Abstract
Himalayan mountains are distinctly characterized for their unique climatic and topographic variations; therefore, unraveling the cold-adaptive mechanisms and processes of native life forms is always being a matter of concern for scientific community. In this perspective, the proteomic response of psychrophilic diazotroph Pseudomonas helmanticensis was studied towards low-temperature conditions. LC-MS-based analysis revealed that most of the differentially expressed proteins providing cold stress resistance were molecular chaperons and cold shock proteins. Enzymes involved in proline, polyamines, unsaturated fatty acid biosynthesis, ROS-neutralizing pathways, and arginine degradation were upregulated. However, proteins involved in the oxidative pathways of energy generation were severalfold downregulated. Besides these, the upregulation of uncharacterized proteins at low temperature suggests the expression of novel proteins by P. helmanticensis for cold adaptation. Protein interaction network of P. helmanticensis under cold revealed that Tif, Tig, DnaK, and Adk were crucial proteins involved in cold adaptation. Conclusively, this study documents the proteome and protein-protein interaction network of the Himalayan psychrophilic P. helmanticensis under cold stress.
Keywords: Cold stress, Psychrophile, Bacterial cold adaptation, LC-MS, Himalaya
Introduction
Cold ecosystems dominate the Earth biosphere (Kuhn 2012), as 80% of Earth’s environments are permanently under cold stress (temperature below 5 °C), particularly in deep oceans, glaciers, polar regions, and alpines (Feller 2017). High-altitude terrestrial ecosystems of Western Indian Himalaya (WIH) are one of such cold stressed ecosystems which undergo frequent freezing-thawing cycles (Awasti et al. 2019). Microorganisms in these Himalayan regions are mostly psychrophilic or psychrotrophic in nature and play critical role in biogeochemical cycle operating under low temperature (Joshi et al. 2017).
Low temperature affects cellular machinery by decreasing membrane permeability, impairing protein folding, and hampering the process of transcription and translation by stabilizing secondary structure of DNA and RNA (Barria et al. 2013; Zhang et al. 2015). Cold-adapted microorganisms have remarkable survival strategies to protect them from detrimental effects of cold stress. Adaptations at the level of cell membrane, RNA metabolism, transcription, translation, and protein degradation/stability are important to carry out the cellular metabolism at low temperature (Tribelli and Lopez 2018). These cold adaptations in bacteria are so effective that the functional low-temperature limit of psychrophilic bacteria is − 12 °C for reproduction and − 20 °C for metabolism (De Maayer et al. 2014).
The degree of adaptations to cold stress varies in different bacterial groups (Barria et al. 2013). In mesophilic organisms, cold shock proteins (CSPs) are transiently induced during cold shock and soon after the acclimatization, their expression is downregulated (Phadtare 2012). These CSPs are crucial for low-temperature growth in mesophiles, as the quadruple csp deletion mutant (cspA, cspB, cspG, and cspE mutant) of E. coli is unable to divide under low temperature (Xia et al. 2001). Intriguingly, in psychrophiles, these proteins are constitutively expressed and act as cold acclimation proteins (CAPs) (Li et al. 2013). Cold-adapted microorganisms also activate complex network of molecular chaperones such as ClpB, DnaJ, DnaK and trigger factor to maintain the protein homeostasis which protects cold-induced protein misfolding (De Maayer et al. 2014). Additionally, low temperature imparts cellular ice crystal formation which causes cellular damage and osmotic imbalance. Antifreeze proteins produced by bacteria control ice crystal formation and recrystallization by lowering the freezing point (De Maayer et al. 2014). Furthermore, compatible solutes viz glycine betaine, sucrose, and mannitol lower the cytoplasmic freezing point thus protecting the cell against freezing, hyper-osmolality, and desiccation (Barria et al. 2013). Trehalose is reported to prevent protein denaturation and facilitate free radicals scavenging under cold stress in Burkholderia pseudomallei and Listeria monocytogenes (Ells and Truelstrup Hansen 2011; Vanaporn et al. 2017).
Besides cold, high-altitude ecosystems also face prolonged radiations, excessive UV, low atmospheric pressure, variable pH, and low nutrient availability (Barria et al. 2013). Osmotic and oxidative stresses are most profound under low temperature. While growing in cold stress, bacteria are reported to alter their metabolic traits to prefer only those metabolic pathways which generate less ROS (reactive oxygen species) (Tribelli and Lopez 2018). Thus, low temperature imparts several changes in global metabolism which provides the major challenges and opportunities to study cold adaptation in bacteria.
Western Indian Himalaya is well known for its rich diversity of cold-adopted microorganisms. Cold-adapted Arthrobacter humicola, Brevibacillus invocatus, Dyadobacter, Pseudomonas palleroniana, Pseudomonas jesenii, Pseudomonas mandelii, Pseudomonas migulae, Pseudomonas helmanticensis, and Rhodococcus qingshengii have been reported from WIH (Kumar et al. 2018; Suyal et al. 2019; Suyal et al. 2017). Despite the huge diversity of cold-adapted bacteria in WIH, there is a limited understanding of cold adaptation in these bacteria. Although previous studies have shown the ubiquitous distribution of genus Pseudomonas in the cold ecological niche of WIH, least is known about the global proteome response under cold stress. In this regard, proteome response of psychrophilic Pseudomonas helmanticensis was studied under simulated cold stress. Pseudomonas helmanticensis was previously isolated from high-altitude Gangotri soil and had optimum growth temperature of 10 °C (Kumar et al. 2019). Gangotri, being the second largest glacier of Himalaya, experiences severe fluctuating cold stress with maximum and minimum annual temperature of 11.1 ± 0.7 °C and − 2.3 ± 0.4 °C, respectively (Kumar et al. 2019). Therefore, liquid chromatography–mass spectrometry (LC-MS)–based differential proteomic study and subsequent protein-protein interaction network analysis would unravel the cold adaptation strategies of psychrophilic diazotroph Pseudomonas helmanticensis.
