Abstract
Background
Ovarian cancer is one of the malignant tumors attacking the female reproductive system. Currently, increasing studies have clearly determined the importance of long non-coding RNAs (lncRNAs) in various human cancers including ovarian cancer. However, the role and in-depth mechanism of ubiquitin specific peptidase 2 antisense RNA 1 (USP2-AS1) in ovarian cancer have been not reported yet.
Purpose
We were absorbed into exploring the character of USP2-AS1 in ovarian cancer.
Methods
RT-qPCR analysis reflected gene expression. The GEPIA database provided further evidences, and bioinformatics tools analyzed the potential molecules downstream USP2-AS1 in ovarian cancer. The changes on ovarian cancer cellular functions were assessed via EdU, TUNEL, JC-1 and transwell assays. RNA pull down, RIP and luciferase reporter assays estimated molecule interactions.
Results
USP2-AS1 was obviously up-regulated in ovarian cancer tissues and cell lines. Inhibiting USP2-AS1 had anti-proliferation, pro-apoptosis, and anti-migration effects on ovarian cancer cells. Furthermore, we confirmed that USP2-AS1 sequestered miR-520d-3p to enhance KIAA1522. In addition, miR-520d-3p silence reversed the effect of depleted USP2-AS1 on ovarian cancer cellular behaviors, while such reversion was then abolished by KIAA1522 knockdown.
Conclusion
USP2-AS1 facilitated ovarian cancer progression via miR-520d-3p/KIAA1522 axis, implying USP2-AS1 as a new perspective for the treatment of ovarian cancer.
Keywords: USP2-AS1, miR-520d-3p, KIAA1522, ovarian cancer
Introduction
Ovarian cancer is one kind of gynecologic malignancy with high mortality.1 Most patients with ovarian cancer are diagnosed at advanced stages, owing to asymptomatic characteristics of this cancer at early stage, and this is also an important factor for high incidence and death rate, except for poor screening tool.2,3 Nowadays, for most cancer types including ovarian cancer, surgery combined with chemotherapy is regarded as effective treatment to eliminate all cancer cells.4,5 Considering ovarian cancer is a heterogeneous disease with complicated molecular and genetic changes, targeted therapies against abnormal molecules have emerged.6–8 Nonetheless, molecules that are suitable for targeted therapies are still limited. Therefore, in this study, we also contributed to explore the underlying molecular mechanism in ovarian cancer, so as to find out the potential biomarkers.
To date, a large proportion of long non-coding RNAs (lncRNAs) have been admittedly considered as promoters or inhibitors in diverse tumors, including ovarian cancer. For example, Zhang et al9 suggested that SUMO1P3 triggered colon cancer cell growth, liver metastasis, and even vascularization. HOTAIR could be regarded as a potential indicator of cell cycle dysregulation in lung cancer.10 MEG3 was down-regulated in ovarian cancer cells and elicited tumor inhibitor role in this disease.11 Besides, lncRNAs have also been reported to affect tumor growth, metastasis, and drug resistance by controlling target gene expression and pathway activation.12,13 For example, TUC338 accelerated the development of lung cancer via targeting MAPK pathway.14 CCAT1 affected breast cancer development via miR-148b.15 Unfortunately, the role and mechanism of ubiquitin specific peptidase 2 antisense RNA 1 (USP2-AS1) in ovarian cancer are still not well understood.
In the current study, we explored the expression of USP2-AS1 in ovarian cancer tissues and cells. Besides, the functional analysis was used to explain the effects of USP2-AS1 interference on the biological behaviors of ovarian cancer cells. Furthermore, we expounded the correlation among USP2-AS1, miR-520d-3p, and KIAA1522, which might help us verify USP2-AS1 as a potential therapeutic target for ovarian cancer.
Materials and Methods
Cell Lines
Human normal ovarian surface epithelial cell line HOSEPiC was bought from Procell Life Science & Technology (Wuhan, China); human ovarian cancer cell lines (ES-2, CAOV3, and SKOV3) were procured from ATCC (Manassas, VA, USA); human ovarian cancer cell line A2780 was purchased from European Collection of Authenticated Cell Cultures (ECACC, Porton Down, Salisbury, UK). HOSEPiC and CAOV3 cells were maintained in DMEM (GIBCO, Grand Island, NY, USA); SKOV3 and ES-2 cells were maintained in McCoy’s 5a medium; A2780 cells were maintained in RPMI 1640 medium (GIBCO). All the culture media were supplemented with 10% fetal bovine serum (FBS; GIBCO). Cell culture was undertaken in a 5% incubator at 37°C.
Total RNA Isolation and RT-qPCR
Total cellular RNA was isolated by TRIzol Reagent (Invitrogen, Carlsbad CA, USA), and synthesis of complementary DNA was carried out via PrimeScript™ RT Master Mix kit (TaKaRa BIO, Shiga, Japan). Gene expressions were detected by use of SYBR Green PCR Kit (Takara) on ABI Prism 7900HT sequence detector (Applied Biosystems, Foster City, CA, USA). Relative expression was processed with 2−ΔΔCt method after normalizing to GAPDH or U6. Three repeated experiments were undertaken.
Cell Transfection
A2780 and SKOV3 cells were cultured in 6-well plates for 48 hours of transfection, in the presence of Lipofectamine 3000 (Invitrogen). The USP2-AS1-specific shRNAs and KIAA1522-specific shRNAs were synthesized by GenePharma (Shanghai, China), as well as the nonspecific shRNAs as the negative control (sh-NC). The miR-520d-3p inhibitor, miR-520d-3p mimics, and NCs (NC inhibitor or NC mimics) were designed and constructed by Ribobio (Guangzhou, China). Three repeated experiments were undertaken.
