Abstract
Background
Circular RNAs (circRNAs) play important roles in tumorigenesis, including lung cancer. However, the expression profile and clinical value of circRNAs in lung adenocarcinoma remain unclear. The purpose of this study was to establish the circRNAs expression profile of lung adenocarcinoma and determine its potential diagnostic and prognostic value.
Materials and Methods
The global expression profile of circRNAs in lung adenocarcinoma tissue was determined from five paired lung adenocarcinoma tissues and adjacent normal tissues. The expression levels of selected candidate circRNA were validated by qRT-PCR. Sequence analysis was used to confirm the specificity of amplified circRNA. The candidate circRNA level was further detected in plasma samples from lung adenocarcinoma patients and healthy controls. The relationships between their levels and clinicopathological factors were explored. Receiver operating characteristic (ROC) curve was constructed to differentiate lung adenocarcinoma from healthy controls. Kaplan–Meier was performed to show survival curves and survival characteristics. The significance of different prognostic factors for overall survival (OS) was analyzed using Cox proportional hazards model.
Results
CircRNA microarray showed 394 circRNAs were differentially expressed, including 215 up-regulated and 179 down-regulated circRNAs. Hsa_circ_0001715 was the most up-regulated circRNA in lung adenocarcinoma tissues. Plasma hsa_circ_0001715 levels were significantly higher in lung adenocarcinoma patients versus healthy controls (P < 0.001). We further found that high plasma hsa_circ_0001715 was significantly correlated with TNM stage (P = 0.039) and distant metastasis (P = 0.030). Furthermore, ROC curve analysis showed that hsa_circ_0001715 had high diagnostic value, and the area under the curve (AUC) was 0.871. Lung adenocarcinoma patients with plasma hsa_circ_0001715 levels over 0.417 had significantly shorter OS than those with lower levels (P = 0.004). Univariate and multivariate survival analysis showed that plasma hsa_circ_0001715 level was an independent prognostic factor for the OS.
Conclusion
Our study revealed an aberrant circRNA expression profile in lung adenocarcinoma, and hsa_circ_0001715 is up-regulated and could act as a novel diagnostic and prognostic biomarker for lung adenocarcinoma.
Keywords: circular RNA, hsa_circ_0001715, lung adenocarcinoma tissues, biomarker, prognosis
Introduction
Lung cancer is the most prevalent malignancy globally and also the leading account of cancer-related death worldwide.1 Non-small-cell lung cancer (NSCLC) accounts for 80–85% of all lung malignancies and lung adenocarcinoma more than 40%.2 Despite remarkable advances in targeted therapy and immunotherapy over the recent years,2,3 the majority of lung adenocarcinoma patients have a dismal prognosis due to the high rate of metastasis and recurrence. The 5-year survival rate of NSCLC staggers at 16%, most patients have progressive disease or distant metastasis upon initial diagnosis.4 Therefore, it is essential to identify novel biomarkers to improve the diagnosis and prognosis of lung adenocarcinoma.
CircRNAs are new type of endogenous noncoding RNAs. Unlike traditional linear RNAs, they have a closed ring structure and are not affected by RNases.5 CircRNAs were previously thought to be lowly expressed and byproducts of splicing errors.6 In addition, their biologically active regulators have not been completely identified. However, with the advent of next-generation sequencing, increasing number of circRNAs have been detected and reported to play important physiological functions. CircRNAs are more stable, abundant and conserved.7 Recent studies have revealed that circRNAs can serve as microRNA sponges, RNA-binding proteins or transcriptional regulators.8–10 Emerging evidence has reported that circRNAs play essential roles in cell proliferation, invasion and metastasis, and apoptosis of cancer cells.11–14
In the present study, we analyzed the global circRNAs expression profile in lung adenocarcinoma through microarrays. We found that hsa_circ_0001715 was the most up-regulated circRNA in lung adenocarcinoma tissue. We further found that plasma hsa_circ_0001715 levels were markedly upregulated and associated with clinicopathological features. On the basis of our data, we conclude that hsa_circ_0001715 plays an important role in lung adenocarcinoma and may serve as a novel potential biomarker for lung adenocarcinoma.
Materials and Methods
Acquisition of Tissues and Plasma Specimens
Forty-eight paired lung adenocarcinoma tissues and adjacent normal tissues and plasma samples of 60 treatment-naive lung adenocarcinoma patients and 57 health controls were acquired from Nanjing Chest Hospital between December 2013 and June 2016. All surgical tissue specimens were snap-frozen and plasma samples were collected at the time of diagnosis before any treatment. The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of Nanjing Chest Hospital. All the study subjects provided written informed consent.