Material and methods
Bacterial strain and growth conditions
Pseudomonas helmanticensis was previously isolated from high-altitude cold climatic Gangotri soil (altitude 3415 m, 30.98° N, 78.93° E) while growing on nitrogen-deficient solid medium (Burk’s medium) at 2 °C for 48 h (Kumar 2018; Kumar et al. 2019). Optimum growth temperature of this bacterium was reported 10 °C in Nutrient Broth medium (HiMedia Laboratories) (Kumar et al. 2019). For proteome extraction, P. helmanticensis was grown separately at 2 °C (T, treatment) and 20 °C (C, control) in 250 mL Erlenmeyer flask with 50 mL of Nutrient Broth medium with shaking at 200 rpm up to mid-log phase.
Bacterial proteome extraction and LC-MS analysis
Bacterial proteome was extracted from mid-log phase culture in triplicates, lyophilized and subsequently analyzed by LC-MS as described earlier (Jain et al. 2010; Suyal et al. 2019). Briefly, lyophilized proteome was dissolved in 1 mL of 40 mM Tris buffer and 100 μL of it was taken for digestion. Samples were diluted in 50 mM NH4HCO3 and treated with 100 mM (dithiothreitol) DTT at room temperature for 1 h, and cysteine residues were subsequently alkylated with 250 mM iodoacetamide at room temperature for 1 h in dark. Protein digestion was carried out with trypsin at 37 °C for 16 h. Peptides were extracted in 0.1% formic acid and incubated at 37 °C for 45 min and centrifuged at 10,000g for 30 min. Supernatant was collected into a separate tube, vacuum dried, and dissolved in 20 μL of 0.1% formic acid in water which was further analyzed using LC-MS.
Identification and quantification of the proteins
After acquisition of raw data, processing and database search was performed using ProteinLynx Global SERVER™ PLGS software 3.0.2. Then, both raw proteomes were matched with bacterial protein database downloaded from Swiss-Prot database. Peptide tolerance was set to 50 ppm. Enzyme specificity was for trypsin, and the number of missed cleavage sites was set to two. The FDR (false discovery rate) was set as < 1% on both protein and peptide levels. Relative protein quantification was performed through spectral counting approach. Only those proteins showing more than 2-fold change in expression were considered upregulated and downregulated.
Protein-protein interaction network analysis
Protein-protein interaction (PPI) network analysis was performed to study the important proteins expressed under cold. Both upregulated and unique proteins were searched in the “STRING: functional protein association networks database” (https://string-db.org/) for finding the possible co-expression interactions among the selected proteins. For higher accuracy, only those interactions were selected from the STRING database for which confidence score was more than 0.7%. PPIs output file from STRING were downloaded and loaded in the Cytoscape to construct the PPI network (Shannon et al. 2003). Topological parameters of the network were analyzed through the Network Analyzer plugin of Cytoscape software as described earlier (Sabetian and Shamsir 2016).
Results
Identification of differentially expressed proteins
LC-MS-based differential proteomics revealed that 1540 proteins were expressed in both the conditions. Of all these proteins, total 952 were common in both, while 430 and 158 proteins were significantly (p ≤ 0.05) unique to control and cold stress, respectively. However, in shared proteins, 153 proteins were significantly (p ≤ 0.05) upregulated, while 159 were significantly (p ≤ 0.05) downregulated (Fig. 1).
Fig. 1.
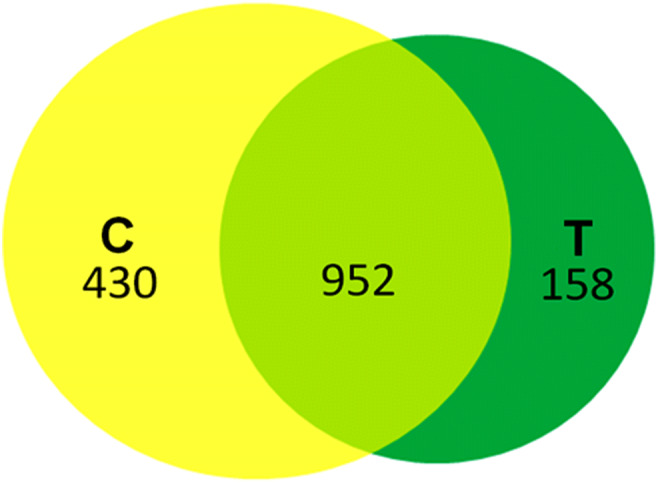
Venn diagram showing the protein expression in control and cold stress, respectively. Overlapped (light green) region represents the common proteins expressed in both conditions; yellow region represents the unique proteins in control while dark green represents the proteins expressed only under cold stress
Comparative protein expression profile showed that ATP-dependent RNA helicase RhlB, cold shock proteins (CspA and CspB), glutathione peroxidase, uncharacterized signaling protein PA1727, and medium-chain-fatty-acid-CoA ligase were major proteins expressed only under low-temperature growth (2 °C) (Table 1). Moreover, gamma-glutamyl phosphate reductase was 41.26-fold upregulated in cold followed by probable 2-ketoarginine decarboxylase (35.51-fold), exodeoxyribonuclease 7 large subunit (29.96-fold), chaperone protein ClpB (6.23-fold), chaperone protein DnaJ (4.43-fold), ribonuclease H (3.86-fold), ribonuclease R (3.71-fold), and 60 kDa chaperonin GroL (3.65-fold) (Table 2). Majority of unique and upregulated proteins expressed under cold were molecular chaperones. ClpB was the most upregulated molecular chaperone with 6.23-fold upregulation. Bifunctional protein HldE was 17.5-fold downregulated under cold followed NADH-quinone oxidoreductase subunit C/D (7.1-fold), oxygen-dependent coproporphyrinogen-III oxidase (6.9-fold), periplasmic trehalase (5.0-fold), phosphoenolpyruvate carboxylase (4.3-fold), quinolinate synthase A (3.7-fold), acetate kinase (2.3-fold), and enolase 2 (2.3-fold) (Table 2). Majority of downregulated proteins were directly or indirectly involved in oxidative pathways of energy generation, heme biosynthesis, and alginate biosynthesis.