EdU Staining Assay
EdU staining assay was implemented in 96-well plates, in the presence of BeyoClick™ EdU Cell Proliferation Kit (Beyotime, Shanghai, China). A pool of 1×104 cells were treated for 2 hours with an EdU kit at 37°C, and with DAPI staining solution for 5 minutes. After washing, the proliferative cells were tested by fluorescence microscope (Olympus, Tokyo, Japan). Three repeated experiments were undertaken.
TUNEL Staining Assay
After fixing in 4% paraformaldehyde, 1×104 cells were prepared in 96-well plates and then subjected to One-Step TUNEL Apoptosis Assay Kit (Beyotime), based on the user guide. Following DAPI staining, the apoptotic cells were measured by use of a fluorescence microscope (Olympus). Three repeated experiments were undertaken.
JC-1 Staining Assay
The transfected cells were rinsed in PBS, and 1 mL of cell suspension was then plated in 6-well plates to analyze the change in mitochondrial membrane potential (Δψm). 2.5 µg/mL of JC-1 dye was employed as guided by the provider (Beyotime). An Olympus fluorescence microscope was used for cell observation and analysis. Three repeated experiments were undertaken.
Cell Migration Assay
Cell migratory potential was determined by transwell chamber (8 μm pore size; Costar, Cambridge, MA, USA). Then 2×104 transfected cells were seeded into the upper chamber with serum-free medium for 24 hours of cultivation, and the complete medium was filled into the lower chamber. Migrated cells were subjected to crystal violet solution after fixing by 4% paraformaldehyde, followed by cell observation under an optical microscope (Olympus). Three repeated experiments were undertaken.
Fish
The fixed A2780 and SKOV3 cells were washed in PBS and then cultured with the USP2-AS1-specific FISH probe (Ribobio) in the hybridization buffer. Following the addition of DAPI solution, the fluorescent analysis was achieved using an Olympus fluorescence microscope, with the images captured as well. Three repeated experiments were undertaken.
Subcellular Fractionation Assay
PARIS™ Kit (Invitrogen) was applied for undertaking subcellular fraction assay, as instructed. A2780 and SKOV3 cells were processed with cell fractionation buffer for the collection of cell cytoplasm, and then centrifuged. Cell nucleus was collected via cell disruption buffer. The content of USP2-AS1 was analyzed by RT-qPCR in both fractions using GAPDH and U6 as the controls. Three repeated experiments were undertaken.
RNA Immunoprecipitation (RIP)
Cell lysates from RIP lysis buffer were subjected to Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA, USA) for RIP assay. The magnetic beads bound with Ago2 antibody or control IgG antibody were mixed with cell lysates to collect the immunoprecipitates. Thereafter, the captured RNAs were subjected to RT-qPCR analysis. Three repeated experiments were undertaken.
RNA Pull Down Assay
Cell lysates from RIPA lysis buffer were employed for RNA pull down assay by use of Pierce Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher Scientific, Waltham, MA, USA). Then 30 μL of magnetic beads was supplemented into samples for capturing the RNA-RNA mixture, followed by RT-qPCR detection of RNA enrichment. Three repeated experiments were undertaken.
Luciferase Reporter Assay
The amplified wild-type fragment of USP2-AS1 or KIAA1522 was inserted into the pmirGLO vector (Promega, Madison, WI, USA) for acquiring USP2-AS1 WT or KIAA1522 WT. Site-direct mutagenesis kit (Takara) was utilized to acquire mutant luciferase vectors (USP2-AS1 MUT and KIAA1522 MUT). Relative luciferase activity was assayed by Dual-Luciferase Reporter Assay System (Promega) after 48 hours of cell co-transfection with indicated reporter vectors and miR-520d-3p mimics or NC mimics. The activity of Renilla luciferase was employed as the internal control. Three repeated experiments were undertaken.
Statistical Analyses
Data from three repeated experiments were analyzed by GraphPad PRISM 6 (GraphPad, San Diego, CA, USA) and expressed as mean±SD. Group difference was examined by Student’s t-test or one-way ANOVA, using P<0.05 as the threshold of statistical significance.
