Abstract
The solute carrier 30 (SLC30) family genes play a fundamental role in various cancers. However, the diverse expression patterns, prognostic value, and potential mechanism of SLC30A family genes in gastric cancer (GC) remain unknown. Herein, we analyzed the expression and survival data of SLC30A family genes in GC patients using multiple bioinformatic approaches. Expression data of SLC30A family genes for GC patients were extracted from the Cancer Genome Atlas (TCGA) and genetic alteration frequency assessed by using cBioportal database. And validated the expression of SLC30A family genes in GC tissues and corresponding normal tissues. The prognostic value of SLC30A family genes in gastric cancer patients were explored using Kaplan–Meier plotter database. Functional enrichment analysis performed using DAVID database and clusterProfiler package. And ssGSEA algorithm was performed to explore the relationship between the SLC30A family genes and the infiltration of immune cells. We found that the median expression levels of SLC30A1-3, 5–7, and 9 were significantly upregulated in gastric cancer tissues compared to non-cancerous tissues, while SLC30A4 was downregulated. Meanwhile, SLC30A1-7, and 9 were significantly correlated with advanced tumor stage and nodal metastasis status, SLC30A5-7, and 9–10 were significantly related to the Helicobacter pylori infection status of GC patients. High expression of five genes (SLC30A1, 5–7, and 9) was significantly correlated with better overall survival (OS), first progression survival (FPS), and post progression survival (PPS). Conversely, upregulated SLC30A2-4, 8, and 10 expression was markedly associated with poor OS, FP and PPS. And SLC30A family genes were closely associated with the infiltration of immune cells. The present study implied that SLC30A5 and 7 may be potential biomarkers for predicting prognosis in GC patients, SLC30A2 and 3 play an oncogenic role in GC patients and could provide a new strategy for GC patients treatment.
Subject terms: Cancer genetics, Gastrointestinal cancer, Oncogenes
Background
Gastric cancer (GC) is one of the most prevalent malignancy worldwide1. According to the latest cancer statistics, GC is considered the second most common cause of cancer-related mortality in the world2. Most GC is induced by a complex interaction between Helicobacter pylori and host factors3. Multiple studies have reported that various environmental elements are considered as gastric cancer risk factors including trace elements4–6. Surgery is the primary therapeutic for GC patients, even with the advances in diagnosis and treatment in the past few years. GC patient prognosis remains unfavorable in that many patients are still initially diagnosed at an advanced stage7. Hence, it is extremely important to seek potential prognostic biomarkers for early diagnosis and novel therapeutic targets.
Cixian and Linxian, located in northern China along the Taihang Mountain chain, are one of the higher-risk areas for upper gastrointestinal cancer both in China and worldwide8–11 (Supplementary Figures S1-2). Previous studies showed that individuals living in Cixian and Linxian have a zinc intake below the recommended daily allowance and higher incidence and mortality rates of GC than that of other regions9,10,12. For zinc to perform its various bioactive roles, many specific systems to transport zinc across the biological membrane are needed13. Therefore, zinc transport proteins are indispensable for facilitating the bioactive roles of zinc. Zinc homeostasis is mostly maintained by the Zn transporter (SLC30A , ZnT) and Irt-related proteins (SLC39A, ZIP), which play critical roles in a wide array of biological processes and cellular functions including growth, endocrine, reproductive, and immune processes14–16.
Emerging evidence indicates that the solute carrier (SLC) 39A family of genes, also known as zinc importer genes, are significantly correlated with prognosis in GC patients17. Therefore, we hypothesized that SLC30A family genes, also known as zinc exporter genes, might also be strongly associated with GC. The SLC30A family, including SLC30A1-10, contribute to the cytoplasmic zinc balance by exporting zinc to the extracellular space or moving cytoplasmic zinc into intracellular compartments when cellular zinc levels are elevated16. Furthermore, multiple studies have revealed that SLC30A family genes are dysregulated and played a critical role in various kinds of cancer, including pancreatic cancer18, invasive breast ductal carcinoma18, and esophageal cell carcinoma20. Previous studies have reported that SLC30A1, 9 and 10 were significantly upregulated in prostate cancer tissues compared to normal tissues, while SLC30A5-6 were strongly downregulated21–23. Upregulated SLC30A5-7 expression might play a critical role in coordinating transcriptional programming associated with the increased activity of the early secretory pathway in colorectal cancer24. Nevertheless, the functional and prognostic significance of SLC30A family genes in GC remains unclear.
To the best of our knowledge, a comprehensive analysis has yet to be applied to clarify the role of SLC30A family genes in GC. Based on the multiple bioinformatics databases, we analyzed the expression and mutation of SLC30A family genes in patients with GC, and evaluated their prognostic value.
Materials and methods
Patients and samples
The present study was performed using data obtained from 40 consecutive patients from Cixian and Cixian, a region in Hebei Province with a high rate of epidemiologically and histologically confirmed GC9,11. All patients were surgically treated at The Fourth Hospital of Hebei Medical University from January 1, 2017 to December 31, 2018. All patients have received pathological diagnosis of primary GC (Supplementary Table 1).
RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from frozen tumor and corresponding non-tumorous tissues using TRIzol reagent (Invitrogen, Thermo Fisher Scientific, Inc.). After the concentration and purity of the total RNA were determined by ultraviolet absorbance spectroscopy, RNA was reverse transcribed into cDNA using RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Lithuania). qRT-PCRs using SuperReal PreMix Plus (SYBR Green) (TianGen, Beijing, China) were performed on ABI7500 Real-Time System (Life Technologies Corp., Foster City, CA, USA). The PCR cycling parameters were as follows: 95 ℃ for 10 min, and 40 cycles of 95 ℃ for 15 s, 60 ℃ for 30 s and 72 ℃ for 30 s. The samples were run in triplicate and the mean value was calculated for each case. The primers for SLC30A family genes are listed in Supplementary Table 2. The human GAPDH gene was employed as an internal control. The relative expression of SLC30A family genes was calculated using the 2−ΔΔCT method according to the previously described protocol25.
TCGA database
TCGA is a large repository of high throughput data of human carcinomas, containing over 30 human tumor cohort studies26. The expression profiling of SLC30A family genes were retrieved from the TCGA-STAD database. In addition, the clinicopathological parameters of GC were downloaded from TCGA in order to assess the diagnostic value of SLC30A family genes in GC patients using receiver operating characteristic (ROC) curve.
UALCAN database
UALCAN is a web resource that provides comprehensive cancer transcriptome data (https://ualcan.path.uab.edu/)27. The expression level of SLC30A family genes in GC tissues and normal gastric tissues were assessed using the UALCAN database.
TIMER database analysis
TIMER (https://cistrome.shinyapps.io/timer/) is an a comprehensive and user-friendly online tool to systematically investigate and visualize the correlation between immune infiltrates and a wide spectrum of factors, including gene expression, clinical outcomes and somatic mutations over 10,897 tumors from 32 cancer types28,29. The differential expression of SLC30A family genes between tumor and normal tissues could be evaluated using Diff Exp module across all the TCGA database tumors and the results were shown with boxplots.
cBioportal database
cBioportal is an interactive open-source platform, that provides large scale cancer genomics data sets (https://www.cbioportal.org/)30,31. The frequency of SLC30A family gene alterations (amplification, deep deletion, and missense mutations) in GC patients was assessed using the cBioportal for Cancer Genomics database and TCGA.
Correlation and functional enrichment analysis of SLC30A family Genes
Correlation between the mRNA expression of SLC30A family genes was evaluated using Pearson’s correlation coefficient and Corrplot32 package in R software. Gene ontology (GO), including biological process (BP), molecular function (MF) and cellular component (CC), is a commonly used bioinformatics tool that provides comprehensive information on gene function of individual genomic data. The Kyoto Encyclopedia of Genes and Genomes (KEGG), a database was used to assign biological function and utilities of target genes. GO and KEGG enrichment analysis and annotations were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) database (https://david.ncifcrf.gov/)33, which provides a user-friendly and comprehensive tools for explore the potential biological meaning of what you are interested gene lists. Enrichment results visualization was performed using ClusterProfiler34 package in R software with criterion false discovery rate (FDR < 0.05). To understand the connections among the SLC30A family genes, STRING database (https://string-db.org/) was used to construct PPI network35,36.
