Abstract
With the number of affected individuals still growing world-wide, the research on COVID-19 is continuously expanding. The deep learning community concentrates their efforts on exploring if neural networks can potentially support the diagnosis using CT and radiograph images of patients’ lungs.
The two most popular publicly available datasets for COVID-19 classification are COVID-CT and COVID-19 Image Data Collection. In this work, we propose a new dataset which we call COVID-19 CT & Radiograph Image Data Stock. It contains both CT and radiograph samples of COVID-19 lung findings and combines them with additional data to ensure a sufficient number of diverse COVID-19-negative samples. Moreover, it is supplemented with a carefully defined split.
The aim of COVID-19 CT & Radiograph Image Data Stock is to create a public pool of CT and radiograph images of lungs to increase the efficiency of distinguishing COVID-19 disease from other types of pneumonia and from healthy chest. We hope that the creation of this dataset would allow standardisation of the approach taken for training deep neural networks for COVID-19 classification and eventually for building more reliable models.
Keywords: COVID-19 classification dataset, CT, Radiograph
Highlights
-
•
The article aims to give a public pool of CT & X-ray lungs images to increase the efficiency of detect COVID-19 disease.
-
•
Binary and multiclass classifiers were trained revealing that precise labels can improve the system performance.
-
•
Models trained on COVID-19 CT & X-ray Image Data Stock are more robust than models trained on other investigated databases.
1. Introduction
At the end of 2019, a new coronavirus SARS-CoV-2 (Severe Acute Respiratory Syndrome Coronavirus 2) appeared in Wuhan, which then triggered a global pandemic. SARS-CoV-2-induced pneumonia has been termed COVID-19 (Coronavirus Disease 2019). The main symptoms of COVID-19 are high fever, dry cough, shortness of breath, muscle pain, diarrhea, myalgia, nasal obstruction and runny nose [1]. As of July 15, 2020, a total of 13,690,108 confirmed cases with COVID-19 pneumonia have been reported globally, including 586,265 deaths (4.28%).
The current diagnostic method for COVID-19 is real time reverse transcription – polymerase chain reaction (RT-PCR) [2]. The main limitation of this method is the insufficient amount and quality of the clinical material from which the nucleic acids are isolated [3]. This can result in false negative Results.
Lung CT and radiograph scans are gradually recognised as an alternative for COVID-19 diagnosis. The lungs of people infected with COVID-19 are characterised by consolidation, ground-glass opacification, bilateral involvement, peripheral and diffuse distribution. Lung CT scans can be used to diagnose COVID-19 in patients in acute and convalescent periods of disease [1]. Only patients with severe or permanent lung damage will show changes in CT after recovery, which makes it impossible to determine the percentage of the population that has undergone the disease based on lung scans [4]. The favorable aspects of CT scanners are their availability in many hospitals and the short amount of time required to obtain the Results estimated to be around 15 min. The use of CT for initial diagnostics might significantly increase testing capabilities. On the other hand, the imaging costs are relatively high, which may limit the use of CT for COVID-19 diagnostics. Moreover, the use of CT for COVID-19 diagnostics requires thorough cleaning of the equipment between examinations and a large surface of contact increases the risk of infection, compared to the RT-PCR method performed in sterile conditions [5]. Despite the large number of publications indicating high sensitivity and specificity of CT, the radiologists’ position from American College of Radiology (ACR) advises against putting lung CT on the first line of COVID-19 diagnostics [6].
The advantages of radiograph scans for COVID-19 diagnostics include greater availability of radiographs, lower radiation doses to which the patient is subjected and a short scanning time.
Recently, both CT and radiograph scans have been shown to enable training models which achieve promising Results in the COVID-19 classification task [7,8].
Considering the advantages and disadvantages of both methods we decided to create a database containing both CT and radiograph images.
Currently, the most popular datasets for COVID-19 classification are COVID-CT [9] and COVID-19 Image Data Collection [10]. These datasets contain images of CT and radiograph chest scans of individuals affected with COVID-19 as well as of patients not affected with COVID-19.
COVID-19 Image Data Collection contains images of both CT and radiograph scans. The number of CT scans is insufficient for training deep models. The number of radiograph images is higher but there is not enough negative samples. Moreover, this dataset does not define a data split.
COVID-CT concentrates on CT scans and defines a data split. However, it provides only a rough categorisation of samples into COVID-19-positive and negative cases, where negative cases can be images of healthy individuals or patients with a different disease.
Training neural networks on these datasets requires including samples from additional data sources such as common bacterial pneumonia [11] or lung nodule analysis [12,13].
Apostolopoulos and Mpesiana [14] used a MobileNet v2 [15] pre-trained on ImageNet [16] for fine-tuning on two datasets which were created using samples from COVID-19 Image Data Collection [10], COVID-19 X-ray collection available on kaggle [17], and a dataset containing radiograph scans of common bacterial pneumonia [11]. They achieved sensitivity of 98% and specificity of 96% on the dataset which included both common bacterial pneumonia and viral pneumonia cases as distractors for the COVID-19 class, and sensitivity of 99% and specificity of 97% on the dataset which included only common bacterial pneumonia cases.
Zhao et al. [9] pre-trained a DenseNet [18] on ChestX-ray14 [19] and fine-tuned it on COVID-CT. They achieve AUC of 0.82.
He et al. [7] used models pre-trained on ImageNet, which were further pre-trained using contrastive self-learning [20] first on LUNA dataset [12,13] and then on COVID-CT, followed by fine-tuning on COVID-CT. This methodology allowed them to achieve AUC of 0.94 with DenseNet-169.
The huge variety of scenarios in which the models are evaluated prevents any comparison between them. As a result, it is difficult to tell which design choices contribute to improved performance of some models and to use this knowledge to build incrementally more reliable solutions.
In this work, we propose COVID-19 CT & Radiograph Image Data Stock, which combines data from multiple sources into a single dataset. The advantages of COVID-19 CT & Radiograph Image Data Stock include:
-
•
a large number of both CT and radiograph scans of COVID-19 class
-
•
a large number of negative samples in both modi
-
•
the exact class of the negative samples is known
-
•
the source of each sample is known
-
•
a data split is defined.
Using COVID-19 CT & Radiograph Image Data Stock does not require employing any additional data sources. We hope that this dataset will allow for better understanding of the influence of individual choices on the final performance of COVID-19 classification models.
To give a better insight into benefits of using COVID-19 CT & Radiograph Image Data Stock for training neural networks, we compare the performance of several popular architectures pre-trained on ImageNet [16] when trained on COVID-CT, COVID-19 Image Data Collection and COVID-19 CT & Radiograph Image Data Stock in multiple scenarios and show that models trained on COVID-19 CT & Radiograph Image Data Stock achieve better Results both in case of CT and radiograph data.
Our main contributions are as follows
-
1.
We build a rich and self-contained database for COVID-19 classification,
-
2.
We train several neural networks using COVID-19 CT & Radiograph Image Data Stock to provide baseline benchmarks,
-
3.
We compare the models trained on COVID-19 CT & Radiograph Image Data Stock with the models trained on COVID-19 Image Data Collection and COVID-CT and show that models trained on COVID-19 CT & Radiograph Image Data Stock are more robust,
-
4.
We show that using a precise class information helps to improve the model's ability to distinguish between COVID-19-positive and negative samples.
The rest of this work is organised as follows: in section 2 we shortly characterize COVID-CT and COVID-19 Image Data Collection, and describe in detail how was COVID-19 CT & Radiograph Image Data Stock created. In section 3, we describe the evaluation of models trained on each of the datasets and in section 4, we present the Results. In section 5, we conclude the paper.
2. COVID-19 CT & Radiograph Image Data Stock
In this section, we briefly characterize COVID-CT [9] and COVID-19 Image Data Collection [10] shortly discussing their strong and weak points and describe in detail how the proposed COVID-19 CT & Radiograph Image Data Stock was created.
COVID-CT. COVID-CT [9] is a dataset containing images derived from over 750 preprints on COVID-19. The images present chest CT scans in axial plane and are in png format. The task is to classify images as belonging to COVID-19-positive or negative class. This dataset has a defined split which allows for comparison between the models and reproducibility of the Results. However, it provides only a rough categorisation of samples into COVID-19-positive and negative cases, where negative cases can be images of healthy individuals or patients with a different disease.
COVID-19 Image Data Collection. COVID-19 Image Data Collection [10] is a dataset containing images of patients with COVID-19, patients with COVID-19 and acute respiratory distress syndrome (ARDS), and images of patients without COVID-19 but with other diseases. The images present CT and radiograph scans of lungs and are in jpg or png format. Each image is accompanied with additional data which describes image characteristic (such as view or modality) and patient characteristic (such as age or survival), however, most of these features are present only for some of the samples. The possible tasks include binary classification of COVID-19-positive and negative patients, and multi-class classification of an exact disease. This dataset does not provide a data split.
COVID-19 CT & Radiograph Image Data Stock. The aim of COVID-19 CT & Radiograph Image Data Stock is to create a public pool of CT and radiograph images of lungs to increase the efficiency of distinguishing COVID-19 from other types of pneumonia and from healthy lungs. We hope this can help to prepare a “ground” for distinguishing between newly discovered and already known viruses and bacteria strains causing pneumonia in order to improve diagnostics in the event of subsequent pandemics. For this reason we included COVID-19-negative samples of several classes which include healthy chest (negative control) and various types of pneumonia (bacterial, fungal, viral).