CircRNA Microarray Assay
GenElute™ Mammalian Total RNA Miniprep Kit (Sigma, USA) was used to extract total cellular RNA according to the manufacturer’s instructions. NanoDrop ND-1000 was used to quantify RNA. We performed sample preparation and microarray hybridization based on the Arraystar’s standard protocols. Briefly, to remove linear RNAs and enrich circRNAs, we treated total RNAs with RNase R (Epicentre Inc., Madison, USA). Then, we amplified and transcribed the enriched circRNAs into fluorescent cRNA utilizing a random priming technique (Arraystar Super RNA Labeling Kit; Arraystar) as instructed by the manufacturer. We hybridized the labeled cRNAs onto the Arraystar Human circRNA Array (8x15K, Arraystar). After slide wash, we scanned the arrays with the Agilent Scanner G2505C. The acquired array images were analyzed with Agilent Feature Extraction software. We used the R software limma package to perform Quantile normalization and subsequent data processing. Volcano Plot filtering was conducted to identify differentially expressed circRNAs between two samples with statistical significance (fold change ≥2.0 or ≤−2.0, P < 0.05). Hierarchical clustering was performed to distinguish circRNAs expression patterns among samples.
Quantitative Real-Time Reverse Transcriptase-Polymerase Chain Reaction (qRT-PCR)
qRT-PCR was performed to validate the result of circRNA microarray assay and detect plasma circRNA levels. We synthesized the primers by Generay Biotech (Shanghai, China) and used GAPDH as an internal control. The divergent primer sequences were as follows: 5ʹ-AGTTCATCGGGGTGCTCTAC-3ʹ (sense) and 5ʹ-CACCTTGATAGCCTGGCCGA-3ʹ (antisense) for hsa_circ_0001715; 5ʹ-GGACCTGACCTGCCGTCTAG-3ʹ (sense) and 5ʹ- GTAGCCCAGGATGCCCTTGA −3ʹ (antisense) for GAPDH. All experiments were independently detected in triple by investigators in our laboratory. CircRNAs expression levels were obtained using the relative level of 2−∆∆Ct method and represented as fold changes.
Statistical Analysis
In the present study, we presented data as mean ± SD and tested continuous variables through Kolmogorov–Smirnov and Shapiro–Wilk. Normally and non-normally distributed continuous variables were measured through two-tailed Student’s t-tests and Mann–Whitney U-tests, respectively. Chi-square test was used to test categorical variables. ROC curve was performed to measure the diagnostic performance of circRNA. The cutoff was calculated according to Youden’s index. Furthermore, with SPSS Statistics 17.0 (IBM, Armonk, NY, USA) and Graphpad Prism 6.0 software (GraphPad Software Inc., La Jolla, CA, USA), we conducted the ANOVA test and ROC analysis. P< 0.05 was considered statistically significant. Kaplan-Meier method and Log rank test were used to perform survival curves and survival characteristics. The significance of different prognostic factors for OS, including age, gender, tissue differentiation, carcinoembryonic antigen (CEA), tumor size, lymph metastasis, distant metastasis, TNM stage, and plasma hsa_circ_0001715 level, was analyzed using univariate and multivariate Cox proportional hazards model.
Results
Expression Profile of circRNA in Lung Adenocarcinoma Patients
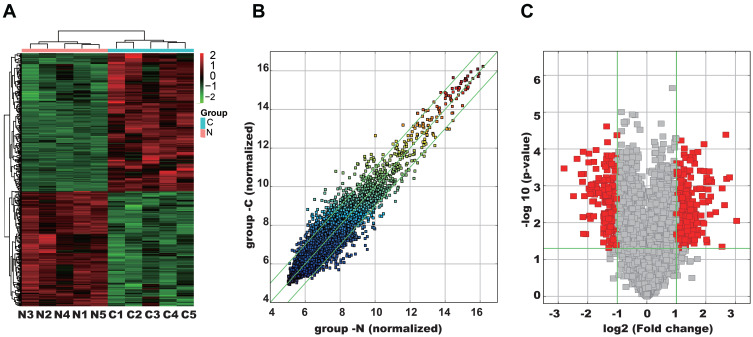
The demographic and clinicopathologic characteristics of patients were shown in Tables 1 and 2. To explore differentially expressed circRNAs in lung adenocarcinoma patients, we performed circRNA microarray assay in 5 paired lung adenocarcinoma tissues and adjacent normal tissue specimens. Hierarchical clustering (Figure 1A) and scatter plot (Figure 1B) showed that circRNAs were variably expressed in lung adenocarcinoma tissues and adjacent normal tissues. Volcano plot filtering (Figure 1C) revealed that 394 circRNAs were differentially expressed (fold-change ≥2.0 and P < 0.05), including 215 up-regulated and 179 down-regulated circRNAs. Among them, hsa_circ_0001715, which is produced at exon 8–11 of the LIMK1 gene locus, was identified to be the most up-regulated circRNA. These results indicate that an aberrant circRNA expression profile in lung adenocarcinoma tissues and hsa_circ_0001715 was filtered for further validation.
Table 1.