Table 1.
List of unique proteins identified in this study involved in cold stress
| Gene | Protein name | Change | Description |
|---|---|---|---|
| rhlB | ATP-dependent RNA helicase RhlB | UCS♦ | DEAD-box RNA helicase - RNA degradation |
| exbB | Biopolymer transport protein | UCS | Transport of various receptor-bound substrates |
| dnaK | Chaperone protein DnaK | UCS | Response to hyperosmotic shock |
| cspA | Cold shock protein A | UCS | Cold-inducible RNA chaperone and antiterminator |
| cspB | Cold shock protein B | UCS | Stress response |
| ycgR | Flagellar brake protein YcgR | UCS | Acts as a flagellar brake, regulating swimming and swarming |
| gpwA | Glutathione peroxidase | UCS | Response to oxidative stress |
| gpuA | Guanidinopropionase | UCS | Hydrolysis of 3-guanidinopropanoate to beta-alanine and urea |
| PA3774 | Histone deacetylase-like amidohydrolase | UCS | Metal ion binding |
| alkK | Medium-chain-fatty-acid--CoA ligase | UCS | Fatty acid metabolic process |
| PputGB1_0956 | Nucleotide-binding protein PputGB1_0956 OS | UCS | ATP-binding, GTP-binding, nucleotide-binding |
| speE2 | Polyamine aminopropyltransferase | UCS | Spermidine synthase activity |
| PA1727 | *Uncharacterized signaling protein PA1727 | UCS | Cellular response to nitric oxide |
♦ Unique to Cold Stress (UCS), *Unannotated proteins
Table 2.
List of major upregulated and downregulated proteins identified in this study involved in cold stress
| Gene | Protein name | Change | Fold change | Description |
|---|---|---|---|---|
| proA | Gamma-glutamyl phosphate reductase | Upregulated | 41.2 | Proline biosynthesis |
| aruI | Probable 2-ketoarginine decarboxylase | Upregulated | 35.5 | Pathway L-arginine degradation |
| xseA | Exodeoxyribonuclease 7 large subunit | Upregulated | 29.9 | Single-stranded DNA degradation |
| ahcY | Adenosylhomocysteinase | Upregulated | 13.5 | Regulate adenosylhomocysteine |
| pepA | Cytosol aminopeptidase | Upregulated | 9.4 | Control intracellular proteins turnover |
| pqqB | Coenzyme PQQ synthesis protein B | Upregulated | 9.2 | Coenzyme PQQ biosynthesis |
| clpB | Chaperone protein ClpB | Upregulated | 6.2 | Part of a stress-induced multi-chaperone system |
| rmuC | DNA recombination protein RmuC | Upregulated | 5.1 | Involved in DNA recombination |
| dnaJ | Chaperone protein DnaJ | Upregulated | 4.4 | Stress-induced multi-chaperone system |
| rnhA | Ribonuclease H | Upregulated | 3.8 | Degradation of RNA in RNA-DNA hybrids |
| rnr | Ribonuclease R | Upregulated | 3.7 | Maturation of structural RNAs |
| groL | 60 kDa chaperonin | Upregulated | 3.6 | Prevents misfolding of unfolded polypeptides |
| tsf | Elongation factor Ts | Upregulated | 3.4 | Associates with the EF-Tu GDP |
| hscA | Chaperone protein HscA | Upregulated | 2.9 | Chaperone involved in the maturation of iron-sulfur cluster-containing proteins |
| lepA | Elongation factor 4 | Upregulated | 2.9 | Protein synthesis under stress conditions |
| htpG | Chaperone protein | Upregulated | 2.8 | Molecular chaperone |
| recA | Protein | Upregulated | 2.7 | Homologous recombination |
| cbpA | Curved DNA-binding protein | Upregulated | 2.4 | Functional analog of DnaJ |
| gyrB | DNA gyrase subunit B | Upregulated | 2.4 | Induce negatively supercoiling in DNA |
| dnaA | Chromosomal replication initiator protein | Upregulated | 2.3 | Regulation of chromosomal replication |
| fusB | Elongation factor G 2 | Upregulated | 2.3 | GTP-dependent ribosomal translocation |
| tig | Trigger factor | Upregulated | 2.0 | Involved in protein export, Acts as a chaperone |
| sodB | Superoxide dismutase | Upregulated | 2.0 | Destroys superoxide anion radicals |
