Abstract
Limitations to successful gene therapy with adeno-associated virus (AAV) can comprise pre-existing neutralizing antibodies to the vector capsid that can block cellular entry, or inefficient transduction of target cells that can lead to sub-optimal expression of the therapeutic transgene. Recombinant serotype 3 AAV (AAV3) is an emerging candidate for liver-directed gene therapy. In this study, we integrated rational design by using a combinatorial library derived from AAV3B capsids with directed evolution by in vitro selection for liver-targeted AAV variants. The AAV3B-DE5 variant described herein was undetectable in the original viral library but gained a selective advantage upon in vitro passaging in human hepatocarcinoma spheroid cultures. AAV3B-DE5 contains 24 capsid amino acid substitutions compared with AAV3B, distributed among all five variable regions, with strong selective pressure on VR-IV, VR-V, and VR-VII. In vivo, AAV3B-DE5 demonstrated improved human hepatocyte tropism in a liver chimeric mouse model. Importantly, this variant exhibited reduced seroreactivity to human intravenous immunoglobulin (i.v. Ig), as well as individual serum samples from 100 healthy human donors. Therefore, molecular evolution using a combinatorial library platform generated a viral capsid with high hepatocyte tropism and enhanced evasion of pre-existing AAV neutralizing antibodies.
Keywords: AAV3, gene therapy, hepatocyte, directed evolution, seroprevalence, neutralizing antibody, capsid library, liver tropism
Graphical Abstract

Biswas and colleagues describe an integrated approach to generate novel AAV3B variants. By integrating a combinatorial AAV3B capsid library platform with directed evolution in a hepatocellular carcinoma cell line, they isolated a candidate AAV3B-DE5, which exhibits improved human hepatocyte tropism and reduced seroreactivity to pre-existing AAV neutralizing antibodies in human serum.
Introduction
Recombinant adeno-associated virus (rAAV) vectors are among the most promising tools for in vivo human gene therapy, as demonstrated by an increasing number of clinical trials as well as treatment approvals being reported worldwide.1 Their favorable safety profile (especially at lower vector doses) due to the episomal localization of the therapeutic gene (non-integrating vector), minimal issues of insertional mutagenesis,2 and lack of pathogenicity in transduced cells makes these small virions the gene delivery vectors of choice. The ability of rAAV vectors to transduce long-lived non-dividing cells such as myocytes or hepatocytes also makes them ideal for sustained expression of transgenes, which would otherwise get diluted upon cell division. Clinical translation of rAAV therapy has so far been successfully applied for hereditary blindness (Luxturna [voretigene neparvovec-rzyl]), neuromuscular disorders (Zolgensma [onasemnogene abeparvovec-xioi]), and coagulation disorders (hemophilia B), among others.3, 4, 5
Wild-type (WT) AAV is a small (~26 nm), non-enveloped parvovirus. It packages a linear single-stranded DNA genome (~4.7 kb), encoding genes necessary for replication (rep) and the viral capsid (cap), flanked by palindromic inverted terminal repeats (ITRs). Except for the ITRs, which are essential, much of the viral DNA genome can be omitted for the purpose of transgene packaging and delivery, allowing for insertion of approximately 4.7 kb of foreign DNA, which altogether forms the transgene expression cassette.6 In some cases, a self-complementary single strand duplex DNA can be packaged,7 although this reduces the transgene capacity to less than half and increases the risk of immune response.8
There are several challenges that impede the successful and broad use of rAAV gene therapy. The first major limitation to systemic or intramuscular administration of AAV is the presence of pre-existing neutralizing antibodies (NAbs) against the vector capsid that can block cellular entry.9 Most of the human population is seropositive for AAV, mostly due to previous subclinical exposure to the WT virus.10 This is a major exclusion criterion for prospective patients, as even very low titers of NAbs in circulation can prevent vector entry and significantly limit effective gene transfer to the target organ. Pre-existing anti-AAV NAbs do not affect rAAV injection into immune-privileged sites such as the eye or brain. Luxturna, the US Food and Drug Administration (FDA)-approved drug for treating inherited retinal disorders, can be successfully delivered into the eye using AAV serotype 2, which is seroprevalent in 40%–70% of the human population.10, 11, 12 The impact of low-titer NAbs (<1:5), particularly on systemic gene transfer, is not accurately known.13, 14, 15 Using a different serotype is complicated because a pattern of cross-reactivity commonly occurs between variants, such as between AAV2, AAV5, and AAV8,16 or AAV1 and AAV6,17 depending on the degree of homology between capsid protein sequences.
A second limitation is transduction efficiency of target cells. Based on specific receptor interaction and post-entry mechanisms, AAV serotypes differ in cell tropism as well as transduction efficiency in the cell type of choice.18 A major challenge to gene therapy is that the functional gene may not transduce the target tissue in high enough numbers to provide therapeutic benefit. Increasing the rAAV dose in this case is not always effective, as a high viral load can induce detrimental capsid-specific T cell immune responses to the transduced cell.14,19
These limitations cannot adequately be addressed by the current limited repertoire of naturally occurring AAV serotypes, and the isolation and characterization of novel variants is time-consuming. The nature of exposed amino acid residues on the capsid surface largely determines receptor attachment, tissue transduction, and antigenicity.20 Therefore, the development of engineered AAV vectors designed either by rational modification of specific amino acids (rational mutagenesis) or in vitro/in vivo selection in the cell type of choice (directed evolution) is an attractive alternative reported by us and others.21,22 These novel rAAV capsid vectors based on different serotypes have been shown to be superior at transducing the cell type of interest and, in some cases, evading pre-existing NAbs in the host.23, 24, 25, 26
In the present study, we integrated a combination of rational design and directed evolution in order to select for an engineered AAV vector derived from the AAV3B capsid backbone. In vitro, this variant showed improved tropism for human hepatocarcinoma cells as compared to WT AAV3B. In vivo, the engineered AAV3B variant showed improved human hepatocyte transduction in a liver chimeric mouse model. Importantly, this variant exhibited reduced seroreactivity to pre-existing NAbs from both pooled human intravenous immunoglobulin (i.v. Ig), as well as in individual serum samples tested from 100 healthy human donors.
Results
Strategies to improve AAV capsid design include (1) a rational approach of mutagenizing known capsid residues critical for binding, entry, and/or intracellular trafficking, and (2) directed evolution to rapidly introduce molecular modifications into the AAV capsids, thus manipulating both diversity and selection. Using detailed knowledge of AAV capsid structures and the sequence of 150 naturally occurring AAV3 variants, as described earlier for AAV2,21 we derived a combinatorial capsid library whereby only variable regions (VRs) on the surface of virion are modified.
AAV Library Generation
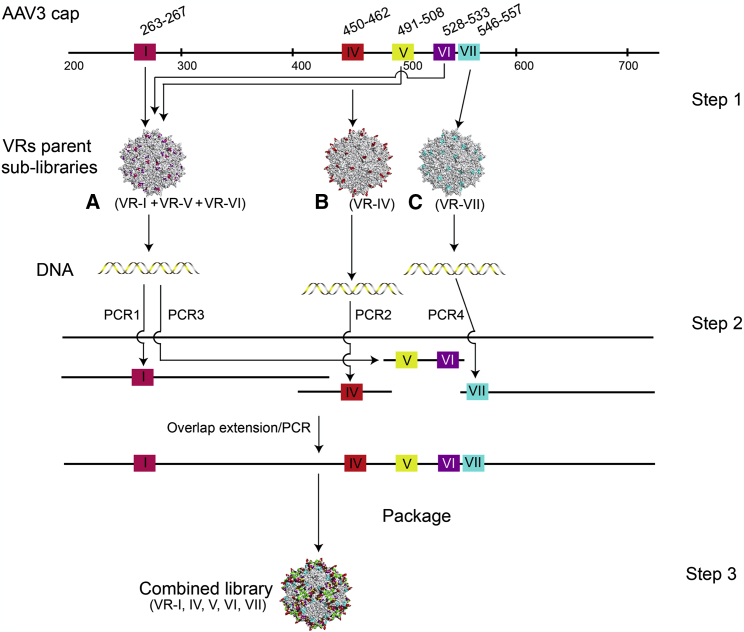
The AAV icosahedral capsid is composed of three structural proteins (VP1, VP2, and VP3), of which VP2 and VP3 are N-terminal truncated versions of VP1.27 VP3 is the most abundant capsid subunit, comprising a major part of the capsid surface, and hence the major determinant of antigenicity and cell tropism. The structure of VP3 contains a β-barrel core, with homologous β strands linked by nine highly variable extended loops called VRs (VR-I to VR-IX).28 Because of their surface location, VRs are predicted to be less critical for capsid assembly but essential for determining cell tropism and antigenicity.29 For this study, a replication-competent AAV3B capsid library was generated by modifying only surface VRs while keeping the backbone sequence unchanged to maintain the integrity of the assembling scaffold. 150 AAV naturally occurring variants in the AAV3B background were aligned as described earlier for AAV2,21 and out of all candidate positions for mutagenesis, residues that were clearly exposed to the surface were selected for modification. Only VR-I (aa residues 263–267 of the AAV3B VP1), VR-IV (aa 450–462), VR-V (aa 491–508), VR-VI (aa 528–533), and VR-VII (aa 546–557) were diversified, because mutagenizing other loops has been previously shown to be detrimental to the library’s assembly and complexity.21 In addition, each amino acid residue substitution was restricted to a subset of residues naturally occurring in this particular position when 150 WT serotypes were aligned as described by Marsic et al.21 Even with the described restrictions, the probabilistic sequence space (also known as theoretical complexity) would amount to 1.29 × 1031 possible permutations, thus exceeding the practical limit of library design. To reduce the complexity and the occurrence of the potential dead-end variants, we applied an approach of a stepwise structural selection library construction, as detailed below.