Results
Knockdown of USP2-AS1 Represses the Progression of Ovarian Cancer
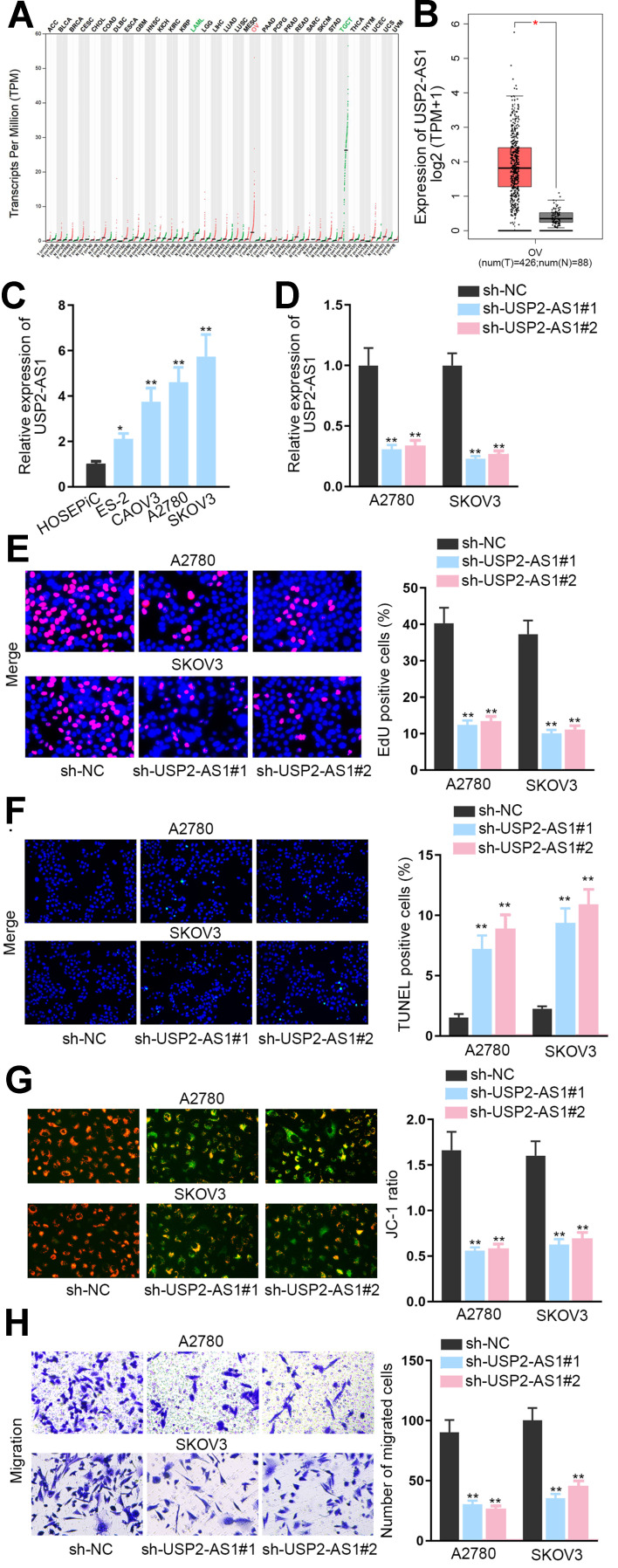
To expose the role of USP2-AS1 in ovarian cancer, data from GEPIA (http://gepia.cancer-pku.cn/detail.php) was analyzed. As a result, we could observe that USP2-AS1 was significantly increased in ovarian cancer (Figure 1A). Besides, ovarian cancer tissues had more abundant USP2-AS1 expression than normal ones (Figure 1B). Moreover, USP2-AS1 was also highly expressed in ovarian cancer cell lines (ES-2, CAOV3, A2780, and SKOV3) compared to normal human ovarian surface epithelial cell line (HOSEPiC) (Figure 1C). Based on these data, we wondered whether such up-regulation of USP2-AS1 had some association with the malignant behaviors of ovarian cancer cells. Therefore, loss-of-function assays were designed via transfecting sh-USP2-AS1#1 or sh-USP2-AS1#2 into A2780 and SKOV3 cells to effectively induce USP2-AS1 silence (Figure 1D). Then EdU assay, TUNEL assay, JC-1 assay, and transwell assay were conducted to observe the effect of USP2-AS1 knockdown on cell biological behaviors. The results of EdU assay demonstrated that USP2-AS1 depletion obviously suppressed ovarian cancer cell proliferation (Figure 1E). However, the outcomes of TUNEL assay and JC-1 assay indicated that cell apoptosis was remarkably strengthened due to USP2-AS1 knockdown (Figure 1F and G). Moreover, transwell assay results revealed that USP2-AS1 deficiency seriously hampered the migration of ovarian cancer cells (Figure 1H). Collectively, USP2-AS1 is up-regulated in ovarian cancer tissues and cells and its down-regulation is effective to depress the malignant phenotypes of ovarian cancer cells.
Figure 1.
Knockdown of USP2-AS1 represses the progression of ovarian cancer. (A) RNA-Seq analysis data from GEPIA detected the expression of USP2-AS1 in diverse cancers including ovarian cancer. (B) GEPIA database analyzed the levels of USP2-AS1 in ovarian cancer tissues compared with normal tissues. (C) USP2-AS1 expression in ovarian cancer cell lines (ES-2, CAOV3, A2780, and SKOV3) and normal human ovarian surface epithelial cell line HOSEPiC was assessed via RT-qPCR. (D) The interference efficiency of USP2-AS1 was assessed by RT-qPCR in A2780 and SKOV3 cells treated with sh-USP2-AS1#1and2. (E) Cell proliferation ability was examined by EdU assay in response to USP2-AS1 silence. (F and G). TUNEL assay and JC-1 assay examined the alteration on the apoptosis of A2780 and SKOV3 cells in the case of silencing USP2-AS1. (H) Transwell assay was used to test cell migration under the context of USP2-AS1 silence. *P<0.05, **P<0.01.