Kaplan–Meier plotter database
ROC curve analysis was conducted using the pROC37 package in R software to explore the sensitivity and specificity of using the SLC30A family genes to distinguish GC patients from healthy individuals. Kaplan–Meier plotter (https://kmplot.com/) is an online database containing microarray gene expression data and survival information extrcated from Gene Expression Omnibus and TCGA database, which contain the gene expression and survival data of 1065 GC patients38. 631 GC patients were included in this study (Supplementary Table 3). Patients missing expression values or lacking complete clinical data, including survival time and status, were exclude. To investigate the underlying prognostic value of SLC30A family genes, we evaluated OS, FPS, and PPS using the Kaplan–Meier plotter database based on median expression (high vs. low). Assessments were made using a Kaplan–Meier survival plot with a hazard ratio with 95% confidence intervals and log rank p-values. Furthermore, the correlation between mRNA expression of SLC30A family genes and different clinicopathological characteristics such as gender, age, HER2 status, clinical stage, Lauren classification, differentiation, perforation, and treatment method were evaluated using this database. Treatment classification in GC patients was divided into surgery alone, 5 FU-based adjuvant, and other adjuvant treatments. Moreover, we performed multivariate Cox regression analysis to determine if SLC30A family genes could serve as prognostic markers based on GSE62254 cohort.
Single-sample gene set enrichment analysis (ssGSEA)
The infiltration levels of immune cell types were quantified by ssGSEA method using gsva package39 in R software. The ssGSEA applies gene signatures expressed by immune cell populations to indivadual cancer samples40. The deconvolution approach used in our study including 24 immune cells that are involved in immunity including B cells, DC, iDC, aDC, pDC, Eosinophils, Macrophages, Mast cells, Neutrophils, NK cells, NK CD56dim cells, NKCD56bright cells, T cell, Cytotoxic cells, CD8 T cells, Tgd, T helper cells, Tcm, Tem, Th1, Th2, Tfh, TReg, Th1741. And we further conducted the ssGSEA algorithm to explore the relationship between the SLC30A family genes and the infiltration of immune cells.
Statistical analysis
All statistical analysis was performed using SPSS 21.0 software (SPSS Company, Chicago, Illinois, USA) and R software. And all methods were performed in accordance with the relevant guidelines and regulations. The real-time RT-PCR results were expressed as the mean ± S.D. Student’s test was used to compare the expression means between different groups. P < 0.05 indicated a statistically significant difference.
Ethics statement
This study was approved by the Institutional Human Ethics Committee of Hebei Medical University Fourth Hospital (ID 2018MEC042), and prior informed consent obtained from all the patients. We confirm that all the methods had been carried out in accordance with the relevant guidelines and regulations of the Declaration of Helsinki.
Consent for publication
All authors have reviewed the manuscript and consented for publication.
Results
Relative transcriptional expression of SLC30A family genes in GC patients using the UALCAN database
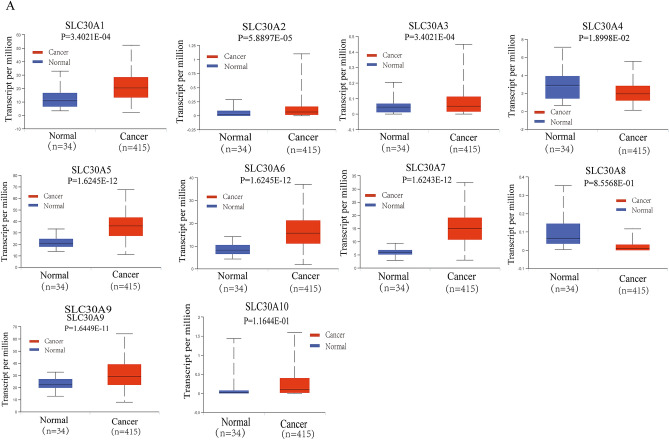
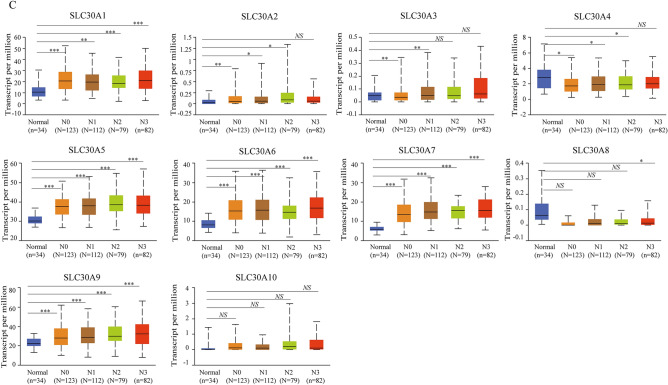
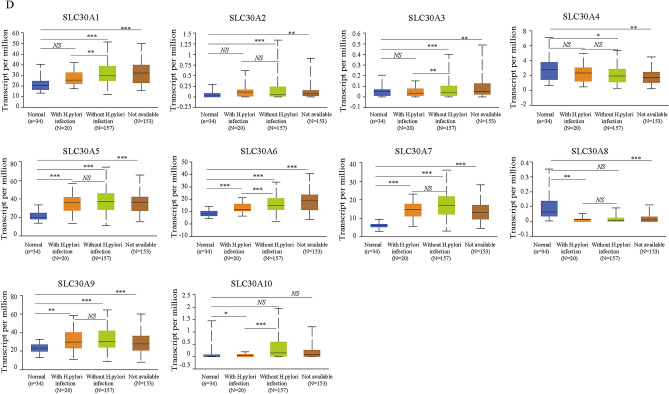
Comparison of the transcriptional expression of SLC30A family genes in gastric tumor tissues and normal tissues indicated that mRNA expression of SLC30A1-3, 5–7, and 9 was significantly upregulated in cancer tissues compared to non-cancerous tissues in GC patients, while SLC30A4 was downregulated in the former compared to the latter (Fig. 1A and Figure S3). Moreover, assessment of the correlation between SLC30A family genes expression levels and the tumor stages of GC patients indicated that the expression levels of most SLC30A family genes, including SLC30A1, 5–7, and 9, were significantly and positively associated with tumor stage in GC patients. Nevertheless, SLC30A8 and 10 expression had no statistical significance (Fig. 1B). We also analyzed the relationship between the expression level of SLC30A family genes and the nodal metastasis status of GC patients. Five genes were positively associated with nodal metastasis for GC patients (SLC30A1, 5–7, and 9). However, SLC30A4 was significantly and negatively correlated with nodal metastasis for GC patients (Fig. 1C). The expression level of most SLC30A family genes was significantly associated with the Helicobacter pylori infection status of GC patients, but the most significant correlation occurred for SLC30A5-10 (Fig. 1D). Furthermore, we validated the expression of SLC30A family genes in 40 GC patients. Most of the expression levels of SLC30A family genes were consistent with those of previous studies, but the expression levels of SLC30A8 and 9 had no significant differences between GC tissues and corresponding non-cancerous tissues (Fig. 1E).
Figure 1.
Relative expression and the correlation between SLC30A family genes in patients with GC. (A) The expression of SLC30A family genes in GC patients (Ualcan database). The P-value was set at 0.05, and most of SLC30A family genes were significantly dysregulated in GC patients. (B) Correlation between expression of SLC30A family genes and tumor stages in GC patients (TCGA data). (C) Expression of SLC30A family genes in GC based on nodal metastasis status (UALCAN database). N0: No regional lymph node metastasis; N1: metastases in 1 to 3 axillary lymph nodes; N2: metastases in 4 to 9 axillary lymph nodes; N3: metastases in 10 or more axillary lymph nodes. (D) expression of SLC30A family genes in GC based on H.pylori infection status (UALCAN database). (E) Relative expression of SLC30A family genes validated in 40 patients with GC. The P-value was set at 0.05. * indicates P-value < 0.05, ** indicates P-value < 0.01, *** indicates P-value < 0.001, NS indicates no statistical significance.
