Abstract

Posttranslational modifications of proteins like citrullination and carbamylation are associated with several diseases. Detailed analytical characterization of citrullinated and carbamylated proteins or peptides could be difficult due to the low concentration of the analytes in complex biological samples. High structural similarity and chemical behavior of citrullinated and carbamylated residues also pose a challenge. We previously reported the “citrulline effect” phenomenon that is manifested in the generation of intense y type ions originating from Cit-Zzz amide bond scissions in collision-induced dissociation tandem mass spectra of citrullinated tryptic peptides. In this study, we created a rigorous tryptic-like model system of both citrulline and homocitrulline-containing peptides that included appropriate and well-defined controls and fragment analogues to quantify the citrulline effect and investigate whether there is an effect for homocitrulline residues as well. Our results show that citrulline residues significantly increased fragmentation at their C-terminus relatively independent of the identity of the following amino acid. In comparison, homocitrulline residues displayed inconclusive results at the same energies. However, the strength of effects was dependent on collision energy and the position of citrulline and homocitrulline in the sequences. As newer software algorithms tend to observe structure–intensity relationships during annotation, this finding increases reliable identification of modified proteins/peptides.
Introduction
Citrullination is an enzymatic posttranslational modification (PTM) of arginine residues leading to citrullines (Cit, X) in proteins.1 Homocitrulline (Hci, B) residues are formed through the nonenzymatic reaction of isocyanic acid with the amino group of lysine residues or with protein N-termini through a process known as carbamylation.2 Both citrullination and carbamylation result in a decreased basicity and thus an altered charge state, activity, or structure compared to the original species. Citrullination plays a role in several physiological functions, including epigenetic regulation3 and skin homeostasis,4 while hypercitrullination is associated with cancer metastasis5 and autoimmune6 and neurodegenerative diseases.7 Carbamylation is normally related to protein aging8,9 and is involved in chronic kidney disease;2 however, it could also be a sample preparation artifact.10,11
Currently—apart from immunodetection techniques, which are unable to determine the exact modification sites—these PTMs could only be characterized at the molecular level using tandem mass spectrometric methods coupled with ultrahigh performance liquid chromatography (UHPLC–MS/MS). Complex samples containing both native and citrullinated peptides may be separated less efficiently using liquid chromatographic methods due to the substoichometric nature of citrullination and the small molecular mass difference between modified and unmodified species. Hyphenation of more charge and structure/ion radius-sensitive techniques like capillary electrophoresis12 and ion mobility spectrometry (IMS)13 with mass spectrometry may be advantageous to overcome these obstacles.
Collision-induced dissociation (CID) and higher energy collision-induced dissociation (HCD)14 are reliable methods for both the identification of citrullinated or carbamylated peptides via the selective loss of isocyanic acid from the side chains and sequencing of the peptides.15 However, determination of modification sites could be sometimes difficult due to differing cleavage preferences of the amide bonds resulting in incomplete fragment ions series.16−19 On the other hand, these phenomena can also yield valuable structural information. Complementary electron-based fragmentation strategies20 are also commonly used, especially in the sequencing of larger sized or highly basic peptides and proteins in MS/MS. However, PTM-specific neutral losses that can be used for identification21 may be, in these cases, of low abundance.22 Another promising technique may be the use of vacuum ultraviolet photodissociation (VUVPD) to randomize the occurrence of amide bond cleavages through the whole sequence.23
We previously reported a neutral loss which was selective to citrulline residues for histone-related pentapeptides in electron-transfer dissociation (ETD).24 In addition, electron-transfer higher energy collision-induced dissociation (EThcD) was proven to be beneficial for both increasing fragmentation efficiency and the incidence of neutral losses.24 We also found that the citrulline effect, which is the enhanced cleavage probability C-terminal to citrulline residues in (higher energy) collision-induced dissociation (HCD, CID), was preserved in EThcD as well. Although several research groups18,25 examined the amide bond scission preferences of tryptic peptides at statistical levels, insufficient data are currently available regarding the effect of special amino acids including citrulline or homocitrulline.
Therefore, our aim was to characterize these effects under well-controlled circumstances, especially concentrating on—but not restricted to—the amide bonds at the C-terminus of citrulline and homocitrulline residues.
We chose the sequences AAXZAAK and AABZAAK, where X and B stand for citrulline or homocitrulline, respectively, and Z denotes any other amino acid, including some special ones like carbamidomethyl cysteine (C*) and oxidized methionine (M′) that are frequently occurring in MS-based experiments. Carbamidomethylation of Cys residues are generally carried out by iodoacetamide; however, this may negatively affect identification rates in proteomics.26 Very recently, Wiśniewski et al. reported a method involving only the reduction of Cys residues without alkylation that resulted in an improved peptide and protein identification.27 Therefore, we included both Cys and carbamidomethylated Cys residues in our experiments.
