Abstract
The faithful segregation of duplicated sister chromatids rely on the remarkable ability of kinetochores to sustain stable load bearing attachments with the dynamic plus ends of kinetochore microtubules (kMTs). The outer layer of the kinetochore recruits several motor and non-motor microtubule-associated proteins (MAPs) that help the kinetochores establish and maintain a load bearing dynamic attachment with kMTs. The primary kinetochore microtubule-binding protein, the Ndc80 complex (Ndc80c), which is highly conserved among diverse organisms from yeast to humans, performs this essential function with assistance from other MAPs. These MAPs are not an integral part of the kinetochore, but they localize to the kinetochore periodically throughout mitosis and regulate the strength of the kinetochore microtubule attachments. Here, we attempt to summarize the recent advances that have been made towards furthering our understanding of this co-operation between the Ndc80c and these MAPs, focusing on the Ska1 complex (Ska1c) and Cdt1 in humans.
Keywords: Microtubules, Kinetochores, Mitosis, Chromosomes, MAPs, Ndc80
Introduction
Kinetochores are mega-protein assemblies formed at the centromeres of chromosomes during mitosis. At the onset of mitosis after the nuclear envelope breakdown, kinetochores embedded within the centromeric region of duplicated sister chromatids initiate attachment to spindle microtubules to drive accurate chromosome segregation (1, 2). Pair of sister chromatids acquire spindle microtubules from opposite spindle poles to form proper bi-oriented attachments during their congression at metaphase. At metaphase, the kinetochores from all sister chromatids acquire end-on attachments with the kMTs. This dynamic attachment is maintained since their inception at prometaphase till the separation of the sister chromatids at anaphase, where they move poleward by harnessing the force of microtubule depolymerization. Until chromosomes form proper bi-oriented attachments, a signaling mechanism called the Spindle Assembly Checkpoint (SAC) remains activated, delaying mitotic exit. Upon the formation of error free kMT attachments, this mitotic surveillance signal turns off allowing the progression of cell cycle from metaphase to anaphase (3, 4). The outer domain of the kinetochore comprising of several MAPs and their binding partners serve as the key interface for kMTs. The Ndc80c, along with a group of proteins that could collectively be referred to as non-motor kinetochore MAPs, are central to this MT-binding function. These human proteins such as Ska1c, Cdt1, chTOG and Astrin-SKAP, along with their homologues have been found to function in concert with the Ndc80c and act as additional couplers of end-on kMT attachments (5, 6).
In this mini-review, we primarily aim to present an overview of the Ska1c and Cdt1 (and their functional yeast modalities) function coupled with the Ndc80c. We direct our readers to recent reviews discussing the role of other MAPs at the kMT interface (5, 6). Additionally, we focus on the mechanism by which these MAPs are targeted to the kMT interface via the Ndc80c.
The Ndc80 complex and the KMN network
During metaphase, the formation of robust kMT attachments or “end-on” kMT attachments are mediated by a highly conserved network of protein complexes referred to as the KMN network (stands for Knl1, Mis12, and Ndc80 complexes) located at mitotic kinetochores. (2, 7). The Ndc80c, an elongated hetero-tetrameric dumbbell-shaped complex composed of four subunits - Hec1, Nuf2, Spc24 and Spc25, is the key component of the KMN network that contributes to robust MT-binding by kinetochores (8, 9). The globular N-terminal regions of Hec1/Nuf2 fold into two Calponin-homology (CH) domains, which serve as the essential MT-binding domains of the Ndc80c (9–12). The ~80 amino acid N-terminal region of the Hec1 subunit composed of intrinsically unstructured and highly positively charged residues has also been shown to be required for high affinity MT-binding (11, 13–16). It has been reported that the Knl1 complex (comprising of Knl1 and Zwint1) of the KMN network is important to form robust kMT attachments, though its precise contribution to this function remains unclear (2, 7). The Ndc80 and Knl1 complexes are targeted to kinetochores by the Mis12 complex. The components of the KMN network are linked to the inner kinetochore domains via interactions with the CENP-C and CENP-T components (Fig. 1A, 1B) of another large network of proteins complexes called the Constitutive Centromere Associated Network (CCAN) (17).
Figure 1. Mechanisms of kinetochore-microtubule coupling in Yeast (1A) and Humans (1B) as summarized in this review.
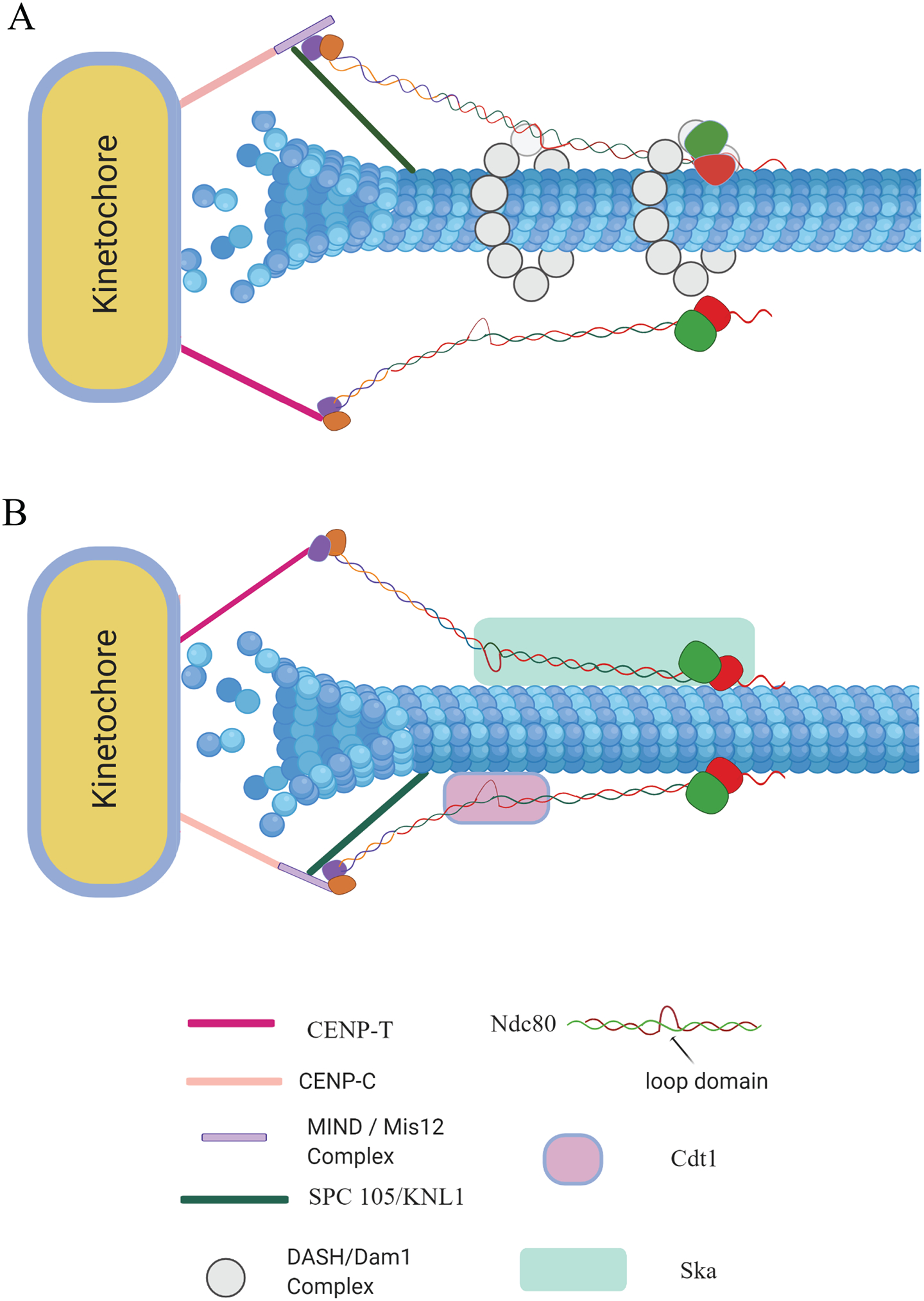
Each model is distinguished by the microtubule‐associated proteins (MAPs) assisting the Ndc80c in the kMT coupling process (1A and 1B). Regions of interaction relevant for each MAP to localize to kinetochores by binding to Ndc80c is also depicted. Proteins or protein complexes are drawn in arbitrary shapes for the purpose of depiction only and are not to a precise scale. The Dam1 ring complex in yeast binds to MT lattice/plus-ends and localizes to the kinetochore via multiple interactions with Ndc80c as described in the main manuscript (1A). Binding of the human Ska1c to kinetochores is also thought to require multiple interaction with the Ndc80c, including those with the loop domain, again, as outlined in the manuscript (1B). As with the Dam1c, Ska1c has been shown to bind both to the MT lattice and to the plus-ends. Human Cdt1 binds MTs directly and its recruitment to kinetochores is dependent on the loop domain of the Ndc80c (1B). The Mis12 complex and the proteins of the CCAN including CENP-C and CENP-T, which are important to localize the Ndc80c to outer kinetochores are indicated with the same colour in both models. These models are not meant to depict that the Ndc80c recruited to kinetochores via a particular component of CCAN (i.e. CENP-C or CENP-T) exhibit any sort of preference for binding to Ska1c or Cdt1. Any such representation is solely meant for the purpose of ease in illustration.