First, the following sub-libraries were constructed (Figure 1): A (VR-I+VR-V+VR-VI), B (VR-IV), and C (VR-VII). These three sub-libraries were constructed from overlapping synthetic oligonucleotides by PCR and isothermal DNA assembly, packaged, and purified separately to select structurally compatible parent viral sub-libraries (Figure 1, step 1). Next, using viral DNAs as PCR templates, the VRs from the sub-libraries A, B, and C were amplified and combined in one ABC library incorporating diversified I, IV, V, VI, and VII loops (Figure 1, step 2). A total of 2.5 × 107 plasmid molecules (extrapolated from colony counts) successfully contributed to creating the plasmid library, which was then used for large-scale packaging of the final viral library.
Figure 1.
Library Construction
Step 1: sub-libraries A, B, and C were first assembled, each with mutations in different regions (VR-I, VR-V, and VR-VI for A; VR-IV for B; VR-VII for C). Step 2: variable regions from the three sub-libraries (from DNA isolated from successfully assembled AAV particles) were recombined to generate the final library that has mutations in all five regions. Step 3: the plasmid library is packaged into the viral library.
AAV Library Characterization
Both plasmid and viral libraries were subjected to next-generation sequencing (NGS). The Illumina platform was chosen because of its higher throughput and lower error rate than PacBio. Due to read length limits, a 393-nt region encompassing VR-IV to VR-VII only (34 of the total 37 diversified amino acid positions) was sequenced from both ends. Reads were subjected to a high stringency filtering to remove sequencing errors and translated into protein sequences. All calculations were performed on the VRs only of the filtered protein sequences (results are shown in Table 1 and Figure 2). A limited sequence bias increase between plasmid and viral libraries was observed (Figure 2A), resulting in only 76.85% of sequences in the viral library being distinct, as opposed to 94.24% in the plasmid library. In both cases, the outlier sequence was WT AAV3b contamination, but even at more than 7,000 times the frequency of most other sequences in the viral library, it only represented 0.69% of the total sequences. The distribution of mutation abundance per sequence was very similar between the plasmid and the viral libraries (Figure 2B) with only a slight decrease (21.56 versus 23.26 mutations per sequence on average, respectively, in the viral and plasmid libraries). Sequence comparison between plasmid and viral libraries shows very little overlap (Figure 2C), suggesting that the actual complexity was much higher than the size of the sequenced sample. Taken together, the NGS data reasonably support a viral complexity on the order of 1 × 107.
Table 1.
Library NGS Summary (VR-IV to VR-VII Only)
| Library | Plasmid | Viral |
|---|---|---|
| Filtered sequences | 1,810,853 | 1,041,321 |
| Distinct sequences (%) | 1,706,527 (94.24%) | 800,244 (76.85%) |
| WT (%) | 381 (0.02%) | 7148 (0.69%) |
| Mutations per sequence | 23.26 | 21.56 |
| % mutant VR-IV | 99.87 | 98.52 |
| % mutant VR-V | 99.03 | 91.91 |
| % mutant VR-VI | 98.11 | 90.25 |
| % mutant VR-VII | 98.03 | 93.95 |
Figure 2.
NGS Analysis of Plasmid and Viral AAV Libraries
(A) Copy number distribution; for each copy number (x axis), the number of distinct sequences present at that copy number is shown (y axis). In the plasmid library, almost all sequences have a single occurrence, while in the viral library there is a wider distribution, with more sequences having diverse levels of abundance; in both cases, the sequence with the highest abundance is WT AAV3B, the library parent, indicated with arrows. (B) Distribution of the number of mutations per sequence; the number of mutations for each sequence varies between 0 (WT AAV3B) and 34 (100% of the 34 variable positions mutated), with a peak at 24 and 23 mutations, respectively, for the plasmid and viral libraries, and an average of 21.56 and 23.26, respectively. (C) Euler diagram showing the overlap between the distinct sequences in the plasmid and the viral libraries.
In Vitro Selection for Liver-Targeted AAV Variants
We generated 3D human hepatocellular carcinoma (HUH-7) spheroid cultures in order to more closely mimic hepatocyte conditions in vivo.30 Discrete spheroids (~100–500 μm) were observed within 5–7 days, and spheres were used on day 7 for serial selection of the AAV3B combinatorial capsid library. The first selection passage (P-1) was carried out using a multiplicity of infection (MOI) of 1. Subsequent passages were carried out using an MOI of 0.01 in order to prevent loss of genotype-phenotype correlation. Altogether, five rounds of in vitro selection were carried out. Ad5 superinfection was performed at each round of selection based on similar studies for AAV library selection.23,31,32
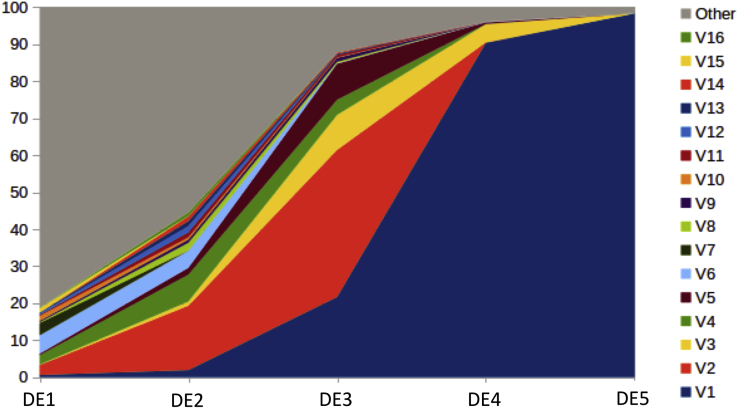
Samples collected after each round were sequenced on the PacBio platform. Evolution of sequence frequencies is displayed in Figure 3 (for clarity, only sequences present at more than 1% in at least one sample are shown individually). One sequence, shown as V1 in Figure 3, was overwhelmingly selected and became predominant at round 4. We refer to this sequence as AAV3B-DE5 (AAV3B-directed evolution 5).
Figure 3.
Evolution of Sequence Frequencies with Selection Rounds
Sequences present at more than 1% after at least one round of selection are displayed with a separate color (V1–V16). All other sequences are combined under a single color (Other). AAV3B-DE5 is shown as sequence V1.
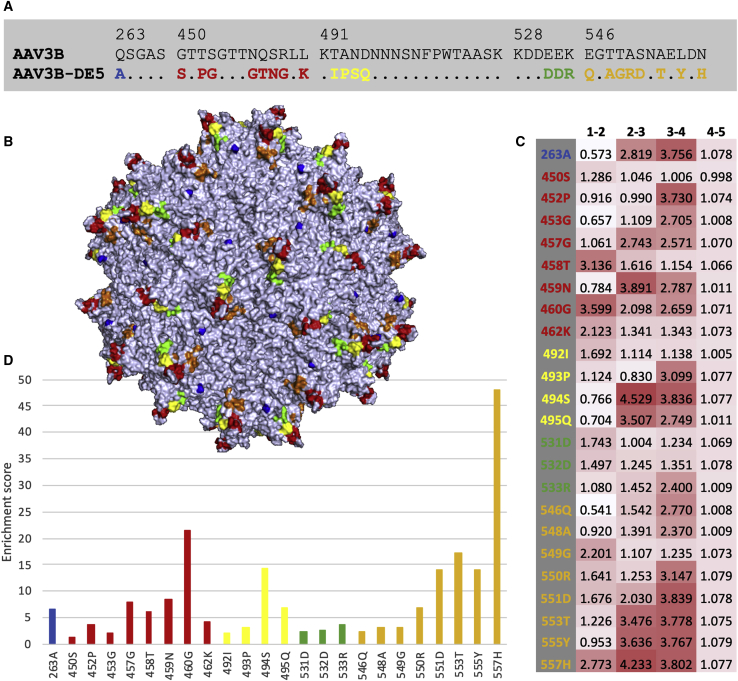
AAV3B-DE5 (more precisely its VR-IV to VR-VII region) was not detected in the original viral library, which means that its frequency was lower than 1 per million. Even the VR-IV region of AAV3B-DE5 itself could not be found in the original viral library, while the VR-V, VR-VI, and VR-VII regions were present at frequencies of 0.11%, 3.43%, and 0.01%, respectively. AAV3B-DE5 contains 24 aa substitutions compared with AAV3B (Figure 4A), distributed among all five VRs, including 23 in VR-IV to VR-VII (slightly more than the library average of 21.56). All substitutions were part of the original library design. The positions of these mutations on the capsid surface are displayed in Figure 4B. Enrichment scores, defined as the products of enrichment factors (Figure 4C) between rounds of selection, were computed for each AAV3B-DE5 mutation (Figure 4D). Mutations N557H, R460G, A553T, N494S, S551D, and L555Y obtained the highest scores, suggesting they were the main drivers for selection, while mutations with lower scores are more likely passengers.
