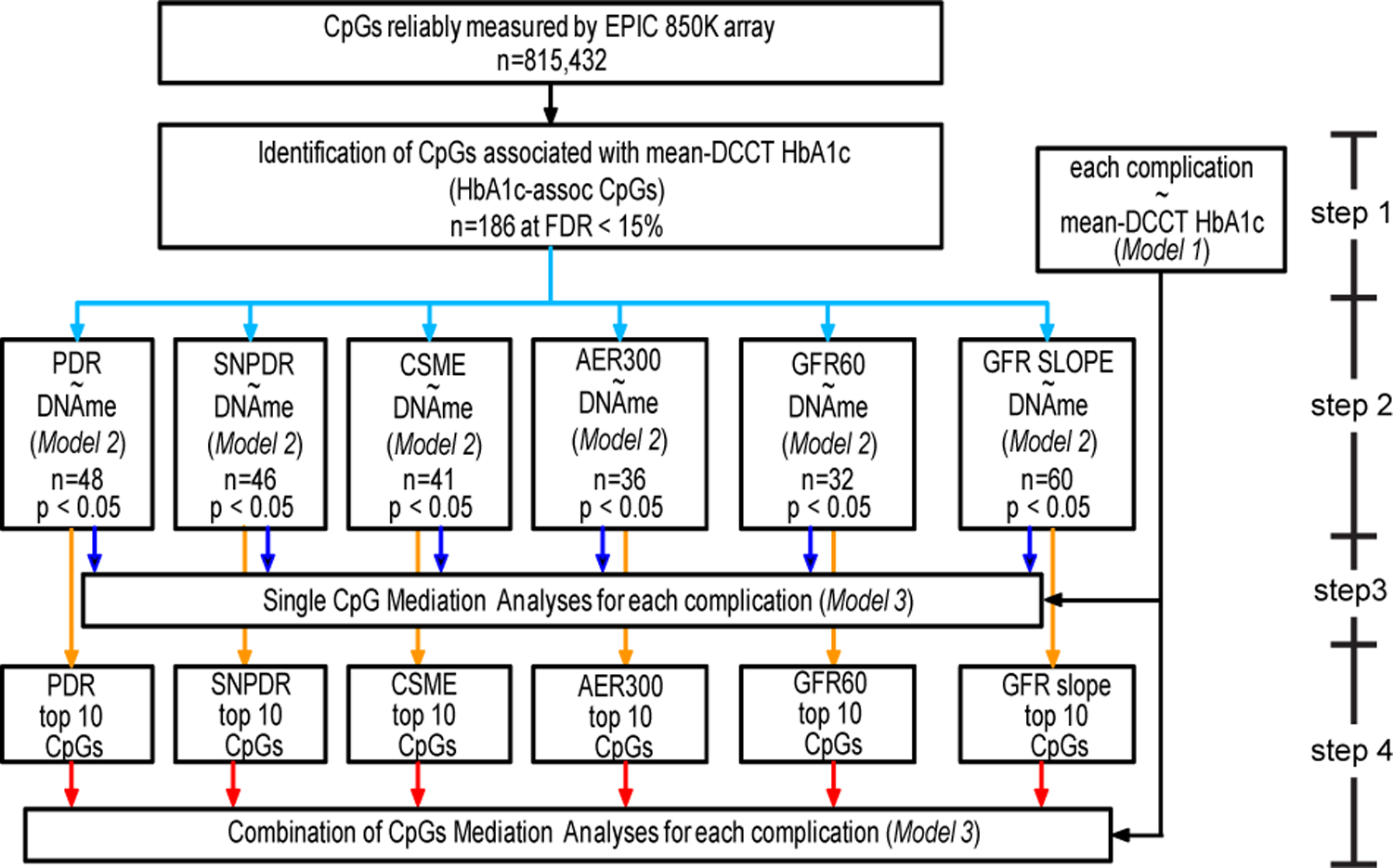
At each selection step, the number of CpGs (n) is shown at the specified cut-off criteria. Arrows with same color reflect the same analytical step using the same model for each complication. The 4 steps as described in the Methods are specified on the right side of the figure. “n” represents number of CpGs identified at each step. In step 1, Model 1 was applied to each complication to analyze the association of mean-DCCT HbA1c with risk of complication development during EDIC (up to EDIC year 18). In step 2, Model 2 was applied to each complication to identify the HbA1c-assoc CpGs whose DNAme is associated with risk of complication development. In step 3, Model 3 was applied to each complication using similar model as Model 1 and Model 2, in which both DNAme and mean-DCCT HbA1c were now included. To identify the best combinations of CpGs to explain the association between HbA1c and complication development in step 4, only the top 10 CpGs (based on the association significance between risk of complication and DNAme identified using model 2 in step2) were considered in order to capture the major mediation effect, while maintaining reasonable computational complexity (which increases dramatically as the number of CpGs included increases using Brute-force approach).