The images which constitute COVID-19 CT & Radiograph Image Data Stock were compiled from public sources. Most of the images come from websites with image collections and about 150 images were collected from online publications. The list of all sources is presented in Table 1 .
Table 1.
Sources of images in COVID-19 CT & Radiograph Image Data Stock.
| diagnosis | CT | radiograph |
|---|---|---|
| healthy | Radiopaedia | [11] |
| COVID-19 | Radiopaedia | SIRM, COVID-19 Resourse site for Imaging and Radiology, EURORAD, Radiopaedia, Radiology Assistant, Cases RSNA, APP Fig. 1, RAD2share, Yxppt, Fig. 1 COVID-19 Chest X-ray Dataset Initiativea [[21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46]], |
| bacterial pneumonia | Radiopaedia [[47], [48], [49], [50]], | Radiopaedia [11], |
| viral pneumonia | Radiopaedia [51,52], | [11,53,54] |
| fungal pneumonia | Radiopaedia [52], | wikipedia, Radiopaedia |
| ARDS | wikipedia, Radiopaedia |
Images were searched by entering the following phrases: COVID-19 lung CT/radiograph images, normal chest CT/radiograph/X-RAY, bacterial pneumonia CT/radiograph images, viral pneumonia CT/radiograph and fungal CT/radiograph images. To qualify the image for the database, it must contain an accurate annotation about the disease (a type of pneumonia). Therefore, images from COVID-CT [9] were rejected for the lack of this information. Most of the images from online publications were downloaded as high quality pictures directly from the web pages.
The current statistics of COVID-19 CT & Radiograph Image Data Stock as of June 25th, 2020 are shown in Table 2 . The database contains over eight thousand COVID-19-positive CT images and few hundred of COVID-19-positive radiographs. Moreover, it includes a substantial number of COVID-19-negative samples in four classes: healthy chest, bacterial pneumonia, viral pneumonia and fungal pneumonia. A few samples of radiographs with acute respiratory distress syndrome (ARDS) are also present.
Table 2.
Detailed COVID-19 CT & Radiograph Image Data Stock description. Given are number of CT and radiograph images in each class and for each section along with the total number of images.
| CT |
radiograph |
|||||
|---|---|---|---|---|---|---|
| coronal | axial | total | sagittal | coronal | total | |
| healthy chest | 417 | 1270 | 1687 | 0 | 1583 | 1583 |
| COVID-19 | 2399 | 5671 | 8070 | 10 | 323 | 333 |
| bacterial pneumonia | 58 | 249 | 307 | 5 | 2801 | 2806 |
| viral pneumonia | 5 | 39 | 44 | 0 | 1509 | 1509 |
| fungal pneumonia | 215 | 1068 | 1283 | 3 | 12 | 15 |
| ARDS | 0 | 0 | 0 | 0 | 4 | 4 |
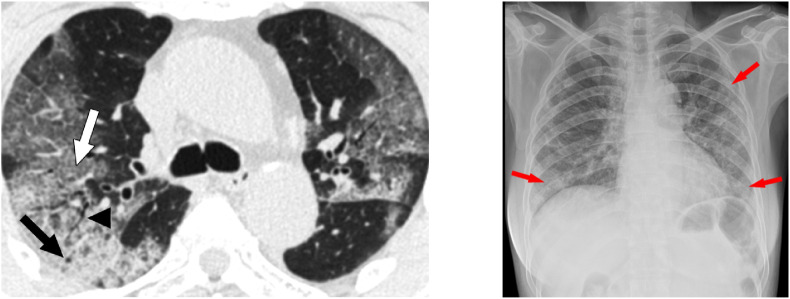
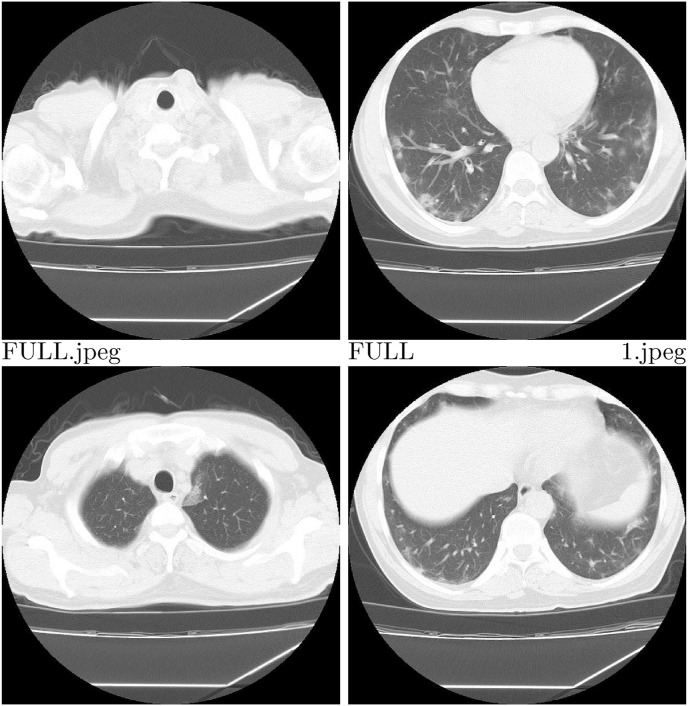
Importantly, each image in the database is accompanied with a precise diagnosis and a source of the image. Less than 1% of the images in the database (90 CTs and 35 radiographs) have markings made by radiologists at the site of lesions characteristic of various types of pneumonia. Information about presence of such markings has been included in the database. Examples of annotated images from COVID-19 CT & Radiograph Image Data Stock are shown in Fig. 1. We categorised CT images into 3 groups: full (whole lungs visible), not full (some part of lungs visible) and empty (no visible lungs). Images described as not full or empty were marked as not suitable for diagnostics. Examples of such images are shown in Fig. 2 . Additional attributes include such information as patient ID or type and section of the image. The full list of attributes is presented in Table 3 .
Fig. 1.
Examples of annotated images from COVID-19 CT & Radiograph Image Data Stock. On the left an example of annotated CT, and on the right an example of annotated radiograph.
Fig. 2.
Examples of CT images categorised as full – upper right corner, not full – lower row, empty – upper left corner.
Table 3.
Description of attributes present in COVID-19 CT & Radiograph Image Data Stock.
| Attribute | Description |
|---|---|
| patient ID | internal identifier |
| file name | name of the file including extension |
| type of image | radiograph or CT |
| section of image | sagittal, axial or coronal |
| diagnosis | healthy chest, COVID-19, bacterial pneumonia, viral pneumonia, fungal pneumonia, or ARDS |
| presence of marks | presence of marks marked by radiologist |
| group train/valid/test | belonging to the train, valid or test group |
| lung presence | the entire surface of the lungs are visible, invisible lungs or only part of the lungs are visible |
| origin | URL of the paper or website where the image came from |
| suitable/not suitable for diagnosis | information if image is suitable or not for diagnosis |
The COVID-19 CT & Radiograph Image Data Stock contains a sequence of images of a full CT lung scan. The number of full CT scans in each class are shown in Table 4 .
Table 4.
Number of full CT lung scans in COVID-19 CT & Radiograph Image Data Stock for each class.
| class | number of full CT scans |
|---|---|
| healthy chest | 10 |
| COVID-19 | 46 |
| bacterial pneumonia | 3 |
| viral pneumonia | 0 |
| fungal pneumonia | 12 |
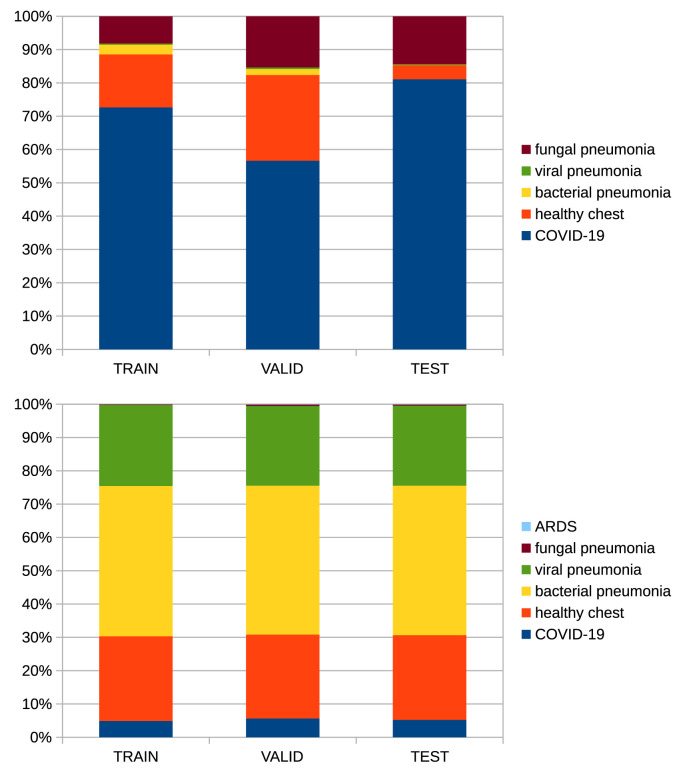
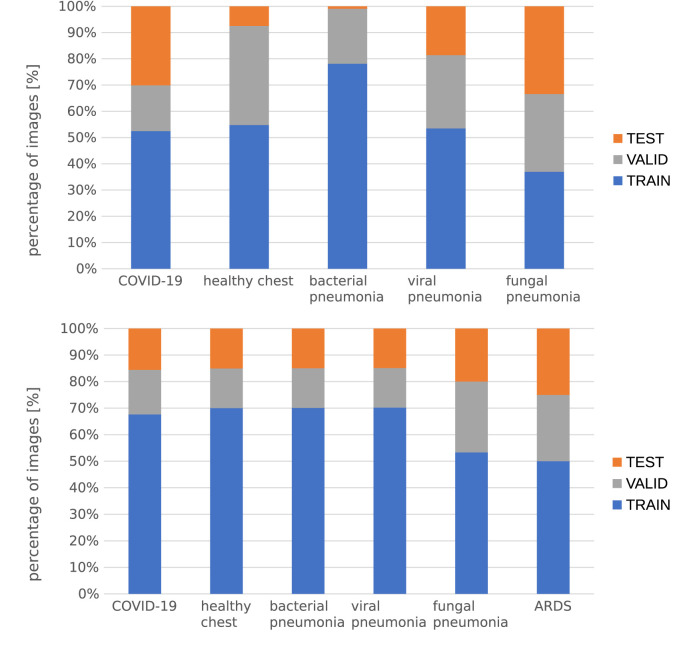
The images were split into train, valid and test sets with 70–15-15 ratio. We ensured that images of the same patient or coming from the same source were included in the same subset. We tried to maintain the class balance and the distributions of different cross-sections (sagittal, axial, and coronal). The details of the data split are shown in Fig. 3, Fig. 4 .