According to Hao et al., citrulline residues are converted to ornithines (Orn, O) using collision-induced dissociation.28 Lee et al. reported that carbamyllysine (Hci) residues are also prone to loss of isocyanic acid upon CID leading to lysine (Lys, K) residues.15 McGee and McLuckey observed that Orn strongly promotes amide bond cleavage at its C-terminus.29 They also proposed a neighboring group reaction mechanism in which the Orn forms a six-membered lactam. We also described a facile cleavage C-terminal to Cit, which could be explained by a loss of isocyanic acid followed by a lactam formation (Scheme 1).30 Thus, we also synthesized peptides where X or Z are substituted for ornithines (O) or lysines (K) to compare the MS/MS behavior of these residues to that of Cit and Hci.
Scheme 1. Literature-Based Two-Step Mechanism for the CID MS/MS Fragmentation of Cit- or Hci-Containing Peptides28−30,
Steps: (1) Conversion of Cit or Hci to Orn or Lys, respectively, by the neutral loss of isocyanic acid. (2) Nucleophilic attack of the carbonyl carbon by the δ- or ε-amino group resulting in the formation of a six- or seven-membered lactam derivative and cleavage of the Cit-Zzz or Hci-Zzz amide bond.
Alanines are reported to have only a small effect on fragmentation;25 therefore, they were incorporated for elongating the peptides. This way, the peptides are relatively large to be successfully separated on C18 columns. The peptides were synthesized with a lysine C-terminus so that they model an enzymatic cleavage by trypsin or LysC, frequently used in bottom-up proteomic experiments.
We also synthesized peptides where X/B/O/K were replaced by alanine residues (AAAZAAK) as controls to examine whether the Z has a contribution in the cleavage increment or solely the X/B/O/K causes the phenomenon as several amino acids, e.g., Gly, Ser, Thr, Cys, Met, Asp, and Asn and especially Pro, have a preference to be cleaved at their N-termini.16,31,32 In addition, Leu, Ile, and Val residues also tend to produce intense peaks corresponding to the cleavage at their C-terminus.32 As Asp and Gly residues commonly occur at the +1 position to Cit residues under physiological conditions,15 alanine was selected as a control again due to its low preference to be cleaved either at its N- or C-terminus to substitute Cit residues. AAZXAAK and AAZBAAK and their above-mentioned control sequences were also designed to check whether the position of X/B has a considerable impact on fragmentation. AA(X/B)ZAAK series were prepared to study the cleavage preference of the third bond, while AAZ(X/B)AAK was prepared to study that of the fourth bond. The latter may offer useful information regarding the dependence of fragmentation characteristics on the position of X residues. Altogether, 225 model peptides were synthesized.
Materials and Methods
Synthesis
Model peptides with a free carboxyl terminus were synthesized using a Multipin Peptide Synthesis Kit (Chiron Technologies Pty Ltd., Clayton VIC, Australia) on macrocrowns (Mimotopes Pty Ltd., Mulgrave VIC, Australia; SynPhase L-series lantern, Wang linker, nominal loading: 15 μmol) with Fmoc/tBu strategy following the manufacturer’s instructions. Heptapeptides containing Cys, Met or Trp were cleaved from the lanterns using a mixture of 82.5% trifluoroacetic acid/5% phenol/5% water/5% thioanisole/2.5% ethanedithiol, all other peptides were cleaved by a mixture consisting of 95% trifluoroacetic acid/2.5% water/2.5% triisopropylsilane for 2 h at room temperature.
Freeze-dried peptide samples were diluted to 1 μM final concentration with water prior to LC–MS analysis. To produce carbamidomethylated cysteine residues, heptapeptides containing Cys were reduced by treatment with dithiothreitol (2 μL, 200 mM in water) for 30 min at 37 °C and alkylated with iodoacetamide (2.5 μL, 200 mM in water) for 30 min at room temperature.
UPLC–MS/MS Conditions
An 8 min long gradient elution was used for separation of the peptides on a Waters Acquity UPLC BEH C18 1.7 μm column (2.1 × 50 mm, Wexford, Ireland). The column temperature was set to 40 °C. Eluent A was composed of 0.1% formic acid, and eluent B was composed of 80% acetonitrile and 0.1% formic acid. The following gradient was used: 0 min 2% B, 1 min 2% B, 4 min 50% B, 4.1 min 90% B, 4.5 min 90% B, 4.6 min 2% B, 8 min 2% B. The flow rate was 300 μL min–1.
Tandem mass spectra were acquired on a Q-Exactive Focus Hybrid Quadrupole-Orbitrap instrument (Thermo Scientific, Bremen, Germany) using higher energy collision-induced dissociation (HCD). Data acquisition and processing were done with Thermo Scientific XCalibur 4.1. Positive mode was used, and a dd-MS2 (Discovery) method was applied. An isolation window of 2 m/z was employed for precursor selection. The same collision energies (10 and 15 eV) were used for the whole peptide set so that the results could be directly compared. The increase of collision energies was tested above 15 eV but was not found to be beneficial due to loss of information regarding fragments with higher m/z. The AGC target was set to the recommended 5e5 value for full MS and 1e5 for MS/MS. Resolution was set to 70000 for both full and tandem MS. The default charge state was 2, as in most proteomic applications these analytes produce the most fragment-rich spectra. Singly charged species are often excluded to enhance selectivity, and triply charged peptides were not present with a reasonable abundance.