USP2-AS1 is Capable of Binding with miR-520d-3p
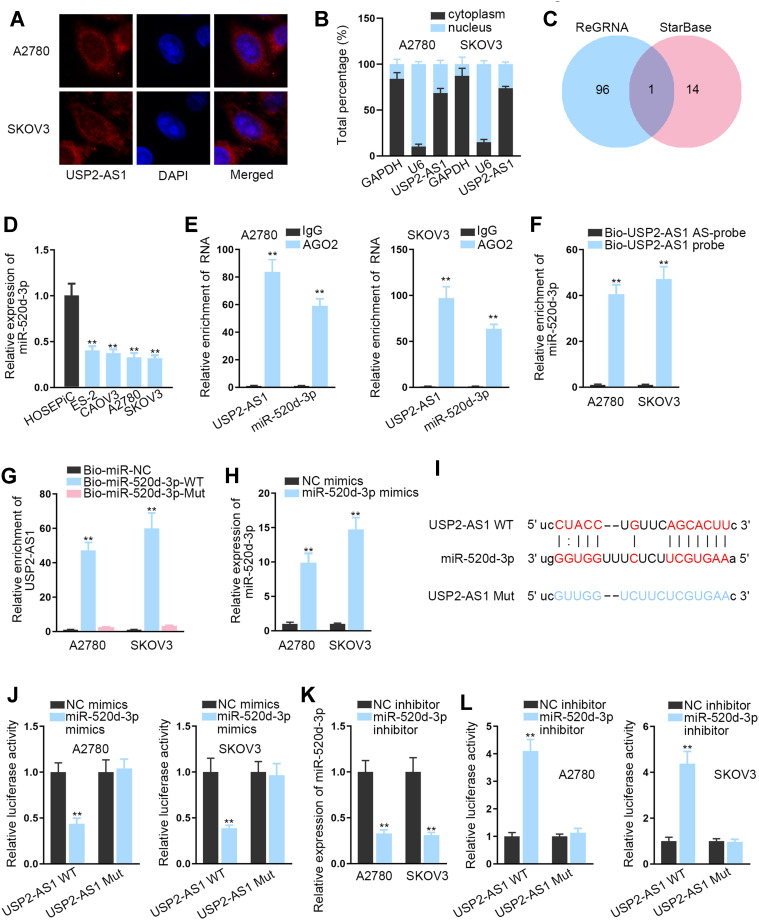
As is well known, lncRNAs have abilities of acting as competitive endogenous RNAs (ceRNAs) to regulate downstream gene expression via sponging microRNAs (miRNAs). Therefore, we aimed to study the in-depth mechanism of USP2-AS1 in ovarian cancer. First, FISH assay and subcellular fractionation assay assisted us to analyze the location of USP2-AS1, and the results indicated the existence of most USP2-AS1 in the cytoplasm of A2780 and SKOV3 cells (Figure 2A and B). Further, miR-520d-3p was shared by the prediction of both ENCORI (http://starbase.sysu.edu.cn/) and ReGRNA (http://regrna2.0.mbc.nctu.edu.tw/index.html) to have the possibility to interact with USP2-AS1 (Figure 2C). Subsequently, RT-qPCR revealed that miR-520d-3p was overtly down-regulated in ovarian cancer cells compared to normal HOSEPiC cells (Figure 2D). Furthermore, RIP assay demonstrated that USP2-AS1 and miR-520d-3p were significantly abundant in the anti-Ago2 group compared with the anti-IgG group (Figure 2E), implying the coexistence of them in RNA-induced silencing complexes (RISCs). Meanwhile, we also discovered the plentiful enrichment of miR-520d-3p in the Bio-USP2-AS1 probe group in A2780 and SKOV3 cells, and the high enrichment of USP2-AS1 captured by Bio-miR-520d-3p-WT (Figure 2F and G). Then, we proved that the expression of miR-520d-3p was effectively up-regulated in ovarian cancer cells treated with miR-520d-3p mimics (Figure 2H). Besides, we exhibited the underlying binding sites of miR-520d-3p and USP2-AS1 that were predicted via ENCORI (Figure 2I). After that, we conducted luciferase reporter assays to verify the interaction between USP2-AS1 and miR-520d-3p. As a result, the luciferase activity of USP2-AS1 WT was overtly declined by miR-520d-3p mimics compared with NC mimics, while that of USP2-AS1 Mut had almost no change (Figure 2J). By contrast, inhibiting miR-520d-3p led to visibly enhanced luciferase activity of USP2-AS1 WT, with no apparent alteration on that of USP2-AS1 Mut (Figure 2K and L). To sum up, USP2-AS1 is able to function as a ceRNA via sponging miR-520d-3p.
Figure 2.
USP2-AS1 is capable of targeting miR-520d-3p. (A and B) FISH assay and subcellular fraction assay were adopted to analyze the distribution of USP2-AS1 in A2780 and SKOV3 cells. (C) The potential miRNAs that could bind to USP2-AS1 were predicted with the aid of ENCORI and ReGRNA. (D) RT-qPCR detected miR-520d-3p expression in ovarian cancer cells and HOSEPiC cells. (E) The co-existence of USP2-AS1 and miR-520d-3p in RISCs was confirmed by RIP assay. (F and G) RNA pull down assays was used to assess the interaction between USP2-AS1 and miR-520d-3p. (H) RT-qPCR analysis detected the overexpression efficiency of miR-520d-3p mimics in ovarian cancer cells. (I) The binding sites between USP2-AS1 and miR-520d-3p were displayed by ENCORI. (J) Under the condition of miR-520d-3p mimics, luciferase reporter assay tested the combination between USP2-AS1 and miR-520d-3p. (K) The interference efficiency of miR-520d-3p inhibitor in ovarian cancer cells was analyzed via RT-qPCR. (L) Under the condition of miR-520d-3p inhibitor, luciferase reporter assay tested combination between USP2-AS1 and miR-520d-3p. **P<0.01.