The Relationships of Hsa_circ_0001715 in Tissues with the Clinicopathological Factors of Lung Adenocarcinoma Patients
| Characteristics | Number | Mean ± SD | P-value |
|---|---|---|---|
| Age (years) | 0.585 | ||
| ≥65 | 18 | 0.529±0.137 | |
| <65 | 25 | 0.555±0.162 | |
| Gender | 0.753 | ||
| Male | 17 | 0.535±0.130 | |
| Female | 26 | 0.550±0.166 | |
| Differentiation | 0.153 | ||
| Moderate – Well | 19 | 0.507±0.159 | |
| Poor | 24 | 0.574±0.141 | |
| Lymph node metastasis | 0.045* | ||
| N0-N1 | 23 | 0.502±0.160 | |
| N2-N3 | 20 | 0.594±0.127 | |
| Tumor size (cm) | 0.348 | ||
| ≤3 | 17 | 0.517±0.175 | |
| >3 | 26 | 0.562±0.134 | |
| TNM stage | 0.028* | ||
| I–II | 21 | 0.493±0.165 | |
| III–IV | 22 | 0.594±0.121 | |
| Distant metastasis | 0.026* | ||
| Absent | 36 | 0.522±0.146 | |
| Present | 7 | 0.660±0.132 |
Note: *Significant difference.
Abbreviation: SD, standard deviation.
Table 2.
The Relationship of Hsa_circ_0001715 in Plasma with the Clinicopathological Factors of Lung Adenocarcinoma Patients
| Characteristics | Number | Mean ± SD | P-value |
|---|---|---|---|
| Age (years) | 0.175 | ||
| ≥65 | 28 | 0.469±0.084 | |
| <65 | 32 | 0.439±0.081 | |
| Gender | 0.260 | ||
| Male | 33 | 0.444±0.087 | |
| Female | 27 | 0.468±0.078 | |
| Differentiation | 0.143 | ||
| Moderate – Well | 30 | 0.439±0.075 | |
| Poor | 30 | 0.471±0.089 | |
| Tumor size (cm) | 0.101 | ||
| ≤3 | 23 | 0.432±0.090 | |
| >3 | 37 | 0.469±0.077 | |
| Lymph node metastasis | 0.056 | ||
| N0-N1 | 35 | 0.437±0.081 | |
| N2-N3 | 25 | 0.479±0.082 | |
| Distant metastasis | 0.030* | ||
| Absent | 44 | 0.440±0.078 | |
| Present | 16 | 0.496±0.086 | |
| TNM stage | 0.039* | ||
| I–II | 31 | 0.433±0.082 | |
| III–IV | 29 | 0.478±0.080 | |
| CEA | 0.022* | ||
| Negative | 34 | 0.433±0.077 | |
| Positive | 26 | 0.482±0.084 |
Note: *Significant difference.
Abbreviations: SD, standard deviation; CEA, carcinoembryonic antigen.
Figure 1.
Analysis of differentially expressed circRNAs in five paired lung adenocarcinoma tissues and adjacent normal tissues. (A) Hierarchical clustering results of circRNA expression profiles between the lung adenocarcinoma tissues and matched normal tissues. The result from unsupervised hierarchical clustering analysis reveals distinguishable circRNAs expression profiling among specimens. Red: higher expression level, green: lower expression level. (B) CircRNAs in the scatter plot above the top green line and below the bottom green line indicate change of circRNAs between the two groups (P< 0.05). (C) Volcano plot of circRNA expression profile. The vertical lines correspond to 2.0-fold upregulation and downregulation, and the horizontal line represents a P-value of 0.05. The red point in the plot represents the differentially expressed circRNAs with statistical significance.
Abbreviations: N, adjacent normal tissues; C, lung adenocarcinoma tissues.