Data Evaluation
Tandem mass spectra originating from doubly charged precursors were extracted from the chromatograms and visualized by mMass 5.5.0.33 Fragments that are usually generated by collision-induced dissociation (bi and yi ions)34 were automatically identified in the spectra using a 5 ppm mass accuracy limit. We calculated the portion of each amide bond cleavage as a relative percentage by the following formula similar to that of Kapp et al.25
where rel%s is the relative percentage of amide bond cleavage at the “sth” position, N is the total number of cleavage sites, k is the charge state of the fragment ion, K is the total number of charge states, and bik+ and yi are the absolute intensities of b- and y-type ions with k charges at the ith cleavage site.
Only these so-called backbone fragments were considered, and the frequently observed peaks corresponding to neutral losses were omitted. Isotope peaks and multiply charged fragments were also taken into account. Spectra were also examined manually to exclude ambiguities of annotation.
Statistical Analysis
As the sample and control data sets did not follow a normal distribution or sometimes had significantly differing variances, robust statistical tests that do not require the equality of population variances and less sensitive to the violation of normality were used to statistically evaluate our results. The equality of expected rank variances was tested by the Welch-type O’Brien35 and Levene tests.36 Robust testing of the hypothesis of stochastic equality was done with a Fligner–Policello test with Welch-like degrees of freedom,37 a Welch test on ranks, and a Brunner–Munzel test.38 In these cases, the confidence intervals for the difference of population means between the sample and control data sets could not be determined; therefore, the stochastic superiority is given instead as a measure of “strength” for the effects
where A12 is the stochastic superiority of group 1 (sample) to group 2 (control), P is the probability, X1 are the data from group 1, and X2 are the data from group 2. Both point and interval estimates were made for the value A12. As an example, an estimated Â12 ∼ 0.8 means that by randomly selecting an X1 and X2 value there is a probability of ∼80% that the X1 has a higher value. One could also use the stochastic difference to characterize the probability by which a randomly selected X1 value is higher than the X2 value. The stochastic difference can be obtained by a scale transformation from Â12:
By definition, δ̂12 can range between [−1;1] or can be given in percentage [−100%;100%]. It may be important to note that these δ̂12 values are only representing a probability of difference and not the strength of the difference between the two population. We used the software ROPstat for the statistical analyses.39
Results and Discussion
We synthesized 225 model peptides to quantify the effect of citrulline and homocitrulline residues on the tandem mass spectra of citrullinated and carbamylated peptides. We previously observed that Cit residues enhance the amide bond scission at their C-terminus in MS/MS.40 To the best of our knowledge, however, there are no data on Hci cleavage preferences. Therefore, in this work, we calculated all of the amide bond cleavage ratios of our new series of peptides containing Cit and Hci residues in the third and fourth position in the sequence and compared them to their variants containing alanines at these positions. We incorporated all the possible natural amino acids as well as oxidized methionine and carbamidomethylated cysteine directly preceding and following Cit, Orn, Hci, and Lys residues to assess the possible differences in cleavage preferences caused by the side chains.
Our results are in great concordance with our previous studies.24,41 As a demonstration of the citrulline effect, the comparison of the tandem mass spectrum of AAXHAAK and its control AAAHAAK can be seen in Figure 1. The intensity of y4-b3 ion corresponding to a cleavage at the Cit C-terminus is 19% higher than that of the control. It is especially interesting that Cit-His cleavage is more favored than that of His-Ala although His residues were described to prefer fragmentation at their C-terminus.32
Figure 1.