Diagnostic value of SLC30A family genes for distinguishing GC patients
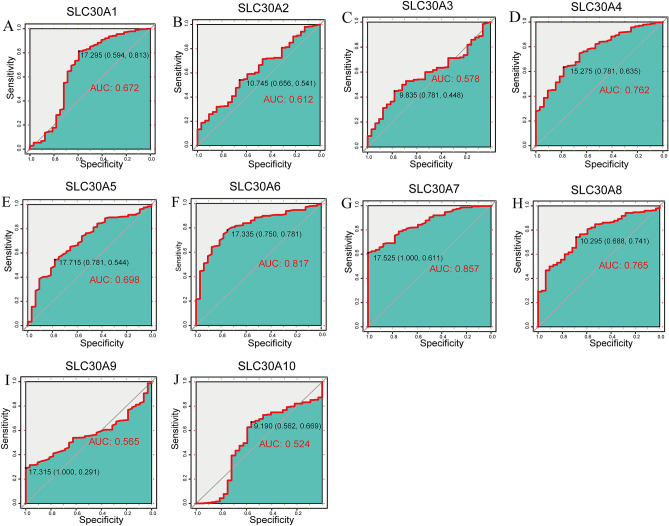
To assess the diagnostic value of SLC30A family genes in GC patients, we performed a receiver operating characteristic (ROC) curve analysis based on data from the Cancer Genome Atlas (TCGA) database. ROC analysis of the predictive efficiency of SLC30A family genes suggested that most of these genes had high diagnostic value for distinguishing GC patients from healthy individuals, including SLC30A1 (0.672), SLC30A2 (0.612), SLC30A4 (0.762), SLC30A5 (0.698), SLC30A6 (0.817), SLC30A7 (0.857), and SLC30A8 (0.765). SLC30A3 (0.578), SLC30A9 (0.565), and SLC30A10 (0.524) had moderate value for distinguishing GC patients (Fig. 2).
Figure 2.
The Receiver operating characteristic (ROC) curve of individual SLC30A family genes. (A) SLC30A1; (B) SLC30A2; (C) SLC30A3; (D) SLC30A4; (E) SLC30A5; (F) SLC30A6; (G) SLC30A7; (H) SLC30A8; (I) SLC30A9; (J) SLC30A10.
Prognostic value of SLC30A family genes in GC patients
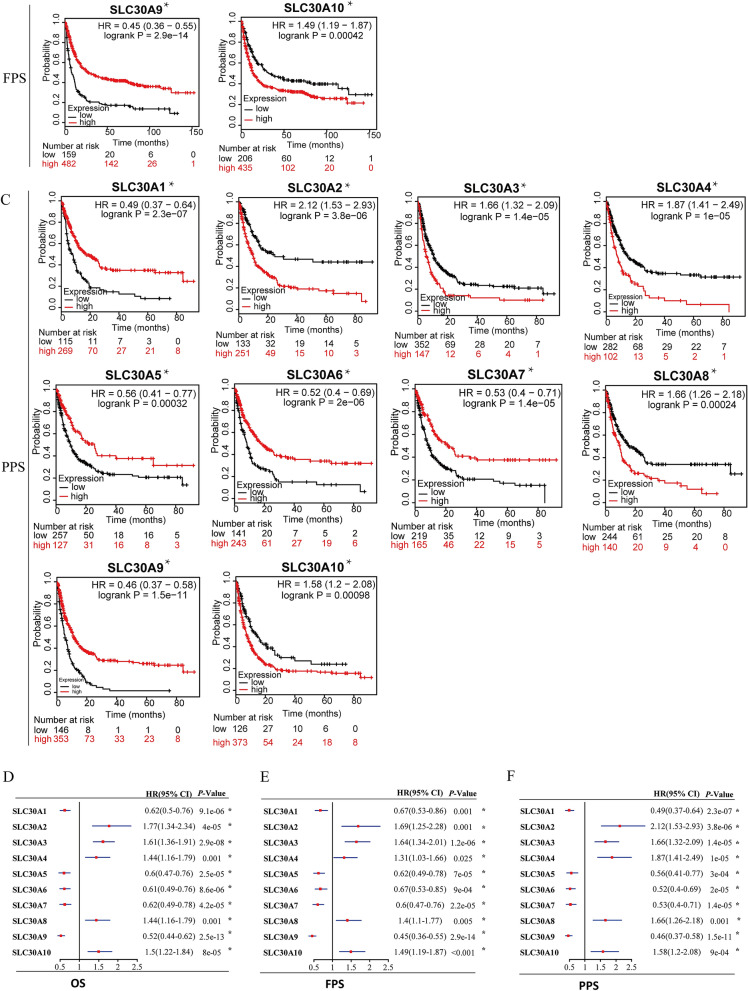
As shown in Fig. 3, all genes were significantly correlated with prognosis in GC patients. Five genes showed a significantly better OS in GC patients when upregulated, (SLC30A1: HR 0.62 [95% CI 0.5–0.76], P = 9.1e−06; SLC30A5: HR 0.6 [95% CI 0.47–0.76], P = 2.5e−05; SLC30A6: HR 0.61 [95% CI 0.49–0.79], P = 8.6e−06; SLC30A7: HR 0.62 [95% CI 0.49–0.78], P = 4.2e−05; and SLC30A9: HR 0.52, [95% CI 0.44–0.62], P = 2.5e−13). Five genes showed a negative correlaion between high expression and significant positive overall survival in GC patients, (SLC3A2: HR 1.77 [95% CI 1.34–2.34], P = 4e−05; SLC30A3: HR 1.61 [95% CI 1.36–1.91], P = 0.9e−08; SLC30A4: HR 1.44 [95% CI 1.16–1.79], P = 0.0010; SLC30A8: HR 1.44 [95% CI 1.16–1.79], P = 0.0008; and SLC30A10: HR 1.5 [95% CI 1.22–1.84], P = 8e−05). Moreover, multivariate Cox regression analysis indicated that SLC30A2, 5 and 7 could serve as OS markers independent of clinicopathological parameters (Table 1).
Figure 3.
Prognostic value of SLC30A family genes in GC patients. (A-C) The correlation between expression level of SLC30A family genes and OS, FPS, and PPS in GC patients (Kaplan–Meier plotter database). (D-F) Forest plot of OS, FPS, PPS and mRNA expression of SLC30A family genes in GC patients. Logrank P was set at 0.05. OS: overall survival. FPS: First Progression Survival; PPS: Post Progression Survival.
Table 1.
Multivariate analysis based on GSE62254.
| Factor | Subgroup | β | SE | Wald | RR (95% CI) | P |
|---|---|---|---|---|---|---|
| TNM stage | T3 | 0.686 | 0.199 | 11.914 | 1.986 (1.345–2.933) | 0.001 |
| N2 | 0.953 | 0.372 | 6.578 | 2.594 (1.252–5.375) | 0.010 | |
| N3 | 1.763 | 0.382 | 21.262 | 5.830 (2.756–12.334) | < 0.001 | |
| M | 1.009 | 0.247 | 16.712 | 2.742 (1.691–4.447) | < 0.001 | |
| SLC30A2 | 0.409 | 0.187 | 4.762 | 1.505 (1.042–2.172) | 0.029 | |
| SLC30A5 | − 0.518 | 0.179 | 8.357 | 0.596 (0.419–0.846) | 0.004 | |
| SLC30A7 | − 0.472 | 0.180 | 6.863 | 0.624 (0.439–0.888) | 0.009 |
Using a forest plot to investigate the potential prognostic value of SLC30A family genes, to reveal the correlation between OS, FPS, PPS, and mRNA expression of SLC30A family genes in GC patients (Fig. 3D–F). The results showed that the high expression of five genes, (SLC30A1, 5–7, and 9), had a positively significant correlation with improved FPS, and PPS. In contrast, upregulated SLC30A2-4, 8, and 10 expression was negatively correlated with favorable FPS, and PPS.
Association of SLC30A family genes prognostic values in GC patients with different clinicopathological features
Investigation of the correlation between clinicopathological features such as gender, clinical stage, Lauren classification, differentiation, HER2 status, treatment types, and perforation and mRNA expression level of SLC30A family genes showed that all SLC30A family gene expression was significantly correlated with gender in GC patients (Table 2). Five genes were promising positive prognostic factors in both male and female patients, including SLC30A1, 5–7, and 9. Nevertheless, SLC30A2-4, 8, and 10 were significantly correlated with poor prognosis in both male and female patients. Upregulated expression of SLC30A1, 5–7, and 9 predicted a favorable prognosis in GC patients with stage III/IV, I/III/IV, I/III/IV, I/III/IV, and I/III/IV, respectively (Table 3). High expression of SLC30A2-4, 8, and 10 was significantly associated with an unfavorable prognosis in stage I/III/IV, III, III/IV, I/III/IV, and I/III GC patients, respectively.
Table 2.