Figure 4.
AAV3-DE5 Mutations
Color code: blue, red, yellow, green, and orange for VR-I, VR-IV, VR-V, VR-VI, and VR-VII, respectively. (A) Alignment between parental capsid AAV3B and evolved capsid AAV3B-DE5; only the four regions that were diversified in the library design are shown (the sequences are identical outside these regions); numbers on top designate the region position in the capsid (VP1 numbering); sequence identity is represented by a dot. (B) Positions of the 24 mutations displayed in AAV3B 3D structure. (C) Heatmap showing enrichment factors between rounds of selection for each mutation; the enrichment factor between two rounds of selection (for example a and b) for a mutation at a particular position is defined as the frequency of the corresponding amino acid at that position in the NGS sample for round b divided by the corresponding frequency for round a. (D) Enrichment scores of AAV3B-DE5 mutations; for each position the score is defined as the product of the four enrichment factors shown in (C).
AAV3B-DE5 was further characterized in vitro and in vivo. Its sequence is deposited in GenBank (GenBank: MT396223).
In Vitro Characterization: Transduction Efficiency
To determine transduction efficiencies, WT AAV3B and AAV3B-DE5 capsids were packaged with the pTR-UF50-BC transgene plasmid to express the firefly luciferase (FLuc) and mApple reporter transgenes (separated by a furin recognition site and self-cleaving 2-A peptide) under control of the chicken β-actin (CBA) promoter, flanked by AAV2 ITRs. We compared transduction efficiencies in spheroid cultures derived from two human hepatocellular carcinoma cell lines, that is, HUH-7 and HEPG2. Increased transduction efficiencies at an MOI of 1 × 104 vector genomes (vg)/cell of the AAV3B-DE5 vector was observed in both cell types as visualized by fluorescence imaging for mApple (Figure 5A). AAV3B-DE5 transduced HUH-7 adherent monolayers at higher frequencies as compared to WT AAV3B at different MOIs. This was confirmed by % mApple+ cells, mApple mean fluorescence intensity (MFI), and relative FLuc activity (Figure 5B). Kinetics of transduction frequencies (% mApple+) as well as AAV genome copies/transduced cell using an MOI of 5 × 105 vg/cell further indicated increased transduction efficiency as well as genome copies per cell for the AAV3B-DE5 variant (Figure 5C). Furthermore, we compared AAV3B-DE5 with AAV8 transduction (both expressing GFP transgene) in primary human, non-human primate, and murine hepatocytes. AAV8 has high tropism for murine livers but transduces human hepatocytes rather poorly.23,33 We confirmed increased in vitro transduction by estimating transgene expression in AAV3B-DE5-transduced human and non-human primate primary hepatocytes, as compared to AAV8. In contrast, AAV3B-DE5 was unable to transduce murine primary hepatocytes, whereas AAV8 displayed high transduction of murine hepatocytes as determined by GFP mRNA transcript levels using real-time RT-PCR (Figure 5D).
Figure 5.
Comparative Transduction of Human Hepatocellular Carcinoma Cell Lines In Vitro
(A) Fluorescence images of HUH-7 and HEPG2 spheroid cultures transduced with WT AAV3B and AAV3B-DE5 vectors, expressing the mApple reporter transgene. (B) Graphical representation of (A) transduction efficiencies (% mApple+ cells), mApple MFI, and FLuc activity levels for HUH-7 adherent cells transduced with either WT AAV3B or AAV3B-DE5 at different MOIs. (C) Kinetics of transduction efficiencies (% mApple+) and AAV genome copy/cell of HUH-7 adherent cells transduced with an MOI of 5 × 105 vg/cell of either WT AAV3B or AAV3B-DE5. (D) GFP transgene expression analysis of primary human hepatocytes, primary monkey hepatocytes, or primary mouse hepatocytes transduced with different MOIs of either AAV8 or AAV3B-DE5 in vitro. Transgene expression was normalized to GAPDH expression levels. Statistical analysis was performed by two-way ANOVA with a Sidak’s multiple comparison test for (B) and (C). Data are represented as mean ± SEM. A p value <0.05 was considered statistically significant (*).
In Vitro Characterization: Determination of NAb Titers
Since pre-existing humoral immunity can block vector transduction efficiency, we tested two different methods to assess vector capsid neutralization by pre-existing NAbs from human sera. WT AAV3B with AAV3B-DE5 vectors expressing the mApple transgene were pre-incubated with serial dilutions of pooled i.v. Ig and then used to transduce HUH-7 cell monolayers at an MOI of 1 × 104 vg/cell. The percent transduced cells (mApple+) as determined by flow cytometry were normalized to AAV-only controls (no i.v. Ig). As compared to WT AAV3B, AAV3B-DE5 was significantly able to evade i.v. Ig neutralization at all concentrations of i.v. Ig tested (Figure 6A). The average reciprocal NAb titer for WT AAV3B was 85.1 ± 0.6 μg/mL i.v. Ig, while for AAV3B-DE5 it was 274.8 ± 0.9 μg/mL i.v. Ig, with an average 3.2-fold difference in NAb titers.
Figure 6.
In Vitro Assays for Neutralization of Transduction
(A) Determination of reciprocal NAb titers to WT AAV3B or AAV3B-DE5 by using increasing concentrations of pooled i.v. Ig (100–1,000 μg/mL). The i.v. Ig concentration at which 50% mApple expression is reduced as compared to no i.v. Ig control is indicated with the red dotted lines. % mApple+ cells are quantified by flow cytometry. (B) Determination of reciprocal NAb titers to WT AAV3B or AAV3B-DE5 by using increasing concentrations of pooled i.v. Ig (100–6,400 μg/mL). The i.v. Ig concentration at which 50% FLuc expression is reduced, as compared to no i.v. Ig control, is indicated with the red dotted lines. (C) Average reciprocal NAb titers for WT AAV3B and AAV3BDE5, determined using the FLuc-based in vitro neutralizing assay. Data are the average of 10 independent experiments. Data are represented as mean ± SD for (A), with statistical significance between WT AAV3B and AAV3B-DE5 conducted by two-way ANOVA with a Sidak’s multiple comparison test. Data are represented as mean ± SEM for (C), with statistical significance between WT AAV3B and AAV3B-DE5 conducted by an unpaired t test with Welch’s correction. A p value <0.05 was considered statistically significant (*).
The sensitivity of anti-AAV NAb titer determination has been shown to depend on the reporter gene used in the assay. We therefore used the more sensitive luciferase reporter-based neutralizing assay as previously described in published reports.34,35 An MOI of 1.36 × 105 vg/cell of WT AAV3B or AAV3B-DE5 expressing the luciferase reporter transgene was pre-incubated with serial dilutions of pooled i.v. Ig and overlaid on HUH-7 cell monolayers as described in Materials and Methods. As observed in Figure 6A, we confirmed substantial evasion by AAV3-DE5 to anti-AAV NAb from pooled i.v. Ig, as detailed in a representative figure (Figure 6B). Average reciprocal NAb titers combined from 10 independent experiments were 793.3 ± 42.7 μg/mL i.v. Ig for WT AAV3B and 1,595 ± 84 μg/mL i.v. Ig for AAV3B-DE5, a 2-fold difference (Figure 6C). The higher NAb titers observed in the luciferase neutralizing assay could be attributed to the higher MOI of AAV used and the increased sensitivity of detection.