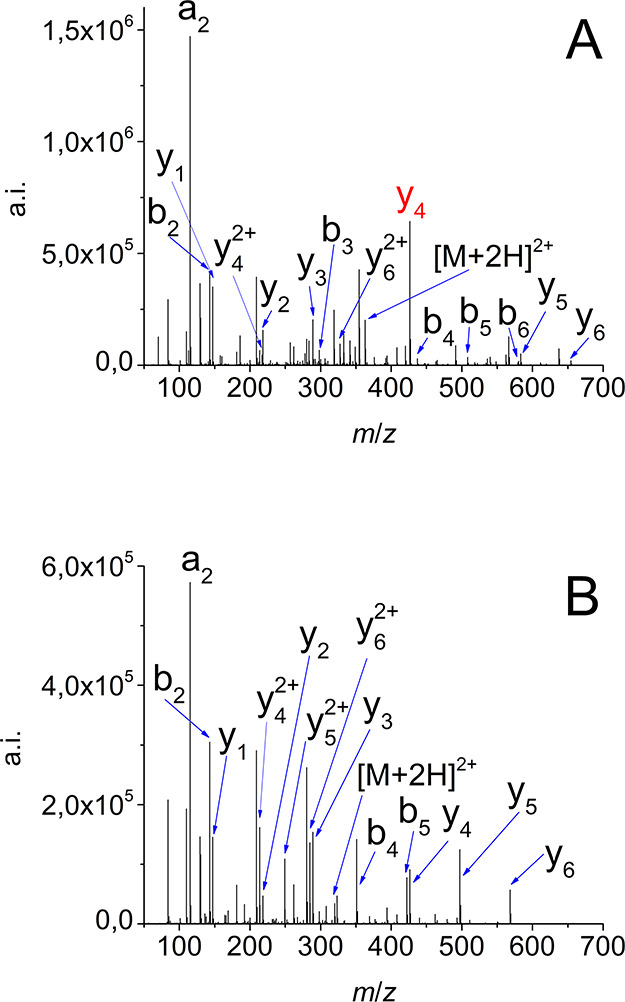
Citrulline effect as demonstrated by the MS/MS fragmentation of AAXHAAK (A) and AAAHAAK (B) model peptides at 15 eV.
Our results demonstrate that Cit residues in the third position substantially and statistically significantly increase the cleavage of the third amide bonds (δ̂12 = 87%) at 10 eV (Table 1). This effect has a very similar strength at 15 eV (δ̂12 = 86%) as it is also summarized in Figure 2A.
Table 1. Stochastic Difference (δ̂12) of Cleavage Products (%) C-Terminal to the Respective Amino Acid Compared to the Alanine-substituted Controla.
| citrulline | ornithine | homocitrulline | lysine | ||
|---|---|---|---|---|---|
| CE = 10 eV | Xxx in 3rd position | 87*** | 98*** | 82*** | 32+ |
| Xxx in 4th position | 42** | 99*** | –48*** | 10 | |
| CE = 15 eV | Xxx in 3rd position | 86*** | 94*** | 80*** | 58*** |
| Xxx in 4th position | 92*** | 100! | 33* | 71*** |
Notation: +, p < 0.10; *, p < 0.05; **, p < 0.01; ***, p < 0.001; !, p-value cannot be determined. Xxx stands for Cit, Orn, Hci or Lys.
Figure 2.

Effects of Cit, Orn, Hci, and Lys on the cleavage of the third amide bond at 15 eV.
Although the increase of cleavage preference of the fourth amide bonds at Cit residues is considerable for the majority of Z (δ̂12 = 92%) (Table 1, Figure 3A) and statistically significant at 15 eV, there are only minor differences at 10 eV (δ̂12 = 42%) compared to the controls. The Cit effect was found to be less strong for the fourth position which could be explained by secondary fragmentation processes that might only be activated at higher collision energies.
Figure 3.

Effects of Cit, Orn, Hci, and Lys on the cleavage of the fourth amide bond at 15 eV.
Similar tendencies could be observed for Hci residues as well (Figure 2C). The values of increase are somewhat lower than that of for Cit residues but not significantly different from Cit effect when the Hci was in the third position. However, the increase was not only absent but there was a slight decrease (δ̂12 = −48%) in cleavage compared to the controls at 10 eV for Hci residues in the fourth position (Figure S2). A minor increase was observed with acceptable statistical significance if the collision energy was set to 15 eV (δ̂12 = 33%), as shown in Figure 3C, but there were several connections where the control displayed higher rel% values.
We also evaluated the data for the peptides that contain an Orn or a Lys as a substitute for Cit and Hci, respectively. These peptides could be derived by a loss of isocyanic acid from the original sequences. Orn residues displayed a statistically significantly higher increase compared to Cit residues (Figure 2B). The Orn effect was also strong when the Orn was at the fourth position (Figure 3B). The decrease of amide bond scission N-terminal to Orn was also stronger than that of the other amino acids.
On the contrary, Lys residues at the third position only produced a slightly significant and very small increase of bond scission preference (δ̂12 = 32%) and not a significant one in the fourth position at 10 eV (δ̂12 = 10%) (Table 1). This may indicate that such a low level of collision energy was not enough to efficiently fragment Lys-containing peptides. Although previous studies implicate that Orn displays a very similar or even higher proton affinity than Lys despite the higher +I effect of the latter one, we found that while Orn residues readily fragmented even at 10 eV, Lys residues showed a lower level of fragmentation (compare Figure S1B with S1D). This may imply a higher proton affinity and, thus, a higher level of proton sequestration by Lys in accordance with both acid–base theoretical considerations and the mobile proton hypothesis of peptide fragmentation. On the other hand, a statistically significant but moderate increase was observed at 15 eV for both the third and fourth position (δ̂12 = 58% and δ̂12 = 71%, respectively), which suggests again that a cleavage C-terminal to Lys residues became slightly favored at a higher collision energy (Table 1, Figures 2D and 3D).