Supplementary Material
ACKNOWLEDGMENTS
A complete list of participants in the DCCT DCCT/EDIC Study Group is presented at the end of the Supplementary Information File. We are deeply grateful to Dr. Andrzej Krolewski (Joslin Diabetes Center, Harvard Medical School, Boston, MA) for generously providing DNA samples and related clinical information from his Joslin Kidney Study cohort of T1D subjects. We are also grateful to the DCCT/EDIC Central Biochemistry Laboratory at the University of Minnesota for assistance in providing the archived DNA samples.
Funding: This study was supported by grants from the National Institutes of Health (NIH): DP3 DK106917-01 and R01 DK065073 (to R.N), and the Wanek family project at the City of Hope (to R.N, ADR and JDT). Research reported in this publication included work performed in the following Campus Cores: Integrative Genomics, and DNA/RNA synthesis (supported by the National Cancer Institute of the NIH under award number P30CA33572), and the Genomics Core at the University of S. California (Dr. Dan Weisenberger, Director). The DCCT/EDIC has been supported by cooperative agreement grants (1982–1993, 2012–2017, 2017–2022), and contracts (1982–2012) with the Division of Diabetes Endocrinology and Metabolic Diseases of the National Institute of Diabetes and Digestive and Kidney Disease (current grant numbers U01 DK094176 and U01 DK094157), and through support by the National Eye Institute, the National Institute of Neurologic Disorders and Stroke, the General Clinical Research Centers Program (1993–2007), and Clinical Translational Science Center Program (2006-present), Bethesda, Maryland, USA.
Industry contributors have had no role in the DCCT/EDIC study but have provided free or discounted supplies or equipment to support participants’ adherence to the study: Abbott Diabetes Care (Alameda, CA), Animas (Westchester, PA), Bayer Diabetes Care (North America Headquarters, Tarrytown, NY), Becton Dickinson (Franklin Lakes, NJ), Eli Lilly (Indianapolis, IN), Extend Nutrition (St. Louis, MO), Insulet Corporation (Bedford, MA), Lifescan (Milpitas, CA), Medtronic Diabetes (Minneapolis, MN), Nipro Home Diagnostics (Ft. Lauderdale, FL), Nova Diabetes Care (Billerica, MA), Omron (Shelton, CT), Perrigo Diabetes Care (Allegan, MI), Roche Diabetes Care (Indianapolis, IN), and Sanofi-Aventis (Bridgewater, NJ).
Footnotes
Reporting Summary
Further information on the research design is available in the Nature Research Reporting Summary linked to this article.
Trial Registration: clinicaltrials.gov NCT00360815 and NCT00360893.
Consortium
DCCT/EDIC Study Group
Barbara H Braffet2, John M Lachin2
DCCT/EDIC Collaborators
Zhuo Chen1, Feng Miao1, Lingxiao Zhang1, Rama Natarajan1, Andrew D Paterson4
Competing interests: All the authors declare that there are no competing interests associated with this manuscript.
Additional Information:
Extended Data Figures, and Supplementary Information (Figures and Tables) accompany this manuscript.
Data Availability
The DNA methylation dataset of 499 DCCT participants from this study has been deposited in the Database of Genotype and Phenotype (dbGaP) (https://www.ncbi.nlm.nih.gov/gap) under the accession # phs002024.v1.p1. Other data that support the findings from this study are available from the corresponding author upon reasonable request.