In another set of experiments, we compared seroprevalence of WT AAV3B or AAV3B-DE5 in individual serum samples using the luciferase-based neutralizing assay. Sera from 100 healthy donors were tested for NAb titers to WT AAV3B or AAV3B-DE5. Using 1:5 serum dilution as a cutoff, pre-existing NAb to WT AAV3B was 37% and to AAV3B-DE5, 25%, an ~1.5-fold lower NAb incidence (p = 0.033, Figure 7A). Since even very low titers of AAV NAbs have been shown to block in vivo transduction in clinical trials and animal studies,14,15 we looked at frequencies of NAb titers >1:2) (Figure 7B). On lowering the detection threshold to ≥1:2, 58% and 42% of healthy donor sera had pre-existing NAb to WT AAV3B and AAV3B-DE5, respectively.
Figure 7.
Neutralization of Individual Serum Samples
(A) Reciprocal NAb titers for WT AAV3B and AAV3B-DE5 obtained from 100 donor serum samples using the in vitro FLuc-based neutralizing assay. (B) Determination of seroprevalence for WT AAV3B and AAV3B-DE5 from (A) using either 1:2 or 1:5 serum dilution as the cutoff. (C) 48 samples with detectable titers were analyzed for fold difference between WT AAV3B and AAV3B-DE5 reciprocal NAb titers. 35% and 40% of these 48 patient samples have 2- to 16-fold and 1.5- to 2-fold higher titers, respectively, for WT AAV3B as compared to AAV3B-DE5. 19% and 6% of patient samples have 1- to 1.5-fold and 0.7- to 1-fold higher titers, respectively, for WT AAV3B as compared to AAV3B-DE5. Statistical analysis for (A) was performed using a two-tailed Mann-Whitney test following a normality distribution determination, and for (B) was conducted by Fisher’s exact test two-tailed test using a 2 × 2 contingency table. A p value <0.05 was considered statistically significant (*).
Of the serum samples that were seropositive for both WT AAV3B and AAV3B-DE5, 33% (16 samples) had 2- to 6-fold higher NAb titers for WT AAV3B as compared to AAV3B-DE5, while one individual sample had a 15-fold higher titer for WT AAV3B (Figure 7C). Most serum samples (40%, 19 samples) had 1.5- to 2-fold higher NAb titers for WT AAV3B. 19% had 1- to 1.5 higher NAb titers for WT AAV3B compared to AAV3B-DE5, while only three samples (6.3%) had a 0.7- to 1-fold difference. Importantly, none of the serum samples had 2-fold or higher titers for AAV3B-DE5 as compared to WT AAV3B. This indicates that original reactivities were maintained or significantly reduced in the AAV3B-DE5 variant, and that new antigenic determinants were not selected for or created.
Serotype Comparison in Human Liver Chimeric Mice
Liver chimeric mouse models allow for in vivo studies of primary human hepatocytes.36 Given the stark species differences in AAV hepatocyte transduction, these human liver chimera models are increasingly applied to test engineered AAV serotypes.23,33,37 In this study, we used Fah−/− non-obese diabetic (NOD) Rag1−/− Il2rgnull (FNRG) mice that were humanized with mouse-passaged primary human hepatocytes (huFNRG mice).38,39 After transplantation, mice were cycled off the protective drug 2-(2-nitro-4-trifluoromethylbenzoyl)-1,3-cyclohexanedione (NTBC or nitisinone) for approximately 3 months to expand the human graft. Chimerism was determined by human albumin (hAlb) quantification in mouse serum, which correlates well with humanization.40, 41, 42 Once mice had reached peak hAlb levels they were challenged 1 day after restarting NTBC, as illustrated by the experimental schematic (Figure 8A). The plateau hAlb levels showed that most animals were >80% humanized (Figure 8B). In order to test transduction of AAV3B-DE5 in vivo, we created three cohorts of huFNRG mice. We compared the performance of the AAV3B-DE5 variant in transducing human hepatocytes with two other AAV vectors, which had earlier been shown to transduce human hepatocytes with high efficiency. LK03 is a bioengineered capsid that is closely related to WT AAV3B, with only 8 aa changes, and it has shown strong tropism for human hepatocytes in humanized mouse livers.23 It is being used in clinical trials for hemophilia A gene therapy (ClinicalTrials.gov: NCT03003533). AAV3B-ST is another engineered AAV3B capsid-modified variant generated with 2 aa changes (S663V+T492V).43 Previous studies by us and others have shown that systemically delivered AAV3B-ST performed better than AAV5, AAV8, AAV9 and WT AAV3B in a human liver xenograft mouse model as well as in non-human primate livers.33,43,44
Figure 8.
In Vivo Transduction Efficiency in Human Liver Chimeric Mice
(A) Schematic representing steps in the generation of the human liver chimeric mouse. Fah−/− NOD Rag1−/−Il2rgnull (FNRG) mice were humanized with mouse-passaged primary human hepatocytes (huFNRG mice). After transplantation, mice were cycled off NTBC to expand the human graft. (B) Chimerism was determined by human albumin (hAlb) quantification in mouse serum, which correlates well with humanization. Once mice had reached peak hAlb levels they were challenged 1 day after restarting NTBC. (C) Flow gating strategy to differentiate between human and mouse hepatocytes, using human HLA-1 and mouse CD81 antibodies, and to quantify the frequencies of GFP-expressing cells. (D) Comparison of transduction frequencies of AAV3-ST, AAV3B-DE5, and LK03 by quantifying percent GFP+ human hepatocytes by flow cytometry. Data for (B) and (D) are shown as median ± SEM.
Animals were intravenously administered with 2 × 1011 vg of either AAV3B-ST, LK03, or AAV3B-DE5, expressing the GFP transgene. Thirteen days later, hepatocytes were isolated from chimeric livers38 and stained for human leukocyte antigen (HLA)-I. Mouse CD81 antibody was used to exclude non-human hepatocytes. After sequential gating (Figure 8C) the GFP+ fraction of human hepatocytes was quantified. AAV3B-DE5 transduced minimally higher fractions of human hepatocytes than did AAV3B-ST, although it did not reach statistical significance. The percentage of GFP+-transduced human hepatocytes was comparable to the previously reported bioengineered LK03 capsid (Figure 8D). These data indicate that AAV3B-DE5 can transduce human hepatocytes in chimeric livers at efficiencies similar to LK03 and possibly slightly better than AAV3B-3ST. Transduction efficiency of mouse hepatocytes cannot be assessed in this highly humanized model because of low numbers, as well as the unhealthy condition of these cells due to NTBC withdrawal.39 However, AAV3B-DE5 failed to transduce primary murine hepatocytes in vitro (Figure 5D), consistent with unpublished observations on failure of in vivo transduction of C57BL/6 mouse liver using this capsid (Vivet Therapeutics, Paris, France, personal communication).
Discussion
The wide and continuous success of AAV as a therapeutic gene delivery vehicle has led to the quest for vectors with improved transduction efficiencies, higher tissue tropism, and lower immunogenicity. These properties are largely dependent on the viral capsid and differ broadly between AAV serotypes. Hepatic gene therapy via systemic AAV administration is an advantageous delivery strategy for long-lived and sustained correction of genetic disorders such as hemophilia, Wilson’s disease, lysosomal storage disorders, or Crigler-Najjar syndrome, among others.45 Identifying an optimal viral capsid with strong liver tropism for efficient transduction of human hepatocytes has thus been under active investigation. While AAV8 and AAV5 serotypes are used in several clinical trials for hepatic gene transfer, emerging data in non-human primates and in “humanized” chimeric liver mice engrafted with human hepatocytes indicate that engineered vectors on the AAV3B backbone show superior transduction efficiencies in human liver.23,33,44 This can be explained by preferential use of the human hepatocyte growth factor receptor as a cellular co-receptor,44 which restricts transduction to the liver. Furthermore, the chimeric AAV capsid (LK03) derived from an AAV shuffled library, found to transduce human primary hepatocytes at a high efficiency in vitro and in vivo, is closely related to AAV3B.23
Methodologies for generating novel AAVs specific for a particular application include generating combinatorial capsid libraries of a high complexity, which is constrained by sequence bias or limited diversity. A rational approach of introducing deliberate mutations into amino acid residues based on our understanding of the capsid structure also faces limits, as in many cases, favorable mutations cannot be combined to produce additive effects. In the current study, we used a naturally existing serotype to generate a library, combining rational mutagenesis and directed evolution to enable us to select a variant specific to our intended application. A common challenge during library generation is contamination with the parental sequence, due to incomplete removal of template DNA after PCR steps. Such contamination can, depending on its extent, seriously limit the library usefulness by reducing its diversity. In this study, parental (WT AAV3B) contamination was limited to 0.69%, which is an acceptable level. Alternatively, the parental sequence can be advantageously used as a control to validate library design, by comparing enrichment of competing variants during selection. Indeed, in our experiment, the frequency of WT AAV3B decreased to 0.49% after the first round of selection, and it was undetectable in subsequent rounds, confirming that the library contained improved variants compared to WT AAV3B.