The relationship between SLC30A family genes and OS in different gender of GC patients (Kaplan–Meier Plotter).
| Gender | Cases | HR (95% CI) | P-value | |
|---|---|---|---|---|
| SLC30A1 | Male | 349 | 0.54 (0.39–0.76) | 0.0004* |
| Female | 187 | 0.52 (0.34–0.8) | 0.0026* | |
| SLC30A2 | Male | 349 | 2 (1.37–2.91) | 0.0002* |
| Female | 187 | 1.95 (1.11–3.4) | 0.0170* | |
| SLC30A3 | Male | 349 | 1.67 (1.34–2.07) | 2.8e−06* |
| Female | 187 | 1.95 (1.36–2.8) | 0.0002* | |
| SLC30A4 | Male | 349 | 1.52 (1.12–2.06) | 0.0065* |
| Female | 187 | 1.68 (1.1–2.59) | 0.0161* | |
| SLC30A5 | Male | 349 | 0.55 (0.4–0.74) | 6.6e−05* |
| Female | 187 | 0.58 (0.36–0.91) | 0.0171* | |
| SLC30A6 | Male | 349 | 0.5 (0.37–0.67) | 2.7e−06* |
| Female | 187 | 0.59 (0.36–0.97) | 0.0370* | |
| SLC30A7 | Male | 349 | 0.45 (0.33–0.61) | 1.3e−07* |
| Female | 187 | 0.64 (0.42–0.99) | 0.0431* | |
| SLC30A8 | Male | 349 | 1.37 (1.02–1.84) | 0.0370* |
| Female | 187 | 2.49 (1.6–3.87) | 2.8e−05* | |
| SLC30A9 | Male | 349 | 0.47 (0.38–0.59) | 3.7e−11* |
| Female | 187 | 0.54 (0.37–0.78) | 0.0010* | |
| SLC30A10 | Male | 349 | 1.73 (1.32–2.25) | 4.8e−05* |
| Female | 187 | 1.35 (0.95–1.92) | 0.0930* |
a: The P-value was set at 0.05 and the * indicate that the results are statistically significant.
b: SLC30, The solute carriers’ families 30; OS, overall survival; HR, hazard ratio; CI, confidence interval.
Table 3.
The relationship between SLC30A family genes and OS in different stages of GC patients (Kaplan–Meier Plotter).
| Stage | Cases | HR (95% CI) | P-value | |
|---|---|---|---|---|
| SLC30A1 | I | 62 | 0.43 (0.14–1.31) | 0.1270 |
| II | 140 | 0.74 (0.37–1.5) | 0.4060 | |
| III | 197 | 0.64 (0.43–0.96) | 0.0280* | |
| IV | 140 | 0.53 (0.34–0.82) | 0.0037* | |
| SLC30A2 | I | 62 | 3,777,800 (0-lnf) | 0.0150* |
| II | 140 | 1.55 (0.82–2.9) | 0.1710 | |
| III | 197 | 1.69 (1.15–2.49) | 0.0067* | |
| IV | 140 | 2 (1.34–3) | 0.0006* | |
| SLC30A3 | I | 62 | 2.34 (0.67–8.22) | 0.1720 |
| II | 140 | 1.66 (0.88–3.13) | 0.1140 | |
| III | 197 | 1.64 (1.23–2.19) | 0.0007* | |
| IV | 140 | 0.75 (0.51–1.12) | 0.1570 | |
| SLC30A4 | I | 62 | 3.06 (0.93–10.1) | 0.0551 |
| II | 140 | 1.56 (0.81–3.01) | 0.1840 | |
| III | 197 | 1.68 (1.14–2.49) | 0.0082* | |
| IV | 140 | 1.6 (1.07–2.38) | 0.0210* | |
| SLC30A5 | I | 62 | 0.21 (0.07–0.64) | 0.0026* |
| II | 140 | 1.65 (0.82–3.33) | 0.1590 | |
| III | 197 | 0.58 (0.4–0.85) | 0.0046* | |
| IV | 140 | 0.56 (0.37–0.84) | 0.0041* | |
| SLC30A6 | I | 62 | 0.14 (0.03–0.64) | 0.0033* |
| II | 140 | 0.55 (0.29–1.05) | 0.0650 | |
| III | 197 | 0.57 (0.38–0.84) | 0.0042* | |
| IV | 140 | 0.48 (0.31–0.74) | 0.0007* | |
| SLC30A7 | I | 62 | 0.19 (0.05–0.7) | 0.0053* |
| II | 140 | 0.67 (0.34–1.35) | 0.2630 | |
| III | 197 | 0.57 (0.39–0.83) | 0.0031* | |
| IV | 140 | 0.63 (0.41–0.98) | 0.0390* | |
| SLC30A8 | I | 62 | 4.6 (1.53–13.82) | 0.0028* |
| II | 140 | 1.8 (0.88–3.69) | 0.1031 | |
| III | 197 | 1.55 (1.07–2.25) | 0.0202* | |
| IV | 140 | 1.68 (1.1–2.57) | 0.0160* | |
| SLC30A9 | I | 62 | 0.22 (0.08–0.6) | 0.0013* |
| II | 140 | 0.58 (0.32–1.08) | 0.0850 | |
| III | 197 | 0.53 (0.39–0.72) | 4.9e−05* | |
| IV | 140 | 0.6 (0.41–0.89) | 0.0100* | |
| SLC30A10 | I | 62 | 2.88 (1.07–7.75) | 0.0280* |
| II | 140 | 1.7 (0.94–3.07) | 0.0780 | |
| III | 197 | 1.54 (1.08–2.18) | 0.0150* | |
| IV | 140 | 0.72 (0.45–1.14) | 0.1550 |
a: The P-value was set at 0.05 and the * indicate that the results are statistically significant.
SLC30A1, 3, 5–7, and 9 were promising favorable prognostic factors in both intestinal and diffuse type GC patients, and high SLC30A5 expression was also significantly correlated with mixed type patients (Table 4). Besides, SLC30A2 and SLC30A8 predicted poorer prognosis in both intestinal and diffuse type patients and high expression of SLC30A3 and SLC30A10 correlated with poor prognosis in intestinal, mixed type GC patients, respectively. High expression of SLC30A2, 4, and 9 were correlated with the improved prognosis in poorly differentiation GC patients (Table 5). Nevertheless, SLC30A1 and 6 were significantly associated with poor OS in moderately differentiation GC patients. Analysis of HER2 status and expression of SLC30A family genes revealed that upregulated expression of SLC30A1, 5–6, and 9 predicted favorable OS in both HER2-positive and HER2-negative patients, while SLC30A2-3, 8, and 10 were associated with a worse prognosis. High expression of SLC30A4 and 7 were significantly associated with unfavorable OS in HER2-negative and improved prognosis in HER2-positive patients, respectively. In this study, treatments in GC patients divided into surgery alone, 5 FU based adjuvant and other treatment (Table 6).
Table 4.
The relationship between SLC30A family genes and OS in different Lauren classification of GC patients (Kaplan–Meier Plotter).
| Lauren classification | Cases | HR (95% CI) | P-value | |
|---|---|---|---|---|
| SLC30A1 | Intestinal | 269 | 0.59 (0.4–0.87) | 0.0065* |
| Diffuse | 240 | 0.66 (0.47–0.94) | 0.0190* | |
| Mixed | 29 | 0.36 (0.08–1.63) | 0.1670 | |
| SLC30A2 | Intestinal | 269 | 1.89 (1.21–2.94) | 0.0043* |
| Diffuse | 240 | 1.97 (1.26–3.06) | 0.0023* | |
| Mixed | 29 | 2.18 (0.66–7.21) | 0.1890 | |
| SLC30A3 | Intestinal | 269 | 1.6 (1.12–2.27) | 0.0086* |
| Diffuse | 240 | 1.28 (0.88–1.85) | 0.1900 | |
| Mixed | 29 | 0.38 (0.14–1.06) | 0.0560 | |
| SLC30A4 | Intestinal | 269 | 1.73 (1.2–2.49) | 0.0028* |
| Diffuse | 240 | 1.34 (0.95–1.89) | 0.0980 | |
| Mixed | 29 | 2.95 (0.97–8.97) | 0.0460* | |
| SLC30A5 | Intestinal | 269 | 0.44 (0.31–0.64) | 7.7e−06* |
| Diffuse | 240 | 0.51 (0.34–0.76) | 0.0009* | |
| Mixed | 29 | 0.56 (0.41–0.79) | 0.0079* | |
| SLC30A6 | Intestinal | 269 | 0.54 (0.37–0.78) | 8e−04* |
| Diffuse | 240 | 0.62 (0.43–0.88) | 0.0070* | |
| Mixed | 29 | 0.53 (0.18–1.58) | 0.245 | |
| SLC30A7 | Intestinal | 269 | 0.53 (0.36–0.78) | 0.0010* |
| Diffuse | 240 | 0.54 (0.38–0.76) | 0.0003* | |
| Mixed | 29 | 1.98 (0.52–7.55) | 0.3100 | |
| SLC30A8 | Intestinal | 269 | 1.51 (1.03–2.2) | 0.0330* |
| Diffuse | 240 | 1.75 (1.24–2.46) | 0.0012* | |
| Mixed | 29 | 2.78 (0.92–8.41) | 0.0590 | |
| SLC30A9 | Intestinal | 269 | 0.42 (0.31–0.58) | 4.2e−08* |
| Diffuse | 240 | 0.46 (0.3–0.71) | 0.0004* | |
| Mixed | 29 | 0.5 (0.17–1.44) | 0.1900 | |
| SLC30A10 | Intestinal | 269 | 1.41 (0.95–2.11) | 0.0880 |
| Diffuse | 240 | 0.7 (0.46–1.05) | 0.0816 | |
| Mixed | 29 | 3.57 (1.23–10.35) | 0.0120* |
a: The P-value was set at 0.05 and the * indicate that the results are statistically significant.