The central region of the Ndc80c consists of dimeric coiled-coil regions of Hec1/Nuf2 and Spc24/Spc25. This complex has a kink or bend in its structure which is thought to be a result of a break in registry of the coil within the Hec1 subunit (18). This kinked region, also referred to as the loop region, may have a unique role in the formation of end-on kMT attachments even though the loop does not seem to impart direct MT-binding activity by itself (19–21). This internal loop is thought to introduce some flexibility into the otherwise rigid coiled-coil region of the complex. Nevertheless, it is still unclear how this motif contributes to the formation of end-on kMT attachments.
Past research in both yeast and higher eukaryotes have provided evidence supporting the notion that the different regions of the Ndc80c including the loop, the coiled-coil, and the CHD/hairpin domain serve as a protein-protein interaction motif that aids in the recruitment of MAPs to kinetochores. The Dam1/DASH complex along with the Dis1 protein and its homologues in yeast, and the Ska1c along with Cdt1 in humans, are all significant MT-binding factors that have been shown to be recruited to kinetochores with the help of the above-mentioned Ndc80c domains [reviewed in Amin et al, 2019 (5)]. Once recruited to kinetochores, these proteins or complexes assist the Ndc80c in forming end-on attachments of optimal strength. In the following sections, we will describe mechanisms mediated by some of these human MAPs including the Ska1c, and Cdt1, and the yeast Dam1 complex that enable coupling of the Ndc80c to dynamic MT plus-ends, which are in a state of flux between assembly and disassembly.
The Dam1/DASH complex
The Dam1/DASHc is a heterodecameric MAP that is found only in fungi and has been shown to tether kinetochores at the plus-ends of spindle MTs (22). The ten-subunit complex consists of Dam1, the key MT binding component of the complex (23), along with nine other subunits; Duo1, Dad1, Dad2, Spc19, Spc34, Ask1, Dad3, Dad4 and Hsk3 (24). It is interesting to note that the Dam1/DASHc is essential for survival in budding yeast, S. cerevisiae, while it is nonessential in fission yeast, S. pombe (25). Mutations in the Dam1/DASH subunits destabilize the kMT attachments and cause spindle disassembly in yeast (26). The Dam1/DASH complexes form stable, closed, ring-like structures around MTs in vitro and are thought to be capable of passive sliding on the MT lattice (Fig. 1A) (27, 28). Cryo-EM structural data from Jenni et al. (29) together with chemical crosslinking experiments from Kim et al. (30) showed that the Dam1/DASHc in C. thermophilum forms rigid ring like structure with flexible extensions that interact with MT and the Ndc80c. Furthermore, Jenni et al. showed that a single Dam1c ring can make necessary contacts with the Ndc80c (29) whereas Kim et al. reported the bridging of two Dam1c rings by the Ndc80c (30).
It is this ability to form rings around MTs after binding preferentially to GTP-tubulin at the plus-ends of dynamic MTs that distinguishes the Dam1/DASHc from many other kinetochore MAPs. It has been shown that the Dam1c complex, unlike the Ndc80c, has an inherent MT plus end-tracking ability. The Dam1c was also shown to influence efficient association of the Ndc80c with dynamic MT plus-ends (31, 32). Flexible extensions from the Dam1/DASHc rings that make multiple weak contact site with binding partners such as the Ndc80c and imparts the load-bearing complex the ability to stochastically detach and reattach, thus facilitating an efficient coupling with the dynamic MT plus end by hand-over hand biased diffusion of the load bearing complex (29, 30).
This supports the notion that the Dam1c is the main source of kinetochore plus end-tracking activity in yeast, while the Ndc80c serve as a structural bridge between MT ends and chromosomes (31). Moreover, the kinetochore recruitment and assembly of the Dam1c requires the Ndc80c (Fig. 1A) (33). Yeast-two hybrid analysis suggest that the Ndc80c loop region is essential for the interaction between the Ndc80c and the Dam1c to facilitate proper loading of the Dam1c onto kinetochores (Fig. 1A) (34). However, this study also found that the Dam1c-Ndc80c interaction does not require the loop in vitro, suggesting loop mutagenesis indirectly affects the Ndc80c-Dam1c interaction, possibly via other proteins. Lampert et al. demonstrated that deletion of the hairpin region of the Ndc80c abrogates the Ndc80c-Dam1c interaction in vitro (35), which is consistent with the observations of the Dam1c-Ndc80c localization by EM of yeast kinetochore particles (36) and cross-linking mass spectrometry studies (30). Localization and two-hybrid studies have yielded evidence supporting a direct interaction between these two complexes (37, 38). It has been demonstrated that the Aurora B kinase homologue in yeast, Ipl1, phosphorylates the Dam1c to reduce its ability to bind to the MT lattice (39). The phosphorylation of some subunits of the Dam1c has also been shown to interfere with its binding to the Ndc80c (32, 38).