Validation of the Specificity of Amplified Hsa_circ_0001715
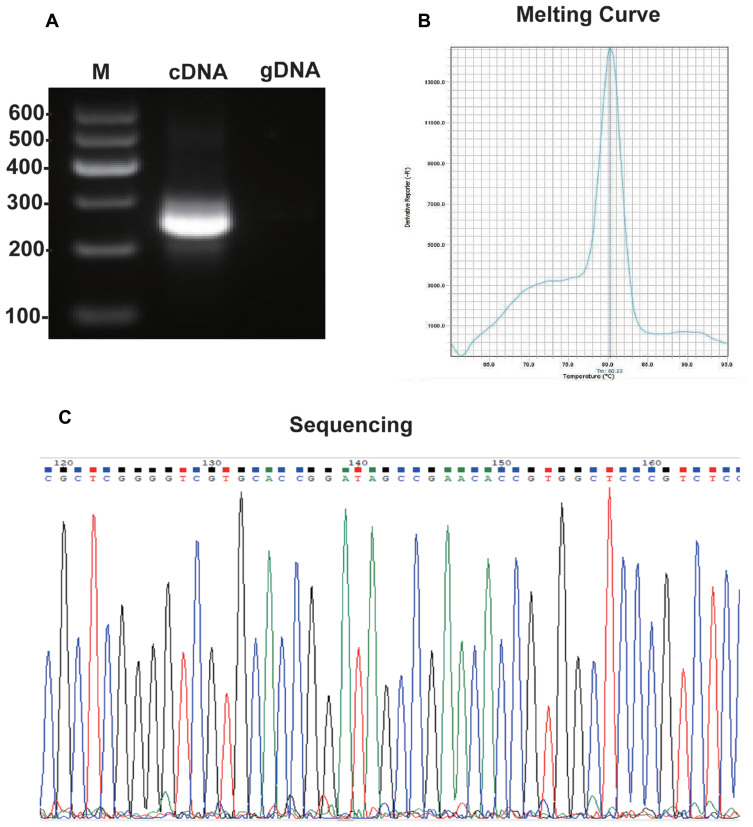
To determine the specificity of hsa_circ_0001715, we amplified hsa_circ_0001715 by qRT-PCR using divergent primers. Agarose electrophoresis revealed a fragment with the expected size of the circRNA fragment (Figure 2A). In addition, melting curve analysis yielded a single peak (Figure 2B). Sequence analysis of the amplified product showed that the sequence of hsa_circ_0001715 was consistent with circBase (Figure 2C). These results show that hsa_circ_0001715 is present in lung adenocarcinoma tissues and could be amplified by qRT-PCR.
Figure 2.
The specificity of amplified hsa_circ_0001715 with divergent primers. (A) Electrophoresis of qRT-PCR product of hsa_circ_0001715. (B) Melting curve analysis of qRT-PCR product of hsa_circ_0001715. (C) Sequencing the amplified product of hsa_circ_0001715.
Upregulated Hsa_circ_0001715 Expression in Lung Adenocarcinoma Tissues
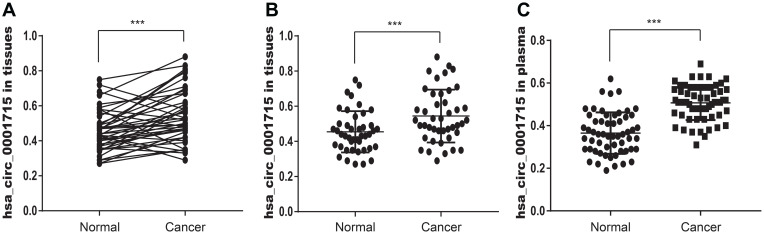
To validate the results of circRNA microarray, we examined the expression of candidate circRNA by qRT-PCR in the other forty-three paired lung adenocarcinoma tissues and adjacent normal tissues. The expression levels of hsa_circ_0001715 in paired lung adenocarcinoma tissues and adjacent normal tissues are shown in Figure 3A. The expression levels of hsa_circ_0001715 in lung adenocarcinoma tissues were significantly higher than those in matched adjacent normal tissues (0.545 ± 0.151 vs 0.454 ± 0.118, P< 0.001, Figure 3B). These results indicate hsa_circ_0001715 is upregulated in lung adenocarcinoma tissues.
Figure 3.
Relative hsa_circ_0001715 expression in lung adenocarcinoma tissues and plasma. (A) The expression levels of hsa_circ_0001715 in each comparison between lung adenocarcinoma and matched-adjacent normal tissue (n = 43). (B) The expression levels of hsa_circ_0001715 in lung adenocarcinoma tissues relative to matched-adjacent normal tissues. (C) The plasma hsa_circ_0001715 levels were significantly higher compared to healthy controls. (***P < 0.001).
Plasma Hsa_circ_0001715 is Elevated in Lung Adenocarcinoma Patients
We further examined plasma hsa_circ_0001715 levels in lung adenocarcinoma patients and healthy controls. Our qRT-PCR assays showed that plasma hsa_circ_0001715 levels were significantly higher in lung adenocarcinoma patients than those in healthy controls (0.455 ± 0.083 vs 0.310 ± 0.096, P< 0.001, Figure 3C). These results indicate the expression of plasma hsa_circ_0001715 is elevated in lung adenocarcinoma patients.
Relationships Between Tissue or Plasma Hsa_circ_0001715 Levels and Clinicopathologic Variables of Lung Adenocarcinoma Patients
We further investigated the association between the expression of hsa_circ_0001715 in lung adenocarcinoma tissues and plasma and clinicopathologic variables of lung adenocarcinoma patients. As shown in Table 1, higher hsa_circ_0001715 levels were observed in patients with N2-N3 versus N0-N1 lung adenocarcinoma (P = 0.045), patients with stage III–IV versus stage I–II lung adenocarcinoma (P = 0.028), and patients with distant metastasis versus no distant metastasis (P = 0.026). However, no statistically significant correlation was found between the expression of hsa_circ_0001715 and other clinical factors, such as age (P = 0.585), gender (P = 0.753), differentiation (P = 0.153) and tumor size (P = 0.348). These results suggest that lymph node metastasis, TNM stage and distant metastasis are significantly associated with hsa_circ_0001715 expression levels in lung adenocarcinoma tissues.