| hldE | Bifunctional protein HldE | Downregulated | 17.5 | Nucleotide-sugar biosynthesis |
| rlmN | Dual-specificity RNA methyltransferase RlmN | Downregulated | 15.3 | Proofreading at the peptidyl transferase center |
| rdgC | Recombination-associated protein | Downregulated | 10.2 | May be involved in recombination |
| nuoC | NADH-quinone oxidoreductase subunit | Downregulated | 7.1 | Electron transport |
| hemF | Oxygen-dependent coproporphyrinogen-III oxidase | Downregulated | 6.9 | Porphyrin-containing compound metabolism. |
| betB | NAD/NADP-dependent betaine aldehyde dehydrogenase | Downregulated | 6.8 | Biosynthesis of the glycine betaine |
| bamD | Outer membrane protein assembly factor BamD | Downregulated | 5.7 | Assembly of proteins in outer membrane |
| PhoR | Phosphate regulon sensor protein PhoR | Downregulated | 5.2 | Phosphate regulon genes expression |
| treA | Periplasmic trehalase | Downregulated | 5.0 | Trehalose degradation |
| ppc | Phosphoenolpyruvate carboxylase | Downregulated | 4.3 | Oxaloacetate generation for the TCA |
| nadA | Quinolinate synthase A | Downregulated | 3.7 | NAD (+) biosynthesis |
| mdcB | Probable 2-(5″-triphosphoribosyl)-3′-dephosphocoenzyme-A synthase | Downregulated | 3.6 | Formation the prosthetic group of the acyl-carrier protein of the malonate decarboxylase |
| alg8 | Mannuronan synthase | Downregulated | 3.3 | Alginate biosynthesis |
| algG | Mannuronan C5-epimerase | Downregulated | 3.3 | Alginate biosynthesis |
| ggpS | Glucosylglycerol-phosphate synthase | Downregulated | 3.1 | Synthesis of the osmolyte glucosylglycerol |
| lysC | Aspartokinase | Downregulated | 2.7 | L-Methionine biosynthesis via de novo pathway |
| aceK | Isocitrate dehydrogenase kinase/phosphatase | Downregulated | 2.6 | Phosphorylate or dephosphorylate isocitrate dehydrogenase |
| metH | Methionine synthase | Downregulated | 2.5 | Synthesizes L-methionine |
| ackA | Acetate kinase | Downregulated | 2.3 | Pathway acetyl-CoA biosynthesis |
| eno2 | Enolase 2 | Downregulated | 2.3 | Part of the pathway glycolysis |
| glgA | Glycogen synthase | Downregulated | 2.2 | Glycogen biosynthesis |
| pgi | Glucose-6-phosphate isomerase | Downregulated | 2.2 | Pathway gluconeogenesis |
Network analysis of the protein-protein interactions
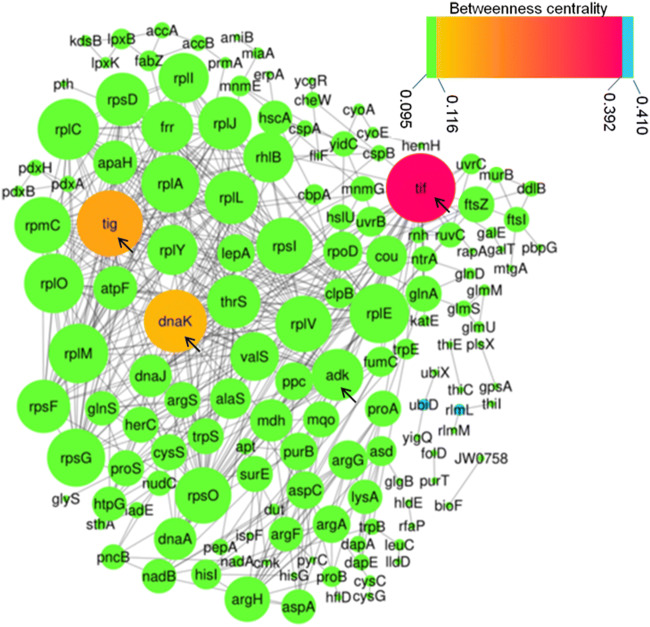
Protein-protein interaction network analysis was performed to investigate the possible interaction among differentially expressed proteins in cold stress. The proteins within PPI network are represented as nodes (circles in the figure) and the interactions among them are represented by edges (lines connecting the nodes). Diameter of nodes represents the degree parameter of the network which indicates number of connections made by the nodes (Sabetian and Shamsir 2016). Thus, nodes with higher degree are more important in the network because removing these nodes will collapse the network. Protein interaction network of P. helmanticensis under cold revealed that Tif, Tig, DnaK, and Adk were hub-bottleneck in this network (Fig. 2). Hub-bottlenecks are the nodes with the highest value of degree and betweenness and are crucial proteins in the PPIs network. Furthermore, DnaJ, DnaA, ProA, and PurB were hub nodes (nodes with higher degree) and UbiD, Frr, and YidC were bottlenecks (high-betweenness proteins). Moreover, CysG, LldD, LpxK, CyoE, and PbpG had high eccentricity, compared with the average eccentricity of the network. Eccentricity in a network represents the easiness of a protein to be functionally reached by all other proteins in the network. Thus, a protein with high eccentricity, compared with the average eccentricity of the network, will be more easily influenced by the activity of other proteins.