MiR-520d-3p Silence Abolishes the Effect of USP2-AS1 Down-Regulation on Ovarian Cancer Cell Proliferation, Apoptosis, and Migration
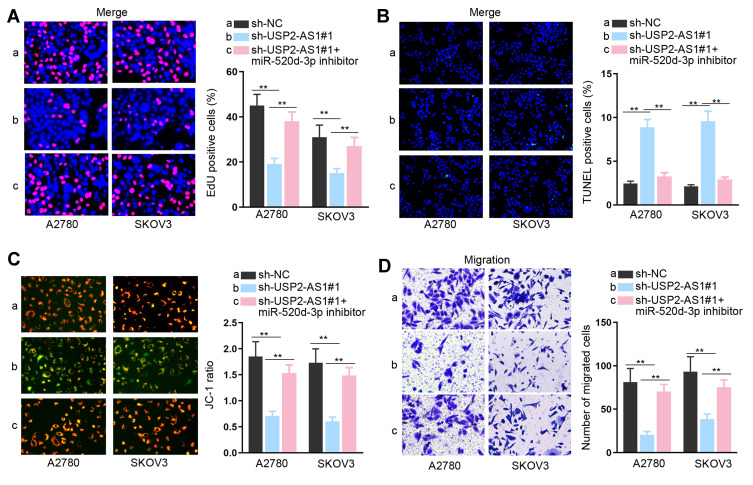
To clear the impact of the USP2-AS1/miR-520d-3p axis on ovarian cancer development, we carried out rescue assays. As indicated by the results of EdU assay, we observed that the EdU positive cell rate that was reduced by silenced USP2-AS1 was re-increased by miR-520d-3p inhibitor (Figure 3A). Also, the elevated apoptosis rates of A2780 and SKOV3 cells due to USP2-AS1 deficiency were reversed under miR-520d-3p inhibition (Figure 3B and C). Furthermore, USP2-AS1 depletion resulted in suppression on ovarian cancer cell migration, which was then absolutely abolished by miR-520d-3p down-regulation (Figure 3D). Taken together, we certified that USP2-AS1 promotes cell proliferation and migration by inhibiting miR-520d-3p in ovarian cancer.
Figure 3.
MiR-520d-3p silence abolishes the effect of USP2-AS1 down-regulation on ovarian cancer cell proliferation, apoptosis and migration. (A) The impact of miR-520d-3p inhibition on the proliferation of USP2-AS1-silence cells was determined by EdU assay. (B and C) TUNEL and JC-1 assays evaluated whether miR-520d-3p inhibitor could regulate the apoptosis of USP2-AS1-depleted A2780 and SKOV3 cells. (D) Transwell assay assessed the migration ability of A2780 and SKOV3 cells transfected with sh-NC, sh-USP2-AS1#1 or sh-USP2-AS1#1+ miR-520d-3p inhibitor. **P<0.01.
USP2-AS1 Acts as a ceRNA to Augment KIAA1522 by Sponging miR-520d-3p
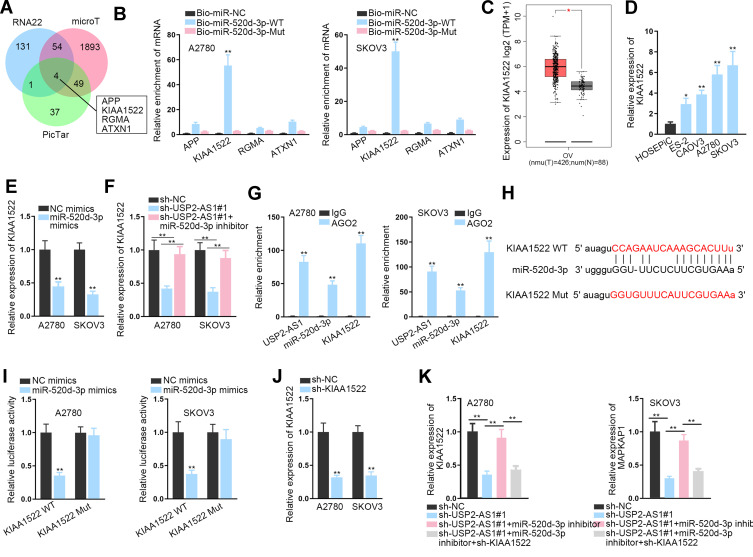
To explore the targets of miR-520d-3p, we utilized online RNA22, microT, and PicTar databases. In result, four underlying target genes, including APP, KIAA1522, RGMA, and ATXN1, were obtained (Figure 4A). Interestingly, only KIAA1522 was obviously harvested by Bio-miR-520d-3p-WT, but not Bio-miR-520d-3p-Mut in both ovarian cancer cells (Figure 4B). In addition, GEPIA data suggested the up-regulation of KIAA1522 in ovarian cancer tissues relative to normal tissues (Figure 4C). Also, KIAA1522 exhibited enhanced expression in ovarian cancer cells by contrast with normal cells (Figure 4D). Importantly, it manifested that miR-520d-3p overexpression remarkably inhibited the expression of KIAA1522 (Figure 4E), and that KIAA1522 expression inhibited by USP2-AS1 silence was rescued by miR-520d-3p depletion (Figure 4F). Furthermore, RIP assay confirmed that USP2-AS1, miR-520d-3p, and KIAA1522 co-existed in Ago2-composed RISCs (Figure 4G). Afterwards, the binding sites were hypothesized between miR-520d-3p and KIAA1522 through ENCORI (Figure 4H). As anticipated, the luciferase activity of KIAA1522-WT was obviously declined by miR-520d-3p elevation while that of KIAA1522-Mut was not affected in both A2780 and SKOV3 cells (Figure 4I). To further reveal the connection among USP2-AS1, miR-520d-3p, and KIAA1522, rescue assays were again performed. It was uncovered that transfection of sh-KIAA1522 apparently knocked down the expression of KIAA1522 in A2780 and SKOV3 cells (Figure 4J). Further, we observed that the decreased KIAA1522 expression resulting from depleted USP2-AS1 was reversed by miR-520d-3p inhibitor, while such recovery was counteracted in response to the absence of KIAA1522 (Figure 4K). Collectively, all data suggested that USP2-AS1 acts as a ceRNA of KIAA1522 by competitively binding to miR-520d-3p.