Fig. 3.
Distribution of classes in data splits – CT on the'top, radiograph on the bottom.
Fig. 4.
Split ratio for each class – CT on the top, radiograph on the bottom.
The images forming the database vary in size and dimension – minimum and maximum width and height are given in Table 5 . All images presented in our database are colour images with three channels. We used 3 channel images because models that we create to evaluate our dataset are pretrained on ImageNet dataset which is also trained on 3 channel images. Images are in png, jpg, and gif formats.
Table 5.
Maximum and minimum width and height of CT and radiograph images given in pixels.
| CT |
radiograph |
|||
|---|---|---|---|---|
| minimum | maximum | minimum | maximum | |
| width/height | 107/85 | 2354/2313 | 156/157 | 4300/4298 |
The COVID-19 CT & Radiograph Image Data Stock is available under the following url: https://is.dicella.com. Until the inhibition of COVID-19 pandemic, the database will be updated monthly.
3. Evaluation methodology
3.1. Data preparation
COVID-CT. COVID-CT was downloaded on May 5, 2020. The dataset contains images of CT scans. We used the data split provided with the dataset, the details of which are presented in Table 6 .
Table 6.
Number of COVID-19-positive and negative samples in each split of COVID-CT dataset.
| COVID-19-positive | COVID-19-negative | total | |
|---|---|---|---|
| train | 191 | 234 | 425 |
| validation | 60 | 58 | 118 |
| test | 98 | 105 | 203 |
COVID-19 Image Data Collection. COVID-19 Image Data Collection was downloaded on May 5, 2020. The dataset contains images of both radiograph and CT scans. Due to insufficient number of CT samples, only the images of radiograph were used during the training and evaluation.
This dataset includes radiograph images in different sections - most of them are front views (posteroanterior and anteroposterior), and there are some in lateral view. The number of samples in lateral view is small and these samples were not used during training. The detailed number of samples in the dataset and samples used for training is presented in Table 7 .
Table 7.
Number of COVID-19-positive and negative images in COVID-19 Image Data Collection for CT and radiograph (full dataset). Number of radiograph images used for training and testing neural networks (after cleaning).
| COVID-19-positive | COVID-19-negative | total | |
|---|---|---|---|
| full dataset CT | 43 | 1 | 44 |
| full dataset radiograph | 253 | 63 | 316 |
| after cleaning radiograph | 233 | 56 | 289 |
The number of COVID-19-negative samples is insufficient for training neural networks. Therefore, following [14], we decided to enrich the COVID-19-negative class with radiograph images from dataset of common bacterial pneumonia [11].
The COVID-19-positive samples were split into train, validation and test sets following 75-15-10 ratio. To achieve class balance 177 samples from bacterial pneumonia dataset were included as COVID-19-negative samples. Subsequently, all the COVID-19-negative samples were split into train, validation and test sets following the same ratio and we ensured that samples coming from the bacterial pneumonia dataset would not be included in the test set. This way, the bacterial pneumonia dataset was used only for choosing the hyperparameters and training neural networks and not for the final evaluation of the models. The data split was performed in such a way to ensure that all the images coming from the same patient are assigned to the same split.
The details of the split sizes can be found in Table 8 . Please note, that the split defined on this dataset is different from the split used in our database.
Table 8.
Description of data split of COVID-19 Image Data Collection. Given are number of COVID-19-positive and negative samples in each split including additional images from bacterial pneumonia dataset.
| COVID-19 Image Data Collection |
bacterial pneumonia |
class balance | total | ||
|---|---|---|---|---|---|
| COVID-19 |
COVID-19 |
COVID-19 |
|||
| positive | negative | negative | |||
| train | 195 | 33 | 162 | 195/195 | 390 |
| validation | 22 | 7 | 15 | 22/22 | 44 |
| test | 16 | 16 | 0 | 16/16 | 32 |
| total | 233 | 56 | 177 | 233/233 | 466 |
COVID-19 CT & Radiograph Image Data Stock. COVID-19 CT & Radiograph Image Data Stock contains images of both CT and radiograph scans, the number of which is presented in Table 9 . CT scans which do not show full lungs were discarded and all radiograph images were used. We used the split provided in the database. We trained separate models for radiograph and CT scans and separate models for binary (distinguishing between COVID-19-positive and negative samples) and multiclass classification (predicting the exact class). For multiclass classification we only used classes with enough samples – in case of CT samples of viral pneumonia were discarded, and in case of radiograph samples of ARDS and fungal pneumonia were discarded. The details of the training, validation and test sets for each scenario are presented in Table 10, Table 11 .
Table 9.
Description of COVID-19 CT & Radiograph Image Data Stock used for training and evaluation of neural networks. Given are number of COVID-19-positive and negative samples of radiograph (dataset radiograph) and CT (dataset CT full) data and number of CT scans used for training and evaluation (CT after cleaning).
| COVID-19-positive | COVID-19-negative | total | |
|---|---|---|---|
| dataset radiograph | 333 | 5917 | 6250 |
| dataset CT full | 8051 | 3228 | 11,279 |
| CT after cleaning | 3980 | 2016 | 5996 |
Table 10.
Data split details of COVID-19 CT & Radiograph Image Data Stock for binary and multiclass classifiers of radiograph scans. Given are number of images in each split for each class.
| binary | COVID-19 |
COVID-19 |
total |
|---|---|---|---|
| positive | negative | ||
| train | 231 | 4143 | 4374 |
| validation | 53 | 887 | 940 |
| test | 49 | 887 | 936 |
| multiclass | COVID-19 |
bacterial |
healthy |
viral |
total |
|---|---|---|---|---|---|
| positive | pneumonia | chest | pneumonia | ||
| train | 231 | 1966 | 1108 | 1059 | 4364 |
| validation | 53 | 420 | 238 | 225 | 932 |
| test | 49 | 420 | 237 | 225 | 935 |
Table 11.
Data split details of COVID-19 CT & Radiograph Image Data Stock for binary and multiclass classifiers of CT scans. Given are number of images in each split for each class.
| binary | COVID-19-positive | COVID-19-negative | total |
|---|---|---|---|
| train | 2749 | 1522 | 4271 |
| validation | 626 | 270 | 896 |
| test | 605 | 320 | 925 |
| multiclass | COVID-19-positive | fungal pneumonia | healthy chest | total |
|---|---|---|---|---|
| train | 2749 | 653 | 773 | 4175 |
| validation | 626 | 127 | 130 | 883 |
| test | 605 | 139 | 151 | 895 |
Pre-processing and normalization. Images from all the datasets were normalised to have zero mean and unit variance. The normalization parameters were calculated on the training datasets only and for each dataset independently and before any other modification. The images present in the datasets have variable aspects and sizes so to simplify the application of neural networks they were all cropped to square ratio and resized to 512 × 512 pixels.
3.2. Models
We trained the following models: ResNet-18, ResNet-50 [55], WideResNet-50 [56], and DenseNet-169 [18]. For these models we used pytorch implementations and ImageNet initialisation. Additionally, we used DenseNet-121 from [57] which is pre-trained on seven datasets: ChestX-ray14 [19], PadChest [58], Chexpert [59], MIMIC-CXR [60], Indiana chest X-ray collection [61], dataset from [62], and the RSNA Pneumonia Detection Challenge dataset1 which is a subset of ChestX-ray8 [19]. We refer to this model as DenseNet-121+ to highlight that it has been pre-trained on medical datasets instead of ImageNet.