Fig. 2.
Protein-protein interaction (PPI) network of differentially expressed proteins under cold stress. The degree (number of neighbors) of nodes is represented by the size of the circle and the interaction with other proteins is represented by edges (lines connecting the nodes). The color of the node represents the betweenness centrality (BC) parameter as indicated on the scale where lawngreen represents low BC and turquoise represents high BC. Crucial proteins in this network are indicated with black arrow
Discussion
The present study aims to investigate the cold adaptation strategies in Himalayan psychrophilic bacterium P. helmanticensis through differential proteomic analysis. Bacterial proteomes at two different temperatures (2 °C and 20 °C) were isolated, characterized through LC-MS, and further validated in silico through PPI network analysis. Presence of several uncharacterized proteins under cold suggests the possibility of novel proteins involved in cold adaptations of P. helmanticensis.
Bacteria respond to low temperature by inducing proteins that facilitate biological process of transcription, translation, and RNA metabolism under cold which are classified as cold-induced proteins. Differential expression of cold-induced proteins under cold stress is reported to be induced by negative supercoiling of DNA (Beckering et al. 2002). Upregulation (2.45-fold) of DNA gyrase subunit B at low temperature could be the possible reason for the differential CSP expression in this study, as inhibition of DNA gyrase in previous study showed decreased expression of cold-induced proteins under cold stress (Prakash et al. 2009). In this study, CspA and CspB were two major cold shock proteins solely expressed under cold stress. CSPs exert its crucial role by facilitating the process of transcription and translation under cold (Phadtare 2012). Previous study on Himalayan cold-adapted Pseudomonas documented that expression of cspA did not change significantly at 5 °C and 30 °C but increased 2.5-fold at 15 °C with respect to cspA levels at 5 °C and 30 °C (Awasti et al. 2019). Therefore, level of cspA expression is different in psychrophilic and psychrotrophic Pseudomonas thus exerting different biological functions in both. While in mesophilic counterpart, CspA is reported to express transiently during cold adaptation, in psychrophilic bacteria, CspA is constitutively expressed. However, above 15 °C, cspA mRNA has the conformation which is not translated and easily recognized by the cellular RNA degrading machinery. Thus, degradation of cspA mRNA at 20 °C could be the possible reason for its absence in this study.
Protein synthesis is significantly impaired under cold shock due to the alteration in structural integrity of ribosomes and formation of RNA secondary structure. DEAD-box RNA helicase genes were reported to play crucial role under cold stress in cold-adapted microorganisms (Kuhn 2012). ATP-dependent RNA helicase, RhlB, was the only DEAD-box RNA helicase expressed under cold stress. Similar to the previous low-temperature differential proteomics studies, several ribosomal proteins, mainly RplM, RplL, RplA, RplI, and RrmA, were upregulated in this study (Dai et al. 2018; Wang et al. 2015). These ribosomal proteins act as chaperones for RNA and proteins under abiotic stress (Aseev and Boni 2011). Thus, overproduction of ribosomal proteins under cold stress could be the cold adaptation strategy in psychrophiles, where one portion of the ribosomal proteins forms the functional ribosomes for protein synthesis and the other portion acts as a molecular chaperon to facilitate cell viability under cold. Furthermore, translation initiation factor IF2 is reported to be involved in ribosome assembly and maturation during cold adaptation (Maillot et al. 2019).
Majority of upregulated and unique proteins expressed under cold were molecular chaperones, including HtpG, ClpB, DnaJ, and trigger factor which were found severalfold upregulated. These molecular chaperones were also found upregulated in previous cold-induced differential proteomics study (Suyal et al. 2019; Zhang et al. 2018). In all these chaperones, caseinolytic protease B (ClpB) was the most upregulated with 6.23-fold upregulation. ClpB has been reported to renature and solubilizes protein aggregates formed under low temperatures (Ito et al. 2014). Two-fold upregulation was observed in trigger factor which is supposed to be the first chaperone to be involved in low-temperature protein stabilization (Hoffmann et al. 2010). Trigger factor controls protein miss-folding by delaying the premature chain compaction and keeping the elongated polypeptide in a non-aggregated state in the absence of structural information for productive folding (Piette et al. 2010). Downregulation of trigger factor in bacterial cells reduces the cell viability during cold stress; however, its overexpression enhances viability under cold stress (Phadtare 2004). Furthermore, DnaK and GroL were slightly upregulated under cold stress. DnaK system with a dedicated J domain protein is reported in cold-adapted Shewanella oneidensis (Brandi et al. 2019).
Other key feature of cold-adapted bacteria growing under cold stress is to tackle with osmotic stress (Mocali et al. 2017). Production of osmoprotectant proline, trehalose, and glycine betaine was reported in bacteria under low-temperature growth. In this study, gamma-glutamyl phosphate reductase was the most upregulated protein (41.26-fold) under cold stress which catalyzes the intermediate step in biosynthesis of proline from glutamate. Therefore, pathways for proline biosynthesis are upregulated under cold stress. The role of proline in osmotic stress is well studied but its exact role in cold adaptation is not well understood. Interestingly, in few studies, proline metabolism is reported to provide oxidative stress resistance in bacteria under cold stress (Zhang et al. 2015). Therefore, proline metabolism plays indispensable role under cold stress for which the exact mechanism is yet to be elucidated.