Another recurring question in library screening is whether the optimal variant has been selected, and whether the same variant would be consistently selected under similar conditions. In other words, the question is whether experimental parameters allow each library variant to compete fairly without selection bias. The risk of eliminating potentially improved variants under our selection conditions (MOI of 1 and 0.01, and multiple rounds of selection) cannot be entirely ruled out.46 Whether the library contains variants with even higher specificity for human hepatocytes than AAV3B-DE5 (e.g., variants from early selection rounds) remains an open question, which will be addressed in future work.
A recent study suggests that library-selected capsids such as LK03 can be further improved by additional site-directed mutagenesis.47 Therefore, it is likely that our newly generated capsid with high tropism for hepatocytes can still be further improved. Earlier studies found that elimination of certain tyrosine residues on the surface of the AAV2 capsid could improve transduction and reduce major histocompatibility complex (MHC) class I presentation of capsid antigen. The rationale for this approach was that the AAV capsid is phosphorylated by tyrosine kinases following cellular entry, which targets the capsid for ubiquitination and proteasomal degradation.25,48, 49, 50, 51 Elimination of phosphorylation sites therefore enhances transfer to the nucleus and reduces presentation of peptides derived from proteasomal degradation, therefore reducing the likelihood of being targeted by capsid-specific CD8+ T cells.9,50,52,53 When attempting to optimize the AAV3B capsid, elimination of a serine and a threonine site (S663V+T492V mutations) was found to be more efficacious33,43,44 for transduction of human and non-human primate hepatocytes. In this study, we diversified the T492 position to include either of the following positions: T/I/A/V, while leaving S663 as in the WT AAV3B. Curiously, the selected AAV3B-DE5 had a T492I mutation, which might have achieved the same purpose as the T492V substitution. This position, as well as S663, and remaining antibody-targeted capsid surface WT residues leave significant room for additional rational engineering of the derived AAV3B-DE5 variant to improve its efficiency and stealth.
The lack of standardized methods to measure vector and neutralizing antibody titers and the variability in testing have made it difficult to predict the lowest NAb titer that can preclude AAV delivery. Using the luciferase-based neutralizing antibody detection assay, and a cutoff of 1:5, the seroprevalence to WT AAV3B in our cohort of 100 individual serum samples was 37%. This was very similar to the 35% previously reported for a UK patient population54 and 36% for a North American population55 respectively. In a separate study, we analyzed a similar number of North American patient samples and found a seroprevalence to AAV3B of 45% (I.Z., unpublished data). While our studies indicate better evasion of pre-existing NAb by AAV3B-DE5 from healthy donors in the 100 serum samples tested, it is noteworthy that in the 48 samples that were seropositive for both WT AAV3B and AAV3B-DE5, ~94% had NAb titers to WT AAV3B that were up to 15-fold higher than for the AAV3B-DE5 variant. In contrast, none of the samples had a titer against AAV3B-DE5 that was even 2-fold higher than for AAV3. Therefore, it appears that no new set of antigenic epitopes was generated by the engineered variant, as patient samples reacted either similarly or less compared to WT AAV3B.
In conclusion, we find that molecular evolution using the combinatorial library platform accomplished at the same time the generation of a viral capsid with high tropism for the desired cell type and reduced neutralization that results from pre-existing immunity to AAV in the human population. This was accomplished without inclusion of human serum or Ig in the selection process but rather resulted from the more extensive changes in the amino acid sequence as compared to site-directed mutagenesis approaches. Nonetheless, rational single amino acid changes can be combined with a library selection approach to achieve superior results.
Materials and Methods
Library Design
The guiding principle behind the library’s design was to modify only the VRs while keeping the backbone sequence unchanged. Following the alignment of 150 AAV naturally occurring variants, candidate positions for mutagenesis were selected from VR-I, VR-IV, VR-V, VR-VI, and VR-VII and diversified. The AAV3B capsid gene fragments incorporating all nucleotide substitutions were assembled from synthetic oligonucleotides and inserted into a plasmid vector containing the AAV3B genome from which the corresponding WT sequence had been removed. The backbone plasmid vector for the library’s design and construction incorporated all AAV3B sequences necessary for replication and packaging, including AAV3 ITRs, rep, and cap.
Three structurally compatible sub-libraries as described in the Results were constructed using synthetic oligonucleotides by PCR and isothermal DNA assembly, packaged, and purified separately. Next, using viral DNAs as PCR templates, the VRs from the sub-libraries A, B, and C were amplified and combined in one ABC library incorporating diversified I, IV, V, VI, and VII loops. Finally, the ABC library was packaged on a large-scale, and amplicons generated from both the plasmid and viral libraries were subjected to Illumina sequencing.
Spheroid Generation and In Vitro Selection
Spheroid cultures were generated by culturing human hepatocellular carcinoma (HUH7) single cells in six-well ultra-low attachment plates (Corning, Corning, NY, USA). Cells were cultured in DMEM/F12K media supplemented with epidermal growth factor (EGF, 20 ng/mL), fibroblast growth factor (FGF, 10 ng/mL), B27 (1×) (Gibco, Gaithersburg, MD, USA). Discrete spheroids were observed within 5–7 days. In order to pre-determine MOIs, cells were seeded on day 0 at 0.5 million cells/mL, dissociated on day 7, counted, and viability was determined (Cellometer Auto T4, Nexcelom Bioscience, Lawrence, MA, USA). On day 7, spheres were centrifuged at 300 × g for 5 min and resuspended in 1 mL of DMEM/F12K medium without FBS. Spheres were first incubated with Ad5 helper virus at an MOI that had been previously determined to have 50% cytopathic effect (CPE). Incubation was carried out for 1 h at 37°C in a humidified CO2 incubator with intermittent manual shaking. Spheres were then centrifuged in 30 mL of medium in order to remove unbound Ad5 virus, resuspended in 1 mL of DMEM/F12K medium. and incubated initially with an MOI of 1 of the recombinant AAV3B library for 1 h at 37°C in a humidified CO2 incubator. Spheres were then extensively washed by centrifugation in 30 mL of medium in order to remove unbound virus and incubated for 72 h. This was passage 1 (P-1) of the recombinant AAV3B library.
For subsequent passages (five total passages), P-1 cells were subjected to three quick freeze-thaw steps. Clarified supernatant from P-1 was then used to infect P-2 of HUH7 spheres, this time at an MOI of 0.01, in order to prevent genome-capsid combinations from occurring. Five selection passages (P-1 to P-5) were thus carried out and subjected to NGS and analysis.
In Vitro Transduction Efficiency
WT AAV3B and AAV3B-DE5 nonreplicating vectors were packaged with the FLuc and mApple double reporter transgene plasmid pTR-UF50-BC, under control of the CBA promoter, flanked by AAV2 ITRs. A furin recognition site and self-cleaving 2-A peptide sequence was cloned downstream of the FLuc gene, and upstream of the mApple gene. Transduction efficiency of WT AAV3B and AAV3B-DE5 in human hepatocyte HUH-7 cells was compared using MOIs of 3 × 104, 6 × 104, 1 × 105, 2.5 × 105, 5 × 105, 1 × 106, and 2 × 106 vg/cell, respectively. Cells were counted and seeded into 96-well plates overnight and the adherent monolayer was incubated with the indicated MOIs of WT AAV3B or AAV3B-DE5 for 1 h at 37°C. Virus was removed by inverting plates, and cells in supplemented media were incubated for 12–72 h at 37°C. For FLuc estimation, cells were treated as above and quantification of luciferase expression was carried out using the Bright-Glo luciferase assay system (Promega, Madison, WI, USA) and measured on an enzyme-linked immunosorbent assay (ELISA) reader with luminescence detection capacity (Synergy HTX, BioTek, Winooski, VT, USA). The same transduction procedure was carried out for human hepatocyte HUH-7, and HEPG2 spheroids with incubation in individual 15-mL conical tubes, following which cells were resuspended in spheroid media and cultured in 48-well ultra-low attachment plates. After 72 h, cells were trypsinized and frequencies of mApple+ cells as well as MFIs were quantified by flow cytometry (LSRFortessa, BD Biosciences, San Jose, CA, USA) and analyzed on FCS Express 7 software.
Real-Time PCR
Genomic DNA was isolated from AAV-transduced HUH-7 cells at indicated time points following either WT AAV3B or AAV3B-DE5 transduction, using the genomic DNA isolation kit (QIAGEN, Germantown, MD, USA) according to the manufacturer’s instructions. 200 ng of input genomic DNA was used to quantitate AAV vg copy numbers using AAV2 ITR-specific primers,56 and a standard curve was generated from serial dilutions of an AAV5 standard of known concentration. A PCR reaction was performed using the 2× SYBR Green supermix (Bio-Rad, Hercules, CA, USA), using the following cycling conditions: 50°C for 2 min, 95°C for 10 min, 40 cycles at 95°C for 15 s, 60°C for 15 s, and 72°C for 30 s, using a CFX96 Touch real-time PCR detection system (Bio-Rad, Hercules, CA, USA). AAV genome copies were normalized to cell number (Countess II, Thermo Fisher Scientific) and number of transduced cells by running flow cytometry in parallel to quantify mApple+ cells, to derive AAV genome copies/cell.