The comparison of the effect of Orn and Lys residues on the fragmentation processes indicates that while the mechanism proposed by our group for the Cit effect which was explained by a Cit to Orn transition followed by Orn effect might be true, it can be conflicting to use it for homocitrulline residues. We propose that the reaction could also follow a one-step mechanism, at least in the case of homocitrulline.
We also investigated the alteration of cleavage products N-terminal to Cit/Hci/Orn/Lys residues and found that an amide bond cleavage at this position is unequivocally unfavored and suppressed compared to the control sequences (Table S1).
To highlight the effect of the Z amino acids on the cleavage preference, we calculated and plotted the difference between the Cit/Hci/Orn/Lys and their alanine-substituted variant for each Z (Figures S3 and S4). These results indicate that for Cit-Zzz connections (Cit in third position) there were no substantial differences between the Z amino acids following the Cit residue in the sequence at neither 10 nor 15 eV. These differences varied between 5–26% and 2–22%, respectively. In the case of Cit in the fourth position, a very low positive increment (0–2%) was measured for the majority of Z compared to the controls at 10 eV. For Z = F, Q, W, and P, a low decrease was observed compared to the controls. At 15 eV, all connections displayed a positive rel% difference (2–15%) with the exception of Z = H.
On the other hand, a very high difference (63%) was found between Orn-His and Orn-Ala connections at 10 eV where Orn was at third position, while other Orn-Zzz connections were not considerably different from each other and the values ranged between 13 and 34%. At 15 eV, this discrepancy was eliminated: 5–32% difference was measured with no remarkable outliers (Figure S3C,D). Orn residues preserved their effect at the fourth position (Figure S4C,D).
For Hci residues, the effect seemed to be occurring only when Hci was at the third position. The cleavage differences varied between 4–18% and 4–21% at 10 and 15 eV, respectively (Figure S3). With Hci at the fourth position, the effect was lacking at 10 eV (only Hci-Glu scored a positive rel% difference). At 15 eV, around half of the amino acid connections were still in the negative range (Figure S4E,F).
As for Lys residues, some connections even showed negative values when Lys was at the third position, with values ranging from −4 to +6% and −5 to +11% at 10 and 15 eV, respectively (Figure S3G,H). With Lys at the fourth positions, 6 out of 25 connections showed a positive rel% difference at 10 eV but 24 out of 25 at 15 eV (Figure S4G,H).
It should also be noted that the lowest values of rel% differences were mostly attributed to Arg and Ala residues (X/B/O/K at third position). Arg is known to “sequestrate” protons and hinder fragmentation, thus giving the lower values. On the other hand, in a previous study, alanine was found to affect the MS/MS fragmentation only in a minor way.25 That is why we selected this amino acid as a control. Our results indicate, however, that the cleavage N-terminal to Ala residues might be rather strongly favored in at least a few cases. When X/B/O/K were at the fourth position, the lowest values of rel% differences were mostly observed for His and Pro residues. For Pro residues, this can be explained by the high preference to be cleaved at its N-terminus in MS/MS. Histidine also has a similar effect that might be comparable to that of Cit and Hci in these cases.
Conclusions
Special amino acids or posttranslational modifications could alter the fragmentation processes of peptides34 in a profound way as previously demonstrated for proline residues.32 Some of these effects could improve or hinder the detection and identification of the peptides. We previously demonstrated an enhanced cleavage preference C-terminal to Cit residues.
Currently, there is a great effort to use the intensity values of the given fragments in the tandem mass spectra of peptides to obtain reliable structural identification and thus reduce false positive identifications41−44
In this work, we quantified the effect of citrulline and homocitrulline residues on the higher energy collision-induced dissociation of tryptic-like model system and found that Cit residues increase the amide bond cleavage ratio at the C-terminus of Cit in a statistically and practically significant manner. The effect was virtually independent of the type of the following natural α amino acid (Z) as opposed to our previous study40 based on a proteomic data set of Lee et al.15 In that work, statistically strong results were only obtained for a small number of Cit-Zzz connections which were the most prevalent ones among native citrullinated peptides. Our results showed that homocitrulline residues produced a considerable effect only in the cases where Hci was in the third position in the sequence, which may indicate that the effect of Hci follows a mechanism that is less favored, in accordance with Scheme 1. Both Cit and Hci effects seemed to be stronger when Cit or Hci were farther from the C-terminus. We also found that Cit and Hci residues (as well as Orn and Lys) always suppress the cleavage at their N-terminus as well.
These results help explain the fragmentation processes of peptides containing a urea moiety and improve tandem mass spectrometry-based proteomic identification by integrating these findings into commercially available software algorithms.