Enzymes involved in glycine betaine production were downregulated under cold in this study and no direct evidence of trehalose production was found. Rather, periplasmic trehalase (trehalose degrading enzyme) was found downregulated under cold stress which is consistent with the previous study which highlighted that the constitutive expression of trehalase abolished the ability of the bacterium to withstand cold shock (Vanaporn et al. 2017). Deletion mutant of trehalase (treA) in Burkholderia pseudomallei and Listeria monocytogenes had shown increased stress tolerance than the wild type (Ells and Truelstrup Hansen 2011; Vanaporn et al. 2017). Therefore, decreased levels of trehalase and consequently increased levels of trehalose could provide cold tolerance in P. helmanticensis.
Oxidative stress is more pronounced at low temperature and cold-adapted bacteria have several strategies to overcome ROS. In this study, glutathione peroxidase and superoxide dismutase were found upregulated under cold stress which are most characterized cryoprotective agents. Spermidine and putrescine have been reported most common cytoprotective polyamines contributing enhanced molecular function by stabilizing nucleic acid and neutralizing ROS (Limsuwun and Jones 2000). SpeE2 was expressed only under cold stress which catalyzes the irreversible transfer of a propylamine group from the amino donor S-adenosylmethioninamine to putrescine to yield spermidine. Putrescine, in turn, is produced from either ornithine or arginine by ornithine decarboxylase or arginine decarboxylase respectively (Koh et al. 2017). In the present investigation, probable 2-ketoarginine decarboxylase was also found 35.51-fold upregulated which suggests the putrescine overproduction under low temperature. Therefore, L-arginine degradation leads to the production of cytoprotective polyamines spermidine and putrescine in P. helmanticensis under cold stress.
Proteins involved in energy conversion (oxygen-dependent coproporphyrinogen-III oxidase, phosphoenolpyruvate carboxylase, quinolinate synthase A, glucosylglycerol-phosphate synthase, acetate kinase, enolase, glycogen synthase, and alcohol dehydrogenase) were found downregulated which was consistent with the previous study (Wang et al. 2015). Most of these downregulated proteins were involved in oxidative pathways of energy generation. As oxidative stress is more pronounced under cold stress, ROS-generating oxidative metabolic pathways have previously been reported to be repressed under low-temperature growth. Intriguingly, preference for less ROS-generating shortened or non-central metabolic pathways has been reported in several cold-adaptive bacteria (Tribelli and Lopez 2018). Several modifications in fatty acids like unsaturation, chain length reduction, and branching of side chain have been reported to provide membrane fluidity at low temperature (Barria et al. 2013). These modifications in this study were highlighted by the upregulation of acetyl-CoA carboxylase, biotin carboxylase, and other genes. Similar modifications were also reported in Pseudomonas species, Psychrobacter sp. PAMC 21119, V. parahaemolyticus, and Shewanella piezotolerans under low-temperature growth (Tribelli and Lopez 2018).
Although, differential proteomics studies could reveal important proteins expressed under certain set of physiological conditions, it does not provide complete interactions of the proteins active under those conditions. Protein-protein interaction network analysis could find out the major proteins which are very crucial under some physiological conditions. In PPI network (Fig. 2), most of the hub-bottleneck, hubs, and bottlenecks were molecular chaperones, ribosomal proteins, and proteins involved in amino acid metabolism. Hub-bottleneck Adk involves in AMP biosynthesis via salvage pathway where it catalyzes the reversible transfer of terminal phosphate group between ATP and AMP (Gutierrez and Csonka 1995). Frr codes for the ribosome-recycling factor which increases the efficiency of translation by recycling ribosomes from one round of translation to another (Janosi et al. 2000). The UbiD is involved in ubiquinone biosynthesis, while YidC is a molecular chaperone which participates in protein folding and protein insertion in the membrane (de Sousa Borges et al. 2015). FabZ is involved in unsaturated fatty acid biosynthesis which is also one of the cold adaptation strategies in psychrophilic bacteria (Liao et al. 2019). These hub-bottleneck, hub, and bottleneck proteins are crucial for low-temperature growth of P. helmanticensis as their deletion will collapse the pathway related to the network.
Therefore, differential proteomics and subsequent protein-protein interaction network analysis suggest that ribosomal proteins assisting in translation, molecular chaperons, cryoprotective polyamines (spermidine and putrescine), proline, unsaturated fatty acid, and ROS-neutralizing proteins had major role in cold adaptation of psychrophilic P. helmanticensis. Furthermore, investigation on protein expression under low-temperature nitrogen stress in the future would unravel the novel proteins/pathways associated with low-temperature diazotrophy in P. helmanticensis.
Concluding remarks
Conclusively, the present study documented the proteome of Himalayan psychrophilic diazotroph P. helmanticensis and elucidated its protein-protein interaction network under cold stress. Most of the proteins providing cold stress resistance were ROS-neutralizing enzymes, molecular chaperons, enzymes involved in cellular cryoprotectant, and unsaturated fatty acid biosynthesis. In conclusion, this study furnishes the complete picture of protein expression profile of P. helmanticensis under cold stress.
Acknowledgments
Author SK acknowledges the Council of Scientific and Industrial Research (CSIR) for granting the Senior Research Fellowship (SRF) and DCS acknowledges the Science and Engineering Research Board (SERB) Young Scientist scheme during the course of this study.
Data availability
All the data is available.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
Not Applicable.
Consent to participate
Not Applicable.
Consent for publication
Not Applicable.