Recent studies reveal that the Ndc80c serves as a bridge between two Dam1c rings in vitro. Cross-linking experiments showed that the interaction domains of the Dam1c subunits were clustered at three separate regions within the Ndc80c that were spread between the hairpin region and the two coiled-coil regions flanking the loop domain (30). Mutation of key residues within the Ndc80c that is responsible for its interaction with the Dam1c result in unusually oblong Ndc80c molecules that can only bind to one Dam1 ring. These mutations translated into defective kMT attachments, improper kinetochore bi-orientation and erroneous chromosomal segregation in vivo (30). This same study further demonstrated that the specific separation between the two Dam1 rings, maintained by the Ndc80c bridges, is critical for proper cell division (30). How we can reconcile these observations with studies showing a dependence of the Dam1c on the Ndc80c loop domain for kinetochore targeting remains an open question.
Since kinetochores of higher eukaryotes are coupled to multiple microtubules that form a bundle as opposed to single microtubule in yeast kinetochores, it necessitates a different mechanism of action for the MT plus end-coupling complex. Various cell biological (40), biochemical (41), and evolutionary (42) analysis suggested that the Ska1c in higher eukaryotes imparts a functional output and also exhibits Ndc80c-dependent kinetochore localization akin to the yeast Dam1c.
The Ska1 complex
The spindle and kinetochore-associated1 (Ska1) complex is a heterotrimeric protein complex formed between three protein subunits (SKA1, SKA2, and SKA3 - also called Rama1). These subunits are paralogs which interact through N terminal coiled-coil regions. The Ska1c further interacts and oligomerizes into a dimer of trimeric Ska1c. The Ska1c is found to localize to spindle microtubules and get enriched selectively at the kinetochores of aligned chromosomes (40, 41, 43, 44). The SKA1 subunit of the Ska1c has also been shown to be involved in SAC silencing (45, 46) as it recruits PP1 phosphatase to facilitate mitotic exit (47, 48). The core of the Ska1c is a W-shaped structure formed by the coil-rich interaction domains of SKA1, SKA2, and SKA3 (49). The N-terminal region of SKA1 facilitates the interaction between SKA2 and SKA3, while the MT-binding domain of the complex resides at the SKA1 C-terminal region. The SKA2/SKA3 subunits thus serve as the structural scaffold for the dimerization interface (49). Although the C-terminal region of the SKA3 subunit has an additional MT-binding property (40), it is not clear precisely how this subunit contributes to the MT-binding activity of the complex. In this context it is worth mentioning that deletion or point mutants of the SKA3 C-terminal substantially reduces the MT-binding affinity of the Ska1c and causes delays in progression of mitotic cells (50).
The critical MT-binding domain of SKA1 adopts a characteristic winged-turn-helix (WTH) fold (49, 51) that has traditionally been recognized as a DNA-binding domain but can also facilitate protein-protein interactions (52, 53). The WTH domain is usually ~110 amino acid consisting of two loops - W1 and W2, three α helices - H1, H2, and H3, and three β-sheets - S1, S2, and S3. These structural components are arranged in the sequence H1-S1-H2-H3-S2-W1-S3-W2 (52) to form a compact α/β structural core (Fig. 2A). The WTH domain of SKA1 has two additional structural modules, apart from the canonical core elements. - Two α helices - α1 and α2 constitute the module I at the N-terminal while three α helices - α4, α5 and α6 constitute the module II in the middle, (Fig. 2B) (51).
Figure 2. Structural comparison of Cdt1 and SKA1 subunit of the Ska1c.
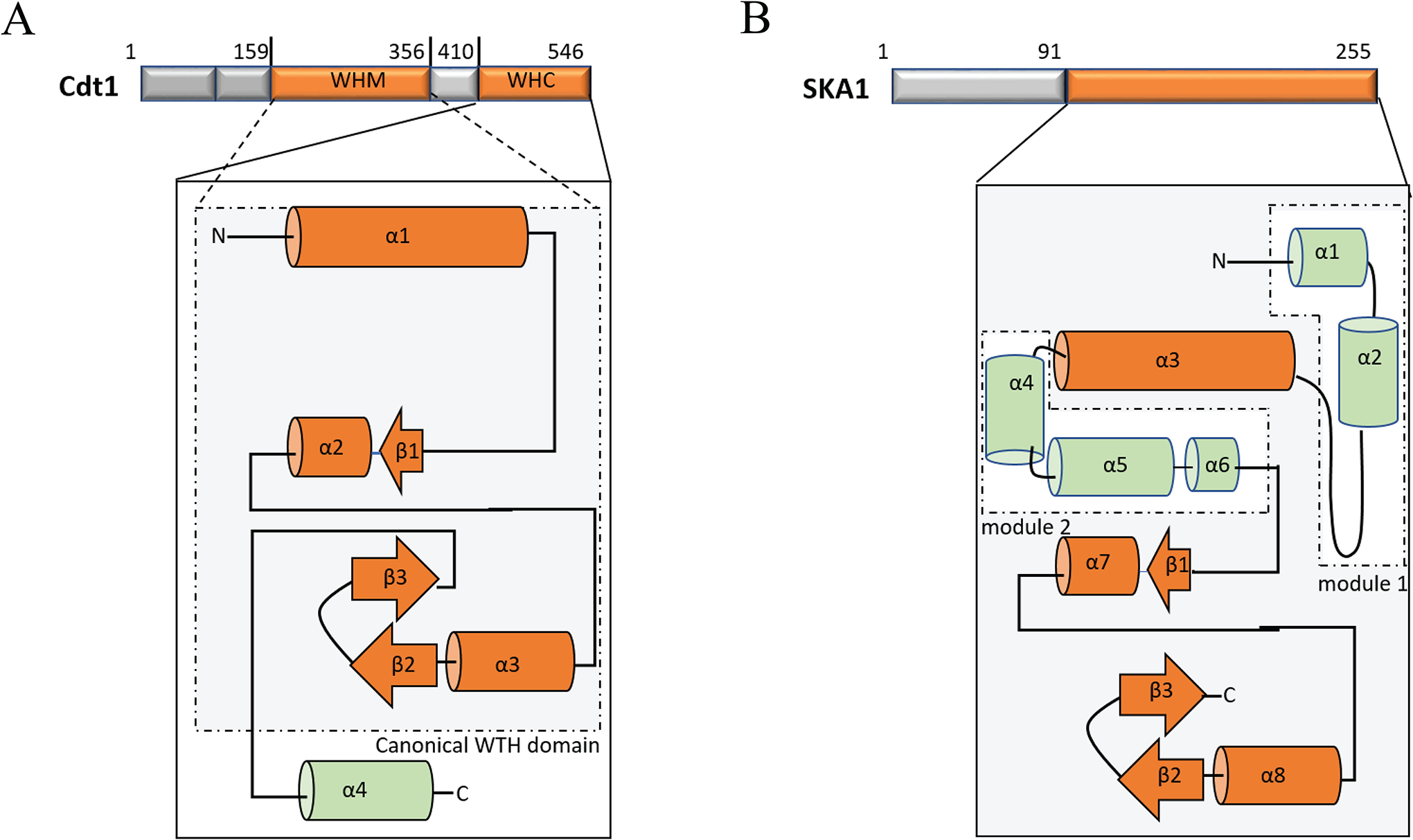
Linear and topology diagram of the canonical WTH domains (shown in orange within the grey area). α and β represent α helix and β sheet structures respectively. Note that Cdt1 (to the left, 2A) contain two WTHs: C-terminal WTH (WHC) is shown under the solid box and the middle WTH (WHM) is shown under the dotted box. In addition to the canonical WTH topology possessed by the WHM of Cdt1, the WHC contains one additional α helical component (α4). The single WTH domain of SKA1 (to the right, 2B) has 5 additional helical elements organized as two separate modules (I and II) as indicated. Secondary structural units that are unique to Cdt1 and SKA1 are shown in green.