RNA Extraction and Gene Expression Analysis
Primary human hepatocytes, primary non-human primate hepatocytes (Macaca fascicularis), and primary mouse hepatocytes were purchased from Innoprot (Bizkaia, Spain). Cells were seeded in MIL130 or MIL600 media with ADD221 or ADD222 additives on PLA137 collagen-coated plates (Biopredic International, Saint Grégoire, France). Cells were transduced after 24 h with AAV3B-DE5-GFP or AAV8-GFP at three different MOIs (104, 3 × 104, and 105 vg/cell) in duplicates. Cells were harvested at 96 h after transduction and RNA was extracted (Maxwell RSC Simply RNA, Promega, Madison, WI, USA) for quantitative RT-PCR analysis of GFP mRNA transcript levels. Extracted RNA was pre-treated with Turbo DNA Free (Ambion, Austin, TX, USA) and retro-transcribed into complementary DNA (cDNA) using Moloney murine leukemia virus (M-MLV) reverse transcriptase (Invitrogen, Carlsbad, CA, USA). Copies of GFP in cDNA were analyzed by qPCR using GoTaq qPCR master mix (Promega, Madison, WI, USA) in a CFX96 real-time detection system (Bio-Rad, Hercules, CA, USA). EGFP expression was analyzed using the following primers: 5′-ATGGTCAGCAAGGGCGAGG-3′ and 5′-TTGCCGGTGGTGCAGATGAA-3′. GFP expression levels were normalized to human, non-human primate, and mouse GAPDH, respectively.
Mice
FNRG female mice were used at the Rockefeller University under Institutional Animal Care and Use Committee (IACUC) protocol 15814.
In Vivo Transduction Efficiency
Cryopreserved pediatric human hepatocytes were purchased from Celsis (Chicago, IL, USA). Fresh human hepatoblasts were isolated from human fetal livers procured from Advanced Bioscience Resources (ABR) as described.57 Human liver cell suspensions were injected intrasplenically (0.5–1 × 106 cells per mouse) under anesthesia into FNRG mice that were generated by a 13-generation backcross of the Fah−/− allele to NOD Rag1−/− Il2rgnull (NRG) animals, respectively, provided by M. Grompe (Oregon Health & Science University)58 or obtained from Jackson Laboratory. Starting on the day of transplantation, mice were cycled off the liver protective drug NTBC (Yecuris, Tualatin, OR, USA) as described by others.37 Human albumin levels in mouse sera were measured by ELISA (Bethyl Laboratories, Montgomery, TX, USA). In vivo transduction efficiency of three vectors were compared, that is, AAV3B-DE5, LK03, and an engineered vector, AAV3-ST, in which one serine and one threonine residue in the AAV3 capsid is eliminated.44 All AAV vectors expressed the GFP transgene and were injected through the tail vein at 1 × 1011 vg/mouse. Two weeks later, human-chimeric mouse livers were removed and processed for flow cytometry and immunohistochemistry.
Human Serum Samples
Individual human serum samples were obtained from Innovative Research (Novi, MI, USA). These were de-identified and had tested negative for hepatitis B surface antigen (HBsAg), hepatitis C virus
(HCV), HIV-1, HIV-2, HIV-1 antigen (HIV-1Ag) or HIV-1 NAT (nucleic acid test), elevated alanine aminotransferase (ALT), and syphilis.
In Vitro Neutralization Assay by Flow Cytometry
An MOI of 1 × 104 vg/cell of either WT AAV3b or AAV3B-DE5 vector-expressing mApple reporter transgene was incubated with varying concentrations of pooled human i.v. Ig (Privigen, CSL Behring, Bradley, IL, USA) ranging from 100 to 1,000 μg/mL for 1 h at 37°C in a humidified CO2 incubator. The vector-i.v. Ig mixture was overlaid on Ad5 pre-treated HUH-7 monolayer cells in a 96-well plate for 1 h at 37°C. The overlay was removed, fresh media were added, and cells were incubated for 72 h at 37°C. Cells were then trypsinized and frequencies of mApple+ events were quantified by flow cytometry. The reciprocal of the highest dilution of i.v. Ig that neutralized 50% of mScarlet expression relative to control wells (transduced in the absence of i.v. Ig) was considered to be the neutralization titer.
In Vitro Neutralization Assay by Luciferase Activity
An MOI of 1.36 × 105 vg/cell of WT AAV3B or AAV3B-DE5 expressing the FLuc and mApple double reporter transgene were incubated with serial 2-fold dilutions of either pooled human i.v. Ig (Privigen, CSL Behring, Bradley, IL, USA) or 100 individual serum samples from donors as described above. HUH-7 cells were transduced as described, except that cells were not pre-treated with Ad5 infection. After 48 h of incubation, quantification of luciferase expression was carried out using the Bright-Glo luciferase assay system (Promega, Madison, WI, USA) and measured on an ELISA reader with luminescence detection capacity (Synergy HTX, BioTek, Winooski, VT, USA). The neutralization titer for each sample is defined as the serum dilution at which the FLuc expression is reduced by 50% compared to the no-serum control.
Statistical Analysis
For neutralization assays, reciprocal NAb titers were extrapolated by non-linear curve fitting methods and analyzed using four- to five-parameter half-maximal inhibitory concentration (IC50) tests or fourth-order polynomial curve fitting in GraphPad Prism 8. For neutralization assays based on luciferase reporter expression, effect size was first measured by a Z factor test. Comparison of two samples was carried out using the Student’s t test with Welch’s correction. For non-normal distribution of data, a non-parametric Mann-Whitney test was carried out. Multiple samples were compared using two-way ANOVA with Sidak’s multiple comparison test. Seroprevalence rates between two treatment groups was calculated by the Fisher’s exact test using a 2 × 2 contingency table (GraphPad Prism).
Author Contributions
M.B., D.M., N.L., C.Z., I.Z., S.R.P.K., J.R., J.S.S.B., and O.K. performed experiments; M.B., D.M., G.G.-A., Y.P.d.J., R.W.H., and S.Z. designed and coordinated studies; M.B., D.M., G.G.-A., Y.P.d.J., R.W.H., and S.Z. analyzed and interpreted data; M.B., D.M., G.G.-A., Y.P.d.J., R.W.H.. and S.Z. wrote the manuscript; M.B. and S.Z. supervised the study.
Conflicts of Interest
R.W.H. serves on the scientific advisory board of Ally Therapeutics. The remaining authors declare no competing interests.
Acknowledgments
This work was supported by National Institutes of Health/National Institute of Allergy and Infectious Diseases grant R01 AI51390 (to R.W.H) and by National Institutes of Health/ National Heart, Lung, and Blood Institute grants R01 HL131093 (to R.W.H. and Y.P.d.J.), R01 HL097088 (to R.W.H.) and R01 HL097088 (to R.W.H. and S.Z.). M.B. was supported by the National Hemophilia Foundation Career Development Award.
Contributor Information
Moanaro Biswas, Email: nbiswas@iu.edu.
Sergei Zolotukhin, Email: szlt@ufl.edu.