As shown in Table 2, higher plasma hsa_circ_0001715 levels were observed in patients with CEA positive versus CEA negative (P = 0.022), patients with TNM stage III–IV versus stage I–II patients (P = 0.039), patients with distant metastasis versus no metastasis (P = 0.030). However, we found no association between plasma hsa_circ_0001715 and other clinicopathological factors, such as age (P = 0.175), gender (P = 0.260), differentiation (P = 0.143), tumor size (P = 0.101) and lymph node metastasis (P = 0.056). Taken together, these results suggest plasma hsa_circ_0001715 levels are significantly associated with plasma CEA, TNM stage and distant metastasis.
Plasma Hsa_circ_0001715 Acted as a Diagnostic Biomarker of Lung Adenocarcinoma
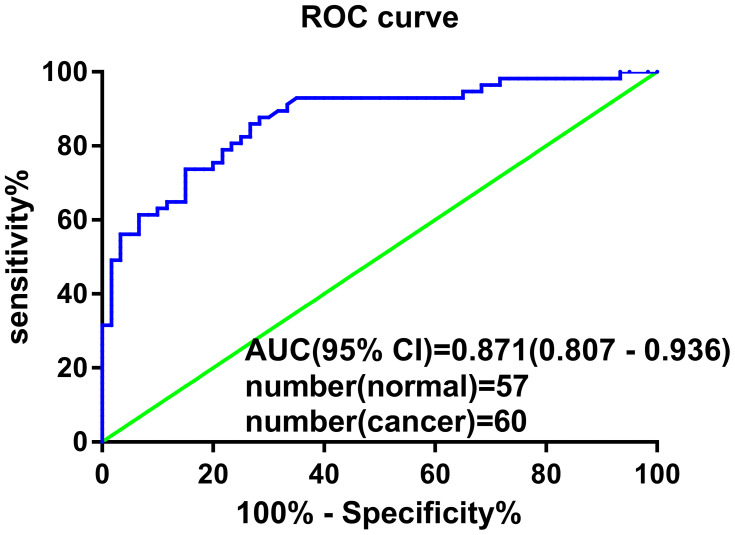
To investigate the diagnostic value of plasma hsa_circ_0001715, we performed ROC curve analysis. Hsa_circ_0001715 had an AUC value of 0.871 (95% CI: 0.807–0.936) (Figure 4). At a cutoff of 0.417, plasma hsa_circ_0001715 had a sensitivity, specificity and accuracy of 87.72%, 71.67% and 79.49%. The positive predictive value was 86.00% and the negative predictive value was 74.63%. These findings indicate that plasma hsa_circ_0001715 could be a promising biomarker for the diagnosis of lung adenocarcinoma.
Figure 4.
ROC curve of plasma hsa_circ_0001715. Plasma hsa_circ_0001715 had an AUC value of 0.871 (95% CI: 0.807–0.936) to discriminate patients with lung adenocarcinoma from healthy controls. With a cutoff of 7.58, the sensitivity, specificity and accuracy were 87.72%, 71.67% and 79.49%, respectively.
Abbreviation: ROC, receiver operating characteristic.