Figure 4.
USP2-AS1 acts as a ceRNA of KIAA1522 by absorbing miR-520d-3p. (A) RNA22, microT and PicTar databases forecasted four latent targets for miR-520d-3p. (B) The binding of miR-520d-3p with the above four candidates was detected via RNA pull down assay. (C) GEPIA data of KIAA1522 expression in ovarian cancer tissues and normal samples. (D) The level of KIAA1522 in ovarian cancer cells was tested by RT-qPCR. (E) RT-qPCR detected the influence of miR-520d-3p on KIAA1522 expression in A2780 and SKOV3 cells. (F) RT-qPCR detected the expression of KIAA1522 in two ovarian cancer cells under indicated conditions. (G) RIP assay confirmed the existence of all USP2-AS1, miR-520d-3p, and KIAA1522 in RISCs. (H) The binding sites between miR-520d-3p and KIAA1522 were exhibited by ENCORI. (I) Luciferase reporter assay was carried out to confirm if miR-520d-3p and KIAA1522 could bind each other. (J) The interference efficiency of KIAA1522 in A2780 and SKOV3 cells was tested by RT-qPCR. (K) RT-qPCR analyzed the alteration of KIAA1522 expression in ovarian cancer cells with appropriate transfection of sh-USP2-AS1#1, miR-520d-3p inhibitor, and sh-KIAA1522. *P<0.05, **P<0.01.
USP2-AS1 Accelerates Ovarian Cancer Development by miR-520d-3p/KIAA1522 Axis
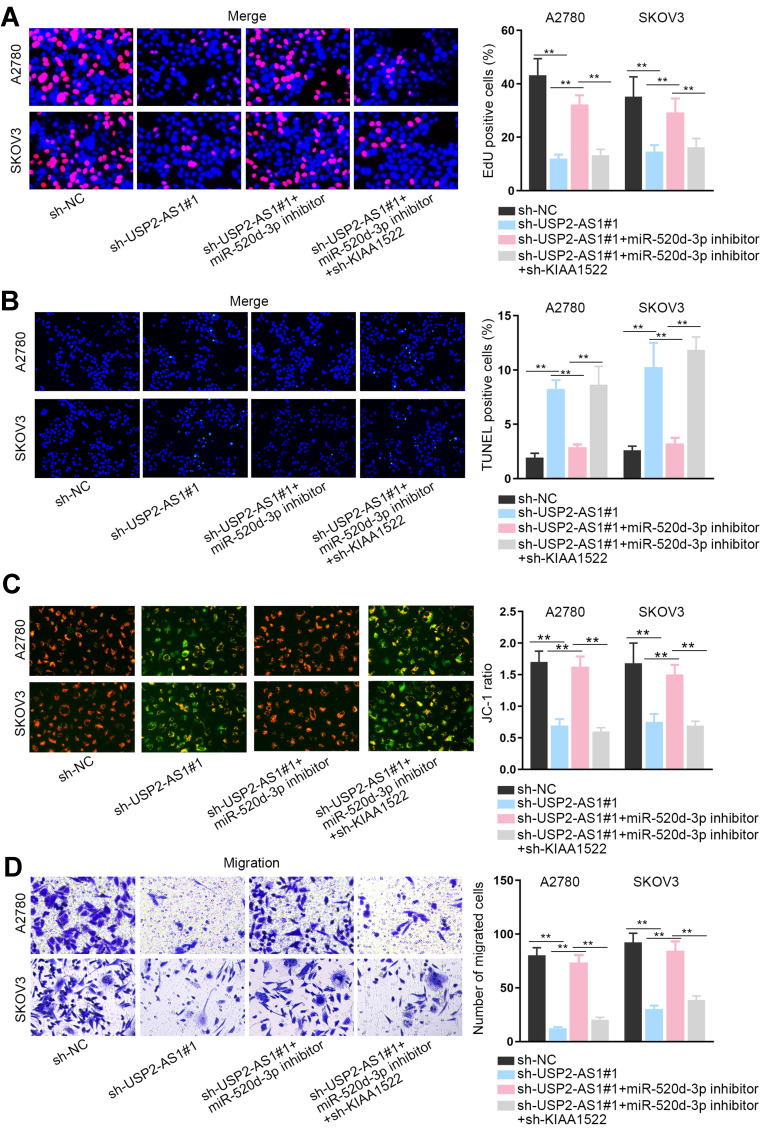
In the last step, a string of rescue assays were designed to confirm the influence of USP2-AS1/miR-520d-3p/KIAA1522 axis on the function of ovarian cancer cells. The results of EdU assay showed that the suppression on ovarian cancer cell proliferation caused by the loss of USP2-AS1 was fully countervailed by miR-520d-3p reduction, while such reversion was then offset by KIAA1522 inhibition (Figure 5A). Subsequently, the normalization of declined miR-520d-3p on the apoptosis of USP2-AS1-silenced ovarian cancer cells was wholly reversed by KIAA1522 down-regulation (Figure 5B and C). Also, the outcomes of transwell assays exhibited that USP2-AS1 silence successfully inhibited ovarian cancer cell migration to a great extent, which was abolished after inhibiting miR-520d-3p; and KIAA1522 depletion further reversed above phenomenon induced by miR-520d-3p inhibition (Figure 5D). In conclusion, USP2-AS1 accelerates ovarian cancer progression by miR-520d-3p/KIAA1522 axis.