References
- 1.Anguela X.M., High K.A. Entering the modern era of gene therapy. Annu. Rev. Med. 2019;70:273–288. doi: 10.1146/annurev-med-012017-043332. [DOI] [PubMed] [Google Scholar]
- 2.Herzog R.W. Encouraging and unsettling findings in long-term follow-up of AAV gene transfer. Mol. Ther. 2020;28:341–342. doi: 10.1016/j.ymthe.2020.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perrin G.Q., Herzog R.W., Markusic D.M. Update on clinical gene therapy for hemophilia. Blood. 2019;133:407–414. doi: 10.1182/blood-2018-07-820720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mendell J.R., Al-Zaidy S., Shell R., Arnold W.D., Rodino-Klapac L.R., Prior T.W., Lowes L., Alfano L., Berry K., Church K. Single-dose gene-replacement therapy for spinal muscular atrophy. N. Engl. J. Med. 2017;377:1713–1722. doi: 10.1056/NEJMoa1706198. [DOI] [PubMed] [Google Scholar]
- 5.Bennett J., Wellman J., Marshall K.A., McCague S., Ashtari M., DiStefano-Pappas J., Elci O.U., Chung D.C., Sun J., Wright J.F. Safety and durability of effect of contralateral-eye administration of AAV2 gene therapy in patients with childhood-onset blindness caused by RPE65 mutations: a follow-on phase 1 trial. Lancet. 2016;388:661–672. doi: 10.1016/S0140-6736(16)30371-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samulski R.J., Muzyczka N. AAV-mediated gene therapy for research and therapeutic purposes. Annu. Rev. Virol. 2014;1:427–451. doi: 10.1146/annurev-virology-031413-085355. [DOI] [PubMed] [Google Scholar]
- 7.Nathwani A.C., Gray J.T., McIntosh J., Ng C.Y., Zhou J., Spence Y., Cochrane M., Gray E., Tuddenham E.G., Davidoff A.M. Safe and efficient transduction of the liver after peripheral vein infusion of self-complementary AAV vector results in stable therapeutic expression of human FIX in nonhuman primates. Blood. 2007;109:1414–1421. doi: 10.1182/blood-2006-03-010181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martino A.T., Suzuki M., Markusic D.M., Zolotukhin I., Ryals R.C., Moghimi B., Ertl H.C., Muruve D.A., Lee B., Herzog R.W. The genome of self-complementary adeno-associated viral vectors increases Toll-like receptor 9-dependent innate immune responses in the liver. Blood. 2011;117:6459–6468. doi: 10.1182/blood-2010-10-314518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verdera H.C., Kuranda K., Mingozzi F. AAV vector immunogenicity in humans: a long journey to successful gene transfer. Mol. Ther. 2020;28:723–746. doi: 10.1016/j.ymthe.2019.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boutin S., Monteilhet V., Veron P., Leborgne C., Benveniste O., Montus M.F., Masurier C. Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: implications for gene therapy using AAV vectors. Hum. Gene Ther. 2010;21:704–712. doi: 10.1089/hum.2009.182. [DOI] [PubMed] [Google Scholar]
- 11.Blacklow N.R., Hoggan M.D., Kapikian A.Z., Austin J.B., Rowe W.P. Epidemiology of adenovirus-associated virus infection in a nursery population. Am. J. Epidemiol. 1968;88:368–378. doi: 10.1093/oxfordjournals.aje.a120897. [DOI] [PubMed] [Google Scholar]
- 12.Calcedo R., Vandenberghe L.H., Gao G., Lin J., Wilson J.M. Worldwide epidemiology of neutralizing antibodies to adeno-associated viruses. J. Infect. Dis. 2009;199:381–390. doi: 10.1086/595830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aronson S.J., Veron P., Collaud F., Hubert A., Delahais V., Honnet G., de Knegt R.J., Junge N., Baumann U., Di Giorgio A. Prevalence and relevance of pre-existing anti-adeno-associated virus immunity in the context of gene therapy for Crigler-Najjar syndrome. Hum. Gene Ther. 2019;30:1297–1305. doi: 10.1089/hum.2019.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manno C.S., Pierce G.F., Arruda V.R., Glader B., Ragni M., Rasko J.J., Ozelo M.C., Hoots K., Blatt P., Konkle B. Successful transduction of liver in hemophilia by AAV-factor IX and limitations imposed by the host immune response. Nat. Med. 2006;12:342–347. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- 15.Jiang H., Couto L.B., Patarroyo-White S., Liu T., Nagy D., Vargas J.A., Zhou S., Scallan C.D., Sommer J., Vijay S. Effects of transient immunosuppression on adenoassociated, virus-mediated, liver-directed gene transfer in rhesus macaques and implications for human gene therapy. Blood. 2006;108:3321–3328. doi: 10.1182/blood-2006-04-017913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Q., Huang W., Zhang H., Wang Y., Zhao J., Song A., Xie H., Zhao C., Gao D., Wang Y. Neutralizing antibodies against AAV2, AAV5 and AAV8 in healthy and HIV-1-infected subjects in China: implications for gene therapy using AAV vectors. Gene Ther. 2014;21:732–738. doi: 10.1038/gt.2014.47. [DOI] [PubMed] [Google Scholar]
- 17.Rutledge E.A., Halbert C.L., Russell D.W. Infectious clones and vectors derived from adeno-associated virus (AAV) serotypes other than AAV type 2. J. Virol. 1998;72:309–319. doi: 10.1128/jvi.72.1.309-319.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Srivastava A. In vivo tissue-tropism of adeno-associated viral vectors. Curr. Opin. Virol. 2016;21:75–80. doi: 10.1016/j.coviro.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ertl H.C.J., High K.A. Impact of AAV capsid-specific T-cell responses on design and outcome of clinical gene transfer trials with recombinant adeno-associated viral vectors: an evolving controversy. Hum. Gene Ther. 2017;28:328–337. doi: 10.1089/hum.2016.172. [DOI] [PubMed] [Google Scholar]
- 20.Agbandje-McKenna M., Kleinschmidt J. AAV capsid structure and cell interactions. Methods Mol. Biol. 2011;807:47–92. doi: 10.1007/978-1-61779-370-7_3. [DOI] [PubMed] [Google Scholar]
- 21.Marsic D., Govindasamy L., Currlin S., Markusic D.M., Tseng Y.S., Herzog R.W., Agbandje-McKenna M., Zolotukhin S. Vector design tour de force: integrating combinatorial and rational approaches to derive novel adeno-associated virus variants. Mol. Ther. 2014;22:1900–1909. doi: 10.1038/mt.2014.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li C., Samulski R.J. Engineering adeno-associated virus vectors for gene therapy. Nat. Rev. Genet. 2020;21:255–272. doi: 10.1038/s41576-019-0205-4. [DOI] [PubMed] [Google Scholar]
- 23.Lisowski L., Dane A.P., Chu K., Zhang Y., Cunningham S.C., Wilson E.M., Nygaard S., Grompe M., Alexander I.E., Kay M.A. Selection and evaluation of clinically relevant AAV variants in a xenograft liver model. Nature. 2014;506:382–386. doi: 10.1038/nature12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paulk N.K., Pekrun K., Zhu E., Nygaard S., Li B., Xu J., Chu K., Leborgne C., Dane A.P., Haft A. Bioengineered AAV capsids with combined high human liver transduction in vivo and unique humoral seroreactivity. Mol. Ther. 2018;26:289–303. doi: 10.1016/j.ymthe.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong L., Li B., Mah C.S., Govindasamy L., Agbandje-McKenna M., Cooper M., Herzog R.W., Zolotukhin I., Warrington K.H., Jr., Weigel-Van Aken K.A. Next generation of adeno-associated virus 2 vectors: point mutations in tyrosines lead to high-efficiency transduction at lower doses. Proc. Natl. Acad. Sci. USA. 2008;105:7827–7832. doi: 10.1073/pnas.0802866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Markusic D.M., Nichols T.C., Merricks E.P., Palaschak B., Zolotukhin I., Marsic D., Zolotukhin S., Srivastava A., Herzog R.W. Evaluation of engineered AAV capsids for hepatic factor IX gene transfer in murine and canine models. J. Transl. Med. 2017;15:94. doi: 10.1186/s12967-017-1200-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rose J.A., Maizel J.V., Jr., Inman J.K., Shatkin A.J. Structural proteins of adenovirus-associated viruses. J. Virol. 1971;8:766–770. doi: 10.1128/jvi.8.5.766-770.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Govindasamy L., DiMattia M.A., Gurda B.L., Halder S., McKenna R., Chiorini J.A., Muzyczka N., Zolotukhin S., Agbandje-McKenna M. Structural insights into adeno-associated virus serotype 5. J. Virol. 2013;87:11187–11199. doi: 10.1128/JVI.00867-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DiMattia M.A., Nam H.J., Van Vliet K., Mitchell M., Bennett A., Gurda B.L., McKenna R., Olson N.H., Sinkovits R.S., Potter M. Structural insight into the unique properties of adeno-associated virus serotype 9. J. Virol. 2012;86:6947–6958. doi: 10.1128/JVI.07232-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kapałczyńska M., Kolenda T., Przybyła W., Zajączkowska M., Teresiak A., Filas V., Ibbs M., Bliźniak R., Łuczewski Ł., Lamperska K. 2D and 3D cell cultures—a comparison of different types of cancer cell cultures. Arch. Med. Sci. 2018;14:910–919. doi: 10.5114/aoms.2016.63743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pekrun K., De Alencastro G., Luo Q.J., Liu J., Kim Y., Nygaard S., Galivo F., Zhang F., Song R., Tiffany M.R. Using a barcoded AAV capsid library to select for clinically relevant gene therapy vectors. JCI Insight. 