Table 5.
The relationship between SLC30A family genes and OS in different differentiation of GC patients (Kaplan–Meier Plotter).
| Differentiation | Cases | HR (95% CI) | P-value | |
|---|---|---|---|---|
| SLC30A1 | Poorly | 121 | 1.55 (0.93–2.6) | 0.0920 |
| Moderately | 67 | 2.41 (1.22–4.77) | 0.0094* | |
| Well | 5 | – | – | |
| SLC30A2 | Poorly | 121 | 0.58 (0.35–0.95) | 0.0290* |
| Moderately | 67 | 1.73 (0.79–3.78) | 0.1680 | |
| SLC30A3 | Poorly | 121 | 0.78 (0.52–1.17) | 0.2240 |
| Moderately | 67 | 1.35 (0.7–2.6) | 0.3770 | |
| SLC30A4 | Poorly | 121 | 0.59 (0.36–0.96) | 0.0320* |
| Moderately | 67 | 1.66 (0.85–3.22) | 0.1334 | |
| SLC30A5 | Poorly | 121 | 1.22 (0.75–1.98) | 0.4230 |
| Moderately | 67 | 0.66 (0.34–1.28) | 0.2150 | |
| SLC30A6 | Poorly | 121 | 1.54 (0.92–2.55) | 0.0950 |
| Moderately | 67 | 2.03 (1.05–3.95) | 0.0330* | |
| SLC30A7 | Poorly | 121 | 1.52 (0.92–2.53) | 0.1020 |
| Moderately | 67 | 1.56 (0.8–3.02) | 0.1850 | |
| SLC30A8 | Poorly | 121 | 1.95 (1.11–3.43) | 0.0180* |
| Moderately | 67 | 0.66 (0.33–1.31) | 0.2340 | |
| SLC30A9 | Poorly | 121 | 0.62 (0.41–0.92) | 0.0180* |
| Moderately | 67 | 0.6 (0.3–1.16) | 0.120 | |
| SLC30A10 | Poorly | 121 | 0.77 (0.51–1.16) | 0.214 |
| Moderately | 67 | 0.75 (0.37–1.52) | 0.4260 |
a: The P-value was set at 0.05 and the * indicate that the results are statistically significant.
Table 6.
The relationship between SLC30A family genes and OS in different HER2 status of GC patients (Kaplan–Meier Plotter).
| HER2 | Cases | HR (95% CI) | P-value | |
|---|---|---|---|---|
| SLC30A1 | Positive | 202 | 0.61 (0.41–0.93) | 0.0200* |
| Negative | 429 | 0.66 (0.52–0.82) | 0.0003* | |
| SLC30A2 | Positive | 202 | 1.46 (1–2.14) | 0.0470* |
| Negative | 429 | 1.67 (1.27–2.18) | 0.0002* | |
| SLC30A3 | Positive | 202 | 1.6 (1.23–2.08) | 0.0004* |
| Negative | 429 | 1.58 (1.25–1.98) | 8.2e−05* | |
| SLC30A4 | Positive | 202 | 1.3 (0.86–1.95) | 0.2120 |
| Negative | 429 | 1.65 (1.26–2.16) | 0.0002* | |
| SLC30A5 | Positive | 202 | 0.6 (0.42–0.88) | 0.0077* |
| Negative | 429 | 0.59 (0.46–0.78) | 0.0001* | |
| SLC30A6 | Positive | 202 | 0.56 (0.36–0.88) | 0.0120* |
| Negative | 429 | 0.53 (0.41–0.69) | 1.9e−06* | |
| SLC30A7 | Positive | 202 | 0.69 (0.45–0.76) | 4.8e−05* |
| Negative | 429 | 0.58 (0.45–1.05) | 0.0830 | |
| SLC30A8 | Positive | 202 | 1.52 (1.05–2.2) | 0.0270* |
| Negative | 429 | 1.54 (1.16–2.05) | 0.0026* | |
| SLC30A9 | Positive | 202 | 0.56 (0.42–0.73) | 2.2e−05* |
| Negative | 429 | 0.48 (0.38–0.61) | 6.2e−10* | |
| SLC30A10 | Positive | 202 | 1.66 (1.2–2.29) | 0.0018* |
| Negative | 429 | 1.29 (1.01–1.65) | 0.0430* |
a: The P-value was set at 0.05 and the * indicate that the results are statistically significant.
b: HER2, human epidermal growth factor receptor 2.
SLC30A6-7 and 9 were strongly related to favorable OS in GC patients based on a surgery only treatment. SLC30A1 and 9–10 were positively associated with other adjuvant treatments, while high expression of SLC30A2 predicted better prognosis in 5 fluorouracil (FU)- based adjuvant treatment. Nevertheless, overexpression of SLC30A2-3 and 8 were correlated with poor prognosis in patients that received surgery alone. SLC30A3, 8, and 10 were strongly negatively associated with OS in patients that received 5-FU based adjuvant treatment (Table 7). Furthermore, analysis of the correlation between mRNA expression of SLC30A family genes and prognosis in patients with no perforation showed that SLC30A9 was a favorable factor in patients without perforation, while overexpression of SLC30A1 and 8 were significantly associated with poor prognosis (Table 8). Taken together, all SLC30A family genes were strongly correlated with clinical characteristics including gender, clinical stage, Lauren classification, differentiation, HER2 status, perforation, and treatment method (Fig. 4).
Table 7.
The relationship between SLC30A family genes and OS in treatment methods of GC patients (Kaplan–Meier Plotter).
| Treatment | Cases | HR (95% CI) | P-value | |
|---|---|---|---|---|
| SLC30A1 | Surgery alone | 380 | 0.79 (0.59–1.06) | 0.1190 |
| 5 FU based adjuvant | 34 | 2.29 (0.75–6.99) | 0.1360 | |
| Other adjuvant | 76 | 0.28 (0.12–0.69) | 0.0030* | |
| SLC30A2 | Surgery alone | 380 | 1.65 (1.16–2.35) | 0.0051* |
| 5 FU based adjuvant | 34 | 0.3 (0.1–0.9) | 0.0230* | |
| Other adjuvant | 76 | 0.61 (0.25–1.49) | 0.2700 | |
| SLC30A3 | Surgery alone | 380 | 1.47 (1.03–2.08) | 0.0300* |
| 5 FU based adjuvant | 34 | 1.99 (1.34–2.95) | 0.0005* | |
| Other adjuvant | 76 | 2.47 (1.02–5.96) | 0.0380* | |
| SLC30A4 | Surgery alone | 380 | 1.26 (0.93–1.7) | 0.1340 |
| 5 FU based adjuvant | 34 | 0.42 (0.16–1.08) | 0.0630 | |
| Other adjuvant | 76 | 2.08 (0.87–5) | 0.0940 | |
| SLC30A5 | Surgery alone | 380 | 0.72 (0.54–0.97) | 0.0300* |
| 5 FU based adjuvant | 34 | 2.22 (0.51–9.66) | 0.2750 | |
| Other adjuvant | 76 | 0.41 (0.14–1.21) | 0.0950 | |
| SLC30A6 | Surgery alone | 380 | 0.74 (0.56–0.99) | 0.0450* |
| 5 FU based adjuvant | 34 | 0.71 (0.29–1.75) | 0.4520 | |
| Other adjuvant | 76 | 1.56 (0.57–4.3) | 0.3810 | |
| SLC30A7 | Surgery alone | 380 | 0.69 (0.51–0.91) | 0.0098* |
| 5 FU based adjuvant | 34 | 0.55 (0.22–1.37) | 0.1930 | |
| Other adjuvant | 76 | 0.3 (0.12–0.75) | 0.0060* | |
| SLC30A8 | Surgery alone | 380 | 1.69 (1.25–2.3) | 0.0006* |
| 5 FU based adjuvant | 34 | 3.17 (1.23–8.16) | 0.0120* | |
| Other adjuvant | 76 | 2.17 (0.89–5.31) | 0.0830 | |
| SLC30A9 | Surgery alone | 380 | 0.68 (0.51–0.91) | 0.0085* |
| 5 FU based adjuvant | 34 | 0.55 (0.38–0.79) | 0.0010* | |
| Other adjuvant | 76 | 0.08 (0.01–0.56) | 0.0010* | |
| SLC30A10 | Surgery alone | 380 | 1.31 (0.93–1.85) | 0.1234 |
| 5 FU based adjuvant | 34 | 1.61 (1.14–2.28) | 0.0067* | |
| Other adjuvant | 76 | 0.17 (0.04–0.75) | 0.0081* |
a: The P-value was set at 0.05 and the * indicate that the results are statistically significant.
b: FU, fluorouracil.