Even though it is established that the Ska1c is dependent on the Ndc80c to localize to kinetochores (Fig. 1B) (40, 44), the finer details of how Ska1c interacts with the Ndc80c are only emerging. Multiple interaction sites of the Ska1c with the Ndc80c have been shown, especially the N-terminal tail region of Hec1 subunit, the tetrameric coiled-coil region and the loop region of the Hec1 subunit. The importance of the loop domain in this interaction is reasonably well recognized (Fig. 1B)(21, 54). One study had shown that Cdk1-mediated phosphorylation of SKA3 promotes its association with the Ndc80c loop domain. Consequently, although the SKA3 mutants defective for Cdk1 phosphorylation localized to MTs, they failed to localize to kinetochores (54). More recently, in vitro reconstitution studies by the same group has demonstrated that these two threonine Cdk1 phosphorylation sites are required for direct interaction with the Ndc80c (55). However, another very recent study has shown that the loop region is dispensable for loading of the Ska1c to kinetochores, but rather the coiled coils region of the Ndc80c subunit, NUF2, is the primary recruiter of the Ska1c to the Ndc80c in vitro (56). This line of investigation is further supported by a previous study that employed cross-linking mass spectrometry to demonstrate that the human Ska1c directly interacts with the Ndc80c (57). Helgeson et al. found that the C-terminal unstructured region of SKA3 formed robust cross-links with the predicted coiled-coil regions of all four subunits of the Ndc80c in the presence of taxol-stabilized MTs (57). A third set of studies proposed an alternative mechanism where the N-terminal unstructured tail of the Ndc80c is involved in the interaction with the Ska1c to recruit it to kinetochores and enable robust kMT attachments (58, 59). The Helgeson et al study, however, found that the Ska1c-mediated stabilization of kMT attachments was independent of the N-terminal tail of the Ndc80c (57). This notion is supported further by evidence from two very recent studies (56, 60). The first study found that, in addition to the loop region, the N-terminal tail also was not required for the Ska1c-binding (56). The 2nd study demonstrates that the N-terminal tail is dispensable for robust kMT attachments and Ska1c recruitment, while being required for kinetochore-force generation (60). One way to reconcile the possibly contradicting observations from all the above mentioned studies is that the N-terminal tail and the CH-domain of the Ndc80c might help to bring the Ska1c in proximity to kinetochores and that the Cdk1 phosphorylation may then promote stable interaction of the Ska1c with the Ndc80c loop and the flanking coiled-coil regions.
The Ska1c-binding increases the affinity of the Ndc80c for MTs which, in turn, contributes to the stabilization of kMT attachments (41, 57). It has been shown that the kinetochore levels of Ska1c increase with the formation of end-coupled KMT attachments (21, 54, 58, 60–63), thereby increasing the strength of the load bearing kMT attachments. This enrichment also seems to be affected by Aurora B-mediated phosphorylation of the unstructured tail domain of the Ndc80c as it was recently shown that cells expressing non-phosphorylable Hec1 mutant have higher level of the Ska1c compared to phosphomimetic Hec1 mutant (60). At non bi-oriented chromosomes on the other hand, this enrichment of SKA1 is antagonized by high levels of SAC protein Bub1, thereby destabilizing the stability of these attachments. Therefore, the Ska1c-Bub1 relationship acts as a sensory mechanism for the bi-oriented kinetochores, stabilizing correct attachments, and destabilizing the incorrect ones. This sensory mechanism also works independently at kinetochores of sister chromatids as varying levels of Ska1c/Bub1 is observed at kinetochores depending on their attachment status (62).
This Ska1c-mediated kMT stabilization is finely regulated by two mitotic kinases, the Aurora B kinase and the Mps1 kinase. Maceiejowski et al. have shown that Mps1 dependent phosphorylation at the hinge region of SKA3 subunit of Ska1c impairs lattice to end-on conversion ability of kinetochores resulting in destabilization of kinetochore microtubule attachment (64). Aurora B phosphorylates both the SKA1 and the SKA3 subunit to regulate the interaction of Ska1c with the components of the KMN network (61). Also, the Ska1c has been shown to directly bind to Aurora B and stimulate Aurora B activity on kinetochore substrates (65). These results demonstrate a significant role of the Ska1c in the correction of erroneous kMT attachments through both Aurora B and Mps1 kinase mediated pathways.
Apart from binding to the straight MT lattice, the Ska1c has been shown to bind equally well to both the polymerizing and depolymerizing MT plus-ends. Conversely, the human Ndc80c is thought to be less versatile in this regard, as it binds only to the MT lattice, and the single Ndc80c molecules lack tracking activity of polymerizing or depolymerizing MT plus-ends (41). The Schmidt et al study demonstrated that the Ska1c can enable the Ndc80c to track dynamic MT plus-ends. More importantly, recent studies using laser-traps have shed light on the mechanism by which the Ska1c contributes to the formation of stable kMT attachments. These studies demonstrate that the Ska1c increases the rupture force of the MT-bound Ndc80c (56, 57). The observation that the yeast Ndc80c can couple to MT-ends both during MT assembly and disassembly (66) either independently or with the assistance of MAPs that are structurally distinct from that of humans, might lead one to believe that there exist substantial differences between yeast and humans in the mechanism of function of the Ndc80c. However, recent studies using laser traps and beads multimerized with the Ndc80c show that the human Ndc80c can indeed track depolymerizing ends in this scenario (56, 67); suggesting that the yeast and human Ndc80c might not be that different in this regard.