Figure 5.
USP2-AS1 accelerates ovarian cancer development by miR-520d-3p/KIAA1522 axis. (A) The proliferation of indicated ovarian cancer cells was measured via EdU assay. (B and C) The apoptosis of transfected cells in different groups was tested by TUNEL and JC-1 assays. (D) The migration of transfected cells from indicated groups was assessed by transwell assay. **P<0.01.
Discussion
The current data have demonstrably proved that a majority of lncRNAs impact the development of ovarian cancer by mediating cell proliferation and apoptosis, even migration and invasion.16,17 For example, TP73-AS1 knockdown hampered ovarian cancer cell growth by modulating cell apoptosis, and also effectively elevated the survival time of patients with this disease.18 MALAT1 stimulated epithelial ovarian cancer cell viability as well as cell migration and invasion.19 Moreover, DUXAP10 was indicated as a potential therapeutic biomarker for ovarian cancer.20 In the present work, we discovered from the GEPIA database that USP2-AS1 was highly-expressed in ovarian cancer tissues. Also, we unveiled the up-regulation of USP2-AS1 in ovarian cancer cells. Furthermore, knocking down USP2-AS1 inhibited cell proliferation and migration, but activated cell apoptosis in ovarian cancer. Our findings demonstrated that USP2-AS1 played as an oncogene in ovarian cancer, and targeting USP2-AS1 might be a novel manner for treating patients with this disease.
Increasing concern has popular hotpot for the interaction among lncRNAs, miRNAs, and mRNAs in diverse tumors. For instance, Chang et al21 suggested that HOTAIR up-regulated CCND1 and CCND2 by sponging miR-206 to facilitate ovarian cancer progression. In the present study, it was unveiled that USP2-AS1 enhanced KIAA1522 expression to aggravate ovarian cancer progression through sequestering miR-520d-3p. As far as miR-520d-3p was concerned, it could availably subdue gastric cancer development by silencing EphA2 expression.22 Besides, SNHG16 acted as a motivator in hemangioma by miR-520d-3p/STAT3 signaling.23 In terms of KIAA1522, some reporters also revealed its role in certain cancers. For instance, Liu et al24 expressed that KIAA1522 was an underlying predictor of the prognosis of patients with non-small cell lung cancer. KIAA1522 targeted by miR-125b-5p contributed to breast cancer development.25 In this research, we disclosed that USP2-AS1 accelerated ovarian cancer development by modulating miR-520d-3p/KIAA1522 axis.
Conclusions
In conclusion, our findings illuminated the positive contribution of USP2-AS1 to the tumorigenesis of ovarian cancer, which was achieved via USP2-AS1/miR-520d-3p/KIAA1522 regulatory mechanism. In this network, USP2-AS1 acted as a ceRNA to enhance KIAA1522 expression by competitively binding with miR-520d-3p. Our work provided the first evidence to demonstrate USP2-AS1 as the promising therapeutic target for ovarian cancer.
Acknowledgment
We appreciate the support of laboratory.
Funding Statement
This study was supported by Anhui Provincial Key Projects of Natural Science Research in Colleges and Universities under grant (No. KJ2017A224 and KJ2018A0215).
Data Sharing Statement
Research Data are included in the manuscript.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising, or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no competing interests in this study.