All of the models were trained with Adam optimiser and used step scheduler with step size 7 and gamma equal to 0.1. ResNet-18 was trained with batch size equal to 32, ResNet-50 with batch size equal to 16, and the remaining networks with batch size equal to 8. The loss function for all the models was cross entropy. The networks were trained for 100 epochs with early stopping with patience equal to 10. The other training parameters were chosen using random search which we describe next.
Random search. Random search was used to find the best hyperparameters for training individual networks. We considered learning rates in range 10e-3 - 10e-6 and the following augmentations: random rotation (in the interval −90 to 90°), brightness (0.11 for COVID-19, 0.2 for COVID-CT), horizontal flip, center crop and contrast (0.2). Out of 32 possible choices of augmentations (each augmentation can be turned on and off which gives 25 possibilities) 5 were sampled and checked with each learning rate. To sum up: 20 models of each family were trained for each dataset. During hyperparameter search the models were trained only for 30 epochs which should be enough to tell apart better and worse performing combinations.
In case of binary classification we have chosen the best performing architectures based on their AUC on the validation set, and in case of multiclass classification using the F1 score. The hyperparameters chosen for each model are presented in Appendix A.
3.3. Metrics
In case of binary classification we report the following metrics: precision, recall, F1 score, accuracy and area under ROC curve (AUC). We assume that the positive class is COVID-19.
In case of multiclass classification we report: precision, recall, F1 score, accuracy, binary AUC calculated by merging COVID-19-negative classes together, and multiclass AUC obtained by calculating binary AUC scores for each class in one-versus-rest regime and taking an unweighted mean of these scores.2 Again, we assume COVID-19 to be the positive class and the remaining classes to be negative.
For all these metrics the higher the score, the better the performance.
4. Results
In this section we present the Results of binary classifiers trained on COVID-CT and COVID-19 Image Data Collection, and binary and multiclass classifiers trained on COVID-19 CT & Radiograph Image Data Stock on their respective test sets. Next, we compare these models by evaluating their performance on an additionally constructed test set. Finally, we compare binary and multiclass classifiers trained on COVID-19 CT & Radiograph Image Data Stock on the task of binary COVID-19 classification and show that precise label information can be used to improve performance on this task.
Results of the binary classifiers trained on CTs on their respective test sets. The results for binary classifiers trained on images of CT scans are shown in Table 12 . In this case, the models trained on COVID-CT usually achieve better performance on their respective test set. The exceptions are ResNet-18 which achieves better performance on its respective test set when trained on COVID-19 CT & Radiograph Image Data Stock, and DenseNet-121+ which in both cases achieves a performance similar to a random classifier. These results are surprising, given that the number of training samples in COVID-19 CT & Radiograph Image Data Stock is much higher than in COVID-CT.
Table 12.
Test Results of binary classifiers on CT data.
| Precision | Recall | F1 score | Accuracy | AUC | |
|---|---|---|---|---|---|
| COVID-CT | |||||
| ResNet-18 | 0.57 | 0.77 | 0.65 | 0.61 | 0.61 |
| ResNet-50 | 0.87 | 0.76 | 0.81 | 0.83 | 0.83 |
| DenseNet-169 | 0.8 | 0.67 | 0.73 | 0.76 | 0.76 |
| WideResNet-50 | 0.85 | 0.80 | 0.82 | 0.83 | 0.83 |
| DenseNet-121+ |
0.67 |
0.04 |
0.08 |
0.53 |
0.51 |
| COVID-19 CT & Radiograph Image Data Stock | |||||
| ResNet-18 | 0.86 | 0.71 | 0.78 | 0.74 | 0.75 |
| ResNet-50 | 0.73 | 0.59 | 0.65 | 0.59 | 0.59 |
| DenseNet-169 | 0.78 | 0.72 | 0.75 | 0.69 | 0.67 |
| WideResNet-50 | 0.77 | 0.99 | 0.87 | 0.81 | 0.72 |
| DenseNet-121+ | 0.69 | 0.67 | 0.68 | 0.59 | 0.55 |
A direct comparison of these models cannot be drawn from Table 12 as they were evaluated on different test sets. To provide a more scrupulous comparison we constructed an additional test set which will be described later in this section.
Results of the binary classifiers trained on radiographs on their respective test sets. In Table 13 we present the results of binary classifiers trained on radiograph images. Models trained on COVID-19 CT & Radiograph Image Data Stock achieve much higher performance on their respective test set than the models trained on COVID-19 Image Data Collection. Since the methodology for optimising models on both datasets was exactly the same, we attribute this to a different number of samples in the respective training sets. It is worth noting, that the models trained on COVID-19 Image Data Collection utilise during training additional samples from a dataset of bacterial pneumonia, which was an arbitrary choice, while in case of the models trained on COVID-19 CT&Radiograph Image Data Stock the additional data was already contained within the dataset and no arbitrary choice had to be made.
Table 13.
Test Results of binary classifiers on radiograph data.
| Precision | Recall | F1 score | Accuracy | AUC | |
|---|---|---|---|---|---|
| COVID-19 Image Data Collection | |||||
| ResNet-18 | 0.6 | 0.75 | 0.67 | 0.62 | 0.63 |
| ResNet-50 | 0.60 | 0.75 | 0.67 | 0.62 | 0.63 |
| DenseNet-169 | 0.64 | 1 | 0.78 | 0.72 | 0.72 |
| WideResNet-50 | 0.62 | 0.94 | 0.75 | 0.69 | 0.69 |
| DenseNet-121+ |
0.5 |
0.88 |
0.64 |
0.5 |
0.5 |
| COVID-19 CT & Radiograph Image Data Stock | |||||
| ResNet-18 | 0.64 | 1.00 | 0.78 | 0.97 | 0.98 |
| ResNet-50 | 0.55 | 1.00 | 0.71 | 0.96 | 0.98 |
| DenseNet-169 | 0.71 | 0.96 | 0.82 | 0.98 | 0.97 |
| WideResNet-50 | 0.73 | 0.96 | 0.83 | 0.98 | 0.97 |
| DenseNet-121+ | 0.36 | 0.94 | 0.52 | 0.91 | 0.92 |
A direct comparison between these models is not possible as they were evaluated on different test sets. Again we provide a more scrupulous comparison later in this section.
Results of the multiclass classifiers. In Table 14 we present results of multiclass classifiers trained on COVID-19 CT & Radiograph Image Data Stock. The binary AUC was calculated by merging COVID-19-negative classes together. Note, that in this case misclassifying a COVID-19-negative sample as a sample of another COVID-19-negative class is not considered as an error – the task is to tell the COVID-19-positive and negative samples apart. The multiclass AUC gives us insight into how well the models separate each class from the others and the resulting mean is not over-influenced by the most common class in the dataset. The confusion matrices for the best performing models are presented in Fig. 5 and the confusion matrices of the remaining models are shown in Appendix B.
Table 14.
Test Results of multiclass classifiers on radiograph and CT data from COVID-19 CT & Radiograph Image Data Stock.
| Precision | Recall | F1 score | Accuracy | Binary |
Multiclass |
|
|---|---|---|---|---|---|---|
| AUC | AUC | |||||
| CT | ||||||
| ResNet-18 | 0.82 | 0.83 | 0.82 | 0.73 | 0.72 | 0.85 |
| ResNet-50 | 0.80 | 0.63 | 0.70 | 0.59 | 0.65 | 0.74 |
| DenseNet-169 | 0.78 | 0.77 | 0.77 | 0.69 | 0.65 | 0.82 |
| WideResNet-50 | 0.74 | 1.00 | 0.85 | 0.64 | 0.99 | 0.87 |
| DenseNet-121+ |
0.76 |
0.82 |
0.79 |
0.70 |
0.64 |
0.83 |
| Radiograph | ||||||
| ResNet-18 | 0.70 | 1.00 | 0.82 | 0.65 | 0.99 | 0.90 |
| ResNet-50 | 0.67 | 1.00 | 0.80 | 0.62 | 0.99 | 0.89 |
| DenseNet-169 | 0.74 | 1.00 | 0.85 | 0.58 | 0.99 | 0.86 |
| WideResNet-50 | 0.74 | 1.00 | 0.85 | 0.64 | 0.99 | 0.87 |
| DenseNet-121+ | 0.47 | 0.94 | 0.63 | 0.62 | 0.94 | 0.85 |
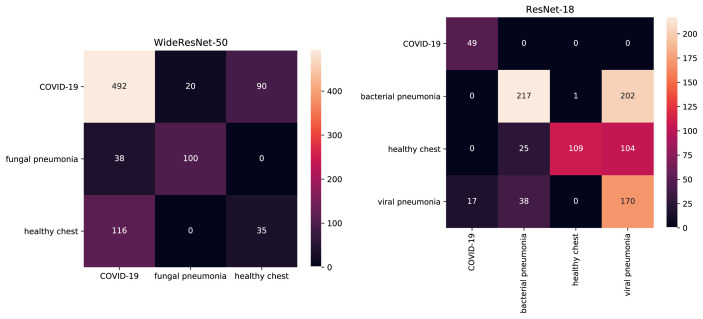
Fig. 5.
Confusion matrices of best performing multiclass models. True labels are in rows, predicted labels in columns. On the left confusion matrix for WideResNet-50 trained on CT and on the right confusion matrix for ReseNet-18 trained on radiograph.