The Ska1c is thought to localize to MT plus-ends via the conserved “microtubule tip localization signal” - SxIP motif (68), “SHLP”, found at the N terminus of SKA1 (69). The C-terminal coiled-coil region of master MT plus-tip interacting protein (+TIP), EB1, has been shown to bind many other +TIPs via their SxIP motifs. It was thus not surprising that the Ska1c recruitment to the plus-ends of MTs and its interaction with EB1 was found to involve the same C-terminal region of EB1 (69). However, recent in vitro studies have indicated that the Ska1c is capable of tracking both polymerizing and depolymerizing MTs independent of EB1 (70). Thus, while the Ska1c is remarkable in its ability to bind both polymerizing and depolymerizing MT plus-ends, more studies are clearly required to delineate the precise details of the Ska1c function in concert with MTs and the Ndc80c.
Cdt1
The protein, Cdt1 (Cdc10-dependent transcript 1), has been recognized to be essential for the process of origin licensing during DNA replication at the G1 phase of the cell cycle. During this licensing process, Cdt1 promotes the loading of mini-chromosome maintenance (MCM) helicase onto DNA-bound ORC (Origin Recognition Complex)/Cdc6 (Cell Division Cycle 6) complexes to generate the pre-RC (pre-replication complex). After MCM loading, the licensed origins can unwind the DNA and recruit the DNA polymerase for DNA replication in S phase (71).
Recent studies have revealed an additional essential role for Cdt1 during mitosis (72). It was found that Cdt1 localized to kinetochores via an interaction with the Hec1 subunit of the Ndc80c (Fig. 1B). This interaction was further found to ensure stable kMT interactions and normal mitotic progression (72). A direct interaction between Cdt1 and the Hec1 subunit of the Ndc80c was confirmed by Two-hybrid analysis and in vivo GST pull-down assay (72). Further, this study demonstrated that the loop region of the Ndc80c was critical for recruiting Cdt1 to kinetochores (Fig. 1B). The loss of Cdt1 function to a great extent phenocopied mutations in the loop domain of the Ndc80c. Further evidence for an interaction between Cdt1 and Hec1was obtained more recently using blot overlay assays (73). Both of the above-mentioned studies together also provided evidence to support the notion that the mitotic function of Cdt1 was independent of its function in DNA replication licensing (72).
A more recent study illuminated the mechanistic details of how Cdt1 contributed to the stabilization of kMT attachments (73). Using multiple in vitro and in vivo assays, it was demonstrated that Cdt1 bound to MTs directly through an extended region (a.a. 92–546) consisting of its central and C-terminal regions (Fig. 1B). This region is comprised of two WTH domains, one in the center and the other at the C-terminus. The middle WTH domain is flanked by two unstructured regions on either side (Fig. 2A). While the details of how Cdt1 binds to MTs is still unclear, it is thought that these WTH domains are critical for Cdt1 MT-binding. The WTH domains of Cdt1, in general, are more similar to the canonical WTH domains as compared to that of Ska1c. However, the C-terminal WTH domain of Cdt1 was found to have an additional α helical component apart from the core elements (Fig. 2A) (73). Cdt1 was also found to be able to bind to dolostatin-induced MT protofilament rings that simulate curved MT protofilaments. Further, it was demonstrated that Cdt1 underwent diffusive motion on MTs in a direction-independent manner (73).
The Agarwal et al. study also finds that Aurora B, negatively regulates the MT-binding activity of Cdt1. This is a phosphorylation mechanism that has been found to regulate MT-binding of multiple other kinetochore proteins, including the Ndc80c. Mitotic cells rescued using an Aurora B phosphomimetic mutants of Cdt1 exhibited defective kMT attachments and severely delayed mitotic progression (73). This finding is in line with previous observations in synchronized cells subjected to Cdt1 depletion specifically during mitosis (72). Surprisingly, the Aurora B sites were found to be distributed over this extended central and C-terminal region of Cdt1 (a.a. 92–546) comprising of the two WTH domains, with no tight grouping of the phosphosites to be discerned. This suggest a mode of regulation of Cdt1-MT binding by Aurora B, different from that observed for the Ndc80c, where the ABK sites are clustered at the N-terminal MT-binding region and influence binding electrostatically. This lead the authors to postulate that phosphorylation by Aurora B possibly regulates Cdt1-MT binding allosterically by inducing a conformational change in Cdt1 (73).
The N-terminal region of Cdt1 is involved in binding to the Ndc80c (72, 73). Cdt1 is thought to serve as a bridge between the loop domain of the Ndc80c and the spindle MTs, thus providing the Ndc80c with an accessory MT-binding site and aiding the formation of robust kMT attachments.
Similarities and differences between the Ska1c and Cdt1 critical for kinetochore-microtubule coupling
In addition to the fact that the ability of both the Ska1c and Cdt1 to form robust kMT attachments is regulated by Aurora B (61, 62, 73), both the Ska1c and Cdt1 seem to use a WTH domain to bind to MTs (Fig. 2A, 2B). It is quite interesting that traditional DNA-binding motifs found in DNA-binding proteins could be repurposed as MT-binding motifs in MT-binding proteins. At the same time, this concept is not entirely surprising, considering that both the DNA and MTs are possibly extended, negatively charged cellular structures likely to be recognized by a diverse array of protein motifs via the use of electrostatic interactions. The WTH domains can thus be deemed as a category of unconventional MAP motifs with the capability to impart MT-binding characteristic to proteins carrying these motifs.
Both the Ska1c and Cdt1 bind to the Ndc80c for recruitment to kinetochores at least in part via the loop domain of the Hec1 subunit. However, the finer details of how these factors dock precisely with the Ndc80c is not clear as has been described in the previous sections. It is also not clear whether these factors exhibit any dependency on one another for their kinetochore or MT-binding. Once recruited to kinetochores, their predominant function is to assist the Ndc80c in forming robust kMT attachments. An interesting point to note here is the ‘cause and effect’ conundrum in the context of the inter-relationship between kinetochore recruitment of the Ska1c or Cdt1 and their function in concert with the Ndc80c. The same domains of the Ndc80c (such as the loop, coiled-coil, N-terminal tail, etc.) that could be deemed essential for kMT attachments and the mutation of which produces a defective kMT function are also the regions that are required for the recruitment of the MAPs. This leads to two possibilities that are not mutually exclusive: (1) A particular Ndc80c domain contributes to the Ska1c or Cdt1 recruitment which in turn is required for stable kMT attachments; or (2) A particular Ndc80c domain promotes stable kMT attachments, which in turn is required for the kinetochore recruitment of the Ska1c or Cdt1. Though hard to distinguish experimentally, future studies will hopefully delineate the distinction between these two possibilities.