2019;4:e131610. doi: 10.1172/jci.insight.131610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grimm D., Zolotukhin S. E pluribus unum: 50 years of research, millions of viruses, and one goal—tailored acceleration of AAV evolution. Mol. Ther. 2015;23:1819–1831. doi: 10.1038/mt.2015.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vercauteren K., Hoffman B.E., Zolotukhin I., Keeler G.D., Xiao J.W., Basner-Tschakarjan E., High K.A., Ertl H.C., Rice C.M., Srivastava A. Superior in vivo transduction of human hepatocytes using engineered AAV3 capsid. Mol. Ther. 2016;24:1042–1049. doi: 10.1038/mt.2016.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meliani A., Leborgne C., Triffault S., Jeanson-Leh L., Veron P., Mingozzi F. Determination of anti-adeno-associated virus vector neutralizing antibody titer with an in vitro reporter system. Hum. Gene Ther. Methods. 2015;26:45–53. doi: 10.1089/hgtb.2015.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mingozzi F., Chen Y., Edmonson S.C., Zhou S., Thurlings R.M., Tak P.P., High K.A., Vervoordeldonk M.J. Prevalence and pharmacological modulation of humoral immunity to AAV vectors in gene transfer to synovial tissue. Gene Ther. 2013;20:417–424. doi: 10.1038/gt.2012.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grompe M., Strom S. Mice with human livers. Gastroenterology. 2013;145:1209–1214. doi: 10.1053/j.gastro.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 37.Bissig-Choisat B., Wang L., Legras X., Saha P.K., Chen L., Bell P., Pankowicz F.P., Hill M.C., Barzi M., Leyton C.K. Development and rescue of human familial hypercholesterolaemia in a xenograft mouse model. Nat. Commun. 2015;6:7339. doi: 10.1038/ncomms8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michailidis E., Vercauteren K., Mancio-Silva L., Andrus L., Jahan C., Ricardo-Lax I., Zou C., Kabbani M., Park P., Quirk C. Expansion, in vivo-ex vivo cycling, and genetic manipulation of primary human hepatocytes. Proc. Natl. Acad. Sci. USA. 2020;117:1678–1688. doi: 10.1073/pnas.1919035117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zou C., Vercauteren K.O.A., Michailidis E., Kabbani M., Zoluthkin I., Quirk C., Chiriboga L., Yazicioglu M., Anguela X.M., Meuleman P. Experimental variables that affect human hepatocyte AAV transduction in liver chimeric mice. Mol. Ther. Methods Clin. Dev. 2020;18:189–198. doi: 10.1016/j.omtm.2020.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vanwolleghem T., Libbrecht L., Hansen B.E., Desombere I., Roskams T., Meuleman P., Leroux-Roels G. Factors determining successful engraftment of hepatocytes and susceptibility to hepatitis B and C virus infection in uPA-SCID mice. J. Hepatol. 2010;53:468–476. doi: 10.1016/j.jhep.2010.03.024. [DOI] [PubMed] [Google Scholar]
- 41.Kawahara T., Toso C., Douglas D.N., Nourbakhsh M., Lewis J.T., Tyrrell D.L., Lund G.A., Churchill T.A., Kneteman N.M. Factors affecting hepatocyte isolation, engraftment, and replication in an in vivo model. Liver Transpl. 2010;16:974–982. doi: 10.1002/lt.22099. [DOI] [PubMed] [Google Scholar]
- 42.Billerbeck E., Mommersteeg M.C., Shlomai A., Xiao J.W., Andrus L., Bhatta A., Vercauteren K., Michailidis E., Dorner M., Krishnan A. Humanized mice efficiently engrafted with fetal hepatoblasts and syngeneic immune cells develop human monocytes and NK cells. J. Hepatol. 2016;65:334–343. doi: 10.1016/j.jhep.2016.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ling C., Wang Y., Zhang Y., Ejjigani A., Yin Z., Lu Y., Wang L., Wang M., Li J., Hu Z. Selective in vivo targeting of human liver tumors by optimized AAV3 vectors in a murine xenograft model. Hum. Gene Ther. 2014;25:1023–1034. doi: 10.1089/hum.2014.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li S., Ling C., Zhong L., Li M., Su Q., He R., Tang Q., Greiner D.L., Shultz L.D., Brehm M.A. Efficient and targeted transduction of nonhuman primate liver with systemically delivered optimized AAV3B vectors. Mol. Ther. 2015;23:1867–1876. doi: 10.1038/mt.2015.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baruteau J., Waddington S.N., Alexander I.E., Gissen P. Gene therapy for monogenic liver diseases: clinical successes, current challenges and future prospects. J. Inherit. Metab. Dis. 2017;40:497–517. doi: 10.1007/s10545-017-0053-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Alencastro G., Pekrun K., Valdmanis P., Tiffany M., Xu J., Kay M.A. Tracking adeno-associated virus capsid evolution by high-throughput sequencing. Hum. Gene Ther. 2020;31:553–564. doi: 10.1089/hum.2019.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ran G., Chen X., Xie Y., Zheng Q., Xie J., Yu C., Pittman N., Qi S., Yu F.X., Agbandje-McKenna M. Site-directed mutagenesis improves the transduction efficiency of capsid library-derived recombinant AAV vectors. Mol. Ther. Methods Clin. Dev. 2020;17:545–555. doi: 10.1016/j.omtm.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhong L., Li B., Jayandharan G., Mah C.S., Govindasamy L., Agbandje-McKenna M., Herzog R.W., Weigel-Van Aken K.A., Hobbs J.A., Zolotukhin S. Tyrosine-phosphorylation of AAV2 vectors and its consequences on viral intracellular trafficking and transgene expression. Virology. 2008;381:194–202. doi: 10.1016/j.virol.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Markusic D.M., Herzog R.W., Aslanidi G.V., Hoffman B.E., Li B., Li M., Jayandharan G.R., Ling C., Zolotukhin I., Ma W. High-efficiency transduction and correction of murine hemophilia B using AAV2 vectors devoid of multiple surface-exposed tyrosines. Mol. Ther. 2010;18:2048–2056. doi: 10.1038/mt.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martino A.T., Basner-Tschakarjan E., Markusic D.M., Finn J.D., Hinderer C., Zhou S., Ostrov D.A., Srivastava A., Ertl H.C., Terhorst C. Engineered AAV vector minimizes in vivo targeting of transduced hepatocytes by capsid-specific CD8+ T cells. Blood. 2013;121:2224–2233. doi: 10.1182/blood-2012-10-460733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Büning H., Srivastava A. Capsid modifications for targeting and improving the efficacy of AAV vectors. Mol. Ther. Methods Clin. Dev. 2019;12:248–265. doi: 10.1016/j.omtm.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shirley J.L., de Jong Y.P., Terhorst C., Herzog R.W. Immune responses to viral gene therapy vectors. Mol. Ther. 2020;28:709–722. doi: 10.1016/j.ymthe.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shirley J.L., Keeler G.D., Sherman A., Zolotukhin I., Markusic D.M., Hoffman B.E., Morel L.M., Wallet M.A., Terhorst C., Herzog R.W. Type I IFN sensing by cDCs and CD4+ T cell help are both requisite for cross-priming of AAV capsid-specific CD8+ T cells. Mol. Ther. 2020;28:758–770. doi: 10.1016/j.ymthe.2019.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perocheau D.P., Cunningham S., Lee J., Antinao Diaz J., Waddington S.N., Gilmour K., Eaglestone S., Lisowski L., Thrasher A.J., Alexander I.E. Age-related seroprevalence of antibodies against AAV-LK03 in a UK population cohort. Hum. Gene Ther. 2019;30:79–87. doi: 10.1089/hum.2018.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang L., Bell P., Somanathan S., Wang Q., He Z., Yu H., McMenamin D., Goode T., Calcedo R., Wilson J.M. Comparative study of liver gene transfer with AAV vectors based on natural and engineered AAV capsids. Mol. Ther. 2015;23:1877–1887. doi: 10.1038/mt.2015.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aurnhammer C., Haase M., Muether N., Hausl M., Rauschhuber C., Huber I., Nitschko H., Busch U., Sing A., Ehrhardt A., Baiker A. Universal real-time PCR for the detection and quantification of adeno-associated virus serotype 2-derived inverted terminal repeat sequences. Hum. Gene Ther. Methods. 2012;23:18–28. doi: 10.1089/hgtb.2011.034. [DOI] [PubMed] [Google Scholar]
- 57.Andrus L., Marukian S., Jones C.T., Catanese M.T., Sheahan T.P., Schoggins J.W., Barry W.T., Dustin L.B., Trehan K., Ploss A. Expression of paramyxovirus V proteins promotes replication and spread of hepatitis C virus in cultures of primary human fetal liver cells. Hepatology. 2011;54:1901–1912. doi: 10.1002/hep.24557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brehm M.A., Cuthbert A., Yang C., Miller D.M., DiIorio P., Laning J., Burzenski L., Gott B., Foreman O., Kavirayani A. Parameters for establishing humanized mouse models to study human immunity: analysis of human hematopoietic stem cell engraftment in three immunodeficient strains of mice bearing the IL2rγnull mutation. Clin. Immunol. 2010;135:84–98. doi: 10.1016/j.clim.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]