References:
- 1.Nathan DM et al. The effect of intensive treatment of diabetes on the development and progression of long-term complications in insulin-dependent diabetes mellitus. N Engl J Med 329, 977–986, doi: 10.1056/NEJM199309303291401 (1993). [DOI] [PubMed] [Google Scholar]
- 2.Nathan DM The Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications study at 30 years: overview. Diabetes Care 37, 9–16, doi: 10.2337/dc13-2112 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lachin JM et al. Effect of intensive diabetes therapy on the progression of diabetic retinopathy in patients with type 1 diabetes: 18 years of follow-up in the DCCT/EDIC. Diabetes 64, 631–642, doi: 10.2337/db14-0930 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Boer IH et al. Intensive diabetes therapy and glomerular filtration rate in type 1 diabetes. N Engl J Med 365, 2366–2376, doi: 10.1056/NEJMoa1111732 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DCCT/EDIC research group. Effect of intensive diabetes treatment on albuminuria in type 1 diabetes: long-term follow-up of the Diabetes Control and Complications Trial and Epidemiology of Diabetes Interventions and Complications study. Lancet Diabetes Endocrinol 2, 793–800, doi: 10.1016/S2213-8587(14)70155-X (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kato M & Natarajan R Epigenetics and epigenomics in diabetic kidney disease and metabolic memory. Nat Rev Nephrol, doi: 10.1038/s41581-019-0135-6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reddy MA, Zhang E & Natarajan R Epigenetic mechanisms in diabetic complications and metabolic memory. Diabetologia 58, 443–455, doi: 10.1007/s00125-014-3462-y (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooper ME & El-Osta A Epigenetics: mechanisms and implications for diabetic complications. Circ Res 107, 1403–1413, doi: 10.1161/CIRCRESAHA.110.223552 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Bird A Perceptions of epigenetics. Nature 447, 396–398, doi: 10.1038/nature05913 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Russo VEA, Martienssen RA & Riggs AD Epigenetic mechanisms of gene regulation. (Cold Spring Harbor Laboratory Press, 1996). [Google Scholar]
- 11.Jirtle RL & Skinner MK Environmental epigenomics and disease susceptibility. Nat Rev Genet 8, 253–262, doi: 10.1038/nrg2045 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simmons R Epigenetics and maternal nutrition: nature v. nurture. Proc Nutr Soc 70, 73–81, doi: 10.1017/S0029665110003988 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Susztak K Understanding the epigenetic syntax for the genetic alphabet in the kidney. J Am Soc Nephrol 25, 10–17, doi: 10.1681/ASN.2013050461 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosen ED et al. Epigenetics and Epigenomics: Implications for Diabetes and Obesity. Diabetes 67, 1923–1931, doi: 10.2337/db18-0537 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sandholm N et al. The Genetic Landscape of Renal Complications in Type 1 Diabetes. J Am Soc Nephrol 28, 557–574, doi: 10.1681/ASN.2016020231 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pollack S et al. Multiethnic Genome-Wide Association Study of Diabetic Retinopathy Using Liability Threshold Modeling of Duration of Diabetes and Glycemic Control. Diabetes 68, 441–456, doi: 10.2337/db18-0567 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hosseini SM et al. The association of previously reported polymorphisms for microvascular complications in a meta-analysis of diabetic retinopathy. Hum Genet 134, 247–257, doi: 10.1007/s00439-014-1517-2 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ko YA et al. Cytosine methylation changes in enhancer regions of core pro-fibrotic genes characterize kidney fibrosis development. Genome Biol 14, R108, doi: 10.1186/gb-2013-14-10-r108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wing MR et al. DNA methylation profile associated with rapid decline in kidney function: findings from the CRIC study. Nephrol Dial Transplant 29, 864–872, doi: 10.1093/ndt/gft537 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chu AY et al. Epigenome-wide association studies identify DNA methylation associated with kidney function. Nat Commun 8, 1286, doi: 10.1038/s41467-017-01297-7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qiu C et al. Cytosine methylation predicts renal function decline in American Indians. Kidney Int 93, 1417–1431, doi: 10.1016/j.kint.2018.01.036 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miao F et al. Evaluating the role of epigenetic histone modifications in the metabolic memory of type 1 diabetes. Diabetes 63, 1748–1762, doi: 10.2337/db13-1251 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen G et al. Aberrant DNA methylation of mTOR pathway genes promotes inflammatory activation of immune cells in diabetic kidney disease. Kidney Int, doi: 10.1016/j.kint.2019.02.020 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Gluck C et al. Kidney cytosine methylation changes improve renal function decline estimation in patients with diabetic kidney disease. Nat Commun 10, 2461, doi: 10.1038/s41467-019-10378-8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salem