Apart from the fact that WTH domains contribute to MT-binding there seems to be no other sequence or structural conservation between the Ska1c and Cdt1. However, there are multiple functional similarities between the Ska1c and Cdt1 with regard to their MT-binding in that both seem to be capable of binding to curved MT protofilaments and also undergo direction-independent diffusion on MTs. Cdt1 is thought to act by bringing conformational change to the Ndc80c suitable for maintaining robust kMT attachment (72). The Ska1c on the other hand works as a direct mediator at the Ndc80c-MT interface, thereby actively regulating the Ndc80c-mediated end-on kMT coupling. Therefore, a plausible mutual co-operation model between the Ska1c and Cdt1 emerges in stabilizing kMT attachments (63). It is possible that at unattached kinetochores, Cdt1 binding to the loop region promotes the Ndc80c conformation to acquire more kMTs which in turn recruits more Ska1c to progressively strengthen kMT attachments as mitotic cells proceed from prometaphase to metaphase. Furthermore, the Ska1c-Bub1 duo form a mutually exclusive kinetochore enrichment pattern to regulate checkpoint signaling (62) whereas, the involvement of Cdt1 might be an evolutionary selection for cells to ascertain that essential replication licensing factors are present before mitotic exit.
Another interesting point of distinction is whether the kinetochore- and spindle MT-binding abilities of the Ska1c or Cdt1 are in anyway inter-dependent. Both the Ska1c and Cdt1 localize to mitotic spindle MTs and the spindle poles, and also readily bind to MTs in vitro (44, 46, 72, 73). These observations suggest that their localization to kinetochores is likely not required for their MT-binding. It has been demonstrated using nocodazole-based assays that the Ska1c localizes to kinetochores at sub-maximal levels in the absence of kMT attachments and that its localization to kinetochores and spindle MT plus-ends is substantially enhanced after the attachments have been formed (21, 54, 58, 60–63). This suggests that spindle MTs are somehow necessary for this typical localization pattern of the Ska1c. It is thus possible that the Ska1c initially localizes to spindle MTs early during mitosis. As kinetochores capture more MTs, the Ska1c might start accumulating at kinetochores via these microtubules as is the case with the loading of Dam1 complexes to kinetochores in budding yeast (74). However, it is not clear if such a distinction exists with regard to Cdt1 kinetochore targeting, as Cdt1 was found to localize normally to kinetochores as well as to the spindle MTs independent of the kMT attachment status (72, 73). Unlike the Ska1c, only a slight increase of Cdt1 levels have been observed between prometaphase and metaphase kinetochores (63). A specific localization to the kMT plus-ends has also not been discerned yet for Cdt1 (72, 73). Further studies are clearly required to dissect out the Ndc80c-loop dependent functions of these kinetochore MAPs in isolation and in concert with each other.
Conclusion
The Ndc80c located at the outer kinetochore is essential for robust kMT interactions. Some of the other outer kinetochore proteins such as Cdt1 and the Ska1c in vertebrates and the Dam1/DASH complex in yeast serve as non-motor kinetochore MAPs while also binding to the Ndc80c to control chromosome alignment and segregation. The Dam/DASH complex interacts with the loop, coiled-coil, and the hairpin/CH domains of the Ndc80c (Fig. 1A). The human Ska1c also exhibits an interaction with parts of the coil region of the Ndc80c in addition to its interactions with the loop region and the unstructured N-terminal tail of the Ndc80c (Fig. 1B). Human Cdt1 has been shown to be dependent only on the loop domain to localize to the kinetochore-bound Ndc80c (Fig. 1B). Other Ndc80c-binding kinetochore MAPs such as human chTOG (and its evolutionarily conserved homologues), the human Astrin-SKAP complex and many other MT plus-binding proteins are currently being actively studied, but the mechanism of how these proteins enable robust kMT attachments is still unclear and is beyond the scope of discussion in this review (5, 6). A key point to add in this context is that the Astrin-SKAP and the Ndc80 complexes bind cooperatively to MTs in vitro (75) while Stu2, the yeast homologue of human chTOG, confers increased rupture force to the MT-bound Ndc80 complexes in an optical trap assay (76). Irrespective of the mechanism by which these MAPs interact with the Ndc80c, their main function is to stabilize the interaction of the Ndc80c to kMTs. In many cases, it is thought that they specifically assist to couple the Ndc80c efficiently to the plus-ends of dynamic microtubules during chromosome segregation. Future work will reveal how these different MAPs could work individually or in tandem to modulate and possibly provide temporal control of kMT attachment stability.
Acknowledgements
The authors would like to acknowledge funding for D.V. (R01GM135391) from the National Institute of Health -National Institute of General Medical Sciences. We would also like to thank Robert Wimbish for his critical comments on the manuscript and Anita Varma for editing the language content.
Abbreviations:
- MT(s)
Microtubule(s)
- kMT
kinetochore-microtubule
- MAP(s)
Microtubule-associated proteins
- WTH
Winged-turn-helix
- CH
Calponin-homology
Footnotes
Conflict of Interest
The authors declare no conflicts of interest.