Prognostic Significance of Plasma Hsa_circ_0001715 for Lung Adenocarcinoma
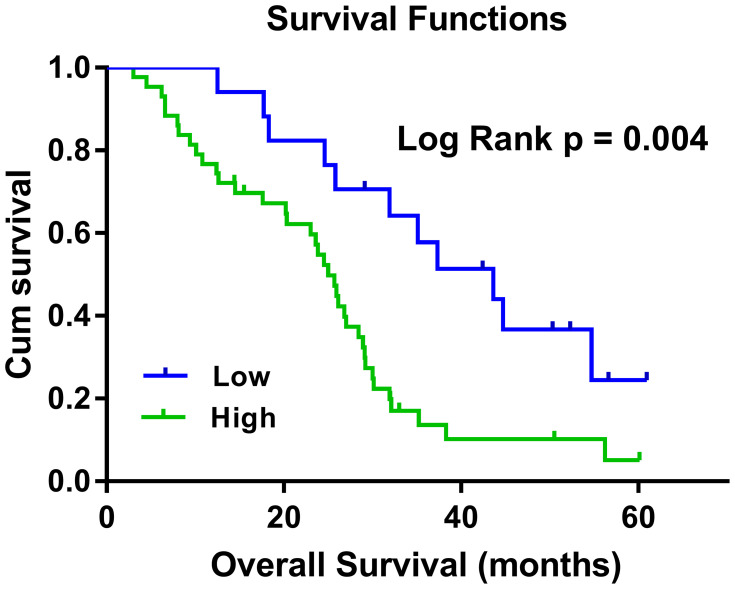
To evaluate the prognostic value of plasma hsa_circ_0001715, lung adenocarcinoma patients were divided into low- and high-concentration of plasma hsa_circ_0001715 groups in accordance with the optimal cutoff value (0.417). Using Kaplan-Meier curves and Log rank test, the OS of lung adenocarcinoma patients with high-level of plasma hsa_circ_0001715 was significantly shorter than those with low-level of hsa_circ_0001715 (24.88 months vs 40.56 months, P = 0.004) (Figure 5). According to univariate analysis (Table 3), OS was associated with tissue differentiation (P = 0.041), lymph node metastasis (P = 0.038), distant metastasis (P = 0.001), TNM stage (P = 0.005) and plasma hsa_circ_0001715 (P = 0.014), but not with age (P = 0.689), gender (P = 0.348), CEA (P = 0.062) and tumor size (P = 0.581). Moreover, to identify the prognostic value of plasma hsa_circ_0001715, we used the Cox proportional hazards model to perform multivariate analysis of prognostic factors. Multivariate Cox regression analysis revealed that plasma hsa_circ_0001715 level (HR 0.807, 95% CI, 0.663–0.982, P = 0.032) and distant metastasis status (HR 2.418, 95% CI, 1.063–5.499, P = 0.035) were independent prognostic biomarkers for OS of lung adenocarcinoma (Table 3). On the whole, our results indicate that hsa_circ_0001715 may act as a novel tumor biomarker for the prognosis of patients with lung adenocarcinoma.
Figure 5.
Kaplan–Meier survival curves for OS of lung adenocarcinoma patients with low- and high-concentration of plasma hsa_circ_0001715. Lung adenocarcinoma patients with high plasma hsa_circ_0001715 expression had a shorter OS than those with low-level of hsa_circ_0001715 (24.88 months vs 40.56 months, P = 0.004).
Table 3.
Univariate and Multivariate Cox Proportional Hazards Analysis of Prognostic Variables for Overall Survival in Lung Adenocarcinoma Patients
| Parameters | Categories | Univariate Analysis HR (95% CI) | P-value | Multivariate Analysis HR (95% CI) | P-value |
|---|---|---|---|---|---|
| Age | <65 vs ≥65 | 0.890 (0.503–1.575) | 0.689 | ||
| Gender | Male vs female | 1.321 (0.739–2.364) | 0.348 | ||
| Tissue differentiation | Poor vs middle-high | 1.893 (1.025–3.498) | 0.041* | 1.303 (0.659–2.575) | 0.447 |
| Tumor size (cm) | >3 vs ≤3 | 1.179 (0.656–2.119) | 0.581 | ||
| Lymph node metastasis | N2-3 vs N0-1 | 1.889 (1.035–3.447) | 0.038* | 0.507 (0.165 −1.559) | 0.236 |
| Distant metastasis | Present vs Absent | 3.038 (1.590–5.804) | 0.001* | 2.418 (1.063–5.499) | 0.035* |
| TNM stage | I–II vs III–IV | 2.391 (1.297–4.408) | 0.005* | 2.393 (0.736–7.783) | 0.147 |
| CEA | Positive vs negative | 1.721 (0.874–3.042) | 0.062 | ||
| hsa_circ_0001715 | High vs Low | 0.778 (0.637–0.950) | 0.014* | 0.807 (0.663–0.982) | 0.032* |
Note: *Significant difference.
Abbreviations: HR, hazard ratio; CI, confidence interval; CEA, carcinoembryonic antigen.
Discussion
CircRNAs are novel class of endogenous noncoding RNAs and characterized by covalently closed continuous loop with back-splicing joined 3′- and 5′-ends.15 CircRNAs were first observed in RNA virus decades ago, but were mistaken as by-products of splicing errors.15,16 With the development of high-throughput RNA sequencing and bioinformatic approaches, many circRNAs were identified as crucial layers of gene regulators.9,17 Increasing evidence proved that circRNAs are implicated in tumor development and can act as microRNA sponge to compete with endogenous RNA. For example, Memczak et al reported that ciRS-7 can sponge miR-7 to sequester and suppress miR-7 activity, and promote overexpression of miR-7 targets, resulting in initiation and development of cancer.18 CircRNAs are widely expressed, highly conserved in sequence, specific expression in tissues and more stable in the presence of RNA exonucleases.19 All these characteristics make circRNA in tumor tissue or peripheral blood an ideal biomarker for diagnosis and prognosis of different tumors.20,21
In the present study, we firstly used 5 paired lung adenocarcinoma and adjacent normal tissue specimens to systematically characterize the circRNA expression profile by microarray analysis. Hsa_circ_0001715 was identified to be the most up-regulated circRNA and the result was validated by qRT-PCR in 43 paired additional lung adenocarcinoma and adjacent normal tissues. Therefore, we selected hsa_circ_0001715 as the candidate circRNA for further study.