Acknowledgments
The research was supported by the MTA Premium Post-Doctorate Research Program of the Hungarian Academy of Sciences (HAS, MTA). This work was completed in the Synthesis+ ELTE Thematic Excellence Programme supported by the Hungarian Ministry for Innovation and Technology. The research within Project No. VEKOP-2.3.3-15-2017-00020 was supported by the European Union and the State of Hungary, cofinanced by the European Regional Development Fund. Project No. 2018-1.2.1-NKP-2018-00005 has been implemented with the support provided from the National Research, Development and Innovation Fund of Hungary, financed under the 2018-1.2.1-NKP funding scheme.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jasms.0c00210.
Supporting figures and tables (PDF)
Author Contributions
All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Nicholas A. P.; Thompson P. R.; Bhattacharya S. K.. Protein Deimination in Human Health and Disease; Springer, 2017. [Google Scholar]
- Jaisson S.; Pietrement C.; Gillery P. Protein Carbamylation: Chemistry, Pathophysiological Involvement, and Biomarkers. Adv. Clin. Chem. 2018, 84, 1–38. 10.1016/bs.acc.2017.12.001. [DOI] [PubMed] [Google Scholar]
- Fuhrmann J.; Thompson P. R. Protein Arginine Methylation and Citrullination in Epigenetic Regulation. ACS Chem. Biol. 2016, 11 (3), 654–668. 10.1021/acschembio.5b00942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méchin M. C.; Takahara H.; Simon M. Deimination and Peptidylarginine Deiminases in Skin Physiology and Diseases. Int. J. Mol. Sci. 2020, 21 (2), 1–15. 10.3390/ijms21020566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzhalin A. E. Citrullination in Cancer. Cancer Res. 2019, 79 (7), 1274–1284. 10.1158/0008-5472.CAN-18-2797. [DOI] [PubMed] [Google Scholar]
- Valesini G.; Gerardi M. C.; Iannuccelli C.; Pacucci V. A.; Pendolino M.; Shoenfeld Y. Citrullination and Autoimmunity. Autoimmun. Rev. 2015, 14, 490–497. 10.1016/j.autrev.2015.01.013. [DOI] [PubMed] [Google Scholar]
- Gallart-Palau X.; Tan L. M.; Serra A.; Gao Y.; Ho H. H.; Richards A. M.; Kandiah N.; Chen C. P.; Kalaria R. N.; Sze S. K. Degenerative Protein Modifications in the Aging Vasculature and Central Nervous System: A Problem Shared Is Not Always Halved. Ageing Res. Rev. 2019, 53, 100909. 10.1016/j.arr.2019.100909. [DOI] [PubMed] [Google Scholar]
- Gorisse L.; Pietrement C.; Vuiblet V.; Schmelzer C. E. H.; Köhler M.; Duca L.; Debelle L.; Fornès P.; Jaisson S.; Gillery P. Protein Carbamylation Is a Hallmark of Aging. Proc. Natl. Acad. Sci. U. S. A. 2016, 113 (5), 1191–1196. 10.1073/pnas.1517096113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas C.; Jaisson S.; Gorisse L.; Tessier F. J.; Niquet-Léridon C.; Jacolot P.; Pietrement C.; Gillery P. Carbamylation and Glycation Compete for Collagen Molecular Aging in Vivo. Sci. Rep. 2019, 9 (1), 18291. 10.1038/s41598-019-54817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollipara L.; Zahedi R. P. Protein Carbamylation: In Vivo Modification or in Vitro Artefact?. Proteomics 2013, 13 (6), 941–944. 10.1002/pmic.201200452. [DOI] [PubMed] [Google Scholar]
- Sun S.; Zhou J. Y.; Yang W.; Zhang H. Inhibition of Protein Carbamylation in Urea Solution Using Ammonium-Containing Buffers. Anal. Biochem. 2014, 446 (1), 76–81. 10.1016/j.ab.2013.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faserl K.; Sarg B.; Maurer V.; Lindner H. H. Exploiting Charge Differences for the Analysis of Challenging Post-Translational Modifications by Capillary Electrophoresis-Mass Spectrometry. J. Chromatogr. A 2017, 1498, 215–223. 10.1016/j.chroma.2017.01.086. [DOI] [PubMed] [Google Scholar]
- Winter D. L.; Wilkins M. R.; Donald W. A. Differential Ion Mobility–Mass Spectrometry for Detailed Analysis of the Proteome. Trends Biotechnol. 2019, 37 (2), 198–213. 10.1016/j.tibtech.2018.07.018. [DOI] [PubMed] [Google Scholar]
- Bayat P.; Lesage D.; Cole R. B. Tutorial: Ion Activation in Tandem Mass Spectrometry Using Ultra-High Resolution Instrumentation. Mass Spectrom. Rev. 2020, 1–23. 10.1002/mas.21623. [DOI] [PubMed] [Google Scholar]