Code availability
Not Applicable.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Aseev LV, Boni IV. Extraribosomal functions of bacterial ribosomal proteins. Mol Biol (Mosk) 2011;45:805–816. doi: 10.1134/S0026893311050025. [DOI] [PubMed] [Google Scholar]
- Awasti S, Anjney S, Saxena P, Yadav J, Pandiyan K, Kumar M, Singh A, Chakdar H, Bhowmik A, Kashyap PL, Srivastava A, Saxena AK. Molecular detection and in silico characterization of cold shock protein coding gene (cspA) from cold adaptive Pseudomonas koreensis. J Plant Biochem Biotechnol. 2019;28(4):405–413. doi: 10.1007/s13562-019-00500-8. [DOI] [Google Scholar]
- Barria C, Malecki M, Arraiano CM. Bacterial adaptation to cold. Microbiology. 2013;159:2437–2443. doi: 10.1099/mic.0.052209-0. [DOI] [PubMed] [Google Scholar]
- Beckering CL, Steil L, Weber MH, Volker U, Marahiel MA. Genomewide transcriptional analysis of the cold shock response in Bacillus subtilis. J Bacteriol. 2002;184:6395–6402. doi: 10.1128/JB.184.22.6395-6402.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandi A, Piersimoni L, Feto NA, Spurio R, Alix JH, Schmidt F, Gualerzi CO. Translation initiation factor IF2 contributes to ribosome assembly and maturation during cold adaptation. Nucleic Acids Res. 2019;47:4652–4662. doi: 10.1093/nar/gkz188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Zhu M, Warren M, Balakrishnan R, Okano H, Williamson JR, Fredrick K, Hwa T (2018) Slowdown of translational elongation in Escherichia coli under hyperosmotic stress. MBio 9(1):e02375-17 [DOI] [PMC free article] [PubMed]
- De Maayer P, Anderson D, Cary C, Cowan DA. Some like it cold: understanding the survival strategies of psychrophiles. EMBO Rep. 2014;15:508–517. doi: 10.1002/embr.201338170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sousa Borges A, de Keyzer J, Driessen AJ, Scheffers DJ. The Escherichia coli membrane protein insertase YidC assists in the biogenesis of penicillin binding proteins. J Bacteriol. 2015;197:1444–1450. doi: 10.1128/JB.02556-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ells TC, Truelstrup Hansen L. Increased thermal and osmotic stress resistance in Listeria monocytogenes 568 grown in the presence of trehalose due to inactivation of the phosphotrehalase-encoding gene treA. Appl Environ Microbiol. 2011;77:6841–6851. doi: 10.1128/AEM.00757-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller G (2017) Cryosphere and psychrophiles: insights into a cold origin of life? Life (Basel) 7(2):25 [DOI] [PMC free article] [PubMed]
- Gutierrez JA, Csonka LN. Isolation and characterization of adenylate kinase (adk) mutations in Salmonella typhimurium which block the ability of glycine betaine to function as an osmoprotectant. J Bacteriol. 1995;177:390–400. doi: 10.1128/JB.177.2.390-400.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Bukau B, Kramer G. Structure and function of the molecular chaperone Trigger Factor. Biochim Biophys Acta. 2010;1803:650–661. doi: 10.1016/j.bbamcr.2010.01.017. [DOI] [PubMed] [Google Scholar]
- Ito F, Tamiya T, Ohtsu I, Fujimura M, Fukumori F. Genetic and phenotypic characterization of the heat shock response in Pseudomonas putida. Microbiologyopen. 2014;3:922–936. doi: 10.1002/mbo3.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Rani A, Marla SS, Goel R. Differential proteomic analysis of psychrotolerant Pseudomonas putida 710A and alkaliphilic Pseudomonas monteilli 97AN for cadmium stress. Int J Biol Med Res. 2010;4:234–241. [Google Scholar]
- Janosi L, Mori H, Sekine Y, Abragan J, Janosi R, Hirokawa G, Kaji A. Mutations influencing the frr gene coding for ribosome recycling factor (RRF) J Mol Biol. 2000;295:815–829. doi: 10.1006/jmbi.1999.3401. [DOI] [PubMed] [Google Scholar]
- Joshi D, Kumar S, Suyal DC, Goel R. Mining of Microbial Wealth and MetaGenomics. Singapore: Springer; 2017. The microbiome of the Himalayan ecosystem; pp. 101–116. [Google Scholar]
- Koh HY, Park H, Lee JH, Han SJ, Sohn YC, Lee SG. Proteomic and transcriptomic investigations on cold-responsive properties of the psychrophilic Antarctic bacterium Psychrobacter sp. PAMC 21119 at subzero temperatures. Environ Microbiol. 2017;19:628–644. doi: 10.1111/1462-2920.13578. [DOI] [PubMed] [Google Scholar]
- Kuhn E. Toward understanding life under subzero conditions: the significance of exploring psychrophilic “cold-shock” proteins. Astrobiology. 2012;12:1078–1086. doi: 10.1089/ast.2012.0858. [DOI] [PubMed] [Google Scholar]
- Kumar S. Documentation of the bacterial and diazotrophic diversity from Garhwal Himalaya through culturable and unculturable approaches. Pantnagar: G.B.P.U.A.&T; 2018. [Google Scholar]
- Kumar S, Suyal DC, Bhoriyal M, Goel R. Plant growth promoting potential of psychrotolerant Dyadobacter sp. for pulses and finger millet and impact of inoculation on soil chemical properties and diazotrophic abundance. J Plant Nutr. 2018;41(8):1035–1046. doi: 10.1080/01904167.2018.1433211. [DOI] [Google Scholar]