The multiclass AUC of the presented models ranges between 0.74 for ResNet-50 and 0.87 for WideResNet-50. When evaluated on the task of binary COVID-19 classification the Results for ResNet-50, DenseNet-169 and DenseNet-121+ are far worse with binary AUC of 0.65, 0.65 and 0.64 respectively. It suggests that these models cannot tell COVID-19-positive and negative samples apart even though they might be able to distinguish between various COVID-19-negative classes. On the other hand, WideResNet-50 achieves binary AUC of 0.99 and multiclass AUC of 0.87 which suggests it might be a good candidate for COVID-19 classification. However, a careful analysis of the confusion matrix for this model shown in Fig. 5 reveals that even though this model is able to correctly classify most of the COVID-19-positive samples, a substantial number them is categorised as being from a healthy patient. It is striking that images of healthy chest are more often classified as being COVID-19-positive (116 cases) than as belonging to its true class (only 35 cases).
In comparison to their CT counterparts, the models trained on radiograph data achieve a higher performance on their respective test set. In case of binary AUC DenseNet-121+ achieves a score of 0.94 and all the other models achieve a score of 0.99. In case of multiclass classification the best performance is achieved by ResNets with AUC of 0.90 and of 0.89 for ResNet-18 and ResNet-50 respectively. The confusion matrix for ResNet-18 is presented in Fig. 5. It can be seen that all COVID-19-positive images are correctly classified and only a small percentage of viral pneumonia images (less then 10%) was classified as COVID-19-positive cases. The separation between the other classes is less accurate – almost a half of bacterial pneumonia images is classified as viral pneumonia cases and over a half of healthy cases are classified as viral (104 cases) or bacterial pneumonia (104 cases). Interestingly, viral pneumonia images are mixed with bacterial pneumonia much less often then the other way around.
Comparison between the models trained on COVID-CT, COVID-19 Image Data Collection and COVID-19 CT & Radiograph Image Data Stock. To compare models trained on COVID-CT, COVID-19 Image Data Collection and COVID-19 CT & Radiograph Image Data Stock we constructed two additional test sets. We used the COVID-19 CT & Radiograph Image Data Stock test sets and removed from them all the images which were present in training or validation sets of models trained on COVID-CT or COVID-19 Image Data Collection. From these reduced test sets we sampled images in such a way to ensure a proper class distribution.
In case of CT data we have not found any images that were present in COVID-CT train or validation sets which is not surprising as this dataset was not used as a source of images for COVID-19 CT & Radiograph Image Data Stock. Since COVID-CT does not provide an exact diagnosis for COVID-19-negative samples we could not ensure the data distribution in the reduced test set to matche the distribution in COVID-CT dataset. The class balance was ensured by sampling 278 COVID-19 images, 139 fungal pneumonia images and 139 images of healthy chest. The resulting test set contains 556 images in total.
In case of Radiograph data, 42 images from COVID-19 CT & Radiograph Image Data Stock test set were found in the COVID-19 Image Data Collection train and validation sets, most of them being COVID-19-positive samples. To imitate the class balance and class distribution in the COVID-19 Image Data Collection all the remaining COVID-19-positive samples were used and an equal number of bacterial pneumonia images was sampled. This resulted in a small test set of 24 images.
We used these datasets to evaluate the performance of the binary classifiers trained on COVID-CT, COVID-19 Image Data Collection and COVID-19 CT & Radiograph Image Data Stock and the multiclass classifiers trained on COVID-19 CT & Radiograph Image Data Stock. The Results of this evaluation are presented in Table 15, Table 16 .
Table 15.
Results of models trained on COVID-CT and COVID-19 CT & Radiograph Image Data Stock on an additional test set.
| Precision | Recall | F1 score | Accuracy | Binary AUC | |
|---|---|---|---|---|---|
| COVID-CT | |||||
| ResNet-18 | 0.88 | 0.05 | 0.10 | 0.28 | 0.52 |
| ResNet-50 | 0.59 | 0.41 | 0.48 | 0.41 | 0.56 |
| DenseNet-169 | 0.12 | 0.02 | 0.04 | 0.21 | 0.43 |
| WideResNet-50 | 0.64 | 0.64 | 0.64 | 0.53 | 0.64 |
| DenseNet-121+ |
0.24 |
0.10 |
0.14 |
0.19 |
0.38 |
| COVID-19 CT & Radiograph Image Data Stock binary | |||||
| ResNet-18 | 0.69 | 0.83 | 0.75 | 0.68 | 0.73 |
| ResNet-50 | 0.66 | 0.61 | 0.63 | 0.57 | 0.65 |
| DenseNet-169 | 0.62 | 0.75 | 0.68 | 0.64 | 0.64 |
| WideResNet-50 | 0.60 | 0.79 | 0.69 | 0.63 | 0.63 |
| DenseNet-121+ |
0.52 |
0.51 |
0.51 |
0.49 |
0.52 |
| COVID-19 CT & Radiograph Image Data Stock multiclass | |||||
| ResNet-18 | 0.84 | 0.72 | 0.78 | 0.57 | 0.79 |
| ResNet-50 | 0.62 | 0.58 | 0.60 | 0.45 | 0.61 |
| DenseNet-169 | 0.68 | 0.71 | 0.69 | 0.67 | 0.74 |
| WideResNet-50 | 0.55 | 0.66 | 0.60 | 0.52 | 0.56 |
| DenseNet-121+ | 0.69 | 0.83 | 0.75 | 0.68 | 0.73 |
Table 16.
Results of models trained on COVID-19 Image Data Collection and COVID-19 CT & Radiograph Image Data Stock on an additional test set.
| Precision | Recall | F1 score | Accuracy | Binary AUC | |
|---|---|---|---|---|---|
| COVID-19 Image Data Collection | |||||
| ResNet-18 | 0.52 | 0.92 | 0.67 | 0.54 | 0.54 |
| ResNet-50 | 0.50 | 1.00 | 0.67 | 0.50 | 0.50 |
| DenseNet-169 | 0.00 | 0.00 | 0.00 | 0.50 | 0.50 |
| WideResNet-50 | 0.00 | 0.00 | 0.00 | 0.38 | 0.38 |
| DenseNet-121+ |
0.50 |
1.00 |
0.67 |
0.50 |
0.50 |
| COVID-19 CT & Radiograph Image Data Stock binary | |||||
| ResNet-18 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| ResNet-50 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| DenseNet-169 | 1.00 | 0.92 | 0.96 | 0.96 | 0.96 |
| WideResNet-50 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| DenseNet-121+ |
0.86 |
1.00 |
0.92 |
0.92 |
0.92 |
| COVID-19 CT & Radiograph Image Data Stock multiclass | |||||
| ResNet-18 | 1.00 | 1.00 | 1.00 | 0.71 | 1.00 |
| ResNet-50 | 1.00 | 1.00 | 1.00 | 0.71 | 1.00 |
| DenseNet-169 | 1.00 | 1.00 | 1.00 | 0.71 | 1.00 |
| WideResNet-50 | 1.00 | 1.00 | 1.00 | 0.75 | 1.00 |
| DenseNet-121+ | 0.91 | 0.83 | 0.87 | 0.54 | 0.87 |
In Table 15 we see that the models trained on COVID-CT achieve AUC similar or lower than a random classifier, with the exception being WideResNet-50 with AUC of 0.64. Both binary and multiclass classifiers trained on COVID-19 CT & Radiograph Image Data Stock achieve better Results. In this case, the worst performing binary classifier is DenseNet-121+ with AUC of 0.52, ResNet-18 achieves AUC of 0.73 and the remaining models score in rage 0.63–0.65.3 out of 5 multiclass classifiers achieve better AUC than their binary counterparts. The lowest AUC of 0.56 belongs to WideResNet-50 and the best performing model in this group is ResNet-18 with AUC of 0.79.
In Table 16 we see that models trained on COVID-19 Image Data Collection achieve very poor Results which are similar to those of a random classifier. On the other hand, models trained on COVID-19 CT & Radiograph Image Data Stock achieve a near maximum or maximal possible AUC. We attribute such a vivid discrepancy to the small size of the testing set, which contains only 24 elements. However, even such a small test set allows to draw a conclusion that models trained on COVID-19 CT&Radiograph Image Data Stock are more robust than models trained on COVID-19 Image Data Collection.
The Results of experiments shown in this section indicate that models trained on COVID-19 CT & Radiograph Image Data Stock are more robust than models trained on COVID-CT or COVID-19 Image Data Collection. This can be a result of a bigger or more diverse training set.
Precise label information improves the model's ability to distinguish between COVID-19-positive and negative samples. In this section, we try to answer the question if including a detailed information about the classes, i.e. not treating all the non-COVID samples as if they were coming from the same class, can help the models to better distinguish between COVID-19-positive and negative samples. We compare models trained in two scenarios: when merging all COVID-19-negative samples into one class and training a binary classifier, and when using the precise label information and training a multiclass classifier.
If the goal of the model is to distinguish between COVID-19-positive and negative samples only, the samples of different COVID-19-negative classes can be merged into a single class and used to train a binary classifier. In this scenario, the task on which the model is trained does not differ from the task of interest. However, the samples in the COVID-19-negative class come from different distributions and as a result models might struggle to treat them as if coming from a single class. Moreover, the exact information about the diagnosis is never presented to the model and so it cannot be used to learn a better representation of the problem.