Table 8.
The relationship between SLC30A family genes and OS of GC patients with no perforation (Kaplan–Meier Plotter).
| Perforation | Cases | HR (95% CI) | P-value | |
|---|---|---|---|---|
| SLC30A1 | No | 169 | 1.52 (1–2.32) | 0.0490* |
| SLC30A2 | No | 169 | 0.71 (0.47–1.06) | 0.0960 |
| SLC30A3 | No | 169 | 0.72 (0.48–1.08) | 0.1080 |
| SLC30A4 | No | 169 | 0.69 (0.45–1.07) | 0.0930 |
| SLC30A5 | No | 169 | 0.82 (0.52–1.27) | 0.3630 |
| SLC30A6 | No | 169 | 1.31 (0.87–1.97) | 0.1980 |
| SLC30A7 | No | 169 | 1.31 (0.86–2.01) | 0.2070 |
| SLC30A8 | No | 169 | 1.76 (1.17–2.64) | 0.0060* |
| SLC30A9 | No | 169 | 0.5 (0.33–0.77) | 0.0012* |
| SLC30A10 | No | 169 | 0.68 (0.41–1.11) | 0.1181 |
a: The P-value was set at 0.05 and the * indicate that the results are statistically significant.
Figure 4.
Forest plot of individuals expression level of SLC30A family genes with OS in different clinicopathological features patients with GC (The P-value was set at 0.05).
Genetic alteration differences of SLC30A family genes in GC patients
To explore the roles of SLC30A family genes in GC patients, genetic alteration of 10 genes was performed using the cBioportal database. A total of 1443 patients from seven GC studies were analyzed. As results showed that mRNA mutation, amplification and deep deletion were the most important factors for alteration in different GC subtypes, including tubular stomach adenocarcinoma, mucinous stomach adenocarcinoma, intestinal type stomach adenocarcinoma, stomach adenocarcinoma, signet ring cell carcinoma of the stomach, diffuse type stomach adenocarcinoma, papillary stomach adenocarcinoma and esophagogastric adenocarcinoma (Fig. 5A). As Fig. 5B shows that SLC30A family genes were altered in 269 samples of 1443 GC patients (19%). The genetic alteration percentages of SLC30A family genes for GC varied from 1.1% to 7% for individual genes (SLC30A1, 2.1%; SLC30A2, 1.1%; SLC30A3, 3%; SLC30A4, 1.6%; SLC30A5, 2.1%; SLC30A6, 1.9%; SLC30A7, 1.8%; SLC30A8, 7%; SLC30A9, 1.9%; SLC30A10, 1.3%). The results of Kaplan–Meier plotter and log-rank test showed no significantly statistical difference in overall survival (OS) and disease-free survival (DFS) in cases with and without SLC30A family genes alterations (P-value was 0.331 and 0.0915, respectively. Figure 5C,D).
Figure 5.
Oncoprint and alteration differences of SLC30A family genes in gastric cancer (cBioportal database). (A) summary of alteration in SLC30Afamily genes. (B) The visual summary Oncoprint based on a query of the SLC30A family genes. (C) Kaplan–Meier plots comparing Overall Survival (OS) in cases with and without SLC30A family genes alterations. (D) Kaplan–Meier plots comparing Disease-free Survival (DFS) in cases with and without SLC30A family genes alterations.
Correlation and functional enrichment analysis of SLC30A family genes
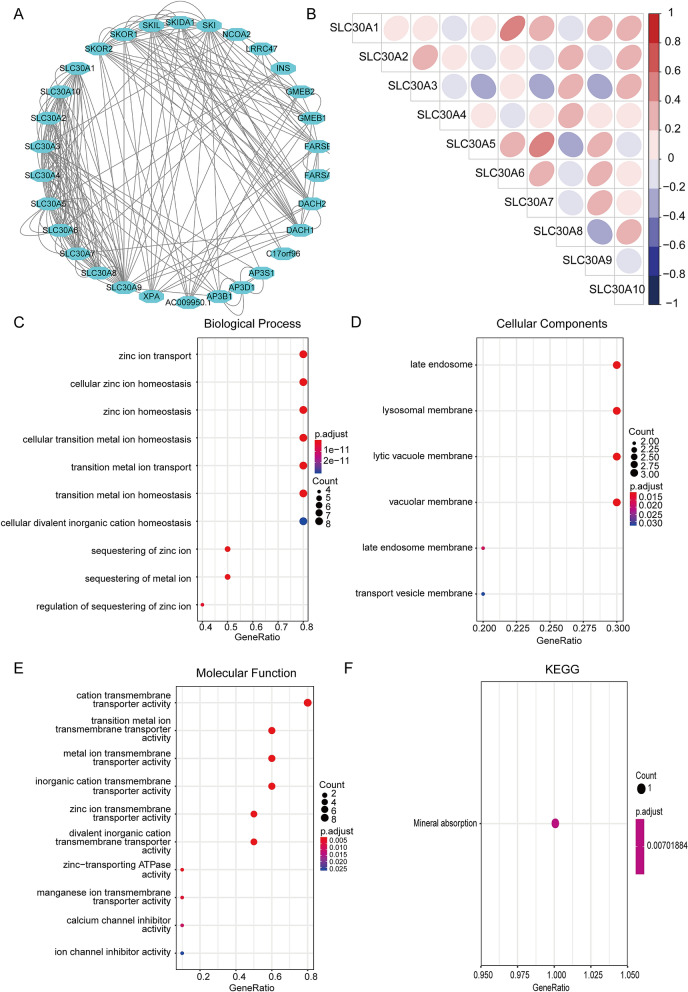
To further reveal the potential functional mechanisms in GC patients, we constructed the correlation between the expression of SLC30A family genes, protein–protein interaction (PPI) network, gene ontology (GO) term analysis, and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis (Fig. 6). The individual mRNA expressions of SLC30A family genes in GC patients were weakly correlated (Fig. 6B). The PPI network showed that 30 genes including XPA, FARSB, DACH1, and DACH2 participated in PPI networks through multiple pathways, physical interactions, genetic interactions, shared protein domains and co-expression (Fig. 6A). SLC30A family genes and their neighboring genes were mainly involved in the zinc transport, cellular zinc ion homeostasis, zinc ion homeostasis, cellular transition metal ion homeostasis, and transition metal ion transport, which are mineral transport related biological processes and mineral absorption pathways analyzed by GO term analysis and KEGG pathway enrichment analysis (Fig. 6C–F).
Figure 6.
Correlation and functional enrichment analysis of SLC30A family genes. (A) Protein–protein interaction network analysis using STRING database. (B) Pearson correlation analysis of individual among SLC30A family genes. (C) Biological process analysis; (D) cellular components; (E) molecular function. (F) Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis. All of terms colored by adjusted P-value and the size of points represent number of genes.
Immune infiltrates in correlation with SLC30A family genes in GC
The complex interactions between solid tumors and their microenvironment remain unclear, and previous studies had shown that immune infiltrates were significantly related to the progression and prognosis of GC41–43. We conducted the ssGSEA algorithm to deconvolve the relative abundance of each cell type based on expression profiling data obtained from GSE62254. The immune phenotype landscape as shown in Fig. 7A. We get further explored the coefficient of the association of SLC30A family genes in immune cell subsets (Fig. 7B). The results showed that SLC30A family genes were closely associated with the infiltration of immune cells, indicating that SLC30A family genes play an important role in GC partly because of immune infiltration.