RM et al. Genome-Wide Association Study of Diabetic Kidney Disease Highlights Biology Involved in Glomerular Basement Membrane Collagen. J Am Soc Nephrol 30, 2000–2016, doi: 10.1681/ASN.2019030218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Z et al. Epigenomic profiling reveals an association between persistence of DNA methylation and metabolic memory in the DCCT/EDIC type 1 diabetes cohort. Proc Natl Acad Sci U S A 113, E3002–3011, doi: 10.1073/pnas.1603712113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shalev A Minireview: Thioredoxin-interacting protein: regulation and function in the pancreatic beta-cell. Mol Endocrinol 28, 1211–1220, doi: 10.1210/me.2014-1095 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Marinis Y et al. Epigenetic regulation of the thioredoxin-interacting protein (TXNIP) gene by hyperglycemia in kidney. Kidney Int 89, 342–353, doi: 10.1016/j.kint.2015.12.018 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Kumar A & Mittal R Mapping Txnip: Key connexions in progression of diabetic nephropathy. Pharmacol Rep 70, 614–622, doi: 10.1016/j.pharep.2017.12.008 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Singh LP Thioredoxin Interacting Protein (TXNIP) and Pathogenesis of Diabetic Retinopathy. J Clin Exp Ophthalmol 4, doi: 10.4172/2155-9570.1000287 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rich SS et al. A genome-wide association scan for acute insulin response to glucose in Hispanic-Americans: the Insulin Resistance Atherosclerosis Family Study (IRAS FS). Diabetologia 52, 1326–1333, doi: 10.1007/s00125-009-1373-0 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neve B et al. Role of transcription factor KLF11 and its diabetes-associated gene variants in pancreatic beta cell function. Proc Natl Acad Sci U S A 102, 4807–4812, doi: 10.1073/pnas.0409177102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fernandez-Zapico ME et al. MODY7 gene, KLF11, is a novel p300-dependent regulator of Pdx-1 (MODY4) transcription in pancreatic islet beta cells. J Biol Chem 284, 36482–36490, doi: 10.1074/jbc.M109.028852 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park IY et al. Dual Chromatin and Cytoskeletal Remodeling by SETD2. Cell 166, 950–962, doi: 10.1016/j.cell.2016.07.005 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kulkarni H et al. Novel epigenetic determinants of type 2 diabetes in Mexican-American families. Hum Mol Genet 24, 5330–5344, doi: 10.1093/hmg/ddv232 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamanouchi M et al. Improved clinical trial enrollment criterion to identify patients with diabetes at risk of end-stage renal disease. Kidney Int 92, 258–266, doi: 10.1016/j.kint.2017.02.010 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toperoff G et al. Genome-wide survey reveals predisposing diabetes type 2-related DNA methylation variations in human peripheral blood. Hum Mol Genet 21, 371–383, doi: 10.1093/hmg/ddr472 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones PA Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13, 484–492, doi: 10.1038/nrg3230 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Kundaje A et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330, doi: 10.1038/nature14248 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thurman RE et al. The accessible chromatin landscape of the human genome. Nature 489, 75–82, doi: 10.1038/nature11232 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Avellino R & Delwel R Expression and regulation of C/EBPalpha in normal myelopoiesis and in malignant transformation. Blood 129, 2083–2091, doi: 10.1182/blood-2016-09-687822 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Javierre BM et al. Lineage-Specific Genome Architecture Links Enhancers and Non-coding Disease Variants to Target Gene Promoters. Cell 167, 1369–1384 e1319, doi: 10.1016/j.cell.2016.09.037 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rickels R & Shilatifard A Enhancer Logic and Mechanics in Development and Disease. Trends Cell Biol 28, 608–630, doi: 10.1016/j.tcb.2018.04.003 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Reynolds LM et al. Age-related variations in the methylome associated with gene expression in human monocytes and T cells. Nat Commun 5, 5366, doi: 10.1038/ncomms6366 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roshandel D et al. Meta-genome-wide association studies identify a locus on chromosome 1 and multiple variants in the MHC region for serum C-peptide in type 1 diabetes. Diabetologia 61, 1098–1111, doi: 10.1007/s00125-018-4555-9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paterson AD et al. A genome-wide association study identifies a novel major locus for glycemic control in type 1 diabetes, as measured by both A1C and glucose. Diabetes 59, 539–549, doi: 10.2337/db09-0653 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hainsworth DP et al. Risk Factors for Retinopathy in Type 1 Diabetes: The DCCT/EDIC Study. Diabetes Care 42, 875–882, doi: 10.2337/dc18-2308 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perkins BA et al. Risk Factors for Kidney Disease in