To combat these issues the models can be trained to perform multiclass classification using exact label information about the diagnosis. In this case, the exact class-information can be used by the model to learn a powerful representation and samples coming from different classes do not need to be squished together which might make the learning process easier. On the other hand, for some classes the number of samples in the dataset might be insufficient for the model to be able to learn a good representation of these classes. If this is the case, such samples can be dropped from the dataset eventually decreasing its size. More importantly, in this scenario the model is trained on a different task than the task of interest.
To answer the question which of these learning paradigms brings better Results in case of COVID-19 classification we compared binary and multiclass models from the previous sections. Precisely, we evaluated the binary classifiers on the multiclass test set after binarising its classes. The results are presented in Table 17 . Comparing this table with the results of multiclass classifiers which were presented in Table 14 we see that in case of radiograph classification the multiclass models consistently achieve a slightly better binary AUC then their binary counterparts. In case of CT classification ResNet-18 and DenseNet-169 achieve slightly better results when trained as binary classifiers. On the other hand, ResNet-50 and DenseNet-121+ achieve slightly better results when trained as multiclass classifiers and WideResNet-50 achieves much better results improving from AUC of 0.74 when trained as a binary classifier to AUC of 0.99 when trained as a multiclass classifier.
Table 17.
Results of binary classifiers trained on COVID-19 CT & Radiograph Image Data Stock and evaluated on COVID-19 CT & Radiograph Image Data Stock multiclass test sets.
| Precision | Recall | F1 score | Accuracy | Binary AUC | |
|---|---|---|---|---|---|
| Radiograph | |||||
| ResNet-18 | 0.76 | 0.96 | 0.85 | 0.50 | 0.97 |
| ResNet-50 | 0.57 | 1.00 | 0.73 | 0.50 | 0.98 |
| DenseNet-169 | 0.75 | 0.96 | 0.84 | 0.50 | 0.97 |
| WideResNet-50 | 0.76 | 0.96 | 0.85 | 0.50 | 0.97 |
| DenseNet-121+ | 0.37 | 0.94 | 0.53 | 0.47 | 0.92 |
| CT | |||||
| ResNet-18 | 0.92 | 0.71 | 0.80 | 0.61 | 0.79 |
| ResNet-50 | 0.77 | 0.59 | 0.67 | 0.50 | 0.60 |
| DenseNet-169 | 0.81 | 0.82 | 0.76 | 0.62 | 0.69 |
| WideResNet-50 | 0.80 | 0.99 | 0.89 | 0.78 | 0.74 |
| DenseNet-121+ | 0.71 | 0.67 | 0.69 | 0.57 | 0.55 |
To conclude, these Results suggest that precise label information can improve the performance of neural networks on the task of binary COVID-19 classification.
5. Conclusion
In this work, we proposed a new self-contained dataset for COVID-19 classification which includes a significant number of both CT and radiograph images from a diverse set of classes. The dataset was constructed ensuring a high quality of samples with each image carefully annotated with its precise label and source. A train-validation-test split is defined. The dataset is publicly available and will be updated monthly until the inhibition of COVID-19 pandemic.
We fine-tuned several neural network architectures pre-trained on ImageNet or medical dataset to provide benchmark Results for the proposed dataset. We compared the performance of models trained on COVID-19 CT & Radiograph Image Data Stock with models trained on COVID-CT and COVID-19 Image Data Collection showing that models trained on COVID-19 CT & Radiograph Image Data Stock perform better. We compared binary and multiclass classifiers trained on COVID-19 CT & Radiograph Image Data Stock revealing that training neural networks with precise label information can improve the performance on binary COVID-19 classification task.
The research we have developed does not allow to compare how 3D CT reconstruction influences classifications compared to 2D dimensional radiograph. Neural networks for CT were trained for each slide separately, which corresponds to the radiograph dimension. In the future we would like to investigate the whole series for patient at ones. Also, we would like to address several remaining questions which include using additional information about the patient to build better-performing models or analysing how the models trained on images of CT and radiograph will perform when used directly on CT or radiograph scans.
Declaration of competing interest
The authors declare that there is no conflict of interest.
Acknowledgement
National Centre for Research and Development, BRIdge Alfa Programme funding within Operational Programme Smart Growth of European Union, grant ID: 4/2017.
Footnotes
In other words, we use scikit-learn implementation of roc_auc_score with parameter multi_class set to ‘ovr’, and parameter average set to ‘macro’.
Appendix A.
Here, we present the hyperparameters chosen for each model presented in the paper. A description of the methodology used to obtain these hyperparametrs is presented in section 3.2.
Table 18.
Hyperparameters chosen with random search for each architecture.
| lr | random rotation | brightness | horizontal flip | center crop | contrast | |
|---|---|---|---|---|---|---|
| COVID-19 Image Data Collection | ||||||
| ResNet-18 | 0.001 | + | + | |||
| ResNet-50 | 0.001 | + | + | + | ||
| DenseNet-169 | 0.0001 | + | + | + | ||
| WideResNet-50 | 0.00001 | + | + | + | ||
| DenseNet-121+ |
0.0001 |
+ |
+ |
|||
| COVID-19 CT & Radiograph Image Data Stock binary X-ray | ||||||
| ResNet-18 | 0.001 | |||||
| ResNet-50 | 0.001 | |||||
| DenseNet-169 | 0.00001 | |||||
| WideResNet-50 | 0.0001 | + | + | + | ||
| DenseNet-121+ | 0.001 | + | + | + | ||
Table 19.
Hyperparameters chosen with random search for each architecture.
| lr | random rotation | brightness | horizontal flip | center crop | contrast | |
|---|---|---|---|---|---|---|
| COVID-19 CT & Radiograph Image Data Stock multiclass Radiograph | ||||||
| ResNet-18 | 0.001 | + | + | |||
| ResNet-50 | 0.001 | |||||
| DenseNet-169 | 0.001 | + | + | |||
| WideResNet-50 | 0.00001 | + | + | + | ||
| DenseNet-121+ | 0.001 | + | ||||
| COVID-CT | ||||||
| ResNet-18 | 0.001 | + | + | + | + | |
| ResNet-50 | 0.0001 | + | + | |||
| DenseNet-169 | 0.0001 | + | ||||
| WideResNet-50 | 0.0001 | + | + | + | ||
| DenseNet-121+ | 0.00001 | + | ||||
Table 20.
Hyperparameters chosen with random search for each architecture.
| lr | random rotation | brightness | horizontal flip | center crop | contrast | |
|---|---|---|---|---|---|---|
| COVID-19 CT & Radiograph Image Data Stock binary CT | ||||||
| ResNet-18 | 0.001 | + | + | |||
| ResNet-50 | 0.001 | |||||
| DenseNet-169 | 0.000001 | + | ||||
| WideResNet-50 | 0.0001 | + | ||||
| DenseNet-121+ |
0.001 |
+ |
||||
| COVID-19 CT & Radiograph Image Data Stock multiclass CT | ||||||
| ResNet-18 | 0.001 | + | + | |||
| ResNet-50 | 0.0001 | + | + | + | ||
| DenseNet-169 | 0.000001 | + | ||||
| WideResNet-50 | 0.000001 | |||||
| DenseNet-121+ | 0.001 | + | ||||
Appendix B.
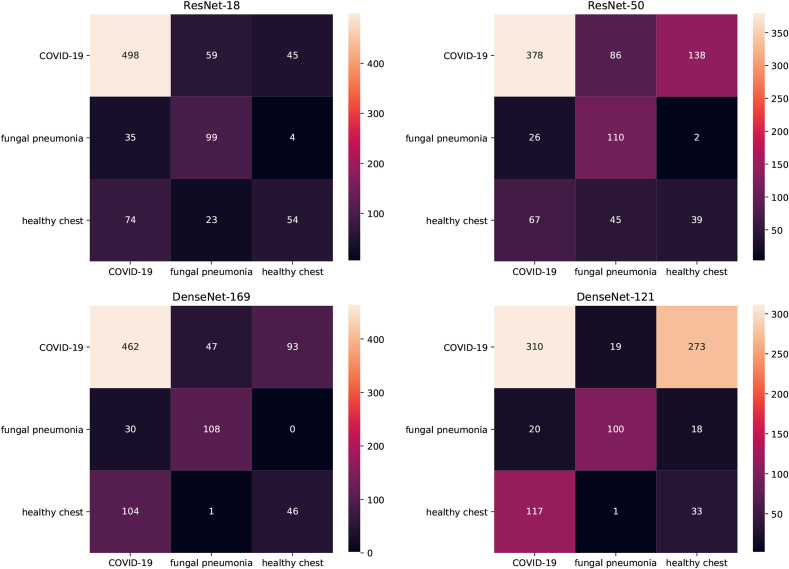
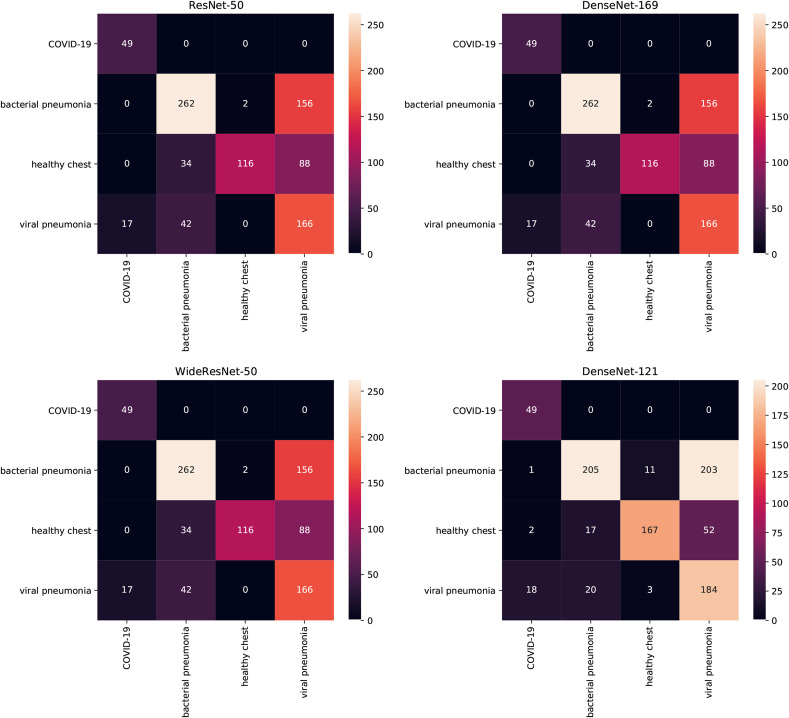
Here, we present the confusion matrices of multiclass classifiers trained on COVID-19 CT & Radiograph Image Data Stock.