Figure 7.
Immune landscape of gastric cancer. (A) Unsupervised clustering of 300 patients from the GSE62254 cohort using single-sample gene set enrichment analysis scores from 24 immune cell types. Molecular subtype, post operation type, number of positive nodes, Lauren classification, stage, T, N, M, age, as well as gender stage were annotated in the higher panel. Hierarchical clustering was performed with Euclidean distance and Ward linkage. (B) SLC30A family genes were associated with immune-cell subset. Red boxes indicate positive correlation and blue boxes indicate negative correlation. *, P < 0.05; **, P < 0.01.
Discussion
In the present study, ROC analysis suggested that most SLC30A family genes had high diagnostic value for distinguishing GC patients from healthy individuals and could play an important role in GC diagnosis. Furthermore, univariate survival analysis showed that upregulated SCL30A1, 5–7, and 9 expression was positively associated with favorable OS, FPS, and PPS. On the contrary, high expression of SLC30A2-4, 8, and 10 were significantly correlated with poor OS, FPS, and PPS in GC patients. Moreover, all SLC30A family genes were strongly correlated with clinical characteristics. Taken together, all members of the SLC30A gene family could be utilized as promising prognostic biomarkers in GC patients.
Zinc is an indispensable trace element that is crucial for the proper function of various cellular proteins and essential for key physiological processes including nucleic acid metabolism, regulation of gene expression, cell division44,45. Furthermore, cancer cells may extract zinc from circulation to promote cancer growth46,47. In this study, to our best knowledge and for the first time, we used various large database, including TCGA, GEO, UALCAN, cBioPortal, STRING, and Kaplan–Meier Plotter, to systematically analyzed the expression level of SLC30A family genes, prognostic values, genetic alterations, and functional enrichment analysis in GC patients.
Aberrant zinc expression levels and regulation of SLC30A family genes have been reported in various kinds of cancer. SLC30A1 is upregulated in bladder cancer and negatively targeted by miR-411 to inhibit the growth and metastasis of bladder cancer cells48. Upregulated SLC30A1 expression of could lead to cytotoxic cell death in human ductal adenocarcinoma cell lines49. Meanwhile, SLC30A1 has high expression in ovarian cancer (OC) cell lines and tissues and a recovery experiment revealed that upregulated SLC30A1 counteracts the effect of miR-8073 mimics on OC cell proliferation and apoptosis to affect the malignant progression of OC50. SLC30A2 is dysregulated in breast cancer lines and SLC30A2-mediated Zn accumulation in mitochondria is associated with increased mitochondrial oxidation51. Meanwhile, SLC30A2 over-expression leads to Zn vesicularization, shifts in cell cycle, enhanced apoptosis, and reduced proliferation and invasion in breast cancer52. SLC30A2-overexpression represses the cytotoxic effects of zinc hyper-accumulation in malignant metallothionein-null T47D breast tumor cells53. SLC30A4 is significantly overexpressed in prostate cancer compared to normal tissues from other organs22. SLC30A5-7 and 9 are significantly upregulated in colorectal cancer and SLC30A9 is involved in the canonical Wnt pathway24. Overexpressed SLC30A7 in esophageal squamous cell carcinoma could be a mechanism adapted by tumor cells to maintain the basal zinc requirement for carrying out vital functions during zinc deficiency54. SLC30A7 is also significantly upregulated in hepatocellular carcinoma55. SLC30A8 is aberrantly expressed in breast cancer and glioblastoma tumors, and decreased expression of SLC30A8 could contribute to the uncontrolled growth, proliferation, and tumor maintenance of glioblastoma multiforme cells56,57. SLC30A9 expression is significantly higher in hepatocellular carcinoma tissues than adjacent non-cancerous tissues, but is not correlated with survival in hepatocellular carcinoma patients58. SLC30A10 is aberrantly expressed in colorectal cancer and is significantly related to the methylation epigenotype and molecular genesis of colorectal cancer59,60. In the present study, mRNA expression of SLC30A1-3, SLC30A5-7, and 9 was significantly upregulated in gastric cancer tissues compared to non-cancer tissues in GC patients, while SLC30A4 was downregulated in cancer tissues.
To further clarify the genetic alteration and carcinogenic mechanism of SLC30A family genes, we found that the percentages of genetic alterations in SLC30A family genes for GC varied from 1.1 to 7% for individual genes. Furthermore, the results of Kaplan–Meier plotter and log-rank test showed no significantly statistical differences in OS and DFS in cases with and without SLC30A family gene alterations. Consistent with previous research, GO term analysis and KEGG pathway enrichment analysis showed that SLC30A family genes contributed to mineral transport related biological processes, including zinc transport, cellular zinc homeostasis, cellular transition metal ion homeostasis, and the mineral absorption pathway and our results showed that SLC30A family genes were closely associated with the infiltration of immune cells,. Therefore, we hypothesized that the action mechanism of SLC30A family genes induced tumorigenesis and progression by regulating zinc homeostasis in tumor cells and partly because of immune infiltration. This may provide a new insight in diagnosis and treatment of GC patients, especially in areas with zinc deficiency such as Cixian and Linxian.
Conclusions
In conclusion, SLC30A family genes were aberrantly expressed in GC tissues. High expression of SLC30A1, 5–7, and 9 as well as low expression of SLC30A2-4, 8, and 10 were significantly associated with favorable prognosis in GC patients. High SLC30A2 expression was significantly correlated with poor OS, FPS, and PPS in in all of GC patients indicating that these genes play an oncogenic role in GC and are markers for improved GC survival and prognostic accuracy.
Supplementary information
Acknowledgements
We would like to thank all patients and investigators who participated in publicly available database.
Abbreviations
- GC
Gastric cancer
- OS
Overall survival
- FPS
First progression survival
- PPS
Post progression survival
- SLC30A
The solute carrier 30
- TCGA
The Cancer Genome Atlas
- FU
Fluorouracil
- HER2
Human epidermal growth factor receptor 2
- ssGSEA
Single-sample gene set enrichment analysis
Author contributions
Y.H. conceived and designed this study; Y.G. analyzed the data and drafted the paper.
Data availability
Publicly available datasets were analyzed in this study. These data can be found here: TCGA, UALCAN, cBioPortal, and Kaplan–Meier Plotter.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-75012-w.