References
- 1.Alushin G, Nogales E. Visualizing kinetochore architecture. Curr Opin Struct Biol. 2011;21(5):661–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9(1):33–46. [DOI] [PubMed] [Google Scholar]
- 3.Bakhoum SF, Compton DA. Kinetochores and disease: keeping microtubule dynamics in check! Curr Opin Cell Biol. 2012;24(1):64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foley EA, Kapoor TM. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat Rev Mol Cell Biol. 2013;14(1):25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amin MA, Agarwal S, Varma D. Mapping the kinetochore MAP functions required for stabilizing microtubule attachments to chromosomes during metaphase. Cytoskeleton (Hoboken). 2019;76(6):398–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monda JK, Cheeseman IM. The kinetochore-microtubule interface at a glance. J Cell Sci. 2018;131(16). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Varma D, Salmon ED. The KMN protein network--chief conductors of the kinetochore orchestra. J Cell Sci. 2012;125(Pt 24):5927–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Umbreit NT, Gestaut DR, Tien JF, Vollmar BS, Gonen T, Asbury CL, et al. The Ndc80 kinetochore complex directly modulates microtubule dynamics. Proc Natl Acad Sci U S A. 2012;109(40):16113–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat Struct Mol Biol. 2007;14(1):54–9. [DOI] [PubMed] [Google Scholar]
- 10.Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Dos Reis G, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133(3):427–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127(5):969–82. [DOI] [PubMed] [Google Scholar]
- 12.DeLuca JG, Musacchio A. Structural organization of the kinetochore-microtubule interface. Curr Opin Cell Biol. 2012;24(1):48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alushin GM, Ramey VH, Pasqualato S, Ball DA, Grigorieff N, Musacchio A, et al. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature. 2010;467(7317):805–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127(5):983–97. [DOI] [PubMed] [Google Scholar]
- 15.Guimaraes GJ, Dong Y, McEwen BF, Deluca JG. Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Curr Biol. 2008;18(22):1778–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zaytsev AV, Mick JE, Maslennikov E, Nikashin B, DeLuca JG, Grishchuk EL. Multisite phosphorylation of the NDC80 complex gradually tunes its microtubule-binding affinity. Mol Biol Cell. 2015;26(10):1829–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Musacchio A, Desai A. A Molecular View of Kinetochore Assembly and Function. Biology (Basel). 2017;6(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang HW, Long S, Ciferri C, Westermann S, Drubin D, Barnes G, et al. Architecture and flexibility of the yeast Ndc80 kinetochore complex. J Mol Biol. 2008;383(4):894–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nilsson J Looping in on Ndc80 - how does a protein loop at the kinetochore control chromosome segregation? Bioessays. 2012;34(12):1070–7. [DOI] [PubMed] [Google Scholar]
- 20.Tang NH, Toda T. Ndc80 Loop as a protein-protein interaction motif. Cell Div. 2013;8(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang G, Kelstrup CD, Hu XW, Kaas Hansen MJ, Singleton MR, Olsen JV, et al. The Ndc80 internal loop is required for recruitment of the Ska complex to establish end-on microtubule attachment to kinetochores. J Cell Sci. 2012;125(Pt 13):3243–53. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka TU, Desai A. Kinetochore-microtubule interactions: the means to the end. Curr Opin Cell Biol. 2008;20(1):53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hofmann C, Cheeseman IM, Goode BL, McDonald KL, Barnes G, Drubin DG. Saccharomyces cerevisiae Duo1p and Dam1p, novel proteins involved in mitotic spindle function. J Cell Biol. 1998;143(4):1029–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McAinsh AD, Tytell JD, Sorger PK. Structure, function, and regulation of budding yeast kinetochores. Annu Rev Cell Dev Biol. 2003;19:519–39. [DOI] [PubMed] [Google Scholar]
- 25.Joglekar AP, Bloom KS, Salmon ED. Mechanisms of force generation by end-on kinetochore-microtubule attachments. Curr Opin Cell Biol. 2010;22(1):57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheeseman IM, Enquist-Newman M, Muller-Reichert T, Drubin DG, Barnes G. Mitotic spindle integrity and kinetochore function linked by the Duo1p/Dam1p complex. J Cell Biol. 2001;152(1):197–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Westermann S, Avila-Sakar A, Wang HW, Niederstrasser H, Wong J, Drubin DG, et al. Formation of a dynamic kinetochore- microtubule interface through assembly of the Dam1 ring complex. Mol Cell. 2005;17(2):277–90. [DOI] [PubMed] [Google Scholar]
- 28.Westermann S, Wang HW, Avila-Sakar A, Drubin DG, Nogales E, Barnes G. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature. 2006;440(7083):565–9. [DOI] [PubMed] [Google Scholar]
- 29.Jenni S, Harrison SC. Structure of the DASH/Dam1 complex shows its role at the yeast kinetochore-microtubule interface. Science. 2018;360(6388):552–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim JO, Zelter A, Umbreit NT, Bollozos A, Riffle M, Johnson R, et al. The Ndc80 complex bridges two Dam1 complex rings. Elife. 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lampert F, Hornung P, Westermann S. The Dam1 complex confers microtubule plus end-tracking activity to the Ndc80 kinetochore complex. J Cell Biol. 2010;189(4):641–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tien JF, Umbreit NT, Gestaut DR, Franck AD, Cooper J, Wordeman L, et al. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J Cell Biol. 2010;189(4):713–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janke C, Ortiz J, Lechner J, Shevchenko A, Shevchenko A, Magiera MM, et al. The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J. 2001;20(4):777–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maure JF, Komoto S, Oku Y, Mino A, Pasqualato S, Natsume K, et al. The Ndc80 loop region facilitates formation of kinetochore attachment to the dynamic microtubule plus end. Curr Biol. 2011;21(3):207–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lampert F, Mieck C, Alushin GM, Nogales E, Westermann S. Molecular requirements for the formation of a kinetochore-microtubule interface by Dam1 and Ndc80 complexes. J Cell Biol. 2013;200(1):21–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gonen S, Akiyoshi B, Iadanza MG, Shi D, Duggan N, Biggins S, et al. The structure of purified kinetochores reveals multiple microtubule-attachment sites. Nat Struct Mol Biol. 2012;19(9):925–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joglekar AP, Bloom K, Salmon ED. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr Biol. 2009;19(8):694–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shang C, Hazbun TR, Cheeseman IM, Aranda J, Fields S, Drubin DG, et al. Kinetochore protein interactions and their regulation by the Aurora kinase Ipl1p. Mol Biol Cell. 2003;14(8):3342–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gestaut DR, Graczyk B, Cooper J, Widlund PO, Zelter A, Wordeman L, et al. Phosphoregulation and depolymerization-driven movement of the Dam1 complex do not require ring formation. Nat Cell Biol. 2008;10(4):407–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaitanos TN, Santamaria A, Jeyaprakash AA, Wang B, Conti E, Nigg EA. Stable kinetochore-microtubule interactions depend on the Ska complex and its new component Ska3/C13Orf3. EMBO J. 2009;28(10):1442–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmidt JC, Arthanari H, Boeszoermenyi A, Dashkevich NM, Wilson-Kubalek EM, Monnier N, et al. The kinetochore-bound Ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Dev Cell. 2012;23(5):968–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Hooff JJE, Snel B, Kops G. Unique Phylogenetic Distributions of the Ska and Dam1 Complexes Support Functional Analogy and Suggest Multiple Parallel Displacements of Ska by Dam1. Genome Biol Evol. 2017;9(5):1295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raaijmakers JA, Tanenbaum ME, Maia AF, Medema RH. RAMA1 is a novel kinetochore protein involved in kinetochore-microtubule attachment. J Cell Sci. 2009;122(Pt 14):2436–45. [DOI] [PubMed] [Google Scholar]