References
- 1.Kujawa KA, Lisowska KM. [Ovarian cancer–from biology to clinic]. Postepy Higieny I Medycyny Doswiadczalnej (Online). 2015;69:1275–1290. doi: 10.5604/17322693.1184451 [DOI] [PubMed] [Google Scholar]
- 2.Rottmann M, Burges A, Mahner S, et al. Cancer of the ovary, fallopian tube, and peritoneum: a population-based comparison of the prognostic factors and outcomes. J Cancer Res Clin Oncol. 2017;143(9):1833–1844. doi: 10.1007/s00432-017-2422-6 [DOI] [PubMed] [Google Scholar]
- 3.Rooth C. Ovarian cancer: risk factors, treatment and management. Br J Nurs. 2013;22(17):S23–30. doi: 10.12968/bjon.2013.22.Sup17.S23 [DOI] [PubMed] [Google Scholar]
- 4.Narod S. Can advanced-stage ovarian cancer be cured? Nat Rev Clin Oncol. 2016;13(4):255–261. doi: 10.1038/nrclinonc.2015.224 [DOI] [PubMed] [Google Scholar]
- 5.Elies A, Riviere S, Pouget N, et al. The role of neoadjuvant chemotherapy in ovarian cancer. Expert Rev Anticancer Ther. 2018;18(6):555–566. doi: 10.1080/14737140.2018.1458614 [DOI] [PubMed] [Google Scholar]
- 6.Grunewald T, Ledermann JA. Targeted therapies for ovarian cancer. Best Pract Res Clin Obstet Gynaecol. 2017;41:139–152. doi: 10.1016/j.bpobgyn.2016.12.001 [DOI] [PubMed] [Google Scholar]
- 7.Pujade-Lauraine E. New treatments in ovarian cancer. Ann Oncol. 2017;28(suppl_8):viii57–viii60. doi: 10.1093/annonc/mdx442 [DOI] [PubMed] [Google Scholar]
- 8.Chiaramonte R, Colombo M, Bulfamante G, et al. Notch pathway promotes ovarian cancer growth and migration via CXCR4/SDF1alpha chemokine system. Int J Biochem Cell Biol. 2015;66:134–140. doi: 10.1016/j.biocel.2015.07.015 [DOI] [PubMed] [Google Scholar]
- 9.Zhang LM, Wang P, Liu XM, Zhang YJ. LncRNA SUMO1P3 drives colon cancer growth, metastasis and angiogenesis. Am J Transl Res. 2017;9(12):5461–5472. [PMC free article] [PubMed] [Google Scholar]
- 10.Liu M, Zhang H, Li Y, et al. HOTAIR, a long noncoding RNA, is a marker of abnormal cell cycle regulation in lung cancer. Cancer Sci. 2018;109(9):2717–2733. doi: 10.1111/cas.13745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Xu W, He Y, Xia Q, Liu S. LncRNA MEG3 impacts proliferation, invasion, and migration of ovarian cancer cells through regulating PTEN. Inflamm Res. 2018;67(11–12):927–936. doi: 10.1007/s00011-018-1186-z [DOI] [PubMed] [Google Scholar]
- 12.Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36(41):5661–5667. doi: 10.1038/onc.2017.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tuo Z, Zhang J, Xue W. LncRNA TP73-AS1 predicts the prognosis of bladder cancer patients and functions as a suppressor for bladder cancer by EMT pathway. Biochem Biophys Res Commun. 2018;499(4):875–881. doi: 10.1016/j.bbrc.2018.04.010 [DOI] [PubMed] [Google Scholar]
- 14.Zhang YX, Yuan J, Gao ZM, Zhang ZG. LncRNA TUC338 promotes invasion of lung cancer by activating MAPK pathway. Eur Rev Med Pharmacol Sci. 2018;22(2):443–449. [DOI] [PubMed] [Google Scholar]
- 15.Lai Y, Chen Y, Lin Y, Ye L. Down-regulation of LncRNA CCAT1 enhances radiosensitivity via regulating miR-148b in breast cancer. Cell Biol Int. 2018;42(2):227–236. doi: 10.1002/cbin.10890 [DOI] [PubMed] [Google Scholar]
- 16.Ren C, Li X, Wang T, et al. Functions and mechanisms of long noncoding RNAs in ovarian cancer. Int J Gynecol Cancer. 2015;25(4):566–569. doi: 10.1097/IGC.0000000000000413 [DOI] [PubMed] [Google Scholar]
- 17.Lu YM, Wang Y, Liu SQ, Zhou MY, Guo YR. Profile and validation of dysregulated long noncoding RNAs and mRNAs in ovarian cancer. Oncol Rep. 2018;40(5):2964–2976. [DOI] [PubMed] [Google Scholar]
- 18.Li X, Wang X, Mao L, Zhao S, Wei H. LncRNA TP73AS1 predicts poor prognosis and promotes cell proliferation in ovarian cancer via cell cycle and apoptosis regulation. Mol Med Rep. 2018;18(1):516–522. [DOI] [PubMed] [Google Scholar]
- 19.Lin Q, Guan W, Ren W, Zhang L, Zhang J, Xu G. MALAT1 affects ovarian cancer cell behavior and patient survival. Oncol Rep. 2018;39(6):2644–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Q, Wang WW, Xu TH, Xu ZF. Highly expressed long non-coding RNA DUXAP10 promotes proliferation of ovarian cancer. Eur Rev Med Pharmacol Sci. 2018;22(2):314–321. [DOI] [PubMed] [Google Scholar]
- 21.Chang L, Guo R, Yuan Z, Shi H, Zhang D. LncRNA HOTAIR regulates CCND1 and CCND2 expression by sponging miR-206 in ovarian cancer. Cell Physiol Biochem. 2018;49(4):1289–1303. doi: 10.1159/000493408 [DOI] [PubMed] [Google Scholar]
- 22.Li R, Yuan W, Mei W, Yang K, Chen Z. MicroRNA 520d-3p inhibits gastric cancer cell proliferation, migration, and invasion by downregulating EphA2 expression. Mol Cell Biochem. 2014;396(1–2):295–305. doi: 10.1007/s11010-014-2164-6 [DOI] [PubMed] [Google Scholar]
- 23.Zhao W, Fu H, Zhang S, Sun S, Liu Y. LncRNA SNHG16 drives proliferation, migration, and invasion of hemangioma endothelial cell through modulation of miR-520d-3p/STAT3 axis. Cancer Med. 2018;7(7):3311–3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu YZ, Yang H, Cao J, et al. KIAA1522 is a novel prognostic biomarker in patients with non-small cell lung cancer. Sci Rep. 2016;6:24786. doi: 10.1038/srep24786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Wang Y, Fan H, Zhang Z, Li N. miR-125b-5p inhibits breast cancer cell proliferation, migration and invasion by targeting KIAA1522. Biochem Biophys Res Commun. 2018;504(1):277–282. doi: 10.1016/j.bbrc.2018.08.172 [DOI] [PubMed] [Google Scholar]