References
- 1.Cavatorta O, et al. Epidemiology of gastric cancer and risk factors. Acta Biomed. 2018;89:82–87. doi: 10.23750/abm.v89i8-S.7966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Tan P, Yeoh KG. Genetics and molecular pathogenesis of gastric adenocarcinoma. Gastroenterology. 2015;149:1153–1162. doi: 10.1053/j.gastro.2015.05.059. [DOI] [PubMed] [Google Scholar]
- 4.Kohzadi S, et al. Evaluation of trace element concentration in cancerous and non-cancerous tissues of human stomach. Chemosphere. 2017;184:747–752. doi: 10.1016/j.chemosphere.2017.06.071. [DOI] [PubMed] [Google Scholar]
- 5.Nakaji S, et al. Relationship between mineral and trace element concentrations in drinking water and gastric cancer mortality in Japan. Nutr. Cancer. 2001;40:99–102. doi: 10.1207/S15327914NC402_4. [DOI] [PubMed] [Google Scholar]
- 6.Navarro Silvera SA, Rohan TE. Trace elements and cancer risk: a review of the epidemiologic evidence. Cancer Causes Control. 2007;18:7–27. doi: 10.1007/s10552-006-0057-z. [DOI] [PubMed] [Google Scholar]
- 7.Chan TH, et al. ADAR-mediated RNA editing predicts progression and prognosis of gastric cancer. Gastroenterology. 2016;151:637–650. doi: 10.1053/j.gastro.2016.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He Y, et al. Cancer incidence and mortality in Hebei province, 2013. Medicine (Baltimore) 2017;96:e7293. doi: 10.1097/MD.0000000000007293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li D, et al. Cancer survival in Cixian of China, 2003–2013: a population-based study. Cancer Med. 2018;7:1537–1545. doi: 10.1002/cam4.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li DJ, et al. Upper gastrointestinal cancer burden in Hebei Province, China: a population-based study. World J. Gastroenterol. 2017;23:2625–2634. doi: 10.3748/wjg.v23.i14.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang D, et al. Gastric cancer burden of last 40 years in North China (Hebei Province): A population-based study. Medicine (Baltimore) 2017;96:e5887. doi: 10.1097/MD.0000000000005887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zou XN, et al. Seasonal variation of food consumption and selected nutrient intake in Linxian, a high risk area for esophageal cancer in China. Int. J. Vitam Nutr. Res. 2002;72:375–382. doi: 10.1024/0300-9831.72.6.375. [DOI] [PubMed] [Google Scholar]
- 13.Kambe T, Tsuji T, Hashimoto A, Itsumura N. The physiological, biochemical, and molecular roles of zinc transporters in zinc homeostasis and metabolism. Physiol. Rev. 2015;95:749–784. doi: 10.1152/physrev.00035.2014. [DOI] [PubMed] [Google Scholar]
- 14.Bai X, Moraes TF, Reithmeier RAF. Structural biology of solute carrier (SLC) membrane transport proteins. Mol. Membr. Biol. 2017;34:1–32. doi: 10.1080/09687688.2018.1448123. [DOI] [PubMed] [Google Scholar]
- 15.Al-Abdulla R, et al. Unraveling 'The Cancer Genome Atlas' information on the role of SLC transporters in anticancer drug uptake. Expert Rev. Clin. Pharmacol. 2019;12:329–341. doi: 10.1080/17512433.2019.1581605. [DOI] [PubMed] [Google Scholar]
- 16.Huang L, Tepaamorndech S. The SLC30 family of zinc transporters - a review of current understanding of their biological and pathophysiological roles. Mol. Aspects Med. 2013;34:548–560. doi: 10.1016/j.mam.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 17.Bisha, D. & Weimin, F. Analysis the prognostic values of solute carrier (SLC) family 39 genes in gastric cancer. Am J Transl Res (2019). [PMC free article] [PubMed]
- 18.Yang J, et al. Gene profile identifies zinc transporters differentially expressed in normal human organs and human pancreatic cancer. Curr. Mol. Med. 2013;13:401–409. [PMC free article] [PubMed] [Google Scholar]
- 19.Farquharson MJ, et al. Zinc presence in invasive ductal carcinoma of the breast and its correlation with oestrogen receptor status. Phys. Med. Biol. 2009;54:4213–4223. doi: 10.1088/0031-9155/54/13/016. [DOI] [PubMed] [Google Scholar]
- 20.Kumar A, Chatopadhyay T, Raziuddin M, Ralhan R. Discovery of deregulation of zinc homeostasis and its associated genes in esophageal squamous cell carcinoma using cDNA microarray. Int. J. Cancer. 2007;120:230–242. doi: 10.1002/ijc.22246. [DOI] [PubMed] [Google Scholar]
- 21.Milosavljevic V, et al. The Zinc-Schiff Base-Novicidin complex as a potential prostate cancer therapy. PLoS ONE. 2016;11:e0163983. doi: 10.1371/journal.pone.0163983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henshall SM, et al. Expression of the zinc transporter ZnT4 is decreased in the progression from early prostate disease to invasive prostate cancer. Oncogene. 2003;22:6005–6012. doi: 10.1038/sj.onc.1206797. [DOI] [PubMed] [Google Scholar]
- 23.Singh CK, et al. Analysis of zinc-exporters expression in prostate cancer. Sci. Rep. 2016;6:36772. doi: 10.1038/srep36772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barresi V, et al. Transcriptome analysis reveals an altered expression profile of zinc transporters in colorectal cancer. J. Cell. Biochem. 2018;119:9707–9719. doi: 10.1002/jcb.27285. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Tomczak K, Czerwinska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp. Oncol. (Pozn) 2015;19:A68–77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chandrashekar DS, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li T, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li T, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020 doi: 10.1093/nar/gkaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013 doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taiyun Wei & Simko., V. R package "corrplot": Visualization of a Correlation Matrix (Version 0.84). (2017).
- 33.Da Huang W, Sherman Brad T, et al. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 34.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szklarczyk D, et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szklarczyk D, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xavier R, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marcell SA, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322–49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barbie DA, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bindea G, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 42.Pan JH, et al. LAYN is a prognostic biomarker and correlated with immune infiltrates in gastric and colon cancers. Front. Immunol. 2019;10:6. doi: 10.3389/fimmu.2019.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiao Z, et al. TGFbeta2 is a prognostic-related biomarker and correlated with immune infiltrates in gastric cancer. J. Cell Mol. Med. 2020 doi: 10.1111/jcmm.15164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwartz JR, Marsh Randall G, Diana DZ. Zinc and skin health: overview of physiology and pharmacology. Dermatol. Surg. 2006;31:837–847. doi: 10.1111/j.1524-4725.2005.31729. [DOI] [PubMed] [Google Scholar]
- 45.Tapiero H, Tew KD. Trace elements in human physiology and pathology: zinc and metallothioneins. Biomed. Pharmacother. 2003;57:399–411. doi: 10.1016/s0753-3322(03)00081-7. [DOI] [PubMed] [Google Scholar]
- 46.Schwartz M K. Role of trace elements in cancer. Cancer Res.35 (1975). [PubMed]
- 47.Xie Y, et al. Higher serum zinc levels may reduce the risk of cervical cancer in Asian women: a meta-analysis. J. Int. Med. Res. 2018;46:4898–4906. doi: 10.1177/0300060518805600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y, et al. MiR-411 suppresses the development of bladder cancer by regulating ZnT1. Onco. Targets Ther. 2018;11:8695–8704. doi: 10.2147/OTT.S173750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jayaraman AK, Jayaraman S. Increased level of exogenous zinc induces cytotoxicity and up-regulates the expression of the ZnT-1 zinc transporter gene in pancreatic cancer cells. J. Nutr. Biochem. 2011;22:79–88. doi: 10.1016/j.jnutbio.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 50.Zhang L, Wang YF, Wang L, Wang L. MiRNA-8073 targets ZnT1 to inhibit malignant progression of ovarian cancer. Eur. Rev. Med. Pharmacol. Sci. 2019;23:6062–6069. doi: 10.26355/eurrev_201907_18419. [DOI] [PubMed] [Google Scholar]
- 51.Bostanci Z, Alam S, Soybel DI, Kelleher SL. Prolactin receptor attenuation induces zinc pool redistribution through ZnT2 and decreases invasion in MDA-MB-453 breast cancer cells. Exp. Cell Res. 2014;321:190–200. doi: 10.1016/j.yexcr.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 52.Chandler P, et al. Subtype-specific accumulation of intracellular zinc pools is associated with the malignant phenotype in breast cancer. Mol. Cancer. 2016;15:2. doi: 10.1186/s12943-015-0486-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lopez V, Foolad F, Kelleher SL. ZnT2-overexpression represses the cytotoxic effects of zinc hyper-accumulation in malignant metallothionein-null T47D breast tumor cells. Cancer Lett. 2011;304:41–51. doi: 10.1016/j.canlet.2011.01.027. [DOI] [PubMed] [Google Scholar]
- 54.Kumar, A., Chatopadhyay T Fau - Raziuddin, M., Raziuddin M Fau - Ralhan, R. & Ralhan, R. Discovery of deregulation of zinc homeostasis and its associated genes in esophageal squamous cell carcinoma using cDNA microarray. (2006). [DOI] [PubMed]
- 55.Peng, X., Fu, H., Yin, J. & Zhao, Q. CHAF1B knockdown blocks migration in a hepatocellular carcinoma model. (2018). [DOI] [PubMed]
- 56.Horvath A, et al. Novel insights into breast cancer genetic variance through RNA sequencing. Sci. Rep. 2013;3:2256. doi: 10.1038/srep02256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lozada-Delgado EL, et al. Targeting MicroRNA-143 leads to inhibition of glioblastoma tumor progression. Cancers (Basel) 2018 doi: 10.3390/cancers10100382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gartmann L, et al. Expression of zinc transporters ZIP4, ZIP14 and ZnT9 in hepatic carcinogenesis—An immunohistochemical study. J. Trace Elem. Med. Biol. 2018;49:35–42. doi: 10.1016/j.jtemb.2018.04.034. [DOI] [PubMed] [Google Scholar]
- 59.Shangkuan W-C, et al. Risk analysis of colorectal cancer incidence by gene expression analysis. PeerJ. 2017;5:e3003. doi: 10.7717/peerj.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yagi K, et al. Three DNA methylation epigenotypes in human colorectal cancer. Clin. Cancer Res. 2010;16:21–33. doi: 10.1158/1078-0432.CCR-09-2006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Publicly available datasets were analyzed in this study. These data can be found here: TCGA, UALCAN, cBioPortal, and Kaplan–Meier Plotter.