- 44.Welburn JP, Grishchuk EL, Backer CB, Wilson-Kubalek EM, Yates JR 3rd, Cheeseman IM. The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev Cell. 2009;16(3):374–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Daum JR, Wren JD, Daniel JJ, Sivakumar S, McAvoy JN, Potapova TA, et al. Ska3 is required for spindle checkpoint silencing and the maintenance of chromosome cohesion in mitosis. Curr Biol. 2009;19(17):1467–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hanisch A, Sillje HH, Nigg EA. Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. EMBO J. 2006;25(23):5504–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sivakumar S, Daum JR, Tipton AR, Rankin S, Gorbsky GJ. The spindle and kinetochore-associated (Ska) complex enhances binding of the anaphase-promoting complex/cyclosome (APC/C) to chromosomes and promotes mitotic exit. Mol Biol Cell. 2014;25(5):594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sivakumar S, Janczyk PL, Qu Q, Brautigam CA, Stukenberg PT, Yu H, et al. The human SKA complex drives the metaphase-anaphase cell cycle transition by recruiting protein phosphatase 1 to kinetochores. Elife. 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jeyaprakash AA, Santamaria A, Jayachandran U, Chan YW, Benda C, Nigg EA, et al. Structural and functional organization of the Ska complex, a key component of the kinetochore-microtubule interface. Mol Cell. 2012;46(3):274–86. [DOI] [PubMed] [Google Scholar]
- 50.Abad MA, Zou J, Medina-Pritchard B, Nigg EA, Rappsilber J, Santamaria A, et al. Ska3 Ensures Timely Mitotic Progression by Interacting Directly With Microtubules and Ska1 Microtubule Binding Domain. Sci Rep. 2016;6:34042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abad MA, Medina B, Santamaria A, Zou J, Plasberg-Hill C, Madhumalar A, et al. Structural basis for microtubule recognition by the human kinetochore Ska complex. Nature Communications. 2014;5:2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gajiwala KS, Burley SK. Winged helix proteins. Curr Opin Struct Biol. 2000;10(1):110–6. [DOI] [PubMed] [Google Scholar]
- 53.Teichmann M, Dumay-Odelot H, Fribourg S. Structural and functional aspects of winged-helix domains at the core of transcription initiation complexes. Transcription. 2012;3(1):2–7. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Q, Sivakumar S, Chen Y, Gao H, Yang L, Yuan Z, et al. Ska3 Phosphorylated by Cdk1 Binds Ndc80 and Recruits Ska to Kinetochores to Promote Mitotic Progression. Curr Biol. 2017. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Q, Hu L, Chen Y, Tian W, Liu H. Multisite Phosphorylation Determines the Formation of Ska-Ndc80 Macro-complexes That Are Essential for Chromosome Segregation during Mitosis. Mol Biol Cell. 2020:mbcE19100569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huis In ‘t Veld PJ, Volkov VA, Stender ID, Musacchio A, Dogterom M. Molecular determinants of the Ska-Ndc80 interaction and their influence on microtubule tracking and force-coupling. Elife. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Helgeson LA, Zelter A, Riffle M, MacCoss MJ, Asbury CL, Davis TN. Human Ska complex and Ndc80 complex interact to form a load-bearing assembly that strengthens kinetochore-microtubule attachments. Proc Natl Acad Sci U S A. 2018;115(11):2740–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Janczyk PL, Skorupka KA, Tooley JG, Matson DR, Kestner CA, West T, et al. Mechanism of Ska Recruitment by Ndc80 Complexes to Kinetochores. Dev Cell. 2017;41(4):438–49 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheerambathur DK, Prevo B, Hattersley N, Lewellyn L, Corbett KD, Oegema K, et al. Dephosphorylation of the Ndc80 Tail Stabilizes Kinetochore-Microtubule Attachments via the Ska Complex. Dev Cell. 2017;41(4):424–37 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wimbish RT, DeLuca KF, Mick JE, Himes J, Jimenez-Sanchez I, Jeyaprakash AA, et al. The Hec1/Ndc80 tail domain is required for force generation at kinetochores, but is dispensable for kinetochore-microtubule attachment formation and Ska complex recruitment. Mol Biol Cell. 2020;31(14):1453–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chan YW, Jeyaprakash AA, Nigg EA, Santamaria A. Aurora B controls kinetochore-microtubule attachments by inhibiting Ska complex-KMN network interaction. J Cell Biol. 2012;196(5):563–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Auckland P, Clarke NI, Royle SJ, McAinsh AD. Congressing kinetochores progressively load Ska complexes to prevent force-dependent detachment. J Cell Biol. 2017;216(6):1623–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Campbell S, Amin MA, Varma D, Bidone TC. Computational model demonstrates that Ndc80-associated proteins strengthen kinetochore-microtubule attachments in metaphase. Cytoskeleton (Hoboken). 2019;76(11–12):549–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maciejowski J, Drechsler H, Grundner-Culemann K, Ballister ER, Rodriguez-Rodriguez JA, Rodriguez-Bravo V, et al. Mps1 Regulates Kinetochore-Microtubule Attachment Stability via the Ska Complex to Ensure Error-Free Chromosome Segregation. Dev Cell. 2017;41(2):143–56 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Redli PM, Gasic I, Meraldi P, Nigg EA, Santamaria A. The Ska complex promotes Aurora B activity to ensure chromosome biorientation. J Cell Biol. 2016;215(1):77–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Powers AF, Franck AD, Gestaut DR, Cooper J, Gracyzk B, Wei RR, et al. The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell. 2009;136(5):865–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Volkov VA, Huis In ‘t Veld PJ, Dogterom M, Musacchio A. Multivalency of NDC80 in the outer kinetochore is essential to track shortening microtubules and generate forces. Elife. 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Honnappa S, Gouveia SM, Weisbrich A, Damberger FF, Bhavesh NS, Jawhari H, et al. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 2009;138(2):366–76. [DOI] [PubMed] [Google Scholar]
- 69.Thomas GE, Bandopadhyay K, Sutradhar S, Renjith MR, Singh P, Gireesh KK, et al. EB1 regulates attachment of Ska1 with microtubules by forming extended structures on the microtubule lattice. Nat Commun. 2016;7:11665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Monda JK, Whitney IP, Tarasovetc EV, Wilson-Kubalek E, Milligan RA, Grishchuk EL, et al. Microtubule Tip Tracking by the Spindle and Kinetochore Protein Ska1 Requires Diverse Tubulin-Interacting Surfaces. Curr Biol. 2017;27(23):3666–75 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pozo PN, Cook JG. Regulation and Function of Cdt1; A Key Factor in Cell Proliferation and Genome Stability. Genes (Basel). 2016;8(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Varma D, Chandrasekaran S, Sundin LJ, Reidy KT, Wan X, Chasse DA, et al. Recruitment of the human Cdt1 replication licensing protein by the loop domain of Hec1 is required for stable kinetochore-microtubule attachment. Nat Cell Biol. 2012;14(6):593–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Agarwal S, Smith KP, Zhou Y, Suzuki A, McKenney RJ, Varma D. Cdt1 stabilizes kinetochore-microtubule attachments via an Aurora B kinase-dependent mechanism. J Cell Biol. 2018;217(10):3446–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li Y, Bachant J, Alcasabas AA, Wang Y, Qin J, Elledge SJ. The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes Dev. 2002;16(2):183–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kern DM, Monda JK, Su KC, Wilson-Kubalek EM, Cheeseman IM. Astrin-SKAP complex reconstitution reveals its kinetochore interaction with microtubule-bound Ndc80. Elife. 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Miller MP, Asbury CL, Biggins S. A TOG Protein Confers Tension Sensitivity to Kinetochore-Microtubule Attachments. Cell. 2016;165(6):1428–39. [DOI] [PMC free article] [PubMed] [Google Scholar]