Our results indicated that hsa_circ_0001715 was significantly up-regulated in lung adenocarcinoma tissues and plasma, and it was positively correlated with TNM stage and distant metastasis. It is of note that increasing TNM stage and present of distant metastasis are associated with the decreased survival time in lung cancer patients, as well as they both are independent prognostic factors for lung cancer,22–24 we speculate that hsa_circ_0001715 play a role in promoting tumor metastasis and may be served as a valuable marker for predicting tumor progression in patients with lung adenocarcinoma.
Increasing evidence proved that plasma circRNAs can be used as efficient molecular biomarkers for the diagnosis of cancer. For instance, Yin et al revealed that plasma hsa_circ_0001785 had better diagnostic accuracy for breast cancer than carbohydrate antigen 153 (CA153) and CEA in breast cancer.25 Qiao et al have demonstrated that plasma hsa_circ_0003998 can be used as an efficient molecular biomarker for hepatocellular carcinoma diagnosis and the combination of hsa_circ_0003988 and AFP can improve sensitivity and specificity.26 Switching to the field of NSCLC, Hang et al reported that the expression of plasma circFARSA was upregulated in NSCLC patients than those in healthy controls and the AUC value was 0.71.27 To investigate the diagnostic value of plasma hsa_circ_0001715 for lung adenocarcinoma, we performed ROC curve analysis in lung adenocarcinoma patients and healthy controls. As shown in Figure 4, an AUC value for plasma hsa_circ_0001715 reached 0.871. Using a cutoff of 7.58, the sensitivity, specificity and accuracy of hsa_circ_0001715 in diagnosis were 87.72%, 71.67% and 79.49%, respectively. These results indicate that plasma hsa_circ_0001715 could be a promising biomarker for the diagnosis of lung adenocarcinoma.
Furthermore, circRNAs have been shown to provide a potential prognostic indicator for survival in many tumors. For example, CircYAP1 in gastric cancer,28 hsa_circ_0078602 in hepatocellular carcinoma,29 and ciRS-7 in colorectal cancer.30 The prognostic value of plasma hsa_circ_0001715 in lung adenocarcinoma patients remains unknown. Using Kaplan-Meier curves and Log rank test, plasma hsa_circ_0001715 level in lung adenocarcinoma patients was conversely correlated with patients’ OS. In other words, compared to low plasma hsa_circ_0001715 levels, lung adenocarcinoma patients with high level had shorter survival time (24.88 months vs 40.56 months, P = 0.004). Multivariate Cox regression analysis revealed that plasma hsa_circ_0001715 level (P = 0.032) and distant metastasis status (P = 0.035) were independent prognostic factors for OS of lung adenocarcinoma.
Our study had some limitations. First, the sample size enrolled in our study is relatively small. Second, when we detect the expression of circRNAs in plasma, we should spike exogenous RNA into the plasma samples before RNA extraction to serve as a reference, the detection of hsa_circ_0001715 in plasma will be more credible. Furthermore, we all know that joint detection can improve the sensitivity and specificity of diagnosis. In the further study, we will validate more other differential expression circRNAs and investigate the value of joint detection of hsa_circ_0001715 and other circRNAs in lung adenocarcinoma.
In conclusion, hsa_circ_0001715 is up-regulated in lung adenocarcinoma tissues and plasma, and its expression level is associated with TNM stage and distant metastasis. Our results reveal that plasma hsa_circ_0001715 may be utilized as a potential non-invasive diagnostic biomarker for lung adenocarcinoma with a favorable degree of sensitivity, specificity and accuracy. Furthermore, plasma hsa_circ_0001715 can serve as a prognostic predictor for lung adenocarcinoma and a high plasma hsa_circ_0001715 is correlated with poor prognosis. Additional studies are necessary to confirm the specific molecular mechanisms of hsa_circ_0001715 in lung adenocarcinoma metastasis.