Type 1 Diabetes. Diabetes Care 42, 883–890, doi: 10.2337/dc18-2062 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kriebel J et al. Association between DNA Methylation in Whole Blood and Measures of Glucose Metabolism: KORA F4 Study. PLoS One 11, e0152314, doi: 10.1371/journal.pone.0152314 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ronn T et al. Impact of age, BMI and HbA1c levels on the genome-wide DNA methylation and mRNA expression patterns in human adipose tissue and identification of epigenetic biomarkers in blood. Hum Mol Genet 24, 3792–3813, doi: 10.1093/hmg/ddv124 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Hidalgo B et al. Epigenome-wide association study of fasting measures of glucose, insulin, and HOMA-IR in the Genetics of Lipid Lowering Drugs and Diet Network study. Diabetes 63, 801–807, doi: 10.2337/db13-1100 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walaszczyk E et al. DNA methylation markers associated with type 2 diabetes, fasting glucose and HbA1c levels: a systematic review and replication in a case-control sample of the Lifelines study. Diabetologia 61, 354–368, doi: 10.1007/s00125-017-4497-7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chambers JC et al. Epigenome-wide association of DNA methylation markers in peripheral blood from Indian Asians and Europeans with incident type 2 diabetes: a nested case-control study. Lancet Diabetes Endocrinol 3, 526–534, doi: 10.1016/S2213-8587(15)00127-8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soriano-Tarraga C et al. Epigenome-wide association study identifies TXNIP gene associated with type 2 diabetes mellitus and sustained hyperglycemia. Hum Mol Genet 25, 609–619, doi: 10.1093/hmg/ddv493 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Cardona A et al. Epigenome-Wide Association Study of Incident Type 2 Diabetes in a British Population: EPIC-Norfolk Study. Diabetes 68, 2315–2326, doi: 10.2337/db18-0290 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ye J et al. Identification of loci where DNA methylation potentially mediates genetic risk of type 1 diabetes. J Autoimmun 93, 66–75, doi: 10.1016/j.jaut.2018.06.005 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Vigorelli V et al. Abnormal DNA Methylation Induced by Hyperglycemia Reduces CXCR 4 Gene Expression in CD 34(+) Stem Cells. J Am Heart Assoc 8, e010012, doi: 10.1161/JAHA.118.010012 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsukada J, Yoshida Y, Kominato Y & Auron PE The CCAAT/enhancer (C/EBP) family of basic-leucine zipper (bZIP) transcription factors is a multifaceted highly-regulated system for gene regulation. Cytokine 54, 6–19, doi: 10.1016/j.cyto.2010.12.019 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Nagareddy PR et al. Hyperglycemia promotes myelopoiesis and impairs the resolution of atherosclerosis. Cell Metab 17, 695–708, doi: 10.1016/j.cmet.2013.04.001 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mitroulis I et al. Modulation of Myelopoiesis Progenitors Is an Integral Component of Trained Immunity. Cell 172, 147–161 e112, doi: 10.1016/j.cell.2017.11.034 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Woroniecka KI et al. Transcriptome analysis of human diabetic kidney disease. Diabetes 60, 2354–2369, doi: 10.2337/db10-1181 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kojima H, Kim J & Chan L Emerging roles of hematopoietic cells in the pathobiology of diabetic complications. Trends Endocrinol Metab 25, 178–187, doi: 10.1016/j.tem.2014.01.002 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.The Diabetes Control and Complications Trial (DCCT). Design and methodologic considerations for the feasibility phase. The DCCT Research Group. Diabetes 35, 530–545 (1986). [PubMed] [Google Scholar]
- 64.Fortin JP, Triche TJ Jr. & Hansen KD Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics 33, 558–560, doi: 10.1093/bioinformatics/btw691 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aryee MJ et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30, 1363–1369, doi: 10.1093/bioinformatics/btu049 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Houseman EA et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13, 86, doi: 10.1186/1471-2105-13-86 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krueger F & Andrews SR Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572, doi: 10.1093/bioinformatics/btr167 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Niewczas MA et al. A signature of circulating inflammatory proteins and development of end-stage renal disease in diabetes. Nat Med 25, 805–813, doi: 10.1038/s41591-019-0415-5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hemani G et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 7, doi: 10.7554/eLife.34408 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The DNA methylation dataset of 499 DCCT participants from this study has been deposited in the Database of Genotype and Phenotype (dbGaP) (https://www.ncbi.nlm.nih.gov/gap) under the accession # phs002024.v1.p1. Other data that support the findings from this study are available from the corresponding author upon reasonable request.