In Fig. 6 the Results of multiclass classifiers trained on CT data is shown. A careful analysis shows that these models struggle to separate between healthy and COVID-19-positive cases – most of the COVID-19-positive samples are well classified but when mistakes occur they are more likely to be categorised as healthy samples than fungal pneumonia samples. At the same time, healthy chest samples are more often classified as COVID-19-positive instances than as members of their own class.
Fig. 6.
Confusion matrices of multiclass models on CT data. True labels are in rows, predicted labels in columns.
In Fig. 7 the Results of multiclass classifiers trained on radiograph data is shown. In this case, the COVID-19-positive samples are always correctly classified and hardly any samples of bacterial pneumonia or healthy chest are categorised as COVID-19-positive samples. The class which is most commonly confused with COVID-19 is viral pneumonia but the number of samples misclassified as COVID-19 never exceeds 10% of samples in this class. Viral and bacterial pneumonia classes are not well separated with bacterial pneumonia cases categorised as viral pneumonia more often than the other way around.
Fig. 7.
Confusion matrices of multiclass models on Radiograph data. True labels are in rows, predicted labels in columns.
The performance metrics for these models are shown in Table 14.
References
- 1.Ding X., Xu J., Zhou J., Long Q. Chest CT findings of COVID-19 pneumonia by duration of symptoms. Eur. J. Radiol. 2020:109009. doi: 10.1016/j.ejrad.2020.109009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3):2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology. 2020:200343. doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang W. Imaging changes of severe COVID-19 pneumonia in advanced stage. Intensive Care Med. 2020:1–3. doi: 10.1007/s00134-020-05990-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.C. Shen, Y. Bar-Yam, Breaking the Testing Logjam: CT Scan Diagnosis .
- 6.ACR Recommendations for the Use of Chest Radiography and Computed Tomography (CT) for Suspected COVID-19 Infection. 2020. https://www.acr.org/Advocacy-and-Economics/ACR-Position-Statements/Recommendations-for-Chest-Radiography-and-CT-for-Suspected-COVID19-Infection [Google Scholar]
- 7.X. He, X. Yang, S. Zhang, J. Zhao, Y. Zhang, E. Xing, P. Xie, Sample-efficient Deep Learning for COVID-19 Diagnosis based on CT Scans, medRxiv doi:\let\@tempa\bibinfo@X@doi10.1101/2020.04.13. 20063941, URL https://www.medrxiv.org/content/early/2020/04/17/2020.04.13.20063941.
- 8.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020:103792. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.J. Zhao, Y. Zhang, X. He, P. Xie, COVID-CT-Dataset: a CT Scan Dataset about COVID-19, arXiv preprint arXiv:2003.13865 .
- 10.J. P. Cohen, P. Morrison, L. Dao, COVID-19 image data collection, arXiv 2003.11597 URL https://github.com/ieee8023/covid-chestxray-dataset.
- 11.Kermany D.S., Goldbaum M., Cai W., Valentim C.C., Liang H., Baxter S.L., McKeown A., Yang G., Wu X., Yan F. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172(5):1122–1131. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 12.van Ginneken B., Jacobs C. 2019. LUNA16 Part 1/2. \let\@tempa\bibinfo@X@doi10.5281/zenodo.3723295, URL. [DOI] [Google Scholar]
- 13.van Ginneken B., Jacobs C. 2019. LUNA16 Part 2/2. \let\@tempa\bibinfo@X@doi10.5281/zenodo.3723299, URL. [DOI] [Google Scholar]
- 14.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020:1. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.A. G. Howard, M. Zhu, B. Chen, D. Kalenichenko, W. Wang, T. Weyand, M. Andreetto, H. Adam, Mobilenets: Efficient Convolutional Neural Networks for Mobile Vision Applications, arXiv preprint arXiv:1704.04861 .
- 16.Russakovsky O., Deng J., Su H., Krause J., Satheesh S., Ma S., Huang Z., Karpathy A., Khosla A., Bernstein M. Imagenet large scale visual recognition challenge. Int. J. Comput. Vis. 2015;115(3):211–252. [Google Scholar]
- 17.A. M. V. Dadario, COVID-19 X rays, doi:\let\@tempa\bibinfo@X@doi10.34740/KAGGLE/DSV/1019469, URL https://www.kaggle.com/dsv/1019469, (????).
- 18.Huang G., Liu Z., Van Der Maaten L., Weinberger K.Q. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Densely connected convolutional networks; pp. 4700–4708. [Google Scholar]
- 19.Wang X., Peng Y., Lu L., Lu Z., Bagheri M., Summers R.M. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Chestx-ray8: hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases; pp. 2097–2106. [Google Scholar]
- 20.Chen T., Kornblith S., Norouzi M., Hinton G. 2002. A Simple Framework for Contrastive Learning of Visual Representations. arXiv preprint arXiv. 05709. [Google Scholar]
- 21.Salehi S., Abedi A., Balakrishnan S., Gholamrezanezhad A. Coronavirus disease 2019 (COVID-19): a systematic review of imaging findings in 919 patients. Am. J. Roentgenol. 2020:1–7. doi: 10.2214/AJR.20.23034. [DOI] [PubMed] [Google Scholar]
- 22.Guan W., Liu J., Yu C. CT findings of coronavirus disease (COVID-19) severe pneumonia. Am. J. Roentgenol. 2020;214(5):W85–W86. doi: 10.2214/AJR.20.23035. [DOI] [PubMed] [Google Scholar]
- 23.Silverstein W.K., Stroud L., Cleghorn G.E., Leis J.A. First imported case of 2019 novel coronavirus in Canada, presenting as mild pneumonia. Lancet. 2020;395(10225):734. doi: 10.1016/S0140-6736(20)30370-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.S.-C. Cheng, Y.-C. Chang, Y.-L. F. Chiang, Y.-C. Chien, M. Cheng, C.-H. Yang, C.-H. Huang, Y.-N. Hsu, First case of coronavirus disease 2019 (COVID-19) pneumonia in taiwan, J. Formos. Med. Assoc. . [DOI] [PMC free article] [PubMed]
- 25.N.-Y. Lee, C.-W. Li, H.-P. Tsai, P.-L. Chen, L.-S. Syue, M.-C. Li, C.-S. Tsai, C.-L. Lo, P.-R. Hsueh, W.-C. Ko, A case of COVID-19 and pneumonia returning from Macau in Taiwan: clinical course and anti-SARS-CoV-2 IgG dynamic, J. Microbiol. Immunol. Infect. . [DOI] [PMC free article] [PubMed]
- 26.W.-H. Hsih, M.-Y. Cheng, M.-W. Ho, C.-H. Chou, P.-C. Lin, C.-Y. Chi, W.-C. Liao, C.-Y. Chen, L.-Y. Leong, N. Tien, et al., Featuring COVID-19 cases via screening symptomatic patients with epidemiologic link during flu season in a medical center of central Taiwan, J. Microbiol. Immunol. Infect. . [DOI] [PMC free article] [PubMed]
- 27.L. B. Adair II, E. J. Ledermann, reportChest CT Findings of Early and Progressive Phase COVID-19 Infection from a US Patient, Radiology Case Reports . [DOI] [PMC free article] [PubMed]
- 28.Thevarajan I., Nguyen T.H., Koutsakos M., Druce J., Caly L., van de Sandt C.E., Jia X., Nicholson S., Catton M., Cowie B. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat. Med. 2020;26(4):453–455. doi: 10.1038/s41591-020-0819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.J.-j. Zhang, X. Dong, Y.-y. Cao, Y.-d. Yuan, Y.-b. Yang, Y.-q. Yan, C. A. Akdis, Y.-d. Gao, Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China, Allergy . [DOI] [PubMed]
- 30.Phan L.T., Nguyen T.V., Luong Q.C., Nguyen T.V., Nguyen H.T., Le H.Q., Nguyen T.T., Cao T.M., Pham Q.D. Importation and human-to-human transmission of a novel coronavirus in Vietnam. N. Engl. J. Med. 2020;382(9):872–874. doi: 10.1056/NEJMc2001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.J. Wu, J. Liu, X. Zhao, C. Liu, W. Wang, D. Wang, W. Xu, C. Zhang, J. Yu, B. Jiang, et al., Clinical Characteristics of Imported Cases of Coronavirus Disease 2019 (COVID-19) in Jiangsu Province: A Multicenter Descriptive Study, Clinical Infectious Diseases . [DOI] [PMC free article] [PubMed]