- Lee C. Y.; Wang D.; Wilhelm M.; Zolg D. P.; Schmidt T.; Schnatbaum K.; Reimer U.; Pontén F.; Uhlén M.; Hahne H.; Kuster B. Mining the Human Tissue Proteome for Protein Citrullination. Mol. Cell. Proteomics 2018, 17 (7), 1378–1391. 10.1074/mcp.RA118.000696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzihradszky K. F.; Chalkley R. J. Lessons In De Novo Peptide Sequencing By Tandem Mass Spectrometry. Mass Spectrom. Rev. 2015, 34, 43–63. 10.1002/mas.21406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.; Triscari J. M.; Pasa-Tolic L.; Anderson G. A.; Lipton M. S.; Smith R. D.; Wysocki V. H. Dissociation Behavior of Doubly-Charged Tryptic Peptides: Correlation of Gas-Phase Cleavage Abundance with Ramachandran Plots. J. Am. Chem. Soc. 2004, 126 (10), 3034–3035. 10.1021/ja038041t. [DOI] [PubMed] [Google Scholar]
- Huang Y.; Triscari J. M.; Tseng G. C.; Pasa-Tolic L.; Lipton M. S.; Smith R. D.; Wysocki V. H. Statistical Characterization of the Charge State and Residue Dependence of Low-Energy CID Peptide Dissociation Patterns. Anal. Chem. 2005, 77 (18), 5800–5813. 10.1021/ac0480949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.; Tseng G. C.; Yuan S.; Pasa-Tolic L.; Lipton M. S.; Smith R. D.; Wysocki V. H. A Data-Mining Scheme for Identifying Peptide Structural Motifs Responsible for Different MS/MS Fragmentation Intensity Patterns. J. Proteome Res. 2008, 7 (1), 70–79. 10.1021/pr070106u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lermyte F.; Valkenborg D.; Loo J. A.; Sobott F. Radical Solutions: Principles and Application of Electron-Based Dissociation in Mass Spectrometry-Based Analysis of Protein Structure. Mass Spectrom. Rev. 2018, 37 (6), 750–771. 10.1002/mas.21560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Q.; Lee M. V.; Rose C. M.; Marsh A. J.; Hubler S. L.; Wenger C. D.; Coon J. J. Characterization and Diagnostic Value of Amino Acid Side Chain Neutral Losses Following Electron-Transfer Dissociation. J. Am. Soc. Mass Spectrom. 2011, 22 (2), 255–264. 10.1007/s13361-010-0029-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermansson M.; Artemenko K.; Ossipova E.; Eriksson H.; Lengqvist J.; Makrygiannakis D.; Catrina A. I.; Nicholas A. P.; Klareskog L.; Savitski M.; Zubarev R. A.; Jakobsson P. J. Mass Spectrometric Analysis of Rheumatoid Arthritic Synovial Tissue Identifies Specific Citrullination Sites on Fibrinogen. Proteomics Clin. Appl. 2010, 4 (5), 511–518. 10.1002/prca.200900088. [DOI] [PubMed] [Google Scholar]
- Shaw J. B.; Robinson E. W.; Paša-Tolić L. Vacuum Ultraviolet Photodissociation and Fourier Transform-Ion Cyclotron Resonance (FT-ICR) Mass Spectrometry: Revisited. Anal. Chem. 2016, 88 (6), 3019–3023. 10.1021/acs.analchem.6b00148. [DOI] [PubMed] [Google Scholar]
- Steckel A.; Uray K.; Kalló G.; Csosz É.; Schlosser G. Investigation of Neutral Losses and the Citrulline Effect for Modified H4 N-Terminal Pentapeptides. J. Am. Soc. Mass Spectrom. 2020, 31 (3), 565–573. 10.1021/jasms.9b00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapp E. A.; Schütz F.; Reid G. E.; Eddes J. S.; Moritz R. L.; O’Hair R. A. J.; Speed T. P.; Simpson R. J. Mining a Tandem Mass Spectrometry Database to Determine the Trends and Global Factors Influencing Peptide Fragmentation. Anal. Chem. 2003, 75 (22), 6251–6264. 10.1021/ac034616t. [DOI] [PubMed] [Google Scholar]
- Müller T.; Winter D. Systematic Evaluation of Protein Reduction and Alkylation Reveals Massive Unspecific Side Effects by Iodine-Containing Reagents. Mol. Cell. Proteomics 2017, 16 (7), 1173–1187. 10.1074/mcp.M116.064048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewski J. R.; Zettl K.; Pilch M.; Rysiewicz B.; Sadok I. ‘Shotgun’ Proteomic Analyses without Alkylation of Cysteine. Anal. Chim. Acta 2020, 1100, 131–137. 10.1016/j.aca.2019.12.007. [DOI] [PubMed] [Google Scholar]