Funding Statement
This work was supported by “Thirteen-Five Plan” the Major Program of Nanjing Medical Science and Technique Development Foundation (grant number ZDX16012).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Smith RA, Cokkinides V, Brawley OW. Cancer screening in the United States, 2009: a review of current American Cancer Society guidelines and issues in cancer screening. CA Cancer J Clin. 2009;59:27–41. doi: 10.3322/caac.20008 [DOI] [PubMed] [Google Scholar]
- 3.Gettinger S, Horn L, Jackman D, et al. Five-year follow-up of nivolumab in previously treated advanced non-small-cell lung cancer: results from the CA209-003 study. J Clin Oncol. 2018;36:1675–1684. doi: 10.1200/JCO.2017.77.0412 [DOI] [PubMed] [Google Scholar]
- 4.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073 [DOI] [PubMed] [Google Scholar]
- 5.Suzuki H, Tsukahara T. A view of pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci. 2014;15:9331–9342. doi: 10.3390/ijms15069331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7:155–160. doi: 10.1096/fasebj.7.1.7678559 [DOI] [PubMed] [Google Scholar]
- 7.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7(2):e30733. doi: 10.1371/journal.pone.0030733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019 [DOI] [PubMed] [Google Scholar]
- 9.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017 [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, Liu H, Hou L, et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. 2017;16:151. doi: 10.1186/s12943-017-0719-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang M, Yu F, Li P. Circular RNAs: characteristics, function and clinical significance in hepatocellular carcinoma. Cancers. 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao-Long M, Kun-Peng Z, Chun-Lin Z. Circular RNA circ_HIPK3 is down-regulated and suppresses cell proliferation, migration and invasion in osteosarcoma. J Cancer. 2018;9:1856–1862. doi: 10.7150/jca.24619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, Lu C, Zhou Y, Zhang Z, Sun L. Circular RNA hsa_circ_0008039 promotes breast cancer cell proliferation and migration by regulating miR-432-5p/E2F3 axis. Biochem Biophys Res Commun. 2018;502(3):358–363. doi: 10.1016/j.bbrc.2018.05.166 [DOI] [PubMed] [Google Scholar]
- 15.Mercer TR, Clark MB, Andersen SB, et al. Genome-wide discovery of human splicing branchpoints. Genome Res. 2015;25:290–303. doi: 10.1101/gr.182899.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Danan M, Schwartz S, Edelheit S, Sorek R. Transcriptome-wide discovery of circular RNAs in archaea. Nucleic Acids Res. 2012;40:3131–3142. doi: 10.1093/nar/gkr1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao MS, Ai Y, Wilusz JE. Biogenesis and functions of circular RNAs come into focus. Trends Cell Biol. 2020;30:226–240. doi: 10.1016/j.tcb.2019.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 19.Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA (New York, NY). 2013;19:141–157. doi: 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen B, Huang S. Circular RNA: an emerging non-coding RNA as a regulator and biomarker in cancer. Cancer Lett. 2018;418:41–50. doi: 10.1016/j.canlet.2018.01.011 [DOI] [PubMed] [Google Scholar]
- 21.Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. Jbiotechnol. 2016;238:42–51. doi: 10.1016/j.jbiotec.2016.09.011 [DOI] [PubMed] [Google Scholar]
- 22.Hwang JK, Page BJ, Flynn D, et al. Validation of the eighth edition TNM lung cancer staging system; a brief report. J Thorac Oncol. 2020;15:649–654. doi: 10.1016/j.jtho.2019.11.030 [DOI] [PubMed] [Google Scholar]
- 23.Luo J, Chen YJ, Narsavage GL, Ducatman A. Predictors of survival in patients with non-small cell lung cancer. Oncol Nurs Forum. 2012;39:609–616. [DOI] [PubMed] [Google Scholar]
- 24.Wao H, Mhaskar R, Kumar A, Miladinovic B, Djulbegovic B. Survival of patients with non-small cell lung cancer without treatment: a systematic review and meta-analysis. Syst Rev. 2013;2:10. doi: 10.1186/2046-4053-2-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yin W-B, Yan M-G, Fang X, Guo -J-J, Xiong W, Zhang R-P. Circulating circular RNA hsa_circ_0001785 acts as a diagnostic biomarker for breast cancer detection. Clin Chimica Acta. 2017;487:363–368. doi: 10.1016/j.cca.2017.10.011 [DOI] [PubMed] [Google Scholar]
- 26.Qiao GL, Chen L, Jiang WH, et al. Hsa_circ_0003998 may be used as a new biomarker for the diagnosis and prognosis of hepatocellular carcinoma. Onco Targets Ther. 2019;12:5849–5860. doi: 10.2147/OTT.S210363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hang D, Zhou J, Qin N, et al. A novel plasma circular RNA circFARSA is a potential biomarker for non-small cell lung cancer. Cancer Med. 2018;7:2783–2791. doi: 10.1002/cam4.1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu H, Liu Y, Bian Z, et al. Circular RNA YAP1 inhibits the proliferation and invasion of gastric cancer cells by regulating the miR-367-5p/p27 (Kip1) axis. Mol Cancer. 2018;17:151. doi: 10.1186/s12943-018-0902-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kou P, Zhang C, Lin J, Wang H. Circular RNA hsa_circ_0078602 may have potential as a prognostic biomarker for patients with hepatocellular carcinoma. Oncol Lett. 2019;17:2091–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weng W, Wei Q, Toden S, et al. Circular RNA ciRS-7-A promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin Cancer Res. 2017;23:3918–3928. doi: 10.1158/1078-0432.CCR-16-2541 [DOI] [PMC free article] [PubMed] [Google Scholar]