- 32.S. Fatima, I. Ratnani, M. Husain, S. Surani, Radiological findings in patients with COVID-19, Cureus 12 (4). [DOI] [PMC free article] [PubMed]
- 33.Giang H.T.N., Shah J., Hung T.H., Reda A., Truong L.N., Huy N.T. The first Vietnamese case of COVID-19 acquired from China. Lancet Infect. Dis. 2020;20(4):408–409. doi: 10.1016/S1473-3099(20)30111-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.J. Lim, S. Jeon, H.-Y. Shin, M. J. Kim, Y. M. Seong, W. J. Lee, K.-W. Choe, Y. M. Kang, B. Lee, S.-J. Park, Case of the index patient who caused tertiary transmission of COVID-19 infection in Korea: the application of lopinavir/ritonavir for the treatment of COVID-19 infected pneumonia monitored by quantitative RT-PCR, J. Kor. Med. Sci. 35 (6). [DOI] [PMC free article] [PubMed]
- 35.Yoon S.H., Lee K.H., Kim J.Y., Lee Y.K., Ko H., Kim K.H., Park C.M., Kim Y.-H. Chest radiographic and CT findings of the 2019 novel coronavirus disease (COVID-19): analysis of nine patients treated in Korea. Korean J. Radiol. 2020;21(4):494–500. doi: 10.3348/kjr.2020.0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi H., Han X., Zheng C. Evolution of CT manifestations in a patient recovered from 2019 novel coronavirus (2019-nCoV) pneumonia in Wuhan, China. Radiology. 2020;295(1) doi: 10.1148/radiol.2020200269. 20–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y.-C., Liao C.-H., Chang C.-F., Chou C.-C., Lin Y.-R. A locally transmitted case of SARS-CoV-2 infection in Taiwan. N. Engl. J. Med. 2020;382(11):1070–1072. doi: 10.1056/NEJMc2001573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song F., Shi N., Shan F., Zhang Z., Shen J., Lu H., Ling Y., Jiang Y., Shi Y. Emerging 2019 novel coronavirus (2019-nCoV) pneumonia. Radiology. 2020;295(1):210–217. doi: 10.1148/radiol.2020200274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wong H.Y.F., Lam H.Y.S., Fong A.H.-T., Leung S.T., Chin T.W.-Y., Lo C.S.Y., Lui M.M.-S., Lee J.C.Y., Chiu K.W.-H., Chung T. Frequency and distribution of chest radiographic findings in COVID-19 positive patients. Radiology. 2020:201160. doi: 10.1148/radiol.2020201160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qian L., Yu J., Shi H. Severe acute respiratory disease in a Huanan seafood market worker: images of an early casualty. Radiology: Cardiothoracic Imaging. 2020;2(1) doi: 10.1148/ryct.2020200033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kong W., Agarwal P.P. Chest imaging appearance of COVID-19 infection. Radiology: Cardiothoracic Imaging. 2020;2(1) doi: 10.1148/ryct.2020200028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ng M.-Y., Lee E.Y., Yang J., Yang F., Li X., Wang H., Lui M.M.-s., Lo C.S.-Y., Leung B., Khong P.-L. Imaging profile of the COVID-19 infection: radiologic findings and literature review. Radiology: Cardiothoracic Imaging. 2020;2(1) doi: 10.1148/ryct.2020200034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.A. Jacobi, M. Chung, A. Bernheim, C. Eber, Portable Chest X-Ray in Coronavirus Disease-19 (COVID-19): A Pictorial Review, Clinical Imaging . [DOI] [PMC free article] [PubMed]
- 45.Abbas A., Abdelsamea M.M., Gaber M.M. 2003. Classification of COVID-19 in Chest X-Ray Images Using DeTraC Deep Convolutional Neural Network; p. 13815. arXiv preprint arXiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.R. Sánchez-Oro, J. T. Nuez, G. Martínez-Sanz, Radiological Findings for Diagnosis of SARS-CoV-2 Pneumonia (COVID-19), Medicina Clinica (English Ed.) . [DOI] [PMC free article] [PubMed]
- 47.Chou D.-W., Wu S.-L., Chung K.-M., Han S.-C., Cheung B.M.-H. Septic pulmonary embolism requiring critical care: clinicoradiological spectrum, causative pathogens and outcomes. Clinics. 2016;71(10):562–569. doi: 10.6061/clinics/2016(10)02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Müller N.L., Franquet T., Lee K.S., Silva C.I.S. Lippincott Williams & Wilkins; 2007. Imaging of Pulmonary Infections. [Google Scholar]
- 49.Okada F., Ando Y., Honda K., Nakayama T., Ono A., Tanoue S., Maeda T., Mori H. Acute Klebsiella pneumoniae pneumonia alone and with concurrent infection: comparison of clinical and thin-section CT findings. Br. J. Radiol. 2010;83(994):854–860. doi: 10.1259/bjr/28999734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garg M., Prabhakar N., Gulati A., Agarwal R., Dhooria S. Spectrum of imaging findings in pulmonary infections. Part 1: bacterial and viral. Pol. J. Radiol. 2019;84 doi: 10.5114/pjr.2019.85812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koo H.J., Lim S., Choe J., Choi S.-H., Sung H., Do K.-H. Radiographic and CT features of viral pneumonia. Radiographics. 2018;38(3):719–739. doi: 10.1148/rg.2018170048. [DOI] [PubMed] [Google Scholar]
- 52.Franquet T. Imaging of pulmonary viral pneumonia. Radiology. 2011;260(1):18–39. doi: 10.1148/radiol.11092149. [DOI] [PubMed] [Google Scholar]
- 53.Paul N.S., Roberts H., Butany J., Chung T., Gold W., Mehta S., Konen E., Rao A., Provost Y., Hong H.H. Radiologic pattern of disease in patients with severe acute respiratory syndrome: the Toronto experience. Radiographics. 2004;24(2):553–563. doi: 10.1148/rg.242035193. [DOI] [PubMed] [Google Scholar]
- 54.Wong K., Antonio G.E., Hui D.S., Lee N., Yuen E.H., Wu A., Leung C., Rainer T.H., Cameron P., Chung S.S. Severe acute respiratory syndrome: radiographic appearances and pattern of progression in 138 patients. Radiology. 2003;228(2):401–406. doi: 10.1148/radiol.2282030593. [DOI] [PubMed] [Google Scholar]
- 55.He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Deep residual learning for image recognition; pp. 770–778. [Google Scholar]
- 56.S. Zagoruyko, N. Komodakis, Wide Residual Networks, arXiv preprint arXiv:1605.07146.
- 57.Cohen J.P., Hashir M., Brooks R., Bertrand H. Medical Imaging with Deep Learning. 2020. On the limits of cross-domain generalization in automated X-ray prediction. [Google Scholar]
- 58.Bustos A., Pertusa A., Salinas J.-M., de la Iglesia-Vayá M. 1901. Padchest: A Large Chest X-Ray Image Dataset with Multi-Label Annotated Reports. arXiv preprint arXiv, 07441. [DOI] [PubMed] [Google Scholar]
- 59.Irvin J., Rajpurkar P., Ko M., Yu Y., Ciurea-Ilcus S., Chute C., Marklund H., Haghgoo B., Ball R., Shpanskaya K. vol. 33. 2019. Chexpert: a large chest radiograph dataset with uncertainty labels and expert comparison; pp. 590–597. (Proceedings of the AAAI Conference on Artificial Intelligence). [Google Scholar]
- 60.Johnson A.E., Pollard T.J., Greenbaum N.R., Lungren M.P., Deng C.-y., Peng Y., Lu Z., Mark R.G., Berkowitz S.J., Horng S. 1901. MIMIC-CXR-JPG, a Large Publicly Available Database of Labeled Chest Radiographs. arXiv preprint arXiv. 07042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Demner-Fushman D., Kohli M.D., Rosenman M.B., Shooshan S.E., Rodriguez L., Antani S., Thoma G.R., McDonald C.J. Preparing a collection of radiology examinations for distribution and retrieval. J. Am. Med. Inf. Assoc. 2016;23(2):304–310. doi: 10.1093/jamia/ocv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Majkowska A., Mittal S., Steiner D.F., Reicher J.J., McKinney S.M., Duggan G.E., Eswaran K., Cameron Chen P.-H., Liu Y., Kalidindi S.R. Chest radiograph interpretation with deep learning models: assessment with radiologist-adjudicated reference standards and population-adjusted evaluation. Radiology. 2020;294(2):421–431. doi: 10.1148/radiol.2019191293. [DOI] [PubMed] [Google Scholar]