- Kumar S, Suyal DC, Yadav A, Shouche Y, Goel R. Microbial diversity and soil physiochemical characteristic of higher altitude. PLoS One. 2019;14:e0213844. doi: 10.1371/journal.pone.0213844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Jiang T, Yu B, Wang L, Gao C, Ma C, Xu P, Ma Y. Escherichia coli transcription termination factor NusA: heat-induced oligomerization and chaperone activity. Sci Rep. 2013;3:2347. doi: 10.1038/srep02347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao JX, Li KH, Wang JP, Deng JR, Liu QG, Chang CQ. RNA-seq analysis provides insights into cold stress responses of Xanthomonas citri pv. citri. BMC Genomics. 2019;20:807. doi: 10.1186/s12864-019-6193-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limsuwun K, Jones PG. Spermidine acetyltransferase is required to prevent spermidine toxicity at low temperatures in Escherichia coli. J Bacteriol. 2000;182:5373–5380. doi: 10.1128/JB.182.19.5373-5380.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillot NJ, Honore FA, Byrne D, Mejean V, Genest O. Cold adaptation in the environmental bacterium Shewanella oneidensis is controlled by a J-domain co-chaperone protein network. Commun Biol. 2019;2:323. doi: 10.1038/s42003-019-0567-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocali S, Chiellini C, Fabiani A, Decuzzi S, de Pascale D, Parrilli E, Tutino ML, Perrin E, Bosi E, Fondi M, Lo Giudice A, Fani R. Ecology of cold environments: new insights of bacterial metabolic adaptation through an integrated genomic-phenomic approach. Sci Rep. 2017;7:839. doi: 10.1038/s41598-017-00876-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadtare S. Recent developments in bacterial cold-shock response. Curr Issues Mol Biol. 2004;6:125–136. [PubMed] [Google Scholar]
- Phadtare S. Escherichia coli cold-shock gene profiles in response to over-expression/deletion of CsdA, RNase R and PNPase and relevance to low-temperature RNA metabolism. Genes Cells. 2012;17:850–874. doi: 10.1111/gtc.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piette F, D'Amico S, Struvay C, Mazzucchelli G, Renaut J, Tutino ML, Danchin A, Leprince P, Feller G. Proteomics of life at low temperatures: trigger factor is the primary chaperone in the Antarctic bacterium Pseudoalteromonas haloplanktis TAC125. Mol Microbiol. 2010;76:120–132. doi: 10.1111/j.1365-2958.2010.07084.x. [DOI] [PubMed] [Google Scholar]
- Prakash JSS, Sinetova M, Zorina A, Kupriyanova E, Suzuki I, Murata N, Los DA. DNA supercoiling regulates the stress-inducible expression of genes in the cyanobacterium Synechocystis. Mol BioSyst. 2009;5:1904–1912. doi: 10.1039/b903022k. [DOI] [PubMed] [Google Scholar]
- Sabetian S, Shamsir MS. Systematic analysis of protein interaction network associated with azoospermia. Int J Mol Sci. 2016;17:1857. doi: 10.3390/ijms17111857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyal DC, Kumar S, Yadav A, Shouche Y, Goel R. Cold stress and nitrogen deficiency affected protein expression of psychrotrophic Dyadobacter psychrophilus B2 and Pseudomonas jessenii MP1. Front Microbiol. 2017;8:430. doi: 10.3389/fmicb.2017.00430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyal DC, Joshi D, Kumar S, Soni R, Goel R. Differential protein profiling of soil diazotroph Rhodococcus qingshengii S10107 towards low-temperature and nitrogen deficiency. Sci Rep. 2019;9:20378. doi: 10.1038/s41598-019-56592-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribelli PM, Lopez NI (2018) Reporting key features in cold-adapted bacteria. Life (Basel) 8(1):8 [DOI] [PMC free article] [PubMed]
- Vanaporn M, Sarkar-Tyson M, Kovacs-Simon A, Ireland PM, Pumirat P, Korbsrisate S, Titball RW, Butt A. Trehalase plays a role in macrophage colonization and virulence of Burkholderia pseudomallei in insect and mammalian hosts. Virulence. 2017;8:30–40. doi: 10.1080/21505594.2016.1199316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Jia Y, Li W. Effects of environmental and biotic factors on carbon isotopic fractionation during decomposition of soil organic matter. Sci Rep. 2015;5:11043. doi: 10.1038/srep11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B, Ke H, Inouye M. Acquirement of cold sensitivity by quadruple deletion of the cspA family and its suppression by PNPase S1 domain in Escherichia coli. Mol Microbiol. 2001;40:179–188. doi: 10.1046/j.1365-2958.2001.02372.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Cong J, Lu H, Li G, Xue Y, Deng Y, Li H, Zhou J, Li D. Soil bacterial diversity patterns and drivers along an elevational gradient on Shennongjia Mountain, China. Microb Biotechnol. 2015;8:739–746. doi: 10.1111/1751-7915.12288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Burkhardt DH, Rouskin S, Li GW, Weissman JS, Gross CA. A stress response that monitors and regulates mRNA structure is central to cold shock adaptation. Mol Cell. 2018;70(274–286):e277. doi: 10.1016/j.molcel.2018.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data is available.