- Hao G.; Wang D.; Gu J.; Shen Q.; Gross S. S.; Wang Y. Neutral Loss of Isocyanic Acid in Peptide CID Spectra: A Novel Diagnostic Marker for Mass Spectrometric Identification of Protein Citrullination. J. Am. Soc. Mass Spectrom. 2009, 20 (4), 723–727. 10.1016/j.jasms.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee W. M.; McLuckey S. A. The Ornithine Effect in Peptide Cation Dissociation. J. Mass Spectrom. 2013, 48 (7), 856–861. 10.1002/jms.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steckel A.; Uray K.; Turiák L.; Gömöry Á.; Drahos L.; Hudecz F.; Schlosser G. Mapping the Tandem Mass Spectrometric Characteristics of Citrulline-Containing Peptides. Rapid Commun. Mass Spectrom. 2018, 32 (11), 844–850. 10.1002/rcm.8105. [DOI] [PubMed] [Google Scholar]
- Zhang Q.; Perkins B.; Tan G.; Wysocki V. H. The Role of Proton Bridges in Selective Cleavage of Ser-, Thr-, Cys-, Met-, Asp-, and Asn-Containing Peptides. Int. J. Mass Spectrom. 2011, 300 (2–3), 108–117. 10.1016/j.ijms.2010.09.025. [DOI] [Google Scholar]
- Tabb D. L.; Smith L. L.; Breci L. A.; Wysocki V. H.; Lin D.; Yates J. R. Statistical Characterization of Ion Trap Tandem Mass Spectra from Doubly Charged Tryptic Peptides. Anal. Chem. 2003, 75 (5), 1155–1163. 10.1021/ac026122m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohalm M.; Kavan D.; Novák P.; Volný M.; Havlíček V. MMass 3: A Cross-Platform Software Environment for Precise Analysis of Mass Spectrometric Data. Anal. Chem. 2010, 82 (11), 4648–4651. 10.1021/ac100818g. [DOI] [PubMed] [Google Scholar]
- Paizs B.; Suhai S. Fragmentation Pathways of Protonated Peptides. Mass Spectrom. Rev. 2005, 24 (4), 508–548. 10.1002/mas.20024. [DOI] [PubMed] [Google Scholar]
- Algina J.; Olejnik S.; Ocanto R. Type I Error Rates and Power Estimates for Selected Two-Sample Tests of Scale. J. Educ. Stat. 1989, 14 (4), 373–384. 10.3102/10769986012001045. [DOI] [Google Scholar]
- Gastwirth J. L.; Gel Y. R.; Miao W. The Impact of Levene’s Test of Equality of Variances on Statistical Theory and Practice. Stat. Sci. 2009, 24 (3), 343–360. 10.1214/09-STS301. [DOI] [Google Scholar]
- Delaney H. D.; Vargha A. Comparing Several Robust Tests of Stochastic Equality with Ordinally Scaled Variables and Small to Moderate Sized Samples. Psychol. Methods 2002, 7 (4), 485–503. 10.1037/1082-989X.7.4.485. [DOI] [PubMed] [Google Scholar]
- Brunner E.; Munzel U. The Nonparametric Behrens-Fisher Problem: Asymptotic Theory and a Small-Sample Approximation. Biom. J. 2000, 42 (1), 17–25. . [DOI] [Google Scholar]
- Vargha A.; Torma B.; Bergman L. R. ROPstat: A General Statistical Package Useful for Conducting Person-Oriented Analysis. J. Pers. Res. 2015, 1 (1–2), 87–98. 10.17505/jpor.2015.09. [DOI] [Google Scholar]
- Steckel A.; Schlosser G. Citrulline Effect Is a Characteristic Feature of Deiminated Peptides in Tandem Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2019, 30 (9), 1586–1591. 10.1007/s13361-019-02271-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degroeve S.; Martens L.; Jurisica I. MS2PIP: A Tool for MS/MS Peak Intensity Prediction. Bioinformatics 2013, 29 (24), 3199–3203. 10.1093/bioinformatics/btt544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degroeve S.; Maddelein D.; Martens L. MS2PIP Prediction Server: Compute and Visualize MS2 Peak Intensity Predictions for CID and HCD Fragmentation. Nucleic Acids Res. 2015, 43 (W1), W326–W330. 10.1093/nar/gkv542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriels R.; Martens L.; Degroeve S. Updated MS2PIP web server delivers fast and accurate MS2 peak intensity prediction for multiple fragmentation methods, instruments and labeling techniques. Nucleic Acids Res. 2019, 47 (W1), W295–W299. 10.1093/nar/gkz299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessulat S.; Schmidt T.; Zolg D. P.; Samaras P.; Schnatbaum K.; Zerweck J.; Knaute T.; Rechenberger J.; Delanghe B.; Huhmer A.; Reimer U.; Ehrlich H. C.; Aiche S.; Kuster B.; Wilhelm M. Prosit: Proteome-Wide Prediction of Peptide Tandem Mass Spectra by Deep Learning. Nat. Methods 2019, 16 (6), 509–518. 10.1038/s41592-019-0426-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



